Nitrate represses the gibberellin pathway and subsequently activates the flowering repressors SMZ and SNZ, delaying flowering time in Arabidopsis.
Keywords: Developmental transition, flowering, gibberellic acid, mineral nutrition, nitrate, nitrate transporter 1.1, Schlafmutze, Schnarchzapfen
Abstract
The reproductive success of plants largely depends on the correct programming of developmental phase transitions, particularly the shift from vegetative to reproductive growth. The timing of this transition is finely regulated by the integration of an array of environmental and endogenous factors. Nitrogen is the mineral macronutrient that plants require in the largest amount, and as such its availability greatly impacts on many aspects of plant growth and development, including flowering time. We found that nitrate signaling interacts with the age-related and gibberellic acid pathways to control flowering time in Arabidopsis thaliana. We revealed that repressors of flowering time belonging to the AP2-type transcription factor family including SCHLAFMUTZE (SMZ) and SCHNARCHZAPFEN (SNZ) are important regulators of flowering time in response to nitrate. Our results support a model whereby nitrate activates SMZ and SNZ via the gibberellin pathway to repress flowering time in Arabidopsis thaliana.
Introduction
Nitrogen (N) is an essential component of many key biological molecules and a limiting factor for plant growth in natural as well as in agricultural systems (Frink et al., 1999). N availability can have profound effects on a variety of developmental programs such as germination, seedling establishment, and flowering (Vidal et al., 2014). Nitrate is one of the main sources of N in the soil. Changes in nitrate concentration are sensed by the transporter and receptor NITRATE TRANSPORTER 1.1 (NRT1.1) (Ho et al., 2009). Nitrate perception is able to trigger signaling events that include an increase in cytoplasmic Ca2+, which acts as a second messenger (Gutiérrez, 2012; Riveras et al., 2015). Nitrate signal transduction produces transcriptional changes in an extensive array of genes that play pivotal roles in N metabolism [e.g. nitrate transporters NRT1.1, NRT2.1, and NRT2.2, nitrate reductase (NIR), and nitrite reductase (NIA1 and NIA2)] as well as in plant ontogeny [e.g. auxin receptor AFB3, bZIP transcription factors TGA1 and TGA4, Arabidopsis Nitrate Regulated 1 (ANR1), lateral organ boundary domain (LBD37/38/39), among others] (Jonassen et al., 2009; Rubin et al., 2009; Gutiérrez, 2012; Alvarez et al., 2014; Vidal et al., 2014; O’Brien et al., 2016).
Flowering is one of the most important developmental transitions during a plant’s life cycle (Koornneef et al., 1998). Floral induction is regulated by an intricate genetic network that integrates both environmental and endogenous signals (Amasino, 2010; Fornara et al., 2010; Srikanth and Schmid, 2011). Major environmental factors known to affect flowering time are the photoperiod (Imaizumi et al., 2003; Imaizumi and Kay, 2006; Giakountis and Coupland, 2008; Michaels, 2009) and prolonged exposure to cold temperatures, a process known as vernalization (Alexandre and Hennig, 2008; Michaels, 2009). On the other hand, endogenous pathways include the gibberellic acid signaling pathway (Mutasa-Göttgens and Hedden, 2009), and the autonomous and age-related pathways that monitor plant developmental state (Simpson, 2004; Wang, 2014). These different pathways converge to regulate expression of a small number of flowering integrator genes that promote flowering, including SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1), FLOWERING LOCUS T (FT), and LEAFY (LFY) (Adrian et al., 2009; Michaels, 2009; Amasino, 2010; Srikanth and Schmid, 2011).
It has been reported that FT encodes the ‘florigen’, a mobile signal that is produced in the leaf tissue and is transmitted to the shoot apical meristem, where it initiates flowering (Kobayashi et al., 1999; Golembeski and Imaizumi, 2015). Due to its importance, FT is subjected to fine transcriptional control. Multiple transcriptional activators, such as GIGANTEA (GI) and CONSTANS (CO), bind to its promoter region; in contrast, repressors such as FLOWERING LOCUS C (FLC) and SHORT VEGETATIVE PHASE (SVP) down-regulate its expression (Sawa and Kay, 2011; Andrés and Coupland, 2012). SCHLAFMUTZE (SMZ) together with its paralog SCHNARCHZAPFEN (SNZ) are floral repressors from the AP2 family of transcription factors. They delay flowering under long-day (LD) conditions and are targets of the microRNA miR172, a pivotal regulator of the ageing pathway. Chromatin immunoprecipitation experiments have demonstrated that SMZ is able to bind directly to the FT locus, down-regulating its expression (Mathieu et al., 2009; Golembeski and Imaizumi, 2015).
Gibberellin (GA) is a plant hormone that regulates flowering time. When bioactive GAs bind to their receptors, they trigger the proteasome-dependent degradation of the DELLA transcription factors (Murase et al., 2008; Shimada et al., 2008; Mutasa-Göttgens and Hedden, 2009). These proteins regulate plant development and physiology by modifying the activity of a myriad of transcription factors, either by inhibiting their DNA binding ability or by acting as co-activators, facilitating their attachment to target promoters (de Lucas et al., 2008; Feng et al., 2008; Hou et al., 2010; Zhang et al., 2011; Hong et al., 2012; Yang et al., 2012; Xu et al., 2014). Application of exogenous GAs promotes the transition from vegetative growth to flowering in a variety of plants (Bernier, 1988; Jacobsen and Olszewski, 1993; Chandler and Dean, 1994). The role of GAs in flowering initiation has been observed primarily in LD plants grown under non-inductive conditions. In Arabidopsis the photoperiodic pathway and its core components CO and FT dominate flowering initiation under inductive conditions (Mutasa-Göttgens and Hedden, 2009). Despite the dominance exerted by the CO-FT module under LD conditions, the GA biosynthesis mutant ga1-3 and the triple GA receptor mutant gid1 show a delayed flowering phenotype when grown under LDs, establishing a role for GAs under inductive conditions (Wilson et al., 1992; Griffiths et al., 2006; Mutasa-Göttgens and Hedden, 2009). Interestingly, there is evidence supporting an interaction between the GA pathway and nitrate nutrition. It has been shown that Arabidopsis plants grown under low-nitrate conditions have higher levels of bioactive GAs. It was proposed that low concentrations of nitrate activate the biosynthesis of GAs, as evidenced by increased expression of the GA biosynthetic enzyme GA1 under low nitrate (Liu et al 2013).
Nitrate and other N-nutrients or metabolites are known to modify flowering time in plants (Klebs, 1913; Dickens and Staden, 1988; Bernier et al., 1993; Loeppky and Coulman, 2001). Arabidopsis plants grown under low-nitrate conditions flower earlier than plants grown under high nitrate (Castro Marín et al., 2011; Yuan et al., 2016). This effect was first attributed to a novel signaling pathway acting directly over floral integrators; however, the identity of the components involved in this pathway is still an open question (Castro Marín et al., 2011; Kant et al., 2011; Liu et al., 2013). Yuan et al. (2016) found that N-signaling affects ferredoxin-NADP+-oxidoreductase (FNR1) and the blue-light receptor cryptochrome 1 (CRY1), causing a delay in flowering time. However, these studies used a mix of N nutrients and metabolites that did not narrow down the contribution of a specific N component to a particular signaling pathway.
In this work, we used molecular genetics approaches in order to find components involved in nitrate-dependent regulation of flowering time. We found that under N-sufficient conditions, nitrate delays flowering time by controlling the expression of the floral repressors SMZ and SNZ. Modulation of SMZ and SNZ gene expression by nitrate requires the GA pathway. Our results support a model whereby NRT1.1-mediated nitrate signaling interacts with the GA pathway and key elements of the ageing pathway in order to control bolting and flowering time in Arabidopsis.
Materials and methods
Plant material
Experiments were performed with Arabidopsis thaliana Columbia-0 (Col-0) and Ler ecotypes as indicated. The following lines have been previously described: chl1-5 (Liu et al., 1999); chl1-9 (Ho et al., 2009); toe1-2 and toe2-1 (Aukerman and Sakai, 2003); smz-2, snz-1, smz-2/snz-1, and toe1-2/toe2-1, smz-2/snz-1/toe1-2/toe2-1 (Mathieu et al., 2009); co (SAIL24H04) (Kim and Michaels, 2006); flc-3 (Michaels and Amasino, 1999); miR156 overexpressor (Schwab et al., 2005); miR172 overexpressor (Mateos et al., 2010); rga-t2/gai-t6/rgl1-1/rgl2-1/rgl3-1 quintuple DELLA mutant (Feng et al., 2008); ft-10 (Yoo et al., 2005); soc1-2 (Lee et al., 2000); the RGA::GFP-RGA line (Silverstone et al., 2001); the overexpressor lines 35S::GNL (35S:YFP:GNL) and 35S::GNC (35S:GNC:GFP), and the gnc-gnl double mutant (gnc, SALK_001778; gnl, SALK_003995) (Richter et al., 2010, 2013a, 2013b).
Growth and treatment conditions
Seeds were stratified at 4 °C for 3 d in complete darkness to synchronize germination, then sterilized and grown in plastic trays with vermiculite. Plants were watered with N-free medium (100 µM H3BO3, 3 mM CaCl2, 100 µM MnSO4, 0.16 µM CuSO4, 0.1 µM Na2MoO4, 1.25 mM KH2PO4, 1.5 mM MgSO4, 50 µM ZnSO4, 10 µM KI, 100 µM FeSO4, 100 µM Na2EDTA, and 0.1 µM CoCl2) supplemented with different concentrations of KNO3 as the only N source. A constant volume of nutrient solution per plant was applied once every week until flowering time. Flowering time was measured as the time between sowing and anthesis (opening of the first flower). Bolting time was recorded when the main inflorescence had reached a height of 0.5 cm. Plants were grown in a growth room, under a controlled environment, with a 16/8 h light/dark cycle, cool white fluorescent illumination of 100 μmol m−2 s−1 and a constant temperature of 22 °C. Leaf production rate was calculated as: number of leaves/days to bolting.
For gene expression assays, seeds were sown on vertical agar plates containing N-free medium supplemented with either 1 or 3 mM KNO3. Seedlings were grown for 7, 9, 11, 13, or 15 d on a Percival incubator (Percival Scientific, Inc.) under a 16/8 light/dark cycle at 22 °C. They were harvested at zeitgeber time 0 (ZT0) on these days.
RNA quantification
Total RNA was isolated from seedlings with the mirVana kit (Life technologies, Carlsbad, CA, catalog no. AM1560) according to the manufacturer’s instructions. For mRNA quantification, reverse transcription was performed using the ImProm-II reverse transcriptase (Promega, Madison, WI). Quantitative real-time PCR was carried out in a StepOne Real time PCR system (Life technologies, Carlsbad, CA). The ADAPTOR PROTEIN-4 MU-ADAPTIN gene (At4g24550) was used as a housekeeping gene (Jonassen et al., 2009; Rubin et al., 2009). Quantification of miR172 levels was performed with the TaqMan microRNA ath-MIR172a assay (Life Technologies, Carlsbad, CA, catalog number 4427975). snoR41Y (Life Technologies, Carlsbad, CA, catalog number 4427975) was used as an internal reference. All experiments were carried out with three independent biological replicates.
GFP-RGA imaging and quantification
Transgenic lines expressing GFP (green fluorescent protein) under the control of the RGA promoter (Silverstone et al., 2001) were grown in N-free medium supplemented with either 1 or 3 mM KNO3 for 7 d. A Zeiss LSM780 confocal microscope was used for imaging. At least eight independent roots were photographed, and the number and relative intensities of GFP-fluorescent particles were automatically calculated using Fiji software (Schindelin et al., 2012).
Statistical analysis
Statistical analyses of bolting/flowering, rosette leaves, and gene expression data were done with one-way ANOVA and Tukey’s HSD tests using the GraphPad scientific software (Prism).
Results
Timing of bolting and flowering are regulated by nitrate in Arabidopsis
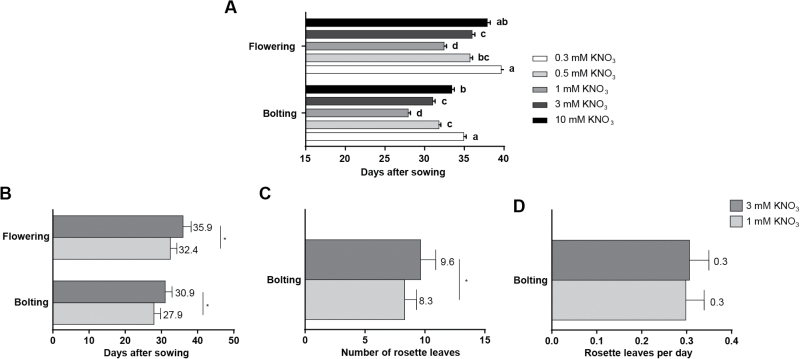
To evaluate the effect of nitrate concentration on flowering time in Arabidopsis, we seeded plants on vermiculite and grew them under a long-day (LD) photoperiod at 22 °C constant temperature. Nitrate concentrations in agricultural soils typically average 6 mM (Crawford and Glass, 1998). Since the ion gets rapidly depleted from the soil solution, the most frequent concentrations to which plants are exposed oscillate between 2 and 5 mM (Crawford and Glass, 1998; Owen and Jones, 2001; Andrews et al., 2013). Plants were fertilized weekly with a nutrient solution lacking nitrogen and supplemented with either 0.1, 0.3, 0.5, 1, 3, or 10 mM KNO3. (Fig. 1A). Plants grown with 0.1 mM KNO3 were unable to complete their life cycle under our experimental conditions. Plants grown with 0.3 or 0.5 mM KNO3 did complete their life cycle, but showed severe signs of N-limitation, including chlorotic leaves and reduced shoot development, as previously described (Bi et al., 2007). Widely used parameters for assessing reproductive phase change and flowering time include recording the number of rosette leaves at bolting (floral stem at 0.1 cm height), or counting the number of days from sowing to bolting, or to flowering (Pouteau and Albertini, 2009). We found that an increased nitrate availability delayed both bolting and flowering time. Plants flowered faster at 1 mM and exhibited increasing delays at 3 mM or 10 mM nitrate concentrations. To further characterize the effect of nitrate on flowering time, we chose to do the rest of our experiments with 1 and 3 mM concentrations, to avoid the confounding effects of severe nutritional stress and to allow plants to complete their life cycle without N excess.
Fig. 1.
Flowering time varies according to nitrate availability. Arabidopsis plants were sown on vermiculite and watered once a week with N-free nutrient solution containing either 0.3, 0.5, 1, 3, or 10 mM KNO3. Number of days to bolting and flowering (A, B), number of rosette leaves at bolting (C), and average number of rosette leaves per day (D) were determined. At least 20 plants were used for each quantification. Data are means and SD of three independent biological replicates. Asterisks and different letters indicate significant differences as determined by Tukey’s Multiple Comparison test (P≤0.01).
As shown in Fig. 1B, Arabidopsis plants grown with 3 mM KNO3 respectively bolted and flowered 3 and 3.5 d later than plants cultivated with 1 mM KNO3. In addition, plants grown at 3 mM KNO3 produced 1.3 more rosette leaves (Fig. 1C). These results are consistent with previous reports, which showed that increased nitrate concentrations delay flowering time as measured by number of rosette leaves or by number of days (Castro Marín et al., 2011; Kant et al., 2011; Liu et al., 2013).
Interestingly, we found no differences in rosette leaf production rate between the two nitrate treatments (Fig. 1D). This suggests that nitrate availability has a direct impact on the phase change, but does not influence the normal rate of plant vegetative growth under our experimental conditions.
Nitrate interacts with the gibberellin pathway and SMZ/SNZ floral repressors to control the reproductive phase transition in Arabidopsis
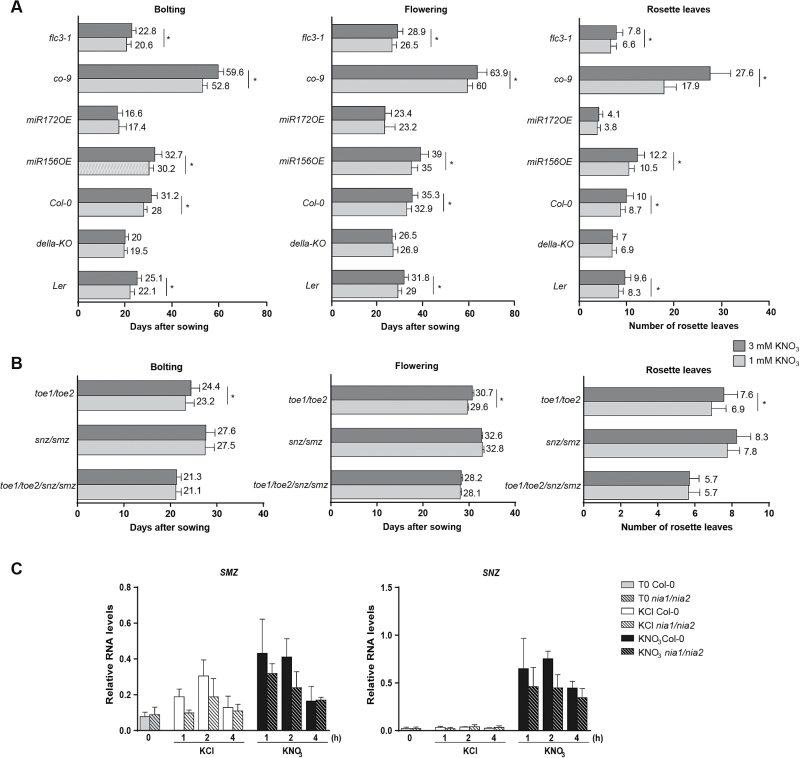
Plants have developed internal and environmentally dependent pathways to finely tune the timing of the reproductive phase transition according to endogenous and exogenous cues (Vidal et al., 2014). In order to determine whether nitrate interacts with any of the previously described flowering pathways, we analysed different mutants on key genes for each pathway. We chose CONSTANS (CO), a central regulator of the photoperiodic pathway that stimulates flowering (Suárez-López et al., 2001), Flowering Locus C (FLC), a strong flowering repressor from the autonomous and vernalization pathways (Koornneef et al., 1994; Lee et al., 2004), and a quintuple-DELLA mutant, della-KO, which lacks all five DELLA proteins and therefore shows constitutive, flowering-promoting GA signaling (Cao et al., 2005). In addition, we evaluated plants that overexpress the miR156 and miR172 microRNAs, regulators of the age-dependent pathway (Wu et al., 2009). We quantified the days to bolting and flowering as well as number of rosette leaves at bolting. We found that the repressive effect of nitrate over bolting and flowering was suppressed in the miR172 overexpressor and in the quintuple DELLA mutant (Fig. 2A). Interestingly, all the other mutants as well as the miR156 overexpressor showed the same bolting and flowering response to nitrate as the wild-type (Col-0) plants, with a delay at 3 mM KNO3 (Fig. 2A). These results indicate that nitrate interacts with the gibberellin pathway and with miR172 or its targets in order to regulate bolting and flowering time.
Fig. 2.
Nitrate-dependent delay in flowering time depends on the gibberellin pathway and the miR172 targets SMZ and SNZ. Plants were sown on vermiculite and watered once a week with N-free nutrient solution containing either 1 mM (light gray) or 3 mM (dark gray) KNO3. Nitrate-dependent flowering time and rosette leaf number of mutants for key genes from different flowering pathways were determined (A). Nitrate-dependent flowering and rosette leaf number of the quadruple toe1/toe2/smz/snz or double toe1/toe2 and smz/snz mutants were also quantified (B). (C) Col-0 (filled bars) and nitrate reductase double-mutant (nia1/nia2) plants (dashed bars) were sown on a N-free hydroponic medium supplemented with 0.5 mM ammonium succinate and then treated with either 5 mM KCl or 5 mM KNO3 for the indicated times. Total RNA was extracted from shoots, and SMZ and SNZ transcript levels were quantified by qRT-PCR. The ADAPTOR PROTEIN-4 MU-ADAPTIN gene (At4g24550) was used as an internal reference. flc3-1, FLOWERING LOCUS C mutant; co-9, CONSTANS mutant; miR172OE, miR172 overexpressor; miR156OE, miR156 overexpressor; della-KO, quintuple DELLA mutant (Ler background); toe1, toe2, TARGET OF EAT mutants; snz, SCHNARCHZAPFEN mutant; smz, SCHLAFMÜTZE mutant. All mutants and overexpressor lines, except della-KO, are in the Col-0 background. At least 20 plants were used for each flowering time measurement. Data are means and SD of three independent biological replicates. Asterisks highlight significant differences as determined by Tukey’s Multiple Comparison test (P≤0.01).
miR172 targets a set of AP2-like transcription factors that act as flowering repressors that down-regulate the floral integrator FT (Jones-Rhoades and Bartel, 2004; Mathieu et al., 2009; Zhu and Helliwell, 2011). We found that a quadruple mutation of the AP2-like floral repressors toe1, toe2, smz, and snz abolished the repressive effect of nitrate over bolting and flowering (Fig. 2B). This effect was observed in smz/snz double- and single-mutants, but not in the toe1 toe2 double-mutant (Fig. 2B, Supplementary Fig. S1 at JXB online). Consistently, we also found that both SMZ and SNZ were induced in nitrate-treated seedlings (Fig. 2C). Therefore, the effect of nitrate on flowering depends on the SMZ and SNZ floral repressors.
Nitrate effect over bolting and flowering time depends on NPF6.3/NRT1.1
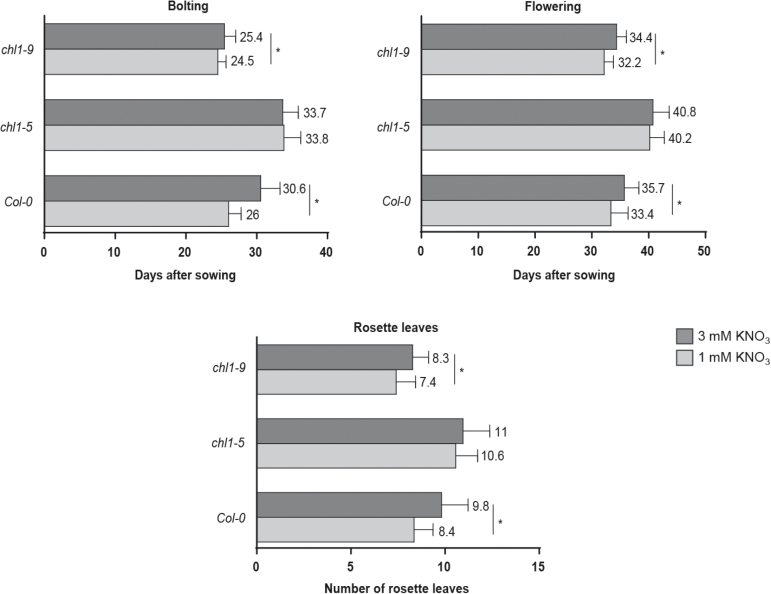
The nitrate transporter and receptor NPF6.3/NRT1.1 is a key factor in the nitrate signaling pathway (Ho et al., 2009; Bouguyon et al., 2015; Riveras et al., 2015). Mutant alleles of NPF6.3/NRT1.1 are known to display a late-flowering phenotype when plants are grown on peat soil under LDs (Guo et al., 2001). In order to determine whether this effect is nitrate-specific, we measured bolting and flowering in the loss-of-function chl1-5 mutant (Liu et al., 1999) grown in 1 and 3 mM nitrate. We found that bolting and flowering were delayed by 3 to 8 d in chl1-5 mutant plants as compared to the wild-type (Figs 1 and 3). This delay was expected because the chl1-5 mutant has impaired nitrate signaling and uptake (Liu et al., 1999) and its flowering phenotype mimics wild-type plants growing under suboptimal nitrate concentrations (Fig. 1A). However, no differences in bolting or flowering times were observed between chl1-5 mutant plants grown with 1 or 3 mM nitrate. This result indicates that NPF6.3/NRT1.1 is important for the effect of nitrate over the timing of the reproductive phase change and flowering. In order to determine whether nitrate repression of bolting and flowering depended on nitrate transport or a different function of NPF6.3/NRT1.1, we measured bolting and flowering in chl1-9 mutant plants (Ho et al., 2009). The chl1-9 mutant has a P492L point-mutation that impacts specific aspects of NPF6.3/NRT1.1 function: it affects nitrate transport capacity in both the high-affinity and low-affinity range without an apparent effect on nitrate sensing and the downstream response of NRT2.1 gene expression (Ho et al., 2009; Bouguyon et al., 2015). As shown in Fig. 3, chl1-9 still had a nitrate-dependent flowering response. When grown with 3 mM KNO3, these plants bolted and flowered 0.9 and 2.2 d later, respectively, than their counterparts grown with 1 mM KNO3. These results indicate that nitrate-dependent repression of bolting and flowering is not dependent on nitrate transport by NPF6.3/NRT1.1 at the plasma membrane. Consistent with a signaling role of nitrate in controlling bolting and flowering, we found that nitrate was able to induce SMZ and SNZ gene expression in wild-type shoots as well as in the nitrate reductase double-mutant nia1/nia2 (Wang et al., 2007) (Fig. 2C).
Fig. 3.
Nitrate repression of flowering is dependent on the NPF6.3/NRT1.1 nitrate transceptor. Nitrate transporter NPF6.3/NRT1.1 mutants chl1-5 and chl1-9 (Col-0 background) were sown on vermiculite and watered once a week with N-free nutrient solution containing either 1 mM (light gray) or 3 mM (dark gray) KNO3. At least 20 plants were used for each measurement. Data are means and SD of three independent biological replicates. Asterisks highlight significant differences as determined by Tukey’s Multiple Comparison test (P≤0.01).
These results indicate that nitrate is the signal that triggers bolting and flowering repression by controlling the transcript levels of the SMZ and SNZ genes.
Nitrate availability controls the developmental expression of the SMZ and SNZ floral repressors and the FT floral integrator
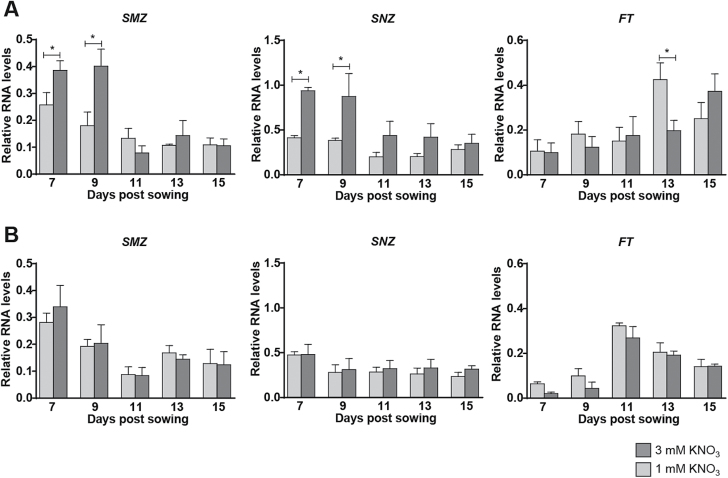
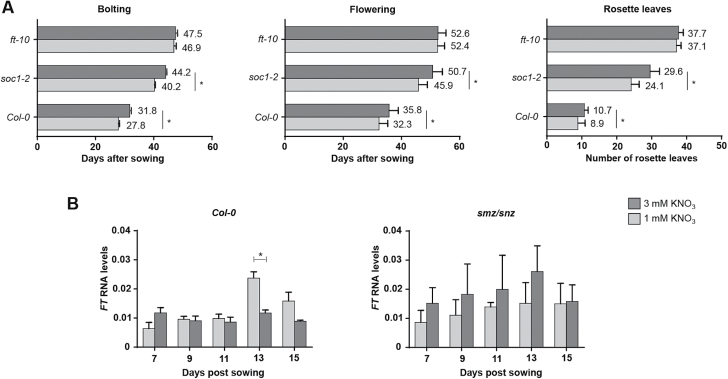
Expression of floral repressors is developmentally regulated, being higher in young seedlings and gradually decreasing throughout the plant’s life cycle. Controlled expression of the floral repressors determines FT accumulation, which is crucial for determining the transition to flowering (Aukerman and Sakai, 2003). In order to determine whether nitrate controls bolting and flowering time by regulating expression of the SMZ and SNZ genes during development, we analysed the levels of these transcription factors over the first 2 weeks of Arabidopsis growth. We found that during early developmental stages, SMZ and SNZ gene expression levels were higher in plants grown with 3 mM KNO3. This difference disappeared on days 11–15, when they showed similar levels when compared to plants grown with 1 mM KNO3 (Fig. 4A). We found that the effect of nitrate concentration on SMZ and SNZ levels was not dependent on post-transcriptional regulation by miR172, since miR172 accumulates in plants grown under 1 mM KNO3 only at later time points (13 and 15 d post-sowing; Supplementary Fig. S2). Consistent with a repressor role of SMZ and SNZ over FT, mRNA levels of this floral integrator showed a peak of induction at day 13 only when plants were grown in 1 mM KNO3 (Fig. 4A).
Fig. 4.
High nitrate availability induces a GA signaling-dependent early accumulation of SMZ and SNZ transcripts that correlates with a delayed increase in FT levels. Arabidopsis wild-type (A) and DELLA quintuple mutants (B) plants were grown on agar plates in N-free nutrient medium supplemented with either 1 mM (light gray) or 3 mM (dark gray) KNO3. At the days indicated, plants were harvested and RNA was extracted and used as a template for qRT-PCR. The ADAPTOR PROTEIN-4 MU-ADAPTIN gene (At4g24550) was used as an internal reference. Data are means and SE for three independent biological replicates of 15 plants. Asterisks highlight statistically different means as determined by Tukey’s Multiple Comparison test (P≤0.05).
It has been established that SMZ directly binds to the promoter of the floral integrators FT and SOC1 (Mathieu et al., 2009). In order to further analyse the role of these integrators over nitrate-dependent flowering time, we tested flowering time of the loss-of-function mutants soc1-2 and ft-10. As shown in Fig. 5A, flowering time of the ft-10 mutant did not respond to different nitrate concentrations. On the contrary, soc1-2 plants flowered later when grown under a higher KNO3 concentration. These results are consistent with a role for FT in nitrate-dependent control of flowering time in a SOC1-independent manner. Furthermore, quantitative RT-PCR showed that the FT transcript increase observed in 13-d-old seedlings grown under 1mM nitrate was suppressed in the smz/snz double-mutants (Fig. 5B). These data suggest that early accumulation of the floral repressors SMZ and SNZ under high nitrate leads to delayed bolting and flowering by controlling the expression of the FT floral integrator.
Fig. 5.
FT is the flowering integrator that mediates nitrate-dependent repression of flowering time. ft-10 and soc1-2 mutants (Col-0 background) were sown on vermiculite and watered once a week with N-free nutrient solution containing either 1 mM (light gray) or 3 mM (dark gray) KNO3. Nitrate-dependent flowering time and rosette leaf number were determined for 20–50 plants of each genotype (A). FT transcript levels of the wild-type (Col-0) and the smz/snz double-mutant (B). Plants were grown on Petri plates containing solid N-free nutrient medium supplemented with either 1 mM or 3 mM KNO3. At the days indicated, plants were harvested, their RNA was extracted and subsequently used for qRT-PCR. The ADAPTOR PROTEIN-4 MU-ADAPTIN gene (At4g24550) was used as an internal reference. Three independent biological replicates of 15 plants were used. Data are means and SD. Asterisks highlight significant differences as determined by Tukey’s Multiple Comparison test (P≤0.05).
Nitrate availability controls the developmental expression of GA biosynthesis genes and DELLA accumulation
Nitrate availability has been shown to control the levels of bioactive GAs to control flowering time (Liu et al., 2013). In Arabidopsis, bioactive GA biosynthesis depends on oxidation of the GA precursors GA12 and GA53 by GIBBERELLIN-20-OXIDASE (GA20OX) into GA9 and GA20. This is followed by an oxidation step performed by GIBERELLIN-3-OXIDASE (GA3OX) family proteins to produce the bioactive forms GA4 and GA1 (Hedden and Phillips, 2000). We analysed the developmental expression of two members of these families, GA20OX1 and GA3OX1, given their predominant role and expression in shoots (Rieu et al., 2008). As shown in Supplementary Fig. S3, the expression of GA20OX1 was lower in the early development of Arabidopsis when plants were grown in 3 mM as compared to 1 mM KNO3. We also found differences in the expression levels of GA3OX1, with an expression pattern under 1 and 3 mM KNO3 during development similar to the pattern of FT transcript accumulation (Figs 4A and 5B).
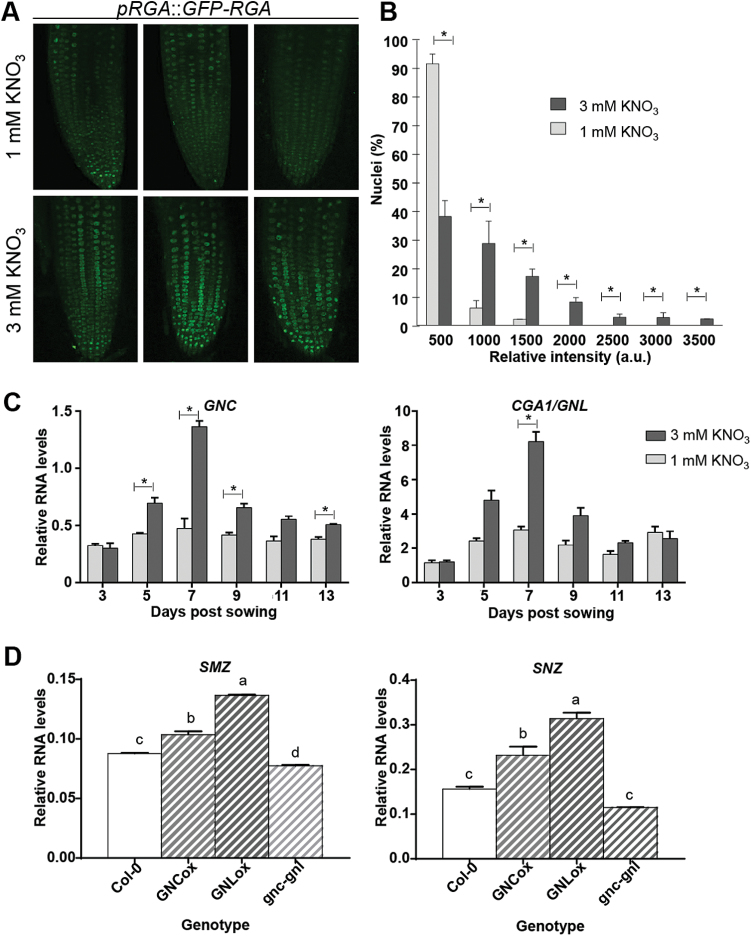
To confirm that nitrate-dependent differences in the expression levels of GA metabolism genes is biologically relevant, we analysed accumulation of the DELLA protein RGA in seedlings grown with 1 mM and 3 mM nitrate concentrations. DELLA proteins have been shown to accumulate when GA synthesis is impaired (Silverstone et al., 2001; Stavang et al., 2009). Consistently, we found that GFP-RGA levels were higher in roots of 7-d-old seedlings grown under 3 mM KNO3 (Fig. 6A, B), suggesting that high nitrate increases the levels of DELLA proteins by controlling the levels of bioactive gibberellins.
Fig. 6.
High nitrate availability promotes the accumulation of DELLA proteins and the induction of the downstream targets GNC and CGA1/GNL, triggering the up-regulation of SMZ and SNZ transcript levels. Representative confocal images showing roots of 7-d-old Arabidopsis RGA::GFP-RGA seedlings grown on agar plates in N-free nutrient medium supplemented with either 1 mM or 3 mM KNO3 (A). Quantification of the relative number of nuclei with specific GFP intensities (n=6) (B). Relative RNA levels for GNC and CGA1/GNL (C). Plants were grown on agar plates in N-free nutrient medium supplemented with either 1 mM (light gray) or 3 mM (dark gray) KNO3. At the days indicated, plants were harvested and RNA was extracted and used as a template for qRT-PCR. (D) Relative levels of SMZ and SNZ. Plants were grown on Petri plates containing solid N-free nutrient medium supplemented with 3 mM KNO3. On day 9, plants were harvested, their RNA was extracted and subsequently used for qRT-PCR. The ADAPTOR PROTEIN-4 MU-ADAPTIN gene (At4g24550) was used as a housekeeping gene. GNCox, GNC overexpressor plants; GNLox, CGA/GNL overexpressor plants; gnc-gnl, gnc-cga/gnl double-mutant plants. Data are means and SE for three independent biological replicates of 15 plants. Asterisks highlight significant differences as determined by Student’s t-test (P≤0.05). Different letters highlight significant differences as determined by Tukey’s Multiple Comparison test (P ≤0.05).
Nitrate controls SMZ and SNZ levels in a GA-dependent manner
Our results are consistent with nitrate regulating the GA and age-related pathways to control bolting and flowering time. These pathways have been previously shown to interact to control flowering time (Porri et al., 2012; Yu et al., 2012; Hyun et al., 2016). As previously shown in Fig. 2, the repressive effect of high nitrate concentration was abolished in the quintuple DELLA mutant, della-KO (Fig. 2A), similar to what was observed in the smz/snz mutant (Fig. 2B). In order to determine a possible crosstalk between the GA and the age-related pathways regarding nitrate-dependent bolting and flowering time, we analysed mRNA levels of the SMZ and SNZ genes in the della-KO mutant. As shown in Fig. 4B, mRNA levels of the SMZ and SNZ genes were similar when these mutant plants were grown in 1 or 3 mM KNO3. Moreover, FT mRNA levels peaked earlier and were similar in plants grown under either nitrate concentration, consistent with the bolting/flowering phenotype of the DELLA mutant. This suggests an interaction between DELLA proteins and SMZ/SNZ floral repressors to control FT levels depending on nitrate availability.
We also measured transcript accumulation of GNC and CGA1/GNL, which are transcription factors that act downstream of DELLA proteins, repressing certain GA responses (Richter et al., 2010). Consistent with our previous findings, the expression levels of both genes were higher under 3 mM KNO3 than 1 mM KNO3 (Fig. 6C). We also found that GNC and CGA1/GNL affected SMZ and SNZ expression, as evidenced by changes in SMZ and SNZ expression in mutants and transgenic lines that had altered levels of GNC and/or GNL (Fig 6D). These results provide additional evidence for a role of GA signaling in the transcriptional control of SMZ and SNZ.
Our findings prompt a model whereby nitrate signaling controls bolting and flowering time by a GA-dependent pathway that leads to changes in the levels of SMZ, SNZ, and FT throughout the development of the plant (Fig. 7).
Fig. 7.
Model for regulation of flowering time by nitrate in Arabidopsis thaliana. A nitrate signal, sensed by the NPF6.3/NRT1.1 transceptor, represses a gibberellin signaling pathway that in time causes an early induction of SMZ and SNZ. Induction of SMZ and SNZ leads to early repression of the floral integrator FT, and a delay of flowering time. This model summarizes our results in a naive lineal model. However, it does not preclude multiple entries stemming from upstream signaling components (e.g. NRT1.1) over the effectors and, similarly, feedback regulation by effectors (e.g. GA pathway) towards upstream components. Dashed lines denote likely indirect regulation. The link between DELLA and the effectors GNC-CGA1/GNL is indirect and has been previously described (Richter et al., 2010).
Discussion
In order to ensure reproductive success, plants have evolved mechanisms to sense environmental and internal cues to tightly control the timing of the transition from vegetative to reproductive growth and their flowering time. Research in the last decade has led to the identification of major pathways controlling this transition, as well as a considerable array of its molecular components. Pathways controlling flowering converge in floral pathway integrators, including FT, which encodes the florigen, a flowering signal that migrates from the leaf to the shoot apex (Corbesier et al., 2007; Mathieu et al., 2007; Andrés and Coupland, 2012). In this work, we have shown that nitrate, the main N source available in agricultural soils, modified the timing of the reproductive phase change and flowering by interacting with two endogenous pathways controlling this process, the GA pathway and the age-related pathway.
Nitrogen is the mineral nutrient that is required in largest amounts by plants (Epstein and Bloom, 2005). Its availability has a direct effect over fitness, as plants grown with higher N concentrations produce higher seed yields (Araus et al., 2016). Nitrate, the main N source for plants, triggers gene expression changes that encompass about 10% of the plant transcriptome. Nitrate-dependent gene networks are extraordinarily complex and have the ability to adjust in response to environmental perturbations. Nitrate assimilation integrates internal signals, such as carbon and energy metabolism, and environmental ones, such as light and N availability (Krouk et al., 2010). It has long been known that N modifies flowering time in plants, with N limitation often inducing early flowering (Klebs, 1913; Dickens and Staden, 1988; Bernier et al., 1993; Loeppky and Coulman, 2001). Other abiotic cues such as salt, drought, heat, cold, and UV radiation alter flowering time as well and this has been interpreted as a strategy that ensures seed production (Martínez et al., 2004; Achard et al., 2006).
Consistent with our findings, previous analyses of nitrate control of flowering have shown that nitrate availability modulates this developmental transition in Arabidopsis, with low nitrate availability accelerating and high nitrate availability delaying flowering time (Castro Marín et al., 2011; Kant et al., 2011; Liu et al., 2013; Yuan et al., 2016). However, these studies did not address whether nitrate interacts with the age-dependent pathway, limiting their results to analyses of the photoperiod, GA, and autonomous pathways (Castro Marín et al., 2011; Kant et al., 2011; Liu et al., 2013). As we have shown in our work, nitrate regulation of SMZ and SNZ levels led to changes in the timing of FT expression, and consequently altered bolting and flowering time. During the plant’s life cycle, the levels of the SMZ and SNZ floral repressors peaked at early stages and diminished as plants aged, partially by the post-transcriptional control exerted by miR172 microRNA (Aukerman and Sakai, 2003). Our results show that nitrate availability was able to control early accumulation of SMZ and SNZ mRNAs. Overexpression of SMZ and SNZ has been shown to repress FT expression, causing a late-flowering phenotype under long-day conditions, with no effect under short days (Mathieu et al., 2009). Accordingly, the timing of FT transcript accumulation was determined by nitrate availability under our experimental conditions, and was delayed under nitrate-sufficient conditions. Furthermore, the influence of nitrate over FT transcript accumulation was lost in the smz/snz double-mutant. Our data suggest that the nitrate-dependent control of SMZ and SNZ expression was mediated by a miR172-independent mechanism. The timing of nitrate-mediated changes in miR172 expression did not support an influence of this microRNA over the nitrate-elicited increase of SMZ and SNZ transcripts. In addition, the other tested targets of this microRNA, TOE1 and TOE2, did not have roles in nitrate-dependent flowering time. Notwithstanding this, our results do not exclude a potential complementary role of miR172 at later time points or under different experimental conditions.
Regulation of SMZ, SNZ, and FT expression by nitrate was similar to what has been described for the control of flowering by trehalose-6-phosphate (T6P). This sugar directly controls transcript levels of FT and SPL family members in leaves, independently from miR156 (van Dijken et al., 2004; Wahl et al., 2013), contrasting with the miR156-dependent pathway, which is controlled by other sugars such as sucrose and glucose (Yang et al., 2013; Yu et al., 2013). Analysis of SMZ and SNZ gene expression in della-KO plants grown under low- and high-nitrate conditions indicated that nitrate controlled SMZ and SNZ mRNA levels by interacting with the GA pathway. A crosstalk between the GA pathway and nitrate-dependent flowering time had previously been suggested from transcript expression studies of genes involved in GA biosynthesis, floral meristem identity, and floral integrators (Kant et al., 2011; Liu et al., 2013). Consistent with a role for GA in controlling nitrate-regulated bolting and flowering, we found that the transcript levels of two key enzymes in active GA biosynthesis depended on nitrate availability. Transcriptomic data from multiple independent studies has also shown down-regulation of the GA biosynthetic genes GA2, GA3OX1, and GA3OX4 in response to nitrate treatments (Canales et al., 2014). Nitrate availability has been shown to control the levels of bioactive GA3, with low and high nitrate causing an increase and a reduction in its levels, respectively, by controlling the expression of GA1, a key enzyme in GA biosynthesis in Arabidopsis (Liu et al., 2013). In agreement, our analysis of downstream effectors from the GA pathway showed that they were also affected by nitrate. First, the DELLA protein RGA accumulated more in the presence of a higher nitrate concentration. Second, the transcript levels of the downstream targets GNC and CGA1/GNL were up-regulated under these conditions. Both results showed that the GA pathway was more active at 1 mM than at 3 mM nitrate, consistent with the flowering time phenotypes observed under these conditions.
The GA and age-dependent flowering pathways are integrated by direct physical interaction between DELLA and SPL proteins (Porri et al., 2012; Yu et al., 2012; Hyun et al., 2016). This interaction down-regulates the SPL-dependent transcription of miR172 and MADS box genes. (Yu et al., 2012). miR172 targets AP2-like flowering repressors, including SMZ and SNZ (Mathieu et al., 2009). We found nitrate-dependent changes in SMZ and SNZ levels, which would suggest a GA-dependent down-regulation through the DELLA-SPL-miR172 module. However, our data also showed that nitrate-dependent flowering time was not affected in plants overexpressing miR156, which targets SPLs. These results strongly suggested that, for nitrate-dependent flowering time, the gibberellin pathway regulates SMZ and SNZ expression levels directly and that this is sufficient to explain the phenotype. Indeed, we obtained evidence identifying a cross-regulatory point between the two pathways. As shown in Fig. 6D, we found that two downstream targets of the GA pathway, GNC and CGA/GNA, had an effect over the expression of the flowering repressors SMZ and SNZ. This evidence was obtained with two transgenic lines overexpressing either GNC or CGA/GNA, and with a gnc-cga/gna double-mutant. Although both overexpressors consistently showed increased SMZ and SNZ levels, the double-mutant only showed a significant decrease of SMZ transcripts. This suggests that additional factors in the GA pathway may also regulate SMZ/SNZ expression.
Castro Marín et al. (2011) reported that nitrate regulates floral induction in Arabidopsis, acting independently of light, GA, and the autonomous pathways. The discrepancy between their conclusions regarding the role of GAs and ours could be attributed to multiple factors. First, their use of a combination of inorganic and organic N sources (nitrate and glutamine) with higher concentrations as compared to our study, where we focused on the effect of nitrate alone at relatively low concentrations. It has been shown that these different N sources can trigger disparate phenotypical effects (Zhang et al., 1999; Alboresi et al., 2005). Second, Castro Marin et al. (2011) tested flowering-time mutants in the gibberellin pathway under a neutral (12 h day/12h night) photoperiod and not LD conditions (16 h day/8 h night). Yuan et al. (2016) reported that N-dependent changes in flowering time are caused by variations in transcript levels of ferredoxin-NADP+-oxidoreductase (FNR1) and the blue-light receptor cryptochrome 1 (CRY1). The experimental conditions used in this study differed from ours in two key aspects. First, the N source used in this study was Murashige and Skoog medium supplemented with different concentrations of nitrate and ammonia. Second, flowering time was assessed in sterile Petri dishes under tissue-culture conditions, an environment that significantly differed from our set-up. These differing experimental settings may explain our disparate conclusions. This evidence, together with the Castro Marin results, strongly suggests that different N metabolites have effects over different pathways in order to control flowering time.
In Arabidopsis, nitrate availability is sensed by the NPF6.3/NRT1.1 nitrate transceptor (Ho et al., 2009). Besides its role as a major nitrate uptake transporter in Arabidopsis roots, NPF6.3/NRT1.1 has been shown to have diverse signaling mechanisms independent of nitrate transport (Bouguyon et al., 2015). According to our results, nitrate regulation of flowering time depended on a signaling function of nitrate that was dependent on NPF6.3/NRT1.1 since we did not find alterations in flowering time control in a NPF6.3/NRT1.1 mutant that was only altered in its nitrate uptake capability (Ho et al., 2009; Bouguyon et al., 2015). As for flowering, the control of seed dormancy by nitrate is also dependent on the NPF6.3/NRT1.1 transceptor (Alboresi et al., 2005), suggesting that signaling by NPF6.3/NRT1.1 might represent a mechanism to coordinate developmental transitions to optimal environmental conditions. A potential connection between NPF6.3/NRT1.1 and the gibberellin pathway is supported by a microarray analysis that was performed with the NPF6.3/NRT1.1 loss-of-function allele nrg1 and its wild-type counterpart (Wang et al., 2009). When treated with 1 mM KNO3, nrg1 mutants showed a 2-fold reduction of the gibberellin biosynthetic enzyme GA3OX1 (At1g15550) and the DELLA target CP1 (At4g36880), suggesting a role for NPF6.3/NRT1.1 in nitrate-dependent regulation of gibberellin (Cao et al., 2006; Wang et al., 2009). Furthermore, the complex relationship between N nutrition and the GA pathway is highlighted by a recent report that showed that the nitrate/nitrite transporter NPF3, a member of the same gene family of the NPF6.3/NRT1.1 transceptor, is a GA transporter (Tal et al., 2016).
Our data are consistent with a model in which nitrate availability controls GA activity, leading to an early regulation of SMZ and SNZ expression levels, which in turn changes the timing of FT induction and bolting and flowering time.
In summary, our phenotypical characterization together with our molecular genetics approach has uncovered a novel mechanism for nitrate-dependent control of flowering time. The shift from vegetative to reproductive development is one of the most important transitions throughout a plant’s ontogeny and, consequently, it is tightly controlled by various genetic and environmental factors. Although many environmental factors such as light, photoperiod, and temperature have been identified and characterized, the influence of mineral nutrients over flowering has been under-explored. Therefore, uncovering the molecular mechanisms of control that N availability exerts over the regulation of flowering time highlights the importance of nutritional status with regards to developmental decisions. Furthermore, it also provides new targets for crop improvement in a key environmental factor that impacts on reproductive success and yield.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Nitrate-dependent delay in flowering time is suppressed in the smz and snz single-mutants.
Fig. S2. miR172 expression is affected by nitrate availability at later stages of plant development.
Fig. S3. Nitrate availability controls the levels of active gibberellin key biosynthetic enzymes.
Acknowledgements
We are grateful to Dr Claus Schwechheimer (Technische Universität München, Germany) for providing 35S::GNC, 35S::GNL, and gnc-gnl seeds. This work was supported by grants from the Howard Hughes Medical Institute, Fondo de Desarrollo de Areas Prioritarias (FONDAP) Center for Genome Regulation (15090007), Millennium Nucleus Center for Plant Systems and Synthetic Biology (NC130030) to RAG; Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT) 1141097 to RAG, 11110083 to DG and 11121225 to EAV; Spanish Ministry of Economy and Competitiveness (grant BIO2013-43184-P) to DA and MAB; Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA) RTA2015-00014-c02-01 to JM; Internationalization Program PUC (PUC1566) to JM; and INIA pre-doctoral fellowship to JDF.
Glossary
Abbreviations:
- GA
Gibberelic acid
- NRT1.1
nitrate transporter 1.1
- SMZ
Schlafmutze
- SNZ
Schnarchzapfen.
Author contributions
Author contributions DG and RAG conceived this study; RAG supervised the research; DG performed most of the experiments; EAV, ER, RAG, JDF, SFU, SM, and DA performed some experiments or analysed data; DG, EAV, ER, SFU, and RAG wrote the paper; MB, DA, JDF, and JM performed specific experiments and helped with writing.
References
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP. 2006. Integration of plant responses to environmentally activated phytohormonal signals. Science 311, 91–94. [DOI] [PubMed] [Google Scholar]
- Adrian J, Torti S, Turck F. 2009. From decision to commitment: the molecular memory of flowering. Molecular Plant 2, 628–642. [DOI] [PubMed] [Google Scholar]
- Alboresi A, Gestin C, Leydecker MT, Bedu M, Meyer C, Truong HN. 2005. Nitrate, a signal relieving seed dormancy in Arabidopsis. Plant, Cell & Environment 28, 500–512. [DOI] [PubMed] [Google Scholar]
- Alexandre CM, Hennig L. 2008. FLC or not FLC: the other side of vernalization. Journal of Experimental Botany 59, 1127–1135. [DOI] [PubMed] [Google Scholar]
- Alvarez JM, Riveras E, Vidal EA, et al. 2014. Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. The Plant Journal 80, 1–13. [DOI] [PubMed] [Google Scholar]
- Amasino R. 2010. Seasonal and developmental timing of flowering. The Plant Journal 61, 1001–1013. [DOI] [PubMed] [Google Scholar]
- Andrés F, Coupland G. 2012. The genetic basis of flowering responses to seasonal cues. Nature Reviews. Genetics 13, 627–639. [DOI] [PubMed] [Google Scholar]
- Andrews M, Raven JA, Lea PJ. 2013. Do plants need nitrate? The mechanisms by which nitrogen form affects plants. Annals of Applied Biology 163, 174–199. [Google Scholar]
- Araus V, Vidal EA, Puelma T, Alamos S, Mieulet D, Guiderdoni E, Gutiérrez RA. 2016. Members of BTB gene family of scaffold proteins suppress nitrate uptake and nitrogen use efficiency. Plant Physiology 171, 1523–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H. 2003. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. The Plant Cell 15, 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier G. 1988. The control of floral evocation and morphogenesis. Annual Review of Plant Physiology and Plant Molecular Biology 39, 175–219. [Google Scholar]
- Bernier G, Havelange A, Houssa C, Petitjean A, Lejeune P. 1993. Physiological signals that induce flowering. The Plant Cell 5, 1147–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi YM, Wang RL, Zhu T, Rothstein SJ. 2007. Global transcription profiling reveals differential responses to chronic nitrogen stress and putative nitrogen regulatory components in Arabidopsis. BMC Genomics 8, 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouguyon E, Brun F, Meynard D, et al. 2015. Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nature Plants 1, 15015. [DOI] [PubMed] [Google Scholar]
- Canales J, Moyano TC, Villarroel E, Gutiérrez RA. 2014. Systems analysis of transcriptome data provides new hypotheses about Arabidopsis root response to nitrate treatments. Frontiers in Plant Science 5, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Cheng H, Wu W, Soo HM, Peng J. 2006. Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiology 142, 509–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Hussain A, Cheng H, Peng J. 2005. Loss of function of four DELLA genes leads to light- and gibberellin-independent seed germination in Arabidopsis. Planta 223, 105–113. [DOI] [PubMed] [Google Scholar]
- Castro Marín I, Loef I, Bartetzko L, Searle I, Coupland G, Stitt M, Osuna D. 2011. Nitrate regulates floral induction in Arabidopsis, acting independently of light, gibberellin and autonomous pathways. Planta 233, 539–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J, Dean C. 1994. Factors influencing the vernalization response and flowering time of late flowering mutants of Arabidopsis-thaliana (L) Heynh. Journal of Experimental Botany 45, 1279–1288. [Google Scholar]
- Corbesier L, Vincent C, Jang S, et al. 2007. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033. [DOI] [PubMed] [Google Scholar]
- Crawford NM, Glass ADM. 1998. Molecular and physiological aspects of nitrate uptake in plants. Trends in Plant Science 3, 389–395. [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S. 2008. A molecular framework for light and gibberellin control of cell elongation. Nature 451, 480–484. [DOI] [PubMed] [Google Scholar]
- Dickens CWS, Staden JV. 1988. The in vitro flowering of Kalanchöe blossfeldiana Poellniz: I. Role of culture conditions and nutrients. Journal of Experimental Botany 39, 461–471. [Google Scholar]
- Epstein E, Bloom AJ. 2005. Mineral nutrition of plants: principles and perspectives. Sunderland, MA: Sinauer Associates, Inc. [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, et al. 2008. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451, 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, de Montaigu A, Coupland G. 2010. SnapShot: Control of flowering in Arabidopsis. Cell 141, 550–550.e2. [DOI] [PubMed] [Google Scholar]
- Frink CR, Waggoner PE, Ausubel JH. 1999. Nitrogen fertilizer: retrospect and prospect. Proceedings of the National Academy of Sciences, USA 96, 1175–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giakountis A, Coupland G. 2008. Phloem transport of flowering signals. Current Opinion in Plant Biology 11, 687–694. [DOI] [PubMed] [Google Scholar]
- Golembeski GS, Imaizumi T. 2015. Photoperiodic regulation of florigen function in Arabidopsis thaliana. The Arabidopsis Book 13, e0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, et al. 2006. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. The Plant Cell 18, 3399–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo FQ, Wang R, Chen M, Crawford NM. 2001. The Arabidopsis dual-affinity nitrate transporter gene AtNRT1.1 (CHL1) is activated and functions in nascent organ development during vegetative and reproductive growth. The Plant Cell 13, 1761–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez RA. 2012. Systems biology for enhanced plant nitrogen nutrition. Science 336, 1673–1675. [DOI] [PubMed] [Google Scholar]
- Hedden P, Phillips AL. 2000. Gibberellin metabolism: new insights revealed by the genes. Trends in Plant Science 5, 523–530. [DOI] [PubMed] [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF. 2009. CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194. [DOI] [PubMed] [Google Scholar]
- Hong GJ, Xue XY, Mao YB, Wang LJ, Chen XY. 2012. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. The Plant Cell 24, 2635–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Lee LY, Xia K, Yan Y, Yu H. 2010. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Developmental Cell 19, 884–894. [DOI] [PubMed] [Google Scholar]
- Hyun Y, Richter R, Vincent C, Martinez-Gallegos R, Porri A, Coupland G. 2016. Multi-layered regulation of SPL15 and cooperation with SOC1 integrate endogenous flowering pathways at the arabidopsis shoot meristem. Developmental Cell 37, 254–266. [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Kay SA. 2006. Photoperiodic control of flowering: not only by coincidence. Trends in Plant Science 11, 550–558. [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA. 2003. FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426, 302–306. [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE. 1993. Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. The Plant Cell 5, 887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen EM, Sévin DC, Lillo C. 2009. The bZIP transcription factors HY5 and HYH are positive regulators of the main nitrate reductase gene in Arabidopsis leaves, NIA2, but negative regulators of the nitrate uptake gene NRT1.1. Journal of Plant Physiology 166, 2071–2076. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP. 2004. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Molecular Cell 14, 787–799. [DOI] [PubMed] [Google Scholar]
- Kant S, Peng M, Rothstein SJ. 2011. Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in arabidopsis. PLoS Genetics 7, e1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Michaels SD. 2006. SUPPRESSOR OF FRI 4 encodes a nuclear-localized protein that is required for delayed flowering in winter-annual Arabidopsis. Development 133, 4699–4707. [DOI] [PubMed] [Google Scholar]
- Klebs G. 1913. Uber das verhaltnis der außenwelt zur entwicklung der pflanze. Sitz-Ber Akad Wiss Heidelberg Ser B 5, 3–47. [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. 1999. A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Peeters AJ, Soppe W. 1998. Genetic control of flowering time in Arabidopsis. Annual Review of Plant Physiology and Plant Molecular Biology 49, 345–370. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Vries H, Hanhart C, Soppe W, Peeters T. 1994. The phenotype of some late‐flowering mutants is enhanced by a locus on chromosome 5 that is not effective in the Landsberg erecta wild‐type. The Plant Journal 6, 911–919. [Google Scholar]
- Krouk G, Crawford NM, Coruzzi GM, Tsay YF. 2010. Nitrate signaling: adaptation to fluctuating environments. Current Opinion in Plant Biology 13, 266–273. [DOI] [PubMed] [Google Scholar]
- Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I. 2000. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes & Development 14, 2366–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kim J, Han JJ, Han MJ, An G. 2004. Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in rice. The Plant Journal 38, 754–764. [DOI] [PubMed] [Google Scholar]
- Liu KH, Huang CY, Tsay YF. 1999. CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. The Plant Cell 11, 865–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Li Y, Ren J, Qian Y, Yang X, Duan W, Hou X. 2013. Nitrate or NaCl regulates floral induction in Arabidopsis thaliana. Biologia 68, 215–222. [Google Scholar]
- Loeppky HA, Coulman BE. 2001. Residue removal and nitrogen fertilization affects tiller development and flowering in meadow bromegrass. Agronomy Journal 93, 891–895. [Google Scholar]
- Martínez C, Pons E, Prats G, León J. 2004. Salicylic acid regulates flowering time and links defence responses and reproductive development. The Plant Journal 37, 209–217. [DOI] [PubMed] [Google Scholar]
- Mateos JL, Bologna NG, Chorostecki U, Palatnik JF. 2010. Identification of microRNA processing determinants by random mutagenesis of Arabidopsis MIR172a precursor. Current Biology 20, 49–54. [DOI] [PubMed] [Google Scholar]
- Mathieu J, Warthmann N, Küttner F, Schmid M. 2007. Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Current Biology 17, 1055–1060. [DOI] [PubMed] [Google Scholar]
- Mathieu J, Yant LJ, Mürdter F, Küttner F, Schmid M. 2009. Repression of flowering by the miR172 target SMZ. PLoS Biology 7, e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD. 2009. Flowering time regulation produces much fruit. Current Opinion in Plant Biology 12, 75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. 1999. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. The Plant Cell 11, 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase K, Hirano Y, Sun TP, Hakoshima T. 2008. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456, 459–463. [DOI] [PubMed] [Google Scholar]
- Mutasa-Göttgens E, Hedden P. 2009. Gibberellin as a factor in floral regulatory networks. Journal of Experimental Botany 60, 1979–1989. [DOI] [PubMed] [Google Scholar]
- O’Brien JA, Vega A, Bouguyon E, Krouk G, Gojon A, Coruzzi G, Gutiérrez RA. 2016. Nitrate transport, sensing, and responses in plants. Molecular Plant 9, 837–856. [DOI] [PubMed] [Google Scholar]
- Owen AG, Jones DL. 2001. Competition for amino acids between wheat roots and rhizosphere microorganisms and the role of amino acids in plant N acquisition. Soil Biology & Biochemistry 33, 651–657. [Google Scholar]
- Porri A, Torti S, Romera-Branchat M, Coupland G. 2012. Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 139, 2198–2209. [DOI] [PubMed] [Google Scholar]
- Pouteau S, Albertini C. 2009. The significance of bolting and floral transitions as indicators of reproductive phase change in Arabidopsis. Journal of Experimental Botany 60, 3367–3377. [DOI] [PubMed] [Google Scholar]
- Richter R, Bastakis E, Schwechheimer C. 2013a. Cross-repressive interactions between SOC1 and the GATAs GNC and GNL/CGA1 in the control of greening, cold tolerance, and flowering time in Arabidopsis. Plant Physiology 162, 1992–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R, Behringer C, Müller IK, Schwechheimer C. 2010. The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes & Development 24, 2093–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R, Behringer C, Zourelidou M, Schwechheimer C. 2013b. Convergence of auxin and gibberellin signaling on the regulation of the GATA transcription factors GNC and GNL in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 110, 13192–13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieu I, Ruiz-Rivero O, Fernandez-Garcia N, et al. 2008. The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. The Plant Journal 53, 488–504. [DOI] [PubMed] [Google Scholar]
- Riveras E, Alvarez JM, Vidal EA, Oses C, Vega A, Gutiérrez RA. 2015. The calcium ion is a second messenger in the nitrate signaling pathway of arabidopsis. Plant Physiology 169, 1397–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G, Tohge T, Matsuda F, Saito K, Scheible WR. 2009. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. The Plant Cell 21, 3567–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M, Kay SA. 2011. GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 108, 11698–11703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nature Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. 2005. Specific effects of microRNAs on the plant transcriptome. Developmental Cell 8, 517–527. [DOI] [PubMed] [Google Scholar]
- Shimada A, Ueguchi-Tanaka M, Nakatsu T, Nakajima M, Naoe Y, Ohmiya H, Kato H, Matsuoka M. 2008. Structural basis for gibberellin recognition by its receptor GID1. Nature 456, 520–523. [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Jung HS, Dill A, Kawaide H, Kamiya Y, Sun TP. 2001. Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. The Plant Cell 13, 1555–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG. 2004. The autonomous pathway: epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Current Opinion in Plant Biology 7, 570–574. [DOI] [PubMed] [Google Scholar]
- Srikanth A, Schmid M. 2011. Regulation of flowering time: all roads lead to Rome. Cellular and Molecular Life Sciences 68, 2013–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavang JA, Gallego-Bartolomé J, Gómez MD, Yoshida S, Asami T, Olsen JE, García-Martínez JL, Alabadí D, Blázquez MA. 2009. Hormonal regulation of temperature-induced growth in Arabidopsis. The Plant Journal 60, 589–601. [DOI] [PubMed] [Google Scholar]
- Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. 2001. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116–1120. [DOI] [PubMed] [Google Scholar]
- Tal I, Zhang Y, Jorgensen ME, et al. 2016. The Arabidopsis NPF3 protein is a GA transporter. Nature Communications 7, 11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijken AJ, Schluepmann H, Smeekens SC. 2004. Arabidopsis trehalose-6-phosphate synthase 1 is essential for normal vegetative growth and transition to flowering. Plant Physiology 135, 969–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Moyano TC, Canales J, Gutiérrez RA. 2014. Nitrogen control of developmental phase transitions in Arabidopsis thaliana. Journal of Experimental Botany 65, 5611–5618. [DOI] [PubMed] [Google Scholar]
- Wahl V, Ponnu J, Schlereth A, Arrivault S, Langenecker T, Franke A, Feil R, Lunn JE, Stitt M, Schmid M. 2013. Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 339, 704–707. [DOI] [PubMed] [Google Scholar]
- Wang JW. 2014. Regulation of flowering time by the miR156-mediated age pathway. Journal of Experimental Botany 65, 4723–4730. [DOI] [PubMed] [Google Scholar]
- Wang R, Xing X, Crawford N. 2007. Nitrite acts as a transcriptome signal at micromolar concentrations in Arabidopsis roots. Plant Physiology 145, 1735–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Xing X, Wang Y, Tran A, Crawford NM. 2009. A genetic screen for nitrate regulatory mutants captures the nitrate transporter gene NRT1.1. Plant Physiology 151, 472–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RN, Heckman JW, Somerville CR. 1992. Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiology 100, 403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. 2009. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Liu Q, Yao T, Fu X. 2014. Shedding light on integrative GA signaling. Current Opinion in Plant Biology 21, 89–95. [DOI] [PubMed] [Google Scholar]
- Yang DL, Yao J, Mei CS, et al. 2012. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proceedings of the National Academy of Sciences, USA 109, E1192–E1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Xu M, Koo Y, He J, Poethig RS. 2013. Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. eLIFE 2, e00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SK, Chung KS, Kim J, Lee JH, Hong SM, Yoo SJ, Yoo SY, Lee JS, Ahn JH. 2005. CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiology 139, 770–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Cao L, Zhou CM, Zhang TQ, Lian H, Sun Y, Wu J, Huang J, Wang G, Wang JW. 2013. Sugar is an endogenous cue for juvenile-to-adult phase transition in plants. eLIFE 2, e00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Galvão VC, Zhang YC, Horrer D, Zhang TQ, Hao YH, Feng YQ, Wang S, Schmid M, Wang JW. 2012. Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. The Plant Cell 24, 3320–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Zhang ZW, Zheng C, et al. 2016. Arabidopsis cryptochrome 1 functions in nitrogen regulation of flowering. Proceedings of the National Academy of Sciences, USA 113, 7661–7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HM, Jennings A, Barlow PW, Forde BG. 1999. Dual pathways for regulation of root branching by nitrate. Proceedings of the National Academy of Sciences, USA 96, 6529–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZL, Ogawa M, Fleet CM, Zentella R, Hu JH, Heo JO, Lim J, Kamiya Y, Yamaguchi S, Sun TP. 2011. SCARECROW-LIKE 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proceedings of the National Academy of Sciences, USA 108, 2160–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QH, Helliwell CA. 2011. Regulation of flowering time and floral patterning by miR172. Journal of Experimental Botany 62, 487–495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.