Abstract
Background
Bone marrow stromal cells (BMSCs) have classically been cultured in media supplemented with fetal bovine serum (FBS). As an alternative to FBS, pooled solvent detergent apheresis platelets, HPGF-C18, was evaluated for BMSC culture.
Methods
A comparison of passage 2 BMSC growth revealed that 10% HPGF-C18 produced similar cell numbers as 20% FBS. Marrow aspirates from 5 healthy subjects were cultured for 4 passages in 10% HPGF-C18 or 20% FBS and were analyzed for proliferation, colony formation efficiency (CFE), surface marker expression, suppression of mixed lymphocyte reactions (MLRs), global gene and microRNA expression analysis. BMSC supernatant cytokine and growth factor concentrations were also compared.
Results
Primary cultures of marrow aspirates in 10% HPGF-C18 and 20% FBS yielded similar numbers and CFE. After 4 passages, 10% HPGF-C18 and 20% FBS yielded similar numbers of BMSCs, surface marker expression patterns and immunosuppression effects. Gene and microRNA expression analysis revealed that BMSCs cultured under the two conditions had distinct expression profiles. Gene Set Enrichment Analysis (GSEA) revealed HPGF-C18-cultured BMSCs were enriched in metabolic processing and biosynthetic pathways, cell proliferation and cell cycle pathways, and immune response pathways. FBS-cultured BMSCs were enriched in MAPK signaling, TGF-beta signaling, cell adhesion and extracellular matrix pathways. Differently expressed microRNAs were related to the osteogenesis of BMSCs. The supernatant of HPGF-C18 BMSCs had higher levels of PEDF and TGFB1 and lower levels of IL6, VEGF, SDF1 and PLGF.
Conclusions
Traditional measures, expansion, surface marker expression and inhibition of MLRs suggest that BMSC cultured in HPGF-C18 and FBS were similar, but analysis at the molecular level revealed many differences. BMSCs cultured in HPGF-C18 should be assessed in specific functional assays that reflect application-specific potency before substituting FBS with HPGF-C18.
Keywords: Bone marrow stromal cells, Mesenchymal stem cells, Fetal bovine serum, Platelet lysate, Potency
Background
Bone marrow stromal cells (BMSCs) are a heterogeneous population of adherent cells obtained from the marrow that have a number of potential clinical applications. Skeletal stem cells found in BMSCs produce bone and can be used for bone repair [1–3]. BMSCs also have immune modulatory properties and can induce angiogenesis and tissue repair. They are being used in early phase clinical trials to treat steroid resistant acute graft-versus-host-disease (GVHD) [4], inflammatory bowel disease [5, 6], ischemic vascular disease [7], acute lung injury [8] and traumatic brain injury [9].
BMSC were first isolated and expanded in media supplemented with fetal bovine serum (FBS) [10] and most laboratories continue to culture BMSCs in FBS. The culture of BMSCs in FBS, however, exposes the recipient of the BMSC product to potential xenogenic infection and immune reaction to bovine proteins [11–13]. In order to avoid the exposure of BMSC recipients to FBS some groups have begun to grow BMSCs in media supplemented with factors derived from human platelets. Preliminary studies have shown that when BMSCs are cultured in media supplemented with the contents of lysed platelets, they express mesenchymal stromal cells (MSC) surface markers CD73, CD90 and CD105; they differentiate in vitro into adipose tissue, chondrocytes, and osteocytes; and, in general, they proliferate faster than cells grown in FBS [14–20].
While the use of platelet lysate avoids exposure to exogenous proteins, platelet lysate is derived from human blood products and has the potential to transmit hepatitis B, hepatitis C, human immune deficiency virus and other transfusion transmitted pathogens. Many transfusion transmitted pathogens can be inactivated by various methods and platelet lysate preparations that have been treated to inactivate pathogens are available. We tested one such product, human platelet growth factor C-18 (HPGF-C18), as a source of platelet factors to support the growth of BMSCs. HPGF-C18 has been subjected to solvent-detergent treatment that inactivates lipid-enveloped viruses and to a lesser extent bacteria which reduces the risk of transmitting an infectious disease to the recipient of the BMSC products [21]. HPGF-C18 is made from a relatively large pool of outdated apheresis platelets, approximately 50 units, and it is produced following Good Manufacturing Practices (GMP) standards, both of which helps keep inter-lot variability minimal.
In this study we found that 10% HPGF-C18 produced similar numbers of BMSCs to 20% FBS in a preliminary experiment using passage 2 BMSCs. We then compared BMSCs manufactured from marrow aspirates with media supplemented with 10% HGPF-C18 or 20% FBS using the same methods that our laboratory has used to produce clinical grade BMSCs. We compared the proliferation, surface marker expression, immunosuppression effects of BMSCs and measured the concentrations of cytokines and growth factors in the supernatant. We also compared the global profiles of the BMSCs cultured by HPGF-C18 and FBS using gene expression and microRNA expression analysis.
Methods
Study design
The study made use of human BMSCs collected from healthy subjects. In order to determine the best concentration of HPGF-C18 for BMSCs growth passage 2 BMSCs were cultured in various concentrations of HPGF-C18 and compared to BMSCs cultured in 20% FBS, the concentration that is used in our laboratory for the production of BMSCs following GMPs [22]. After the best concentration of HPGF-C18 was identified, bone marrow aspirates were obtained from 5 healthy subjects and BMSCs were isolated and cultured for 4 passages, the cellular expansion, surface markers, and immunosuppressive activities were compared between HPGF-C18 cultured BMSCs and 20% FBS cultured BMSCs. We also measured a variety of cytokines and growth factors in the supernatant and compared the gene expression profiles and microRNA profiles of the BMSCs cultured under both conditions. These studies were approved by an NHLBI committee on the use of human subjects in research.
HPGF-C18
HPGF-C18 was prepared from 52 outdated units of apheresis platelets each of which contained approximately 4 × 1011 platelets (GwoWei Technology Co, Ltd., Taipei, Taiwan). The pooled platelets were solvent/detergent (S/D)-treated (1% tri-n-butyl phosphate and 1% Triton X-45), extracted with oil, purified by C18 hydrophobic interaction chromatography and sterile filtered as previously described [23].
Determination of the optimal HPGF-C18 concentration for BMSC growth
Cryopreserved passage 2 BMSCs were cultured to determine the best concentration of HPGF-C18 for BMSC growth. These passage 2 BMSCs were isolated from marrow aspirates of 3 healthy subjects using media supplemented in 20% FBS and were cryopreserved in 5% DMSO and 6% HES and stored in the vapor phase of liquid nitrogen. The passage 2 BMSCs were thawed, washed and suspended in alpha MEM with 2 mM glutamine (Lonza, Walkersville, MD), supplemented with 10 µg/mL Gentamicin and 20% lot-selected FBS (Hyclone, Thermo Fischer Scientific, Waltham, MA) or 5, 10, 15 and 20% HPGF-C18 and were seeded on T75 flasks at a density of 3000 cells/cm2. The cells were cultured and harvested with 5 mL TrypLE Express (Invitrogen, Life Technologies) when they reached 70–80% confluence and were re-seeded on T75 flasks at a density of 3000 cells/cm2 for expansion. Passage 4 cells were harvested and surface marker of passage 4 BMSCs was compared. The number of BMSCs at passage 3 and 4 was manually counted and the population doubling (PD) for each passage was calculated.
Culture of BMSCs from marrow aspirates
The marrow aspiration and BMSC culture were performed according to standard operating procedures (SOP) established in our laboratory [22]. After obtaining informed consent, marrow was collected from the posterior iliac crest of 5 healthy donors. A total of 5–10 mL of marrow was collected in Bone Marrow Prep Syringes (Pharmacy Department, NIH, Bethesda, MD) and then washed with 2.5 × volume of HBSS (Lonza, Walkersville, MD). A single cell suspension was made with BMSC culture media (BMSC CM) [alpha MEM with 2 mM glutamine (Lonza), supplemented with 20% lot-selected FBS (Hyclone, Thermo Fischer Scientific, Waltham, MA) or 10% HPGF-C18 and 10 µg/mL gentamicin] and was plated at a density of 2 × 105/cm2 in T-75 flasks (Corning Life Sciences, Corning, NY) and incubated at 37 °C in 5% CO2. Non-adherent cells were removed after 24 h; the media was changed every 3 days until the colonies reached 70–80% confluence.
The primary BMSCs were washed with 10 mL HBSS twice and lifted with 5 mL TrypLE Express (Invitrogen, Life Technologies, Grand Island, NY), the cells were then centrifuged at 406×g for 10 min and the cell number was counted. The cells harvested at this stage were designated as Passage 1. The BMSCs were then seeded on plastic surface at a density of 3000 cells/cm2, cultured and harvested as described above when they reached 70–80% confluence.
Passage 4 cells were harvested for evaluation of surface marker expression, suppression on the proliferation of mixed lymphocytes, global gene expression profiling, and microRNA expression analysis. Cytokine and growth factor levels were measured in the supernatant of passage 4 BMSCs. The number of BMSCs at passage 3 and 4 was manually counted and the population doublings (PDs) for each passage was calculated. Cumulative PDs were calculated in relation to the number of cells at the first passage.
Primary colony-forming efficiency (CFE) enumeration
Bone marrow aspirates were diluted in culture media with 20% FBS or 10% HPGF respectively, and then plated at a density of 1 × 105 per T25 flask and cultured for 13 days without changing culture medium. The colonies were fixed with methanol for 30 min and stained with saturated methyl-violet water solution for 20 min. Colonies were observed under low magnitude light microscope field (25×). Colonies containing 50 or more cells of fibroblastic morphology were counted, and CFE (number of BMSC colonies/plating nucleated cells) was calculated.
Surface marker expression
BMSC surface markers were analyzed by flow cytometry. The cells were incubated with antibodies CD90-FITC, CD73-PE, CD146-PE, CD106-APC, CD45-FITC, CD14-PE, CD19-FITC, CD34-APC, HLA-DR-APC (BD Bioscience, San Diego, CA), CD11b-FITC and CD105-APC (eBioscience, San Diego, CA) for 20 min at 4 °C, washed on Lyse Wash Assistant (LWA, BD Bioscience) and acquired 30,000 events on a FACSCanto (BD Bioscience). The data were analyzed using FACSDiva 6.0 software (BD Bioscience).
Cytokine and growth factor analysis of BMSC culture supernatant
The cytokine and growth factor concentrations in BMSC culture supernatant were evaluated using SearchLight Protein Array Analysis (Aushon Biosystems, Billerica, MA). Culture supernatant was collected, centrifuged for 10 min at 1400 rpm to remove cell debris and then stored at − 80 °C. The supernatants of BMSC from 5 healthy donors were evaluated for IL4, IL6, IL8, IL10, KGF, LIF, PEDF, TGFB1, FGF2, HGF, PDGFBB, VEGF, SDF1, PLGF, ANG2, and Endoglin.
Mixed lymphocyte reaction (MLR)
The immunosuppressive properties of BMSCs were compared using MLR assay (SAIC-Frederic, Frederic, MD). Ficoll-separated peripheral blood mononuclear cells were plated in 96-well plates at 1 × 105 responders per well. Responders were co-cultured with 2500 cGy irradiated stimulator peripheral blood mononuclear cells at a concentration of 1 × 105 cells per well. BMSCs cultured by either 10% HPGF-C18 or 20% FBS were added at concentrations of 1 × 104 and 4 × 104 cells/well. Culture plates were incubated for 6 days in a humidified 5% CO2 incubator at 37 °C. On the day of harvest, 0.5 μ Ci of 3H-thymidine (3H-TdR) was added to each well for 4 h with lymphocyte proliferation measured using a liquid scintillation counter. The effect of BMSCs on MLR was calculated as the percentage of the suppression compared with the proliferative response of the positive control without BMSC, where the positive control was set to 0% suppression. The experiments were performed three times for each variable described.
Microarray gene expression analysis
Total RNA was extracted using miRNeasy Mini Kit (Qiagen, Hilden, Germany) and assessed using Nano Drop 2000 (Thermo Scientific, Wilmington, DE). Microarray expression experiments were performed on 4 × 44 K Whole Human Genome Microarray (Agilent technologies, Santa Clara, CA, USA) according to our protocols [24]. Generally, 0.5 µg of BMSC RNA was labeled with Cyanine 5-CTP and Universal Human Reference RNA (Stratagene, Santa Clara, CA, USA) was labeled with Cyanine 3-CTP using a Quick Amp Labeling kit (Agilent). After purification, 825 ng of labeled cRNA from BMSC and reference RNA was pooled, fragmented and then hybridized on 4 × 44 K microarrays for 17 h at 65 °C. Images of the arrays were acquired using a microarray scanner G2505B (Agilent technologies) and image analysis was performed using feature extraction software version 9.5 (Agilent Technologies). The Agilent GE2-v5_95 protocol was applied using default settings.
RT-qPCR analysis of gene expression
For verification of the gene expression profiling results, RT-qPCR gene expression analysis was performed using custom made PCR arrays (Qiagen). The data analysis was conducted using the ΔΔCt method. HPRT1 was used as housekeeping gene and its Ct values were used to normalize the data. The normalized ΔCt for each gene of interest (GOI) was calculated by deducting the averaged Ct of HPRT1 from the Ct of each GOI: ΔCt = (CtGOI − CtHPRT1). The ΔΔCt for each GOI was calculated by deducting the average ΔCt of GOI in the HPGF group from the ΔCt of each GOI in the FBS group: ΔΔCt = average ΔCt (HPGF group)—average ΔCt (FBS group). The fold-change of each GOI in HPGF group compared to the FBS group was calculated as: fold-change = 2(−ΔΔCt).
MicroRNA expression analysis
The expression of MicroRNAs was measured by using the Human miRNome miScript miRNA PCR Array (Qiagen) following the manufacturer’s instructions. In brief, 1 µg of RNA was reverse-transcribed with miScript reverse transcription (RT) master mix for 60 min at 37 °C, then heated at 95 °C for 5 min and diluted with water. The diluted RT product was loaded into miRNA PCR Array for amplification using the following parameters, 95 °C for 1 min, then 40 cycles of amplification: 94 °C for 15 s, 55 °C for 30 s, 70 °C for 30 s, followed by the dissociation curve stage. The data analysis was conducted using ΔΔCt method. RNU6-2 was used to normalize the data. The normalized ΔCt for each miRNA of interest (MOI) was calculated by deducting the averaged Ct of RNU6-2 from the Ct of each MOI: ΔCt = (CtMOI − CtRNU6-2). The ΔΔCt for each MOI was calculated by deducting the average ΔCt of MOI in the HPGF group from the ΔCt of each MOI in the FBS group: ΔΔCt = average ΔCt (HPGF group)—average ΔCt (FBS group). The fold-change of each MOI compared to the FBS group was calculated as: fold-change = 2(−ΔΔCt).
Data processing and statistical analyses
Global gene expression analysis was performed according to a standard procedure. Raw data was uploaded into mAdb database (http://madb.nci.nih.gov/) and then imported into BRB-ArrayTools [25] (http://linus.nci.nih.gov/BRB-ArrayTools.html). Tests for differences between HPGF-C18 and FBS were conducted for individual genes using paired two-sided t tests, considering P values of < 0.001 as significant. In a class prediction model, genes significantly different between the classes (HPGF-C18 vs FBS) at 0.001 significance level were used for class prediction, and the leave-one-out cross-validation method was used to compute misclassification rate. The Benjamini and Hochberg method was used to estimate the false discovery rate. Gene set enrichment analysis (GSEA) was conducted by following the instructions of Broad Institute [26] (http://www.broadinstitute.org/gsea/index.jsp).
Results
Optimization of HPGF-C18 concentration
In order to determine the optimum concentration of HPGF-C18 to support the growth of BMSCs, cryopreserved passage 2 BMSCs prepared from 3 healthy subjects were thawed and cultured for 2 additional passages in αMEM supplemented with 5, 10, 15 and 20% HPGF-C18 and 20% FBS. For all 3 donors, BMSCs cultured in 10% HPGF-C18 exhibited similar growth rate to those cultured in 20% FBS, and faster than those cultured in 5, 15 and 20% HPGF, except that 15% HPGF-C18 was more supportive for BMSC growth from donor 09FC49 (Fig. 1a). In summary, the proliferative rate of BMSCs as measured by populations doublings (PD) per day was greatest in BMSCs grown with 10% HPGF-C18, which was similar to that of BMSCs cultured in 20% FBS (Fig. 1b).
Fig. 1.
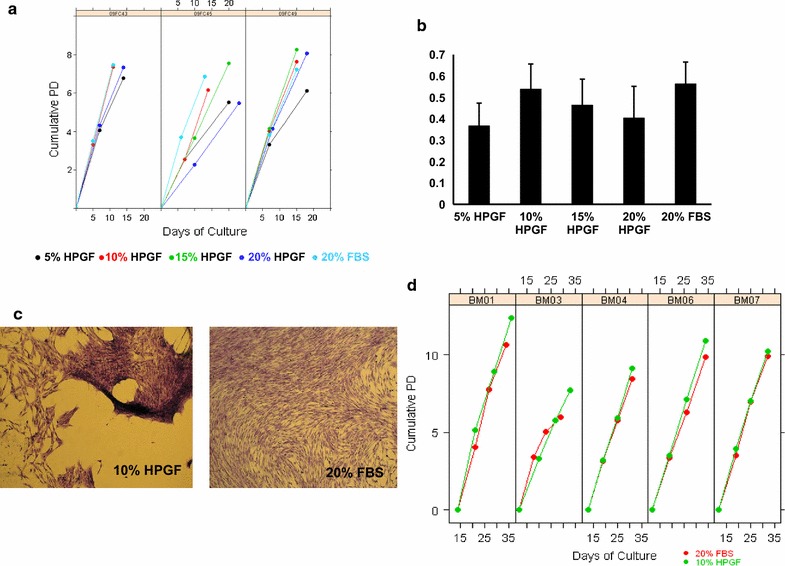
Proliferation of BMSCs in HPGF-C18 and FBS. Cryopreserved passage 2 BMSCs from 3 healthy subjects were thawed and cultured for 2 additional passages in αMEM supplemented with 5, 10, 15 and 20% HPGF-C18 and 20% FBS. The cumulative population doublings (PDs) at passages 3 and 4 are shown in a. The mean (± 1 SD) cumulative PDs normalized by total number of days in culture at passage 4 for each culture condition is show in b. Primary cultures of marrow aspirates cultured in αMEM supplemented with 10% HPGF-C18 showed irregular spacing and piling up of BMSCs while primary cultures of the same marrow aspirated cultured in 20% FBS formed more uniform monolayers (c). BMSCs from marrow aspirates of 5 healthy subjects were cultured for 4 passages in 10% HPGF-C18 and 20% FBS. The cumulative population doubling for BMSCs grown in 10% HPGF-C18 are shown in green and of 20% FBS in red. Each panel represents BMSCs isolated and expanded from one healthy subject. Each circle represents one BMSC passage (d)
The proportion of BMSCs expressing CD73, CD90, CD105, CD146 and CD166 was over 90% and similar for all culture conditions, while the expression of CD106 was variable. When the mean fluorescence intensity (MFI) was compared, CD73, CD90, CD105, CD146 and CD166 had similar expression levels in BMSCs cultured in HPGF-C18 and 20% FBS. The expression of CD105 was slightly brighter in BMSCs cultured in 20% FBS than those cultured in HPGF-C18; and that of CD106 was slightly dimmer in 20% FBS cultured BMSCs (data not shown). Since BMSCs cultured in media supplemented with 10% HPGF-C18 were most similar to those cultured in 20% FBS, all further studies were performed with media supplemented with 10% HPGF-C18.
Comparison of BMSCs Cultured in 20% FBS and 10% HPGF-C18
Primary BMSC Cultures
Marrow aspirates were collected from 5 healthy subjects and BMSCs were cultured for 4 passages in αMEM supplemented with 10% HPGF-C18 or 20% FBS. The primary BMSCs cultured with 10% HPGF-C18 and 20% FBS were harvested on the same day, ranging from day 12–14. Compared with BMSCs cultured in 20% FBS, the number of BMSCs harvested from 10% HPGF-C18 was similar from one subject, less from 2 subjects, and greater from 2 subjects (Table 1). It was noted that the morphology of the colonies grown in the two media supplements differed slightly. BMSCs cultured with 20% FBS formed uniform monolayers, while some colonies cultured with 10% HPGF-C18 were uneven and clumped (Fig. 1c). The media with HPGF-C18 was found to contain particles and the BMSCs appeared to clump around the particles. For the culture of the last two marrow aspirates, BM06 and BM07, the particles were allowed to settle and only the particle-reduced supernatant was used. This resulted in more uniform cell growth, but some clumping still occurred. However, this difference disappeared with cell passaging. The CFE was also evaluated for each aspirate in media supplemented with 10% HPGF-C18 and 20% FBS, and it was similar for the two media supplements for all 5 aspirates (Table 1).
Table 1.
Comparison of primary BMSCs cultured with 10% HPGF-C18 and 20% FBS from marrow aspirates of 5 healthy subjects
| Media supplement | Number of nucleated cell from marrow | Total number of BMSCs harvested | Days in culture | CFE per 1 × 105 cells | |
|---|---|---|---|---|---|
| BM01 | 10% HPGF-C18 | 10.0 × 106 | 1.20 × 105 | 14 | 2 |
| 20% FBS | 10.0 × 106 | 1.14 × 106 | 14 | 1 | |
| BM03 | 10% HPGF-C18 | 6.10 × 106 | 3.00 × 105 | 12 | 4a |
| 20% FBS | 6.10 × 106 | 4.20 × 105 | 12 | 3 | |
| BM04 | 10% HPGF-C18 | 55.2 × 106 | 7.8 × 105 | 13 | 5a |
| 20% FBS | 55.2 × 106 | 5.94 × 106 | 13 | 4 | |
| BM06b | 10% HPGF-C18 | 5.8 × 106 | 2.2 × 106 | 12 | 9a |
| 20% FBS | 5.8 × 106 | 3.78 × 106 | 12 | 11 | |
| BM07b | 10% HPGF-C18 | 21.7 × 106 | 2.4 × 106 | 12 | 30a |
| 20% FBS | 21.7 × 106 | 1.14 × 106 | 12 | 25 |
aColonies with atypical features
bParticles were allowed to settle for the preparation of HPGF-C18
BMSC Expansion from Passages 2 through 4
The proliferation rate of BMSCs cultured in 10% HPGF was similar to those cultured in 20% FBS but the accumulative population doublings was greater in media supplemented with 10% HPGF-C18 (Fig. 1d). The expression of stromal cell and hematopoietic cell surface markers were similar on passage 4 BMSCs cultured in 10% HPGF-C18 and 20% FBS; over 90% of cells expressed CD73, CD90, CD105, CD146 and CD166, while the expression of CD11b, CD14, CD19, CD34, CD45 and HLA-DR was almost undetectable in all BMSC samples. The expression of CD106 was higher in BMSCs cultured with 10% HPGF-C18 than those cultured with 20% FBS (Table 2).
Table 2.
The expression of surface markers by flow cytometry
| Donor | Supplement | CD90 | CD73 | CD105 | CD146 | CD166 | CD106 | CD11b | CD34 | CD45 | CD14 | HLA-DR | CD19 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BM01 | 10% HPGF | 99.5 | 99.2 | 99.4 | 99.1 | 99.6 | 14.4 | 0.6 | 2.1 | 0.9 | 0.9 | 0.9 | 0.6 |
| 20% FBS | 99.9 | 99.9 | 99.9 | 99.9 | 99.6 | 12.3 | 0.8 | 1.4 | 0.9 | 0.3 | 0.6 | 0.7 | |
| BM03 | 10% HPGF | 97.6 | 97.2 | 97 | 92.1 | 97.3 | 58.8 | 1 | 4 | 1 | 0.4 | 1.2 | 0.7 |
| 20% FBS | 99 | 99 | 98.9 | 98.5 | 98.9 | 18.3 | 1.1 | 1.3 | 1.1 | 0.3 | 1.3 | 1.3 | |
| BM04 | 10% HPGF | 99.3 | 99.2 | 99 | 98.6 | 99.1 | 49.5 | 1 | 5.7 | 1.2 | 1 | 1.2 | 0.9 |
| 20% FBS | 99.9 | 99.9 | 99.9 | 99.8 | 100 | 26.1 | 1.1 | 9.6 | 0.8 | 1 | 15.6 | 0.7 | |
| BM06 | 10% HPGF | 99.5 | 99.4 | 99.3 | 93.8 | 99.5 | 32.6 | 1.5 | 3.7 | 1.2 | 1.4 | 1.6 | 1.2 |
| 20% FBS | 100 | 100 | 100 | 99.4 | 100 | 8.5 | 1.4 | 2.9 | 1.2 | 1 | 1.2 | 1.5 | |
| BM07 | 10% HPGF | 99.3 | 99.2 | 99.3 | 98.8 | 99.3 | 47.5 | 1.2 | 2.1 | 1.3 | 0.9 | 1 | 0.8 |
| 20% FBS | 98.7 | 98.8 | 98.8 | 99 | 99.3 | 31.1 | 1.2 | 3.6 | 1 | 0.1 | 1.1 | 1.3 |
Suppression of MLRs by BMSCs
To evaluate the immunosuppressive activity of BMSCs cultured with the 2 media supplements, their ability to suppress MLRs was evaluated using two cell doses; 10,000 BMSCs per well and 40,000 BMSCs per well with BMSC: lymphocyte ratios of 1:10 and 1:2.5 respectively. At the 10,000 BMSCs per well dose level (BMSC: lymphocyte ratio is 1:10) BMSCs cultured in HPGF-C18 displayed lower immunosuppression activity than those cultured in FBS, but the difference is not statistically significant (FBS vs HPGF, 0.56 vs 0.49, p = 0.23). At the 40,000 BMSCs per well dose level (BMSC: lymphocyte ratio is 1:2.5) BMSCs cultured in HPGF-C18 displayed similar immunosuppression activity to those cultured in FBS (FBS vs HPGF, 0.62 vs 0.61, p = 0.90) (Fig. 2).
Fig. 2.
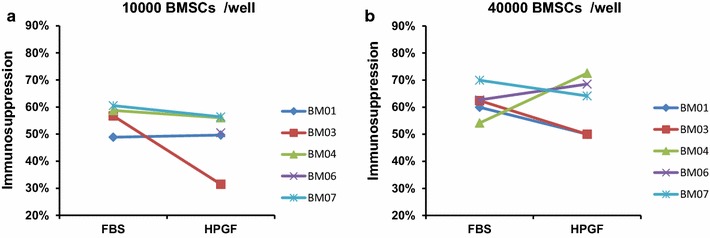
Inhibition of mixed lymphocyte reactions by BMSCs cultured with 10% HPGF-C18 and 20% FBS on the proliferation of mixed lymphocytes. BMSC suppression of mixed lymphocyte reactions was measured by H3-thymdine incorporation methods; the percent suppression was calculated by normalizing the values to the mixed lymphocyte reaction without BMSCs. Two doses of BMSCs were evaluated, 1 × 104 BMSCs per well (a, data for BM06 is missing due to technical issues) and 4 × 104 per well (b). There was no significant difference on their suppressive activity
Concentrations of Cytokines and Growth Factors in the Culture Supernatant
The concentrations of several cytokines and growth factors were measured in BMSC culture supernatants. The specific proteins that were evaluated were those related to BMSC immunosuppressive functions and other beneficial effects of BMSCs. Among these factors IL4, IL10, bFGF and ANG2 were not detected in either HPGF-C18 or FBS BMSC supernatants (data not shown). The levels of PEDF and TGFB1 were significantly increased (p < 0.05) in the supernatant of HPGF-C18-cultured BMSCs compared to FBS-cultured BMSCs, while IL6, VEGF, SDF1 and PLGF were significantly decreased (p < 0.05) (Fig. 3). The levels of other factors did not differ significantly including IL8 (FBS vs HPGF; 300.69 vs 216.54 pg/ml, p = 0.21); KGF (FBS vs HPGF; 124.66 vs 25.77 pg/ml, p = 0.11), LIF (FBS vs HPGF; 92.80 vs 100.34 pg/ml, p = 0.77), HGF (FBS vs HPGF; 49.75 vs 38.97 pg/ml, p = 0.50). Interestingly, endoglin (CD105), which is a BMSC surface marker, was detected in the supernatant from both culture conditions, and it was significantly increased in the supernatant of HPGF-C18-cultured BMSCs (FBS vs HPGF; 544.97 vs 2497.79 pg/ml, p = 0.0028).
Fig. 3.

Concentration of proteins in BMSC culture supernatants. The supernatants of BMSCs cultured in 10% HPGF-C18 or 20% FBS were collected and the concentrations of proteins were measured by SearchLight. The p-values were calculated by paired t-test
Global Gene Expression Analysis
Passage 3 and 4 BMSCs were analyzed by global gene expression analysis. Principal Component Analysis (PCA) revealed that the BMSCs cultured with 10% HPGF-C18 clustered separately from those cultured with 20% FBS (Fig. 4a) indicating that BMSCs cultured in the two media supplements represent two different cell types. We also used 6 class prediction methods to compare BMSCs cultured with 2 media supplements. All class prediction methods including Compound Covariate Predictor, Diagonal Linear Discriminant Analysis, 1-Nearest Neighbor, 3-Nearest Neighbors, Nearest Centroid, and Support Vector Machines separated the BMSCs cultured with 10% HPGF-C18 and 20% FBS with 100% accuracy providing further evidence that BMSCs cultured in the two media supplements represent two different cell types.
Fig. 4.
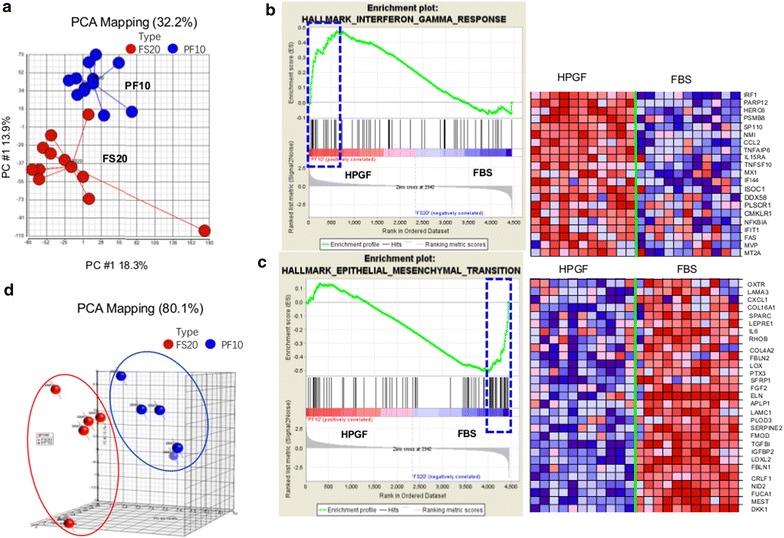
Global transcriptome and microRNA analysis of BMSCs cultured in 10% HPGF-C18 and 20% FBS. BMSCs were prepared from marrow aspirates of 5 healthy subjects and were cultured for 4 passages in 10% HPGF-C18 or 20% FBS. Passage 3 and 4 BMSCs from all 5 subjects were analyzed by global gene expression analysis. The results of the principal component analysis (PCA) analysis of the gene expression data are shown, the blue circles represent BMSCs cultured in 10% HPGF-C18 and the red circles BMSCs cultured in 20% FBS (a). Microarray data were analyzed using Gene Set Enrichment Analysis (GSEA) software to identify functionally related groups of genes (gene sets) with statistically significant enrichment. The enrichment plot for interferon gamma response and the 21 genes on the leading edge that are positively correlated with 10% HPGF-C18 (indicated by the blue rectangle) are shown in b. The enrichment plot for epithelial–mesenchymal transition and the 29 genes on the leading edge that are negatively correlated with HPGF-C18 culture (as indicated by the blue rectangle) were shown in c. In both b, c the plot on the left shows the distribution of genes in the set that are positively and negatively correlated with the HPGF-C18 phenotype. The plot on the right shows the relative gene expression (red = high, blue = low) for each gene for the indicated samples. Passage 4 BMCSs were also analyzed by microRNA PCR array, the PCA results on the microRNAs were shown, the blue circles represent BMSCs cultured in 10% HPGF-C18 and the red circles BMSCs cultured in 20% FBS (d)
We ran Gene Set Enrichment Analysis (GSEA) for functional annotation of the difference between HPGF-C18 and FBS cultured BMSCs. We found that distinct gene sets were enriched by the two cell types; specifically, 70 of 404 gene sets were upregulated by HPGF-C18-cultured BMSCs, while 12 gene sets were significantly enriched by FBS-cultured BMSC (FDR < 25%, Table 3). Interestingly, the gene sets enriched by HPGF-C18 were more likely to include cell proliferation and cell cycle related pathways. These results were consistent with our observed proliferation kinetics. A number of metabolic processing and biosynthetic pathways were also enriched by HPGF-C18 such as Cholesterol homeostasis and Lipid Biosynthetic process. Several signaling pathways such as MTORC1 Signaling and Neurotrophin Signaling pathways were also enriched by HPGF-C18 (Table 3). Moreover, a few immune response gene sets were enriched in HPGF-C18-cultured BMSCs; for example, Interferon-Gamma Response was positively correlated with HPGF-18 culture (Fig. 4b). Among the genes that contributed most to the enrichment of the Interferon-Gamma Response pathway were: CCL2, TNFAIP6, TNFSF10, NFKBIA and FAS suggesting that the immune modulatory functions of HPGF-C18 cultured BMSCs may different from those cultured with FBS. The gene sets enriched in FBS-cultured BMSC included canonical BMSC related signaling pathways, such as Activation of MAPK activity and TGF-beta signaling pathway as well as cell adhesion and ECM pathways including Cell Adhesion Molecules—CAMs and ECM-Receptor Interaction. This implies that lower expression of cell adhesion molecules by HPGF-C18-cultured BMSCs may account for the abnormal “clumping” phenomena. Another interesting gene set enriched by FBS was Epithelial-Mesenchymal transition; the genes contributing most to the enrichment of this pathway included CXCL1, SPARC, IL6, SFRP1, FGF2, TGFBI and DKK1 (Fig. 4c), all of which are representative markers of BMSCs cultured in FBS. Clearly, the results of GSEA analysis indicated that the BMSCs cultured with HPGF-C18 and FBS have different biological activities.
Table 3.
Gene set enrichment analysis (GSEA) on the global genes
| NAME | SIZE | ES | NES | FDR |
|---|---|---|---|---|
| Enriched gene sets in HPGF-C18 group (FDR < 0.25) | ||||
| HALLMARK_CHOLESTEROL_HOMEOSTASIS | 24 | 0.87 | 2.21 | 0.00 |
| HALLMARK_MTORC1_SIGNALING | 59 | 0.60 | 1.99 | 0.02 |
| HALLMARK_ANDROGEN_RESPONSE | 20 | 0.56 | 1.87 | 0.02 |
| KEGG_NEUROTROPHIN_SIGNALING_PATHWAY | 26 | 0.53 | 1.86 | 0.02 |
| HALLMARK_MITOTIC_SPINDLE | 49 | 0.71 | 1.88 | 0.02 |
| RESPONSE_TO_DNA_DAMAGE_STIMULUS | 38 | 0.69 | 1.89 | 0.02 |
| KEGG_P53_SIGNALING_PATHWAY | 20 | 0.69 | 1.89 | 0.02 |
| LIPID_BIOSYNTHETIC_PROCESS | 32 | 0.52 | 1.90 | 0.03 |
| NUCLEOCYTOPLASMIC_TRANSPORT | 20 | 0.69 | 1.83 | 0.03 |
| KEGG_PROGESTERONE_MEDIATED_OOCYTE_MATURATION | 25 | 0.72 | 1.91 | 0.03 |
| HALLMARK_INTERFERON_ALPHA_RESPONSE | 31 | 0.59 | 1.81 | 0.03 |
| M_PHASE | 38 | 0.72 | 1.74 | 0.04 |
| DNA_METABOLIC_PROCESS | 65 | 0.59 | 1.75 | 0.04 |
| CELL_CYCLE_PROCESS | 62 | 0.69 | 1.75 | 0.04 |
| HALLMARK_ADIPOGENESIS | 39 | 0.47 | 1.73 | 0.04 |
| DNA_REPAIR | 26 | 0.72 | 1.75 | 0.04 |
| CELL_CYCLE_GO_0007049 | 95 | 0.57 | 1.77 | 0.04 |
| HALLMARK_APOPTOSIS | 57 | 0.44 | 1.73 | 0.04 |
| DNA_RECOMBINATION | 15 | 0.80 | 1.75 | 0.04 |
| CELL_CYCLE_PHASE | 56 | 0.67 | 1.76 | 0.04 |
| HALLMARK_INTERFERON_GAMMA_RESPONSE | 63 | 0.48 | 1.77 | 0.05 |
| CHROMOSOME_ORGANIZATION_AND_BIOGENESIS | 24 | 0.63 | 1.78 | 0.05 |
| NUCLEAR_TRANSPORT | 21 | 0.66 | 1.77 | 0.05 |
| RESPONSE_TO_ENDOGENOUS_STIMULUS | 50 | 0.55 | 1.71 | 0.05 |
| KEGG_CELL_CYCLE | 45 | 0.73 | 1.70 | 0.05 |
| MITOTIC_CELL_CYCLE | 50 | 0.69 | 1.71 | 0.05 |
| BIOCARTA_HIVNEF_PATHWAY | 17 | 0.59 | 1.70 | 0.05 |
| MICROTUBULE_BASED_PROCESS | 25 | 0.62 | 1.70 | 0.05 |
| HALLMARK_DNA_REPAIR | 22 | 0.66 | 1.69 | 0.06 |
| CELL_CYCLE_CHECKPOINT_GO_0000075 | 20 | 0.74 | 1.68 | 0.06 |
| ESTABLISHMENT_OF_CELLULAR_LOCALIZATION | 71 | 0.42 | 1.66 | 0.07 |
| KEGG_OOCYTE_MEIOSIS | 31 | 0.56 | 1.66 | 0.07 |
| GAMETE_GENERATION | 37 | 0.47 | 1.66 | 0.07 |
| PROTEIN_TARGETING | 22 | 0.59 | 1.64 | 0.07 |
| ESTABLISHMENT_OF_PROTEIN_LOCALIZATION | 40 | 0.47 | 1.63 | 0.07 |
| KEGG_APOPTOSIS | 21 | 0.53 | 1.64 | 0.07 |
| HALLMARK_G2M_CHECKPOINT | 68 | 0.78 | 1.64 | 0.07 |
| M_PHASE_OF_MITOTIC_CELL_CYCLE | 26 | 0.75 | 1.64 | 0.07 |
| MACROMOLECULE_LOCALIZATION | 47 | 0.47 | 1.64 | 0.07 |
| ORGANELLE_ORGANIZATION_AND_BIOGENESIS | 118 | 0.41 | 1.62 | 0.07 |
| REGULATION_OF_CELL_CYCLE | 57 | 0.53 | 1.62 | 0.07 |
| MITOSIS | 26 | 0.75 | 1.64 | 0.07 |
| CELLULAR_LOCALIZATION | 74 | 0.41 | 1.61 | 0.08 |
| DNA_DEPENDENT_DNA_REPLICATION | 15 | 0.70 | 1.61 | 0.08 |
| PROTEIN_TRANSPORT | 35 | 0.47 | 1.60 | 0.08 |
| CYTOSKELETON_ORGANIZATION_AND_BIOGENESIS | 71 | 0.40 | 1.58 | 0.10 |
| DNA_REPLICATION | 26 | 0.63 | 1.58 | 0.10 |
| KEGG_PEROXISOME | 21 | 0.46 | 1.57 | 0.10 |
| CELL_MIGRATION | 26 | 0.43 | 1.56 | 0.11 |
| HALLMARK_ESTROGEN_RESPONSE_EARLY | 63 | 0.40 | 1.56 | 0.11 |
| PROTEIN_LOCALIZATION | 42 | 0.46 | 1.56 | 0.11 |
| DETECTION_OF_STIMULUS | 19 | 0.55 | 1.55 | 0.11 |
| HALLMARK_IL2_STAT5_SIGNALING | 59 | 0.37 | 1.54 | 0.12 |
| INTRACELLULAR_PROTEIN_TRANSPORT | 30 | 0.49 | 1.53 | 0.12 |
| INTERPHASE | 22 | 0.60 | 1.53 | 0.12 |
| CELL_PROLIFERATION_GO_0008283 | 185 | 0.34 | 1.53 | 0.12 |
| HALLMARK_E2F_TARGETS | 60 | 0.72 | 1.52 | 0.13 |
| INTERPHASE_OF_MITOTIC_CELL_CYCLE | 19 | 0.61 | 1.51 | 0.13 |
| SEXUAL_REPRODUCTION | 48 | 0.46 | 1.51 | 0.13 |
| ORGAN_MORPHOGENESIS | 54 | 0.37 | 1.50 | 0.14 |
| ANATOMICAL_STRUCTURE_MORPHOGENESIS | 118 | 0.33 | 1.50 | 0.14 |
| HALLMARK_UV_RESPONSE_UP | 49 | 0.37 | 1.49 | 0.15 |
| KEGG_PYRIMIDINE_METABOLISM | 22 | 0.55 | 1.48 | 0.16 |
| NUCLEOBASENUCLEOSIDENUCLEOTIDE_AND_NUCLEIC_ACID_METABOLIC_PROCESS | 263 | 0.34 | 1.48 | 0.16 |
| RESPONSE_TO_STRESS | 140 | 0.32 | 1.46 | 0.17 |
| TRANSLATION | 30 | 0.42 | 1.46 | 0.17 |
| HALLMARK_SPERMATOGENESIS | 47 | 0.45 | 1.45 | 0.18 |
| ACTIN_CYTOSKELETON_ORGANIZATION_AND_BIOGENESIS | 38 | 0.36 | 1.45 | 0.18 |
| KEGG_BASAL_CELL_CARCINOMA | 16 | 0.47 | 1.41 | 0.22 |
| HALLMARK_TNFA_SIGNALING_VIA_NFKB | 69 | 0.38 | 1.39 | 0.24 |
| Enriched gene sets in FBS group (FDR < 0.25) | ||||
| HALLMARK_GLYCOLYSIS | 54 | 0.56 | 1.97 | 0.02 |
| ACTIVATION_OF_MAPK_ACTIVITY | 15 | 0.65 | 1.73 | 0.14 |
| POSITIVE_REGULATION_OF_MAP_KINASE_ACTIVITY | 17 | 0.63 | 1.72 | 0.14 |
| KEGG_CELL_ADHESION_MOLECULES_CAMS | 48 | 0.49 | 1.68 | 0.15 |
| KEGG_ECM_RECEPTOR_INTERACTION | 33 | 0.54 | 1.74 | 0.15 |
| KEGG_FOCAL_ADHESION | 61 | 0.43 | 1.66 | 0.16 |
| HALLMARK_HYPOXIA | 66 | 0.51 | 1.68 | 0.16 |
| KEGG_TGF_BETA_SIGNALING_PATHWAY | 25 | 0.55 | 1.68 | 0.17 |
| EXTRACELLULAR_STRUCTURE_ORGANIZATION_AND_BIOGENESIS | 16 | 0.73 | 1.74 | 0.18 |
| FEMALE_PREGNANCY | 21 | 0.59 | 1.75 | 0.23 |
| HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION | 79 | 0.51 | 1.78 | 0.23 |
| TRANSMEMBRANE_RECEPTOR_PROTEIN_SERINE_THREONINE_KINASE_SIGNALING_PATHWAY | 18 | 0.54 | 1.60 | 0.25 |
ES enrichment score, NES normalized enrichment score, FDR false discovery rate
RT-qPCR Analysis on Differentially Expressed Genes
We also used RT-qPCR to compare the expression of 82 genes among HPGF-C18- and FBS-cultured BMSCs; and the expression 52 genes differed significantly (p < 0.05). Among these genes 22 were up-regulated by HPGF-C18 culture while 30 were up-regulated by FBS-culture (Table 4). Consistent with our microarray data, the expression of a few of the BMSC function related genes did not differ significantly, including CXCL12, IGF1, IL6, IL10, RUNX2 and TGFB1. However, the expression of some genes differed by microarray analysis but did not differ significantly by RT-qPCR analysis including EDN1, FGF2, HGF and VEGFA. Interestingly, TNFSF10 which was confirmed to be up-regulated by HPGF-C18 was expressed by MSC spheres when cultured in chemically defined xeno-free media [27]. Another gene, MMP1, that was up-regulated by HPGF-C18 is critical to MSC migration [28]. Among the genes confirmed to be up-regulated by FBS; HOXA3, FOXF2 and BEX2 were transcription factors; PTGS2, PTGES and PTGIS were immune response modulatory genes; DKK1, SFRP1, and BMP6 were involved in Wnt signaling pathway.
Table 4.
qRT-PCR analysis of differentially expressed genes
| Genes | p value | FDR | FS20/PF10 fold change |
|---|---|---|---|
| PDPN | 0.00 | 0.00 | − 28.57 |
| TNFSF10 | 0.00 | 0.00 | − 27.78 |
| TLR5 | 0.00 | 0.00 | − 13.89 |
| MMP1 | 0.00 | 0.00 | − 12.66 |
| BMPR1B | 0.00 | 0.00 | − 12.05 |
| TP63 | 0.02 | 0.04 | − 11.11 |
| CTSC | 0.00 | 0.00 | − 9.09 |
| INSIG1 | 0.00 | 0.00 | − 8.33 |
| CTSK | 0.00 | 0.00 | − 8.33 |
| EGR2 | 0.00 | 0.00 | − 7.69 |
| KLF8 | 0.00 | 0.00 | − 6.25 |
| PLXDC1 | 0.00 | 0.01 | − 5.56 |
| COMP | 0.00 | 0.01 | − 5.56 |
| IL16 | 0.01 | 0.03 | − 5.56 |
| ROBO4 | 0.01 | 0.03 | − 5.26 |
| TGFB3 | 0.00 | 0.01 | − 4.55 |
| IL27RA | 0.00 | 0.01 | − 4.35 |
| CHRD | 0.01 | 0.02 | − 3.70 |
| WNT7B | 0.02 | 0.04 | − 3.70 |
| MMP13 | 0.06 | 0.10 | − 3.03 |
| TNFAIP6 | 0.02 | 0.04 | − 2.63 |
| SMAD1 | 0.04 | 0.08 | − 2.44 |
| SRGN | 0.04 | 0.08 | 2.13 |
| TGFBI | 0.05 | 0.09 | 2.32 |
| IL6 | 0.04 | 0.08 | 2.54 |
| HMOX1 | 0.05 | 0.09 | 2.62 |
| CD40 | 0.03 | 0.06 | 2.88 |
| LTBP1 | 0.04 | 0.08 | 3.36 |
| HOXA3 | 0.01 | 0.03 | 3.53 |
| ALDH1A3 | 0.02 | 0.04 | 3.56 |
| SMAD6 | 0.03 | 0.06 | 3.72 |
| STC2 | 0.01 | 0.03 | 3.74 |
| ID2 | 0.00 | 0.01 | 3.86 |
| MFGE8 | 0.01 | 0.02 | 3.94 |
| CCL26 | 0.00 | 0.01 | 4.32 |
| DKK1 | 0.00 | 0.01 | 4.51 |
| HAS2 | 0.03 | 0.05 | 5.48 |
| PTGS2 | 0.00 | 0.00 | 5.66 |
| ID4 | 0.00 | 0.00 | 6.63 |
| BMP6 | 0.00 | 0.00 | 6.96 |
| TGFB2 | 0.00 | 0.00 | 7.21 |
| KRT18 | 0.00 | 0.00 | 7.96 |
| IL1B | 0.00 | 0.01 | 8.44 |
| SFRP1 | 0.00 | 0.00 | 8.64 |
| LEPR | 0.00 | 0.00 | 14.92 |
| PGF | 0.00 | 0.00 | 19.59 |
| SERPINE2 | 0.00 | 0.01 | 20.77 |
| HAS3 | 0.00 | 0.01 | 21.80 |
| SERPING1 | 0.00 | 0.01 | 32.69 |
| PTGES | 0.00 | 0.00 | 42.11 |
| PTGIS | 0.00 | 0.00 | 53.63 |
| BEX2 | 0.00 | 0.00 | 53.81 |
Expression of MicroRNAs
We also evaluated the expression of microRNAs using RT-PCR assays. After Quality Control, 256 microRNAs passed the selection criteria and were used for further analysis. Unsupervised clustering using the 256 microRNAs clearly separated all the BMSCs into 2 clusters, one with all the BMSCs cultured with FBS and the other with BMSCs cultured in HPGF-C18 (Fig. 4d), again suggesting that the BMSCs cultured with two media supplements have distinct biological properties. A total of 44 microRNAs were significantly changed (p < 0.05). Among these 44 microRNAs 22 were up-regulated by HPGF-C18 and 22 were up-regulated by FBS (Table 5). Interestingly, microRNAs with the highest changes were related to the osteogenesis of BMSC, including has-miR-146a, has-miR-135b, miR-378, miR-335-5p and miR-210. Hsa-miR-146a regulates the expression of JMJD3 and RUNX2 and thus affects osteogenic differentiation [29]. Hsa-miR-135b is abnormally up-regulated in MSCs from multiple myeloma patients, and it negatively regulates MSCs osteogenesis [30] and the over-expression of hsa-miR-135b results in decreased mineralization [31]. Over-expression of miR-378 attenuates high glucose-suppressed osteogenic differentiation through the targeting CASP3 and activating the PI3 K/Akt pathway [32]. The functional roles of miR-335-5p are, however, controversial. For example, it may activate Wnt signaling and promote osteogenic differentiation by downregulating DKK1 [33], but its over-expression may inhibit the osteogenic and adipogeneic potential of MSC. In addition, miR-335 may also directly target RUNX2 [34] or increase the expression of chondrogenic marker genes [35]. Over-expression of miR-210 significantly reduces MSC apoptosis under oxidative stress, increases cell viability and superoxide dismutase activity [36].
Table 5.
microRNA identified by RT-qPCR
| miRNA | p value | FDR | FS20/PF10 fold change |
|---|---|---|---|
| hsa-miR-146a | 0.00 | 0.07 | − 7.69 |
| hsa-miR-135b | 0.00 | 0.07 | − 7.69 |
| hsa-miR-378 | 0.00 | 0.07 | − 4.35 |
| hsa-miR-146b-5p | 0.00 | 0.07 | − 4.00 |
| hsa-miR-33a | 0.02 | 0.15 | − 3.57 |
| hsa-miR-126* | 0.02 | 0.15 | − 3.13 |
| hsa-miR-218 | 0.01 | 0.13 | − 3.03 |
| hsa-miR-135a | 0.00 | 0.07 | − 2.78 |
| hsa-miR-542-5p | 0.01 | 0.09 | − 2.50 |
| hsa-miR-15b* | 0.01 | 0.09 | − 2.00 |
| hsa-miR-142-3p | 0.00 | 0.07 | − 1.96 |
| hsa-miR-450a | 0.01 | 0.11 | − 1.89 |
| hsa-miR-29b | 0.00 | 0.07 | − 1.79 |
| hsa-miR-424 | 0.00 | 0.07 | − 1.67 |
| hsa-miR-149 | 0.01 | 0.13 | − 1.64 |
| hsa-miR-132 | 0.03 | 0.17 | − 1.61 |
| hsa-miR-20a | 0.02 | 0.14 | − 1.59 |
| hsa-miR-455-5p | 0.01 | 0.13 | − 1.43 |
| hsa-miR-484 | 0.01 | 0.08 | − 1.41 |
| hsa-miR-576-5p | 0.02 | 0.14 | − 1.33 |
| hsa-miR-15a | 0.03 | 0.18 | − 1.18 |
| hsa-miR-28-5p | 0.00 | 0.07 | 1.19 |
| hsa-let-7i* | 0.04 | 0.24 | 1.69 |
| hsa-miR-493 | 0.00 | 0.07 | 1.76 |
| hsa-miR-485-3p | 0.02 | 0.15 | 1.77 |
| hsa-let-7i | 0.02 | 0.15 | 1.80 |
| hsa-miR-380 | 0.01 | 0.14 | 1.82 |
| hsa-miR-337-3p | 0.01 | 0.11 | 1.83 |
| hsa-miR-127-3p | 0.01 | 0.09 | 1.86 |
| hsa-miR-495 | 0.03 | 0.17 | 1.90 |
| hsa-miR-299-3p | 0.00 | 0.07 | 2.04 |
| hsa-miR-431 | 0.02 | 0.15 | 2.04 |
| hsa-miR-411 | 0.00 | 0.07 | 2.09 |
| hsa-miR-1 | 0.02 | 0.14 | 2.14 |
| hsa-miR-487a | 0.01 | 0.13 | 2.21 |
| hsa-miR-376c | 0.02 | 0.14 | 2.21 |
| hsa-miR-487b | 0.00 | 0.07 | 2.23 |
| hsa-miR-329 | 0.01 | 0.14 | 2.30 |
| hsa-miR-127-5p | 0.00 | 0.07 | 2.62 |
| hsa-miR-379 | 0.04 | 0.24 | 2.76 |
| hsa-miR-210 | 0.02 | 0.14 | 3.04 |
| hsa-miR-432 | 0.02 | 0.15 | 3.46 |
| hsa-miR-452 | 0.03 | 0.21 | 3.79 |
| hsa-miR-335 | 0.00 | 0.07 | 12.76 |
Asterick (*) is a part of the microRNA name, indicating the microRNA arises from the 3′ arm of a hairpin
Discussion
In this study we compared BMSCs cultured from marrow aspirates in media supplemented with HPGF-C18 and FBS. Using the same methods that we used to produce clinical grade BMSCs we found that HPGF-C18 supported the growth of BMSCs from marrow aspirates. BMSC proliferation, CFE, cell surface marker expression and immunosuppression activities were similar among BMSCs grown in HPGF-C18 and FBS. These results are similar to another study which found that HPGF-C18 supported the growth of adipose tissue derived MSCs and surface makers expression was similar for adipose tissue-derived MSC cultured in HPGF-C18 and FBS [23].
BMSCs cultured in HPGF-C18 and FBS were, however, not identical. Differences were found in gene expression profiles, microRNA profiles and cytokines/growth factor concentrations. The most striking differences in gene expression were in pathways involved with cell proliferation, metabolic pathway, immune response, MAPK signaling pathway, TGF-β signaling pathway and cell adhesion pathway. Since many clinical applications of BMSCs involve immune modulation, we evaluated the immune modulatory functions of BMSCs cultured in HPGF-C18 and FBS. We found that BMSCs grown in HPGF-C18 and FBS suppressed lymphocyte proliferation to a similar extent. We then assessed the expression of genes that are involved in the immune modulation using RT-qPCR and found that the expression of many BMSC function related genes did not differ significantly including CXCL12 (SDF1), IGF1, IL6, IL10, RUNX2 and TGFB1. However, the culture supernatant protein levels of SDF1, IL6, TGFB1 and VEGFA differed significantly among HPGF-C18- and FBS-cultured BMSCs. This discrepancy may be due to the post-transcriptional regulation of these genes or technical differences in the two platforms that were used for the analysis. While our results indicate that there are functional differences between BMSCs cultured in HPGF-C18 and FBS, it is not certain if these differences are clinically relevant. Further comparison studies that make use of application specific function assays and models that reflect BMSC potency should be performed prior to substituting HPGF-C18 for FBS for the manufacture of clinical BMSC products.
While the proliferation of BMSCs was similar when grown in media supplemented with 10% HPGF and 20% FBS, we noticed that the BMSCs cultured with HPGF-C18 displayed different morphologies and formed abnormal colonies. The growth of BMSCs in FBS resulted in uniform colonies and monolayer, while growth in HPGF-C18 resulted in some areas where no cells grew and other areas where cells grew in multiple layers. The HPGF-C18 contained some particles and we hypothesized that the irregularities might be due partially to cell attachment to these particles. For the last 2 marrow aspirates we allowed HPGF-C18 to settle and used the supernatant for making cell culture medium. This removed some but not all of the particles. While this reduced the number of areas of irregular growth, they were not completely eliminated and it is possible that complete removal of particles by filtration may further improve BMSC growth. Other mechanisms may also account for the irregularities, differences in BMSC adhesion and mobility may have caused or contributed to the abnormal BMSC growth, for example, HPGF-C18 cultured BMSCs expressed lower levels of cell adhesion molecules such as LAMA3, COL4A4, ITGA8, COL4A2, LAMB2, COL11A2, THBS3, LAMC1 and ITGA8; and expressed high level of motility related gene MMP1. Therefore, it will be interesting to investigate the cell adhesion and motility regulation activities of BMSCs cultured in HPGF-C18 and FBS.
We also noticed differences in the expression of genes belonging to metabolism pathways, and this may be due to differences in the concentrations of growth factors in the two media supplements. Compared to FBS, HPGF-C18 has greater concentrations of Leptin, adiponectin, and immune related proteins, such as EGF, MIP-1, PDGF-BB and CCL1 [23].
Platelet growth factor preparations provide an alternative source of the limited reagent FBS since the growth of cellular therapies will soon result in a shortage of FBS [37]. Besides, there are some advantages in regard to the use of HPGF-C18 rather than FBS; the major advantage is that the use of platelet factor preparations avoids exposure of the recipient of the BMSC products to xenogenic proteins and xenogenic infection.
There are some important differences between HPGF-C18 and other media supplements made from platelets. A relatively large number of apheresis platelet components are used to manufacture each lot of HPGF, approximately 40–50; more than can be easily made in-house by cell processing facilities located in academic health care centers. The use of this relatively large number of apheresis components ensures that the variability among lots is kept to a minimum. In addition, the solvent-detergent treatment used in the manufacture of HPGF-C18 inactivates pathogens. The apheresis platelets used to make platelet-derived media supplements are collected from healthy donors who meet all blood donor healthy history screening and transfusion transmitted disease marker criteria and consequently the possibility that they would transmit a pathogen is low. Solvent-detergent treatment of the pooled apheresis platelets reduces the risk of transmission of a lipid-enveloped virus and inactivates some bacteria. In addition, the final HPGF-C18 product is passed through a sterilizing filter [21], all these measures were taken to make HPGF-C18 a safe alternative to FBS. There may be also functional differences between BMSCs cultured in media supplemented with HPGF-C18 and other platelet-derived media supplements. The quantities of factors released from platelets by freeze/thaw and other methods used to lyse platelets may differ from those released by solvent-detergent treatment. In addition, the process of treating pooled platelet products with solvent-detergent and removing the solvent-detergent by oil extraction and chromatography can also result in the loss of some factors. For example the recovery of TGF-b1, EGF and IGF is over 90% during the hydrophobic interactive chromatography, but the recovery of PDGF-AA, PDGF-AB and PDGF and VEGF was less than 40% [38].
Conclusions
We found that BMSCs from marrow aspirates could be grown in HPGF-C18 supplemented culture medium; when BMSCs cultured in HPGF-C18 were compared to those cultured in FBS using traditional assays such as proliferation, surface marker expression and inhibition of MLRs, the two types of BMSCs appeared very similar. However, a more comprehensive analysis revealed significant differences at the gene and microRNA expression level and in the composition of the cell supernatants. If HPGF-C18 is to be used in place of FBS for BMSC culture, comparison of BMSCs grown in the two types of media supplements using application specific functional assays that reflect BMSC potency will be necessary.
Authors’ contributions
JR, DW, PJ, MS, PGR and DFS designed the studies. DW, SC, KT, RJ and PJ preformed assays and analyzed the data. JR, DW, PJ, PGR and DFS prepared the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets generated for analysis during the current study are available in Gene Expression Omnibus (GEO) under accession number GSE111495.
Consent for publication
Not applicable.
Ethics approval and consent to participate
These studies were approved by the NHLBI Institutional review board and all subjects who donated cells for manufacturing BMSCs gave informed consent.
Funding
This research was supported by the Intramural Research Programs of the NIH Clinical Center and National Institute of Dental and Craniofacial Research.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- BMSCs
bone marrow stromal cells
- FBS
fetal bovine serum
- CFE
colony forming efficiency
- MLR
mixed lymphocyte culture
- GSEA
gene set enrichment analysis
- GVHD
graft versus host disease
- HPGF-C18
human platelet growth factor C-18
- GMP
good manufacturing practices
- SOP
standard operating procedure
- PDs
population doublings
- PCA
principal component analysis
Footnotes
Jiaqiang Ren and Dawn Ward contribute equally to this work
Contributor Information
Jiaqiang Ren, Email: Jiaqiang.ren@cellbiomedgroup.com.
Dawn Ward, Email: DWard@mednet.ucla.edu.
Steven Chen, Email: stchen17@gmail.com.
Katherine Tran, Email: katherine3tran@gmail.com.
Ping Jin, Email: PJin@mail.cc.nih.gov.
Marianna Sabatino, Email: msabatino@kitepharma.com.
Pamela G. Robey, Email: probey@dir.nidcr.nih.gov
David F. Stroncek, Phone: 301-402-3314, Email: dstroncek@cc.nih.gov
References
- 1.Kuznetsov SA, Mankani MH, Robey PG. In vivo formation of bone and haematopoietic territories by transplanted human bone marrow stromal cells generated in medium with and without osteogenic supplements. J Tissue Eng Regen Med. 2013;7(3):226–235. doi: 10.1002/term.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuznetsov SA, et al. Long-term stable canine mandibular augmentation using autologous bone marrow stromal cells and hydroxyapatite/tricalcium phosphate. Biomaterials. 2008;29(31):4211–4216. doi: 10.1016/j.biomaterials.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mankani MH, et al. Canine cranial reconstruction using autologous bone marrow stromal cells. Am J Pathol. 2006;168(2):542–550. doi: 10.2353/ajpath.2006.050407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yin F, et al. Bone marrow mesenchymal stromal cells to treat tissue damage in allogeneic stem cell transplant recipients: correlation of biological markers with clinical responses. Stem Cells. 2014;32(5):1278–1288. doi: 10.1002/stem.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duijvestein M, et al. Autologous bone marrow-derived mesenchymal stromal cell treatment for refractory luminal Crohn’s disease: results of a phase I study. Gut. 2010;59(12):1662–1669. doi: 10.1136/gut.2010.215152. [DOI] [PubMed] [Google Scholar]
- 6.Ciccocioppo R, et al. Autologous bone marrow-derived mesenchymal stromal cells in the treatment of fistulising Crohn’s disease. Gut. 2011;60(6):788–798. doi: 10.1136/gut.2010.214841. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Y, et al. Direct injection of autologous mesenchymal stromal cells improves myocardial function. Biochem Biophys Res Commun. 2009;390(3):902–907. doi: 10.1016/j.bbrc.2009.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson JG, et al. Mesenchymal stem (stromal) cells for treatment of ARDS: a phase 1 clinical trial. Lancet Respir Med. 2015;3(1):24–32. doi: 10.1016/S2213-2600(14)70291-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Menge T, et al. Mesenchymal stem cells regulate blood-brain barrier integrity through TIMP3 release after traumatic brain injury. Sci Transl Med. 2012;4(161):161ra150. doi: 10.1126/scitranslmed.3004660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedenstein AJ, Chailakhjan RK, Lalykina KS. The development of fibroblast colonies in monolayer cultures of guinea-pig bone marrow and spleen cells. Cell Tissue Kinet. 1970;3(4):393–403. doi: 10.1111/j.1365-2184.1970.tb00347.x. [DOI] [PubMed] [Google Scholar]
- 11.Selvaggi TA, Walker RE, Fleisher TA. Development of antibodies to fetal calf serum with arthus-like reactions in human immunodeficiency virus-infected patients given syngeneic lymphocyte infusions. Blood. 1997;89(3):776–779. [PubMed] [Google Scholar]
- 12.Macy E, et al. Anaphylaxis to infusion of autologous bone marrow: an apparent reaction to self, mediated by IgE antibody to bovine serum albumin. J Allergy Clin Immunol. 1989;83(5):871–875. doi: 10.1016/0091-6749(89)90099-7. [DOI] [PubMed] [Google Scholar]
- 13.Mackensen A, et al. Presence of IgE antibodies to bovine serum albumin in a patient developing anaphylaxis after vaccination with human peptide-pulsed dendritic cells. Cancer Immunol Immunother. 2000;49(3):152–156. doi: 10.1007/s002620050614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jonsdottir-Buch SM, Lieder R, Sigurjonsson OE. Platelet lysates produced from expired platelet concentrates support growth and osteogenic differentiation of mesenchymal stem cells. PLoS ONE. 2013;8(7):e68984. doi: 10.1371/journal.pone.0068984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffiths S, et al. Human platelet lysate stimulates high-passage and senescent human multipotent mesenchymal stromal cell growth and rejuvenation in vitro. Cytotherapy. 2013;15(12):1469–1483. doi: 10.1016/j.jcyt.2013.05.020. [DOI] [PubMed] [Google Scholar]
- 16.Ben Azouna N, et al. Phenotypical and functional characteristics of mesenchymal stem cells from bone marrow: comparison of culture using different media supplemented with human platelet lysate or fetal bovine serum. Stem Cell Res Ther. 2012;3(1):6. doi: 10.1186/scrt97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bieback K, et al. Human alternatives to fetal bovine serum for the expansion of mesenchymal stromal cells from bone marrow. Stem Cells. 2009;27(9):2331–2341. doi: 10.1002/stem.139. [DOI] [PubMed] [Google Scholar]
- 18.Schallmoser K, et al. Human platelet lysate can replace fetal bovine serum for clinical-scale expansion of functional mesenchymal stromal cells. Transfusion. 2007;47(8):1436–1446. doi: 10.1111/j.1537-2995.2007.01220.x. [DOI] [PubMed] [Google Scholar]
- 19.Lange C, et al. Accelerated and safe expansion of human mesenchymal stromal cells in animal serum-free medium for transplantation and regenerative medicine. J Cell Physiol. 2007;213(1):18–26. doi: 10.1002/jcp.21081. [DOI] [PubMed] [Google Scholar]
- 20.Bieback K, et al. Altered gene expression in human adipose stem cells cultured with fetal bovine serum compared to human supplements. Tissue Eng Part A. 2010;16(11):3467–3484. doi: 10.1089/ten.tea.2009.0727. [DOI] [PubMed] [Google Scholar]
- 21.Chou ML, et al. Impact of solvent/detergent treatment of plasma on transfusion-relevant bacteria. Vox Sang. 2012;102(4):277–284. doi: 10.1111/j.1423-0410.2011.01560.x. [DOI] [PubMed] [Google Scholar]
- 22.Sabatino M, et al. The establishment of a bank of stored clinical bone marrow stromal cell products. J Transl Med. 2012;10:23. doi: 10.1186/1479-5876-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shih DT, et al. Expansion of adipose tissue mesenchymal stromal progenitors in serum-free medium supplemented with virally inactivated allogeneic human platelet lysate. Transfusion. 2011;51(4):770–778. doi: 10.1111/j.1537-2995.2010.02915.x. [DOI] [PubMed] [Google Scholar]
- 24.Ren J, et al. Intra-subject variability in human bone marrow stromal cell (BMSC) replicative senescence: molecular changes associated with BMSC senescence. Stem Cell Res. 2013;11(3):1060–1073. doi: 10.1016/j.scr.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon R, et al. Analysis of gene expression data using BRB-array tools. Cancer Inform. 2007;3:11–17. doi: 10.1177/117693510700300022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ylostalo JH, et al. Unique characteristics of human mesenchymal stromal/progenitor cells pre-activated in 3-dimensional cultures under different conditions. Cytotherapy. 2014;16(11):1486–1500. doi: 10.1016/j.jcyt.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho IA, et al. Matrix metalloproteinase 1 is necessary for the migration of human bone marrow-derived mesenchymal stem cells toward human glioma. Stem Cells. 2009;27(6):1366–1375. doi: 10.1002/stem.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huszar JM, Payne CJ. MIR146A inhibits JMJD3 expression and osteogenic differentiation in human mesenchymal stem cells. FEBS Lett. 2014;588(9):1850–1856. doi: 10.1016/j.febslet.2014.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu S, et al. Upregulation of miR-135b is involved in the impaired osteogenic differentiation of mesenchymal stem cells derived from multiple myeloma patients. PLoS ONE. 2013;8(11):e79752. doi: 10.1371/journal.pone.0079752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaap-Oziemlak AM, et al. MicroRNA hsa-miR-135b regulates mineralization in osteogenic differentiation of human unrestricted somatic stem cells. Stem Cells Dev. 2010;19(6):877–885. doi: 10.1089/scd.2009.0112. [DOI] [PubMed] [Google Scholar]
- 32.You L, et al. MiR-378 overexpression attenuates high glucose-suppressed osteogenic differentiation through targeting CASP3 and activating PI3K/Akt signaling pathway. Int J Clin Exp Pathol. 2014;7(10):7249–7261. [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, et al. Effects of miR-335-5p in modulating osteogenic differentiation by specifically downregulating Wnt antagonist DKK1. J Bone Miner Res. 2011;26(8):1953–1963. doi: 10.1002/jbmr.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tome M, et al. miR-335 orchestrates cell proliferation, migration and differentiation in human mesenchymal stem cells. Cell Death Differ. 2011;18(6):985–995. doi: 10.1038/cdd.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin X, et al. MiR-335-5p promotes chondrogenesis in mouse mesenchymal stem cells and is regulated through two positive feedback loops. J Bone Miner Res. 2014;29(7):1575–1585. doi: 10.1002/jbmr.2163. [DOI] [PubMed] [Google Scholar]
- 36.Xu J, et al. miR-210 over-expression enhances mesenchymal stem cell survival in an oxidative stress environment through antioxidation and c-Met pathway activation. Sci China Life Sci. 2014;57(10):989–997. doi: 10.1007/s11427-014-4725-z. [DOI] [PubMed] [Google Scholar]
- 37.Brindley DA, et al. Peak serum: implications of serum supply for cell therapy manufacturing. Regen Med. 2012;7(1):7–13. doi: 10.2217/rme.11.112. [DOI] [PubMed] [Google Scholar]
- 38.Su CY, et al. A virally inactivated functional growth factor preparation from human platelet concentrates. Vox Sang. 2009;97(2):119–128. doi: 10.1111/j.1423-0410.2009.01180.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated for analysis during the current study are available in Gene Expression Omnibus (GEO) under accession number GSE111495.


