GmPHR25, which is up-regulated by phosphate starvation, is a vital regulator in the phosphorus signaling network, and controls phosphate homeostasis in soybean.
Keywords: Expression pattern, PHR, Pi homeostasis, Pi starvation, soybean
Abstract
As an essential nutrient element, phosphorus (P) plays an important role in plant growth and development. Low P availability is a limiting factor for crop production, especially for legume crops (e.g. soybean), which require additional P to sustain nitrogen fixation through symbiotic associations with rhizobia. Although PHOSPHATE STARVATION RESPONSE 1 (PHR1) or PHR1-like is considered as a central regulator of phosphate (Pi) homeostasis in several plant species, it remains undefined in soybean. In this study, 35 GmPHR members were cloned from the soybean genome and expression patterns in soybean were assayed under nitrogen (N) and P deficiency conditions. GmPHR25, which is up-regulated in response to Pi starvation, was then overexpressed in soybean hairy roots in vitro and in vivo to investigate its functions. The results showed that overexpressing GmPHR25 increased Pi concentration in transgenic soybean hairy roots under normal conditions, accompanied with a significant decrease in hairy root growth. Furthermore, transcripts of 11 out of 14 high-affinity Pi transporter (GmPT) members as well as five other Pi starvation-responsive genes were significantly increased in soybean hairy roots with GmPHR25 overexpression. Taken together, this study suggests that GmPHR25 is a vital regulator in the P signaling network, and controls Pi homeostasis in soybean.
Introduction
Phosphorus (P) is an essential macronutrient in plants that is not only a major constituent in plant cells, but is also involved in metabolic processes (Raghothama, 1999; Richardson, 2009). Furthermore, meeting crop P needs with supplemental additions is problematic due to the fact that applied phosphate (Pi) fertilizers are easily fixed by soil particles into unavailable forms (e.g. aluminum, iron, and calcium phosphates), which results in low soil Pi availability (Beardsley, 2011; Veneklaas et al., 2012). Equally concerning is the prediction that the rock phosphate sources used in fertilizers will be largely depleted within a number of decades (Vance et al., 2003; Cordell et al., 2009). Therefore, development of ‘smart’ crop cultivars with superior P-use efficiency and optimization of field P management are imperative for the future of sustainable agriculture (Shen et al., 2011; Tian et al., 2012; Veneklaas et al., 2012; Wu et al., 2013).
Plants have evolved a range of morphological, physiological, and molecular strategies in adaptation to P deficiency, including changes of root morphology and architecture, increased exudation of organic acids and purple acid phosphatases, and formation of symbiotic interactions with arbuscular mycorrhizal (AM) fungi (Veneklaas et al., 2012). Many of these adaptive strategies enhance soil P mobility or plant acquisition of this limiting resource, and thus increase P efficiency. In recent years, identification of the multiple genes and proteins that regulate the relevant adaptive processes has significantly contributed to an emerging picture of the complex signaling network involved in plant responses to P deficiency.
Among the genes and proteins identified, several are considered as vital regulators, including a plant small ubiquitin-like modifier E3 ligase (SIZ1), PHR1, microRNA399, and proteins containing the SYG1/PHO81/XPR1 (SPX) domain (Chiou and Lin, 2011; Veneklaas et al., 2012; Wu et al., 2013; Liang et al., 2014). As a MYB-CC type transcription factor, PHR1 and its homologs appear to play central roles in P signaling networks (Chiou and Lin, 2011; Liang et al., 2013; Sun et al., 2016). Phosphate Starvation Response 1 (CrPSR1) is an ancestral MYB-CC type transcription factor that may be critical for the acclimation of the unicellular green alga Chlamydomonas reinhardtii to P deficiency (Wykoff et al., 1999). In plants, the identification and functional analysis of the CrPSR1 homolog AtPHR1 was a milestone accomplishment along the path to elucidating the P signaling network in Arabidopsis (Arabidopsis thaliana) (Rubio et al., 2001). Over time, phr1 mutations have been associated with decreases in anthocyanin accumulation and Pi concentration, as well as lower root-to-shoot ratios (Rubio et al., 2001; Bustos et al., 2010). Furthermore, transcription of multiple Pi starvation-responsive genes is impaired in phr1 mutants (Rubio et al., 2001; Bustos et al., 2010). At the opposite extreme, overexpression of AtPHR1 in Arabidopsis leads to significant increases in Pi concentration, accompanied by increased transcription of Pi starvation-responsive genes, such as miR399, RNase 1, and PHOSPHATE TRANSPORTER 1–7 (Nilsson et al., 2007). One of these Pi starvation-responsive genes, miR399, is suggested to repress expression levels of a ubiquitin-conjugating E2, PHO2, which regulates the abundance of PHOSPHATE TRANSPORTER (PHT), and thereby modulates Pi acquisition and accumulation (Fujii et al., 2005). Thus, it is well known that PHR1, miR399, PHO2, and PHT form a branch of the P signaling network controlling Pi homeostasis in plants (Bari et al., 2006; Chiou and Lin, 2011; Liang et al., 2014).
Recently, PHR1 homologs have also been documented to play important roles in regulating Pi homeostasis, including PHL1 and PHL2 in Arabidopsis, OsPHR1, OsPHR2, OsPHR3, and OsPHR4 in rice (Oryza sativa), BnPHR1 in rape (Brassica napus), TaPHR1 in wheat (Triticum aestivum), ZmPHR1 in maize (Zea mays), and PvPHR1 in bean (Phaseolus vulgaris) (Valdés-lópez et al., 2008; Zhou et al., 2008; Bustos et al., 2010; Ren et al., 2012; Wang et al., 2013a, 2013b; Guo et al., 2015; Sun et al., 2015; Ruan et al., 2017). Furthermore, binding of AtPHR1 and OsPHR2 to the P1BS site (PHR1-binding sequence: GNATATNC) is inhibited by Pi-dependent interactions with AtSPX1 and AtSPX2 in Arabidopsis and with OsSPX1, OsSPX2, and OsSPX4 in rice, which suggests the presence of another layer of complexity in the P signaling network in plants (Lv et al., 2014; Puga et al., 2014; Wang et al., 2014; Zhou et al., 2015).
Although much of the complex P signaling network has been elucidated in model plants (e.g. rice and Arabidopsis), genome-wide analysis of PHR members responsive to Pi starvation and their functions in controlling Pi homeostasis remain scarce and fragmentary in other plants, notably in legume crops. Soybean (Glycine max) is an important oil-bearing legume with high nutritional value (Herridge et al., 2008). It has been demonstrated that soybean exhibits multiple adaptive strategies to P deficiency, including formation of a shallower root system, increases of organic exudation and acid phosphatase (APase) activity, and alterations in symbiotic associations with AM fungi and rhizobia (Tian et al., 2003; Zhao et al., 2004; Liao et al., 2006; Liu et al., 2008; Qin et al., 2011; Liang et al., 2013).
With the availability of soybean genome sequences, expression analysis for soybean responses to Pi starvation has been conducted for several gene families, including expansin (EXPB), purple acid phosphatase (PAP), phosphate transporter (PT), and SPX (Wu et al., 2011; Li et al., 2012; Qin et al., 2012a; Fan et al., 2013; Li et al., 2014; Yao et al., 2014). Furthermore, functional analysis of several Pi starvation-responsive genes has led to elucidation of the molecular mechanisms underlying soybean adaptations to P deficiency. For example, a β-expansin gene, GmEXPB2, is highly induced in soybean roots by P deficiency, and overexpressing GmEXPB2 in Arabidopsis leads to enhanced root growth and Pi uptake (Guo et al., 2011). In addition, GmPT5, a high-affinity phosphate transporter, is mainly expressed in nodules and plays an important role in maintenance of Pi homeostasis in soybean nodules (Qin et al., 2012b). Furthermore, GmSPX3 has recently been suggested as a critical regulator in the P signaling network because it regulates transcription of a group of Pi starvation-responsive genes in soybean, including GmEXPB2 and GmPT5 (Yao et al., 2014).
Despite this progress in elucidating P signaling networks in soybean, the role of GmPHR members in these networks remains unclear. In the present study, genome-wide analysis of 35 GmPHR members was conducted. Beyond identification and phylogenetic analysis, expression patterns of GmPHR members were examined in different soybean tissues in response to P deficiency. Furthermore, functional analysis of a GmPHR member up-regulated by Pi starvation, GmPHR25, suggests that it is a key regulator in the P signaling network controlling Pi homeostasis in soybean.
Materials and methods
Identification of the GmPHR family in soybean
BLAST searches were performed, firstly using the AtPHR1 (AT4G28610) sequence as a query sequence. Then, using all identified GmPHR sequences as query sequences at the phytozome website (http://www.phytozome.net), a total of 35 GmPHR members were identified in the soybean genome that harbor two conserved domains (i.e. MYB and Coiled-Coil) and exhibit more than 24% similarity with AtPHR1. The members of the GmPHR family were named GmPHR1 to GmPHR35 based on their positions on the chromosomes. General information for each GmPHR member (e.g. numbers of exons and introns, length of open reading frame) was extracted from the same website. Protein molecular weights were predicted using the ExPASy web server (http://www.expasy.org/). Phylogenetic tree analysis of the PHR proteins was conducted using a ClustalX multiple-sequence alignment and the neighbor-joining method with 1000 bootstrap replicates in MEGA 5.05, as described previously (Tamura et al., 2007).
Plant growth conditions
The soybean genotype YC03-3 was used in these experiments. For expression analysis of GmPHR members in various soybean tissues, from 7 d after seed germination seedlings were grown in a full-strength nutrient solution containing 1500 μM KNO3, 1200 μM Ca(NO3)2, 400 μM NH4NO3, 500 μM KH2PO4, 500 μM MgSO4, 25 μM MgCl2, 300 μM K2SO4, 300 μM (NH4)2SO4, 1.5 μM MnSO4, 1.5 μM ZnSO4, 0.5 μM CuSO4, 0.16 μM (NH4)6Mo7O24, 2.5 μM NaB4O7, and 40 μM Fe-EDTA. On day 25 of growth in the nutrient solution, entirely expanded young leaves, roots, and flowers were harvested separately. Pods of 1 cm length and immature seeds were harvested separately on days 30 and 40 after transferring seedlings into the nutrient solution. All samples were stored at −80 °C prior to RNA extraction.
For the nutrient deficiency experiment, 7 d after seed germination, soybean seedlings were grown for 14 d in complete nutrient solution as a control, or in nutrient solution without nitrogen (N) or phosphorus (P). Then, entirely expanded young leaves and roots were harvested separately for further analysis, as described previously (Yao et al., 2014). Briefly, in the N deficiency (–N) solution, K2SO4 and CaCl2 were used to replace the KNO3 and Ca(NO3)2, respectively. In the P deficiency (–P) solution, K2SO4 replaced KH2PO4. For the rhizobium inoculation experiment, 7 d after seed germination, soybean seedlings were inoculated with rhizobium, Bradyrhizobium sp. BXYD3, for 1 h, and then transferred into low-nitrogen (100 μM total N) nutrient solution containing 5 μM or 500 μM KH2PO4, as described previously (Yao et al., 2014). Nodules were harvested at 30 d after inoculation for further analysis. All experiments included four biological replicates.
RNA extraction and quantitative real-time PCR analysis
Total RNA was extracted from various soybean tissues using TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions. Subsequently, RNA samples were treated with RNase-free DNase I (TaKaRa, Japan) to remove genomic DNA. The first cDNA strand was synthesized using MMLV-reverse transcriptase (Promega, USA) according to the given protocol. Quantitative real-time PCR (qPCR) was performed and analysed using SYBR Green PCR master mix (Promega, USA) and a Rotor-Gene 3000 system (Corbett Research, Australia). Expression levels of the soybean housekeeping gene, EF1-α (Glyma.17G186600) or ACTIN (Glyma.18G290800) were used as an endogenous control to normalize the samples, as described previously (Li et al., 2015; Liu et al., 2016). The specific primer sequences used in the study are listed in Supplementary Data Table S2 at JXB online.
Subcellular localization of GmPHR25
To determine the subcellular localization of GmPHR25, the coding region of GmPHR25 was amplified with specific primers (Supplementary Table S3), and cloned into the pMDC43 vector and the pBEGFP vector for fusion with green fluorescent protein (GFP) at its N- and C-terminus, respectively, according to the manufacturer’s instructions (Invitrogen, USA). The plasma membrane marker AtPIP2A-mCherry was used for co-localization analysis. Each of 35S:GFP, 35S:GFP-GmPHR25, 35S:GmPHR25-GFP, and 35S:AtPIP2A-mCherry fusion vectors was separately introduced into Agrobacterium tumefaciens strain GV3101, and then transformed into tobacco (Nicotiana benthamiana) leaves for transient expression, as described previously (Liu et al., 2012). After 3 d, transformed tobacco leaf epidermal cells were imaged on a Zeiss LSM7 DUO confocal microscope (Zeiss, Germany). Fluorescence of GFP and mCherry was stimulated at 488 and 543 nm, respectively.
In vitro overexpression of GmPHR25 in soybean hairy roots
The coding region of GmPHR25 was amplified with specific primers (Supplementary Table S3), and the PCR product was ligated into the pYLRNAi vector after digestion by Sac I and Pst I. GmPHR25-OE or empty vector constructs were separately transformed into Agrobacterium rhizogenes strain K599, which was further used to infect soybean cotyledons to obtain transgenic hairy roots in vitro, as described previously (Guo et al., 2011). Transgenic hairy roots were grown on Murashige and Skoog (MS) medium supplied with carbenicillin, and were then confirmed by PCR and qPCR analysis. Two independent GmPHR25 overexpression lines and the control line were selected, and established for further experiments. About 0.2 g (fresh weight) of hairy roots from each of the three independent lines was sub-cultured in MS medium supplied with 1.25 mM KH2PO4 (+P) or without KH2PO4 (–P). After 14 d of growth, hairy roots were harvested for dry weight and Pi concentration analysis, as described previously (Liang et al., 2010). Each independent transgenic line had three biological replicates.
GmPHR25 overexpression and suppression in soybean composite plants
To construct the GmPHR25-RNAi vector, a 360-bp specific fragment from the GmPHR25 coding region was amplified with specific primers (Supplementary Table S3), and PCR products were ligated into the pYLRNAi vector after digestion by BamH I and Hind III, Mlu I and Pst I. Subsequently, GmPHR25-OE, GmPHR25-RNAi, or empty vector constructs were separately transformed into Agrobacterium rhizogenes strain K599, which was also used to infect soybean seedlings in order to obtain composite soybean plants with transgenic hairy roots, as described previously (Guo et al., 2011). When transgenic hairy roots grew to approximately 10 cm long, a small portion was harvested for PCR and qPCR analysis. The transgenic composite soybean plants were grown in nutrient solution supplied with 500 µM KH2PO4 (+P) or 25 µM KH2PO4 (–P). For each P treatment, six independent transgenic lines were included for GmPHR25-OE, GmPHR25-RNAi, or control lines. After 14 d of growth, entirely expanded young leaves, shoots, and hairy roots were separately harvested to determine dry weight, along with total P and soluble Pi concentration, as described previously (Liang et al., 2010). Small portions of hairy roots were also harvested for further qPCR analysis. One independent transgenic line originating from a composite soybean plant with transgenic hairy roots was considered as a semi-biological replicate. A total of six replicates were included in this experiment.
Measurement of soluble Pi and total P concentration
For the soluble Pi concentration assay, about 0.1 g samples of fresh plant tissue were ground and extracted by distilled water. After centrifugation, the supernatant was assayed as described previously (Murphy and Riley, 1962). For the plant total P concentration assay, whole plants were heated at 75 °C until completely dry, then shoots and roots were ground into powder and, after digestion by H2SO4, Pi concentration was determined as above.
Expression analysis of downstream genes in soybean composite plants
Total RNA was extracted from transgenic hairy roots in soybean composite plants, and then qPCR was conducted to analyse transcription of downstream genes, including GmHAD1-2 (Glyma07g01410), GmSPX5 (Glyma10g40820), GmEXPB2 (Glyma10g24080), GmPAP14 (Glyma08g09880), GmPAP21 (Glyma10g08300), 14 soybean high-affinity phosphate transporter (GmPT) members (Qin et al., 2012a), and the other 34 GmPHR members (i.e. except GmPHR25). All qPCR primers were designed according to sequences from the phytozome website (http://www.phytozome.net).
Transcriptional activity and DNA-binding affinity analysis of GmPHR25
To detect transcriptional activity of GmPHR25, the full-length GmPHR25 coding region was amplified with specific primers (Supplementary Table S3) and inserted into pGBKT7 fused with GAL4 DNA-BD using the Matchmaker yeast two-hybrid system (Clontech, USA). The constructs were transformed into yeast strain AH109, and screened on the minimal medium SD/-Trp and SD/-Trp-His-A to examine the reporter gene expression. Yeast transformed with the empty pGBKT7 (BD) vector was used as a negative control.
For detection of DNA-binding affinity of GmPHR25, three soybean high-affinity Pi transporters (GmPT9, GmPT10, and GmPT12) were selected because their expression levels were up-regulated by GmPHR25 and their promoter region contains one or two PHR1 biding sites (P1BS; 5′-GNATATNC-3′). Therefore, fragments were separately amplified from the promoter regions of GmPT9, GmPT10, and GmPT12 using specific primers containing at least one P1BS element (Supplementary Table S3), which were subsequently cloned into pABAi vectors. The constructs were transformed into yeast strain Y1HGold (Clontech, USA). Meanwhile, a fragment containing four P1BS (5′-GAATATTC-3′) elements was synthesized and cloned into the pABAi vector as the positive control, as described previously (Sun et al., 2016; Ruan et al., 2017). The full-length GmPHR25 coding region was amplified and cloned into the pGADT7 (AD) vector and transformed into yeast bait strain. Yeast transformed with pGADT7 was used as a negative control. Modified medium without uracil (Ura) or leucine (Leu) was used for selection.
Statistical analyses
All data were analysed using Microsoft Excel 2003 (Microsoft Company, USA) to calculate means and standard errors, and SPSS 10.1 (SPSS Institute Inc., Cary, NC, USA) was used to conduct Student’s t-tests.
Results
Identification and characterization of GmPHR members in soybean
A total of 35 putative GmPHR members were identified through BLAST searching of the soybean genome database at http://www.phytozome.net, And general information on them is summarized in Table 1. The GmPHR members were unevenly distributed on soybean chromosomes 1–3, 7–13, 15, 16, and 18–20 (Table 1). Based on their positions on these chromosomes, the 35 GmPHR members were named from GmPHR1 to GmPHR35. As shown in Table 1, open reading frames of the GmPHR members ranged from 642 to 1455 bp in length, which was predicted to encode proteins containing 213–484 amino acids, and exhibiting 41–61% sequence identity with AtPHR1 (Table 1).
Table 1.
General information for the 35 GmPHR members
| Gene | Locus | Chromosomal location | Exon/Intron number | Length of ORF (bp) | Number of amino acids (aa) | Protein size (kD) | Identity to AtPHR1 |
|---|---|---|---|---|---|---|---|
| GmPHR1 | Glyma01g01300 | 1 | 6/5 | 813 | 270 | 30.3 | 48% |
| GmPHR2 | Glyma01g05920 | 1 | 6/5 | 1035 | 334 | 38.2 | 45% |
| GmPHR3 | Glyma02g07790 | 2 | 6/5 | 1251 | 416 | 46.9 | 46% |
| GmPHR4 | Glyma02g12070 | 2 | 6/5 | 1056 | 351 | 39.2 | 44% |
| GmPHR5 | Glyma02g30714 | 2 | 7/6 | 642 | 213 | 24.3 | 48% |
| GmPHR6 | Glyma02g30800 | 2 | 6/5 | 1269 | 422 | 47.1 | 54% |
| GmPHR7 | Glyma03g00590 | 3 | 6/5 | 804 | 267 | 29.0 | 58% |
| GmPHR8 | Glyma03g29940 | 3 | 6/5 | 1284 | 427 | 47.7 | 58% |
| GmPHR9 | Glyma03g32350 | 3 | 7/6 | 1446 | 481 | 53.2 | 50% |
| GmPHR10 | Glyma03g41040 | 3 | 8/7 | 1308 | 435 | 48.6 | 43% |
| GmPHR11 | Glyma07g35700 | 7 | 6/5 | 996 | 331 | 37.3 | 43% |
| GmPHR12 | Glyma08g17400 | 8 | 6/5 | 1122 | 373 | 41.4 | 42% |
| GmPHR13 | Glyma09g02030 | 9 | 6/5 | 945 | 314 | 34.3 | 42% |
| GmPHR14 | Glyma09g02040 | 9 | 6/5 | 990 | 329 | 35.7 | 53% |
| GmPHR15 | Glyma09g17404 | 9 | 6/5 | 1275 | 424 | 47.0 | 61% |
| GmPHR16 | Glyma09g34461 | 9 | 6/5 | 798 | 265 | 30.0 | 48% |
| GmPHR17 | Glyma10g04540 | 10 | 7/6 | 1446 | 481 | 53.0 | 54% |
| GmPHR18 | Glyma10g34050 | 10 | 6/5 | 1164 | 387 | 43.1 | 41% |
| GmPHR19 | Glyma11g18990 | 11 | 6/5 | 1245 | 414 | 46.2 | 44% |
| GmPHR20 | Glyma12g09490 | 12 | 6/5 | 1218 | 405 | 45.2 | 45% |
| GmPHR21 | Glyma12g31020 | 12 | 6/5 | 1263 | 420 | 47.4 | 47% |
| GmPHR22 | Glyma13g18805 | 13 | 7/6 | 1440 | 479 | 53.0 | 54% |
| GmPHR23 | Glyma13g39290 | 13 | 6/5 | 1203 | 400 | 45.0 | 47% |
| GmPHR24 | Glyma15g12930 | 15 | 6/5 | 942 | 313 | 34.2 | 42% |
| GmPHR25 | Glyma15g12940 | 15 | 6/5 | 990 | 329 | 35.6 | 54% |
| GmPHR26 | Glyma15g29620 | 15 | 6/5 | 1068 | 355 | 39.6 | 49% |
| GmPHR27 | Glyma15g41740 | 15 | 7/6 | 1164 | 387 | 43.2 | 45% |
| GmPHR28 | Glyma16g26820 | 16 | 6/5 | 1251 | 416 | 46.9 | 47% |
| GmPHR29 | Glyma18g43130 | 18 | 6/5 | 714 | 237 | 26.8 | 47% |
| GmPHR30 | Glyma19g30220 | 19 | 6/5 | 819 | 272 | 29.6 | 58% |
| GmPHR31 | Glyma19g32850 | 19 | 6/5 | 1206 | 401 | 45.2 | 52% |
| GmPHR32 | Glyma19g35080 | 19 | 7/6 | 1455 | 484 | 53.3 | 50% |
| GmPHR33 | Glyma19g43690 | 19 | 8/7 | 1209 | 402 | 44.7 | 44% |
| GmPHR34 | Glyma20g04630 | 20 | 6/5 | 1005 | 334 | 37.4 | 43% |
| GmPHR35 | Glyma20g33540 | 20 | 6/5 | 1182 | 393 | 43.2 | 42% |
Gene locus, exon and intron number, length, and protein size were extracted from the Phytozome website (http://www.phytozome.net). The identity between AtPHR1 and each GmPHR member was determined by BLAST analysis (https://blast.ncbi.nlm.nih.gov/Blast.cgi). ORF, open reading frame.
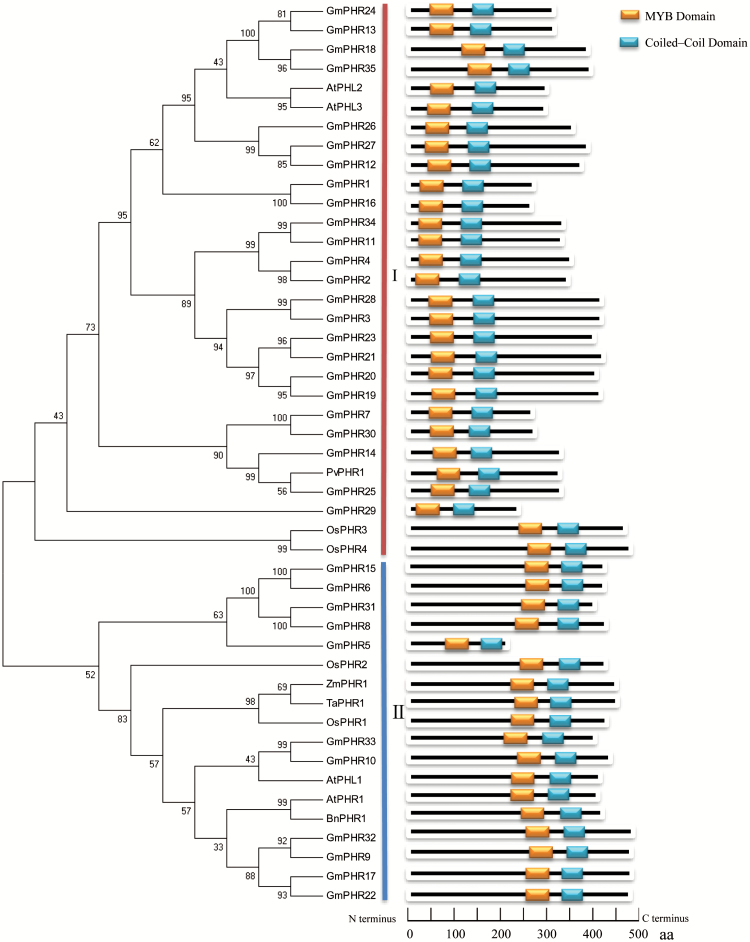
In order to determine evolutionary relationships among PHR members in soybean, Arabidopsis, rice, bean, wheat, maize, and rape, a phylogenetic tree was constructed. The results showed that plant PHR proteins can be divided into two groups, labeled as group I and group II in Fig. 1. Group I consisted of 24 GmPHR members together with AtPHL2 and AtPHL3 from Arabidopsis, PvPHR1 from bean, OsPHR3 and OsPHR4 from rice; however, 11 other GmPHR members were classified into group II, including GmPHR5, 6, 8, 9, 10, 15, 17, 22, 31, 32, 33, together with AtPHR1 and AtPHL1 from Arabidopsis, OsPHR1 and OsPHR2 from rice, ZmPHR1 from maize, TaPHR1 from wheat, and BnPHR1 from rape, (Fig. 1). Furthermore, two typical domains of PHR members, MYB binding and Coiled-Coil, were closely localized on the C-terminus for all PHR members in group II, but on the N-terminus for all PHR members in group I, except for OsPHR3 and OsPHR4 (Fig. 1).
Fig. 1.
Phylogenetic analysis of PHR proteins. For information on the soybean proteins see Table 1. The GenBank accession numbers of the proteins or gene loci for other species are as follows: AtPHR1 (At4g28610), AtPHL1 (At5g29000), AtPHL2 (At3g24120), AtPHL3 (At4g13640), OsPHR1 (Os03g21240), OsPHR2 (Os07g25710), OsPHR3 (Os02g04640), OsPHR4 (Os06g49040), BnPHR1 (JN806156), TaPHR1 (KC218925), ZmPHR1 (JF831533), PvPHR1 (EU500763). At, Arabidopsis thaliana; Gm, Glycine max; Os, Oryza sativa; Pv, Phaseolus vulgaris; Bn, Brassica napus; Zm, Zea mays; Ta, Triticum aestivum. The phylogenetic tree was created using the Mega 5.05 program.
Tissue-specific expression of GmPHR members
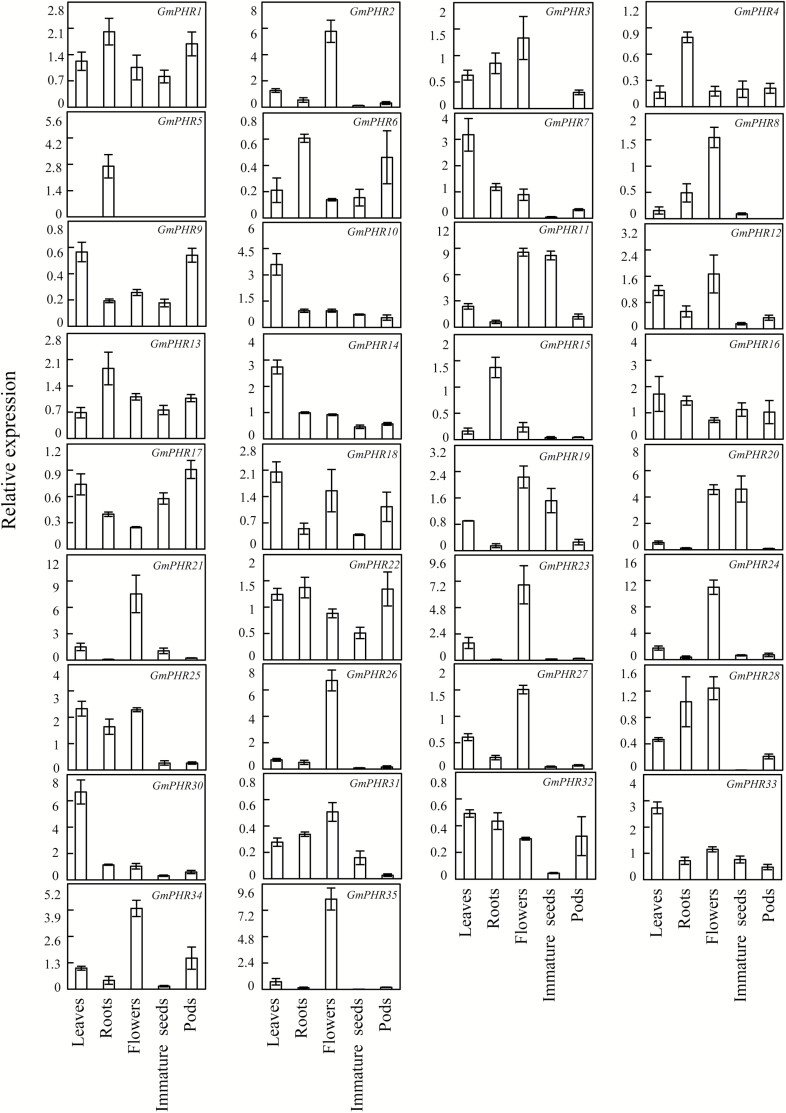
Expression patterns of GmPHR members were determined by qPCR analysis of soybean leaves, roots, flowers, pods, and seeds. The results showed that transcripts could be detected for all GmPHR members except for GmPHR29 (Fig. 2), and that expression patterns varied throughout the tissues that were tested . For example, GmPHR7, GmPHR10, GmPHR14, GmPHR30, and GmPHR33 were most highly expressed in leaves, GmPHR2, GmPHR8, GmPHR21, GmPHR23, GmPHR24, GmPHR26, GmPHR27, GmPHR34, and GmPHR35 were most highly expressed in flowers, and GmPHR4, GmPHR5, and GmPHR15 were mainly expressed in roots. The expressions of GmPHR11, GmPHR19, and GmPHR20 were higher in flowers and immature seeds than in other tissues. For GmPHR32, expression levels were similar in leaves and other tissues, except for immature seeds (Fig. 2).
Fig. 2.
Tissue-specific expression patterns of GmPHR members. Soybean seedlings were grown in complete nutrient solution and entirely expanded young leaves, roots, flowers, 1-cm pods, and immature seeds were separately harvested for qPCR analysis. Data are means of four replicates ±SE.
Transcriptional responses of GmPHR to nutrient deficiencies
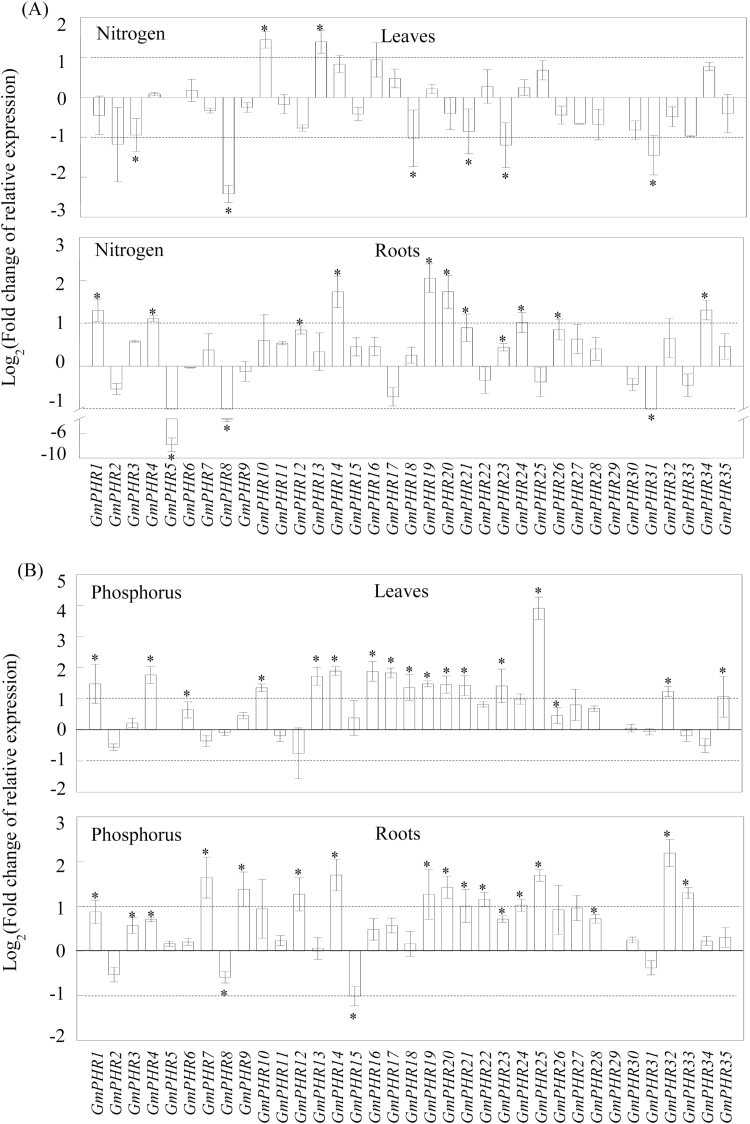
Expression patterns of GmPHR members in both leaves and roots were further examined under nitrogen (N) or phosphorus (P) nutrient deficiency conditions. The results showed that GmPHR members exhibited diverse responses to N and P deficiencies (Fig. 3).
Fig. 3.
Expression patterns of GmPHR members in response to nitrogen and phosphorus deficiency in leaves and roots. Soybean seedlings were grown for 14 d in complete nutrient solution as a control, or in nutrient solution without nitrogen (A) or phosphorus (B). Data are means of four replicates ±SE, expressed as the binary logarithm of fold-changes of relative expression of GmPHR members under nutrient deficiency compared with normal conditions. *, Significant difference between normal and nutrient deficiency treatments (Student’s t-test, P<0.05).
In the N deficiency treatment, the transcription of most GmPHR members remained largely unchanged, with fold-changes below 2 compared with controls observed in both soybean leaves and roots for all genes, except for six members in leaves and ten members in roots (Fig. 3A). In leaves, only GmPHR10 and GmPHR13 were found to be significantly up-regulated by more than 2-fold, while GmPHR8, GmPHR18, GmPHR23, and GmPHR31 were significantly down-regulated by more than 2-fold by nitrogen starvation (Fig. 3A). In roots, the expressions of seven GmPHR members (GmPHR1, 4, 14, 19, 20, 24, 34) were significantly up-regulated by more than 2-fold under nitrogen deficiency conditions, while the expressions of GmPHR5, GmPHR8, and GmPHR31 were significantly down-regulated by more than 2-fold (Fig. 3A).
In contrast to N deficiency, expression levels of most GmPHR members were significantly up-regulated by Pi starvation in soybean leaves and roots (Fig. 3B). In leaves, transcription of 15 members (GmPHR1, 4, 10, 13, 14, 16–21, 23, 25, 32, 35) was significantly increased by more than 2-fold in the P deficiency treatment, especially for GmPHR25, which exhibited a 16-fold increase of transcript levels in response to P deficiency (Fig. 3B). Similarly, in roots, except for GmPHR15 where expression was significantly down-regulated by more than 2-fold, expression levels were significantly up-regulated by more than 2-fold by Pi starvation in 12 members (GmPHR7, 9, 12, 14, 19–22, 24, 25, 32, 33), particularly for GmPHR32, for which transcription increased 5-fold in responses to P deficiency (Fig. 3B).
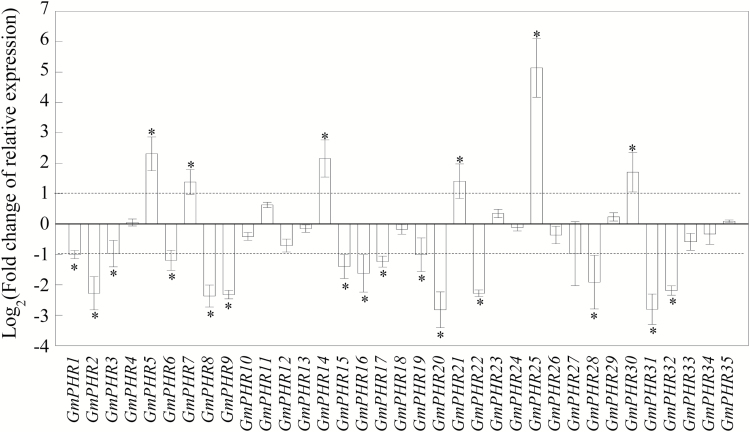
Effects of P deficiency on transcription of GmPHR members in soybean nodules
Since soybean can form a special organ, the nodule, with rhizobium symbionts, the effects of P deficiency on the expression of GmPHR members in nodules were investigated by qPCR analysis. The results showed that transcripts of 21 members were significantly up-regulated or down-regulated by more than 2-fold in nodules under Pi deficiency conditions relative to those under P sufficient conditions (Fig. 4). Among them, expression levels were up-regulated for GmPHR5, GmPHR7, GmPHR14, GmPHR21, GmPHR25, and GmPHR30, especially for GmPHR25 where transcripts were 30-fold higher in nodules under P deficiency. On the other hand, expression levels of 15 members (GmPHR1–3, 6, 8, 9, 15–17, 19, 20, 22, 28, 31, 32) decreased dramatically in response to P deficiency, with GmPHR20 and GmPHR31 transcripts decreased by 6-fold (Fig. 4). Expression levels of the other GmPHR members in nodules were relatively unaltered by P deficiency (Fig. 4). Using the housekeeping gene GmACTIN as a reference, similar expression patterns of eight GmPHR members in nodules were observed (Supplementary Fig. S6).
Fig. 4.
Expression patterns of GmPHR members in root nodules, expressed as the binary logarithm of fold-changes of relative expression of GmPHR members under high-P and low-P treatments. Data are mean of four replicates ±SE. *, Significant difference between high-P and low-P treatments (Student’s t-test, P<0.05).
Subcellular localization of GmPHR25
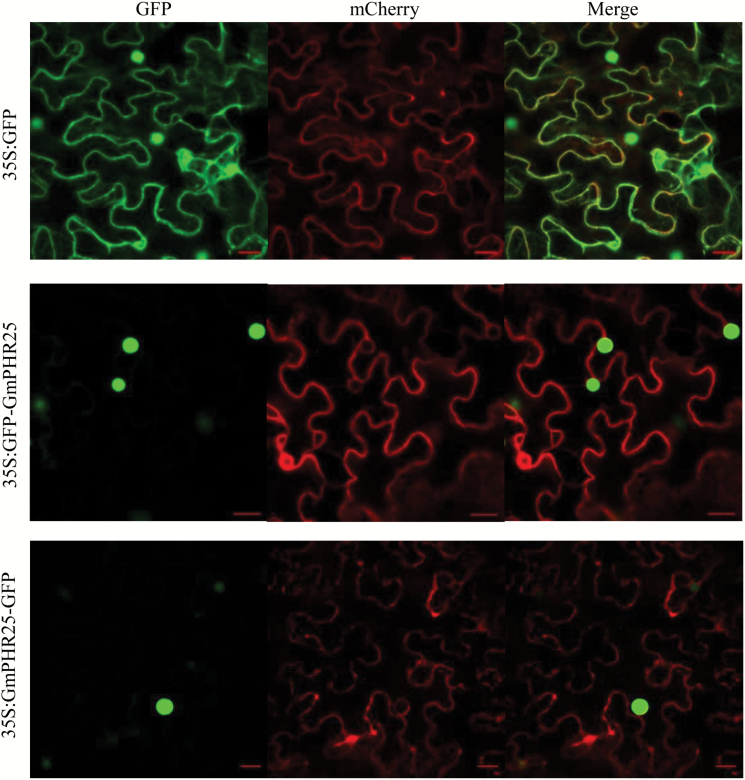
Since transcription of GmPHR25 exhibited the most responses to P deficiency in all the soybean organs tested (i.e. leaves, roots, and nodules), and its MYB and Coiled-Coil domains were closely located on the N-terminus, which was different from most PHR members with well-known functions in plants (Fig. 1), GmPHR25 was selected for further analysis. To investigate the subcellular localization of GmPHR25, its encoding region was fused to GFP at either its N-terminus (GFP-GmPHR25) or C-terminus (GmPHR25-GFP), and the constructs were transiently expressed in tobacco leaves. Subcellular localization was examined by the detection of GFP signals (Fig. 5). Signals of the empty vector control were observed in the plasma membrane, cytoplasm, and nucleus (Fig. 5), whereas signals from fusion with GmPHR25 were only detected in the nucleus (Fig. 5), strongly suggesting that this is where GmPHR25 predominantly localizes.
Fig. 5.
Subcellular localization of GmPHR25 fused to GFP protein in tobacco mesophyll cells: 35S:GFP, 35S:GFP-GmPHR25, 35S:GmPHR25-GFP, and 35S:AtPIP2A-mCherry fusion vectors are shown. GFP fluorescence and mCherry fluorescence were observed using confocal microscopy. Scale bars are 20 µm.
Overexpressing GmPHR25 increases Pi concentration in soybean hairy roots in vitro
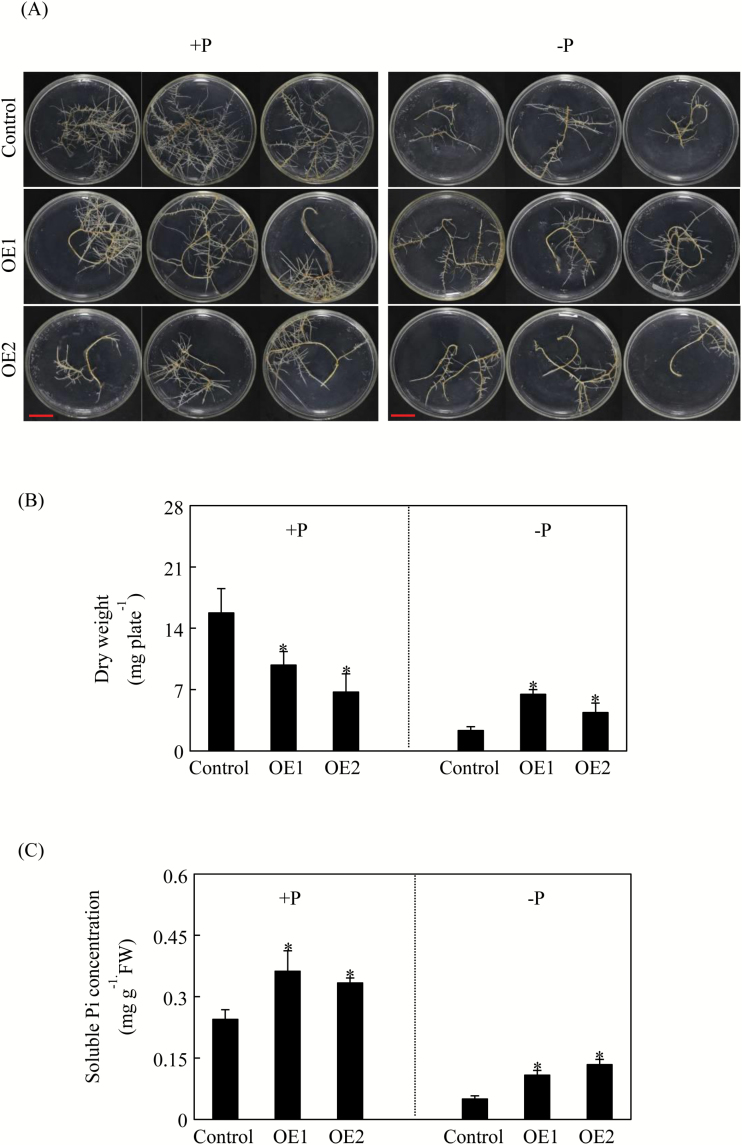
In order to assess GmPHR25 functions in the control of plant Pi homeostasis, soybean hairy roots overexpressing GmPHR25 in vitro were generated. Increased expression of GmPHR25 was verified through qPCR analysis, with GmPHR25 expression levels being increased by more than 3-fold over empty vector control hairy roots (Supplementary Fig. S1A). Furthermore, GmPHR25 overexpression significantly affected soybean hairy root growth and Pi concentration (Fig. 6). Overexpression of GmPHR25 inhibited hairy root growth under P-sufficient conditions, as reflected by 37% and 57% decreases in root dry weights of the GmPHR25 overexpression lines relative to the controls (Fig. 6B). However, overexpression of GmPHR25 enhanced hairy root growth under P-deficient conditions, as reflected by 170% and 80% increases in hairy root dry weights (Fig. 6B). In contrast to changes in dry weight, relative to the control line, soluble Pi concertation in GmPHR25 overexpression lines was increased by more than 30% (OE2 compared to control) and 110% (OE1 compared to control) under Pi-sufficient and -deficient conditions, respectively (Fig. 6C). Taken together, these results suggest that GmPHR25 regulates soybean hairy root growth and Pi homeostasis.
Fig. 6.
Dry weight and soluble Pi concentration in soybean hairy roots of control and GmPHR25-overexpressing plants. (A) Phenotype of hairy roots, (B) dry weight, and (C) soluble Pi concentration. Hairy roots were grown in MS medium for 14 d prior to transferring into MS medium containing 1.25 mM (+P) or 0 mM (–P) phosphorus. After a further 14 d, roots were harvested for analysis. Control represents hairy roots transformed with the empty vector; OE indicates transgenic hairy roots overexpressing GmPHR25. FW, fresh weight. Data are means of three replicates +SE. *, Significant difference between OE and control (Student’s t-test, P<0.05). Scale bars in (a) are 1 cm.
Functional analysis of GmPHR25 in soybean composite plants
Functions of GmPHR25 were further investigated in overexpressing soybean transgenic composite plants. Increased expression of GmPHR25 in transgenic hairy roots was verified through qPCR analysis (Supplementary Fig. S1B). Under P-sufficient conditions, compared to control lines, overexpression of GmPHR25 in soybean composite plants also resulted in a 56% decrease in plant dry weight (Fig. 7B), while total P concentration increased by 23% (Fig. 7C). More precisely, the soluble Pi concentration rose by 38% in leaves and by 52% in roots under Pi-sufficient conditions (Fig. 7D, E). However, under Pi-deficient condition, overexpression of GmPHR25 only resulted in increased dry weight and soluble Pi concentration in leaves, compared with the control lines (Fig. 7B, D). These results further reinforce the suggestion that GmPHR25 affects Pi homeostasis in plants. However, it was observed that suppressed GmPHR25 expression did not affect dry weight and P concentration for transgenic composite plants, except for decreased soluble Pi concentrations in leaves under P-sufficient conditions (Supplementary Fig. S2), suggesting that function redundancy might be present for GmPHR25 in soybean.
Fig. 7.
Dry weight and P concentration of control and GmPHR25-overexpressing composite soybean plants. (A) Phenotype of composite soybean plants, (B) dry weight, (C) total P concentration in the whole plant, (D) soluble Pi concentration of leaves, and (E) soluble Pi concentration of roots. Composite soybean plants with transgenic hairy roots were grown in normal nutrient solution for 14 d, then plants were transferred to nutrient solution containing 500 μM (+P) or 25 μM (–P) KH2PO4. After a further 14 d, shoots and roots were separately harvested for analysis. Control represents hairy roots transformed with the empty vector; OE indicates transgenic hairy roots overexpressing GmPHR25. DW, dry weight; FW, fresh weight. Data are means of six replicates ±SE. *, Significant differences between OE and control (Student’s t-test, P<0.05). Scale bars in (A) are 10 cm.
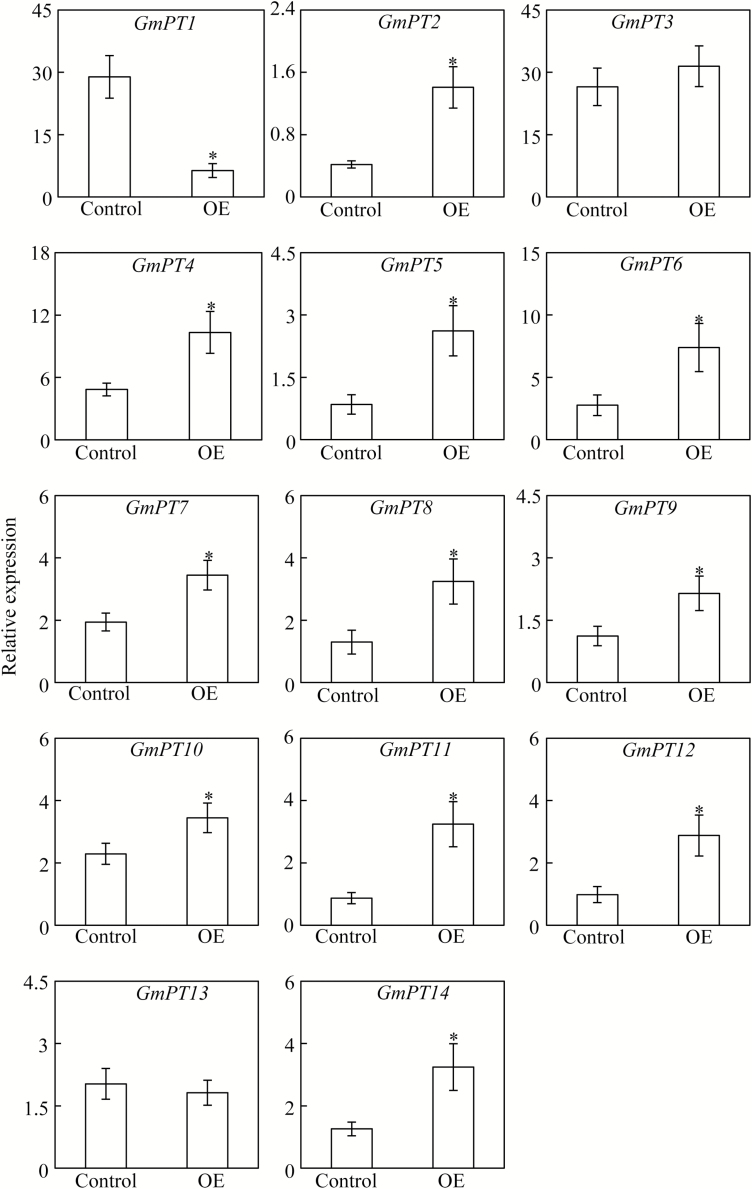
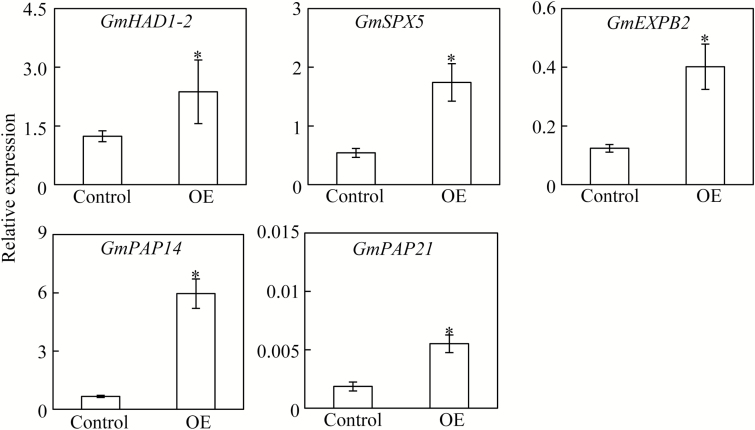
To further elucidate the regulatory roles of GmPHR25 in soybean, transcription of 14 high-affinity Pi transporters (GmPT), five Pi starvation-responsive genes, and 34 other GmPHR members were analysed in hairy roots of transgenic composite plants. Except for GmPT1, GmPT3, and GmPT13, transcripts of other GmPT members were significantly increased in composite plant hairy roots overexpressing GmPHR25, with expression of GmPT2 increased over 2-fold (Fig. 8). Consistently, at least one P1BS element could be detected in the promoter regions of GmPT members, including GmPT2, GmPT5, GmPT8, GmPT9, GmPT10, and GmPT12 (Supplementary Table S4). These results indicate that GmPHR25 regulates GmPT expression patterns, and thus controls Pi homeostasis in soybean. In addition, overexpression of GmPHR25 significantly increased the expression levels of five Pi starvation-responsive genes, namely GmHAD1-2, GmSPX5, GmEXPB2, GmPAP14, and GmPAP21 (Fig. 9). However, except for significant increases of GmPHR8 and GmPHR22 expression levels (Supplementary Fig. S7), overexpression of GmPHR25 had no effect on the transcription levels of the other 32 GmPHR members (data not shown). These results strongly suggest that GmPHR25 plays an important role in the P signaling network in soybean.
Fig. 8.
Transcripts of GmPTs in GmPHR25-overexpressing composite soybean plants. Plants were grown in nutrient solution containing 500 μM KH2PO4 for 14 d, and transcripts in hairy roots were determined by qPCR. Control represents soybean hairy roots transformed with the empty vector; OE indicates transgenic soybean hairy roots overexpressing GmPHR25. Data are means of six replicates ±SE. *, Significant differences in downstream gene expression between OE and control plants (Student’s t-test, P<0.05).
Fig. 9.
Expression of five Pi starvation-responsive genes in GmPHR25-overexpressing composite soybean plants. Plants were grown in nutrient solution containing 500 μM KH2PO4 for 14 d, and transcripts of candidate genes in hairy roots were determined by qPCR. Control represents soybean hairy roots transformed with the empty vector; OE indicates transgenic soybean hairy roots overexpressing GmPHR25. Data are means of six replicates ±SE. *, Significant differences in gene expression between OE and control plants (Student’s t-test, P<0.05).
Discussion
The critical roles of PHR1 and PHR1-like genes in the P signaling network have been well elucidated in several plant species, including Arabidopsis, rice, bean, wheat, maize, and rape (Rubio et al., 2001; Valdés-López et al., 2008; Zhou et al., 2008; Ren et al., 2012; Wang et al., 2013a, 2013b; Sun et al., 2016; Ruan et al., 2017). Furthermore, all PHR1 members with known functions have been characterized as having the MYB and Coiled-Coil domains localization at their C-terminus, except for PvPHR1 in bean, AtPHL2 and AtPHL3 in Arabidopsis (Fig. 1). However, expression patterns of PHR family members in soybean and functions of GmPHR members exhibiting the MYB and Coiled-Coil domains at the N-terminus remain unclear. With the release of soybean genome sequences, it is now possible to characterize GmPHR members and dissect their involvement in adaptations to P deficiency. In the current study, gene structures and expression patterns of GmPHR members were analysed in soybean for the first time. Furthermore, the roles of GmPHR25 in controlling Pi homeostasis were investigated through functional analysis in transgenic hairy roots in vitro and in vivo.
In total, 35 GmPHR members were identified in the soybean genome via BLAST searches on the phytozome website. All GmPHR members could be further divided into two groups through phylogenetic analysis (Fig. 1). It was found that 11 soybean GmPHR members belong to group II, which also contains representatives from other species known to function as key regulators in P signaling pathways, including AtPHR1 and AtPHL1 in Arabidopsis, OsPHR2 in rice, BnPHR1 in rape, ZmPHR1 in maize, and TaPHR1 in wheat (Rubio et al., 2001; Valdés-López et al., 2008; Zhou et al., 2008; Ren et al., 2012; Wang et al., 2013a, 2013b; Guo et al., 2015). The other 24 GmPHR members were more homologous to PvPHR1 in common bean (Fig. 1). Among two the GmPHR groups, each PHR member contains at least one copy, and presents as duplicated pairs, such as GmPHR1 and GmPHR16, GmPHR7 and GmPHR30, GmPHR6 and GmPHR15, GmPHR8 and GmHR31 (Fig. 1). Consistent with this, it has been suggested that soybean has experienced at least two rounds of whole-genome duplication, thus resulting in approximately 75% of its genes being present in multiple copies (Shoemaker et al., 2006; Schmutz et al., 2010). Furthermore, diverse functions of duplicated genes in soybean have been suggested, such as GmCHLI controlling chlorophyll biosynthesis and GmTfl1 controlling growth habit (Tian et al., 2010; Li et al., 2016). It was observed that several GmPHR duplicated pairs exhibit different expression patterns, suggesting diverse functions present in GmPHR paralogs in soybean. For example, GmPHR6 exhibited high expression levels in both roots and pods (Fig. 2); however, its duplicated paralog GmPHR15 only exhibited relatively high transcripts in roots. GmPHR7 expression was significantly increased by P deficiency in roots, but this was not the case for its duplicated paralog GmPHR30 (Fig. 3B).
Genome-wide transcriptomic analysis in soybean has revealed transcripts of GmPHR members in various tissues, including leaves, roots, nodules, flowers, pods, and seeds, which are summarized in Supplementary Table S1 (Libault et al., 2010; Severin et al., 2010). Consistent with these published results, transcripts of GmPHR members were also detected in the tissues tested in the current study through qPCR analysis using their specifc primers, including leaves, roots, flowers, pods, and seeds (Fig. 2, Supplementary Fig. S8). However, relative expression levels of several GmPHR members differed from previous transcriptomics results. For example, the expression of GmPHR10 and GmPHR14 in the current study was highest in leaves (Fig. 2), whereas previous studies indicate it was highest in roots (Supplementary Table S1). These inconsistencies might be the result of differences in experimental materials, growth conditions, or analytical techniques. Despite these differences, the observation that GmPHR members are widely expressed throughout soybean, with variations among tissues, remains valid and is largely consistent with published reports.
In this study, P deficiency significantly increased expression levels of most GmPHR members (Fig. 3B), and demonstrated that responses vary among members. Similar results have also been observed in other plant species. For example, in Arabidopsis, expression levels of two PHR members (AtPHL2 and AtPHL3) were significantly increased by Pi starvation, while two other members (AtPHR1 and AtPHL1) exhibited no responses (Rubio et al., 2001; Sun et al., 2016). In rice, P deficiency resulted in significantly increased transcription of OsPHR3 and OsPHR4, but not OsPHR1 or OsPHR2 (Zhou et al., 2008; Guo et al., 2015; Ruan et al., 2017). These results strongly suggest that regulatory mechanisms underlying PHR member responses to P deficiency vary among these members, which warrants further study of the underlying mechanisms.
Further analysis of expression patterns of GmPHR members in both leaves and roots under N deficiency conditions revealed that several Pi starvation-responsive GmPHR members also respond to N deficiency in both leaves and roots (Fig. 3A). For example, in roots, expression of Pi starvation-responsive GmPHR14 and GmPHR19 increased by 2.3 and 3.1-fold, respectively, under N deficiency conditions (Fig. 3). These results indicate potential crosstalk in plant responses to P and N deficiencies. Recently, global transcriptome profiling has demonstrated that a large number of genes are responsive to both N and P deficiencies (Cai et al., 2013; Takehisa et al., 2015). For example, it was found that 159 genes in roots and 101 genes in shoots were up-regulated after 7 d of both N and P deficiencies, with one notable example being MYB101 (a MYB transcription factor) in rice (Cai et al., 2013). Furthermore, several key regulators have been suggested to play crucial roles in both N and P signaling in plants. For example, AtNLA is considered a key regulator of Arabidopsis adaptations to N-limiting conditions, because the nla mutant fails to develop essential adaptive responses to N limitation, and thus exhibits early and rapid senescence (Peng et al., 2007). Meanwhile, AtNLA can recruit PHOSPHAT2 (PHO2) to degrade PT2, and thus control Pi homeostasis in Arabidopsis (Kant et al., 2011; Park et al., 2014). Therefore, it seems that soybean responses to P and N deficiencies share common signaling pathway elements, which might be regulated by GmPHR members.
Soybean can interact with rhizobia, and thus form symbiotic associations, which significantly affects N and P acquisition and utilization by the plant. Recently, it has been shown that rhizobium inoculation not only improves N nutrition, but also influences P acquisition and utilization in soybean (Cheng et al., 2009; Qin et al., 2011; Ding et al., 2012). Rhizobium inoculation resulted in significant increases in exudation of protons and organic acids, and thus enhanced the capability of soybean to mobilize Ca-P and Al-P (Qin et al., 2011; Ding et al., 2012). Furthermore, Pi starvation-responsive GmPT5 and GmEXPB2 have been documented to control soybean Pi homeostasis and nodule development, respectively (Qin et al., 2012b; Li et al., 2015). It is therefore suggested that nodules contain adaptive strategies for responding to Pi starvation. Consistent with this hypothesis, expression levels of six GmPHR members (GmPHR5, GmPHR7, GmPHR14, GmPHR21, GmPHR25, and GmPHR30) were significantly increased by P deficiency in nodules, with GmPHR25 being particularly notable (Fig. 4). Furthermore, expression levels of GmEXPB2 and GmPT5 were up-regulated by GmPHR25 overexpression in soybean hairy roots (Figs 8, 9), which strongly suggests that GmPHR25 is involved in regulating Pi homeostasis in the nodules.
The role of AtPHR1 and its orthologues in P signaling and homeostasis has been well established in several plant species, which indicates that members of this gene family share a conserved function as central regulators in plant Pi homeostasis (Rubio et al., 2001; Valdés-López et al., 2008; Zhou et al., 2008; Bustos et al., 2010; Ren et al., 2012; Wang et al., 2013a, 2013b). In this study, GmPHR25 was selected as a candidate for further functional analysis because it exhibits the highest sequence similarity to PvPHR1, higher Pi starvation responses in leaves, roots, and nodules than other GmPHR members, and it is localized to the nucleus (Fig. 5). It was observed that GmPHR25 overexpression increased soluble Pi concentration in transgenic soybean hairy roots in vivo and in vitro under Pi-sufficient conditions (Figs 6, 7), which strongly supports the hypothesis that GmPHR25 acts in controlling Pi homeostasis. Furthermore, accompanied by increased Pi concentration, decreased growth in transgenic lines with GmPHR25 overexpression was observed under Pi-sufficient conditions (Figs 6, 7), strongly suggesting excessive Pi accumulation might inhibit plant growth. Consistent with this, similar results have also been found in rice through overexpressing OsPHR2 and OsPHR4 (Zhou et al., 2008; Ruan et al., 2017). However, the molecular mechanisms underlying a significant decrease in plant growth with excessive Pi accumulation remain unknown.
Since Pi transporters play important roles in Pi uptake and translocation in plants, expression patterns of soybean Pi transporters (GmPT) were assayed by qPCR analysis in hairy roots overexpressing GmPHR25. As a result of GmPHR25 overexpression, transcription increased for most GmPT members, except GmPT1, GmPT3, and GmPT13 (Fig. 8), which strongly suggests that GmPHR25 regulates GmPT expression, and thus controls Pi homeostasis (Figs 6, 7). Within plant signaling networks, it has been well documented that several PT members act downstream of PHR1 (Chiou and Lin, 2011; Liang et al., 2014). For example, transcript levels of AtPht1;4, AtPht1;7, AtPht1;8, and AtPht1;9 were all significantly increased by AtPHR1 overexpression in Arabidopsis (Nilsson et al., 2007). In rice, OsPT1, OsPT5, OsPT7, OsPT9, and OsPT12 have also been placed downstream of OsPHR2, because overexpression or suppression OsPHR2 leads to respective increases or decreases in their expression levels (Zhou et al., 2008). Although the functions of most GmPT members remain largely unknown, with the exception of GmPT5, it is reasonable to conclude that their expression levels influence Pi homeostasis in soybean. Beyond Pi transporters, transcripts of other Pi starvation-responsive genes were also increased by GmPHR25 overexpression in soybean hairy roots. The Pi-responsive genes tested here included GmEXPB2, GmSPX5, GmHAD1-2, GmPAP14, and GmPAP21 (Fig. 9). Among them, GmEXPB2 has been suggested to play a critical role in soybean root and nodule growth, possibly through cell wall modification, thereby affecting Pi acquisition and accumulation (Guo et al., 2011; Li et al., 2015). Recently, GmPAP21 has been suggested to be involved in P utilization in soybean nodules (Li et al., 2017). Therefore, it is possible that GmPHR25 is a critical regulator in controlling Pi acquisition and uptake in soybean through effects on the transcription of Pi starvation-responsive genes. However, unlike AtPHR1 or OsPHR2, GmPHR25 did not harbor any transcriptional activity in yeast (Supplementary Fig. S3). Since OsPHR2 transcription activity is determined by 230 amino acids at its N-terminus in the yeast analysis system (Zhou et al., 2008), which are lacking in GmPHR25 (Fig. 1), it might be plausible that GmPHR25 exhibits no transcription activity, and thus is different from OsPHR2. Furthermore, GmPHR25 exhibited binding activity against the synthetic fragment containing four P1BS elements despite having no binding activity against the promoters of GmPT9, GmPT10, and GmPT12 (Supplementary Figs S4, S5). These results strongly suggest that GmPHR25 might regulate transcription of its downstream genes through interaction with other regulators. Consistent with this, it has recently been reported that OsPHR4 could interact with other OsPHR members to regulate transcription of downstream genes in rice (Ruan et al., 2017).
In summary, GmPHR members were systematically characterized, including expression patterns among tissues, and responses to nutrient deficiencies (i.e. N and P). In addition, evidence placing GmPHR25 as a regulator in the P signaling network has also been presented. These data provide not only a comprehensive list of GmPHR members in soybean, but also information on their properties, as well as results confirming their roles in the soybean P signaling network.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Expression of GmPHR25 in soybean hairy roots.
Fig. S2. Dry weight, total P concentration and soluble Pi concentration of control and RNAi-GmPHR25 in composite soybean plants.
Fig. S3. Transcriptional activity analysis of GmPHR25 in yeast.
Fig. S4. Yeast one-hybrid analysis of the DNA-binding affinity of GmPHR25 for the 4xP1BS module.
Fig. S5. Yeast one-hybrid analysis of the DNA-binding affinity of GmPHR25 for the GmPT9, GmPT10, and GmPT12 promoters.
Fig. S6. Expression patterns of eight GmPHR members and GmEF1-α in nodules at two P levels.
Fig. S7. Transcripts of GmPHR8 and GmPHR22 in overexpressing GmPHR25 composite soybean plants.
Fig. S8. Amplification efficiency for each pair of of GmPHR-specific primers for qRT-PCR analysis.
Table S1. The expression profiles of GmPHR members from the SoyBase (http://soybase.org/soyseq/).
Table S2. Primer sequences used in this study for qPCR.
Table S3. Primer sequences used in this study for vector construction.
Table S4. Analysis of the putative PHR1-binding site in a 3.0-kb sequence upstream of the start codon of each GmPT member.
Supplementary Material
Acknowledgements
This work was supported by grants from the National Key Research and Development Program of China (2016YFD0100700), the National Natural Science Foundation of China (31372119 and 31422046), the Natural Science Foundation of Guangdong Province (2015A030306034), the High Level Talents Special Support Plan of Guangdong Province (2015TX01N042 and 2015TQ01N078), and the Research Team Project of the Natural Science Foundation of Guangdong Province (2016A030312009). The authors thank Dr Thomas Walk for critical reading of the original manuscript. The authors have no conflict of interest to declare.
References
- Beardsley TM. 2011. Peak phosphorus. Bioscience 61, 91. [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible WR. 2006. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiology 141, 988–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, Solano R, Leyva A, Paz-Ares J. 2010. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genetics 6, e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Xie W, Lian X. 2013. Comparative analysis of differentially expressed genes in rice under nitrogen and phosphorus starvation stress conditions. Plant Molecular Biology Reporter 31, 160–173. [Google Scholar]
- Cheng F, Cao G, Wang X, Zhao J, Yan X, Liao H. 2009. Isolation and application of effective nitrogen fixation rhizobial strains on low-phosphorus acid soils in South China. Chinese Science Bulletin 54, 412–420. [Google Scholar]
- Chiou TJ, Lin SI. 2011. Signaling network in sensing phosphate availability in plants. Annual Review of Plant Biology 62, 185–206. [DOI] [PubMed] [Google Scholar]
- Cordell D, Drangert JO, White S. 2009. The story of phosphorus: global food security and food for thought. Global Environmental Change 19, 292–305. [Google Scholar]
- Ding X, Sui X, Wang F, Gao J, He X, Zhang F, Yang J, Feng G. 2012. Synergistic interactions between Glomus mosseae and Bradyrhizobium japonicum in enhancing proton release from nodules and hyphae. Mycorrhiza 22, 51–58. [DOI] [PubMed] [Google Scholar]
- Fan C, Wang X, Hu R, Wang Y, Xiao C, Jiang Y, Zhang X, Zheng C, Fu YF. 2013. The pattern of Phosphate transporter 1 genes evolutionary divergence in Glycine max L. BMC Plant Biology 13, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK. 2005. A miRNA involved in phosphate-starvation response in Arabidopsis. Current Biology 15, 2038–2043. [DOI] [PubMed] [Google Scholar]
- Guo M, Ruan W, Li C et al. . 2015. Integrative comparison of the role of the PHOSPHATE RESPONSE1 subfamily in phosphate signaling and homeostasis in rice. Plant Physiology 168, 1762–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Zhao J, Li X, Qin L, Yan X, Liao H. 2011. A soybean β-expansin gene GmEXPB2 intrinsically involved in root system architecture responses to abiotic stresses. The Plant Journal 66, 541–552. [DOI] [PubMed] [Google Scholar]
- Herridge DF, Peoples MB, Boddey RM. 2008. Global inputs of biological nitrogen fixation in agricultural systems. Plant & Soil 311, 1–18. [Google Scholar]
- Kant S, Peng M, Rothstein SJ. 2011. Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genetics 7, e1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Gui S, Yang T, Walk T, Wang X, Liao H. 2012. Identification of soybean purple acid phosphatase genes and their expression responses to phosphorus availability and symbiosis. Annals of Botany 109, 275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Li C, Zhang H, Liao H, Wang X. 2017. The purple acid phosphatase GmPAP21 enhances internal phosphorus utilization and possibly plays a role in symbiosis with rhizobia in soybean. Physiologia Plantarum 159, 215–227. [DOI] [PubMed] [Google Scholar]
- Li Q, Fang C, Duan Z, Liu Y, Qin H, Zhang J, Sun P, Li W, Wang G, Tian Z. 2016. Functional conservation and divergence of GmCHLI genes in polyploid soybean. The Plant Journal 88, 584–596. [DOI] [PubMed] [Google Scholar]
- Li X, Zhao J, Tan Z, Zeng R, Liao H. 2015. GmEXPB2, a cell wall β-expansin, affects soybean nodulation through modifying root architecture and promoting nodule formation and development. Plant Physiology 169, 2640–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhao J, Walk TC, Liao H. 2014. Characterization of soybean β-expansin genes and their expression responses to symbiosis, nutrient deficiency, and hormone treatment. Applied Microbiology and Biotechnology 98, 2805–2817. [DOI] [PubMed] [Google Scholar]
- Liang C, Tian J, Lam HM, Lim BL, Yan X, Liao H. 2010. Biochemical and molecular characterization of PvPAP3, a novel purple acid phosphatase isolated from common bean enhancing extracellular ATP utilization. Plant Physiology 152, 854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Tian J, Liao H. 2013. Proteomics dissection of plant responses to mineral nutrient deficiency. Proteomics 13, 624–636. [DOI] [PubMed] [Google Scholar]
- Liang C, Wang J, Zhao J, Tian J, Liao H. 2014. Control of phosphate homeostasis through gene regulation in crops. Current Opinion in Plant Biology 21, 59–66. [DOI] [PubMed] [Google Scholar]
- Liao H, Wan H, Shaff J, Wang X, Yan X, Kochian LV. 2006. Phosphorus and aluminum interactions in soybean in relation to aluminum tolerance. Exudation of specific organic acids from different regions of the intact root system. Plant Physiology 141, 674–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M, Farmer A, Joshi T, Takahashi K, Langley RJ, Franklin LD, He J, Xu D, May G, Stacey G. 2010. An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. The Plant Journal 63, 86–99. [DOI] [PubMed] [Google Scholar]
- Liu J, Li Y, Wang W, Gai J, Li Y. 2016. Genome-wide analysis of MATE transporters and expression patterns of a subgroup of MATE genes in response to aluminum toxicity in soybean. BMC Genomics 17, 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Liao H, Wang XR, Yan XL. 2008. Regulation effect of soil P availability on mycorrhizal infection in relation to root architecture and P efficiency of Glycine max. Chinese Journal of Applied Ecology 19, 564–568. [PubMed] [Google Scholar]
- Liu TY, Huang TK, Tseng CY, Lai YS, Lin SI, Lin WY, Chen JW, Chiou TJ. 2012. PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. The Plant Cell 24, 2168–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Q, Zhong Y, Wang Y et al. . 2014. SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. The Plant Cell 26, 1586–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy J, Riley J. 1962. A modifed single solution method for the determination of phosphate in natural water. Analytica Chimica Acta 27, 31–35. [Google Scholar]
- Nilsson L, Müller R, Nielsen TH. 2007. Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant, Cell & Environment 30, 1499–1512. [DOI] [PubMed] [Google Scholar]
- Park BS, Seo JS, Chua NH. 2014. NITROGEN LIMITATION ADAPTATION recruits PHOSPHATE2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. The Plant Cell 26, 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M, Hannam C, Gu H, Bi YM, Rothstein SJ. 2007. A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts the adaptability of Arabidopsis to nitrogen limitation. The Plant Journal 50, 320–337. [DOI] [PubMed] [Google Scholar]
- Puga MI, Mateos I, Charukesi R et al. . 2014. SPX1 is a phosphate-dependent inhibitor of PHOSPHATE STARVATION RESPONSE 1 in Arabidopsis. Proceedings of the National Academy of Sciences, USA 111, 14947–14952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Guo Y, Chen L, Liang R, Gu M, Xu G, Zhao J, Walk T, Liao H. 2012a. Functional characterization of 14 Pht1 family genes in yeast and their expressions in response to nutrient starvation in soybean. PLoS ONE 7, 543–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Jiang H, Tian J, Zhao J, Liao H. 2011. Rhizobia enhance acquisition of phosphorus from different sources by soybean plants. Plant & Soil 349, 25–36. [Google Scholar]
- Qin L, Zhao J, Tian J, Chen L, Sun Z, Guo Y, Lu X, Gu M, Xu G, Liao H. 2012b. The high-affinity phosphate transporter GmPT5 regulates phosphate transport to nodules and nodulation in soybean. Plant Physiology 159, 1634–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama KG. 1999. Phosphate acquisition. Annual Review of Plant Physiology and Plant Molecular Biology 50, 665–693. [DOI] [PubMed] [Google Scholar]
- Ren F, Guo QQ, Chang LL, Chen L, Zhao CZ, Zhong H, Li XB. 2012. Brassica napus PHR1 gene encoding a MYB-like protein functions in response to phosphate starvation. PLoS ONE 7, e44005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AE. 2009. Regulating the phosphorus nutrition of plants: molecular biology meeting agronomic needs. Plant and Soil 322, 17–24. [Google Scholar]
- Ruan W, Guo M, Wu P, Yi K. 2017. Phosphate starvation induced OsPHR4 mediates Pi-signaling and homeostasis in rice. Plant Molecular Biology 93, 327–340. [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J. 2001. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes & Development 15, 2122–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J et al. . 2010. Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. [DOI] [PubMed] [Google Scholar]
- Severin AJ, Woody JL, Bolon YT et al. . 2010. RNA-Seq Atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biology 10, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Yuan L, Zhang J, Li H, Bai Z, Chen X, Zhang W, Zhang F. 2011. Phosphorus dynamics: from soil to plant. Plant Physiology 156, 997–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker RC, Schlueter J, Doyle JJ. 2006. Paleopolyploidy and gene duplication in soybean and other legumes. Current Opinion in Plant Biology 9, 104–109. [DOI] [PubMed] [Google Scholar]
- Sun L, Song L, Zhang Y, Zheng Z, Liu D. 2016. Arabidopsis PHL2 and PHR1 act redundantly as the key components of the central regulatory system controlling transcriptional responses to phosphate starvation. Plant Physiology 170, 499–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehisa H, Sato Y, Antonio B, Nagamura Y. 2015. Coexpression network analysis of macronutrient deficiency response genes in rice. Rice 8, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24, 1596–1599. [DOI] [PubMed] [Google Scholar]
- Tian J, Liao H, Wang X, Yan X. 2003. Phosphorus starvation-induced expression of leaf acid phosphatase isoforms in soybean. Acta Botanica Sinica 45, 1037–1042. [Google Scholar]
- Tian J, Wang X, Tong Y, Chen X, Liao H. 2012. Bioengineering and management for efficient phosphorus utilization in crops and pastures. Current Opinion in Biotechnology 23, 866–871. [DOI] [PubMed] [Google Scholar]
- Tian Z, Wang X, Lee R, Li Y, Specht JE, Nelson RL, McClean PE, Qiu L, Ma J. 2010. Artificial selection for determinate growth habit in soybean. Proceedings of the National Academy of Sciences, USA 107, 8563–8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés-López O, Arenas-Huertero C, Ramírez M, Girard L, Sánchez F, Vance CP, Luis Reyes J, Hernández G. 2008. Essential role of MYB transcription factor: PvPHR1 and microRNA: PvmiR399 in phosphorus-deficiency signalling in common bean roots. Plant, Cell & Environment 31, 1834–1843. [DOI] [PubMed] [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DL. 2003. Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytologist 157, 423–447. [DOI] [PubMed] [Google Scholar]
- Veneklaas EJ, Lambers H, Bragg J et al. . 2012. Opportunities for improving phosphorus-use efficiency in crop plants. New Phytologist 195, 306–320. [DOI] [PubMed] [Google Scholar]
- Wang J, Sun J, Miao J et al. . 2013a. A phosphate starvation response regulator Ta-PHR1 is involved in phosphate signalling and increases grain yield in wheat. Annals of Botany 111, 1139–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Bai J, Liu H, Sun Y, Shi X, Ren Z. 2013b. Overexpression of a maize transcription factor ZmPHR1 improves shoot inorganic phosphate content and growth of Arabidopsis under low-phosphate conditions. Plant Molecular Biology Reporter 31, 665–677. [Google Scholar]
- Wang Z, Ruan W, Shi J et al. . 2014. Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proceedings of the National Academy of Sciences, USA 111, 14953–14958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Shou H, Xu G, Lian X. 2013. Improvement of phosphorus efficiency in rice on the basis of understanding phosphate signaling and homeostasis. Current Opinion in Plant Biology 16, 205–212. [DOI] [PubMed] [Google Scholar]
- Wu Z, Zhao J, Gao R, Hu G, Gai J, Xu G, Xing H. 2011. Molecular cloning, characterization and expression analysis of two members of the Pht1 family of phosphate transporters in Glycine max. PLoS ONE 6, e19752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff DD, Grossman AR, Weeks DP, Usuda H, Shimogawara K. 1999. Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. Proceedings of the National Academy of Sciences, USA 96, 15336–15341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Tian J, Liao H. 2014. Comparative characterization of GmSPX members reveals that GmSPX3 is involved in phosphate homeostasis in soybean. Annals of Botany 114, 477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Fu J, Liao H, He Y, Nian H, Hu Y, Qiu L, Dong Y, Yan X. 2004. Characterization of root architecture in an applied core collection for phosphorus efficiency of soybean germplasm. Chinese Science Bulletin 49, 1611–1620. [Google Scholar]
- Zhou J, Jiao F, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P. 2008. OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiology 146, 1673–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Wang Z, Lv Q, Shi J, Zhong Y, Wu P, Mao C. 2015. SPX proteins regulate Pi homeostasis and signaling in different subcellular level. Plant Signaling & Behavior 10, e1061163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.