Abstract
Metabotropic glutamate 1 (mGlu) receptor has been proposed as a target for the treatment of metastatic melanoma. Studies have demonstrated that inhibiting the release of glutamate (the natural ligand of mGlu1 receptors), results in a decrease of melanoma tumor growth in mGlu1 receptor-expressing melanomas. Here, we demonstrate that mGlu1 receptors, which have been previously characterized as oncogenes, also behave like dependence receptors by creating a dependence on glutamate for sustained cell viability. In the mGlu1 receptor-expressing melanoma cell lines, SK-MEL-2 (SK2) and SK-MEL-5 (SK5), we show that glutamate is both necessary and sufficient to maintain cell viability, regardless of underlying genetic mutations. Addition of glutamate increased DNA synthesis while removal of glutamate not only suppressed DNA synthesis, but also promoted cell death in SK2 and SK5 melanoma cells. Using genetic and pharmacological inhibitors we established that this effect of glutamate is mediated by activation of mGlu1 receptors. The stimulatory potential of mGlu1 receptors was further confirmed in vivo in a melanoma cell xenograft model. In this model, subcutaneous injection of SK5 cells with shRNA targeted downregulation of mGlu1 receptors resulted in a decrease in the rate of tumor growth relative to control. We also demonstrate for the first time, that a selective mGlu1 receptor antagonist, JNJ16259685 [(3,4-Dihydro-2H-pyrano[2,3-b]quinolin-7-yl)-(cis-4-methoxycyclohexyl)-methanone], slows SK2 and SK5 melanoma tumor growth in vivo. Taken together, these data suggest that pharmacological inhibition of mGlu1 receptors may be a novel approach for the treatment of metastatic melanoma.
Keywords: melanoma, metabotropic glutamate receptor 1, dependence receptor, glutamate, JNJ16259685
Introduction
Melanoma accounts for less than 5% of skin cancers, but is responsible for the majority of skin cancer-related deaths1. The 5-year survival rate is 98% for localized melanomas, but if diagnosed once distant metastases have occurred, the survival rate decreases to 15%1. This aggressive disease is often resistant to conventional therapy2,3, and as a result, there is an urgent need for the development of better therapies.
Previous studies have identified metabotropic glutamate 1 (mGlu1) receptor as a potentially novel target for the treatment of metastatic melanoma4–11 . mGlu1 receptors belong to a family of G-protein coupled receptors (GPCRs), which includes eight subtypes categorized into three groups based on sequence homology and pharmacology12,13. A role for mGlu receptors has been suggested in cancers in addition to melanoma14, including triple negative breast cancer15,16, colorectal adenocarcinoma17, glioblastoma18, astrocytoma18, medulloblastoma18, and oral squamous cell carcinoma19. All mGlu receptor subtypes are activated by glutamate, which is released at elevated concentrations by many cancer cells4,20–23. This increase in glutamate release may act in an autocrine loop4,22–23, involving mGlu1 receptors, which can promote the growth4,23 and invasion22 of tumors.
Glutamate release inhibitors have been used to selectively slow the growth of melanomas expressing mGlu1 receptors4,7,10,11, suggesting that there may be an important role played by glutamate in melanoma cell proliferation. However, a direct link between glutamate availability and the status of mGlu1 receptor expression has not been shown. In the present study, we show that the expression of mGlu1 receptors in SK-MEL-2 (SK2) and SK-MEK-5 (SK5) melanoma cells confers a dependence on glutamate for sustained viability and DNA synthesis, regardless of the presence of other common mutations. In addition, both targeted knockdown of mGlu1 receptors using shRNA and pharmacological inhibition with a non-competitive mGlu1 receptor antagonist, selectively inhibited viability of melanoma cells expressing mGlu1 receptors in vitro. In a xenograft model, SK5 melanoma cells with down-regulated mGlu1 receptor expression grew slower than native SK5 melanoma cells or those with a scrambled shRNA control plasmid. Finally, we showed for the first time that an mGlu1 receptor antagonist decreased the rate of tumor growth in two xenograft mouse models of melanoma. From these studies, we conclude that mGlu1 receptors are a plausible therapeutic target for the treatment of metastatic melanoma.
Results
Glutamate is necessary and sufficient to sustain the viability of melanoma cells that express mGlu1 receptors
There are increasing data suggesting that glutamate and mGlu1 receptors may play a role in the growth and proliferation of melanoma cells4–11. However, due to the heterogeneity of melanoma, further studies are necessary to determine if there is a direct link between glutamate-stimulated proliferation and mGlu1 receptor protein expression. In the present study, SK2 and SK5 cells were used to determine if the presence of glutamate is necessary and/or sufficient to maintain the viability of melanoma cells expressing mGlu1 receptors. These specific cell lines were selected because SK2 cells have an NRAS mutation11,23, while SK5 cells contain a BRAF mutation11,24, both common mutations that might alter the effects of mGlu1 receptors on viability and growth.
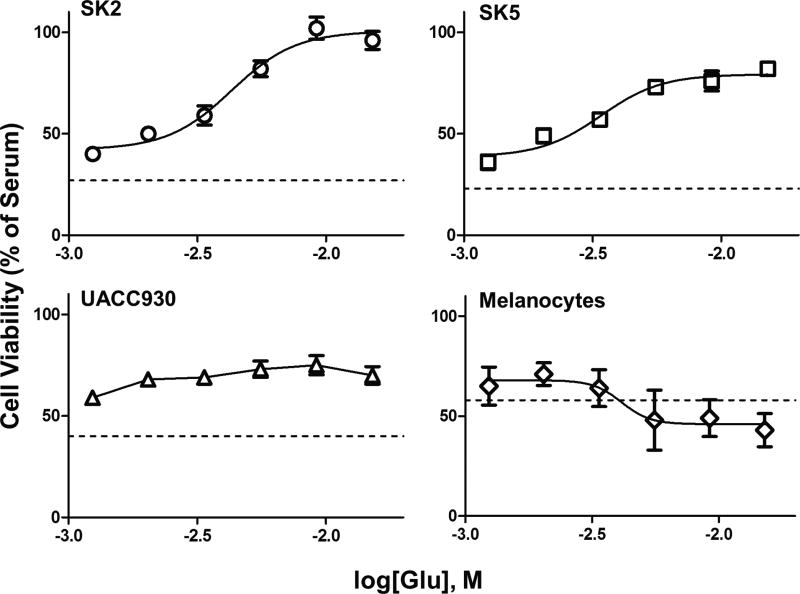
To begin assessing whether depletion of glutamate affects melanoma cell growth, the cell lines were transferred to a medium containing dialyzed serum, which lacks all low molecular weight (MW) compounds (MW<10,000) including glutamate. Under these conditions SK2 and SK5 melanoma cell viability decreased by 74% and 77%, respectively, as compared with growth in full serum (Fig. 1). Replacement of glutamate increased cell viability of SK2 (EC50=4.3mM) and SK5 (EC50=3.4mM) human melanoma cells (both of which express mGlu1 receptors11,23,25) in a dose-dependent manner (Fig. 1). In contrast, the effect of glutamate on UACC930 melanoma cell viability (which do not express mGlu1 receptors4,11,23) was ambiguous and did not fit to a dose-response curve (Fig. 1). Human melanocytes, which also lack expression of mGlu1 receptors 4–6,26,27 were tested, and in those cells high concentrations of glutamate not only failed to sustain cell viability, but instead, induced cellular toxicity (EC50=4.1mM) (Fig. 1).
Figure 1. Glutamate dose-dependently increases cell viability of melanoma cells expressing mGlu1 receptors.
A, Dose-response curves of glutamate-induced cell viability in SK2 cells, SK5 cells, UACC930 cells, and melanocytes. All values are normalized to full serum (set at 100% cell viability). Dashed line represents growth in dialyzed serum lacking glutamate. Addition of glutamate to dialyzed serum dose-dependently increased cell viability (as measured by MTT assay) after 7 days, in SK2 (EC50=4.3mM) and SK5 cells (EC50=3.4mM) as calculated by the four-parameter logistic equation in GraphPad Prism. The four-parameter equation did not fit the UACC930 data. In melanocytes, glutamate decreased cell viability (EC50=4.1mM). All data points are means from at least two independent experiments performed in triplicate with error bars representing standard error of mean (SEM).
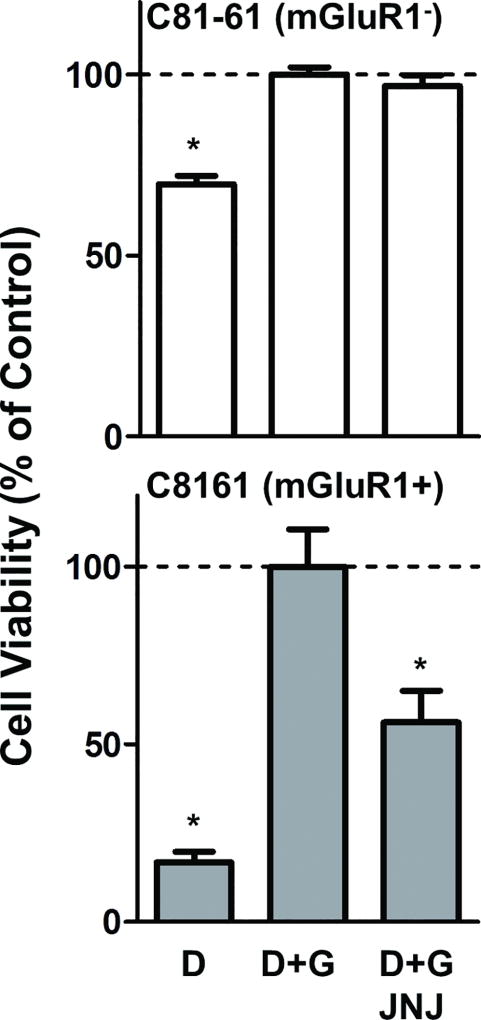
The effect of glutamate depletion and addition on cell viability was also tested in two melanoma cell lines that originated from the same patient, but differ in their level of expression of mGlu1 receptors: C81-61 and C816128. Like SK2 and SK5 cells, C8161 melanoma cells express mGlu1 receptors; however, C8161 cells are wild type for BRAF and NRAS. C8161 cell viability was reduced 83% in dialyzed serum relative to dialyzed serum containing 10mM glutamate (Fig. 6 bottom). The effect of glutamate depletion was less pronounced in C81-61 melanoma cells, which lack mGlu1 receptors. In these cells, viability was only reduced 30% in dialyzed serum relative to dialyzed serum containing 10mM glutamate (Fig. 6 top).
Figure 6. Glutamate withdrawal and pharmacological antagonism preferentially inhibits viability in mGlu1 receptor expressing melanoma cells.
Viability of C81-61 (no mGluR1) and C8161 (mGluR1+) melanoma cells in dialyzed serum (D), dialyzed serum containing 10mM glutamate (D+G), and dialyzed serum containing 10mM glutamate and 40 µM JNJ16259685 (D+G+JNJ). Values were normalized to dialyzed serum containing 10mM glutamate (dashed line- set at 100% cell viability). All data points are means from at least two independent experiments performed in triplicate with error bars representing SEM. *p<0.01 compared to dialyzed serum containing 10mM glutamate as measured by one-way ANOVA and Bonferroni post-hoc test.
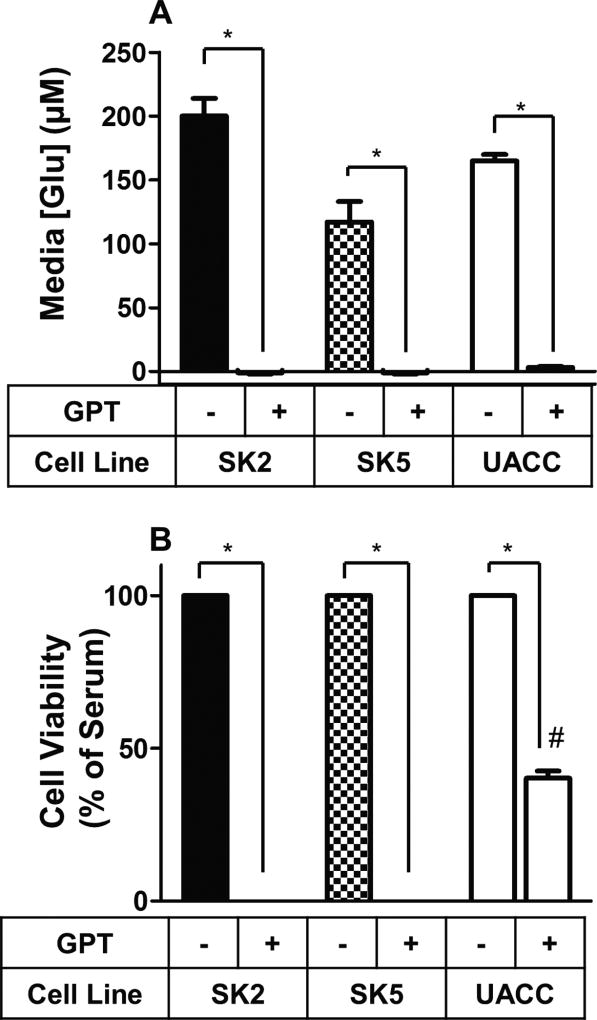
Because dialyzed serum lacks many factors in addition to glutamate, we assessed whether selective enzymatic depletion of glutamate using glutamate pyruvate transaminase (GPT)29 decreased melanoma cell viability. The levels of glutamate were measured by the Amplex Red Glutamic Acid/Glutamate Oxidase Assay Kit in cultures of SK2, SK5, and UACC930 melanoma cells in the presence and absence of GPT. As shown in Fig. 2A, GPT effectively removed all measurable traces of glutamate from SK2, SK5, and UACC930 growth media. Consistent with the results using dialyzed serum, which showed that melanoma cells expressing mGlu1 receptors are more sensitive to glutamate, this selective removal of glutamate significantly decreased SK2 and SK5 melanoma cell viability to approximately zero, as measured by MTT assay (Fig 2B). In contrast, GPT only reduced the mGlu1 receptor negative melanoma cell line (UACC930) to 40% relative to full serum (Fig. 2B). Taken together, the glutamate removal assays (either using dialyzed serum or full serum with GPT) suggest that melanoma cells that express mGlu1 receptors become dependent on glutamate and as a result exhibit increased sensitivity to both its withdrawal and addition.
Figure 2. Glutamate is necessary for SK2 and SK5 melanoma cell viability.
A, Glutamate pyruvate transaminase (GPT) treatment for 7 days completely depleted glutamate from serum-containing media in cultures of SK2, SK5, and UACC930 melanoma cells. B, Glutamate removal decreased SK2, SK5, and UACC930 cell viability at 7 days after GPT treatment as measured by an MTT assay. Values were normalized to full serum without GPT treatment (set at 100% cell viability) for each cell line. All data points are means from at least two independent experiments performed in triplicate with error bars representing SEM. [Glu], glutamate concentration, *p<0.01 assessed by 2-sample t-test. #p<0.01 (different from 0) assessed by 1-sample t-test.
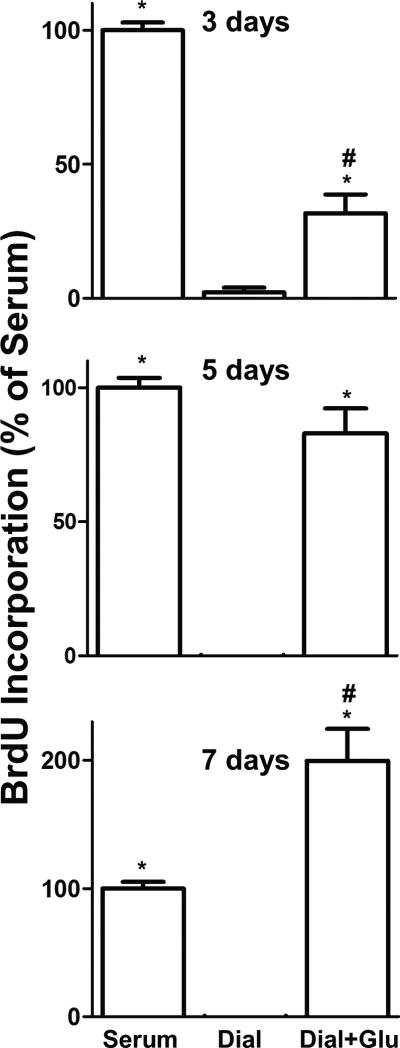
Therefore, to determine how glutamate affects the proliferation index of melanoma cells, we performed a 5-bromo-2'-deoxyuridine (BrdU) incorporation assay in the presence and absence of glutamate. In the BrdU assay, cells were incubated with BrdU, which is incorporated instead of thymidine into newly synthesized DNA. The amount of BrdU incorporation over time, which is an indicator of the rate of DNA synthesis, was measured by chemiluminescence. SK5 cell BrdU incorporation in dialyzed serum, which lacks glutamate, was significantly reduced relative to full serum at 3, 5, and 7 days post-treatment (Fig. 3). Addition of 10mM glutamate to dialyzed serum resulted in an increase in DNA synthesis relative to dialyzed serum alone, which became more pronounced over time (Fig. 3).
Figure 3. Addition of glutamate to dialyzed serum increases DNA synthesis in SK5 melanoma cells.
BrdU incorporation is reduced in dialyzed serum (Dial) relative to full serum, and is stimulated by the addition of 10mM glutamate (Dial+Glu) at 3, 5, and 7 days post-treatment. Values were normalized to full serum (set at 100% cell viability). All data points are means from at least two independent experiments performed in triplicate with error bars representing SEM. *p<0.01 compared to dialyzed serum and #p<0.01 as compared to serum as measured by one-way ANOVA and Bonferroni post-hoc test.
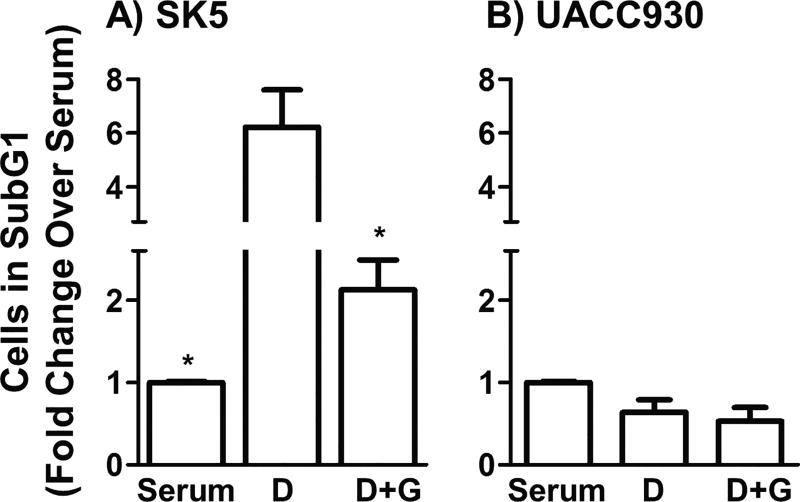
Using flow cytometry we next assessed the effects of glutamate depletion on apoptosis in SK5 and UACC930 melanoma cells. SubG1 analysis using ethanol fixation and propidium iodide was used to estimate the fraction of apoptotic cells, identified by increases in DNA fragmentation and decreased DNA content30,31. Incubation of SK5 melanoma cells in dialyzed serum led to an increase in the fraction of cells undergoing apoptosis, relative to full serum, and the addition of 10mM glutamate to dialyzed serum reversed this effect (Fig. 4A). In contrast, in UACC930 melanoma cells (which lack mGlu1 receptors) neither incubation in dialyzed serum nor the addition of glutamate to dialyzed serum had an effect on apoptosis (Fig. 4B). These results are fully consistent with the hypothesis that mGlu1 receptor positive melanoma cells become reliant on glutamate for survival and that increased levels of glutamate can promote proliferation by stimulating DNA synthesis.
Figure 4. Addition of glutamate to dialyzed serum suppresses apoptosis in SK5 melanoma cells.
Effect of dialyzed serum (D) and dialyzed serum containing 10mM glutamate (D+G) for 5 to 7 days on the fraction of apoptotic cells in the subG1 phase of the cell cycle. All values are means from at least two independent experiments with at least 20 000 cells per experiment with error bars representing SEM, and are expressed as fold changes of the fraction of cells in SubG1 relative to full serum. p<0.01 as measured by one-way ANOVA for SK5 melanoma cells, but not UACC930 cells. *p<0.01 as compared to dialyzed serum by Bonferroni post-hoc test.
Glutamate activation of mGlu1 receptors is required for sustained SK5 melanoma cell viability in vitro
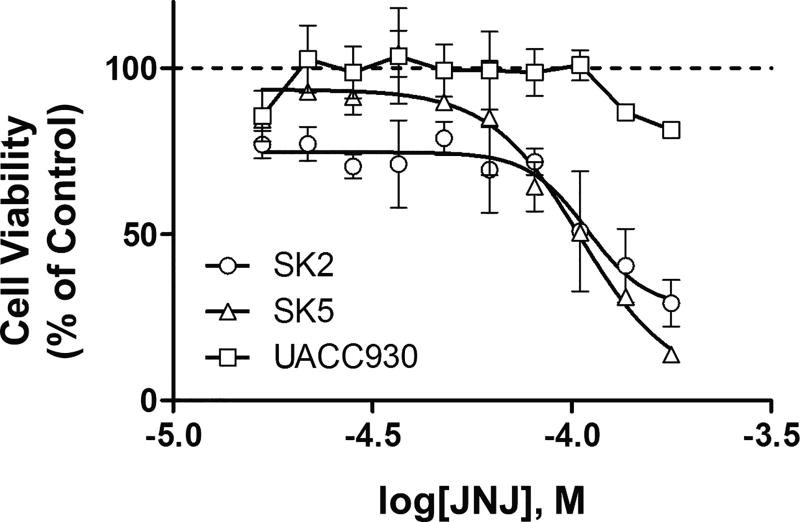
Since glutamate dose-dependently increased viability only in melanoma cells expressing mGlu1 receptors, we next tested whether or not an mGlu1 receptor antagonist could inhibit this effect. JNJ16259685 [(3,4-Dihydro-2H-pyrano[2,3-b]quinolin-7-yl)-(cis-4-methoxycyclohexyl)-methanone], a selective32 non-competitive32 mGlu1 receptor antagonist, that to our knowledge has never been used in cancer cells, dose-dependently decreased cell viability in response to stimulation with glutamate in SK2 (IC50=109µM) and SK5 (IC50=105µM) melanoma cells. Identical concentrations of the antagonist only partially inhibited the viability of UACC930 melanoma cells (Fig. 5). At the highest concentration tested (178 µM), JNJ16259685 decreased cell viability of SK2, SK5, and UACC930 cells by 71%, 86%, and 19% respectively, relative to viability in dialyzed serum containing 10 mM glutamate. This suggests that JNJ16259685 is decreasing SK2 and SK5 melanoma cell viability primarily through blockade of mGlu1 receptors. To confirm these results, experiments were repeated using two melanoma cells lines originating from the same patient: C81-61 (no mGlu1 receptors) and C8161 (moderate mGlu1 receptor expression)28. Treatment of these cell lines with JNJ16259685 significantly inhibited C8161 melanoma cell viability, without affecting C81-61 melanoma cell viability (Fig. 6). These results demonstrate that JNJ16259685 effectively decreases melanoma cell viability in vitro by selective inhibition of mGlu1 receptors.
Figure 5. Pharmacological inhibition of mGlu1 receptors in vitro, effectively decreases viability of melanoma cells that express mGlu1 receptors.
A, Dose-response curves of mGlu1 receptor antagonist, JNJ16259685 in dialyzed serum in the presence of 10mM glutamate in SK2, SK5 and UACC930 melanoma cells. JNJ16259685 significantly decreased cell viability in SK2 (IC50=109µM) and SK5 cells (IC50=105µM) (p<0.01), but not in UACC930 cells (p=0.31) relative to dialyzed serum containing 10mM of glutamate and 1% DMSO (set at 100% viability). All data points are means from at least three experiments performed in triplicate with error bars representing SEM. Statistical significance was assessed using linear regression to determine if the slope was different from zero.
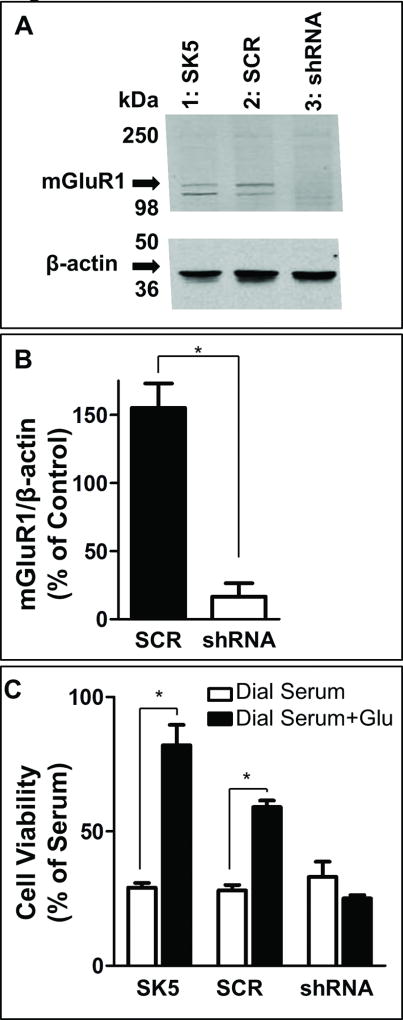
To further confirm the role of mGlu1 receptor activation in maintaining melanoma cell viability, we specifically downregulated mGlu1 receptor expression in SK5 cells using shRNA (SK5-shRNA-mGluR1). shRNA targeted to mGlu1 receptors effectively decreased mGlu1 receptor protein expression (Fig. 7A), and quantification from two independent experiments performed in duplicate (Fig. 7B) confirmed a significant reduction in mGlu1 receptor protein by 83% in SK5-shRNA-mGluR1 cells relative to native SK5 cells. The scrambled shRNA control plasmid (SK5-SCR-shRNA) did not reduce mGlu1 receptor protein levels. Moreover, shRNA targeted to mGlu1 receptors, but not scrambled shRNA, abolished the stimulatory effects of glutamate on SK5 melanoma cell viability (Fig. 7C). These data are consistent with the pharmacological inhibition experiments performed with JNJ16259685 and further indicate that mGlu1 receptor positive cells are highly dependent on both the expression of mGlu1 receptors and glutamate for proliferation.
Figure 7. Knockdown of mGlu1 receptors abolishes the ability of glutamate to stimulate SK5 melanoma cell growth in vitro.
A, Representative Western blot of mGlu1 receptor expression (130kDa), with β-actin as a loading control (48kDa), in native SK5 cells (lane 1), SK5 cells transfected with a scrambled shRNA control plasmid (SCR) (lane 2), and SK5 cells transfected with a combination of shRNA plasmids targeted to mGlu1 receptors (shRNA) (lane 3). Images were cropped to improve clarity. B, Quantification of mGlu1 receptor protein, expressed as a percent of mGlu1 receptor protein present in native SK5 cells, from 2 independent experiments performed in duplicate with error bars representing SEM. C, Viability of native SK5 cells, SK5 scrambled shRNA (SCR) cells, and SK5 shRNA (shRNA) cells after a 7 day incubation in the absence or presence of 25mM glutamate. Data points are means from at least three independent experiments with error bars representing SEM. *p<0.01, as assessed by 2 sample t-test.
mGlu1 receptor activity is required for melanoma growth in vivo
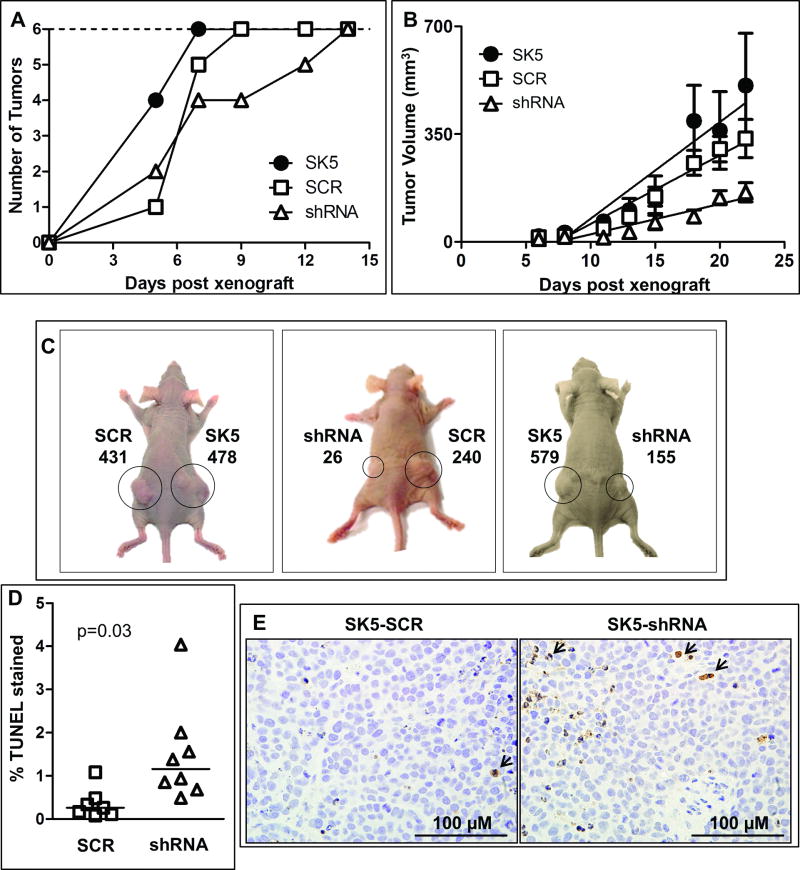
To test whether mGlu1 receptor activity was important for melanoma cell growth in vivo, a xenograft mouse model was used whereby athymic nude mice were injected subcutaneously in the flanks with either native SK5 cells, SK5-shRNA-mGluR1 cells, or SK5-SCR-shRNA cells. The time to tumor formation was assessed by manual palpation and tumor growth was quantified by caliper measurements (n=6 tumors). Tumor initiation was delayed for the SK5-shRNA-mGluR1 cells as it took 14 days for all of the tumors to become palpable, versus 7 days for native SK5 cells and 9 days for SK5-SCR-shRNA cells (Fig. 8A). Furthermore, SK5-shRNA-mGluR1 tumors grew significantly slower than untransfected SK5 tumors and SK5-SCR-shRNA tumors, while the rate of growth between native SK5 and SK5-SCR-shRNA tumors was not significantly different (Fig. 8B). Representative photographs of final tumors volumes (22 days post-injection) are shown in Fig. 8C. dUTP nick end labeling (TUNEL) staining of histologic sections of the tumor masses revealed a significant increase in apoptosis in SK5-shRNA-mGluR1 tumors versus SK5-SCR tumors (Fig. 8D-E). Taken together, these results demonstrate that knockdown of mGlu1 receptors decreases the rate of SK5 tumor initiation (Fig. 8A) and SK5 tumor growth (Fig. 8B), which can be explained, at least partially through an increase in apoptosis (Fig 8D-E).
Figure 8. Knockdown of mGlu1 receptors suppresses SK5 melanoma tumor growth in vivo.
A, Tumor formation measured by palpation was delayed after knockdown of mGlu1 receptors. All melanoma cell xenografts (n=6) of SK5, SK5-SCR-shRNA (SCR) or SK5-shRNA-mGluR1 (shRNA) cells formed tumors 7, 9 or 14 days after melanoma cell injection, respectively. B, Rate of tumor growth, as calculated by linear regression, is significantly decreased in SK5 cells with downregulated mGlu1 receptors (shRNA) relative to native SK5 cells and SK5 cells transfected with a scrambled shRNA control plasmid (SCR). P-values on the slope were SK5 vs. SCR (p=0.14), SK5 vs. shRNA (p<0.001), and SCR vs. shRNA (p<0.0001). Average tumor volumes on final day of the experiment (day 22) were 508, 336, and 161 mm3 in SK5, SCR, and shRNA tumors, respectively. Data points represent average tumor volumes with error bars representing SEM (n=6 tumors). C, Representative photographs of mice (day 22) with tumors labeled with cell-type and volume of tumor (mm3). D, Quantification of TUNEL staining from SK5-SCR-shRNA (SCR) and SK5-shRNA-mGluR1 (shRNA) tumor tissue at final day (day 22) of the experiment. Data points represent average percentage of TUNEL stained cells at 60X magnification of at least 400 cells. Horizontal lines represent median percentage of TUNEL staining, which is increased in SK5-shRNA-mGluR1 (shRNA) tumors relative to SK5-SCR-shRNA (SCR) tumors (p=0.03), as measured by 2-sample t-test. E, Representative photographs of TUNEL stained tissue with arrows indicating examples of apoptotic, TUNEL stained cells.
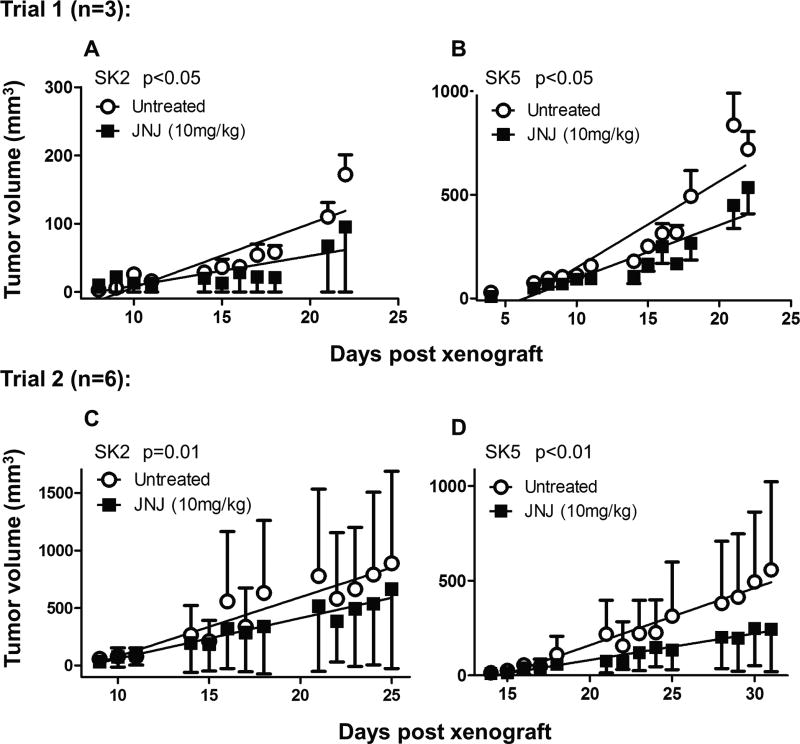
Next, we tested whether pharmacological inhibition of these receptors using the non-competitive mGlu1 receptor antagonist JNJ16259685 would decrease the rate of SK2 and SK5 tumor growth in mice. The first trial (n=3) showed a statistically significant decrease in the rate of SK2 (Fig. 9A) and SK-5 (Fig. 9B) tumor growth in the 10mg/kg JNJ16259685 treated group relative to the DMSO control group. We repeated this study with additional animals (n=6) and JNJ16259685 again significantly decreased the rate of SK2 (Fig. 9C) and SK5 (Fig. 9D) tumor growth. These in vivo results identify mGlu1 receptors as a possible pharmacological target for the treatment of metastatic melanoma.
Figure 9. Pharmacological inhibition of mGlu1 receptors significantly decreases SK2 and SK5 melanoma tumor growth in mice.
JNJ16259685 (10mg/kg dissolved in DMSO and diluted in PBS) or vehicle control (DMSO in PBS) was subcutaneously injected into the flanks of athymic mice and tumors were measured 5 times/week. JNJ16259685 treated mice had a significantly decreased rate of tumor growth relative to vehicle treatment, as calculated by linear regression, in trial 1 (n=3 tumors) in (A) SK2 cells (p<0.05) and (B) SK5 cells (p<0.05) and in Trial 2 (n=6 tumors) in (C) SK2 cells (p=0.01) and (D) SK5 cells (p<0.0001). R2 values: Trial 1; SK2: untreated=0.78, 10mg/kg JNJ=0.60. SK5: untreated=0.83, 10mg/kg JNJ=0.82. Trial 2; SK2: untreated=0.89, 10 mg/kg JNJ=0.94. SK5: untreated=0.96, 10mg/kg JNJ=0.96. Data points represent average tumor volumes with error bars representing SEM.
4. Discussion
Deregulation of glutamate signaling has been implicated in the growth and invasion of various cancers33,34. This deregulation can manifest as an elevation in glutamate release, which is seen in melanomas4,6,35, gliomas20–22,36, breast cancers34, and prostate cancers34,37 or through genetic alterations of glutamate receptors as seen in melanomas5,38–39 and colorectal carcinomas17. In this study we determined that sustained glutamate signaling is required for the maintenance of mGlu1 receptor positive human melanoma cell viability in vitro, as selective glutamate depletion severely decreased cell viability and addition of glutamate dose-dependently restored this suppression. Additionally, reduction of mGlu1 receptor signaling in vivo reduced melanoma xenograft growth, demonstrating potential clinical significance of these results.
mGlu1 receptors have been shown to play a role in both the transformation of melanocytes to melanoma and the proliferation of melanomas in vitro and in vivo4,6,9. Human melanocytes and benign nevi do not express mGlu1 receptors, however mGlu1 receptors are expressed in 80% of human melanoma cell lines and 65% of primary to metastatic biopsies9. The genetic and epigenetic mechanisms that suppress mGlu1 receptor expression in melanocytes can be deregulated in melanomas, leading to the expression of mGlu1 receptors26. This regulation appears to have a direct impact on tumorigenesis, as transfection with mGlu1 receptor cDNA was shown to transform mouse melanocytes into melanoma cells, characterized by TPA independence, anchorage independent growth, glutamate release, and tumorigenic potential in vivo6. Xenografts of mouse melanocytes expressing mGlu1 receptors, formed tumors that were dependent for growth on the continued expression of mGlu1 receptors6. In a separate study, induced knockdown of mGlu1 receptors in vivo, decreased melanoma tumor volume relative to control9. Additionally, pharmacological antagonism of mGlu1 receptors using BAY36-76204,11 and LY3673854, blocked the growth of mGlu1 expressing melanoma cells in vitro4,11. These published data are consistent with our results where downregulation of mGlu1 receptor signaling, using either shRNA or pharmacological inhibition with a selective32, non-competitive32 mGlu1 receptor antagonist (JNJ16259685), decreased SK2 and SK5 melanoma cell viability in vitro and reduced tumor growth in vivo.
To our knowledge, the non-competitive mGlu1 receptor antagonist, JNJ16259685 has never been used to treat cancer cells; however, it has been tested for behavioral effects on anxiety40 and memory41 in vivo. Based on the doses used in these studies, we chose to use 10mg/kg of JNJ16259685. This non-competitive antagonist was used, instead of a competitive antagonist, to prevent the need for extremely high, and possibly toxic concentrations that would be needed to compete with the reported high levels of glutamate released by melanoma cells4,6,35.
A previous study from our group has demonstrated that, in neurons, mGlu1 receptors exhibit the properties of dependence receptors, inducing death in the absence of the endogenous agonist, glutamate, and promoting survival in its presence42. This pro-apoptotic, signaling pathway is often inhibited during tumor progression43. One mechanism for this inhibition, which is seen in breast cancer43,44, non-small-cell lung cancer43,45, and neuroblastomas43,46 is to constantly produce and release the ligand of the dependence receptor in order to prevent apoptosis43. Melanomas have been reported to release elevated levels of glutamate4,6,35, which would inhibit the pro-apoptotic, negative signaling cascade through mGlu1 receptors. When SK5 melanoma cells were grown in dialyzed serum, which lacks glutamate, this not only led to a reduction in DNA synthesis, but also resulted in an increase in the percentage of apoptotic cells, relative to full serum. Addition of glutamate to dialyzed serum, not only re-activated the proliferative signaling pathway leading to an increase in DNA synthesis, but also blocked the negative, pro-apoptotic signaling pathway. In contrast, removal of glutamate did not promote apoptosis in melanoma cells lacking mGlu1 receptors. Further evidence that mGlu1 receptors may act as dependence receptors in melanoma comes from comparing the effect of selective glutamate depletion on melanoma cells that express mGlu1 receptors (SK2 and SK5) to those that lack mGlu1 receptor expression (UACC930 cells). SK2 and SK5 cells had no viability in the absence of glutamate, while melanoma cells lacking mGlu1 receptors were viable; however, they exhibited an overall slowing of growth rates without significant increases in apoptosis. This decrease in proliferation following extracellular glutamate depletion in melanoma cells that lack mGlu1 receptors may result from reduced stimulation of other glutamate receptors, such as ionotropic glutamate receptors, which have been shown to contribute to proliferation in other cancers47. Moreover, while GPT treatment depletes extracellular glutamate, it cannot be excluded that under these conditions intracellular glutamate concentrations may be affected, which could alter cellular metabolism resulting in decreased cellular viability.
This stimulatory effect of glutamate, seen in mGlu1 receptor-expressing melanoma cell lines, was not observed in immortalized human melanocytes. Instead, glutamate significantly decreased cell viability in a dose-dependent manner (Fig. 1). It has been shown in neurons that high concentrations of glutamate can induce toxicity through activation of ionotropic glutamate receptors48. Because melanocytes originate from the neural crest2,27,33, express ionotropic glutamate receptors27,49, and do not express mGlu1 receptors4–6,26,27, it is possible that in melanocytes glutamate is promoting toxicity through NMDA and/or AMPA receptor activation. In contrast, in UACC930 melanoma cells, which also do not express mGlu1 receptors, addition of high glutamate concentrations had no toxic effects. As NMDA and AMPA receptor activation has been shown to increase proliferation in various cancers47, it is possible that in transformed cells ionotropic glutamate receptors may lose their toxic potential and acquire the ability to sustain cell growth.
In conclusion, our results demonstrate that specific melanomas are dependent on the presence of glutamate and mGlu1 receptors for proliferation. While the mGlu1 receptor antagonist JNJ16259685 did not fully inhibit melanoma tumor growth, our results suggest that JNJ16259685 or other non-competitive mGlu1 receptor antagonists may be useful as adjuvants in the context of other melanoma treatments. However, we do not expect that JNJ16259685, as such, would be a useful drug for the treatment of melanomas since its pharmacodynamic properties, shown here, and the reported pharmacokinetics32 are not optimal. Nevertheless, JNJ16259685 further identifies mGlu1 receptors as a potential therapeutic target and could serve as a lead compound for the synthesis of new therapeutic molecules. Additionally, success has been achieved in melanoma treatment through combination of MAPK pathway inhibitors to delay resistance to monotherapy. For example, in an open-label study two inhibitors of the MAPK pathway were combined (dabrafenib 150mg 2X/day with trametinib 2mg/day) and increased progression free survival and rate of complete or partial response, relative to dabrafenib monotherapy50. Our laboratory has demonstrated using transfected CHO cells that glutamate activation of mGlu1 receptors causes sustained stimulation of the MAPK pathway51, and as a result aberrant expression of mGlu1 receptors in melanomas may be an additional resistance mechanism to current MAPK inhibitors. In addition, because the mGlu1 receptor antagonist JNJ16259685 slows the rate of growth in BRAF (SK5) and NRAS (SK2) mutant melanomas, and BRAF inhibitor resistance often develops through mutations in NRAS and expression of mutant BRAF splice variants52, we hypothesize that mGlu1 receptor antagonists could be useful for the treatment of melanomas that are BRAF inhibitor resistant. Therefore, it will be important to test in the future, both preclinically and clinically, whether combining mGlu1 receptor antagonists with current MAPK inhibitors will enhance progression free survival in melanomas that express mGlu1 receptors. Furthermore, because mGlu1 receptors are involved in the growth of other cancers15,16,53, our results suggest that mGlu1 receptor antagonists may not only be useful for the treatment of melanoma, but may also be useful in the treatment of additional types of cancer.
2. Materials and Methods
Materials
DMEM, RPMI 1640, PBS, fetal bovine serum, dialyzed fetal bovine serum, and antibiotic-antimycotic for cell cultures were purchased from Life Technologies (Carlsbad, CA). Glutamate and JNJ16259685, were purchased from Tocris Bioscience (Briston, United Kingdom). Compounds for melanocyte media including 12-O-tetradecanoylphorbol-13-acetate (TPA), 3-Isobutyl-1-methylxanthine (IBMX), endothelin 1, and human stem cell factor were purchased from Sigma-Aldrich (St. Louis, MO).
Cell Cultures
SK2 and SK5 human melanoma cell lines were obtained from Lombardi Comprehensive Cancer Center Tissue Culture Shared Resource (Georgetown University, Washington, DC). UACC930, C81-61, and C8161 melanoma cells were generously provided by Dr. Suzie Chen (Rutgers University, Piscataway, NJ). HERMES 2 immortalized human melanocytes were purchased from the Wellcome Trust Functional Genomics Cell Bank (University of London, London, United Kingdom). Cells were cultured in 6% CO2 at 37 °C on 60 mm dishes from MidSci (St. Louis, MO). Melanoma cells were cultured in DMEM (high glucose) containing 10% fetal bovine serum, 2 mM glutamine and antibiotic–antimycotic. Melanocytes were cultured in RPMI 1640 growth media supplemented with antibiotic–antimycotic,10 mM HCl, 200 nM TPA, 300 µM IBMX, 10 nM endothelin 1, 10 ng/ml human stem cell factor, 10% fetal bovine serum, and 2 mM glutamine (Life Technologies).
Pharmacological treatments
Cells were plated at 2 000 cells/well on a 96-well plate in a final volume of 100 µl. The next day, the medium was replaced with fresh complete medium or medium containing dialyzed serum and glutamate with or without antagonist was added to the cultures. The selective, non-competitive, mGlu1 receptor antagonist JNJ16259685 was dissolved in DMSO and equal amounts of DMSO were added to controls (1% final). Glutamate was dissolved in equilmolar solution of NaOH and pH was adjusted to 7.4.
Assessment of Cell Viability (MTT)
After 7 days of treatment, medium was removed and cells were incubated for 40 minutes at 37 °C with 0.2 mg/ml of MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide; Life Technologies). The solution was removed and 70 µl of DMSO was added to dissolve the formazan product, which was measured colorimetrically on a plate reader (Envision, Perkin-Elmer, Waltham, MA) as described previously51.
Glutamate depletion
Cells were plated at a density of 2 000 cells/well in 96-well plates in complete media and after 24 hours cells were treated with glutamate pyruvate transaminase (GPT) (Roche, Indianapolis, IN) (35µg/ml) in the presence of 10 mM pyruvate (Life Technologies). GPT converts glutamate and pyruvate into α-ketoglutarate and alanine, which effectively depletes glutamate from the medium.
Glutamate release assay
Cells were plated in 96-well plates, in 100 µl of complete medium. Concentrations of glutamate in medium were measured using fluorescence-based Amplex Red Glutamic Acid/Glutamate Oxidase Assay Kit (Life Technologies) without amplification (without GPT and alanine addition). In short, 50 µl of solution were combined with 50 µl of an Amplex Red working solution (100µM Amplex Red + 0.25U/mL HRP, 0.08 U/mL of L-glutamate oxidase in 10mM Tris-HCl, pH 7.5). A standard curve of glutamate was used to calculate the concentration of glutamate in each well. MTT assay was used to immediately assess cell viability.
BrdU Incorporation
DNA synthesis was measured in SK5 cells using the Cell Proliferation ELISA, BrdU kit (Roche). In short, cells were plated in 96-well plates in complete media and 24 hours later treated with dialyzed serum with or without glutamate. Three, five, or seven days later BrdU labeling solution was added for two hours. Cells were fixed and denatured before addition of anti-BrdU-POD working solution. Chemiluminescence was measured on a plate reader (Envision).
Cell-cycle analysis
SK5 cells were plated at a density of 53 000 cells per well on 6-well plates and 24 hours later treated with fresh serum, dialyzed FBS, or dialyzed FBS containing 10mM of glutamate. After 7 days, cells were collected, washed with PBS, fixed in 75% ethanol, treated with RNase A solution (Sigma-Aldrich), and labeled with prodidium iodide (Sigma-Aldrich) for 30 minutes at room temperature and then 1 hour at 4°C as previously described30. Cell cycle analysis was done by the Flow Cytometry & Cell Sorting Shared Resource at the Lombardi Cancer Center (Georgetown University, Washington, DC) using a LSRFortessa (Becton Dickinson, Franklin Lakes, NJ). Data was modeled using ModFit (Verity Software, Topsham, ME).
shRNA plasmid to mGlu1 receptors
Four different plasmids encoding shRNA to mGlu1 receptors as well as a scrambled control plasmid were purchased from GeneCopoeia (Rockville, MD). shRNA was targeted to sequences (starting base listed) 722 (cacgttggataagatcaac), 1507 (aggtcaggtcatttgatga), 1978 (gagtgctgaacattgatga), and 2690 (ggaagtctaccttatctgc).
Transfection of plasmids
SK5 cells were plated at 50% confluency in 24-well plates. 24 hours later, cells were transfected using Lipofectamine 2000 (Life Technologies) with either a mixture of four shRNA plasmids to mGlu1 receptors or the scrambled control plasmid (total 500ng/well). Stable cell lines were created using 2 µg/mL of puromycin dihydrochloride (Life Technologies).
Western blotting
SK5 cells, SK5 cells expressing a scrambled plasmid, and SK5 cells expressing shRNA to mGlu1 receptors were grown in 10-cm dishes until they reached 90% confluency. Cells were washed with ice-cold PBS and then harvested into 3 mL of 10 mM Tris, 5 mM EDTA (Fisher Scientific, Pittsburgh, PA), 5 mM EGTA (Sigma-Aldrich) (TEE), and 1X protease inhibitor cocktail (Sigma-Aldrich). Samples were sonicated and centrifuged (30,000 g for 15 minutes) at 4°C and the pellet was resuspended in 500 µl of TEE. Samples were sonicated again and centrifuged (30,000gx for 30 minutes) at 4°C and the pellet was resuspended in 60 µl of TEE. A Bradford assay (Bio-Rad, Hercules, CA) was used to measure protein concentration. All samples were diluted to 50 µg/20µl in 10% glycerol (Life Technologies), 62.5 mM Tris HCL (pH 6.8), 2% SDS (Sigma-Aldrich), 0.01 mg/mL bromophenol blue (Fisher Scientific), and 20 mM dithiothreitol (Sigma-Aldrich). 50 µg of protein sample were added to each well in a 4–20% Tris-Glycine Gel (Life Technologies) and proteins separated at 125 V for 90 minutes. Proteins were transferred to Immobilin-FL membranes (Millipore, Billerica, MA) at 100 mA for 3 hours. Membranes were blocked for 1 hour in Odyssey blocking buffer (LI-COR BioSciences, Lincoln, NE) and antibodies were added against mGlu1 receptor (Upstate/Millipore) and β-actin (Sigma-Aldrich) for 24 hours at 4°C. Proteins were visualized using fluorescently labeled secondary antibodies (LI-COR BioSciences), and molecular weights and protein levels were calculated using the Odyssey infrared imaging system software.
Growth of SK5 Cell Xenografts in vivo
The animal protocols were approved by the Institutional Review Board for the Use and Care of Animals and the Animal Care and Facilities Committee of Georgetown University. Approximately 106 melanoma cells (SK5, SK5-SCR or SK5-shRNA) were subcutaneously injected into the flanks of 4 month old nude C57/BL6 mice (n=6 tumors). Tumor length (a), width (b), and height (c) were measured by a blinded researcher three times per week with an electronic vernier caliper. Tumor volume was calculated using, V = (π/6)(a*b*c).
Immunohistochemistry
The Histopathology and Tissue Shared Resource at The Lombardi Cancer Center (Georgetown University, Washington, DC) performed the standard dUTP nick end labeling (TUNEL) immunohistochemistry. Tumor slices were imaged with the Olympus BX61 microscope with Olympus D70 camera and acquired using the DP controller software (Olympus, Center Valley, PA) at a resolution of 144 pixels per inch. Image resolution was enhanced to 300 pixels per inch using Adobe Photoshop and quantitative assessment was performed on 3 random fields from each tumor.
Pharmacological treatment in vivo
Approximately 106 SK2 or SK5 melanoma cells were subcutaneously injected into the flanks of 4 week old nude C57/BL6 mice. The next day (day 1) mice were split into two groups and subcutaneously injected (starting on day 1) five times per week with either (1) JNJ16259685 (dissolved in DMSO and diluted in PBS) or (2) vehicle control (5% DMSO in PBS) (Trial 1: n=3, Trial 2, n=6). Five times per week a blinded researcher measured tumor volume as above.
Calculations and Statistics
For dose-response data, curves were fitted by nonlinear regression using a four-parameter logistic equation and GraphPad Prism was used to calculate EC50’s and IC50’s. For antagonist data, statistical significance was assessed using linear regression to determine if the slope from shown data points was different from zero. One-sample Student’s t-test was used when comparing data to a set-value of zero, while two-sample Student’s t-test was used when comparing two groups. One-way ANOVA followed by Bonferroni post-hoc test was used when comparing more than two groups. For in vivo data, average tumor size and standard error of mean were plotted as a function of time. The day of melanoma cell injection was set as day zero. Once measureable tumors were present, the slopes of the lines were determined by linear regression. GraphPad Prism was used to test whether the slope was affected by treatment. Statistical significance was deemed as p<0.05.
Acknowledgments
We thank the Flow Cytometry & Cell Sorting Shared Resource (partially supported by NIH/NCI grant P30-CA051008) particularly Karen Creswell, and the Histopathology & Tissue Shared Resource (partially supported by NIH/NCI grant P30-CA051008) at The Georgetown-Lombardi Comprehensive Cancer Center (Georgetown University, Washington, DC) for their expertise. Also, we thank Dr. Suzie Chen from Rutgers University for her guidance and generosity and Maria Salinas (Georgetown) for her technical expertise. This work was partially supported by the Pharmaceutical Research and Manufacturers of America Foundation Pre Doctoral Fellowship in Pharmacology/Toxicology to TG, The National Institutes of Health Grant NS37436 to JTW, and CA129003 to CA.
Footnotes
Conflict of Interest: The authors declare no conflict of interest.
References
- 1.American Cancer Society. Cancer Facts & Figures 2013. American Cancer Society; Atlanta: 2013. [Google Scholar]
- 2.Yajima I, Kumasaka MY, Thang ND, Goto Y, Takeda K, Yamanoshita O, et al. RAS/RAF/MEK/ERK and PI3K/PTEN/AKT Signaling in Malignant Melanoma Progression and Therapy. Dermatol Res Pract. 2012;2012:1–5. doi: 10.1155/2012/354191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maira F, Catania A, Candido S, Russo AE, McCubrey JA, Libra M, et al. Molecular targeted therapy in melanoma: a way to reverse resistance to conventional drugs. Curr Drug Deliv. 2012;9:17–29. doi: 10.2174/156720112798376032. [DOI] [PubMed] [Google Scholar]
- 4.Namkoong J, Shin SS, Lee HJ, Marín YE, Wall BA, Goydos JS, et al. Metabotropic glutamate receptor 1 and glutamate signaling in human melanoma. Cancer Res. 2007;67:2298–2305. doi: 10.1158/0008-5472.CAN-06-3665. [DOI] [PubMed] [Google Scholar]
- 5.Pollock PM, Cohen-Solal K, Sood R, Namkoong J, Martino JJ, Koganti A, et al. Melanoma mouse model implicates metabotropic glutamate signaling in melanocytic neoplasia. Nat Genet. 2003;34:108–12. doi: 10.1038/ng1148. [DOI] [PubMed] [Google Scholar]
- 6.Shin SS, Namkoong J, Wall BA, Gleason R, Lee HJ, Chen S. Oncogenic activities of metabotropic glutamate receptor 1 (Grm1) in melanocyte transformation. Pigment Cell Melanoma Res. 2008;21:368–378. doi: 10.1111/j.1755-148X.2008.00452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khan AJ, Wall B, Ahlawat S, Green C, Schiff D, Mehnert JM, et al. Riluzole Enhances Ionizing Radiation-induced Cytotoxicity in Human Melanoma Cells that Ectopically Express Metabotropic Glutamate Receptor 1 In Vitro and In Vivo. Clin Cancer Res. 2011;17:1807–1814. doi: 10.1158/1078-0432.CCR-10-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shin SS, Martino JJ, Chen S. Metabotropic glutamate receptors (mGluRs) and cellular transformation. Neuropharmacology. 2008;55:396–402. doi: 10.1016/j.neuropharm.2008.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wangari-Talbot J, Wall BA, Goydos JS, Chen S. Functional effects of GRM1 suppression in human melanoma cells. Mol Cancer Res. 2012;10:1440–1450. doi: 10.1158/1541-7786.MCR-12-0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohtani Y, Harada T, Funasaka Y, Nakao K, Takahara C, Abdel-Daim M, et al. Metabotropic glutamate receptor subtype-1 is essential for in vivo growth of melanoma. Oncogene. 2008;27:7162–7170. doi: 10.1038/onc.2008.329. [DOI] [PubMed] [Google Scholar]
- 11.Le MN, Chan JL, Rosenberg SA, Nabatian AS, Merrigan KT, Cohen-Solal KA, et al. The glutamate release inhibitor riluzole decreases migration, invasion, and proliferation of melanoma cells. J Invest Dermatol. 2010;130:2240–2249. doi: 10.1038/jid.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conn PJ, Pin JP. Pharmacology and functions of metabotropic glutamate receptors. Annu Rev Pharmacol Toxicol. 1997;37:205–237. doi: 10.1146/annurev.pharmtox.37.1.205. [DOI] [PubMed] [Google Scholar]
- 13.Pin JP, Duvoisin R. The metabotropic glutamate receptors: structure and functions. Neuropharmacology. 1995;34:1–26. doi: 10.1016/0028-3908(94)00129-g. [DOI] [PubMed] [Google Scholar]
- 14.Teh J, Chen S. Metabotropic glutamate receptors and cancerous growth. Wiley Interdiscip Rev Membr Transp Signal. 2012;1:211–220. doi: 10.1002/wmts.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Speyer CL, Smith JS, Banda M, DeVries JA, Mekani T, Gorski DH. Metabotropic glutamate receptor-1: a potential therapeutic target for the treatment of breast cancer. Breast Cancer Res Treat. 2012;132:565–573. doi: 10.1007/s10549-011-1624-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banda M, Speyer CL, Semma SN, Osuala KO, Kounalakis N, Torres KE, et al. Metabotropic glutamate receptor-1 contributes to progression in triple negative breast cancer. PLoS One. 2014;9(1):1–12. doi: 10.1371/journal.pone.0081126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang HJ, Yoo BC, Lim SB, Jeong SY, Kim WH, Park JG. Metabotropic glutamate receptor 4 expression in colorectal carcinoma and its prognostic significance. Clin Cancer Res. 2005;11:3288–3295. doi: 10.1158/1078-0432.CCR-04-1912. [DOI] [PubMed] [Google Scholar]
- 18.Brocke KS, Staufner C, Luksch H, Geiger KD, Stepulak A, Marzahn J, et al. Glutamate receptors in pediatric tumors of the central nervous system. Cancer Biol Ther. 2010;9:455–468. doi: 10.4161/cbt.9.6.10898. [DOI] [PubMed] [Google Scholar]
- 19.Park SY, Lee SA, Han IH, Yoo BC, Lee SH, Park JY, et al. Clinical significance of metabotropic glutamate receptor 5 expression in oral squamous cell carcinoma. Oncol Rep. 2007;17:81–87. [PubMed] [Google Scholar]
- 20.de Groot J, Sontheimer H. Glutamate and the biology of gliomas. Glia. 2011;59:1181–1189. doi: 10.1002/glia.21113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye ZC, Sontheimer H. Glioma Cells Release Excitotoxic Concentrations of Glutamate. Cancer Res. 1999;59:4383–4391. [PubMed] [Google Scholar]
- 22.Lyons SA, Chung WJ, Weaver AK, Ogunrinu T, Sontheimer H. Autocrine glutamate signaling promotes glioma cell invasion. Cancer Res. 2007;67:9463–9471. doi: 10.1158/0008-5472.CAN-07-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee HJ, Wall BA, Wangari-Talbot J, Shin SS, Rosenberg S, Chan JL, et al. Glutamatergic pathway targeting in melanoma: single-agent and combinatorial therapies. Clin Cancer Res. 2011;17:7080–7092. doi: 10.1158/1078-0432.CCR-11-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468(7326):968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DiRaddo JO, Pshenichkin S, Gelb T, Wroblewski JT. Two newly identified exons in human GRM1 express a novel splice variant of metabotropic glutamate 1 receptor. Gene. 2013;519:367–373. doi: 10.1016/j.gene.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee HJ, Wall BA, Wangari-Talbot J, Chen S. Regulation of mGluR1 expression in human melanocytes and melanoma cells. BBA-Gene Regulatory Mechanisms. 2012;1819:1123–31. doi: 10.1016/j.bbagrm.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frati C, Marchese C, Fisichella G, Copani A, Nasca MR, Storto M, et al. Expression of functional mGlu5 metabotropic glutamate receptors in human melanocytes. J Cell Physiol. 2000;183:364–372. doi: 10.1002/(SICI)1097-4652(200006)183:3<364::AID-JCP9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 28.Wen Y, Li J, Koo J, Shin SS, Lin Y, Jeong BS, et al. Activation of the Glutamate Receptor GRM1 Enhances Angiogenic Signaling to Drive Melanoma Progression. Cancer Res. 2014;74:2499–2509. doi: 10.1158/0008-5472.CAN-13-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng W, Yue Q, Rosenberg PA, Volpe JJ, Jensen FE. Oligodendrocyte excitotoxicity determined by local glutamate accumulation and mitochondrial function. J Neurochem. 2006;98:213–222. doi: 10.1111/j.1471-4159.2006.03861.x. [DOI] [PubMed] [Google Scholar]
- 30.Darzynkiewicz Z, Bruno S, Del Bino G, Gorczyca W, Hotz MA, Lassota P, et al. Features of apoptotic cells measured by flow cytometry. Cytometry. 1992;13:795–808. doi: 10.1002/cyto.990130802. [DOI] [PubMed] [Google Scholar]
- 31.Ringer L, Sirajuddin P, Heckler M, Ghosh A, Suprynowicz F, Yenugonda VM, et al. VMY-1-103 is a novel CDK inhibitor that disrupts chromosome organization and delays metaphase progression in medulloblastoma cells. Cancer Biol Ther. 2011;12:818–826. doi: 10.4161/cbt.12.9.17682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lavreysen H, Wouters R, Bischoff F, Nóbrega Pereira S, Langlois X, Blokland S, et al. JNJ16259685 , a highly potent , selective and systemically active mGlu1 receptor antagonist. Neuropharmacology. 2004;47:961–972. doi: 10.1016/j.neuropharm.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 33.Prickett TD, Samuels Y. Molecular pathways: dysregulated glutamatergic signaling pathways in cancer. Clin Cancer Res. 2012;18:4240–4246. doi: 10.1158/1078-0432.CCR-11-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willard SS, Koochekpour S. Glutamate, glutamate receptors, and downstream signaling pathways. Int J Biol Sci. 2013;9:948–959. doi: 10.7150/ijbs.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seidlitz EP, Sharma MK, Saikali Z, Ghert M, Singh G. Cancer cell lines release glutamate into the extracellular environment. Clin Exp Metastasis. 2009;26:781–787. doi: 10.1007/s10585-009-9277-4. [DOI] [PubMed] [Google Scholar]
- 36.Takano T, Lin JH, Arcuino G, Gao Q, Yang J, Nedergaard M. Glutamate release promotes growth of malignant gliomas. Nat Med. 2001;7:1010–1015. doi: 10.1038/nm0901-1010. [DOI] [PubMed] [Google Scholar]
- 37.Koochekpour S, Majumdar S, Azabdaftari G, Attwood K, Scioneaux R, Subramani D, et al. Serum glutamate levels correlate with Gleason score and glutamate blockade decreases proliferation, migration, and invasion and induces apoptosis in prostate cancer cells. Clin Cancer Res. 2012;18:5888–5901. doi: 10.1158/1078-0432.CCR-12-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei X, Walia V, Lin JC, Teer JK, Prickett TD, Gartner J, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011;43:442–446. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ortiz P, Vanaclocha F, López-Bran E, Esquivias JI, López-Estebaranz JL, Martín-González M, et al. Genetic analysis of the GRM1 gene in human melanoma susceptibility. Eur J Hum Genet. 2007;15:1176–82. doi: 10.1038/sj.ejhg.5201887. [DOI] [PubMed] [Google Scholar]
- 40.Steckler T, Lavreysen H, Oliveira AM, Aerts N, Van Craenendonck H, Prickaerts J, et al. Effects of mGlu1 receptor blockade on anxiety-related behaviour in the rat lick suppression test. Psychopharmacology (Berl) 2005;179:198–206. doi: 10.1007/s00213-004-2056-7. [DOI] [PubMed] [Google Scholar]
- 41.Steckler T, Oliveira AF, Van Dyck C, Van Craenendonck H, Mateus AM, Langlois X, et al. Metabotropic glutamate receptor 1 blockade impairs acquisition and retention in a spatial Water maze task. Behav Brain Res. 2005;14:52–60. doi: 10.1016/j.bbr.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 42.Pshenichkin S, Dolińska M, Klauzińska M, Luchenko V, Grajkowska E, Wroblewski JT. Dual neurotoxic and neuroprotective role of metabotropic glutamate receptor 1 in conditions of trophic deprivation - possible role as a dependence receptor. Neuropharmacology. 2008;55:500–508. doi: 10.1016/j.neuropharm.2008.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goldschneider D, Mehlen P. Dependence receptors: a new paradigm in cell signaling and cancer therapy. Oncogene. 2010;29:1865–1882. doi: 10.1038/onc.2010.13. [DOI] [PubMed] [Google Scholar]
- 44.Fitamant J, Guenebeaud C, Coissieux MM, Guix C, Treilleux I, Scoazec JY, et al. Netrin-1 expression confers a selective advantage for tumor cell survival in metastatic breast cancer. Proc Natl Acad Sci USA. 2008;105:4850–4855. doi: 10.1073/pnas.0709810105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delloye-Bourgeois C, Brambilla E, Coissieux MM, Guenebeaud C, Pedeux R, Firlej V, et al. Interference with netrin-1 and tumor cell death in non-small cell lung cancer. J Natl Cancer Inst. 2009;101:237–247. doi: 10.1093/jnci/djn491. [DOI] [PubMed] [Google Scholar]
- 46.Delloye-Bourgeois C, Fitamant J, Paradisi A, Cappellen D, Douc-Rasy S, Raquin MA, et al. Netrin-1 acts as a survival factor for aggressive neuroblastoma. J Exp Med. 2009;206:833–847. doi: 10.1084/jem.20082299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rzeski W, Ikonomidou C, Turski L. Glutamate antagonists limit tumor growth. Biochem Pharmacol. 2002;98:1195–1200. doi: 10.1016/s0006-2952(02)01218-2. [DOI] [PubMed] [Google Scholar]
- 48.Blaabjerg M, Fang L, Zimmer J, Baskys A. Neuroprotection against NMDA excitotoxicity by group I metabotropic glutamate receptors is associated with reduction of NMDA stimulated currents. Exp Neurol. 2003;183:573–580. doi: 10.1016/s0014-4886(03)00204-8. [DOI] [PubMed] [Google Scholar]
- 49.Hoogduijn MJ, Hitchcock IS, Smit NP, Gillbro JM, Schallreuter KU, Genever PG. Glutamate receptors on human melanocytes regulate the expression of MiTF. Pigment Cell Res. 2006;19:58–67. doi: 10.1111/j.1600-0749.2005.00284.x. [DOI] [PubMed] [Google Scholar]
- 50.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK Inhibition in Melanoma with BRAF V600 Mutations. NEJM. 2012;367:1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emery AC, Pshenichkin S, Takoudjou GR, Grajkowska E, Wolfe BB, Wroblewski JT, et al. The protective signaling of metabotropic glutamate receptor 1 is mediated by sustained, beta-arrestin-1-dependent ERK phosphorylation. J Biol Chem. 2010;285:26041–26048. doi: 10.1074/jbc.M110.139899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edward HJ, Kevin Basile K, Aplin AE. Beneficial Effects of RAF Inhibitor in Mutant BRAF Splice Variant-expressing Melanoma. Mol Cancer Res. 2014:OF1–OF8. doi: 10.1158/1541-7786.MCR-13-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martino JJ, Wall BA, Mastrantoni E, Wilimczyk BJ, La Cava SN, Degenhardt K, et al. Metabotropic glutamate receptor 1 (Grm1) is an oncogene in epithelial cells. Oncogene. 2013;32:4366–4376. doi: 10.1038/onc.2012.471. [DOI] [PMC free article] [PubMed] [Google Scholar]