Abstract
Healthcare delivery has advanced due to the implementation of point-of-care testing, which is often performed within minutes to hours in minimally equipped laboratories or at home. Technologic advances are leading to point-of-care kits that incorporate nucleic acid–based assays, including polymerase chain reaction, isothermal amplification, ligation, and hybridization reactions. As a limited number of single-nucleotide polymorphisms are associated with clinically significant human immunodeficiency virus (HIV) drug resistance, assays to detect these mutations have been developed. Early versions of these assays have been used in research. This review summarizes the principles underlying each assay and discusses strategic needs for their incorporation into the management of HIV infection.
Keywords: HIV, drug resistance, point-of-care tests
Point-of-care tests (POCTs) are assays performed near the patient with rapid turnaround times that allow patient management during the same clinical encounter [1–3]. POCTs generally bypass transport of specimens to central laboratories, reduce specimen processing, and require less-skilled laboratory technicians. These simplifications can reduce the turnaround time for test results, and, in regions with limited laboratory infrastructure, can increase access to diagnostic tests (eg, human immunodeficiency virus [HIV] antibody detection and plasma HIV RNA quantification) [4]. POCTs to detect single-nucleotide polymorphisms (SNPs) associated with HIV drug resistance (HIVDR) mutations are desirable because these assays would (1) allow for faster institution of appropriate antiretroviral therapy (ART), permitting the associated improvements in patient’s health [5]; (2) likely cost less than Sanger sequencing, the most widely used method to detect HIVDR; and (3) be performed within the laboratory infrastructure in low-resource settings.
The new antiretroviral dolutegravir (DTG) rarely selected HIVDR in early clinical trials [6–8], leading to speculation that, with increased distribution and lower costs, pretreatment testing for HIVDR would not be needed in low-resource settings. However, HIVDR mutations are selected in individuals taking DTG monotherapy [9–14], which suggests that (1) for maximal efficacy of DTG-based ART regimens, HIV must be susceptible to coadministered nucleosides, and (2) testing for HIVDR to these nucleosides may be needed to sustain effectiveness of first-line DTG-based regimens. Moreover, testing for HIVDR at virologic failure (VF) may inform the decisions of clinicians prescribing subsequent ART regimens. Assays that combine viral load and testing for HIVDR could minimize the time from recognizing VF to assessing HIVDR and to selecting the next ART regimens.
Several assays in development for the rapid detection of HIVDR mutations in decentralized laboratories are described here. The developers of these tests are simplifying the methods to facilitate assay performance by minimally trained personnel and minimize the cost of POCTs.
OLA_SIMPLE V.1 FOR PRETREATMENT HIV DRUG RESISTANCE
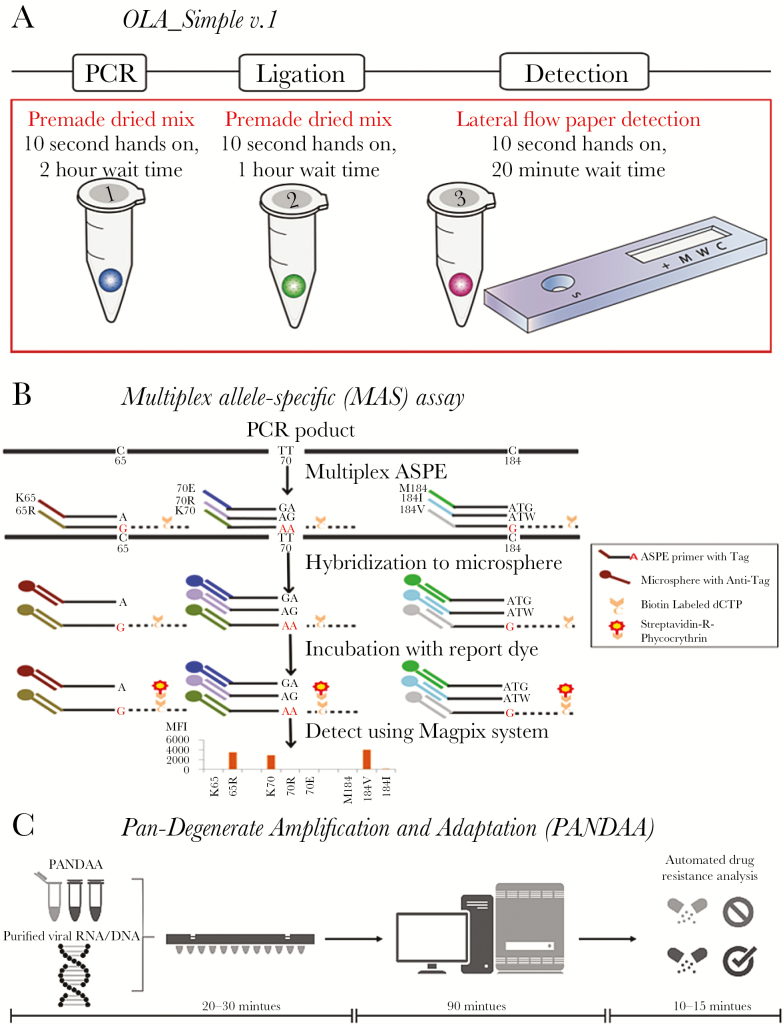
An oligonucleotide ligation-based assay (OLA) that detects HIVDR mutations K65R, K103N, Y181C, M184V, and G190A in HIV pol encoding reverse transcriptase was proven to detect pre-ART drug resistance (PDR) associated with virologic failure to nonnucleoside reverse transcriptase inhibitor (NNRTI)–based ART in Kenya [15–17]. The OLA can use patient-derived DNA or RNA from whole blood, plasma, or dried blood spot specimens. Nucleic acids are specifically amplified by polymerase chain reaction (PCR), annealed to labeled probes that are ligated and then detected by an enzyme-linked immunoassay [18–21]. Recently, OLA_Simple v.1 was developed for laboratories in low-resource settings. The kit shortens and simplifies workflow by combining lyophilized reagents and lateral flow paper detection (Figure 1A) using inexpensive instruments (ie, thermocycler, microfuge). OLA_Simple v.1 uses negatively immunoselected CD4 cells from whole blood, followed by cell lysis and PCR amplification of a region of HIV pol DNA (120 minutes). The mutant codons above, plus V106M, are detected after ligation of labeled probes (60 minutes). The dried reagent mixtures are stable at approximately 24°C and allow quick single-tube preparation (10 seconds). The paper detection (similar to pregnancy tests), read both visually and by a camera, has been benchmarked against the plate-based enzyme-linked immunoassay [22]. To detect resistance at VF, the next generation of OLA_Simple will isolate virion RNA from whole blood, reverse-transcribe the viral RNA, and employ isothermal or more rapid PCR amplification, all contained within plasticware to preclude amplicon cross-contamination.
Figure 1.
Point-of-care test to detect human immunodeficiency virus drug resistance. A, Simplified kit with single-use reagents to test one specimen for drug resistance mutations prior to nonnucleoside reverse transcriptase inhibitor–based antiretroviral therapy. Kit detects mutant codons predictive of virologic failure [17]. The kit amplifies DNA using premade dried polymerase chain reaction (PCR) mixture. The product is added to a dried ligation mix and subsequently detected in a paper-strip test cartridge. B, Targets are PCR amplified, then the multiplex allele-specific (MAS) assay uses allele-specific primer extension (ASPE) with specific primers mixed together in one reaction tube containing reaction reagent mixture and a template. When the primer complementary to the 3ʹ-terminal nucleotide of the target, primer extension occurs and biotinylated deoxycytodine triphosphates (dCTPs) are incorporated into the extended products. ASPE products are hybridized to microspheres through the specificity of “TAG”/”Anti-TAG” recognition and read with the suspension array system. C, Premixed pan-degenerate amplification and adaptation (PANDAA) with quantitative PCR (qPCR) enzymes, buffer, primers, and probes labeled with 3 distinct fluorophores to detect 2 drug resistance mutations and quantify total viral nucleic acid. Viral RNA from plasma, DNA from whole blood, or PCR amplicon previously generated for Sanger sequencing can be used as the input template. PANDAA is a one-step reaction that does not require a first-round cDNA synthesis or PCR step prior to qPCR. PANDAA can be run on any qPCR machine that can distinguish the FAM, VIC, and NED fluorophores (or equivalent fluorophores with a similar emission spectra). Automated data analysis allows the relative abundance of each drug resistance mutation to be quantified with additional data handling by the user.
ALLELE-SPECIFIC PCR
Allele-specific PCR (ASPCR) uses laboratory-based quantitative PCR (qPCR) to detect HIVDR mutations [23–27]. These assays rely on the 3ʹ-terminal nucleotide of primers for specificity that is enhanced by a mismatch at the adjacent base to discriminate between a HIVDR vs wild-type base. One such assay was recently developed for K65R, K103N, Y181C, and M184V in HIV type 1 (HIV-1) subtype C at YRG-CARE, Chennai, India. In preliminary analyses of 46 patients failing tenofovir, lamivudine, and NNRTI ART, ASPCR identified K65R not detected by Sanger sequencing in 4% of RNA and 13% of DNA samples in >5% of the HIV quasispecies [28].
MULTIPLEX ALLELE-SPECIFIC ASSAY
Multiplex allele–specific (MAS) assays use reverse-transcription PCR products spanning the protease and reverse transcriptase regions of HIV pol to perform allele-specific primer extension (ASPE) in a single well. Forty-five primers simultaneously detect the varied genotypes that encode 20 HIVDR amino acids to NNRTIs, nucleoside reverse transcriptase inhibitors (NRTIs), and protease inhibitors. Primers matching the 3ʹ-terminal nucleotide initiate primer extension with biotinylated deoxycytidine triphosphates. The ASPE products are uniquely annealed to microspheres through the specificity of “TAG”/”anti-TAG” recognition and are detected within a suspension array system (US$24000) linked to each microsphere by its internal dye, recording the dye intensity as mean fluorescence intensity (Figure 1B). The primers can detect all major HIVDR mutations associated with World Health Organization–recommended first- and second-line ART regimens (except integrase strand transfer inhibitors) in HIV subtype B or C viruses using plasma or dried blood spot specimens [29, 30]. The subtype C assay was implemented for a survey of transmitted HIVDR in Swaziland in 2011 and detected PDR at a sensitivity to comparable to Sanger sequencing [31]. An HIV-1 group M multisubtype MAS assay was also developed to identify DRM against tenofovir and emtricitabine used for preexposure HIV prophylaxis in high-risk populations [32].
PAN-DEGENERATE AMPLIFICATION AND ADAPTATION
Pan-degenerate amplification and adaptation (PANDAA) is an HIV subtype-independent assay that overcomes the HIV genomic heterogeneity that has previously precluded implementing probe-based qPCR to discriminate SNPs associated with drug resistance [33] (Figure 1C). With traditional qPCR, secondary polymorphisms within a probe-binding site, which are proximal to a DRM, prevent probe hybridization and generate false-negative results. Using highly degenerate primers that overlap with the probe-binding site, PANDAA adapts the targeted genomic region through site-directed mutagenesis during the initial qPCR cycles. This generates a homogeneous amplicon population whereby the only point of nucleotide variation within the probe-binding site is at the HIVDR mutation. PANDAA is under commercialization for both NNRTI/NRTI [34] and protease inhibitor–based ART failure management, and for ultrasensitive detection of low frequency (≥1%) mutations in all antiretroviral drug classes. PANDAA uses either RNA or DNA to detect 2 distinct mutations, as well as total viral copy number, in a multiplex one-step qPCR. The automated data analysis returns a percentage abundance of each mutation in the virus population within 2 hours of sample purification.
LIGATION ON RNA AMPLIFICATION
This assay-in-development uses one-step ligation to detect SNPs from RNA templates. In a single-tube assay, ligase, DNA polymerase and oligonucleotide probes are combined and subjected to qPCR of specific probes [35]. The RNA sample is initially denatured to open any secondary structures. Subsequently, ligation occurs during cycling between 25°C and 48°C, whereas the DNA polymerase remains in its chemically inactivated state. This is followed by heat activation of the polymerase and inactivation of the T4 DNA ligase, allowing DNA polymerase extension and endonuclease activity. Linear amplification of ligated K103N plasmid sensitively detected 1% mutant, with essentially 100% specificity conferred by the ligase. The theoretical advantage of ligation on RNA amplification over ASPCR assays is that the lower annealing temperature allows testing of more polymorphic clinical specimens.
SIMPLE METHOD TO AMPLIFY RNA TARGETS
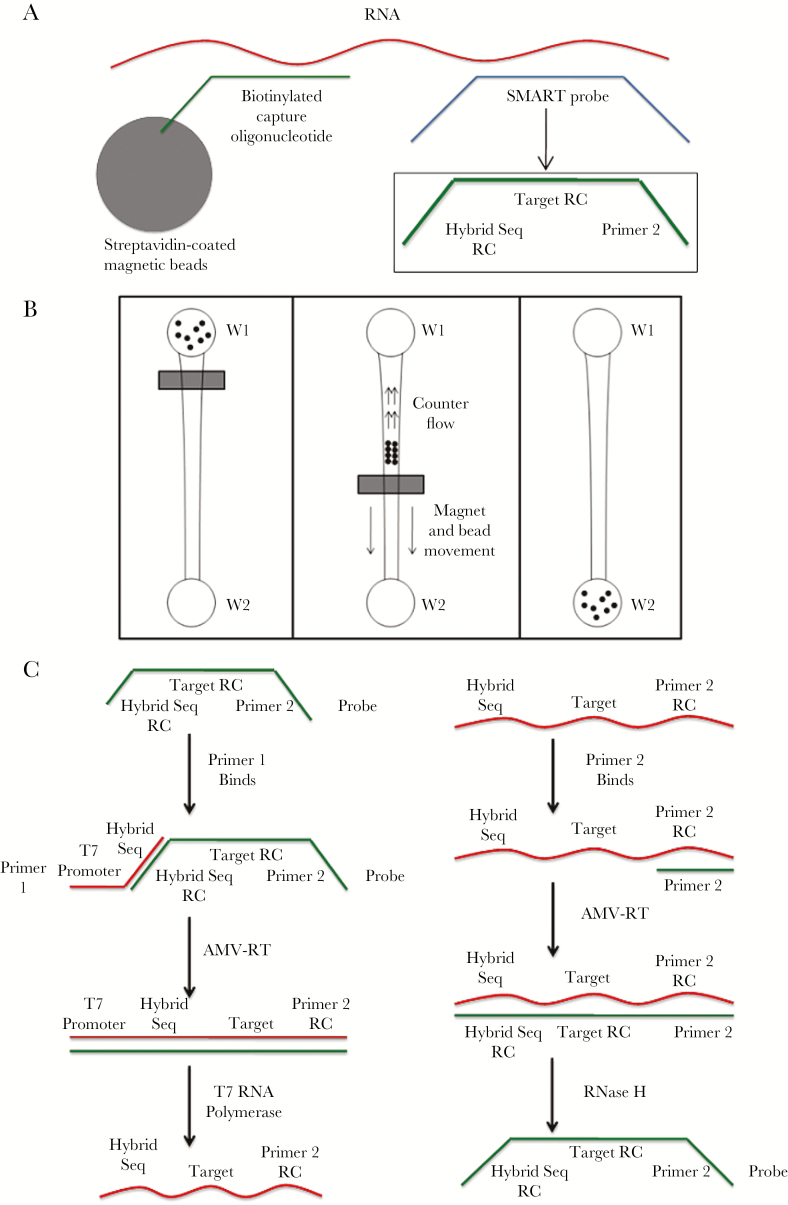
The simple method to amplify RNA targets (SMART), another early-stage method (Figure 2), combines HIV RNA in a solution with streptavidin-coated beads conjugated with biotinylated capture oligonucleotides and SMART probes. The solution is introduced into a microchip well followed by magnetic separation of the bead-bound complex. SMART probes are amplified isothermally [36]. In a preliminary experiment using synthetic DNA sequences with K103N, 6000 copies/mL were detected within 180 minutes [37], which is a relatively short duration for reactions designed to detect SNPs.
Figure 2.
Overview of the simple method to amplify RNA targets (SMART). A, The RNA sequences, isolated from a clinical sample, are put in a solution containing 2 probes that bind to specific sequences within the target RNA, allowing testing of RNA mutations. The first probe (capture probe) is attached to a magnetic bead, which hybridizes RNA via a general consensus sequence. The 5ʹ end of the biotinylated capture probe readily binds to a streptavidin-coated magnetic bead. Simultaneously a mutation-specific probe molecule (~25 nucleotides, SMART or amplification probe) that hybridizes with the RNA. The center sequence of the SMART probe molecule is the reverse complement (RC) of the target strain sequence. The sequence of the 2 flanking ends (called “hybrid seq RC” and “primer 2”) of the SMART probe can be adjusted by the user to optimize amplification reaction kinetics. At the conclusion of (A), a chain-linked molecule complex of streptavidin-coated magnetic beads—biotinylated oligo–RNA—SMART probe is centered about the target region of the RNA. B, The magnetic bead bound complex is microfluidically separated from the unbound SMART probes and/or other molecules in reservoir W1 to reservoir W2. C, Amplification of the SMART probe is performed via an isothermal scheme that utilizes the designed primer sequences for optimal reaction kinetics. Here, various enzymes (AMV-RT, RNase, and T7 polymerase) are used for transcription and amplification. Subsequently, molecular beacons or other fluorescent molecules can be used for detection of amplified SMART probes. The SMART scheme employs an isothermal and exponential amplification of SMART probes, which is suitable for point-of-care testing.
IMPLEMENTATION OF POINT-OF-CARE TESTING
Given the technical complexity and equipment needed for Sanger (or next-generation) sequencing, some experts have advocated for performing large-scale HIVDR testing in centralized laboratories [38]. However, off-site testing leads to delays in clinical decisions compared to on-site POCTs. Implementing POCTs on a large-scale with rapid turnaround times in resource-limited settings would require hiring sufficient staff to perform the diagnostic tests in the clinics, training healthcare workers to properly interpret these tests, and ensuring clinics have adequate infrastructure, including necessary equipment and reliable electricity [3, 39]. Operational research would be needed to incorporate POCTs into daily workflows [40]. Reliable supply chains would be needed to ensure that health centers do not experience POCT stockouts, which has been a challenge for ART medications and laboratory supplies in some settings [41]. POCTs should be regulated for reliable manufacturing, technicians’ skill in conducting the assay should be monitored by a proficiency testing program, and the POCTs should be proven to offer clinically meaningful data. In short, there is a need for efficient and reliable POCT programs, not simply innovative POCT technology [3]. The large-scale implementation of GeneXpert MTB/RIF in South Africa for the diagnosis of tuberculosis provides several years of experience in strengthening health systems, and relevant lessons learned can be applied to HIVDR POCTs [42].
CONCLUSIONS
Given that HIVDR mutations have historically developed to all ART regimens, resistance will likely continue to diminish the long-term success of ART programs. Accessible testing for HIVDR can enable an evidence-based approach to medical care. With further development, the assays described here could offer POCTs that may improve clinical outcomes. However, as additional ART regimens become available, ongoing surveillance will be needed to monitor HIVDR mutations, and relevant mutations will need to be added to POCTs. Implementation science, outcomes research, and mathematical modeling can help evaluate the use of POCTs to optimize use of limited resources to address HIVDR and improve health outcomes.
Notes
Disclaimer. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Financial support. This work was supported by the National Institutes of Health (grant numbers R01 AI110375 to N. P., I. A. B., B. L., J. L., R. M. K., L. M. F.; R01 AI100037 to I. A. B., M. H. C., L. M. F.; R21AI106392 to R. K., S. S.; R44AI118441 to I. J. M.; R44AI128974 to I. J. M.; 272201600022C-0-0-1, P30AI042853 to I. J. M.; UM1 AI106716 to I. A. B., L. M. F.; K12 HD000850 to H. A. D.); Massachusetts General Hospital (to N. P.); P30AI042853 and Brown University Dean’s Award (to R. K. and A. T.); Centers for Disease Control and Prevention (to G. Z. and C. Y.); and President’s Emergency Plan for AIDS Relief (PEPFAR) through the US Centers for Disease Control and Prevention (to G. Z. and C. Y.).
Supplement sponsorship. This work is part of a supplement sponsored by the National Institute of Allergy and Infectious Disease, NIH, and the Centers for Disease Control and Prevention.
Potential conflicts of interest. J. L. reports grants from the National Institute of Allergy and Infectious Diseases (NIAID), during the conduct of the study; other from Nexgenia, outside the submitted work. In addition, J. L. has a 2 patents issued (US9429570 and US9080933). I. J. M. reports grants from the National Institutes of Health, and personal fees and other from Aldatu Biosciences. In addition, L. J. M. has a patent (US20160244817 A1) licensed to Aldatu Biosciences. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Peeling RW, Holmes KK, Mabey D, Ronald A. Rapid tests for sexually transmitted infections (STIs): the way forward. Sex Transm Infect 2006; 82(suppl 5):v1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schito M, Peter TF, Cavanaugh S et al. . Opportunities and challenges for cost-efficient implementation of new point-of-care diagnostics for HIV and tuberculosis. J Infect Dis 2012; 205(suppl 2):S169–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pai NP, Vadnais C, Denkinger C, Engel N, Pai M. Point-of-care testing for infectious diseases: diversity, complexity, and barriers in low- and middle-income countries. PLoS Med 2012; 9:e1001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yager P, Domingo GJ, Gerdes J. Point-of-care diagnostics for global health. Annu Rev Biomed Eng 2008; 10:107–44. [DOI] [PubMed] [Google Scholar]
- 5. Lundgren JD, Babiker AG, Gordin F et al. ; for the Insight Start Study Group Initiation of antiretroviral therapy in early asymptomatic HIV infection. N Engl J Med 2015; 373:795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Walmsley SL, Antela A, Clumeck N et al. ; SINGLE Investigators Dolutegravir plus abacavir-lamivudine for the treatment of HIV-1 infection. N Engl J Med 2013; 369:1807–18. [DOI] [PubMed] [Google Scholar]
- 7. Raffi F, Jaeger H, Quiros-Roldan E et al. ; Extended SPRING-2 Study Group Once-daily dolutegravir versus twice-daily raltegravir in antiretroviral-naive adults with HIV-1 infection (SPRING-2 study): 96 week results from a randomised, double-blind, non-inferiority trial. Lancet Infect Dis 2013; 13:927–35. [DOI] [PubMed] [Google Scholar]
- 8. Clotet B, Feinberg J, van Lunzen J et al. ; ING114915 Study Team Once-daily dolutegravir versus darunavir plus ritonavir in antiretroviral-naive adults with HIV-1 infection (FLAMINGO): 48 week results from the randomised open-label phase 3b study. Lancet 2014; 383:2222–31. [DOI] [PubMed] [Google Scholar]
- 9. Brenner BG, Thomas R, Blanco JL et al. . Development of a G118R mutation in HIV-1 integrase following a switch to dolutegravir monotherapy leading to cross-resistance to integrase inhibitors. J Antimicrob Chemother 2016; 71:1948–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blanco JL, Oldenbuettel C, Thomas R et al. . Pathways of resistance in subjects failing dolutegravir monotherapy. In: Conference on Retroviruses and Opportunistic Infections, Seattle, WA, 2017. [Google Scholar]
- 11. Quashie PK, Mesplède T, Han YS et al. . Biochemical analysis of the role of G118R-linked dolutegravir drug resistance substitutions in HIV-1 integrase. Antimicrob Agents Chemother 2013; 57:6223–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Quashie PK, Oliviera M, Veres T et al. . Differential effects of the G118R, H51Y, and E138K resistance substitutions in different subtypes of HIV integrase. J Virol 2015; 89:3163–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Malet I, Gimferrer Arriaga L, Artese A et al. . New raltegravir resistance pathways induce broad cross-resistance to all currently used integrase inhibitors. J Antimicrob Chemother 2014; 69:2118–22. [DOI] [PubMed] [Google Scholar]
- 14. Munir S, Thierry E, Malet I et al. . G118R and F121Y mutations identified in patients failing raltegravir treatment confer dolutegravir resistance. J Antimicrob Chemother 2015; 70:739–49. [DOI] [PubMed] [Google Scholar]
- 15. Chung MH, Beck IA, Dross S et al. . Oligonucleotide ligation assay detects HIV drug resistance associated with virologic failure among antiretroviral-naive adults in Kenya. J Acquir Immune Defic Syndr 2014; 67:246–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chung M, Beck I, Levine M et al. . Prospective randomized HIV drug resistance testing of Kenyans before first-line ART. In: Conference on Retroviruses and Opportunistic Infections, Boston, MA, 2016. [Google Scholar]
- 17. Beck IA, Levine M, Milne R et al. . Impact of pre-treatment HIV-drug resistance on virologic outcome of first-line art. In: Conference on Retroviruses and Opportunistic Infections, Seattle, WA, 2017. [Google Scholar]
- 18. Frenkel LM, Wagner LE 2nd, Atwood SM, Cummins TJ, Dewhurst S. Specific, sensitive, and rapid assay for human immunodeficiency virus type 1 pol mutations associated with resistance to zidovudine and didanosine. J Clin Microbiol 1995; 33:342–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Beck IA, Mahalanabis M, Pepper G et al. . Rapid and sensitive oligonucleotide ligation assay for detection of mutations in human immunodeficiency virus type 1 associated with high-level resistance to protease inhibitors. J Clin Microbiol 2002; 40:1413–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Beck IA, Crowell C, Kittoe R et al. . Optimization of the oligonucleotide ligation assay, a rapid and inexpensive test for detection of HIV-1 drug resistance mutations, for non-North American variants. J Acquir Immune Defic Syndr 2008; 48:418–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Micek MA, Blanco AJ, Beck IA et al. . Nevirapine resistance by timing of HIV type 1 infection in infants treated with single-dose nevirapine. Clin Infect Dis 2010; 50:1405–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Panpradist N, Beck IA, Chung MH, Kiarie JN, Frenkel LM, Lutz BR. Simplified paper format for detecting HIV drug resistance in clinical specimens by oligonucleotide ligation. PLoS One 2016; 11:e0145962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Loubser S, Balfe P, Sherman G, Hammer S, Kuhn L, Morris L. Decay of K103N mutants in cellular DNA and plasma RNA after single-dose nevirapine to reduce mother-to-child HIV transmission. AIDS 2006; 20:995–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Palmer S, Boltz V, Martinson N et al. . Persistence of nevirapine-resistant HIV-1 in women after single-dose nevirapine therapy for prevention of maternal-to-fetal HIV-1 transmission. Proc Natl Acad Sci U S A 2006; 103:7094–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Paredes R, Cheng I, Kuritzkes DR, Tuomala RE; Women and Infants Transmission Study (WITS) Group Postpartum antiretroviral drug resistance in HIV-1-infected women receiving pregnancy-limited antiretroviral therapy. AIDS 2010; 24:45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Metzner KJ, Rauch P, Braun P et al. . Prevalence of key resistance mutations K65R, K103N, and M184V as minority HIV-1 variants in chronically HIV-1 infected, treatment-naïve patients. J Clin Virol 2011; 50:156–61. [DOI] [PubMed] [Google Scholar]
- 27. Boltz VF, Bao Y, Lockman S et al. ; OCTANE/A5208 Team Low-frequency nevirapine (NVP)-resistant HIV-1 variants are not associated with failure of antiretroviral therapy in women without prior exposure to single-dose NVP. J Infect Dis 2014; 209:703–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Saravanan S, Dinesha T, Poongulali S et al. . A point mutation resistance assay to optimize HIV-1 subtype C antiretroviral therapy in India. In: 21st International AIDS Conference, Durban, South Africa, 2016. [Google Scholar]
- 29. Zhang G, Cai F, Zhou Z et al. . Simultaneous detection of major drug resistance mutations in the protease and reverse transcriptase genes for HIV-1 subtype C by use of a multiplex allele-specific assay. J Clin Microbiol 2013; 51:3666–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang G, Cai F, de Rivera IL et al. . Simultaneous detection of major drug resistance mutations of HIV-1 subtype B viruses from dried blood spot specimens by multiplex allele-specific assay. J Clin Microbiol 2016; 54:220–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Beard R, Duong Y, Maphalala G et al. . National prevalence of transmitted HIV drug resistance in Swaziland in 2011. In: 20th International AIDS Conference, Melbourne, Australia, 2014. [Google Scholar]
- 32. Zhang G, Guo H, DeVos J et al. . A sensitive and single-tube detection assay for identifying HIV-1 drug-resistance mutations associated with pre-exposure prophylaxis. In: 20th Conference on Retroviruses and Opportunistic Infections, Atlanta, GA, 2013. [Google Scholar]
- 33. Clutter DS, Rojas Sanchez P, Rhee SY, Shafer RW. Genetic variability of HIV-1 for drug resistance assay development. Viruses 2016; 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rhee SY, Jordan MR, Raizes E et al. . HIV-1 drug resistance mutations: potential applications for point-of-care genotypic resistance testing. PLoS One 2015; 10:e0145772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang L, Wang J, Coetzer M, Angione S, Kantor R, Tripathi A. One-step ligation on RNA amplification for the detection of point mutations. J Mol Diagn 2015; 17:679–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McCalla S. Kinetics, mass transport, and adsorption in diagnostic microfluidic devices. PhD dissertation, Brown University, 2012. [Google Scholar]
- 37. Morabito K, Kantor R, Tai W, Schreier L, Tripathi A. Detection of HIV-1 minority variants containing the K103N drug-resistance mutation using a simple method to amplify RNA targets (SMART). J Mol Diagn 2013; 15:401–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Inzaule SC, Ondoa P, Peter T et al. . Affordable HIV drug-resistance testing for monitoring of antiretroviral therapy in sub-Saharan Africa. Lancet Infect Dis 2016; 16:e267–75. [DOI] [PubMed] [Google Scholar]
- 39. Pai NP, Wilkinson S, Deli-Houssein R et al. . Barriers to implementation of rapid and point-of-care tests for human immunodeficiency virus infection: findings from a systematic review (1996–2014). Point Care 2015; 14:81–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Palamountain KM, Baker J, Cowan EP et al. . Perspectives on introduction and implementation of new point-of-care diagnostic tests. J Infect Dis 2012; 205(suppl 2):S181–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Médicins Sans Frontières. Empty shelves come back tomorrow: ARV stockouts undermine efforts to fight HIV 2015. http://cdn.msf.mx/sites/mexico/files/attachments/empty_shelves_report_msf-01-12-2015_0.pdf. Accessed April 2017.
- 42. Stevens WS, Scott L, Noble L, Gous N, Dheda K. Impact of the GeneXpert MTB/RIF technology on tuberculosis control. Microbiol Spectr 2017; 5. [DOI] [PubMed] [Google Scholar]