The genetic basis of tomato fruit variability in cuticular lipid composition is analyzed using the Solanum pennellii introgression line population. A QTL map for cuticular lipids and associated genes is presented.
Keywords: Cuticle, cuticular waxes, cutin monomers, introgression line population, tomato fruit, quantitative trait loci
Abstract
The cuticle is a specialized cell wall layer that covers the outermost surface of the epidermal cells and has important implications for fruit permeability and pathogen susceptibility. In order to decipher the genetic control of tomato fruit cuticle composition, an introgression line (IL) population derived from a biparental cross between Solanum pennellii (LA0716) and the Solanum lycopersicum cultivar M82 was used to build a first map of associated quantitative trait loci (QTLs). A total of 24 cuticular waxes and 26 cutin monomers were determined. They showed changes associated with 18 genomic regions distributed in nine chromosomes affecting 19 ILs. Out of the five main fruit cuticular components described for the wild species S. pennellii, three of them were associated with IL3.4, IL12.1, and IL7.4.1, causing an increase in n-alkanes (≥C30), a decrease in amyrin content, and a decrease in cuticle thickness of ~50%, respectively. Moreover, we also found a QTL associated with increased levels of amyrins in IL3.4. In addition, we propose some candidate genes on the basis of their differential gene expression and single nucleotide polymorphism variability between the introgressed and the recurrent alleles, which will be the subjects of further investigation.
Introduction
Modern tomato (Solanum lycopersicum) cultivars are the result of several rounds of domestication and improvement (Frary et al., 2010, Gur et al., 2011; Blanca et al. 2012). Wild relatives of cultivated tomato have often been used in modern breeding as a source of genes for abiotic stress tolerance (Pineda et al., 2012), disease resistance (Sandbrink et al., 1995), and increased yield (Gur and Zamir, 2004). The wild species Solanum pennellii is particularly well adapted to conditions of extreme environmental stress (Yeats and Rose, 2013; Bolger et al., 2014) and has been an important germplasm donor for S. lycopersicum. Special characteristics of the S. pennellii leaf cuticle, including those involved in regulating cuticle-related genes (Bolger et al., 2014), enable it to adapt to these conditions. These characteristics probably make S. pennellii an ideal source of genes for cuticle reinforcement. One of the genetic resources most frequently used to identify genomic regions associated with quantitative traits of interest is the S. lycopersicum cv. M82 × S. pennellii interspecific introgression line (IL) population (Eshed and Zamir 1995). In this set of lines, the full genome of the wild species is represented as small introgressed fragments in 75 nearly-isogenic lines of the cultivar M82 genetic background. Of the more than 3000 quantitative trait loci (QTLs) reported (Alseekh et al., 2013), nearly 2000 control fruit quality traits, for example, Brix, sugar content, volatiles, firmness, or flavonoid content (Causse et al., 2002; Chapman et al., 2012; Alseekh et al., 2013). However, to date, the S. pennellii population has not been used to study QTLs involved in fruit cuticle lipid composition.
The cuticle has been described as the outermost layer covering all aerial plant organs. It is largely composed of acyl lipids, polyphenols, polysaccharides, and proteins (Riederer and Müller, 2006; Segado et al., 2016a). The acyl lipids found in the cuticle include a small fraction of soluble waxes (Samuels et al., 2008; Yeats and Rose, 2013) embedded in an insoluble matrix of the polymer cutin. Cutin represents the major component of the cuticle and is an amorphous, viscoelastic layer consisting of C16 and C18 polyhydroxy and epoxyhydroxy fatty acids (Pollard et al., 2008; Beisson et al., 2012). Additionally, plant cuticles can contain non-acyl lipid components such as pentacyclic triterpenols or polyphenols (Buschhaus and Jetter, 2011). All these compounds define a layered structure in which, schematically, the cuticle is placed on the top, the cell wall at the base, and an interconnected region with both type of polymers is found in between. This layered structure, which was described almost a half century ago and is extensively reviewed by Jeffree (2006), has been questioned in the past few years. Some scientists have started to consider the plant cuticle as a non-layered structure derived from a modified epidermal cell wall (Guzman-Puyol et al., 2015; Segado et al., 2016a, 2016b) and to study the interactions between the cuticle components from a new perspective. For example, in the embryo there were cell wall components, mostly polysaccharides and pectins, partially merged with the cutin matrix in an amorphous structure during early stages of development (Javelle et al., 2011). It is accepted that the cuticular membrane is deposited on and within the cell wall scaffold, creating a single, efficient barrier that plays important roles in plant physiology, ecology, development, and fitness (Riederer and Müller, 2006; Hen-Avivi et al., 2014; Martin and Rose, 2014). Hence, the study of the cuticle represents an interesting goal for breeding, considering that any modification in the cuticle will be related to important traits such as yield, by preventing water loss or pathogen infections (Hovav et al., 2007; Isaacson et al., 2009; Buxdorf et al., 2014), and quality, by modifying glossiness or firmness (Domínguez et al., 2011; Chapman et al., 2012; Lara et al., 2014).
In recent years, the tomato cuticle has received increased attention that has focused on molecular dissection (Bourdenx et al., 2011, Girard et al., 2012, Hen-Avivi et al., 2014, Lashbrooke et al., 2015), biomechanical properties (Dominguez et al., 2011), water loss (Hovav et al., 2007; Isaacson et al., 2009), fruit cracking (Domínguez et al., 2012), pathogen infection, and glossiness (Kunst and Samuels, 2009). Most of these studies resulted from the characterization of a small number of mutants and/or transgenic lines displaying different molecular, biomechanical, or visual phenotypes. However, the biosynthesis of cuticular lipids and their regulatory mechanisms remain unclear (Yeats and Rose, 2013; Hen-Avivi et al., 2014; Martin and Rose, 2014). The use of genetic variability derived from wild tomato relatives or breeding/mapping populations to identify the genetic basis of natural variation in cuticle traits has been very limited. A number of tomato fruit cuticle studies in wild species (Yeats et al., 2012) have revealed substantial variation in cuticle composition in genetically distant wild relatives, including S. pennellii. The availability of ILs, including global gene expression patterns for each population line, and the recent release of the S. pennellii genome constitute an excellent opportunity to investigate the genetic basis of tomato cuticle variability (Bolger et al., 2014). Therefore, the aim of the present study was to screen QTLs involved in cuticular wax and cutin monomer composition in tomato fruit cuticle using the S. pennellii IL population resource, and to propose candidate genes associated with them.
Materials and methods
Plant material
We used the interspecific IL population derived from the cross between the cultivated tomato S. lycopersicum cv. M82 and the wild tomato S. pennellii, LA716. Of the 75 ILs (Eshed and Zamir, 1995), 72 were analyzed for cutin monomer composition (the missing lines were IL3.1, IL5.2, and the subILs 4.3.2 and 6.2.2) and 63 were studied for cuticular waxes (the excluded lines were IL1.2, IL3.1, IL3.2, IL4.4, IL5.3, IL5.5, IL7.3, and the subILs 2.4, 5.2, 6.2.2, 9.1.3, and 9.3.1). The population was cultivated in a single-block trial, each block consisting of four consecutive plants, except for M82, which consisted of 10 plants, under standard growth conditions (natural light and controlled temperature, 24 °C during the day and 18 °C at night) in a greenhouse during the 2010 summer season in Rehovot, Israel. Four fruits per plant were harvested at the red stage, except for lines producing small fruits, for which eight red fruits were collected. To validate the genetic effects, in 2011 a number of selected lines were grown at the same facilities and under the same conditions. In this case, plants were randomized in the greenhouse.
GC-FID and GC-MS combined profiling for cuticular lipids and data analysis
An efficient method recently described for wax screening (Fernandez-Moreno et al., 2016) was used to screen the S. pennellii IL population for variability of cuticular lipid composition in fruit. Cuticular waxes were extracted as described previously (Fernandez-Moreno et al., 2016). Briefly, discs of ~19 cm2 from tomato fruit pericarp were excised from the equatorial region of each fruit and enzymatically treated to remove the non-cutinized cell wall components. Then, the material was dried for 2 days and then dipped twice into 4 ml chloroform for 30 s at room temperature to extract both epicuticular and major intracuticular waxes. The internal standard n-tetracosane (0.2 mg/ml) was added to each extracted sample. Next, dewaxed tissue discs were exhaustively delipidated in methanol:chloroform (1:1 v/v) for 15 days and then air-dried for 12 hours to proceed with the cutin extraction. To analyze cutin monomer variability, we used a well-known method that involves cutin depolymerization by a methyl-esterification reaction followed by cutin monomer extraction in an organic solvent (Franke et al., 2005; Shi et al., 2012; Lashbrooke et al., 2015). In brief, a portion of the initial dewaxed sample (~13 cm2) was used for depolymerization of the cutin matrix by incubating the samples in 2 ml methanol/BF3 (Aldrich catalogue no.: B1252-250MLl) for 16 hours at 70 °C. Then, n-tetracosane was added to the reaction mix before adding 2 ml of saturated NaHCO3–water solution to stop the reaction. Next, cutin monomers were extracted in 2 ml chloroform three times, transferring the organic fraction to an empty vial each time. The pooled organic fraction was washed twice with 1 ml distilled water and the remaining water was removed by anhydrous Na2SO4 salt. Finally, extracts were concentrated to 1 ml using a stream of N2 gas and stored at −20 °C until further analysis.
Cuticular lipid profiling was performed by gas chromatography–flame ionization detector (GC-FID) using fruit tissue from three different plants per IL as previously described (Adato et al., 2009). Additionally, a reference sample was prepared containing a mix of the extracts (1:1:1 v/v) from the three plants per IL and used to identify cuticular waxes and cutin monomers using gas chromatography–mass spectrometry (GC-MS) as described by Mintz-Oron et al. (2008) and Fernandez-Moreno et al. (2016). For the validation analysis of the best QTL obtained during the initial screening, a second-year analysis was performed on selected ILs using the more informative GC-MS technology to quantify the cuticular lipids instead of the GC-FID used in the first-year screening.
Wax constituents were identified in the total ion chromatogram using their Kovats indices and by comparing their mass spectra with those of authentic standards as well as data from the literature as detailed by Fernandez-Moreno et al. (2016). Compound quantification was calculated as the ratio of the peak area of each compound and that of the internal standard. These data were normalized on the basis of the total amount of internal standard (5 μg per mixture) and the total extraction area (18.84 cm2 for cuticular waxes and 12.56 cm2 for cutin monomers) to produce the final dataset (μg cm−2). Before statistical analysis, data were pretreated by centering , where i=1…I rows referring to the metabolites and j=1…J columns referring to the different ILs, and scaling using range scaling to facilitate the observation of minor cuticular lipids from the total set of components (van den Berg et al., 2006).
Hierarchical clustering analysis
Phenotype variation in the S. pennellii IL population for both cuticular wax and cutin monomer metabolites was represented using a two-way hierarchical clustering analysis (HCA). A normalized, pre-treated (centered and range-scaled) dataset was used for the HCA. Statistical analysis was performed with JMP® 10.0.0 software (SAS Institute Inc., 2012).
QTL analysis and mapping
Data were recorded for each single plant and the mean was calculated for each IL and used for subsequent analysis. Plants were not considered as biological replicates as they were grown in the same block. Cuticular trait QTL analysis was conducted by single nucleotide polymorphism (SNP) marker–trait association by one-way ANOVA. A set of 216 Solcap_SNP genetic markers previously genotyped by Ofner et al. (2016), distributed across the 72 ILs (see Supplementary Table S1 at JXB online), were used. These genetic markers allowed the identification of a total of 72 bins, each defined by the recombination points in the IL collection. Significant bins were selected for marker genotype comparison by a Tukey’s honest significant difference test, adjusting the statistical threshold to declare significant effects to α=0.05/72, following Bonferroni correction.
For QTL verification, selected ILs containing S. pennellii introgressions on genomic regions associated with cutin/wax composition were evaluated in 2011. IL means were compared with the M82 control mean by a Dunnett’s or Student’s t test (P<0.05), depending on whether multiple mean or pairwise mean comparisons were performed, respectively; in addition, effects of IL, year, and the interaction IL × year were assessed by two-way ANOVA. Statistical analysis was done using JMP® 10.0.0 software (SAS Institute Inc., 2012).
Candidate gene selection
Candidate genes for each significant metabolite QTL (mQTL) were selected on the basis of: (i) the corresponding gene annotation being related to the biosynthetic process affecting the metabolite differentially accumulated in the QTL; (ii) the differential gene expression level in the IL of interest being consistent with its participation. Expression values were obtained from the transcriptome profiling of the same S. pennellii IL population in the experiment, available at http://ted.bti.cornell.edu/. In this experiment, RPKM (reads per kilobase per million mapped reads) values were obtained from this website and correspond to Illumina RNA-sequencing analysis on samples of red tomato fruits; and (iii) SNP differences between S. lycopersicum and S. pennellii producing non-conserved protein changes. To determine this, we used the S. lycopersicum mRNA sequence of each candidate gene to perform a BLASTx (translated nucleotide to protein database) search against the S. pennellii protein database (http://www.solgenomics.net/tools/blast/). The protein alignment resulting from this BLASTx was inspected carefully to identify non-conserved amino acid substitutions. Based on the above, we named candidate genes as any gene in the QTL interval that satisfied two or three of these criteria.
Histological staining and electron microscopy
Thin sections of fixed and embedded pericarp fruit tissue samples were stained for lipids with Sudan IV as previously described (Isaacson et al., 2009; Shi et al., 2012). For transmission electron microscopy (TEM), 9 mm2 fruit exocarp pieces were fixed using paraformaldehyde, glutaraldehyde, and osmium tetroxide solutions, and embedded in EPON resin following the protocol developed at the Electron Microscopy Unit of the Weizmann Institute of Science (Rehovot, Israel), available online at the website of the Electron Microscopy Unit of the Weizmann Institute of Sciences http://www.weizmann.ac.il/Chemical_Research_Support/EM_Unit/links. Imaging was performed using a Tecnai T12 transmission electron microscope (FEI). Ultra-thin sections were also used for fluorescence microscopy (Nikon fluorescence microscope, Nikon Eclipse e-800, Japan) and to measure cuticle thickness using ImageJ software.
Results
Cuticular lipids identified in the IL population
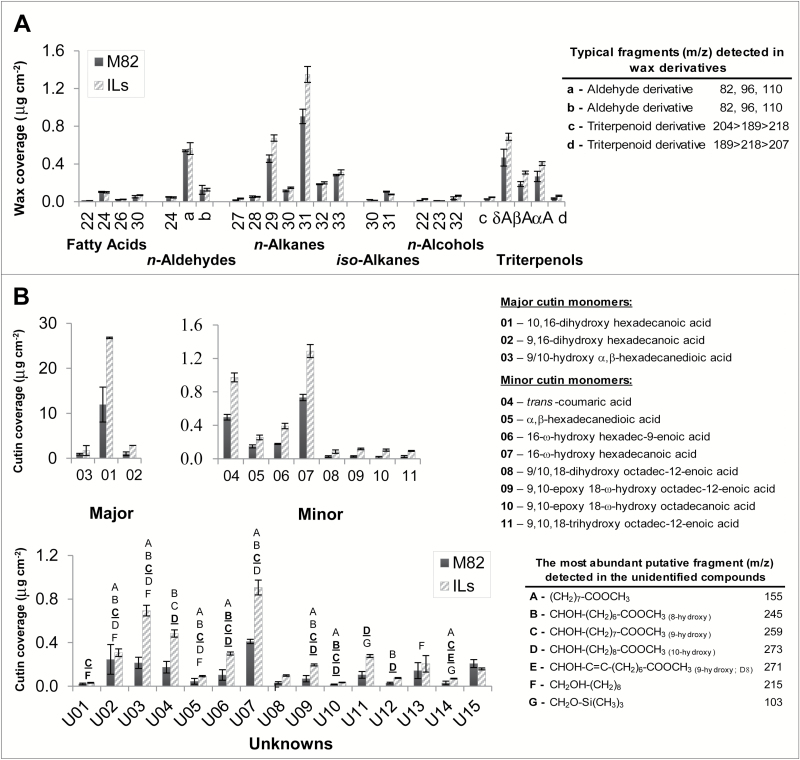
A set of 24 cuticular waxes, including both epicuticular and the most abundant intracuticular waxes, was identified and used for the initial screening. The cuticular waxes identified and quantified were the following (Fig. 1A): even-numbered fatty acids C22, C24, C26, and C30; n-aldehyde C24; odd-numbered n-alkanes C27–C33 and even-numbered n-alkanes C28–C32; iso-alkanes C30 and C31; n-alcohols C22, C23, and C32; α-, β-and -amyrins (pentacyclic triterpenols). We detected a further two waxes with spectra similar to those of the n-aldehydes (labelled a, b in Fig. 1A) and others with spectra typical of pentacyclic triterpenoids (labelled c, d in Fig. 1A). The major waxes detected were the odd n-alkanes C29, C31, and C33, and the three amyrins (Fig. 1A).
Fig. 1.
Changes in cuticular lipid coverage in the S. pennellii IL population. A. Wax coverage. B. Cutin monomer coverage. For the bars representing the parental S. lycopersicum cultivar M82, n=3; bars for ILs represent pooled data (n=63 and n=73 for wax and cutin monomer datasets, respectively). In A, aldehyde derivatives that were detected in the population are shown as a and b; triterpenol derivatives are shown as c and d. In B, uncharacterized cutin monomers are represented by ‘U’ and some of their structural information is provided. Data are presented as mean±SD.
For the cutin screening, a set of 11 cutin monomers was used (Fig. 1B): six C16:X monomers (labelled #01–#03 and #05–#07 in Fig. 1B), including 10/9,16 dihydroxyhexadecanoic acid, the major cutin monomer accumulating in this population, (#01 in Fig. 1B); four C18:X monomers (compounds #08–#11 in Fig. 1B); and one phenolic compound, trans-coumaric acid (#04 in Fig. 1B). An additional set of 15 cutin monomers was also detected (Fig. 1B). Their complete structures were not identified but they were characterized by the presence of structural features such as the 9(10)-midchain-hydroxyl group or some saturation. In total, a set of 50 cuticular lipid components (CLCs; i.e. 24 cuticular waxes and 26 cutin monomers) was detected in the IL population.
Screening for cuticular lipid composition in the S. pennellii IL population
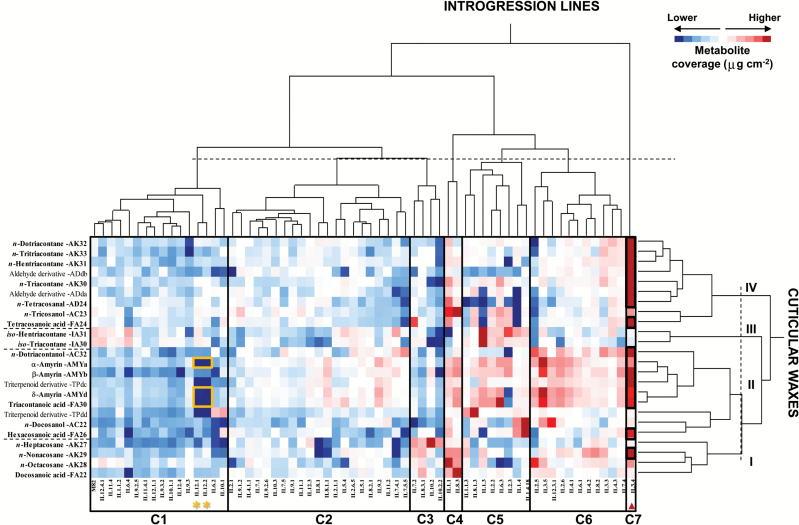
To determine the variability of the 50 CLCs across the population and the parental S. lycopersicum M82, two independent two-way hierarchical clustering analyses (HCAs) were performed, one for ILs and cuticular waxes (Fig. 2) and the other for ILs and cutin monomers (Fig. 3). The first HCA indicated that the ILs were grouped into seven clusters on the basis of wax coverage (Fig. 2), ranging from lines with lower wax levels in cluster C1 to those with higher levels in cluster C7. HCA results also showed that cuticular waxes could be organized according to the profiles obtained across the IL population, and this produced four clusters based on their abundance but also according to the class of wax. Thus, wax clusters II and IV comprised the most abundant cuticular waxes in the population and included amyrins and n-alkanes with more than C30 carbons, respectively, whereas wax clusters I and III contained the least abundant waxes and included n-alkanes with less than C30 carbons and iso-alkanes, respectively. In general, the IL population showed limited variability in fruit cuticular wax coverage (Fig. 2): just over half of the lines (58%) had similar wax coverage to the parental S. lycopersicum M82 (see clusters 1 and 2 in Fig. 2). The remaining lines showed higher differences in specific groups of waxes or, even, in particular wax compounds, when compared with the parental M82 (see clusters 3–7 in Fig. 2). For example, ILs in clusters C3 and C4 accumulated higher levels of n-alkanes with shorter chain length (<C30) than the rest of the population, whereas IL3.4 from cluster C7 accumulated higher levels of n-alkanes with more than C30 carbons. In addition, the triterpenoid wax class was found to be the most variable group in the entire population (see cluster II in Fig. 2), ranging from nearly no amyrin accumulation in IL12.1 and IL12.2 (in cluster C1) to almost a three-fold increase in amyrin levels in lines IL3.5 and IL3.4 (in clusters C6 and C7, respectively) when compared with the parental M82. In contrast, one of the least variable wax classes in the IL population was the iso-alkane class (see cluster III in Fig. 2). Here, the only exception was IL1.3 in cluster C5, which accumulated unusually high levels for both C30 and C31iso-alkanes by comparison with both the entire population and the parental M82.
Fig. 2.
Two-way hierarchical clustering heatmap of cuticular waxes and S. pennellii ILs. The ILs were grouped into seven clusters (C1–C7); cuticular waxes were classified into four clusters (I–IV). Wax coverage (μg cm−2) for each compound in every line is represented by a colored gradient, the intensity of which ranges from dark blue, corresponding to lower wax accumulation levels, to dark red, corresponding to higher wax accumulation levels. The vlcfa3.4 and amy3.4 QTLs in IL3.4, containing cuticular waxes with different levels of accumulation relative to the parental S. lycopersicum M82 (black squares) in the second-year experiments (Student’s t test, α<0.05) are highlighted with a red triangle. Asterisks indicate the amy12.1 QTL also described in previous reports (Yeats et al., 2012; Bolger et al., 2014; Ofner et al., 2016). (This figure is available in colour at JXB online.)
Fig. 3.
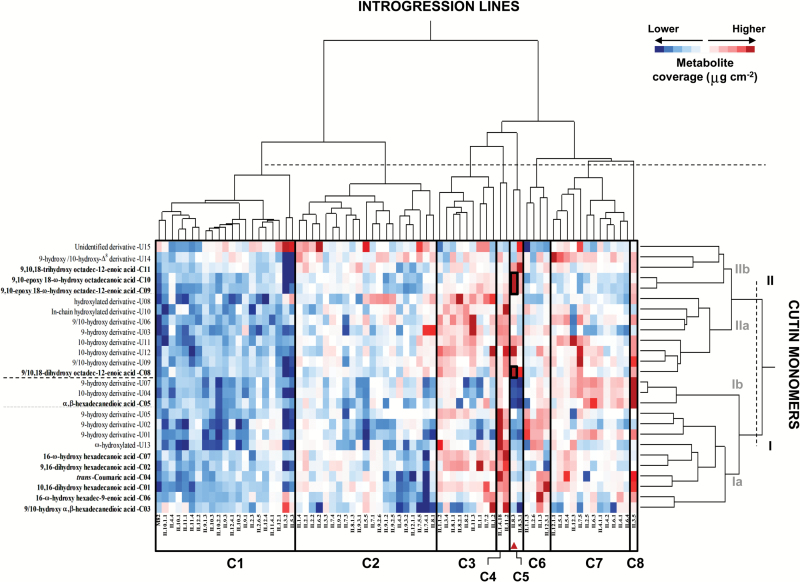
Two-way hierarchical clustering heatmap of cutin monomers and S. pennellii ILs. The ILs were classified into eight clusters (C1–C8); cutin monomers were classified into two main clusters (I and II) and four subclusters (Ia, Ib, IIa, and IIb). Cutin coverage (μg cm−2) for each monomer in every line is represented by a colored gradient, the intensity of which ranges from dark blue, corresponding to lower wax accumulation levels, to dark red, corresponding to higher wax accumulation levels. The ehfa8.3 QTL in IL8.3 containing cutin monomers with different levels of accumulation relative to the parental S. lycopersicum M82 (black squares) in the second-year experiments (Dunnett’s test, P<0.05) is highlighted with a red triangle. (This figure is available in colour at JXB online.)
HCA of cutin monomer composition organized the ILs into eight clusters (Fig. 3). ILs were grouped according to their cutin coverage, from lower levels in cluster C1 to higher levels in cluster C8 (Fig. 3). Interestingly, the 11 cutin monomers were grouped by their chain length, with C16:X monomers in cluster I and C18:X monomers in cluster II (Fig. 3). However, the subgrouping of two monomers—α,β-hexadecanedioic acid in cluster I and 9/10, 18-dihydroxy octadec-12-enoic acid in cluster II—differed from that of the other monomers in each respective cluster (see subclusters Ib and IIa in Fig. 3, respectively). The unidentified cutin monomers were also distributed across the four subclusters, probably by chain length and hydroxylation and/or saturation levels. For example, a putative mid-chain hydroxylated C16:X monomer (labelled U13 in Fig. 3) was clustered together with other C16:X monomers; and a putative 9,10,18-trihydroxy octadecanoic acid (U15) was grouped together with its Δ12-unsaturated counterpart, the 9,10,18-trihydroxy octadec-12-enoic acid in the same subcluster (Fig. 3). Based on the cutin monomer profiles, the IL population also showed cluster associations. In fact, five ILs displayed three clearly different patterns of cutin accumulation: IL3.5 over-accumulated mostly cutin monomers from cluster I (C16:X); IL8.3 and IL8.3.1 showed over-accumulation only of monomers belonging to cluster II (C18:X), particularly those in subcluster IIb; and IL1.4.18 and IL11.2 displayed over-accumulation of almost every cutin monomer in clusters I and II (Fig. 3).
A draft cuticular lipid composition QTL map for tomato fruit in the S. pennellii IL population
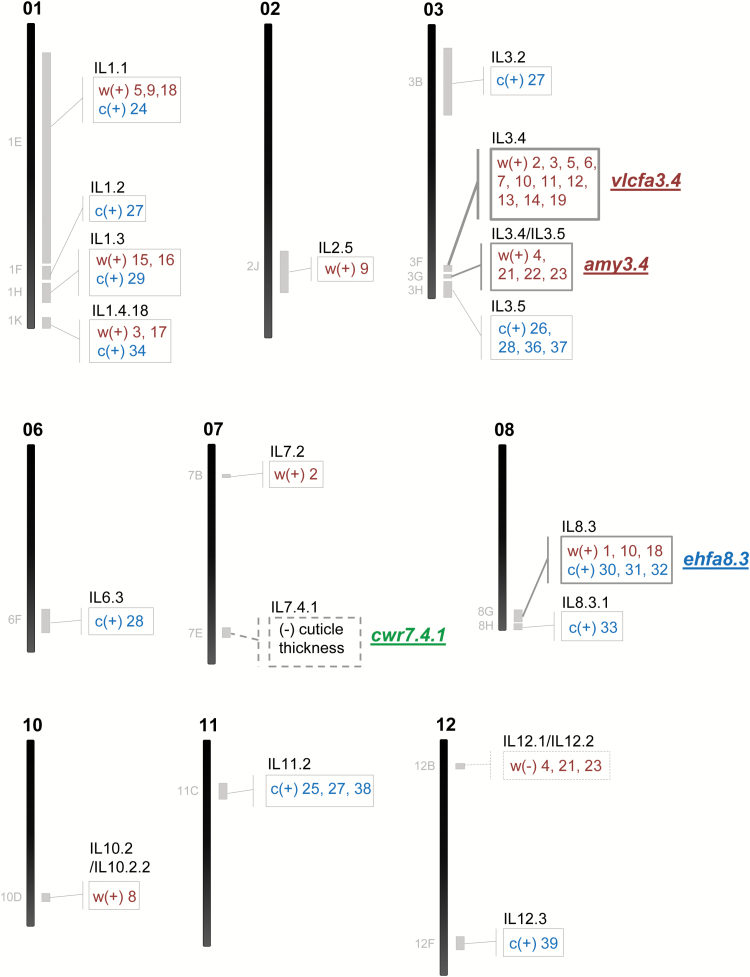
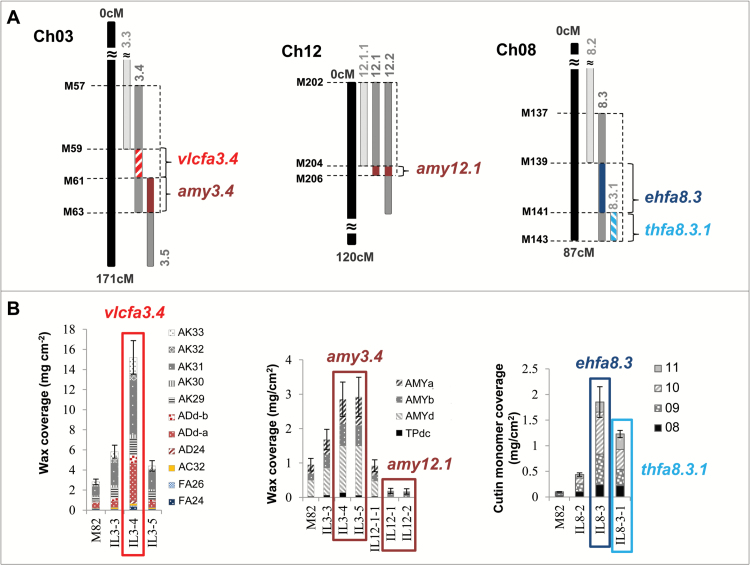
Cuticular waxes and cutin monomers were associated to SNPs by means of one-way ANOVAs using the IL genotype matrix from Ofner et al. (2016). SNPs located in nine different chromosomes (except chromosomes 4, 5, and 9), which were included in the introgressed S. pennellii regions of 19 ILs (~26% of the population), were associated to cuticular wax and/or cutin monomer variations. Table 1 summarizes the ILs containing genomic regions associated with differential metabolite accumulations; these metabolites are also mapped in Fig. 4 and indicated in Figs 2 and 3. Most of these genomic regions were associated with just one or two differentially accumulated cuticular lipid compounds. However, five of these genomic regions were associated with changes in a larger set of cuticular lipids. We focused on these five genomic regions, which were located in four ILs belonging to three chromosomes, affecting the composition of both cuticular waxes and cutin monomers. Accumulation of the major cuticular waxes detected in the population (i.e. n-alkanes with >C30 carbons in length and amyrins) was significantly altered in IL3.4 and IL12.1. Thus, whereas IL3.4 exhibited increased accumulation of both types of major cuticular waxes, IL12.1 showed reduced levels for just one type (amyrins). These changes were associated with three different genomic regions: two of them mapped to IL3.4 and the third to IL12.1 (Fig. 5A). The first of these regions mapped to a 600 kb interval in IL3.4 not shared by either IL3.3 or IL3.5 (Fig. 5B), which was associated with increased levels of n-aldehydes and n-alkanes (Fig. 5B). The second region in IL3.4 mapped to a 410 kb interval shared with IL3.5 (Fig. 5A) and also associated with increased levels of the three amyrins (Fig. 5B). The third region mapped to a 0.98 Mb interval in IL12.1 shared with IL12.2 (Fig. 5A), which was associated with a dramatic reduction in levels two of the three amyrins: δ- and α-amyrins (Fig. 5B). The metabolic phenotype observed in this last region has been reported previously in tomato fruit using both the S. pennellii and Solanum habrochaites IL populations (Yeats et al., 2012), and more recently in tomato leaves using the same S. pennellii IL population (Bolger et al., 2014; Ofner et al., 2016). In fact, two TRITERPENOID SYNTHASE genes, SlTTS1 and SlTTS2 (Solyc12g006530 and Solyc12g006520, respectively; Table 2, Supplementary Tables S2 and S3) involved in the conversion of epoxysqualene to amyrins and other triterpenoids were described in this interval. SlTTS1 was described as a specific β-amyrin synthase, whereas SlTTS2 seemed to be a general amyrin synthase, producing mostly δ-amyrin and then α-amyrin (Wang et al., 2011; Yeats et al., 2012). As the metabolic phenotype for neither of the other two regions in IL3.4 had been reported previously, we wanted to check whether they would be also a consistent QTL. With the purpose of validating these two QTLs, IL3.4 was analyzed and evaluated in a second experiment the following year. Similar to what we observed in the first season, the three amyrins and most of the n-alkanes and n-aldehydes were differentially over-accumulated in IL3.4 (Supplementary Fig. S1A). On the basis of these results, two wax-related QTLs in IL3.4, a very-long-chain fatty acid QTL (vlcfa3.4) and an amyrin QTL (amy3.4), were defined.
Table 1.
QTLs for cuticular lipid composition in tomato fruit uncovered in the S. pennellii introgression line population
| # | Chain length | Other name | M82spa | Introgressed region affectedb |
| Waxes | ||||
| Fatty acids | ||||
| 1 | 22 | 0.007 ± 0.0008 | IL8.2/IL8.3 | IL8.3 (0.04 ± 0.034) | IL8.3.1 (0.02 ± 0.015) | |
| 2 | 24 | 0.1 ± 0.01 | IL3.4 (0.3 ± 0.11) | IL7.2 (0.3 ± 0.13) | |
| 3 | 26 | 0.02 ± 0.002 | IL1.4.18 (0.1 ± 0.01) | IL3.3/IL3.4 (0.1 ± 0.02) | |
| 4 | 30 | 0.05 ± 0.021 | IL3.4 (0.1 ± 0.02)/IL3.5 (0.1 ± 0.03) | IL12.1 (ND)/IL12.2 (0.005 ± 0.0001)↓ | |
| n-Aldehydes | ||||
| 5 | 24 | 0.05 ± 0.007 | IL1.1 (0.1 ± 0.02) | IL3.3/IL3.4 (0.3 ± 0.12) | IL3.4 | IL3.4/IL3.5 | |
| 6 | derivative a | 0.5 ± 0.02 | IL3.3/IL3.4 (3.7 ± 1.68) | IL3.4 | IL3.4/IL3.5 | |
| 7 | derivative b | 0.1 ± 0.08 | IL3.3/IL3.4 (1.0 ± 0.28) | IL3.4 | IL3.4/IL3.5 | |
| n-Alkanes | ||||
| 8 | 27 | 0.02 ± 0.005 | IL10.2 (0.2 ± 0.09) | IL10.2.2 (0.1 ± 0.01) | |
| 9 | 28 | 0.05 ± 0.015 | IL1.1 (0.2 ± 0.06) | IL2.5 (0.2 ± 0.02) | |
| 10 | 29 | 0.5 ± 0.07 | IL3.4 (1.6 ± 0.29) | IL8.2 (1.3 ± 0.77) / IL8.3 (1.3 ± 0.82) | |
| 11 | 30 | 0.1 ± 0.01 | IL3.4 (0.4 ± 0.09) | |
| 12 | 31 | 0.9 ± 0.13 | IL3.4 (5.3 ± 1.97) | |
| 13 | 32 | 0.2 ± 0.01 | IL3.4 (0.7 ± 0.28) | |
| 14 | 33 | 0.3 ± 0.01 | IL3.3 (0.8 ± 0.30)/IL3.4 (1.6 ± 0.70) | |
| iso-Alkanes | ||||
| 15 | 30 | 0.02 ± 0.002 | IL1.3 (0.04 ± 0.001) | IL1.3/IL1.4 | |
| 16 | 31 | 0.1 ± 0.01 | IL1.3 (0.2 ± 0.01) | IL1.3/IL1.4 | |
| n-Alcohols | ||||
| 17 | 22 | 0.01 ± 0.002 | IL1.4.18 (0.2 ± 0.01) | |
| 18 | 23 | 0.01 ± 0.002 | IL1.1 (0.02 ± 0.011) | IL8.3 (0.02 ± 0.018) | |
| 19 | 32 | 0.04 ± 0.025 | IL3.3 (0.2 ± 0.02)/IL3.4 (0.2 ± 0.05) | IL3.4 | IL3.4/IL3.5 (0.1 ± 0.03) | |
| Triterpenols | ||||
| 20 | derivative c | 0.03 ± 0.002 | IL3.3 (0.06 ± 0.006)/IL3.4 (0.1 ± 0.01) | IL3.4 | IL3.4/IL3.5 (0.06 ± 0.052) | |
| 21 | δ-Amyrin | 0.5 ± 0.16 | IL3.4 (1.4 ± 0.024)/IL3.5 (1.4 ± 0.032) | IL12.1 (0.01 ± 0.002)/IL12.2 (ND)↓ | |
| 22 | β-Amyrin | 0.2 ± 0.05 | IL3.4 (0.6 ± 0.06)/IL3.5 (0.6 ± 0.20) | |
| 23 | α-Amyrin | 0.3 ± 0.09 | IL3.4 (0.7 ± 0.11)/IL3.5 (0.8 ± 0.20) | IL12.1 (0.01 ± 0.001)/IL12.2 (0.01 ± 0.006)↓ | |
| Cutin monomers | ||||
| Major cutin monomers | ||||
| 24 | 16 | 9,16-dihydroxy hexadecanoic acid | 1.0 ± 0.68 | IL1.1 (7.7 ± 1.51) |
| 25 | 16 | 9(10)-hydroxy α,β-hexadecanedioic acid | 0.9 ± 0.36 | IL11.2 (4.2 ± 0.71) |
| Minor cutin monomers | ||||
| 26 | trans-coumaric acid | 0.5 ± 0.06 | IL3.5 (2.4 ± 1.02) | |
| 27 | 16 | α,β-hexadecanedioic acid | 0.2 ± 0.03 | IL1.2 (1.1 ± 0.74) | IL3.2 (1.0 ± 0.34) | IL11.2 (1.1 ± 0.04) |
| 28 | 16 | 16-ω-hydroxy hexadec-9-enoic acid | 0.2 ± 0.01 | IL3.5 (1.5 ± 0.87) | IL6.3 (1.3 ± 0.92) |
| 29 | 16 | 16-ω-hydroxy hexadecanoic acid | 0.7 ± 0.07 | IL1.3 (3.9 ± 2.61) |
| 30 | 18 | 9,18-dihydroxy octadec-12-enoic acid | 0.03 ± 0.017 | IL8.3 (0.2 ± 0.09) | IL8.3.1 (0.2 ± 0.09) |
| 31 | 18 | 9,10-epoxy 18-ω-hydroxy octadec-12-enoic acid | 0.03 ± 0.012 | IL8.3 (0.6 ± 0.16) | IL8.3.1 (0.3 ± 0.18) |
| 32 | 18 | 9,10-epoxy 18-ω-hydroxy octadecanoic acid | 0.02 ± 0.018 | IL8.3 (0.8 ± 0.19) | IL8.3.1 (0.4 ± 0.26) |
| 33 | 18 | 9,10,18-trihydroxy octadec-12-enoic acid | 0.03 ± 0.024 | IL8.3 (0.2 ± 0.04) | IL8.3.1 (0.3 ± 0.05) |
| Unidentified cutin monomers | ||||
| 34 | Unknown 02 | 0.3 ± 0.24 | IL1.4.18 (1.6 ± 0.26) | |
| 35 | Unknown 03 | 0.2 ± 0.09 | IL11.2 (1.6 ± 0.54)/IL11.3 (2.1 ± 0.44) | IL11.3 | |
| 36 | Unknown 04 | 0.2 ± 0.09 | IL3.5 (1.7 ± 0.86) | |
| 37 | Unknown 07 | 0.4 ± 0.03 | IL3.5 (3.3 ± 1.71) | |
| 38 | Unknown 10 | 0.02 ± 0.005 | IL11.2 (0.1 ± 0.02) | IL11.2/IL11.3 | |
| 39 | Unknown 11 | 0.1 ± 0.05 | IL12.3 (0.3 ± 0.05) | |
a Wax coverage (μg cm−2) and standard deviation (n=3) are provided for parental M82sp.
b wax coverage (μg cm−2) and standard deviation (n=3) are provided for those lines showing values significant (P<0.0005) by one-way ANOVA.
ND Non-detected metabolites.
Only negative QTLs are in bold type and indicated with an arrow (↓).
Fig. 4.
QTL map for cuticular lipid composition in tomato fruit uncovered in the S. pennellii IL population. The ILs containing significant changes for some cuticular lipids during the screening are indicated along the chromosomes, on the right side (see # numbers in Table 1). Waxes (w) are shown in red and cutin monomers (c) in blue. Most QTLs were positive (+) (cuticular lipid accumulation was higher in the IL than in the parental M82); negative (–) QTLs are represented with a dashed line in the map. The bins associated with the metabolic changes are also shown, on the left side of each chromosome (see Supplementary Table S1). The four QTLs confirmed by the second-year experiments (vlcfa3.4, amy3.4, cwr7.4.1, and ehfa8.3) are indicated on the map. See also details in Figs 5 and 6. (This figure is available in colour at JXB online.)
Fig. 5.
QTLs associated with the main cuticular lipid traits of S. pennellii fruits. A. Chromosome intervals. B. CLC metabolites with differential accumulation in association with each of the intervals in A. AC, alcohol; AD, aldehyde; ADd, aldehyde derivative; AK, alkane; amy, amyrins; ehfa, epoxyhydroxy fatty acids; FA, fatty acid; thfa, trihydroxy fatty acids; TPd, triterpenoid derivative; vlcfa, very-long-chain fatty acids. See cutin monomers #08–#11 in Fig. 1. For more genetic marker information, see Supplementary Table S1. Data are presented as mean±SD. (This figure is available in colour at JXB online.)
Table 2.
Candidate genes proposed for the five QTLs associated with the three of the main traits in S. pennellii fruit cuticle
| QTLa | ILs | Chr | Bin (Mb)b | Phenotype | Gene ID | Annotationc |
| Waxes | ||||||
| vlcfa3.4 | IL3.4 | 3 | M60d (0.68) | Increased VLCFAs from decarbonylation pathway | Solyc03g117800 | Homologous to AtCER3 (ECERIFERUM 3) |
| amy3.4 | IL3.4/IL3.5 | 3 | M61–M63 (0.41) | Increased α-, β- and δ-amyrins | Solyc03g118540 | Homologous to AtJAZ3 (JASMONATE-ZIM-DOMAIN PROTEIN 3) |
| amy12.1 | IL12.1/IL12.2 | 12 | M205–M206 (0.98) | Decreased δ- and α-amyrins | Solyc12g006530 | SlTTS1 (TRITERPENOID SYNTHASE 1) |
| Solyc12g006520 | SlTTS2 (TRITERPENOID SYNTHASE 2) | |||||
| Cutin monomers | ||||||
| ehfa8.3 | IL8.3 | 8 | M140e (0.54) | Increased epoxyhydroxy fatty acids | Solyc08g081220 | Homologous to AtCYP86A8/LCR (LACERATA) |
| Cuticle thickness | ||||||
| cwr7.4.1 | IL7.4.1 | 7 | M122–M123 (2.18) | Reduced cuticle thickness | Solyc07g056000 | Homologous to AtXTR7 |
| Solyc07g055990 | Homologous to AtXTR7 | |||||
| Solyc07g056290 | Uncharacterized tomato POLYGALACTURONASE 28 | |||||
| Solyc07g055920 | Homologous to tomato TAGL1 (SHATTERPROOF 1) | |||||
amy, amyrins; Chr, chromosome; ehfa, epoxyhydroxy fatty acids; thfa, trihydroxy fatty acid, vlcfa, very-long-chain fatty acids.
a The IL number is included for each QTL.
b Genetic markers are described in Supplementary Table S1. The size of the interval in million bases (Mb) is provided.
c Gene annotations were obtained from the SL2.40 tomato database (http://www.solgenomics.net/).
d The phenotype observed for vlcfa3.4 was stronger in the non-overlapping interval of IL3.4. However, it was also observed for the overlapping regions IL3.3/IL3.4 (M57–M59) and IL3.4/IL3.5 (M61–M63).
e The phenotype observed for ehfa8.3 was stronger in the non-overlapping interval of IL8.3. However, it was also observed for the overlapping regions IL8.2/IL8.3 (M137–M139) and IL8.3/IL8.3.1 (M141–M143).
Most of the minor cutin monomers were found to be differentially accumulated in the IL population, but not the major cutin monomer, 10,16-dihydroxyhexadecanoic acid. Similar to what was observed for cuticular waxes, most of the ILs affected in their cutin monomer composition showed differential accumulation of one or two monomers. Only two genomic regions belonging to two overlapping ILs in chromosome 8 showed alterations in more than two cutin monomers (Table 1). The first of these regions mapped to a 540 kb interval in IL8.3 (Fig. 5A), which was associated with an increase in three C18 cutin monomers: 9,10-epoxy 18-ω-hydroxy octadecanoic acid, 9,10-epoxy 18-ω-hydroxy octadec-12-enoic acid, and 9/10,18-dihydroxy octadec-12-enoic acid (Fig. 5B). The second region mapped to a 360 kb interval shared by both IL8.3 and IL8.3.1 (Fig. 5A), which was associated with an increase in 9,10,18-trihydroxy octadec-12-enoic acid (Fig. 5B). In a similar way as for the promising regions in IL3.4, we also checked both IL8.3 and IL8.3.1 in a second-year experiment. In the second year, only two cutin monomers in IL8.3 were validated, while the other two were more variable for the parental M82 than in the first year. This explained why they were not significant enough in any of the IL8.3 and IL8.3.1 even when they were accumulated in higher levels in both ILs (Supplementary Fig. S1). Thus, we validated one of the two QTLs in chromosome 8: the epoxyhydroxy fatty acid QTL in IL8.3 (ehfa8.3). Further analysis should be carried out to further elucidate the trihydroxy fatty acid QTL in IL8.3.1 (thfa8.3.1).
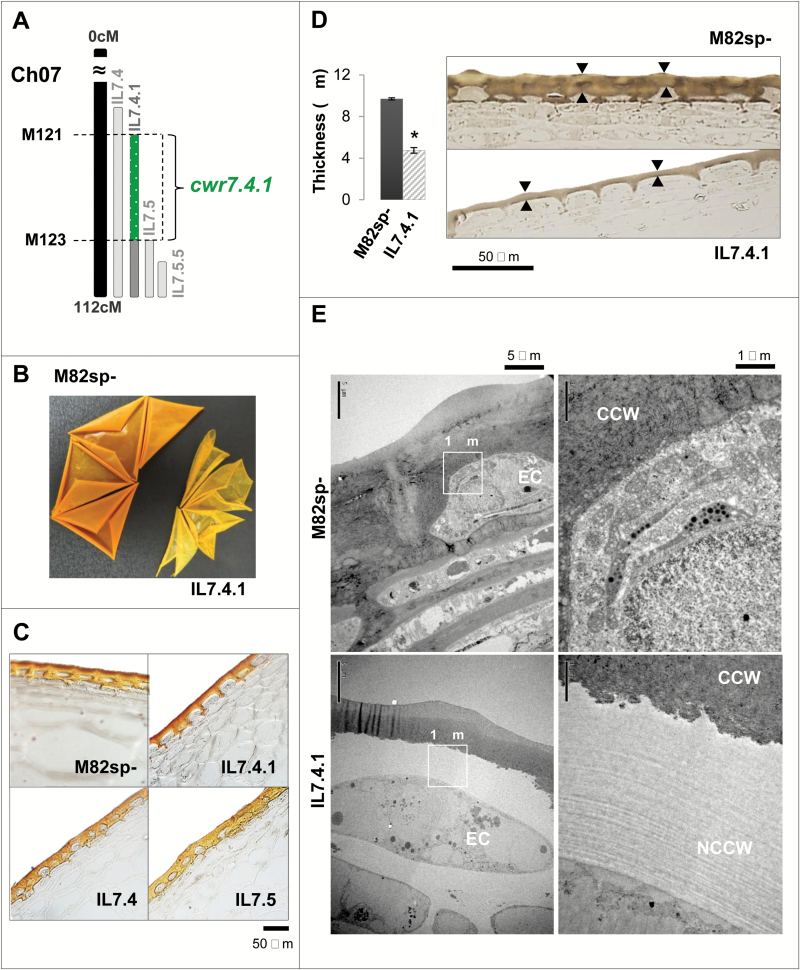
A QTL associated with cuticle thickness was mapped to chromosome 7
The IL population was also phenotyped for cuticle thickness as a proxy for possible changes in fruit cuticular lipids. A significant marker trait association in the SNPs located in the introgression harbored by IL7.4.1 was found that exhibited a very thin, fragile isolated cuticle (Fig. 6B). In fact, IL7.4.1—which in the S. pennellii introgression is contained completely within IL7.4 and partially within IL7.5 and IL7.5.5 (Fig. 6A)—displayed a much thinner cuticle than the other overlapping lines, even thinner than the cuticle from parental M82 (Fig. 6C), as revealed by staining with the lipid-specific dye Sudan IV. More precisely, IL7.4.1 showed an almost two-fold reduction in cuticle thickness compared with the parental line (Fig. 6D). However, our analysis revealed that IL7.4.1 fruit did not accumulate fewer waxes or cutin monomers compared with the parental line M82. To investigate other causes of this cuticular phenotype, TEM was used to examine structural differences in more detail. TEM showed that IL7.4.1 contains a layer of non-cutinized cell wall beneath the cuticle, whereas the parental line M82 showed a completely cutinized cell wall (Fig. 6E). This phenotype was also observed in a second-year experiment, suggesting that changes in some cell wall components may influence the effect of IL7.4.1 cuticle thickness without altering its lipid composition.
Fig. 6.
Putative cell wall-related QTL identified in IL7.4.1 fruits. A. Chromosome intervals. B. Macroscopic changes in cuticular membrane phenotype. C. Cuticle phenotype under light microscopy using the lipid-specific dye Sudan IV as stain. D. Reduction in cuticle thickness (black arrows). Ultrathin sections for TEM stained with lipid-specific osmium tetroxide were used for cuticle thickness measurements. Data are presented as mean±SD. E. TEM appearance of changes in the cell wall of the epidermal cell (EC) in mature red tomatoes. CCW, cutinized cell wall; NCCW, non-cutinized cell wall. (This figure is available in colour at JXB online.)
Candidate genes for the validated QTLs
Using the online resources available for tomato, a search for candidate genes in the four validated QTLs was performed. Out of the 606 genes found in the four intervals (a total of 3.73 Mb), nine candidate genes can be proposed (Table 2) on the basis of their gene annotation, their changes in gene expression levels in ripe fruit samples from informative ILs containing the significant QTLs compared with the reference background, and their SNP variability between S. pennellii and S. lycopersicum, which would produce non-conservative protein changes (Supplementary Tables S2 and S3). Thus, in vlcfa3.4 an ortholog of the Arabidopsis ECERIFERUM3 gene (AtCER3), Solyc03g117800, was found. This gene has been described as being involved in the wax biosynthetic decarbonylation pathway (Rowland et al., 2007; Lam et al., 2012) producing the different n-aldehydes and n-alkanes. The expression levels for this gene on the published databases for the IL population and for the parental S. pennellii revealed that it was increased in IL3.4 compared with the parental S. lycopersicum M82, but no change was observed for the wild species S. pennellii (Supplementary Table S2). In addition to this, six non-conserved amino acid substitutions, including three amino acid insertions (Supplementary Table S3), were found when the protein sequences for both S. lycopersicum and S. pennellii alleles were compared. For the second QTL in IL3.4, amy3.4, no annotated biosynthetic gene likely involved in the production of the amyrins was found. Incidentally, an ortholog of the Arabidopsis JASMONATE-ZIM-DOMAIN PROTEIN 3 gene (AtJAZ3), Solyc03g118540 (Table 2), was found in this region. This gene has been described as an inhibitor of methyl jasmonate biosynthesis (Valenzuela et al., 2016), involved in the activation of pentacyclic triterpenoid biosynthesis in Arabidopsis (Misra et al., 2014). This gene was found to be down-regulated in IL3.4 compared with the parental M82 (Supplementary Table S2), which fitted with the expected biological effect and, in addition, it contained a non-conservative change in its protein sequence (Supplementary Table S3). Moving on to the next QTL in chromosome 8, ehfa8.3, an ortholog of the Arabidopsis CYP86A8 gene (AtLCR), Solyc08g081220, (Table 2) was found. This gene has been described as a fatty acid (ω-1)-hydroxylase, which uses non-epoxidated fatty acids to incorporate a hydroxyl group at the end of the molecule (Kandel et al., 2006). Although no changes in gene expression pattern could be detected (Supplementary Table S2), a non-conservative amino acid change was found in the wild protein (Supplementary Table S3). Finally, when looking for candidate genes in cwr7.4.1, four genes with annotations that could be associated with cell wall remodeling processes (Table 2) were identified: two homologs of the Arabidopsis XYLOGLUCAN ENDOTRANSGLYCOSYLASE 7 gene (AtXTR7), Solyc07g055990 and Solyc07g056000, which is involved in secondary cell wall development and organization (Bourquin et al., 2002; Han et al., 2015); an uncharacterized tomato POLYGALACTURONASE 28, Solyc07g056290, with pectinase activity; and a homolog of the Arabidopsis SHATTERPROOF 1 gene (AtSHP1/AGL1), the SlTAGL1 Solyc07g055920, which affects pericarp thickness (Vrebalov et al., 2009). Both xyloglucan endotransglycosilases and the SHATTERPROOF 1 homolog, but not POLYGALACTURONASE 28, showed a reduction in their gene expression levels in IL7.4.1 compared with the expression level in the parental M82 (Supplementary Table S2). For the four candidate genes, non-conservative changes in their predicted amino acid sequence were observed (Supplementary Table S3), especially in POLYGALACTURONASE 28 and SHATTERPROOF1, which contained several deletions in addition to point amino acid substitutions.
Discussion
Given its complexity, studies aimed at deciphering the genetic basis of cuticular lipid composition in tomato using large populations are scarce (Yeats et al., 2012; Bolger et al., 2014; Ofner et al., 2016), and the situation is not much better for other plants. To date, only one study in tomato has centered on the fruit cuticle and this was mainly focused on wax triterpenoid composition (Yeats et al., 2012); all other studies have targeted leaves. Here, we propose the first screening done for a large set of cuticular lipids (not only amyrins) in tomato fruit using the S. pennellii IL population. The wild species S. pennellii exhibits great differences in cuticular lipid composition, ranging from higher amounts of wax to reduced cutin monomer coverage, compared with the fruit of cultivated S. lycopersicum cv. M82 (Yeats et al., 2012). In fact, S. pennellii fruit is characterized by the presence of a thin cuticle showing reduced levels of the major cutin monomer 10-ω-hydroxy hexadecanoic acid but increased levels of the minor cutin monomer 9,10,18-trihydroxy octadecanoic acid. Additionally, the S. pennellii cuticle contains higher levels of n-alkanes, in particular C29 and C30n-alkanes, but very low levels of α- and δ-amyrins (Yeats et al., 2012). Of these five special cuticular traits, described for the wild accession LA716, in the present study we found that three of them showed significant variation in the IL population: reduced amyrins, reduced cuticle thickness, and increased n-alkane levels.
The biosynthesis of n-alkanes occurs by the decarbonylation pathway (Riederer and Müller, 2006). CER3 has been described in Arabidopsis as the main biosynthetic gene involved in this pathway, in association with CER1 (Rowland et al., 2007; Lam et al., 2012). In vlcfa3.4 we found the tomato ortholog of the AtCER3 gene, associated with an increase in n-aldehyde and n-alkane composition. The putative protein sequence for the S. pennellii wild allele contains some non-conservative changes that may affect substrate specificity when compared with the cultivated allele, which might result in stronger activation of the wax decarbonylation pathway and may explain the phenotype observed. Thus, vlcfa3.4 could represent a strong candidate in the study of CER3 function in the decarbonylation pathway for tomato fruit.
Although the reduction in amyrin content has been described for S. pennellii, and we found an already-reported QTL associated with this trait in IL12.1 (amy12.1), we also found an additional QTL involved in the contrasting phenotype, that is, the over-accumulation of the three amyrins in amy3.4. The metabolic changes observed in amy3.4 were shared by both IL3.4 and IL3.5, suggesting that the molecular basis associated with this phenotype would be present only in this shared region. As mentioned above, we could not find any amyrin biosynthetic gene in this region. However, we propose the ortholog of AtJAZ3, a negative regulator of methyl jasmonate biosynthesis (Valenzuela et al., 2016), as the putative gene involved in the over-accumulation of amyrins. Misra et al. (2014) described how methyl jasmonate was involved in the accumulation of amyrins in Arabidopsis. Taking these two reports in mind, we postulate that JAZ3 could also regulate amyrin biosynthesis by regulating methyl jasmonate biosynthesis in tomato fruit. Moreover, the wild allele version of this gene showed a decrease in expression levels in tomato fruit (Supplementary Table S2), together with important changes in its protein sequence (Supplementary Table S3) that could be the reason for the increased amyrin content observed in IL3.4. Further studies of methyl jasmonate content in association with the biosynthesis of amyrins could clarify this type of regulation.
The thinner cuticular phenotype observed in the wild species S. pennellii has been associated with changes in the accumulation of the major cutin monomer (Yeats et al., 2012). In contrast with this, here we found a very similar cuticular phenotype, which was independent from the cuticular lipid composition in cwr7.4.1. These results differ from those obtained in cuticular mutants in which cutin composition or polymerization was affected (Hovav et al., 2007; Isaacson et al., 2009; Girard et al., 2012; Shi et al., 2012; Petit et al., 2014; Yeats et al., 2014). The most recent studies on tomato fruit cuticle consolidate the idea of the cuticle as a cell wall scaffold over which the different cuticular lipids are distributed (Guzman-Puyol et al., 2015; Segado et al., 2016a, 2016b). In this context, having a genomic region associated with the interaction between cell wall and CLCs represents a great opportunity to consolidate this working model. One of the genes mapped to cwr7.4.1 was the tomato gene SlTAGL1, which is involved in pericarp thickness (Vrebalov et al., 2009). One possibility could be that this gene might affect cell wall composition not only in pericarp cells but also in epidermal cells. As the cwr7.4.1 QTL was mapped to IL7.4.1, which completely overlaps with IL7.4, it could be that SlTAGL1 requires the function of another gene mapped on to the IL7.4.1 region, and only when the alleles of both genes belong to the same species (i.e. both cultivated alleles or both wild alleles) do they show activity. In this context, the presence of several indels and point non-conservative amino acid changes in the wild allele could be essential in the interaction with other proteins encoded by alleles from the same species. Another three genes involved in cell wall remodeling were also mapped on to this QTL. These genes affect the xyloglucan and pectin links with the cellulose and hemicellulose scaffold (Bourquin et al., 2002; Han et al., 2015). Thus, another hypothesis to explain the phenotype observed in cwr7.4.1 could give details on how defective alleles for these genes might affect the formation of the cell wall scaffold, which would also affect the deposition of cuticular lipids on it. As a result, part of the cell wall surrounding the epidermal cells would be incompletely cutinized, as we saw for IL7.4.1 (Fig. 6E). For either or both hypotheses the detailed characterization of this QTL might be of great interest to elucidate the means by which both types of cuticular components are connected and regulated.
Moreover, we also found a fourth QTL, ehfa8.3, which affects the accumulation of epoxyhydroxy fatty acids. Very little is known about the biosynthetic pathway of the epoxyhydroxy fatty acids, and no related biosynthetic gene or regulator has been described in tomato fruit. These types of compounds have been described as being involved in plant defenses against biotic stress such as fungal infection or wounding (Aghofack-Nguemezi et al., 2011). Thus, more in-depth studies on IL8.3 could not only provide clues to decipher such pathways in tomato fruit but also could help to improve traits associated with yield and post-harvest behavior. In ehfa8.3 we mapped the ortholog to AtLCR, whose product which hydroxylates the last carbon of a C18:X fatty acid and rarely uses epoxy fatty acids as a substrate in Arabidopsis (Kandel et al., 2006). One possibility could be that this gene in tomato could take both epoxy- and non-epoxidated fatty acids as a substrate. Another possibility might be that the tomato allele hydroxylates 9,10-dihydroxy fatty acids, which are considered to be the precursors of the epoxyhydroxy fatty acids (Meesapyodsuk and Qiu, 2011). Although we found a non-conservative change in the amino acid sequence of this gene in the wild allele, we did not find a differential expression of this gene for either IL8.3 or S. pennellii. One explanation for this finding could be that this gene acts in tomato fruit earlier than the red stage. However, it will be necessary to conduct molecular analyses to clarify the function of this gene in tomato fruit.
In addition to all the above-mentioned traits, the increased levels of trihydroxy fatty acids described for the wild species S. pennellii was hardly observed in our experiments with the IL population. Thus, although we observed this phenotype in IL8.3.1 during the first year of experiments, it was not confirmed during the second year as a result of a large variation in the parental line M82. This could suggest that, contrary to the other validated QTLs, this QTL in IL8.3.1 was not strong enough. However, this IL could be a starting point for further studies on trihydroxy fatty acid biosynthesis and/or regulation [we found some promising candidate genes in this IL (data not shown)]. Finally, we were unable to identify any QTL associated with the major cutin monomer, 9,16-dihydroxy hexadecanoic acid, although this monomer was drastically reduced in the S. pennellii cuticles. Possible explanations for this finding could be strict homeostatic regulation and/or that the alteration of only one gene in the pathway does not affect the synthesis of this monomer (perhaps some of the genes in the pathway are duplicated), or even that two or more genes (or copies) distantly linked or located on different chromosomes could be altered to produce the diminished accumulation observed in S. pennellii. The analysis of populations such as RILs or MAGIC—in which epistatic interactions can be investigated—could shed some light on this trait.
In conclusion, here we present a first comprehensive screening of cuticular lipid composition in a large population. The S. pennellii IL population has been shown to be an excellent resource to study the molecular basis of cuticular composition, as suggested by the QTLs mapped and confirmed in this study. Furthermore, some candidate genomic regions and associated genes have been reported in association with some of the main cuticular traits observed for the wild species S. pennellii. We therefore believe that our results represent the starting point for future studies using different inbreeding populations, as well as for conducting associated molecular studies to advance our understanding of the molecular basis underlying fruit cuticular phenotypes.
Supplementary data
Supplementary data are available at JXB online.
Table S1. DNA markers used to screen cuticular lipid components in the S. pennellii IL population.
Table S2. Gene expression levels for the selected candidate genes.
Table S3. Gene and protein information for the selected candidate genes.
Fig. S1. Validated QTLs associated with the main cuticular lipid traits of S. pennellii fruits during the second season.
Supplementary Material
Acknowledgements
Research at the IBMCP was supported by the Spanish Ministry of Education and Culture (BIO2013-42193-R) and H2020 TRADITOM (634561). AA, AG, and J-PF-M thank COST FA1106 Quality Fruit for STSM and networking activities. This work was supported by the Israel Science Foundation (ISF) personal grant to AA (grant no. 646/11). We would like to thank the Adelis Foundation, the Leona M. and Harry B. Helmsley Charitable Trust, the Jeanne and Joseph Nissim Foundation for Life Sciences, Tom and Sondra Rykoff Family Foundation Research, and the Raymond Burton Plant Genome Research Fund for supporting AA’s laboratory activity. AA is the incumbent of the Peter J. Cohn Professorial Chair. We are very grateful to Prof. Dani Zamir for providing us the S. pennellii IL collection and to Prof. Antonio Heredia for his valuable advice in preparing the manuscript for publication. We would like to acknowledge the help offered by the Electron Microscopy Unit at the WIS (Israel) for the TEM sample preparation and imaging, especially Elena Kartvelishvily, Eugenia Klein, and Eyal Shimoni. Finally, we would also like to thank Calanit Raanan and Tamara Berkutzki (Department of Veterinary Resources, WIS) for their help in tissue fixation and embedding, as well as Hanna Levanony (Department of Plant Sciences, WIS) for her help in tissue staining for the light microscopy studies.
References
- Adato A, Mandel T, Mintz-Oron S, et al. 2009. Fruit-surface flavonoid accumulation in tomato is controlled by a SlMYB12-regulated transcriptional network. PLoS Genetics 5, e1000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghofack-Nguemezi J, Fuchs C, Yeh SY, Huang FC, Hoffmann T, Schwab W. 2011. An oxygenase inhibitor study in Solanum lycopersicum combined with metabolite profiling analysis revealed a potent peroxygenase inactivator. Journal of Experimental Botany 62, 1313–1323. [DOI] [PubMed] [Google Scholar]
- Alseekh S, Ofner I, Pleban T, Tripodi P, Di Dato F, Cammareri M, Mohammad A, Grandillo S, Fernie AR, Zamir D. 2013. Resolution by recombination: breaking up Solanum pennellii introgressions. Trends in Plant Science 18, 536–538. [DOI] [PubMed] [Google Scholar]
- Beisson F, Li-Beisson Y, Pollard M. 2012. Solving the puzzles of cutin and suberin polymer biosynthesis. Current Opinion in Plant Biology 15, 329–337. [DOI] [PubMed] [Google Scholar]
- Bird D, Beisson F, Brigham A, Shin J, Greer S, Jetter R, Kunst L, Wu X, Yephremov A, Samuels L. 2007. Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. The Plant Journal 52, 485–498. [DOI] [PubMed] [Google Scholar]
- Blanca J, Cañizares J, Cordero L, Pascual L, Diez MJ, Nuez F. 2012. Variation revealed by SNP genotyping and morphology provides insight into the origin of the tomato. PLoS One 7, e48198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A, Scossa F, Bolger ME, et al. 2014. The genome of the stress-tolerant wild tomato species Solanum pennellii. Nature Genetics 46, 1034–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdenx B, Bernard A, Domergue F, et al. 2011. Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiology 156, 29–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourquin V, Nishikubo N, Abe H, Brumer H, Denman S, Eklund M, Christiernin M, Teeri TT, Sundberg B, Mellerowicz EJ. 2002. Xyloglucan endotransglycosylases have a function during the formation of secondary cell walls of vascular tissues. The Plant Cell 14, 3073–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschhaus C, Jetter R. 2008. Sealing plant surface: cuticular wax formation by epidermal cells. Annual Review of Plant Biology 59, 683–707. [DOI] [PubMed] [Google Scholar]
- Buschhaus C, Jetter R. 2011. Composition differences between epicuticular and intracuticular wax substructures: how do plants seal their epidermal surfaces?Journal of Experimental Botany 62, 841–853. [DOI] [PubMed] [Google Scholar]
- Buxdorf K, Rubinsky G, Barda O, Burdman S, Aharoni A, Levy M. 2014. The transcription factor SlSHINE3 modulates defense responses in tomato plants. Plant Molecular Biology 84, 37–47. [DOI] [PubMed] [Google Scholar]
- Causse M, Saliba-Colombani V, Lecomte L, Duffé P, Rousselle P, Buret M. 2002. QTL analysis of fruit quality in fresh market tomato: a few chromosome regions control the variation of sensory and instrumental traits. Journal of Experimental Botany 53, 2089–2098. [DOI] [PubMed] [Google Scholar]
- Chapman NH, Bonnet J, Grivet L, et al. 2012. High-resolution mapping of a fruit firmness-related quantitative trait locus in tomato reveals epistatic interactions associated with a complex combinatorial locus. Plant Physiology 159, 1644–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez E, Cuartero J, Heredia A. 2011. An overview on plant cuticle biomechanics. Plant Science 181, 77–84. [DOI] [PubMed] [Google Scholar]
- Domínguez E, Fernández MD, Hernández JC, Parra JP, España L, Heredia A, Cuartero J. 2012. Tomato fruit continues growing while ripening, affecting cuticle properties and cracking. Physiologia Plantarum 146, 473–486. [DOI] [PubMed] [Google Scholar]
- Eshed Y, Zamir D. 1995. An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141, 1147–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Moreno J-P, Malitsky S, Lashbrooke J, Biswal AK, Racovita RC, Mellerowicz EJ, Jetter R, Orzaez D, Aharoni A, Granell A. 2016. An efficient method for medium throughput screening of cuticular wax composition in different plant species. Metabolomics 12, 73–68. [Google Scholar]
- Franke R, Briesen I, Wojciechowski T, Faust A, Yephremov A, Nawrath C, Schreiber L. 2005. Apoplastic polyesters in Arabidopsis surface tissues—a typical suberin and a particular cutin. Phytochemistry 66, 2643–2658. [DOI] [PubMed] [Google Scholar]
- Frary A, Göl D, Keleş D, Okmen B, Pinar H, Siğva HO, Yemenicioğlu A, Doğanlar S. 2010. Salt tolerance in Solanum pennellii: antioxidant response and related QTL. BMC Plant Biology 10, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard AL, Mounet F, Lemaire-Chamley M, et al. 2012. Tomato GDSL1 is required for cutin deposition in the fruit cuticle. The Plant Cell 24, 3119–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur A, Semel Y, Osorio S, et al. 2011. Yield quantitative trait loci from wild tomato are predominately expressed by the shoot. Theoretical and Applied Genetics 122, 405–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur A, Zamir D. 2004. Unused natural variation can lift yield barriers in plant breeding. PLoS Biology 2, e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman-Puyol S, Benítez JJ, Domínguez E, Bayer IS, Cingolani R, Athanassiou A, Heredia A, Heredia-Guerrero JA. 2015. Pectin-lipid self-assembly: influence on the formation of polyhydroxy fatty acids nanoparticles. PLoS One 10, e0124639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Zhu Q, Zhang Z, Meng K, Hou Y, Ban Q, Suo J, Rao J. 2015. Analysis of xyloglucan endotransglycosylase/hydrolase (XTH) genes and diverse roles of isoenzymes during persimmon fruit development and postharvest softening. PLoS One 10, e0123668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hen-Avivi S, Lashbrooke J, Costa F, Aharoni A. 2014. Scratching the surface: genetic regulation of cuticle assembly in fleshy fruit. Journal of Experimental Botany 65, 4653–4664. [DOI] [PubMed] [Google Scholar]
- Höfer R, Briesen I, Beck M, Pinot F, Schreiber L, Franke R. 2008. The Arabidopsis cytochrome P450 CYP86A1 encodes a fatty acid ω-hydroxylase involved in suberin monomer biosynthesis. Journal of Experimental Botany 59, 2347–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovav R, Chehanovsky N, Moy M, Jetter R, Schaffer AA. 2007. The identification of a gene (Cwp1), silenced during Solanum evolution, which causes cuticle microfissuring and dehydration when expressed in tomato fruit. The Plant Journal 52, 627–639. [DOI] [PubMed] [Google Scholar]
- Isaacson T, Kosma DK, Matas AJ, et al. 2009. Cutin deficiency in the tomato fruit cuticle consistently affects resistance to microbial infection and biomechanical properties, but not transpirational water loss. The Plant Journal 60, 363–377. [DOI] [PubMed] [Google Scholar]
- Javelle M, Vernoud V, Rogowsky PM, Ingram GC. 2011. Epidermis: the formation and functions of a fundamental plant tissue. New Phytologist 189, 17–39. [DOI] [PubMed] [Google Scholar]
- Jeffree C. 2006. The fine structure of the plant cuticle. In: Riederer M, Müller C, eds. Biology of the plant cuticle. Oxford: Blackwell,11–125. [Google Scholar]
- Kandel S, Sauveplane V, Olry A, Diss L, Benvenist I, Pinot F. 2006. Cytochrome p450-dependent fatty acid hydroxylases in plants. Phytochemistry Review 5, 359–372. [Google Scholar]
- Koenig D, Jimenez-Gomez JM, Kimura S, et al. 2013. Comparative transcriptomic reveals patterns of selection in domesticated and wild tomato. Proceedings of the National Academy of Sciences of the United States of America 110, E2655–E2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L, Samuels L. 2009. Plant cuticles shine: advances in wax biosynthesis and export. Current Opinion in Plant Biology 12, 721–727. [DOI] [PubMed] [Google Scholar]
- Lam P, Zhao L, McFarlane HE, Aiga M, Lam V, Hooker TS, Kunst L. 2012. RDR1 and SGS3, components of RNA-mediated gene silencing, are required for the regulation of cuticular wax biosynthesis in developing inflorescence stems of Arabidopsis. Plant Physiology 159, 1385–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara I, Belge B, Goulao LF. 2014. The fruit cuticle as a modulator of postharvest quality. Postharvest Biology and Technology 87, 103–112. [Google Scholar]
- Lashbrooke J, Adato A, Lotan O, et al. 2015. The tomato MIXTA-like transcription factor coordinates fruit epidermis conical cell development and cuticular lipid biosynthesis and assembly. Plant Physiology 169, 2553–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin LB, Rose JK. 2014. There’s more than one way to skin a fruit: formation and functions of fruit cuticles. Journal of Experimental Botany 65, 4639–4651. [DOI] [PubMed] [Google Scholar]
- Meesapyodsuk D, Qiu X. 2011. A peroxygenase pathway involved in the biosynthesis of epoxy fatty acids in oat. Plant Physiology 157, 454–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz-Oron S, Mandel T, Rogachev I, et al. 2008. Gene expression and metabolism in tomato fruit surface tissues. Plant Physiology 147, 823–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra RC, Maiti P, Chanotiya CS, Shanker K, Ghosh S. 2014. Methyl jasmonate-elicited transcriptional responses and pentacyclic triterpene biosynthesis in sweet basil. Plant Physiology 164, 1028–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofner I, Lashbrooke J, Pleban T, Aharoni A, Zamir D. 2016. Solanum pennellii backcross inbred lines (BILs) link small genomic bins with tomato traits. The Plant Journal 87, 151–160. [DOI] [PubMed] [Google Scholar]
- Pineda B, García-Abellán JO, Antón T, et al. 2012. Tomato: genomic approaches for salt and drought stress tolerance. In: Tuteja N, GiII SS, Tiburcio AF, Tuteja R, eds. Improving crop resistance to abiotic stress, Vol. 1 & 2 Weinheim: Wiley-VCH, 1085–1120. [Google Scholar]
- Pollard M, Beisson F, Li Y, Ohlrogge JB. 2008. Building lipid barriers: biosynthesis of cutin and suberin. Cell Review 13, 1360–1385. [DOI] [PubMed] [Google Scholar]
- Petit J, Bres C, Just D, Garcia V, Mauxion JP, Marion D, Bakan B, Joubès J, Domergue F, Rothan C. 2014. Analyses of tomato fruit brightness mutants uncover both cutin-deficient and cutin-abundant mutants and a new hypomorphic allele of GDSL lipase. Plant Physiology 164, 888–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riederer M, Müller C. 2006. Biology of the plant cuticle. Oxford: Blackwell. [Google Scholar]
- Rowland O, Lee R, Franke R, Schreiber L, Kunst L. 2007. The CER3 wax biosynthetic gene from Arabidopsis thaliana is allelic to WAX2/YRE/FLP1. FEBS Letters 581, 3538–3544. [DOI] [PubMed] [Google Scholar]
- Samuels L, Kunst L, Jetter R. 2008. Sealing plant surfaces: cuticular wax formation by epidermal cells. Annual Review Plant of Biology 59, 683–707. [DOI] [PubMed] [Google Scholar]
- Sandbrink JM, van Ooijen JW, Purimahua CC, Vrielink M, Verkerk R, Zabel P, Lindhout P. 1995. Localization of genes for bacterial canker resistance in Lycopersicon peruvianum using RFLPs. Theoretical and Applied Genetics 90, 444–450. [DOI] [PubMed] [Google Scholar]
- Segado P, Domínguez E, Heredia A. 2016a. Ultrastructure of the epidermal cell wall and cuticle of tomato fruit (Solanum lycopersicum L.) during development. Plant Physiology 170, 935–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segado P, Domínguez E, Heredia A. 2016b. Cutinisation of tomato fruit epidermis: structural and morphological details. Plant Signaling & Behavior 11, e1168558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi JX, Adato A, Alkan A, et al. 2012. The tomato SlSHINE3 transcription factor regulates fruit cuticle formation and epidermal patterning. New Phytologist 197, 468–480. [DOI] [PubMed] [Google Scholar]
- Valenzuela CE, Acevedo-Acevedo O, Miranda GS, Vergara-Barros P, Holuigue L, Figueroa CR, Figueroa PM. 2016. Salt stress response triggers activation of the jasmonate signaling pathway leading to inhibition of cell elongation in Arabidopsis primary root. Journal of Experimental Botany 67, 4209–4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg RA, Hoefsloot HC, Westerhuis JA, Smilde AK, van der Werf MJ. 2006. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics 7, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Pan IL, Arroyo AJ, et al. 2009. Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. The Plant Cell 21, 3041–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Guhling O, Yao R, Li F, Yeats TH, Rose JK, Jetter R. 2011. Two oxidosqualene cyclases responsible for biosynthesis of tomato fruit cuticular triterpenoids. Plant Physiology 155, 540–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Buda GJ, Wang Z, Chehanovsky N, Moyle LC, Jetter R, Schaffer AA, Rose JK. 2012. The fruit cuticles of wild tomato species exhibit architectural and chemical diversity, providing a new model for studying the evolution of cuticle function. The Plant Journal 69, 655–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Huang W, Chatterjee S, Viart HM, Clausen MH, Stark RE, Rose JK. 2014. Tomato cutin deficient 1 (CD1) and putative orthologs comprise an ancient family of cutin synthase-like (CUS) proteins that are conserved among land plants. The Plant Journal 77, 667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Rose JK. 2013. The formation and function of plant cuticles. Plant Physiology 163, 5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.