Abstract
The diverse responses of critically ill patients to infection with multi-drug resistant (MDR) bacteria are determined by many complex factors. These include the nature of the immune response activated by specific organisms. Properties unique to each organism such as adherence proteins, microvesicle formation, toxin production and the propensity to form biofilms are important factors in pathogenesis. Equally important is the variability in the host immune response, whether due to genetic or iatrogenic factors, including the presence of major comorbidities, treatment with immunomodulatory therapy and disruption of the microbiome. Future approaches in treating infections caused by MDR bacteria will be heavily influenced by a precision medicine approach, with rapid diagnostic techniques of both bacterial and host factors and high throughput screening of novel therapeutics becoming the mainstay of treatment.
Keywords: MDR-bacteria, pathogenesis, ICU, therapeutics
Infections due to multidrug-resistant (MDR) bacteria are a common complication for patients in the intensive care unit (ICU), increasing morbidity and mortality rates, and length of stay and imposing a significant cost to hospitalization [1–3]. Reducing healthcare-associated infection such as ventilator-associated pneumonia and catheter-related bloodstream infection has become a major objective for hospital regulatory organizations to improve patient outcome and reduce the financial burden [4–6]. Recent reports of isolates resistant to all antibiotics highlight the urgency of ongoing research to develop therapies and to prevent and treat thes infections [7]. Novel and more efficient diagnostic tools are now widely available to rapidly identify such MDR pathogens. These advances have provided insights into the molecular epidemiology of clusters of infection in ICUs and have helped dissect the molecular features of the most highly successful pathogens [8, 9].
There is increasing appreciation of the remarkable heterogeneity of the pathogens that infect ICU patients as well as fundamental differences in the quality of the hosts that they infect [10]. This is best illustrated by the newly developed guidelines for the management of sepsis and septic shock. It attempts to take into account the impact of the profound immune dysregulation that accompanies sepsis, as well as comorbid conditions and immunosuppressive medications that affect the individual's unique clinical phenotype [11]. Substantial data document the multiple factors that influence this host response to infection, such as host genetics, alterations in the microbiome, and various mechanisms of host immunosuppression [10, 12]. The multiple variables surrounding the key properties of both the host and pathogen contribute to the difficulty in establishing and validating diagnostic criteria and stratification of risk for critically ill patients, such as those with suspected ventilator-associated pneumonia [13, 14].
The articles in this supplement highlight many of the similarities as well as the differences in the epidemiology and pathogenesis of the most frequent MDR pathogens associated with pneumonia and sepsis in the ICU setting. The impact of the MDR gram-negative pathogens and limited availability of effective antimicrobial therapy are a growing concern worldwide [2, 8, 15]. However, antibiotic resistance does not completely explain the success of such pathogens, perhaps best exemplified by the global spread of methicillin-resistant Staphylococcus aureus (MRSA), as discussed by Planet in this supplement [16]. Despite the availability of several antimicrobial agents with good in vitro activity against MRSA, the associated morbidity and mortality rates remain high, especially after influenza, as recently documented [17–19]. The selection of organisms resistant to antibiotics as well as to innate immune clearance mechanisms contribute to the success of these healthcare-associated infections. This may not necessarily impose a major burden in bacterial fitness, as reviewed herein by Geisinger and Isberg [20]. Understanding the nature of the host response elicited by these organisms and how they differ may ultimately help clinicians to use therapies targeting pathological immune responses.
BACTERIAL FACTORS
Pathogen-Specific Immune Signaling in the Airway
Critical care reports refer to “ventilator-associated pneumonias” as a single entity, assuming common pathogenesis and a homogeneous host population. However, given the diversity of the pathogens associated with this clinical entity and the nature of the host responses elicited, optimal clinical management of these infections may require a more personalized approach. What unites these patients is a shared predilection for pneumonia, namely, intubation, critical illness in and of itself, and the complications of ICU care. The diagnosis alone does not predict either the nature or quality of the host immune response or the natural history of the infecting organisms, though many predictive measures have tried [13]. Many ICU opportunists do share common microbiological properties, such as a propensity to form biofilms that thwart antimicrobial and phagocytic clearance (Table 1). Strategies to prevent biofilm formation by changing the material of foreign bodies, such as endotracheal tubes, are an avid area of study [28, 29].
Table 1.
Characteristics of Multidrug-Resistant Pathogens Commonly Isolated in the ICU
| Organism | Biofilms | Clonal Diversity | Carbapenemase Production | Human Reservoir | Source |
|---|---|---|---|---|---|
| Pseudomona aeruginosa | Y | +++ | Y | ++ | Jorth et al [21] |
| Klebsiella pneumoniae | Y | ++ | ++ | ++ | Gomez-Simmonds et al [22]; Chen et al [23] |
| Acinetobacter sp. | Y | UNK | ++ | Y | Murray et al [24] |
| Enterobacter sp. | Y | ++++ | ++ | Y | Gomez-Simmonds et al [25]; Girlich et al [26] |
| Staphylococcus aureus (MRSA) | Y | Y | − | ++ | Uhlemann et al [27] |
Abbreviations: −, none; +, low; ++, medium; +++, high; ++++, very high; ICU, intensive care unit; MRSA, methicillin resistant S. aureus; UNK, unknown; Y, yes.
Some organisms, such as the small colony variants of S. aureus, adopt a low metabolic profile, replicating intracellularly at a reduced rate but providing a nidus for recurrent and disseminated infection [30]. Other pathogens may be associated with exaggerated pathological proinflammatory signaling, often achieved through activation of the inflammasome [31]. Whereas other organisms flourish as “stealth” pathogens, replicating to high densities without stimulating a sufficient inflammatory response to affect clearance, a problem compounded by their resistance to multiple antimicrobial agents and host organ dysfunction [32]. Moreover, mixed populations of the same species of bacteria with varying antimicrobial susceptibility and replication rates heterogeneous phenotype may be selected out during the course of infection [21].
MULTIPLE MECHANISMS OF PATHOGEN-HOST ACTIVATION
The pathogenesis of most lower respiratory tract infections in the ICU is through aspiration of upper airway flora, presumably planktonic organisms released from biofilms associated with endotracheal tubes or aspiration of the upper airway microbiota after loss of airway reflexes [33, 34]. These organisms interact with mucosal epithelial cells as well as immune cells recruited into the airway. Most aspirated organisms wind up enmeshed in mucins though direct attachment to epithelial surfaces is not necessary to elicit proinflammatory signaling [35]. Some opportunists, such as Klebsiella pneumoniae (fimbrial proteins) [36] and S. aureus (fibronectin-binding protein) [37], express adherence factors that contribute to their pathogenic properties. Shed bacterial cell wall components, such as microvesicles, harbor multiple components and are readily released from intact bacteria, associated with cytotoxicity and immune stimulation [38]. Host cells take up these microvesicles, facilitating the activation of intracellular receptors such as those involved in initiating inflammasome and interferon activation. Furthermore, bacterial production of pathogen-associated molecular patterns, such as lipopolysaccharide, enables stimulation of both superficial and cytoplasmic receptors to initiate host signaling.
Bacterial targeting of specific components of innate immunity has a major impact on the pathogenesis of infection with complex and often redundant signaling pathways for immune cellular recruitment. The induction of the types I and III interferons, which is mediated by bacterial ligation of intracellular receptors in both epithelial and immune cells, is a major host response to airway pathogens [39, 40], as discussed by Parker in this supplement [41]. Moreover, the interferons regulate the cytokine milieu of the airway, which influence the density of pathogen colonization, and hence the relative risk of aspiration of organisms such as MRSA [42]. Surveillance of the airway contents by immune cells is also an important component of airway defense. In their resting state, alveolar macrophages have a predominantly anti-inflammatory phenotype to prevent unnecessary inflammatory responses [43]. These cells can be targeted by bacterial toxins, such as those produced by MRSA, and destroyed by pyroptosis or necroptosis resulting in excessive immune activation [44]. T cells in the airway are also readily activated by bacterial components, in addition to superantigens, contributing to the inflammatory damage stimulated by a brisk host-response to infection [45]. Several lines of evidence indicate that the redundancy of signaling pathways resulting in proinflammatory gene expression, especially the generation of interleukin 1β, leads to impaired bacterial clearance and airway damage in murine models of infection [46].
Pseudomonas aeruginosa Infections
Pseudomona aeruginosa is a common cause of severe ICU infections, reviewed in this volume by Oliver and colleagues [47]. It is perhaps the best-studied airway pathogen, in part because of its association with airway infection in cystic fibrosis and the concerted research effort to understand the pathogenesis of pulmonary infection associated with CFTR mutations [48]. With its large genome and tremendous metabolic flexibility, P. aeruginosa efficiently adapts to the human airway, scavenges iron through its expression of siderophores to support growth, and rapidly adapts to microenvironments throughout the lung, including areas with low oxygenation [21]. Its production of pyocyanin acts as an antioxidant for further protection against phagocytic clearance [49]. These organisms express multiple pathogen-associated molecular patterns, including flagella that activate proinflammatory signaling through Toll-like receptor 5 [50] and the NLRC4 inflammasome [51, 52] and provide motility, a key factor in invasive infection. P. aeruginosa associated with ICU pneumonias express the type 3 secreted toxins, including ExoU, a patatin-like protease associated with tissue destruction [53]. Components of the type III secretion system also contribute to inflammasome activation [54]. In addition to the planktonic, flagellated P. aeruginosa associated with acute infection, mixed populations of these organisms exist in chronically infected lungs. A dynamic equilibrium exists between the less immunostimulatory mutants associated with biofilms and the more virulent organisms that can be genotypically similar but phenotypically different. In the setting of chronic airway infection in cystic fibrosis, substantiated by whole-genome sequencing, numerous clonally distinct isolates with different antimicrobial susceptibility and phenotypes exist within different areas of the lung [21], a scenario that is likely to occur in selected ICU patients.
S. aureus Infections
Of all the ICU pathogens, S. aureus is the most common cause of pneumonia and in some ways the best understood [4, 55]. In contrast to the gram-negative pathogens, S. aureus is less genetically heterogeneous, as detailed by Planet in this supplement [16]. However, its repertoire of genes designed specifically to thwart human immune mediated clearance is sufficient to assure its continued success as a pathogen [56]. Several antimicrobials with substantial in vitro activity have been developed since the emergence of MRSA [57]. However, the persistence of both MRSA and methicillin-susceptible S. aureus as major ICU pathogens, despite the availability of effective antibiotics, suggests adaptations to immune clearance mechanisms. This indicates that therapeutic strategies to assist the host clear these organisms would be useful.
It is also evident that an excessive proinflammatory response to S. aureus infection is injurious, associated with respiratory failure and sepsis. S. aureus is a major cause of “pyogenic” infections, requiring neutrophils for bacterial clearance at the expense of proinflammatory sequelae. Other mechanisms of inflammation, such as the contribution of α-toxin (Hla), are reviewed in this issue by Bubeck-Wardenburg and colleagues [58]. This toxin facilitates invasive infection by targeting ADAM10 in the pulmonary epithelium [59]. It has a major role in the immunotoxicity of this organism by activating both necroptosis, a proinflammatory form of cell death and pyroptosis, the consequence of caspase 1 activity mediated by the NLRP3 inflammasome [44]. S. aureus clearance is significantly improved in murine models of pneumonia in which activation of the inflammasome is prevented [60]. Numerous other toxins, including the human-specific bicomponent toxins Panton-Valentine leukocidin and LukAB, also contribute to pathogenicity, but are difficult to evaluate in murine models of infection [61]. Moreover, S. aureus effectively thwarts phagocytosis and is capable of intracellular persistence within phagocytes and stromal cells, further contributing to its persistence within the infected lung [62]. The innumerable effects of protein A on the generation of effective B cell responses, as well as interference with opsonization, also promote persistence and repeated infection [63, 64]. The immunological correlates of resistance to S. aureus infection remain to be defined.
K. pneumoniae Infections
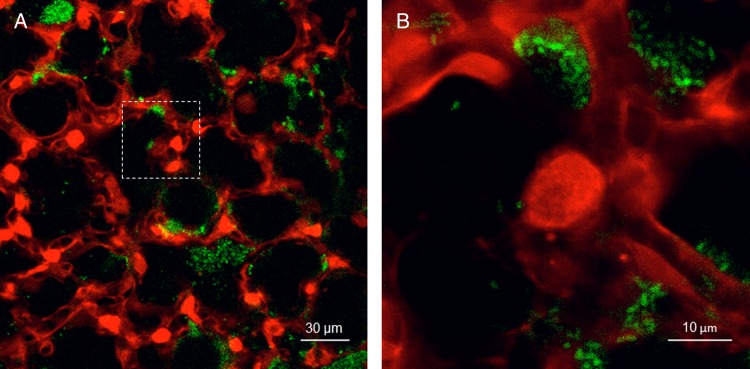
K. pneumoniae, especially the carbapenem-resistant isolates, have emerged as a major clinical problem worldwide (Figure 1). It is evident that K. pneumoniae strains are highly diverse and have substantially different clinical behaviors [65]. Best studied are the highly virulent strains, as typified by the prototypic American Type Culture Collection 43816 (KPPR1), whose metabolic properties are characterized by Mandel and colleagues in a novel model of infection in this supplement [66] . Also highly virulent are the hyperencapsulated K. pneumoniae associated with sepsis and liver abscesses primarily in Asia [67]. More commonly seen in ICUs in the United States are the ST258 strains, now responsible for >70% of K. pneumoniae isolates causing healthcare-associated infections and the predominant clone of carbabenemase-resistant K. pneumoniae (CRKP). These CRKP strains also have been recently found as common components of the fecal flora, with nearly a 5% carriage rate in nursing home residents [68].
Figure 1.
(A) Murine model of green fluorescent protein (GFP)–labeled carbapenemase-producing Klebsiella pneumoniae ST258 in the alveoli as seen with in vivo confocal microscopy. (B) Magnified image of the dashed box in Panel A. Red represents calcein red; green, GFP-labeled K. pneumoniae. (Courtesy of Jaime Hook, MD, Columbia University Medical Center.)
The genetic diversity of CRKP strains is detailed by Uhlemann and coworkers in this supplement [69]. They have been a major clinical problem in ICU patients, particularly in immunocompromised transplant recipients, in whom they cause prolonged, relatively indolent pulmonary infection associated with frequent bacteremias, necessitating toxic combinations of antimicrobial agents with limited efficacy [22]. The heterogeneity of the host response to these organisms has been modeled in mice [70]. There are clearly major differences in the types of host responses activated by different K. pneumoniae, and even different ST258 strains. Specific ST258 isolates are highly resistant to neutrophil-mediated killing [71] but are relatively susceptible to uptake and killing by monocytes [72].
The K. pneumoniae strains that are now considered a global threat [73, 74] exhibit clinically significant genomic and phenotypic heterogeneity. They are resistant to phagocytic clearance by neutrophils, evade inflammatory signaling, and are resistant to almost all available antibiotics. These qualities lead to persistence and promote their rapid selection in a critically ill patient. Whether other major gram-negative ESKAPE pathogens, such as Acinetobacter or Enterobacter, will be as genetically heterogeneous remains to be fully appreciated, though promiscuity in the exchange of genetic material between species would suggest that this is likely [75]. Nonetheless, based on the data accrued from analyses of P. aeruginosa and K. pneumoniae, it is evident that the acquisition of antimicrobial resistance and genes that foster proliferation in the setting of innate immunity also promote positive selection of these bacterial populations.
HOST FACTORS
Sources of Infection – Colonization and the Microbiome
A major question underlying the diagnosis and management of ventilator-associated pneumonias and sepsis is how and when host colonization with MDR pathogens becomes active infection with a pathological immune response. This is particularly important because the reservoirs for antibiotic resistance expands in the community owing to overutilization of broad-spectrum antibiotics, as documented for MRSA [76] and extended-spectrum β-lactamase Enterobacteriaceae [77]. Multiple studies indicate that the endogenous flora of these patients, the respiratory tract for gram-positive infections such as S. aureus, and the gut microbiome for the MDR gram-negatives, such as the carbapenem-resistant isolates [55, 78], are major sources of serious infection for ICU patients, and not patient-to-patient spread. Many factors influence how and when organisms colonize the lower airways, bloodstream, and urinary tract. In the example of ventilator-associated pneumonia, loss of normal protective reflexes due to sedation and paralysis, alterations in the acidity of the stomach by medications, and dysbiosis as a consequence of broad-spectrum antibiotic use influence which organisms colonize the lower airways and have the potential for invasion.
Unrecognized until recently is the importance of the gut microbiome in regulating the immunological tone of the entire host, especially the lung [79]. Alterations in the microbiome alone in the murine models of S. aureus pneumonia can significantly alter the inflammatory and cytokine response to antigenic challenge, ultimately influencing bacterial clearance [80]. The phenotype of dysbiosis can be so striking that refractory episodes of septic shock have been “cured” with fecal transplant in critically ill patients [81]. Studies to understand the influence of the microbiome on the human immune system are in progress, and there is increasing appreciation for its role in establishing the immunological tone of the host response in the defense against bacterial pathogens [82].
Despite the emphasis placed on hand hygiene and careful attention to the prevention of patient-to-patient spread of MDR pathogens, local outbreaks still occur [8]. However, multiple studies of ICU outbreaks using whole bacterial genome sequencing techniques document the heterogeneity, particularly of the gram-negative pathogens, even of the same species that cause healthcare-associated infections [8]. The most obvious phenotypic marker is antimicrobial resistance, but these strains must also harbor genes that facilitate proliferation despite a local antimicrobial milieu consisting of antimicrobial peptides, lactoferrin, mucociliary clearance, phagocytic clearance, and the cytokine responses of the T cells recruited in response to infection.
Status of the Host Immune System
Although there are occasional ICU patients without serious underlying illnesses, most have significant impairment in innate immune clearance mechanisms. Long-standing debilitating illnesses, poor nutrition, or a major disruption in their normal microbiome all contribute to increased susceptibility to serious infection through suppression of the immune system [79, 83]. The increased susceptibility to life-threatening illness of neutropenic patients is widely accepted, and the use of broad-spectrum antimicrobials in these hosts remains the standard of care, with increasing appreciation of negative effects on the microbiota and its contribution to immune function [79]. The pathological effects of immune signaling on host susceptibility to infection is also well appreciated, as illustrated in the setting of bacterial superinfection post influenza [84]. Excessive activation of interleukin 17–mediated responses [85] and the participation of the types I and III interferon cascade significantly increase host susceptibility to serious bacterial pneumonia post influenza. Virally induced interferon responses also influence susceptibility to subsequent bacterial infection by altering the composition of the upper airway microbiome [42].
Many ICU patients receive immunosuppressant drugs to prevent inflammatory damage due to autoimmune diseases or to prevent rejection of solid organ or hematopoietic stem cell transplants. Increasing numbers of patients receive drugs that specifically target T-cell signaling, the so-called biologics used to treat autoimmune and oncological processes. These include drugs such as the check point inhibitors that activate T cells in the setting of malignancy, or drugs that block T-cell activation in the setting of autoimmunity, tumor necrosis factor, interleukin 6, or interleukin 1 antagonists. Dysregulated T-cell signaling is a major component of pathological proinflammatory responses, as well illustrated in the case of S. aureus superantigens [45]. T cells also potentiate innate immune responses to improve clearance of airway pathogens [85] and are essential to the coordination of adaptive immunity. Exactly how these potent immunomodulators change susceptibility to pneumonia and sepsis remains to be established.
A theme common to several articles presented in this supplement is the importance of the regulation of the inflammatory responses activated by airway infection and how this may differ depending on the nature of both the host and the specific pathogen. In this era of “personalized medicine,” it would be useful to have a more detailed characterization of the organisms directly associated with ICU infections, not only their genus and species, but their whole-genomic repertoire, as can be readily accomplished [86]. In addition to the epidemiological importance, the identification of specific biomarkers that correlate with invasion or immune tolerance could be useful in guiding therapy, as monoclonal antibodies directed at specific virulence factors become available. Identifying patients who need amplified inflammatory responses, as opposed to those who need anti-inflammatory modifiers, could be difficult in the setting of severe pneumonia, although new diagnostic strategies such as focused RNA array, cytokine array, or blast proteomics [87, 88] may be far superior to conventional and newer biomarkers, such as C-reactive protein, lactate, procalcitonin, and soluble triggering receptor expressed on myeloid cells (sTREM) 1 [89, 90]. It is also conceivable that markers for pathological inflammatory responses in the lung may become available to identify patients most likely to benefit from immunomodulatory therapy, with specific reagents already developed for other diseases characterized by pathological proinflammatory responses, such as autoimmune disorders.
IDENTIFICATION OF THERAPEUTIC TARGETS: NOVEL STRATEGIES
The search for a universal therapeutic target for bacterial infection has been fruitless, mostly owing to the reductionist approach to severe infection [91]. Promising therapies in animal models have failed to make a major impact on the outcome in critically ill patients [10, 92]. Despite these shortcomings, ongoing trials to study biologics for heterogeneous disease states continue as in the case of global TNF or interleukin 1β neutralization [93–95]. A straightforward approach currently under study targets bacterial toxins that are important in pathogenesis. A monoclonal antibody targeting the α-toxin has been shown to enhance bacterial clearance from the lung, acting through several discrete mechanisms [96]. Neutralization of Hla could also potentially prevent pathological consequences of inflammasome activation. Clinical trials are already in progress to test safety and efficacy of a humanized pegylated anti-P. aeruginosa V-antigen targeting a component of the type III secretion system of P. aeruginosa in the setting of ventilator-associated pneumonia [97]. This would also have the potential of multiple beneficial effects, both neutralizing the effects of the damaging exotoxins necessary for invasion and blocking activation of the NLRC4 inflammasome. Therapeutic strategies to control apparently excessive immune activation in the human lung by counteracting the interleukin 1 cascade have been used in patients with underlying immune diseases and infection, such as chronic granulomatous disease or severe sepsis with features of macrophage activation syndrome [95, 98].
THE FUTURE OF THERAPEUTIC STRATEGIES: PRECISION MEDICINE
Several advances in the field of medicine will probably have a major impact on the treatment of critically ill patients with MDR infections. Epidemiological strategies, such as upper airway and skin decontamination, hand hygiene, antibiotic stewardship, and strategies to reduce healthcare-associated infections, will continue to reduce the incidence of these infections. From a diagnostic approach, the early detection and identification of pathogenic bacteria continues to improve with advances in culture-independent methods [99, 100]. Technologies in the rapid characterization of the host phenotype in severe infection have also advanced, with improvements in focused RNA-sequencing arrays [87, 101] and whole-exome sequencing [102].
For therapeutics, the motivation for the discovery of novel antimicrobials is outweighed by the high economic burden of drug development and likelihood of rapid bacterial adaptation [73]. As discussed above, few novel strategies for specific pathogens have already been identified, and universal therapeutics have a limited effect because of the heterogeneity of disease. More exciting is the idea that host- and pathogen-specific immunomodulatory therapy can be used as adjunctive therapies, similar to host- and disease-specific therapeutics in oncology and rheumatology. An example of how personalized medicine could affect clinical outcome is the use of high-throughput RNA sequencing to identify phenotypically silent mutations, such as interferon regulatory factor 7 deficiency, in a critically ill patient with life-threatening influenza infection. Ciancanelli et al [103] used this approach combined with confirmatory testing in pluripotent immune cells with the patient's DNA to characterize the deficient pathway and identify potential therapeutics. This approach is becoming the standard of care in oncology, with the now routine and rapid next-generation sequencing of tumor tissue [104], and development of high-throughput screening to identify targeted therapies. The adaptation of such novel interventions into infectious diseases could provide a paradigm shift in our understanding of the intricate interactions between host and pathogen.
Notes
Financial support. This work was supported by the National Institutes of Health (grants R01 HL079395 and R01 HL1073989 to A. P. and K12HD047349 to D. A.).
Potential conflict of interest. Both authors: No reported conflicts. Both authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Klevens RM, Edwards JR, Richards CL Jr et al. Estimating health care-associated infections and deaths in U.S. hospitals, 2002. Public Health Rep 2007; 122:160–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vardakas KZ, Rafailidis PI, Konstantelias AA, Falagas ME. Predictors of mortality in patients with infections due to multi-drug resistant gram negative bacteria: the study, the patient, the bug or the drug? J Infect 2013; 66:401–14. [DOI] [PubMed] [Google Scholar]
- 3. Parker CM, Kutsogiannis J, Muscedere J et al. Ventilator-associated pneumonia caused by multidrug-resistant organisms or Pseudomonas aeruginosa: prevalence, incidence, risk factors, and outcomes. J Crit Care 2008; 23:18–26. [DOI] [PubMed] [Google Scholar]
- 4. Magill SS, Edwards JR, Bamberg W et al. Multistate point-prevalence survey of health care-associated infections. N Engl J Med 2014; 370:1198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Agency for Healthcare Research and Quality. AHRQ's Healthcare-Associated Infections Program. http://www.ahrq.gov/professionals/quality-patient-safety/hais/index.html Accessed 9 June 2016.
- 6. Slayton RB, Toth D, Lee BY et al. Vital signs: estimated effects of a coordinated approach for action to reduce antibiotic-resistant infections in health care facilities—United States. MMWR Morb Mortal Wkly Rep 2015; 64:826–31. [PMC free article] [PubMed] [Google Scholar]
- 7. McGann P, Snesrud E, Maybank R et al. Escherichia coli harboring mcr-1 and blaCTX-M on a novel IncF plasmid: first report of mcr-1 in the USA. Antimicrob Agents Chemother 2016; 60:4420–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Snitkin ES, Zelazny AM, Thomas PJ et al. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med 2012; 4:148ra16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koser CU, Holden MT, Ellington MJ et al. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N Engl J Med 2012; 366:2267–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Iskander KN, Osuchowski MF, Stearns-Kurosawa DJ et al. Sepsis: multiple abnormalities, heterogeneous responses, and evolving understanding. Physiol Rev 2013; 93:1247–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Singer M, Deutschman CS, Seymour CW et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016; 315:801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol 2016; 16:341–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nair GB, Niederman MS. Ventilator-associated pneumonia: present understanding and ongoing debates. Intensive Care Med 2015; 41:34–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kett DH, Cano E, Quartin AA et al. Implementation of guidelines for management of possible multidrug-resistant pneumonia in intensive care: an observational, multicentre cohort study. Lancet Infect Dis 2011; 11:181–9. [DOI] [PubMed] [Google Scholar]
- 15. Peleg AY, Hooper DC. Hospital-acquired infections due to gram-negative bacteria. N Engl J Med 2010; 362:1804–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Planet P. Life after USA300: the rise and fall of a superbug. J Infect Dis 2017; 215 (suppl 1); S71–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Randolph AG, Vaughn F, Sullivan R et al. Critically ill children during the 2009–2010 influenza pandemic in the United States. Pediatrics 2011; 128:e1450–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hageman JC, Uyeki TM, Francis JS et al. Severe community-acquired pneumonia due to Staphylococcus aureus, 2003–04 influenza season. Emerg Infect Dis 2006; 12:894–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McDanel JS, Perencevich E, Storm J et al. Increased mortality rates associated with Staphylococcus aureus and influenza co-infection, Maryland and Iowa, USA. Emerg Infect Dis 2016; 22:1253–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Geisinger E, Isberg R. Interplay between antibiotic resistance and virulence during disease promoted by multidrug-resistant bacteria. J Infect Dis 2017; 215 (suppl 1); S9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jorth P, Staudinger BJ, Wu X et al. Regional isolation drives bacterial diversification within cystic fibrosis lungs. Cell Host Microbe 2015; 18:307–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gomez-Simmonds A, Greenman M, Sullivan SB et al. Population structure of Klebsiella pneumoniae causing bloodstream infections at a New York City tertiary care hospital: diversification of multidrug-resistant isolates. J Clin Microbiol 2015; 53:2060–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol 2014; 22:686–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Murray GL, Tsyganov K, Kostoulias XP et al. Global gene expression profile of Acinetobacter baumannii during bacteremia. J Infect Dis 2017; 215 (suppl 1); S52–7. [DOI] [PubMed] [Google Scholar]
- 25. Gomez-Simmonds A, Hu Y, Sullivan SB, Wang Z, Whittier S, Uhlemann AC. Evidence from a New York City hospital of rising incidence of genetically diverse carbapenem-resistant Enterobacter cloacae and dominance of ST171, 2007–14. J Antimicrob Chemother 2016; 71:2351–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Girlich D, Poirel L, Nordmann P. Clonal distribution of multidrug-resistant Enterobacter cloacae. Diagn Microbiol Infect Dis 2015; 81:264–8. [DOI] [PubMed] [Google Scholar]
- 27. Uhlemann AC, Otto M, Lowy FD, DeLeo FR. Evolution of community- and healthcare-associated methicillin-resistant Staphylococcus aureus. Infect Genet Evol 2014; 21:563–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fernandez JF, Levine SM, Restrepo MI. Technologic advances in endotracheal tubes for prevention of ventilator-associated pneumonia. Chest 2012; 142:231–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. May RM, Hoffman MG, Sogo MJ et al. Micro-patterned surfaces reduce bacterial colonization and biofilm formation in vitro: Potential for enhancing endotracheal tube designs. Clin Transl Med 2014; 3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Proctor RA, von Eiff C, Kahl BC et al. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat Rev Microbiol 2006; 4:295–305. [DOI] [PubMed] [Google Scholar]
- 31. Rathinam VA, Fitzgerald KA. Inflammasome complexes: emerging mechanisms and effector functions. Cell 2016; 165:792–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Merrell DS, Falkow S. Frontal and stealth attack strategies in microbial pathogenesis. Nature 2004; 430:250–6. [DOI] [PubMed] [Google Scholar]
- 33. Chevret S, Hemmer M, Carlet J, Langer M. European Cooperative Group on Nosocomial Pneumonia. Incidence and risk factors of pneumonia acquired in intensive care units. Results from a multicenter prospective study on 996 patients. Intensive Care Med 1993; 19:256–64. [DOI] [PubMed] [Google Scholar]
- 34. Torres A, Serra-Batlles J, Ros E et al. Pulmonary aspiration of gastric contents in patients receiving mechanical ventilation: the effect of body position. Ann Intern Med 1992; 116:540–3. [DOI] [PubMed] [Google Scholar]
- 35. Whitsett JA, Alenghat T. Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat Immunol 2015; 16:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sebghati TA, Korhonen TK, Hornick DB, Clegg S. Characterization of the type 3 fimbrial adhesins of Klebsiella strains. Infect Immun 1998; 66:2887–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Menzies BE. The role of fibronectin binding proteins in the pathogenesis of Staphylococcus aureus infections. Curr Opin Infect Dis 2003; 16:225–9. [DOI] [PubMed] [Google Scholar]
- 38. Brown L, Wolf JM, Prados-Rosales R, Casadevall A. Through the wall: extracellular vesicles in gram-positive bacteria, mycobacteria and fungi. Nat Rev Microbiol 2015; 13:620–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Parker D, Planet PJ, Soong G, Narechania A, Prince A. Induction of type I interferon signaling determines the relative pathogenicity of Staphylococcus aureus strains. PLoS Pathog 2014; 10:e1003951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cohen TS, Parker D. Microbial pathogenesis and type III interferons. Cytokine Growth Factor Rev 2016; 29:45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Parker D. Impact of type I and III interferons on respiratory superinfections due to multidrug-resistant pathogens. J Infect Dis 2017; 215 (suppl 1); S58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Planet PJ, Parker D, Cohen TS et al. Lambda interferon restructures the nasal microbiome and increases susceptibility to Staphylococcus aureus superinfection. mBio 2016; 7:e01939–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hussell T, Bell TJ. Alveolar macrophages: plasticity in a tissue-specific context. Nat Rev Immunol 2014; 14:81–93. [DOI] [PubMed] [Google Scholar]
- 44. Kitur K, Parker D, Nieto P et al. Toxin-induced necroptosis is a major mechanism of Staphylococcus aureus lung damage. PLoS Pathog 2015; 11:e1004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Parker D, Ryan CL, Alonzo F III, Torres VJ, Planet PJ, Prince AS. CD4+ T cells promote the pathogenesis of Staphylococcus aureus pneumonia. J Infect Dis 2015; 211:835–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cohen TS, Prince AS. Activation of inflammasome signaling mediates pathology of acute P. aeruginosa pneumonia. J Clin Invest 2013; 123:1630–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Juan C, Peña C, Oliver A. Host and Pathogen biomarkers for severe Pseudomonas aeruginosa infections. J Infect Dis 2017; 215 (suppl 1); S44–51. [DOI] [PubMed] [Google Scholar]
- 48. Pier GB. The challenges and promises of new therapies for cystic fibrosis. J Exp Med 2012; 209:1235–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Price-Whelan A, Dietrich LE, Newman DK. Pyocyanin alters redox homeostasis and carbon flux through central metabolic pathways in Pseudomonas aeruginosa PA14. J Bacteriol 2007; 189:6372–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Morris AE, Liggitt HD, Hawn TR, Skerrett SJ. Role of Toll-like receptor 5 in the innate immune response to acute P. aeruginosa pneumonia. Am J Physiol Lung Cell Mol Physiol 2009; 297:L1112–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Miao EA, Mao DP, Yudkovsky N et al. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc Natl Acad Sci U S A 2010; 107:3076–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sutterwala FS, Mijares LA, Li L, Ogura Y, Kazmierczak BI, Flavell RA. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J Exp Med 2007; 204:3235–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Howell HA, Logan LK, Hauser AR. Type III secretion of ExoU is critical during early Pseudomonas aeruginosa pneumonia. mBio 2013; 4:e00032–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhao Y, Yang J, Shi J et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 2011; 477:596–600. [DOI] [PubMed] [Google Scholar]
- 55. Jones RN. Microbial etiologies of hospital-acquired bacterial pneumonia and ventilator-associated bacterial pneumonia. Clin Infect Dis 2010; 51(suppl 1):S81–7. [DOI] [PubMed] [Google Scholar]
- 56. Thammavongsa V, Kim HK, Missiakas D, Schneewind O. Staphylococcal manipulation of host immune responses. Nat Rev Microbiol 2015; 13:529–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Burkhardt O, Pletz MW, Mertgen CP, Welte T. Linezolid—the first oxazolidinone in the treatment of nosocomial MRSA pneumonia. Recent Pat Antiinfect Drug Discov 2007; 2:123–30. [DOI] [PubMed] [Google Scholar]
- 58. Sampedro GR, Bubeck Wardenburg J. Staphylococcus aureus in the ICU: are these golden grapes ripe for a new approach? J Infect Dis 2017; 215 (suppl 1); S64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Inoshima I, Inoshima N, Wilke GA et al. A Staphylococcus aureus pore-forming toxin subverts the activity of ADAM10 to cause lethal infection in mice. Nat Med 2011; 17:1310–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kebaier C, Chamberland RR, Allen IC et al. Staphylococcus aureus alpha-hemolysin mediates virulence in a murine model of severe pneumonia through activation of the NLRP3 inflammasome. J Infect Dis 2012; 205:807–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. DuMont AL, Yoong P, Liu X et al. Identification of a crucial residue required for Staphylococcus aureus LukAB cytotoxicity and receptor recognition. Infect Immun 2014; 82:1268–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kuipers A, Stapels DA, Weerwind LT et al. The S. aureus polysaccharide capsule and Efb-dependent fibrinogen shield act in concert to protect against phagocytosis. Microbiology 2016; 162:1185–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Viau M, Longo NS, Lipsky PE, Zouali M. Staphylococcal protein a deletes B-1a and marginal zone B lymphocytes expressing human immunoglobulins: an immune evasion mechanism. J Immunol 2005; 175:7719–27. [DOI] [PubMed] [Google Scholar]
- 64. Goodyear CS, Silverman GJ. Staphylococcal toxin induced preferential and prolonged in vivo deletion of innate-like B lymphocytes. Proc Natl Acad Sci U S A 2004; 101:11392–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lawlor MS, Hsu J, Rick PD, Miller VL. Identification of Klebsiella pneumoniae virulence determinants using an intranasal infection model. Mol Microbiol 2005; 58:1054–73. [DOI] [PubMed] [Google Scholar]
- 66. Henry CS, Rotman E, Lathem WW, Tyo KEJ, Hauser AR, Mandel MJ. Generation and validation of the iKp1289 metabolic 1 model for Klebsiella pneumoniae KPPR1. J Infect Dis 2017; 215 (suppl 1); S37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Siu LK, Fung CP, Chang FY et al. Molecular typing and virulence analysis of serotype K1 Klebsiella pneumoniae strains isolated from liver abscess patients and stool samples from noninfectious subjects in Hong Kong, Singapore, and Taiwan. J Clin Microbiol 2011; 49:3761–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cunha CB, Kassakian SZ, Chan R et al. Screening of nursing home residents for colonization with carbapenem-resistant Enterobacteriaceae admitted to acute care hospitals: incidence and risk factors. Am J Infect Control 2016; 44:126–30. [DOI] [PubMed] [Google Scholar]
- 69. Gomez-Simmonds A, Uhlemann AC. Clinical implications of genomic adaptation and evolution of carbapenem-resistant Klebsiella pneumoniae. J Infect Dis 2017; 215 (suppl 1); S18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Xiong H, Carter RA, Leiner IM et al. Distinct contributions of neutrophils and CCR2+ monocytes to pulmonary clearance of different Klebsiella pneumoniae strains. Infect Immun 2015; 83:3418–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kobayashi SD, Porter AR, Dorward DW et al. Phagocytosis and killing of carbapenem-resistant ST258 Klebsiella pneumoniae by human neutrophils. J Infect Dis 2016; 213:1615–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Xiong H, Keith JW, Samilo DW, Carter RA, Leiner IM, Pamer EG. Innate lymphocyte/Ly6Chi monocyte crosstalk promotes Klebsiella pneumoniae clearance. Cell 2016; 165:679–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. The grim prospect. The Economist, 2016. [Google Scholar]
- 74. World Health Organization. Antimicrobial resistance: global report on surveillance. http://apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf Accessed 27 June 2016.
- 75. Munoz-Price LS, Poirel L, Bonomo RA et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 2013; 13:785–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gaze W, O'Neill C, Wellington E, Hawkey P. Antibiotic resistance in the environment, with particular reference to MRSA. Adv Appl Microbiol 2008; 63:249–80. [DOI] [PubMed] [Google Scholar]
- 77. Stoesser N, Xayaheuang S, Vongsouvath M et al. Colonization with Enterobacteriaceae producing ESBLs in children attending pre-school childcare facilities in the Lao People's Democratic Republic. J Antimicrob Chemother 2015; 70:1893–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Scholte JB, van der Velde JI, Linssen CF et al. Ventilator-associated pneumonia caused by commensal oropharyngeal flora: a retrospective analysis of a prospectively collected database [published erratum appears in BMC Pulm Med 2015; 15:104]. BMC Pulm Med 2015; 15:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Becattini S, Taur Y, Pamer EG. Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol Med 2016; 22:458–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gauguet S, D'Ortona S, Ahnger-Pier K et al. Intestinal microbiota of mice influences resistance to Staphylococcus aureus pneumonia. Infect Immun 2015; 83:4003–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Li Q, Wang C, Tang C et al. Successful treatment of severe sepsis and diarrhea after vagotomy utilizing fecal microbiota transplantation: a case report. Crit Care 2015; 19:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bernasconi E, Pattaroni C, Koutsokera A; SysCLAD Consortium. Airway microbiota determines innate cell inflammatory or tissue remodeling profiles in lung transplantation. Am J Respir Crit Care Med 2016; PMID: 27248293. [DOI] [PubMed] [Google Scholar]
- 83. van Vught LA, Klein Klouwenberg PM, Spitoni C et al. Incidence, risk factors, and attributable mortality of secondary infections in the intensive care unit after admission for sepsis. JAMA 2016; 315:1469–79. [DOI] [PubMed] [Google Scholar]
- 84. Shahangian A, Chow EK, Tian X et al. Type I IFNs mediate development of postinfluenza bacterial pneumonia in mice. J Clin Invest 2009; 119:1910–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. McAleer JP, Kolls JK. Directing traffic: IL-17 and IL-22 coordinate pulmonary immune defense. Immunol Rev 2014; 260:129–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zankari E, Hasman H, Cosentino S et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 2012; 67:2640–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sweeney TE, Wong HR. Risk stratification and prognosis in sepsis: what have we learned from microarrays? Clin Chest Med 2016; 37:209–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Cao Z, Robinson RA. The role of proteomics in understanding biological mechanisms of sepsis. Proteomics Clin Appl 2014; 8:35–52. [DOI] [PubMed] [Google Scholar]
- 89. Brenner T, Uhle F, Fleming T et al. Soluble TREM-1 as a diagnostic and prognostic biomarker in patients with septic shock: an observational clinical study. Biomarkers 2016:1–7. [DOI] [PubMed] [Google Scholar]
- 90. Trasy D, Tanczos K, Nemeth M et al. Early procalcitonin kinetics and appropriateness of empirical antimicrobial therapy in critically ill patients: a prospective observational study. J Crit Care 2016; 34:50–5. [DOI] [PubMed] [Google Scholar]
- 91. Marshall JC. Why have clinical trials in sepsis failed? Trends Mol Med 2014; 20:195–203. [DOI] [PubMed] [Google Scholar]
- 92. Seok J, Warren HS, Cuenca AG et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A 2013; 110:3507–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Abraham E, Laterre PF, Garbino J et al. Lenercept (p55 tumor necrosis factor receptor fusion protein) in severe sepsis and early septic shock: a randomized, double-blind, placebo-controlled, multicenter phase III trial with 1,342 patients. Crit Care Med 2001; 29:503–10. [DOI] [PubMed] [Google Scholar]
- 94. Bernard GR, Francois B, Mira JP et al. Evaluating the efficacy and safety of two doses of the polyclonal anti-tumor necrosis factor-alpha fragment antibody AZD9773 in adult patients with severe sepsis and/or septic shock: randomized, double-blind, placebo-controlled phase IIb study. Crit Care Med 2014; 42:504–11. [DOI] [PubMed] [Google Scholar]
- 95. Shakoory B, Carcillo JA, Chatham WW et al. Interleukin-1 receptor blockade is associated with reduced mortality in sepsis patients with features of macrophage activation syndrome: reanalysis of a prior phase III trial. Crit Care Med 2016; 44:275–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Cohen TS, Hilliard JJ, Jones-Nelson O et al. Staphylococcus aureus alpha toxin potentiates opportunistic bacterial lung infections. Sci Transl Med 2016; 8:329ra31. [DOI] [PubMed] [Google Scholar]
- 97. Sawa T, Ito E, Nguyen VH, Haight M. Anti-PcrV antibody strategies against virulent Pseudomonas aeruginosa. Hum Vaccin Immunother 2014; 10:2843–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. de Luca A, Smeekens SP, Casagrande A et al. IL-1 receptor blockade restores autophagy and reduces inflammation in chronic granulomatous disease in mice and in humans. Proc Natl Acad Sci U S A 2014; 111:3526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Douglas IS, Price CS, Overdier KH et al. Rapid automated microscopy for microbiological surveillance of ventilator-associated pneumonia. Am J Respir Crit Care Med 2015; 191:566–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. May AK, Brady JS, Romano-Keeler J et al. A pilot study of the noninvasive assessment of the lung microbiota as a potential tool for the early diagnosis of ventilator-associated pneumonia. Chest 2015; 147:1494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Davenport EE, Burnham KL, Radhakrishnan J et al. Genomic landscape of the individual host response and outcomes in sepsis: a prospective cohort study. Lancet Respir Med 2016; 4:259–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Schulert GS, Zhang M, Fall N et al. Whole-exome sequencing reveals mutations in genes linked to hemophagocytic lymphohistiocytosis and macrophage activation syndrome in fatal cases of H1N1 influenza. J Infect Dis 2016; 213:1180–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ciancanelli MJ, Huang SX, Luthra P et al. Infectious disease. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science 2015; 348:448–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Joseph L. The clinical utility of molecular genetic cancer profiling. Expert Rev Mol Diagn 2016; 16:827–38. [DOI] [PubMed] [Google Scholar]