Identification of all MATE family members in Populus and characterization of PtrMATE1 mediated citrate exudation from the roots of Populus in response to Al stress.
Keywords: Aluminum stress, citrate exudation, MATE transporter, Populus, root apex
Abstract
Ionic aluminum (Al) in acidic soils, comprising approximately 50% of arable land globally, is highly toxic to most plant species. Populus grow naturally in acidic soils and tolerate high concentrations of Al. Multidrug and toxic compound extrusion (MATE) family genes in plants are involved in responses to Al tolerance. To date, however, the functional roles of the MATE genes in Populus remain unclear. In the present study, 71 putative MATE transporters were predicted in the genome of Populus trichocarpa. The chromosome distribution, phylogenetic relationships, and expression level analysis revealed that four candidate MATE genes belonging to subgroup IIIc might contribute to high Al tolerance in poplar. Further, the expression levels of two subgroup IIIc members, PtrMATE1 and PtrMATE2, were induced by Al stress. Transient expression in onion epidermal cells showed that PtrMATE1 was localized to the plasma membrane. Overexpression of PtrMATE1 increased Al-induced secretion of citrate from the root apex of transgenic plants. Al-induced inhibition of root growths were alleviated in both PtrMATE1 overexpression lines in Populus and in Arabidopsis compared with wild-type plants. In addition, PtrMATE1 expression was induced at 12 h after exposure to Al stress whereas PtrMATE2 expression was induced at 24 h, indicating that these proteins coordinately function in response to Al stress in poplar. Taken together, these results provide important insights into the molecular mechanisms involved in Al tolerance in poplar.
Introduction
Aluminum (Al) toxicity is a major limitation to plant growth in acidic soils, namely with a pH<5.5, in which the Al complex in aluminosilicate clays is solubilized as the most toxic trivalent cation, Al3+ (Kochian et al., 2004). Al3+ toxicity primarily targets the root apex, thereby inhibiting root growth (Liu et al., 2009). In most plant species, two main types of Al resistance mechanisms have been described: Al exclusion mechanisms, which prevent Al from entering the root apex, and Al tolerance mechanisms, which detoxify and sequester Al in plants (reviewed by Kochian et al., 2015). To date, the most well-characterized Al exclusion mechanism involves Al-induced exudation of organic acids (OAs), including malate, citrate, and oxalate efflux from roots (Delhaize and Craig, 1993; Delhaize and Ryan, 1995; Kinraide et al., 2005; Delhaize et al., 2007). These OA anions are transported from root cells into rhizospheres, in which Al3+ is effectively chelated, forming non-toxic complexes (Kinraide et al., 2005). However, the regulatory mechanism of Al-induced oxalate secretion remains largely unknown (Feng et al., 1998) and studies of functional genes related to the oxalate synthesis pathway did not reveal the regulation mechanism of oxalate excretion induced by Al in plants (Franceschi and Nakata, 2005; Xu and Peng, 2006).
Al3+ concentrations, even as low as 50 μM, affect root growth in herbaceous plants (Brauer, 2001; Kumari et al., 2008; Chandran et al., 2008). By contrast, woody plants generally tolerate high concentrations of Al and grow naturally in acidic soil (Schaedle et al., 1989). For example, seedlings of Norway spruce (Picea abies) and birch (Betula pendula) showed no changes in root growth at Al concentrations ranging from 0.3 mM to 3 mM (Göransson and Eldhuset, 1987; Göransson and Eldhuset, 1991). Woody plants have evolved an adaptive mechanism that enables tolerance to high Al conditions (Grisel et al., 2010). Elaboration of Al tolerance in woody plants will therefore broaden the current understanding of the Al tolerance mechanism in plants.
Currently, homologs of the malate transporter gene ALMT1 have been isolated from wheat (Sasaki et al., 2004), Arabidopsis (Hoekenga et al., 2006) and oilseed rape (Ligaba et al., 2006). Many Al-induced citrate efflux genes have also been identified and characterized in herbaceous plants, which are all members of the MATE family, such as HvAACT1 in barley (Furukawa et al., 2007), SbMATE in sorghum (Magalhaes et al., 2007), AtMATE in Arabidopsis (Liu et al., 2009), TaMATE in wheat (Ryan et al., 2009), ScFRDL1 in rye (Secale cereale L.; Yokosho et al., 2011), and ZmMATE1 in maize (Maron et al., 2010). The encoded proteins of all these homologous genes are primarily localized to root epidermis cells (Furukawa et al., 2007) and are required for external Al resistance (Yokosho et al., 2011). The MATE proteins are a large family of multidrug efflux transporters (Kuroda and Tsuchiya, 2009) that are widely distributed in plants, bacteria, fungi, and mammals (Omote et al., 2006). Many putative MATE transporters have been predicted in Arabidopsis (Li et al., 2002), Oryza sativa (Tiwari et al., 2013), and Medicago truncatula (Zhao and Dixon, 2009). Recently, some plant MATE transporters have been implicated in controlling diverse developmental and physiological processes, including transport of secondary metabolites, such as alkaloids, flavonoids, and anthocyanins (Shoji et al., 2009; Pérez-Díaz et al., 2014), detoxification of toxic compounds or heavy metals (Diener et al., 2001; Li et al., 2002), improvement of abiotic resistance (Sun et al., 2011), and transport of phytohormones (Zhang et al., 2014), iron (Yokosho et al., 2009; Zhang et al., 2014), and Al (Furukawa et al., 2007; LiLiu et al., 2009; Fujii et al., 2012; Zhou et al., 2013; Liu et al., 2016). Screening the entire MATE gene family is therefore helpful to identify putative Al-induced citrate transporters from plant species.
Early phytoremediation studies showed that Populus, a fast-growing tree species, could be a suitable candidate for phytoremediation of metal-polluted soils (Cunningham and Ow, 1996; Migeon et al., 2010). A recent study showed that Populus tolerates high concentrations of Al and that one of the aspen MATE homologous genes may be related to Al tolerance in the roots of aspen according to transcriptomic predictions (Grisel et al., 2010) with the completion of the poplar genome sequence (Tuskan et al., 2006). To date, however, the characterization of MATE genes in Populus remains lacking. In the present study, we performed a genome-wide search for all putative MATE transporters in P. trichocarpa according to whole genome sequence data from P. trichocarpa Torr. & A. Gray (http://genome.jgi-psf-org/Poptrl_l/Poptrl_l.home.html). The chromosome distribution, gene duplication, phylogenetic relationships, and gene and protein structures of these poplar putative MATE transporters were analyzed and the expression levels of 10 candidate MATE genes in the shoots and roots of Populus after Al treatment were characterized using qRT-PCR analysis. Furthermore, the function of PtrMATE1 in transgenic poplar and Arabidopsis was identified and characterized. Finally, the coordinated roles of PtrMATE1 and PtrMATE2 in response to long-term Al stress were also investigated. These results provide insights into the Al tolerance mechanism in Populus under acidic soil conditions and will be helpful for further understanding the roles of MATE genes in Populus.
Materials and methods
Plant material and treatments
Plant materials and growth conditions
P. trichocarpa Torr. & A. Gray and P. tomentosa Carr. (clone 741) (Chinese white poplar) were cultivated in a greenhouse at 24◦C under a 14 h/10 h light/dark cycle with 5,000 lux of light and maintained in sterile woody plant medium (WPM) containing 0.8% (w/v) agar. The WPM medium contained the following macro- and micronutrients: NH4NO3 (400mg/L), Ca(NO3)2·4H2O (556mg/L), K2SO4 (990mg/L), CaCl2·2H2O (96mg/L), KH2PO4 (170mg/L), Na2MoO4·2H2O (0.25mg/L), MgSO4·7H2O (370mg/L), MnSO4·H2O (22.4mg/L), ZnSO4·7H2O (8.6mg/L), CuSO4·5H2O (0.25mg/L), FeSO4·7H2O (27.8mg/L), Na2-EDTA (37.3mg/L), vitamin B1 (1.0mg/L), vitamin B6 (0.5 mg/L), inositol (100 mg/L), nicotinic acid (0.5 mg/L), and glycine (2.0 mg/L) (Mccown and Lloyd, 1981). The normal pH was 5.2. In the present study, for the Al treatment experiments, the pH was 4.0. The plants were transplanted every 6–8 weeks.
Al and La treatments
In general, 4-week-old plant roots were immersed in WPM medium containing 500 µM AlCl3 or 500 µM LaCl3 at pH 4.0 during the treatment, and subsequently, the shoots and roots were collected and rinsed in WPM at pH 4.0. Plants treated with WPM medium, again at pH 4.0, were used as a control.
For the time-course experiments, 10 mm root apices were rinsed three times for 10 min each time with the 500 µM CaCl2 solution at pH 4.0 (Ca solution) to avoid potential ion leakage around root apices. The root apices were subsequently treated with a CaCl2 solution containing either 500 µM AlCl3 (Al solution) or 500 µM LaCl3 in 1.5 ml centrifuge tubes at 25◦C every 3h. The appropriate solutions and root samples were collected at each time point. Plants treated with the CaCl2 solution were used as controls. At least three biological replicates were performed for each treatment. All samples were immediately frozen in liquid nitrogen and stored at -80◦C until further analysis.
Identification of MATE transporters in Populus
The genomic and protein sequences of 57 MATE transporters in Arabidopsis were obtained from Phytozome v11.0 (Goodstein et al., 2012; http://phytozome.jgi.doe.gov/pz/portal.html). P. trichocarpa putative MATE protein sequences were retrieved by BLASTP searches against the target (P. trichocarpa v3.0) proteome in Phytozome v11.0 using the Arabidopsis MATE protein sequences as queries, with E-value≤1e-7). These MATE sequences were further filtered using Pfam (Marshall, 2003; http://pfam.xfam.org/) and the Simple Modular Architecture Research Tool (Letunic et al., 2015; http://smart.embl-heidelberg.de/smart/batch.pl) based on the presence of a conserved MATE domain (Pfam: PF01554).
Chromosomal locations and gene duplication analysis
The online tool PopGenIE (http://www.popgenie.org/) was used to determine and plot the chromosomal localization of all PtrMATE genes. Genes separated by five gene loci within a 100 kb distance were considered tandem duplicates (Hu et al., 2010). Segmental duplications resulting from salicoid genome-wide duplications were identified based on the duplication coordinates from the Populus genome assembly. Blocks in the same colors represent the homologous chromosomal segments. Segmental and tandem duplication events of the MATE family were conducted according to Tuskan et al. (2006).
Phylogenetic and structural analyses of MATE transporters in Populus
The full protein sequences of 71 Populus MATE proteins and 30 previously reported MATE proteins from other plant species (see Table S1 available at the Dryad Digital Repository http://dx.doi.org/10.5061/dryad.vb047) were used for multiple sequence alignments using ClustalW in MEGA 6.0 (Tamura et al., 2013). The maximum likelihood (ML) was constructed by MEGA 6.0 using an algorithm with 1000 bootstraps, based on the equal input model, using partial deletion of 95% site coverage for gaps and missing data. The online program Gene Structure Display Server (GSDS) with default settings (Hu et al., 2015; http://gsds.cbi.pku.edu.cn) was used to analysis the MATE gene structure. The online tool Multiple EM for Motif Elicitation (MEME; Bailey and Elkan, 1994; http://meme-suite.org/) was used to predict the motifs in MATE proteins. The maximum number of motifs was set at 10.
RNA isolation and real-time quantitative PCR
Total RNA was extracted using the RNA RNeasy Plant Mini Kit (Qiagen, Duesseldorf, Germany). First-strand cDNA synthesis was performed using the PrimeScript™ RT reagent kit (Perfect Real Time; Takara, Dalian, China). GoTaq® qPCR Master Mix (Promega, Madison, WI USA) was used to perform qRT-PCR to determine the transcript levels of MATE genes in P. trichocarpa. (See gene-specific primers used for qRT-PCR analysis in Table S2 at the Dryad). At least two biological replicates of each sample and three technical replicates of each biological replicate were performed to ensure the accuracy of the results. The reference gene UBQ (FJ438462) was used as an internal control. A relative quantification method was used to evaluate quantitative variation among replicates. The PCR conditions and relative gene expression calculations were conducted as previously described (Zhang et al., 2014).
Citrate measurement
Citrate concentrations were determined based on the citric acid (CA) content of the test box (Jiancheng Bioengineering Institute, Nanjing, China) according to the manufacturer’s instructions. Briefly, citrate reduces Cr (VI) in acid solutions, and produces Cr3+, which has a specific absorption peak at 545 nm. By measuring the increase in absorbance at A540, the CA content in the sample can be calculated.
Cloning of PtrMATE1 and transformation of poplar and Arabidopsis
The full open-reading frame of PtrMATE1 was amplified with gene-specific primers. The amplification products were cloned into the plant binary vector pCAMBIA1302. Agrobacterium tumefaciens strain GV3101 containing p35S:PtrMATE1 was used to transform plants.
Poplar transformation was performed according to Jia et al. (2010). Briefly, poplar leaves were excised from in vitro plantlets, cut into disks, and dipped in the diluted Agrobacterium culture for 8–10min; the leaf disks were then transferred to WPM medium. The infected disks were co-cultivated in the dark for 2 d and subsequently transferred to callus-inducing medium. After 2–3 weeks, leaf disks with induced calli were subcultured on screening medium to induce adventitious buds. Regenerated shoots were transferred to rooting medium. The rooted plantlets were acclimatized in pots placed inside a humid chamber for 2 weeks and finally transferred to the greenhouse.
A. tumefaciens harboring the p35S:PtrMATE1 construct was also transformed into wild-type Arabidopsis (Columbia ecotype, Col-0) and the T-DNA insertion mutants AtMATE-KO (SALK_081671; Liu et al., 2009) by the floral dip method (Clough & Bent, 1998). Selection of transformants was performed on ½ Murashige and Skooge (MS) medium supplied with 50 mg·L-1 hygromycin. Homozygote progeny from T2 transformants with a single copy of the transgene were screened. The PtrMATE1-OX and PtrMATE1-R transgenic plants were found using qRT-PCR. PtrMATE1-OX was the overexpression of PtrMATE1 in wild-type Arabidopsis, PtrMATE1-R was the overexpression of PtrMATE1 in AtMATE-KO mutants.
Evaluation of transgenic plants for resistance against Al stress
When the roots of wild-type and transgenic plants grew to 1 cm, the nutrient solution was replaced with autoclaved WPM medium at pH 4.0. After 2 d, the nutrient solution was replaced with a treatment solution comprising autoclaved WPM medium at pH 4.0, supplemented with AlCl3 up to 500 μM. The pH of the Al treatment solutions was adjusted to 4.0 with KOH.
Root growth was monitored photographically prior to the treatment at 2 d and during the entire treatment at 12 h intervals. We used a digital camera focused on a 0.1 mm grid of the graph paper. The pictures were cropped and normalized using the grid on the graph paper with IMAGEJ 1.50i. The normalized pictures were used to measure the increase in root length during particular time intervals. The root growth rate was estimated by dividing each increment by the time elapsed. Following these treatments, the roots were separated from the shoots and rinsed in WPM at pH 4.0. The root samples were collected and transferred to a sterile 1.5 ml tubes. The pooled leaves and stems were separately collected. All tissues were frozen in liquid nitrogen and stored at -80 °C until RNA extraction.
Results
Identification of MATE family genes in Populus
Genome-wide analysis was performed to predict all MATE genes in P. trichocarpa Torr. & A. Gray using genome sequence data (http://genome.jgi-psf-org/Poptrl_l/Poptrl_l.home.html). Initially, we identified 77 putative full-length protein sequences encoding MATE genes from the Populus whole genome using a BLASTP search with 57 MATE protein sequences in Arabidopsis (collected from Phytozome v11.0) as queries. These putative MATE sequences were filtered based on the presence of the conserved MATE domain (Pfam: PF01554) using the Pfam database. Eventually, a total of 71 genes encoding unique poplar MATE genes were identified and designated with consecutive nomemclature as PtrDTX1-71 (for Populus detoxification 1-71) based on their physical location (Li et al., 2002). These MATE genes encoded proteins that varied in length from 120 to 608 amino acids with an average of 480 amino acids, a molecular weight range of 13.14 to 65.49 kDa, and predicted isoelectric point values between 5.05 and 9.50. Detailed information for all 71 Populus MATE proteins is listed in Table S1 at Dryad.
Chromosomal location and gene duplication of Populus MATE genes
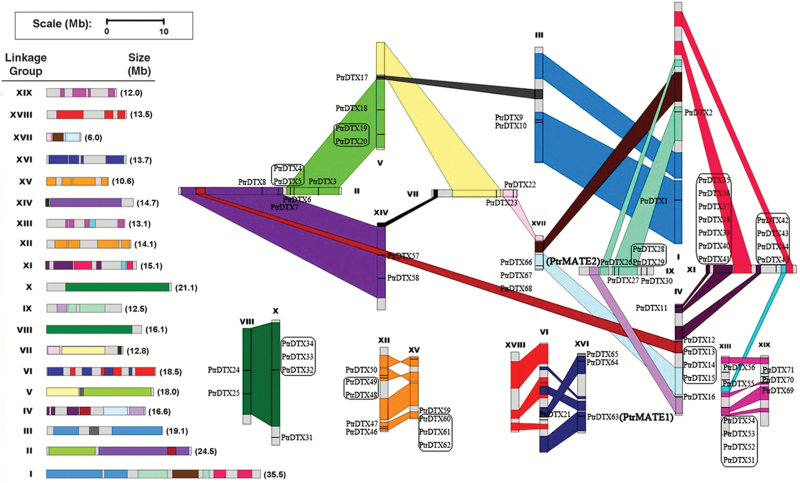
To examine the physical relationship of PtrMATE genes in the chromosomes of Populus, we performed chromosomal location and gene duplication analyses. As shown in Fig. 1, in silico mapping of the gene locus showed that 71 Populus MATE genes were mapped to 18 linkage groups (LG), except LG XVIII. LG XI harbored the highest number of PtrMATE genes at 11 genes, followed by LG II and LG XIII with 6 genes. Only one PtrMATE gene was observed on LG VI, and two PtrMATE genes were observed on each of LG I, III, VII, VIII, and XIV. Substantial clustering of PtrMATE genes was observed on LG XI (Fig. 1).
Fig. 1.
Chromosomal location of Populus MATE genes. A total of 71 MATE genes are mapped to the 18 linkage groups (LG). Schematic view of chromosome reorganization through recent whole genome duplication in Populus is shown. Segmentally duplicated homologous blocks are indicated with the same color. The scale represents megabases (Mb). The LG numbers are indicated at the top of each bar.
Recent studies have shown that the Populus genome has undergone at least three rounds of genome-wide duplications, followed by multiple segmental duplication, tandem duplication, and transposition events (Tuskan et al., 2006; Hu et al., 2012). The segmental duplication associated with a salicoid duplication event in particular contributed to the expansion of many multi-gene families (Barakat et al., 2009; Rogers et al., 2009; Hu et al., 2010). We mapped Populus MATE genes to the duplicated blocks established in previous studies to determine the potential relationships between MATE genes and putative segmental duplications. The distributions of MATE genes relative to the corresponding duplicate blocks are illustrated in Fig. 1. Among these 71 genes, 36 genes were preferentially retained duplicates located in both duplicated regions and 18 duplicated blocks only contained MATE genes on one of the blocks and lacked duplicates on the corresponding block. Almost half of the genes, 35 of 71, were represented in distinct tandem duplicate gene clusters. These results revealed that tandem repeats and duplication events contributed to the expansion of the MATE gene family in the Populus genome.
Phylogenetic analyses of PtrMATE proteins
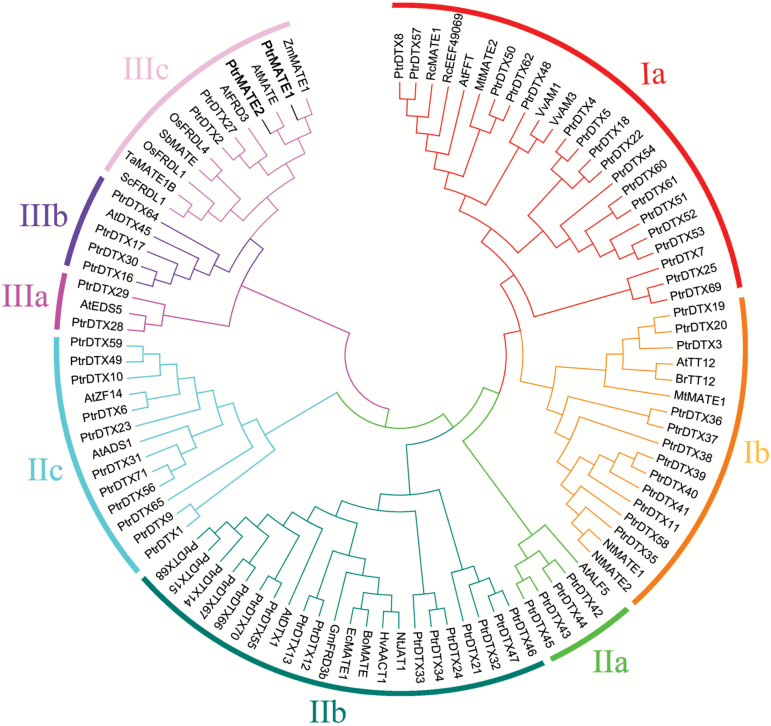
To characterize the phylogenetic relationships among Populus MATE proteins, we constructed a phylogenetic tree using the full-length proteins of the 71 Populus MATE members and 30 previously reported plant MATE proteins from 15 other plant species using Clustal W and MEGA 6.0 (Fig. 2; Table S1 at Dryad). These PtrMATE proteins can be classified into three subfamilies with eight smaller subgroups, namely subfamily I (subgroups Ia and Ib), II (subgroups IIa, IIb, and IIc), and III (subgroups IIIa, IIIb and IIIc). The functions of the PtrMATE proteins were inferred from functionally characterized MATE proteins from other plant species according to their phylogenetic relationships.
Fig. 2.
The phylogenetic tree of MATE proteins from P. trichocarpa and other plant species. The phylogenetic tree was constructed using MEGA 6.0 with the Maximum Likelihood (ML) method. Bootstrap values in percentages (1000 replicates) are indicated on the nodes. Different subgroups are highlighted using different colors and marked with arcs outside the cycle tree.
Subfamily I contains 24 transporters in subgroup Ia and 17 in subgroup Ib. Among these proteins, many MATE members have previously been implicated in the transport of flavonoids, alkaloids, and other phenolics in Arabidopsis, Medicago truncatula, grapevine, and tobacco (Zhao and Dixon, 2009; Shoji et al., 2009; Gomez et al., 2009; Thompson et al., 2010; Gomez et al., 2011), suggesting that this MATE subfamily might be involved in the transport and accumulation of secondary metabolites. There are three subgroups in subfamily II: IIa, IIb and IIc. The IIa subgroup comprised four PtrMATE proteins and AtALF5, which confers Arabidopsis resistance to toxins (Diener et al., 2001). Subgroup IIb, comprised 16 PtrMATEs and six characterized MATE proteins involved in the efflux of plant-derived antibiotics and other toxic compounds, and detoxification of heavy metals, among other functions (Li et al., 2002; Morita et al., 2009). In addition, subgroup IIc includes 11 PtrMATEs and AtADS1 and AtZF14. Interestingly, the functions of AtADS1 and AtZF14 are diverse, including the regulation of plant disease resistance, increasing the leaf initiation rate, and the regulation of hypocotyl cell elongation and iron homeostasis (Sun et al., 2011; Seo et al., 2012; Zhang et al., 2014; Wang et al., 2015). However, most of these identified CA exporters belong to subfamily III. These transporters secrete OAs to the apoplast to chelate Al3+ in the rhizosphere and alleviate Al3+ toxicity in acidic soils. At least eight known MATE transporters, including ScFDRL1, TaMATE1B, OsFRDL1, SbMATE, OsFRDL4, AtFRD3, AtMATE, and ZmMATE1, have been associated with Al detoxification and/or iron translocation (Yokosho et al., 2009; Tovkach et al., 2009; Liu et al., 2009; Maron et al., 2010; Sivaguru et al., 2013; Charlier et al., 2015). Subgroup IIIc also contains four Populus members, PtrMATE1, PtrMATE2, PtrDTX2, and PtrDTX27, suggesting that these PtrMATEs might also be involved in Al detoxification or iron translocation in Populus.
Structure and motif distribution analysis of Populus MATE genes
To better understand their functional characteristics, the structural diversity of all of the Populus MATE genes was analyzed by comparing the corresponding coding sequences with their genomic DNA sequences. The PtrMATE genes contained 1–14 exons, and these intron-exon structures are plotted in Fig. 3A. Consistent with the characteristics shown in the phylogenetic analysis, closely related genes were more structurally similar, differing in the lengths of introns and exons in the same subfamily. For example, almost all of the PtrMATE genes in subfamily I contained seven exons and the members in subfamily II had one (PtrDTX23, PtrDTX31) to nine (PtrDTX12) exons, while the PtrMATE genes in subfamily III exhibited the largest number of exons, between 10 to 14.
Fig. 3.
The gene structures and conserved motifs of PtrMATE family members. (A) Exons and introns of PtrMATE genes are plotted using green boxes and black lines, respectively. The blue boxes indicate upstream/downstream sequences. (B) Protein motifs of the PtrMATE family. The motifs of Populus MATE proteins are shown as colored boxes, each motif is represented as a number in the colored box. The genes are listed according to the order of subfamily I to III from the phylogenetic tree and different subfamilies are highlighted with lines.
We also predicted conservative motifs in PtrMATE proteins using MEME software. A total of 10 conserved motifs were identified as shown in Fig. 3B and Table S3 at Dryad. The types and sequences of the motifs were similar among subfamilies I and II but significantly different from subfamily III. Fewer motifs were observed among subfamily III proteins compared with the other subfamily members. All of the MATE family members contained motif 3 except for PtrDTX10 (Fig. 3B).
Expression patterns of the PtrMATE genes in response to Al stress
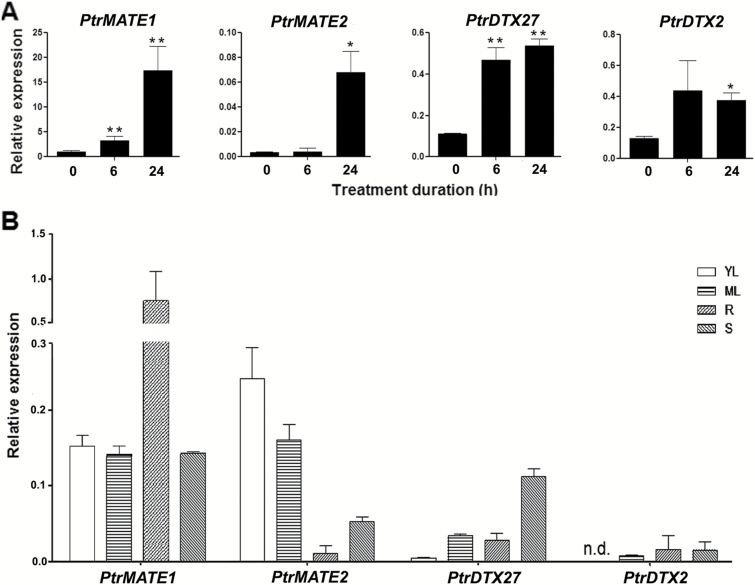
To investigate the function of the PtrMATE genes in subfamily III, we determined the expression patterns of these genes in response to Al exposure using quantitative real-time PCR (qRT-PCR) with the 4-week-old poplars immersed in the WPM medium containing 500 µM Al3+ at pH 4.0) for 12 h. Poplar ubiquitin (UBQ) expression was used as a control and gene-specific primers were used for qRT-PCR analysis of PtrMATE genes. The results revealed that the expression levels of all of the subgroup IIIc genes were significantly increased in roots treated with 500 μM Al for 12 h, while other subgroup III members were not induced (see Fig. S1 at Dryad). A time-course experiment showed that PtrMATE1 was activated 6 h after the exposure to Al and at 24 h the expression increased by approximately 18 times compared with the level detected prior to Al treatment (Fig. 4A). Both PtrDXT2 and PtrDXT27 were also upregulated at 6 h under Al stress conditions. By contrast, PtrMATE2 was not induced at 6 h after Al treatment but was significantly upregulated at 24 h. These results suggest that these PtrMATE genes might be involved in the response to Al stress.
Fig. 4.
Expression analysis of subgroup III MATE genes using qRT-PCR. (A) Expression of four PtrMATE members in poplar roots under 500 μM Al3+ for 6 h and 24 h. (B) Relative quantities of four members in roots, stems, young leaves, and mature leaves are illustrated. Expression of PtrMATE1 in wild-type roots was arbitrarily fixed at one. The results are shown as the mean expression ± standard deviation (SD) of three independent experiments. Poplar ubiquitin (UBQ) expression was used as a control and gene-specific primers were used for qRT-PCR analysis of PtrMATE genes. Student’s t-test, *P<0.05, **P<0.01.
We also examined the expression patterns of these four subgroup IIIc genes in different tissues of Populus, including the roots, stems, young leaves, and mature leaves. As shown in Fig. 4B, these PtrMATE genes had different tissue-specific expression patterns. PtrMATE1 was highly expressed in the roots and its expression was more abundant than that of other PtrMATE genes, suggesting that PtrMATE1 might play a critical role in response to Al stress in poplar.
Sequence analysis and subcellular localization of PtrMATE1
To investigate the function of PtrMATE1 in response to Al stress, we further focused on the characterization and analysis of PtrMATE1 in Populus. In the Populus MATE gene family, PtrMATE1 and PtrMATE2 shared 64% amino acid sequence identity with each other; these genes are the closest sequences to AtMATE, a citrate transporter, with 65% and 66% identity, respectively. Multiple alignments of PtrMATE1 and PtrMATE2 with the AtMATE protein also revealed that they share strong sequence homology with each other (see Fig. S2 at Dryad). Consistent with the structure of AtMATE, PtrMATE1 and PtrMATE2 were predicted to have similar topology using the CCTOP program (Liu et al., 2009; http://cctop.enzim.ttk.mta.hu/).
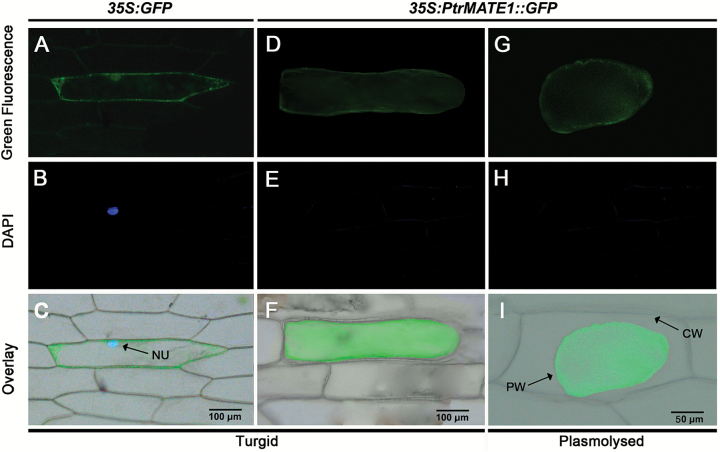
To determine the subcellular location of PtrMATE1, the p35S:PtrMATE1-GFP construct, in which the PtrMATE1-GFP fusion gene was under the control of the CaMV 35S promoter, was transformed into onion epidermal cells using Agrobacterium-mediated transformation. The PtrMATE1-GFP signal was observed in the plasma membrane of onion epidermal cells (Fig. 5D, E, F), compared with the control that expressed GFP alone (Fig. 5A, B, C). To distinguish localization in the plasma membrane from that in the cell wall, we added 0.1 M sucrose to induce plasmolysis. In plasmolyzed cells, the GFP signal was exclusively detected in the plasma membrane after transformation of the p35S:PtrMATE1-GFP construct (Fig. 5G, H, I), indicating that PtrMATE1 is localized to the plasma membrane, consistent with other citrate permeable plant MATE homologs.
Fig. 5.
Subcellular localization of PtrMATE1. GFP alone (A–C) or fusion protein PtrMATE1::GFP (D–F) were transiently expressed in onion epidermal cells, respectively. The images were acquired before (D–F) and after (G–I) plasmolysing the cells with 0.1 M sucrose. The overlay images of the brightfield and fluorescence images are shown (C, F, I). Scale bar, 100 μm (C, F) or 50 μm (I). NU, nucleus; PM, plasma membrane; CW, cell wall.
Expression patterns of PtrMATE1
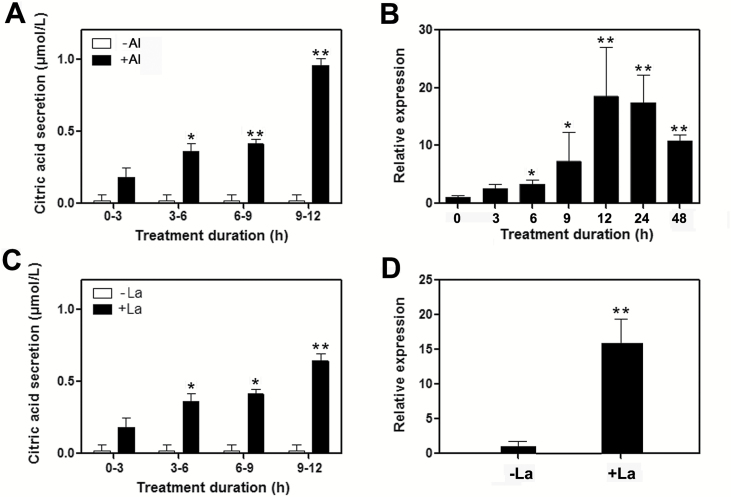
There are two different OA secretion patterns in response to Al treatment based on the timing of secretion in plants (Ma et al., 2001). To establish the OA release pattern of Populus and the connection between citrate and gene inducible secretion, we performed a time-course analysis of citrate secretion and PtrMATE1 expression in the root apex of poplar. The citrate secretion rates remained unchanged for the duration of the treatment without Al stress. However, with Al treatment, citrate secretion increased at 3 h, continued to increase, and at 9-12 h the secretion rate rapidly increased (Fig. 6A). In parallel, the transcript abundance of PtrMATE1 increased with Al treatment, reached a peak at 12 h and subsequently gradually decreased (Fig. 6B). These results indicated that Populus shows the typical pattern of OA release in response to Al exposure.
Fig. 6.
Citrate release and PtrMATE1 expression in poplar roots in response to Al and La treatments. (A) Induction of citrate secretion from the root apices of poplar in response to Al treatment. (B) Time course of PtrMATE1 expression in the roots of poplar in response to Al treatment. (C) Induction of citrate secretion from root apices of poplar in response to La treatment. (D) PtrMATE1 expression in the roots of poplar in response to 12 h La treatment. Excised root apices, 10 mm in length, were placed in the solution containing 0 μM or 500 μM Al or La, respectively. Data are presented as the means ± standard deviation (SD) of three independent experiments. Poplar ubiquitin expression was used as a control. The results are shown as the mean expression ± standard deviation (SD) of three independent experiments. Student’s t-test, *P<0.05, **P<0.01.
Several studies have shown that several lanthanides such as La3+, have ionic properties similar to Al3+ (Kataoka et al., 2002; Liu et al., 2013) but citrate secretion in rice bean specifically occurs in response to Al stress not La stress (Liu et al., 2013). To determine whether both citrate secretion and PtrMATE1 expression in Populus were specific to Al stress, we analyzed PtrMATE1 expression and citrate secretion under La treatment. The results showed that La enhanced the expression of PtrMATE1 in Populus (Fig. 6D) and that La-induced secretion of citrate from the roots was also increased (Fig. 6C), although the expression level and citrate secretion were relatively lower in La-stressed roots than that in Al-stressed roots. Interestingly, PtrMATE2 expression was not induced after 12 h of La treatment (see Fig. S3B at Dryad).
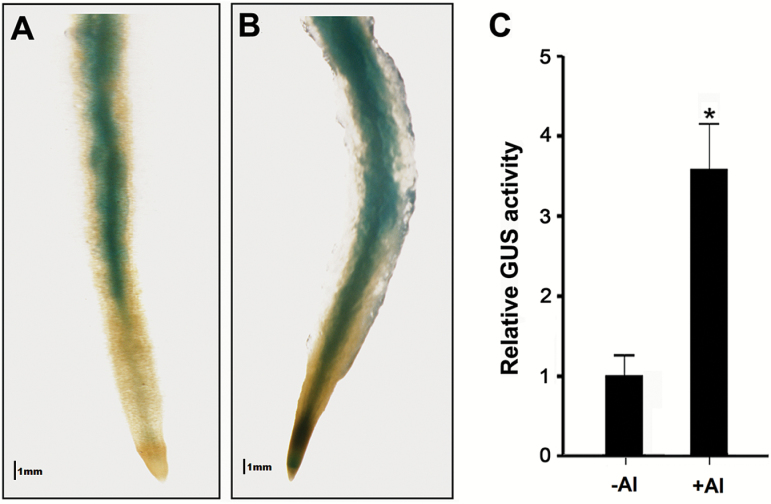
To further confirm the expression pattern of PtrMATE1, the 2 kb promoter fragment of PtrMATE1 was fused to a GUS reporter gene and transformed into wild-type poplar. GUS staining was observed in the central cylinder of mature roots but not detected in the root tips without Al (Fig. 7A). After exposure to Al, GUS activity was not only observed in the central cylinder but also extended to the entire root apex (Fig. 7B). GUS activity was quantitatively measured using a F-7000 fluorospectrophotometer (Hitachi, Japan) with fluorospectrophotometry (Jefferson et al., 1987). The results showed the GUS activity in the 2 cm root apices was increased by approximately 3.5 times under Al stress conditions (Fig. 7C).
Fig. 7.
GUS activity in roots of transgenic PtrMATE1p::GUS plants. The PtrMATE1 gene promoter-driven GUS expression vector was introduced into P. tomentosa Carr. Transgenic plant seedlings were treated with 500 μM Al3+ for 0 h (A) and 12 h (B), respectively. GUS staining was observed in the mature roots of transgenic poplar. Scale bars, 1 mm. (C) Quantitative GUS activity in the roots of transgenic plants. Student’s t-test, *P<0.05. Results are shown as mean expression ± standard deviation (SD) of three independent experiments.
Overexpression of PtrMATE1 in transgenic poplar confers citrate efflux and Al tolerance
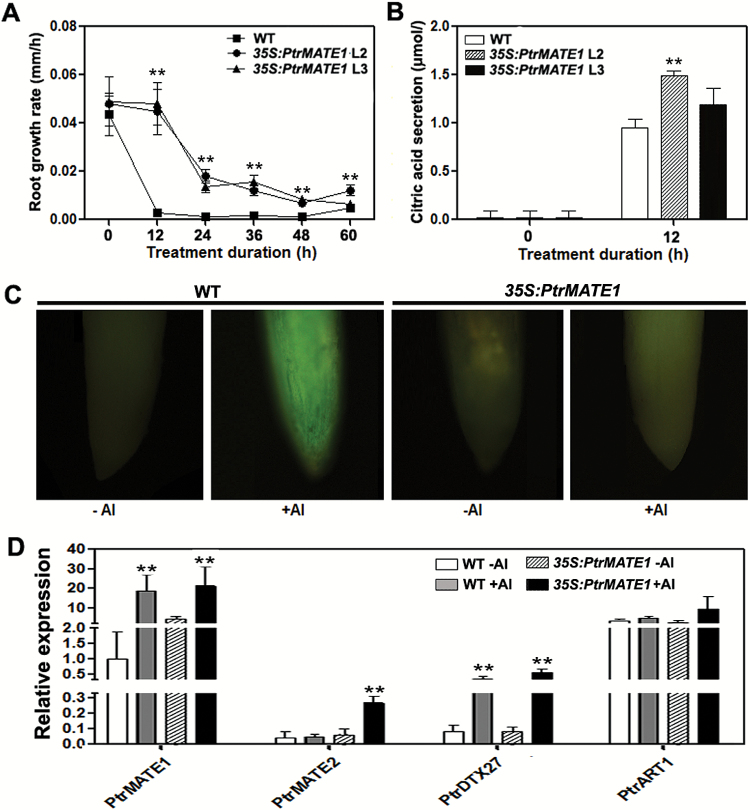
To further investigate the function of PtrMATE1 in mediating Al transport, transgenic poplar plants overexpressing PtrMATE1, named 35S::PrtMATE1, were generated and grown in the greenhouse. The transcript levels of PtrMATE1 were determined in these transgenic lines using qRT-PCR (see Fig. S4 at Dryad) and two independent lines, L2 and L3, with high PtrMATE1 expression were selected for further analysis. We first compared Al tolerance between wild-type and transgenic plants. A time-course experiment showed that, at 12 h after the exposure to Al, strong inhibition of root growth was observed in wild-type, with a reduction in growth rate of ≥90% (Fig. 8A). By contrast, the root growth rate of the transgenic lines, L2 and L3, was only reduced by 10% and 15% at the same time point, respectively. Up to 48 h under Al treatment, the root growth rates of the transgenic lines were significantly higher than that of the control (P<0.01). Furthermore, citrate secretion in transgenic poplar overexpressing PtrMATE1 was enhanced in the presence of Al compared with the wild-type control (Fig. 8B). This in vivo result consistently indicated that PrtMATE1 is a citrate transporter induced by Al stress in poplar. Additionally, the PtrMATE1 gene driven by the 35S promoter was also transformed into the wild-type and AtMATE knockout mutant, AtMATE-KO, Arabidopsis to generate the overexpression lines, named PtrMATE1-OX, and the functional complementation line, named PtrMATE1-R, respectively. Overexpression of PtrMATE1 in Arabidopsis strengthened the response to Al stress (Fig. 6S at Dryad). The results also indicated that PtrMATE1 complements the function of AtMATE in response to Al stress.
Fig. 8.
Overexpression of PtrMATE1 in transgenic poplar confers citrate efflux and Al tolerance. (A) Root growth of wild-type and transgenic plants overexpressing PtrMATE1 in nutrient solution containing 500 μM Al3+ for 60 h. All results represent the means ±standard deviation (SD) of three independent experiments. (B) Induction of citrate secretion from root apices of wild-type and transgenic poplar in response to 500 μM Al3+ treatment for 12 h. Data are represented as the means ± standard deviation (SD) of three independent experiments. (C) Callose accumulation in the root tips following Al treatment. Poplar seedlings were exposed to a nutrient solution containing 0 M (-Al3+) or 500 μM AlCl3 (+Al3+) for 12 h. Seedlings were subsequently fixed, stained with 0.1% aniline blue at pH 9.0, and observed using fluorescence microscopy. The fluorescence images indicate callose accumulation. WT-Al3+, wild-type plants in a solution without Al3+; WT+Al3+, wild-type plants in a solution containing 500 μM Al3+; 35S:PtrMATE1-Al3+, transgenic plants in a solution without Al3+; 35S:PtrMATE1+Al3+, transgenic plants in a solution containing 500 μM Al3+. (D) Relative expression levels of PtrMATE1, PtrMATE2, PtrDTX27, and PtrART1 in roots of wild-type and transgenic 35S:PtrMATE1 plants under 500 μM Al3+ treatment for 0 h and 12 h. Student’s t-test, **P<0.01.
To further confirm that PtrMATE1 correlated with Al tolerance in Populus, we detected the callose content in root apex stained with aniline blue, which is an indicator of Al toxicity and accumulates in plants upon Al exposure (Kochian et al., 2005). In the absence of Al, the root apex of both wild-type and transgenic poplar exhibited a weak fluorescence signal (Fig. 8C). Intense fluorescence, indicating callose accumulation, was observed in the root tips of wild-type plants under Al stress conditions. By contrast, even in the presence of Al, callose deposition in the root tips of the PtrMATE1 overexpression lines was slightly higher than in the untreated control. Additionally, with Al treatment, PtrMATE2 expression was also upregulated in the PtrMATE1 overexpression lines, but the transcript level of PtrART1 (Al resistance transcription factor 1, Yamaji et al., 2009) was not changed (Fig. 8D). This result indicated that Al toxicity was significantly ameliorated in the PtrMATE1 overexpression lines and further confirmed that PrtMATE1 is a citrate exporter in response to Al stress in poplar.
Functional coordination of PtrMATE1 and PrtMATE2 in response to long-term exposure to Al
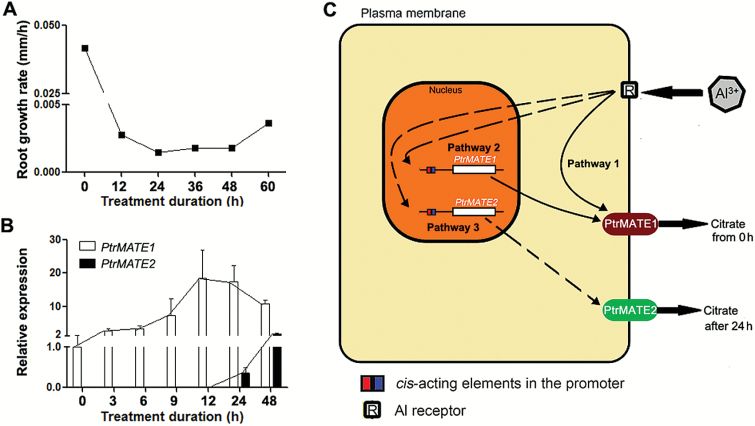
Phylogenetic analysis of MATE proteins showed that Arabidopsis AtMATE corresponds to PtrMATE1 and PtrMATE2 in Populus (Fig. 2). To determine the functional diversity of these duplicated pairs in response to Al stress, we analyzed the relationship between the root growth rate of Populus and expression levels of PtrMATE1 and PtrMATE2 under Al stress. The results showed that, although the transcript level of PrtMATE1 was decreased at 24 h after Al treatment (Fig. 9B), the root growth rate was stable between 24 h to 48 h during Al treatment (Fig. 9A). Time-course analysis of PtrMATE2 expression during the 48 h Al treatment showed that the transcript levels of PtrMATE2 increased 10 times at 24 h and 150 times at 48 h after exposure to Al, compared with the untreated control (Fig. 9B; Fig. S3A at Dryad). The different expression patterns of PtrMATE1 and PtrMATE2 suggested functional diversification and potential function redundancy in Al tolerance in poplar.
Fig. 9.
Hypothetical model for PtrMATE transporter-mediated citrate secretion in response to Al stress in roots of Populus. (A) Root growth of wild-type plants in nutrient solution containing 500 μM Al3+ for 60 h. (B) Expression levels of PtrMATE1 and PtrMATE2 in poplar roots during 48 h under Al treatment. Expression of the poplar ubiquitin (UBQ) gene was used as a control. All results are shown as the mean expression ± standard deviation (SD) of three independent experiments. (C) Hypothetical model for Al-induced citrate secretion from roots of Populus. There are at least three pathways involved in the process. Pathway 1: the receptor (R) on the plasma membrane binds Al3+ and activates PtrMATE1 to transport citrate immediately out of the cell through the plasma membrane. Pathway 2: PtrMATE1 expression is induced by Al3+ and subsequently PtrMATE1 transports citrate out of the cell. Pathway 3: PtrMATE2 expression is induced by Al3+ at 24 h after treatment and subsequently PtrMATE2 transports citrate in cooperation with PtrMATE1.
Discussion
Characterization of the Populus MATE gene family
In the present study, a total of 71 MATE genes, classified into three subfamilies, were identified in the P. trichocarpa genome (Fig. 2). Each subfamily contains at least one MATE motif (Fig. 3). The lengths of the Populus MATE proteins, comprising 120 to 608 amino acids, were significantly varied, whereas in Arabidopsis, the lengths of the MATE proteins ranged from 400 to 700 amino acids (Li et al., 2002). These PtrMATE genes displayed marked variation in gene structure and protein motifs, implying a high degree of complexity among the Populus MATE family. The integration and realignment of the gene fragments induced exon-intron increases or decreases (Xu et al., 2012). Gene structure variations therefore play a major role in the evolution of gene families (Xu et al., 2012). In addition, the number of exons in the PtrMATE genes divided into phylogenetic groups were nearly the same, but varied between different subfamilies. Obviously, the PtrMATE genes in subfamily III contained more exons than the other members (Fig. 3).
Previous studies have shown that the Populus genome undergoes at least three rounds of genome duplication, followed by multiple segmental duplications, tandem duplications, and transposition events (Tuskan et al., 2006; Hu et al., 2012). The segmental duplications associated with salicoid duplication events in particular contributed to the expansion of many multi-gene families (Barakat et al., 2009; Rogers et al., 2009; Hu et al., 2010). In the present study, almost 80% of the PtrMATE genes were located in duplicated regions and approximately half of the PtrMATE members were represented in distinct tandem duplicate gene clusters (Fig. 1). These results suggest that the expansion of the PtrMATE gene family in the Populus genome largely reflected duplication events and tandem repeats. We also observed that most of the Arabidopsis MATE genes have at least two pairs of orthologous genes in the Populus genome (Fig. 2). The loss of a few MATE orthologous genes in poplar might reflect dynamic rearrangements following segmental duplication (Li et al., 2015).
PtrMATE proteins respond to Al stress under acidic conditions
Approximately 50% of the potential arable soil worldwide is acidic soil, in which Al toxicity is one of the major factors limiting plant growth and crop yields (reviewed by Kochian et al., 2015). Al-induced citrate and malate exclusion are the two major Al resistance processes in both dicots and monocots, mediated by members of the MATE and ALMT families, respectively (Kochian et al., 2015). In recent decades, many Al-induced citrate transporter genes of the MATE family have been identified and characterized, including SbMATE from sorghum (Magalhaes et al., 2007), HvMATE from barley (Furukawa et al., 2007), AtMATE from Arabidopsis (Liu et al., 2009), VuMATE from rice bean (Yang et al., 2011) and ZmMATE from maize (Maron et al., 2010). Interestingly, many tree species grow normally in acidic soil and often exhibit natural tolerance to Al toxicity. Previous studies have shown that aspen (Populus tremula) tolerates high concentrations of Al by releasing citrate and oxalate from roots in response to Al stress (Grisel et al., 2010). In this biological process, the citrate efflux transporter gene MATE is an important component of the Al tolerance mechanism in aspen.
To elucidate the molecular mechanism of Al tolerance in poplar, in the present study, we isolated an Al-inducible MATE gene PtrMATE1 from P. trichocarpa. As shown in Fig. S2 (available at Dryad), PtrMATE1 with a conserved transmembrane domain structure shares 65% amino acid identity with AtMATE, a major contributor to Arabidopsis Al tolerance (Liu et al., 2009). PtrMATE1 expression was detected after exposure to Al stress, while citrate exudation from roots could also be detected as early as 3 h after Al exposure (Fig. 6). GUS staining showed that PtrMATE1 expression was specifically high in the central cylinder of mature roots of poplar and was significantly increased under Al treatment (Fig. 7). Overexpression of PtrMATE1 in Populus resulted in an associated increase in Al tolerance and root citrate exudation (Fig. 8). Notably, the timing of PtrMATE1 upregulation by Al is closely correlated with the onset of Al-induced citrate exudation from roots. This rapid regulation of PtrMATE1 expression and citrate exudation by Al is consistent with that in maize, but different from that in sorghum and rice bean (Magalhaes et al., 2007; Maron et al., 2010; Yang et al., 2011).
Although the functional and structural characteristics of PtrMATE1 are similar to those of reported citrate-permeable MATE proteins, the mechanism for Al resistance mediated by PtrMATE1 in poplar is different from other MATEs in herbaceous plants. First, in Arabidopsis and crop plants, Al concentrations as low as 50 µM are capable of inhibiting root growth (Kumari et al., 2008; Chandran et al., 2008), whereas poplar plants grow normally in acidic soil with high Al concentrations of 200 µM (Qin et al., 2007), implying that Populus has developed adaptive mechanisms to tolerate high Al stress. Most citrate-permeable MATE genes from other plant species are activated in response to a low Al concentration of 50 µM (Yokosho et al., 2009; 2011; Wang et al., 2015). However, expression of PtrMATE1 was induced by 500 µM and even 1000 µM Al treatment (Figs 4A and 7). These results confirmed that PtrMATE1-mediated citrate secretion is responsible for resistance to high Al concentration stress in poplar. Also transformed into Arabidopsis Atmate knockout mutants was 35S:PtrMATE1 and here we observed that PtrMATE1 strengthened the activity of Al-induced citrate exudation in transgenic Arabidopsis (see Fig. S6 at Dryad). Second, expression of PrtMATE1 remained relatively constant, peaking at 12–24 h after exposure to Al (Fig. 6), a period almost two times longer than that of other MATE homologs in rice bean and maize (Maron et al., 2010; Liu et al., 2013). Third, PtrMATE1 expression is not only upregulated by Al stress but also highly induced by La3+ treatment (Fig. 6D). Unlike in rice bean, citrate secretion specifically occurs in response to Al stress but not La stress (Liu et al., 2013). These differences suggest that the citrate exudation response in Populus and herbaceous species may be regulated by different mechanisms. Additionally, ART1, a C2H2-type zinc finger transcription factor in rice, regulates expression of the MATE gene OsFRDL2 in Al tolerance (Yamaji et al., 2009; Yokosho et al., 2011). Thus, high Al tolerance is achieved through an ART1-regulated pathway in rice. However, PtrART1 expression was not regulated by Al stress in wild-type or transgenic 35S:PtrMATE1 poplar plants (Fig. 8D), suggesting that other unknown components, such as transcription factors, might be involved in the response to Al in Populus.
Coordinated roles of PtrMATE1 and PtrMATE2 enhanced resistance to long-term Al stress
Previous studies have shown that the Populus genome has undergone genome-wide duplications, followed by multiple segmental and tandem duplications (Tuskan et al., 2006). Phylogenetic analysis showed that the Arabidopsis AtMATE gene corresponds to PtrMATE1 and PtrMATE2 in Populus (Fig. 2). Interestingly, PtrMATE1 expression was induced by Al, peaking at 12 h after Al treatment (Fig. 6B), whereas the expression level of PtrMATE2 was low before 24 h of Al exposure, but rapidly upregulated after 24–48 h (Fig. S3 at Dryad). The root growth rate of Populus was rapidly reduced at 12 h after Al exposure, but remained constant between 12–48 h. We therefore speculated that PtrMATE1 might play a major role in Al-induced citrate exudation from poplar roots at the early stage and that PtrMATE1 and PtrMATE2 coordinately mediate citrate release after 24 h of Al exposure. Future studies should include a more detailed analysis of PtrMATE2 function to clarify the mechanism underlying PtrMATE-mediated exudation of citrate in poplar roots in response to Al stress.
Finally, we propose a hypothetical model for Al-induced citrate secretion from roots of Populus. As illustrated in Fig. 9C, there should be at least three different pathways mediating citrate efflux in response to Al stress in Populus. In pathway 1, the Al-stress signal receptor (R) directly activates PtrMATE1 to export citrate out of the roots without post-transcriptional or translational level regulation. As shown in Fig. 6A, CA was detected in the root in response to treatment with Al. In pathway 2, transcriptional factor(s) or upstream control elements are synthesized de novo after sensing the receptor and further regulate the expression and translation of PtrMATE1. Subsequently PtrMATE1 continues to transport citrate out of the cell. In Fig. 6A, after Al treatment, expression of PtrMATE1 and CA secretion both generally increased with time. In pathway 3, PtrMATE2 expression was induced after 24 h of Al treatment; PtrMATE2 may transport citrate in cooperation with PtrMATE1 (Fig. 9C). As described above, the expression level of PrtMATE2 was significantly upregulated after Al treatment for 24 h, while PtrMATE1 expression smoothly decreased (Figs 9A, B and S3A). Taken together, these results not only reveal the functional diversity of PtrMATE transporters in Populus against Al stress but also provide insights into Al tolerance mechanisms in Populus grown in acidic soil. The present study will be helpful to enhance the current understanding of the roles of MATE genes in Populus and also provides an important resource for the generation of tree or crop varieties more suitable for growth on acidic soils.
Data deposition
The following data are available at Dryad Data Repository: http://dx.doi.org/10.5061/dryad.vb047
Sequences of all MATE proteins from Populus, Arabidopsis, rice, and 13 plant species.
Supplementary Data
Supplementary data are available at JXB online.
Fig. S1. Expression of 10 PtrMATE genes in poplar shoots and roots under 500 μM Al3+ for 12 h.
Fig. S2. Multiple sequence alignment of PtrMATE1, PtrMATE2 and AtMATE.
Fig. S3. Time course of PtrMATE2 expression in the roots of Populus in response to Al or La treatment.
Fig. S4. qRT-PCR analysis of transgenic poplar plants.
Fig. S5. PtrMATE1 expression in the roots of poplar under Al treatments with different concentration.
Fig. S6. Overexpression of PtrMATE1 in transgenic AtMATE knockout mutant (ATMATE-KO) and wild-type Arabidopsis confers Al tolerance.
Table S1. Details of MATEs from different species.
Table S2. Primers used for qRT-PCR and gene cloning in the present study.
Table S3. Motifs of PtrMATE proteins.
Supplementary Material
Acknowledgements
The authors thank Prof. Guoliang Wang (Hunan Agricultural University, Changsha 410128, China) for providing the plant binary vector pCXSN. This work was supported by National Key Research and Development Program (2016YFD0600105), the National Natural Science Foundation of China (31370672, 31500216, 31400063, 31500544 and 31370317) and Fundamental Research Funds for the Central Universities (XDJK2016B032; XDJK2017B030).
Glossary
Abbreviations:
- Al
aluminium
- CA
citric acid
- MATE
multidrug and toxic compound extrusion
- OAs
organic acids
- PtrDTX1-71
Populus detoxification 1-71
- WPM
woody plant medium.
References
- Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings. International Conference on Intelligent Systems for Molecular Biology 2, 28–36. [PubMed] [Google Scholar]
- Barakat A, Bagniewska-Zadworna A, Choi A, Plakkat U, DiLorreto DS, Yellanki P, Carlson JE. 2009. The cinnamyl alcohol dehydrogenase gene family in Populus: phylogeny, organization, and expression. BMC Plant Biology 9, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran D, Sharopova N, VandenBosch KA, Garvin DF, Samac DA. 2008. Physiological and molecular characterization of aluminum resistance in Medicago truncatula. BMC Plant Biology 8, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlier JB, Polese C, Nouet C, Carnol M, Bosman B, Krämer U, Motte P, Hanikenne M. 2015. Zinc triggers a complex transcriptional and post-transcriptional regulation of the metal homeostasis gene FRD3 in Arabidopsis relatives. Journal of Experimental Botany 66, 3865–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cunningham SD, Ow DW. 1996. Promises and Prospects of Phytoremediation. Plant Physiology 110, 715–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Craig S, Beaton CD, Bennet RJ, Jagadish VC, Randall PJ. 1993. Aluminum tolerance in wheat (Triticum aestivum L.) (I. Uptake and distribution of aluminum in root apices). Plant Physiology 103, 685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Gruber BD, Ryan PR. 2007. The roles of organic anion permeases in aluminium resistance and mineral nutrition. FEBS Letters 581, 2255–2262. [DOI] [PubMed] [Google Scholar]
- Delhaize E, Ryan PR. 1995. Aluminum toxicity and tolerance in plants. Plant Physiology 107, 315–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener AC, Gaxiola RA, Fink GR. 2001. Arabidopsis ALF5, a multidrug efflux transporter gene family member, confers resistance to toxins. The Plant Cell 13, 1625–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Ma J, Hiradate S, Matsumoto H. 1998. High aluminum resistance in buckwheat. Ii. Oxalic acid detoxifies aluminum internally. Plant Physiology 117, 753–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi VR, Nakata PA. 2005. Calcium oxalate in plants: formation and function. Annual Review of Plant Biology 56, 41–71. [DOI] [PubMed] [Google Scholar]
- Fujii M, Yokosho K, Yamaji N, Saisho D, Yamane M, Takahashi H, Sato K, Nakazono M, Ma JF. 2012. Acquisition of aluminium tolerance by modification of a single gene in barley. Nature Communications 3, 713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa J, Yamaji N, Wang H, Mitani N, Murata Y, Sato K, Katsuhara M, Takeda K, Ma JF. 2007. An aluminum-activated citrate transporter in barley. Plant & Cell Physiology 48, 1081–1091. [DOI] [PubMed] [Google Scholar]
- Göransson A, Eldhuset TD. 1987. Effects of aluminium on growth and nutrient uptake of Betula pendula seedlings. Physiologia Plantarum 69, 193–199. [Google Scholar]
- Göransson A, Eldhuset TD. 1991. Effects of aluminium on growth and nutrient uptake of small Picea abies and Pinus sylvestris plants. Trees 5, 136–142. [Google Scholar]
- Gomez C, Conejero G, Torregrosa L, Cheynier V, Terrier N, Ageorges A. 2011. In vivo grapevine anthocyanin transport involves vesicle-mediated trafficking and the contribution of anthoMATE transporters and GST. The Plant Journal 67, 960–970. [DOI] [PubMed] [Google Scholar]
- Gomez C, Terrier N, Torregrosa L et al. . 2009. Grapevine MATE-type proteins act as vacuolar H+-dependent acylated anthocyanin transporters. Plant Physiology 150, 402–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R et al. . 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Research 40, D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisel N, Zoller S, Künzli-Gontarczyk M, Lampart T, Münsterkötter M, Brunner I, Bovet L, Métraux JP, Sperisen C. 2010. Transcriptome responses to aluminum stress in roots of aspen (Populus tremula). BMC Plant Biology 10, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekenga OA, Maron LG, Piñeros MA et al. . 2006. AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proceedings of the National Academy of Sciences, USA 103, 9738–9743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. 2015. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Chi X, Chai G, Kong Y, He G, Wang X, Shi D, Zhang D, Zhou G. 2012. Genome-wide identification, evolutionary expansion, and expression profile of homeodomain-leucine zipper gene family in poplar (Populus trichocarpa). PLOS One 7, e31149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Qi G, Kong Y, Kong D, Gao Q, Zhou G. 2010. Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biology 10, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara T, Sekine KT, Hase S et al. . 2008. Overexpression of the Arabidopsis thaliana EDS5 gene enhances resistance to viruses. Plant Biology 10, 451–461. [DOI] [PubMed] [Google Scholar]
- Jefferson RA. 1987. Assaying chimeric genes in plants: The GUS gene fusion system. Plant Molecular Biology Reporter 5, 387–405. [Google Scholar]
- Jia Z, Sun Y, Yuan L, Tian Q, Luo K. 2010. The chitinase gene (Bbchit1) from Beauveria bassiana enhances resistance to Cytospora chrysosperma in Populus tomentosa Carr. Biotechnology Letters 32, 1325–1332. [DOI] [PubMed] [Google Scholar]
- Kataoka T, Stekelenburg A, Nakanishi TM, Delhaize E, Ryan PR. 2002. Several lanthanides activate malate efflux from roots of aluminium-tolerant wheat. Plant Cell & Environment 25, 453–460. [Google Scholar]
- Kinraide TB, Parker DR, Zobel RW. 2005. Organic acid secretion as a mechanism of aluminium resistance: a model incorporating the root cortex, epidermis, and the external unstirred layer. Journal of Experimental Botany 56, 1853–1865. [DOI] [PubMed] [Google Scholar]
- Kochian LV Piñeros MA, Liu J, Magalhaes JV. 2015. Plant adaptation to acid soils: the molecular basis for crop aluminum resistance. Annual Review of Plant Biology 66, 571–598. [DOI] [PubMed] [Google Scholar]
- Kochian LV, Hoekenga OA, Piñeros MA. 2004. How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annual Review of Plant Biology 55, 459–493. [DOI] [PubMed] [Google Scholar]
- Kochian LV, Piñeros MA, Hoekenga OA. 2005. The physiology, genetics and molecular biology of plant aluminum resistance and toxicity. Plant and Soil 274, 175–195. [Google Scholar]
- Kumari M, Taylor GJ, Deyholos MK. 2008. Transcriptomic responses to aluminum stress in roots of Arabidopsis thaliana. Molecular Genetics and Genomics 279, 339–357. [DOI] [PubMed] [Google Scholar]
- Kuroda T, Tsuchiya T. 2009. Multidrug efflux transporters in the MATE family. Biochimica Et Biophysica Acta 1794, 763–768. [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. 2015. SMART: recent updates, new developments and status in 2015. Nucleic Acids Research 43, D257–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Xu X, Hu X et al. . 2015. Genome-wide analysis and heavy metal-induced expression profiling of the HMA gene family in Populus trichocarpa. Frontiers in Plant Science 6, 1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, He Z, Pandey GK, Tsuchiya T, Luan S. 2002. Functional cloning and characterization of a plant efflux carrier for multidrug and heavy metal detoxification. The Journal of Biological Chemistry 277, 5360–5368. [DOI] [PubMed] [Google Scholar]
- Li N, Meng H, Xing H, Liang L, Zhao X, Luo K. 2017. Data from: Genome-wide analysis of MATE transporters and molecular characterization of aluminum resistance in Populus trichocarpa. Dryad Digital Repository. http://dx.doi.org/10.5061/dryad.vb047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligaba A, Katsuhara M, Ryan PR, Shibasaka M, Matsumoto H. 2006. The BnALMT1 and BnALMT2 genes from rape encode aluminum-activated malate transporters that enhance the aluminum resistance of plant cells. Plant Physiology 142, 1294–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Li Y, Wang W, Gai J, Li Y. 2016. Genome-wide analysis of MATE transporters and expression patterns of a subgroup of MATE genes in response to aluminum toxicity in soybean. BMC Genomics 17, 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Magalhaes JV, Shaff J, Kochian LV. 2009. Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. The Plant Journal 57, 389–399. [DOI] [PubMed] [Google Scholar]
- Liu MY, Chen WW, Xu JM, Fan W, Yang JL, Zheng SJ. 2013. The role of VuMATE1 expression in aluminium-inducible citrate secretion in rice bean (Vigna umbellata) roots. Journal of Experimental Botany 64, 1795–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JF, Ryan PR, Delhaize E. 2001. Aluminium tolerance in plants and the complexing role of organic acids. Trends in Plant Science 6, 273–278. [DOI] [PubMed] [Google Scholar]
- Magalhaes JV, Liu J, Guimarães CT et al. . 2007. A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nature Genetics 39, 1156–1161. [DOI] [PubMed] [Google Scholar]
- Maron LG, Piñeros MA, Guimarães CT, Magalhaes JV, Pleiman JK, Mao C, Shaff J, Belicuas SN, Kochian LV. 2010. Two functionally distinct members of the MATE (multi-drug and toxic compound extrusion) family of transporters potentially underlie two major aluminum tolerance QTLs in maize. The Plant Journal 61, 728–740. [DOI] [PubMed] [Google Scholar]
- Mccown BH, Lloyd G. 1981. Woody Plant Medium (Wpm) - a mineral nutrient formulation for microculture of woody plant-species. Hortscience 16, 453–453. [Google Scholar]
- Migeon A, Blaudez D, Wilkins O, Montanini B, Campbell MM, Richaud P, Thomine S, Chalot M. 2010. Genome-wide analysis of plant metal transporters, with an emphasis on poplar. Cellular and Molecular Life Sciences 67, 3763–3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Shitan N, Sawada K et al. . 2009. Vacuolar transport of nicotine is mediated by a multidrug and toxic compound extrusion (MATE) transporter in Nicotiana tabacum. Proceedings of the National Academy of Sciences, USA 106, 2447–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Heck S, Parinthawong N, Métraux JP. 2002. EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. The Plant cell 14, 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omote H, Hiasa M, Matsumoto T, Otsuka M, Moriyama Y. 2006. The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends in Pharmacological Sciences 27, 587–593. [DOI] [PubMed] [Google Scholar]
- Ryan PR, Raman H, Gupta S, Horst WJ, Delhaize E. 2009. A second mechanism for aluminum resistance in wheat relies on the constitutive efflux of citrate from roots. Plant Physiology 149, 340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Díaz R, Ryngajllo M, Pérez-Díaz J, Peña-Cortés H, Casaretto JA, González-Villanueva E, Ruiz-Lara S. 2014. VvMATE1 and VvMATE2 encode putative proanthocyanidin transporters expressed during berry development in Vitis vinifera L. Plant Cell Reports 33, 1147–1159. [DOI] [PubMed] [Google Scholar]
- Rogers EE, Wu X, Stacey G, Nguyen HT. 2009. Two MATE proteins play a role in iron efficiency in soybean. Journal of Plant Physiology 166, 1453–1459. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Yamamoto Y, Ezaki B, Katsuhara M, Ahn SJ, Ryan PR, Delhaize E, Matsumoto H. 2004. A wheat gene encoding an aluminum-activated malate transporter. The Plant Journal 37, 645–653. [DOI] [PubMed] [Google Scholar]
- Schaedle M, Thornton FC, Raynal DJ, Tepper HB. 1989. Response of tree seedlings to aluminum. Tree Physiology 5, 337–356. [DOI] [PubMed] [Google Scholar]
- Seo PJ, Park J, Park MJ, Kim YS, Kim SG, Jung JH, Park CM. 2012. A Golgi-localized MATE transporter mediates iron homoeostasis under osmotic stress in Arabidopsis. The Biochemical Journal 442, 551–561. [DOI] [PubMed] [Google Scholar]
- Shoji T, Inai K, Yazaki Y et al. . 2009. Multidrug and toxic compound extrusion-type transporters implicated in vacuolar sequestration of nicotine in tobacco roots. Plant Physiology 149, 708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Gilroy EM, Chini A et al. . 2011. ADS1 encodes a MATE-transporter that negatively regulates plant disease resistance. New Phytologist 192, 471–482. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson EP, Wilkins C, Demidchik V, Davies JM, Glover BJ. 2010. An Arabidopsis flavonoid transporter is required for anther dehiscence and pollen development. Journal of Experimental Botany 61, 439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari M, Sharma D, Singh M, Tripathi RD, Trivedi PK. 2014. Expression of OsMATE1 and OsMATE2 alters development, stress responses and pathogen susceptibility in Arabidopsis. Scientific Reports 4, 3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S et al. . 2006. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604. [DOI] [PubMed] [Google Scholar]
- Wang R, Liu X, Liang S, Ge Q, Li Y, Shao J, Qi Y, An L, Yu F. 2015. A subgroup of MATE transporter genes regulates hypocotyl cell elongation in Arabidopsis. Journal of Experimental Botany 66, 6327–6343. [DOI] [PubMed] [Google Scholar]
- Warty VS, Busch RP, Virji MA. 1984. A kit for citrate in foodstuffs adapted for assay of serum and urine. Clinical Chemistry 30, 1231–1233. [PubMed] [Google Scholar]
- Xu G, Guo C, Shan H, Kong H. 2012. Divergence of duplicate genes in exon-intron structure. Proceedings of the National Academy of Sciences, USA 109, 1187–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu HW, Ji XM, He ZH, Shi WP, Zhu GH, Niu JK, Li BS, Peng XX. 2006. Oxalate accumulation and regulation is independent of glycolate oxidase in rice leaves. Journal of Experimental Botany 57, 1899–1908. [DOI] [PubMed] [Google Scholar]
- Yang XY, Yang JL, Zhou Y, Piñeros MA, Kochian LV, Li GX, Zheng SJ. 2011. A de novo synthesis citrate transporter, Vigna umbellata multidrug and toxic compound extrusion, implicates in Al-activated citrate efflux in rice bean (Vigna umbellata) root apex. Plant, Cell & Environment 34, 2138–2148. [DOI] [PubMed] [Google Scholar]
- Yokosho K, Yamaji N, Ueno D, Mitani N, Ma JF. 2009. OsFRDL1 is a citrate transporter required for efficient translocation of iron in rice. Plant Physiology 149, 297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokosho K, Yamaji N, Ma JF. 2011. An Al-inducible MATE gene is involved in external detoxification of Al in rice. The Plant Journal 68, 1061–1069. [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhu H, Pan Y, Yu Y, Luan S, Li L. 2014. A DTX/MATE-type transporter facilitates abscisic acid efflux and modulates ABA sensitivity and drought tolerance in Arabidopsis. Molecular Plant 7, 1522. [DOI] [PubMed] [Google Scholar]
- Zhao J, Dixon RA. 2009. MATE transporters facilitate vacuolar uptake of epicatechin 3’-O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and Arabidopsis. The Plant Cell 21, 2323–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Delhaize E, Zhou M, Ryan PR. 2013. The barley MATE gene, HvAACT1, increases citrate efflux and Al(3+) tolerance when expressed in wheat and barley. Annals of Botany 112, 603–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.