Abstract
Klebsiella pneumoniae poses a major challenge to healthcare worldwide as an important cause of multidrug-resistant infections. Nosocomial clones, including epidemic sequence type 258 (ST258), have shown an affinity for acquiring and disseminating resistance plasmids, particularly variants of the K. pneumoniae carbapenemase. By comparison, the resurgence of severe community-associated K. pneumoniae infections has led to increased recognition of hypervirulent strains belonging to the K1 and K2 capsular serotypes, predominantly in eastern Asia. Genomic and functional studies suggest that a variety of virulence and immune evasive factors contribute to the success of nosocomial and community-associated clonal lineages, aided by mechanisms of genetic plasticity that contribute to uptake of genes associated with antimicrobial resistance and pathogenicity. While there currently appears to be limited overlap between resistant and hypervirulent lineages, specific bacterial and host factors contributing to the emergence of dominant clones remain incompletely understood. This review summarizes recent advances in our understanding of the molecular epidemiology, virulence potential, and host-pathogen interactions of K. pneumoniae.
Keywords: K. pneumoniae, carbapenem resistance, bacterial genomics, immune evasion
Klebsiella pneumoniae has been recognized as an urgent threat to human health owing to its widespread accumulation of multiclass antimicrobial resistance (AMR) in nosocomial settings [1], as well as its capacity to cause severe community-associated (CA) infections [2]. While ubiquitous in the environment, in humans K. pneumoniae is a common intestinal commensal that causes infections of the respiratory tract, urinary tract, and other sites, particularly during periods of immunosuppression and debility [3, 4]. Carbapenem-resistant K. pneumoniae (CRKP) associated with plasmid-encoded carbapenemases poses distinct clinical challenges due to its ability to acquire multiclass antibiotic resistance, producing invasive infections with high mortality [3–6]. A second clinical pathotype, hypervirulent K. pneumoniae (HvKP), known to cause liver abscesses and metastatic infections in healthy individuals in the community, has similarly emerged over the past 2 decades but occupies distinct clonal lineages [7, 8].
A variety of virulence factors have been implicated in the success of K. pneumoniae, including capsular polysaccharide (CPS), other outer membrane proteins [9–12], and metabolic factors, such as those enabling iron acquisition [13–15]. A range of adaptations may also allow K. pneumoniae to evade or overcome host immune system detection and clearance [14, 16–19]. While CRKP and HvKP isolates have distinct clonal backgrounds and virulence gene profiles [7, 20], whole-genome sequencing (WGS) studies increasingly demonstrate remarkable potential for K. pneumoniae genetic plasticity [20–22]. Focusing on our current knowledge of factors associated with the successful emergence of CRKP, here we summarize the spectrum of virulence among major K. pneumoniae clinical strains, its interactions with the host immune system, and recent advances in our understanding of the genetic factors enabling the success of this organism in diverse settings.
MOLECULAR EPIDEMIOLOGY OF HIGH-RISK K. PNEUMONIAE CLONES
Assessments of K. pneumoniae population structure have been enabled by genotyping techniques, including multilocus sequence typing (MLST) and, more recently, WGS [20, 21, 23]. While these studies show considerable genetic variability in K. pneumoniae, CRKP and HvKP isolates largely demonstrate clonal restriction. However, CRKP strains appear to be diversifying, and transfer of mobile genetic elements may enable dissemination of resistance and virulence genes.
Capsular Typing
Traditionally, typing and classification of K. pneumoniae was based on capsular serotyping, or K typing. However, this technique has several limitations, including high labor intensity, frequent cross-reactivity between capsular groups, and inability to type unencapsulated strains [24]. CRKP strains are frequently nontypable, and relatively few studies have attempted to use traditional serotyping methods to classify these isolates [25]. Newer molecular typing methods based on the capsular antigen gene cluster (cps), including wzc or wzi gene sequencing and phage-depolymerase typing of the polymerase chain reaction (PCR)–amplified cps locus, have facilitated classification of isolates [26–29]. To date, 79 K. pneumoniae capsule types have been reported and reflect substantial differences between community and nosocomial isolates, as well as limited overlap between hypervirulent and resistant strains [7]. Generally speaking, however, homologous recombination of cps appears to allow for substantial redistribution of capsule types among K. pneumoniae clones [8, 30].
MLST and K. pneumoniae Population Structure
In 2005, a validated MLST scheme was developed for K. pneumoniae [23] and largely replaced pulsed-field gel electrophoresis as the preferred genotyping method for broadly assessing isolate relatedness. MLST has since facilitated assessment of K. pneumoniae population structure on a global scale [8] and shows remarkable restriction of isolates harboring carbapenemase genes or distinct virulence phenotypes to a narrow range of STs [4, 7, 8, 20].
The clonal complex (CC) that includes ST258 and its single-locus variants ST11 and ST340 dominates among CRKP isolates [31]. ST258 in particular has been credited with the global dissemination of K. pneumoniae carbapenemase (KPC) over the past 2 decades, predominantly subtypes KPC-2 and KPC-3 in association with the plasmid-associated Tn4401 transposon and its variants [32]. The epidemiology of blaKPC-harboring K. pneumoniae (KPC-Kp) [3, 5] has recently been reviewed in detail and is not discussed further here. Internationally, the rapid spread of carbapenemase genes OXA-48 and the metallo-β-lactamases VIM and NDM-1 contribute increasingly to carbapenem resistance in Europe, the Indian subcontinent, the Middle East, and East Asia [6, 33, 34], although they have less commonly been associated with specific lineages. This presages the continued proliferation of CRKP strains and growing complexity of K. pneumoniae clonal and resistance plasmid relationships.
Although CC258 dominates among KPC-Kp, non-CC258 strains may be emerging, presumably as a result of transmission of blaKPC-harboring plasmids to susceptible strains [35]. Several centers with high ST258 endemicity, including our tertiary care center in New York City, have reported an increased proportion of polyclonal KPC-Kp to up to 60% CRKP isolates [4, 36, 37]. In areas with a low prevalence of CRKP, diverse non-ST258 K. pneumoniae are the primary strains in local hospital outbreaks [35, 38]. It is worth noting that CRKP is most often reported in outbreak settings and that the full spectrum of genotypes is likely underreported. Little is also known about the clonal background of susceptible K. pneumoniae isolates. Limited evidence suggests that these are composed of substantial clonal heterogeneity [4, 39]. We reported 127 STs among 194 consecutive susceptible bloodstream isolates [4], while a WGS study including 29 susceptible isolates of 24 STs demonstrated no evidence of clustering [39]. Minimal overlap was detected between carbapenem-susceptible and CRKP clones in both studies. Taken together, these studies point to a need for additional investigations elucidating the interplay between carbapenem-resistant and -susceptible lineages.
By comparison, CA-HvKP strains have also emerged over the past 20 years, particularly in association with the K1 and K2 capsular serotypes and epidemiologically with pyogenic liver abscess (PLA) and metastatic infection in eastern Asia [2]. Like CRKP, these K1 and, to a lesser degree, K2 strains demonstrate clonal restriction, suggesting a high degree of genetic specialization. Hypervirulent K1 isolates almost universally belong to CC23, which includes ST23 and ST57 and geographically diverse isolates, although CC82 also comprises K1 isolates [7, 8, 40]. Among K2 isolates, at least 3 different CCs predominate, CC65, CC86, and CC14 [7, 8, 41, 42]. Minimal clonal overlap has been observed between CRKP and HvKP isolates [7].
WGS Studies
Advances in bacterial WGS have enabled the reconstruction of many critical events in the evolutionary history of CRKP. This has confirmed and extended earlier observations based on capsule and MLST typing, which may underestimate clinically relevant variation in gene content between samples. Initial comparative sequencing efforts were largely focused on ST258 and its role in hospital outbreaks [43]. DeLeo et al subsequently fully closed 2 ST258 genomes and demonstrated that ST258 comprises at least 2 clades defined by differences in a 215-kb region including the cps locus [21]. Contrary to prior presumptions that carbapenem resistance in CRKP is due to frequent uptake of various blaKPC-encoding plasmids, a KPC-3 plasmid was stably associated with clade I [21], suggesting dissemination of a successful clone.
Subsequent studies revealed that recombination of large genomic fragments contributed to the emergence of both ST258 clades. Clade II is a hybrid of ST442 and ST11 fragments [22], while clade I likely originated from the hybrid clade II after a second recombination event involving the capsule locus from ST42. It has since been suggested that the CC258 cps locus [44] may comprise as many as 11 distinct loci [30]. PCR-based wzi typing suggests a stable association between blaKPC-harboring plasmids and select subclones: clade I isolates are characterized by wzi29 and KPC-2 and clade II by wzi154 and KPC-3 carriage [4, 25, 28]. The clinical implications of capsule variability in CC258 have yet to be fully determined.
WGS of 300 geographically and clinically diverse K. pneumoniae isolates derived from humans, animals, and the environment has helped to more clearly define the population structure of K. pneumoniae [20]. Four distinct Klebsiella phylogroups, likely representing 3 distinct species (K. pneumoniae, K. quasipneumoniae, and K. varicola), were identified, with overall little evidence of homologous recombination between them. K. pneumoniae belonged to a distinct phylogroup (KpI) containing opportunistic hospital-associated (HA) and CA invasive isolates. While a set of common genes was present in almost all genomes, isolates harbored thousands of additional accessory genes, suggesting that not lineage per se but rather gene content determines virulence [20]. In pan–genome-wide association analyses, rmpA/2, siderophore genes, and 5 additional predicted iron-metabolism genes were significantly associated with invasive infection as compared to carriage or noninvasive disease. Hypervirulent clones such as ST23 were significantly enriched for siderophores and other virulence genes, while acquisition of these genes within individual clones was associated with invasive disease and was generally limited to CA isolates. Acquired AMR genes were most common in HA isolates, particularly nosocomial carriage isolates, which harbored the highest number of these per isolate. This suggests that the accumulation of AMR genes in HA K. pneumoniae clones was a consequence of their successful spread through asymptomatic colonization, rather than a result of resistance [20]. Although biased or incomplete sampling of the colonizing reservoir may limit this interpretation, this study approach clearly demonstrates the strength of analyzing the breadth of K. pneumoniae isolates, rather than CRKP alone, to identify key genomic determinants of virulence and clonal success [20].
Given the frequency of resistance plasmid uptake among K. pneumoniae, concerns have been raised about the possible emergence of hypervirulent CRKP clones. In another WGS study [7], CCs associated with CA invasive infections (CC23, CC86, CC375, and CC380) harbored known virulence factors, including siderophores and rmpA, but lacked AMR. However, in 2 CC23 isolates both virulence and AMR genes (blaCTX-M and blaTEM) were detected. In addition, a small but increasing number of blaKPC-harboring HvKp isolates have been reported from China and Brazil, including those with additional resistance to polymyxin [45, 46]. While these are still isolated cases, extended-spectrum β-lactamase (ESBL)–positive isolates accounted for up to 12.5% of HvKp bloodstream isolates from a large geographic area [47].
Difficulties with plasmid assembly from short-read sequencing data have limited the contribution of WGS to analyses of plasmid-mediated dissemination of virulence and AMR genes in K. pneumoniae. As an alternative, a plasmid typing strategy based on plasmid replicon sequencing has been developed to facilitate the classification and tracking of plasmids [48]. This has allowed detection of plasmids associated with blaKPC and led to the identification of highly promiscuous plasmids associated with multistrain CRKP outbreaks. In a WGS-based assessment of KPC-Kp at a single institution, more than half of the 37 isolates collected >5 years belonged to 10 different STs harboring one of 2 novel conjugative plasmids [35]. New sequencing technologies, such as Pacific Biosciences and Oxford Nanopore sequencing, are capable of producing long sequencing reads and are likely to simplify plasmid analyses. These methods have recently been used to produce plasmid scaffolds for subsequent assembly [49, 50] and to reconstruct plasmid-associated outbreaks [51, 52].
K. PNEUMONIAE VIRULENCE MECHANISMS
K. pneumoniae uses a range of virulence factors to enhance invasiveness and overcome host defenses. Notably, studies of virulence factors in K. pneumoniae have been largely based on a number of well-defined reference strains characterized by virulence rather than drug resistance. Prominent reference strains include susceptible K. pneumoniae isolates ATCC 43816, a K2 clinical pneumonia isolate, and its rifampin-resistant derivative KPPR1; NTUH-K2044, a K1 isolate from a patient with PLA and meningitis [53, 54]; and MGH 78578, a K52 pneumonia isolate resistant to third- and fourth-generation cephalosporins but not carbapenems [55]. Both ATCC 43816 and NTUH-K2044 are highly virulent (median lethal dose [LD50], < 100 colony-forming units [CFU]) in a mouse pneumonia model [9, 54]. In contrast, MGH 78578 is relatively avirulent and is internalized at higher levels than ATCC 43816 and NTUH-K2044 [54]. The role of specific virulence determinants in K. pneumoniae pathogenesis have yet to be defined in a broader array of CA and nosocomial strains, and few have included CRKP and ST258 isolates. While CRKP strains likely lack strong virulence factors, they may harbor immune-evasive mechanisms that are only beginning to be comprehensively studied and understood.
Capsular Serotypes and Regulatory Genes
One of K. pneumoniae's most highly investigated virulence mechanism is the thick polysaccharide capsule that surrounds the organism and is encoded by the cps locus. The K. pneumoniae capsule has been shown to mediate resistance to antimicrobial peptides such as β-defensins, bind to and neutralize K antigen–specific antibodies, and mediate antiphagocytosis by human neutrophils and macrophages [56–58]. It may also play a role in biofilm formation [59]. Polysaccharide overproduction is associated with increased mucoviscosity and also produces a distinct virulence phenotype [60]. Mucoviscosity is mediated by a distinct genetic locus including rmpA/2 [61], while magA encodes a component of the CPS polymerase and is also required for capsule biosynthesis [62].
As described, the cps locus is highly variable and has been implicated in distinct clinical phenotypes. The epidemiological association of the K1 and K2 capsule types with invasive infection has been corroborated using mouse models [2]. In a mouse bacteremia model, capsule-switched recombinants were constructed from K2 and K21a parent strains [63]. Recombination of the K21a cps locus into a K2 background led to higher clearance from the bloodstream and decreased macrophage binding in the K2 recombinant, regardless of strain background. However, K2 cps locus recombination into a K21a parent strain produced isolates with intermediate virulence, suggesting that the K2 capsule is highly protective against phagocytosis, but it does not fully explain lethality in mice. Conversely, CRKP clones harbor an array of highly variable, non-K1/K2 capsular types exhibiting differential virulence potential in animal models. A Galleria mellonella wax worm model of ST258 isolates representing 3 different capsule types demonstrated variability in macrophage killing and virulence, although all strains were nonlethal in mice [25]. While the success of ST258 may be explained in part by capsular variability, which may potentiate evasion of the host immune response [21], this requires experimental validation.
CPS also plays a role in establishing infection and activating host immune responses [9]. In an intranasal inoculation mouse model, cps mutants generated from a wild-type (WT) KPPR1 strain were rapidly cleared from all tissues within 48 hours at an inoculum equivalent to the WT LD50 (104 CFU). At a high inoculum (107 CFU), cps mutants failed to disseminate and were gradually cleared from the trachea, although small numbers persisted in the lungs for days. In a follow-up study, cps mutants elicited an altered cytokine response, including increased interferon γ (IFN-γ) production, suggesting that the K. pneumoniae capsule dampens the immune response [64]. Histologically, WT infection induced a much more dramatic response in the lungs, although cps mutant–infected lung tissue was notable for the absence of neutrophils and large areas of lymphocyte infiltration, reflecting either increased influx or loss in WT infection.
CPS may also facilitate intestinal colonization with K. pneumoniae. In a mouse model, a WT LM21 strain persisted at high levels for up to 20 days, whereas capsule-defective mutants achieved only low-level colonization and appeared clumped [65]. In mixed infections, cps mutants were rapidly outcompeted by WT. In contrast, isogenic nonencapsulated variants of K. pneumoniae C105 and C3019 were as efficient as WT in colonizing the intestines of mice [66] but showed lower bacterial counts in bladders of transurethrally inoculated mice, suggesting complex site-specific interactions between CPS and host that require further study.
Alterations in Fimbriae, Porins, and Lipopolysaccharide (LPS)
Several membrane proteins play critical roles at the intersection of K. pneumoniae virulence and AMR. LPS is found on the outer bacterial membrane and is important in host immune system interactions. Its O antigen protrudes from the bacterial surface and has been shown to interfere with macrophage and neutrophil phagocytosis and cytokine production [11]. In murine models, mutants lacking the LPS O antigen had substantially decreased lethality as compared to WT animals and failed to produce invasive infection or showed rapid clearance of bacteremia [10, 11]. The LPS O antigen also contributed to virulence in CA strains causing PLA in an intraperitoneal mouse model [67]. Although O1-deficient strains were resistant to serum killing and C3 complement deposition, the O1 mutants showed decreased local proliferation and dissemination.
The lipid-anchoring component of LPS, lipid A, contains patterns that are recognized by the innate immune system and can trigger the host immune response [68]. Modification of lipid A in response to different host microenvironments is thought to help bacteria evade immune detection, promote antimicrobial peptide resistance, and alter outer membrane properties [68]. K. pneumoniae with lipid A modification had diminished activation of inflammatory responses, including less tumor necrosis factor α (TNF-α) secretion and activation of the nuclear factor κB (NF-κB) signaling cascade by macrophages in a mouse pneumonia model [68]. Interestingly, here K. pneumoniae was found to remodel its lipid A in a tissue-dependent manner. Lipid A species found in the lungs were consistent with a 2-hydroxyacyl–modified lipid A dependent on the PhoPQ-regulated oxygenase LpxO, leading to reduction in the activation of inflammatory responses and mediating colistin resistance.
Type 1 and 3 fimbriae are components of the K. pneumoniae cell membrane and play an important role in bacterial interaction with host cells, tissues, and environmental surfaces by mediating binding to both biological and nonbiological surfaces [69–71]. The type 3 fimbrial gene cluster includes mrkA, encoding the major fibrial subunit, and mrkD, encoding an adhesin polypeptide that mediates binding to epithelial cells and basement membrane components, including collagen [69]. Biofilm-deficient ATCC 43816 transposon mutants were able to express type 3 fimbriae and form biofilms following transformation with a plasmid containing the mrk gene cluster [70], with its structural (MrkA) component primarily responsible for biofilm formation. Here the transposon insertion site was a gene encoding a hypothetical transport protein of unclear significance. Expression of type 3 fimbriae and biofilm formation were regulated by the cyclic di-GMP–dependent transcriptional activator MrkH, likely in response to environmental factors [72]. Type 1 fimbriae are similarly expressed in a phase-variable manner mediated by an invertible DNA element [71] and are particularly important in establishing urinary tract infections, where expression appears highly upregulated as a result of enhanced on switching [71]. Type 1 fimbriae also potentiate adherence to host cells and contribute to biofilm formation, including on urinary catheters [73]. By facilitating interaction with host tissues and efficient outgrowth and biofilm formation, these cell membrane structures may potentiate K. pneumoniae colonization and persistence in vivo.
OmpA is a major integral protein of the outer membrane and also plays a role in adhesion to epithelial and other host cells and immune evasion [12]. In a series of in vitro and mouse pneumonia models, K. pneumoniae ompA mutants as compared to WT K. pneumoniae 52145 (K2) showed similar levels of binding to lung epithelial cells but induced higher levels of proinflammatory signaling and bacterial clearance from the lungs [12]. OmpK35 and OmpK36, the most important outer membrane porins in K. pneumoniae, also contribute to antiphagocytosis and virulence and have been widely implicated in AMR [18, 19]. Mutations in K. pneumoniae porin genes occur frequently in clinical isolates and increase carbapenem minimum inhibitory concentrations to the resistant range, particularly in combination with ESBL or AmpC β-lactamases [74]. AraC transcriptional proteins, the best studied of which is RamA, modulate bacterial permeability by regulating expression of several K. pneumoniae outer membrane proteins and have also been linked to virulence [75]. Overexpression of ramA, resulting from inactivating mutations of its repressor ramR, is associated with efflux pump modulation, porin downregulation, and modifications in the anchoring lipid A moiety of LPS, leading to reduced macrophage uptake and clearance in an intranasal mouse model [75]. Importantly, these alterations have been linked to increased resistance to the polymyxins and tigecycline [75–77], which are critical antibiotics of last resort used to treat CRKP.
Enhanced virulence or drug resistance frequently exerts a major fitness cost on organisms. Porin mutations are widespread in K. pneumoniae, and both ompK36 and ompK35/ompK36 double mutants showed increased susceptibility to neutrophil phagocytosis and reduce bacterial fitness [18, 19]. Similarly, laboratory-derived colistin-resistant mutants on the background of hypervirulent clinical ST23 isolates showed significantly reduced fitness as compared to parental strains [78]. Mutants also had decreased formation of CPS and expression of regulatory genes and a decrease in their hypervirulent phenotype. Matrix-assisted laser desorption/ionization time-of-flight analysis showed addition of aminoarabinose or palmitate residues to the lipid A moiety of LPS [78, 79].
Metabolic Alterations and Siderophores
Siderophores enable iron uptake into bacterial cells during infection and are thought to contribute substantially to the virulence potential of K. pneumoniae [13]. To date, 3 major siderophores have been implicated in K. pneumoniae virulence: yersiniabactin, enterobactin, and aerobactin [13, 14]. Yersiniabactin is among the most common K. pneumoniae virulence-associated genes and is associated with invasive respiratory tract infections in humans [20] and lethality in mouse pneumonia models [13, 14]. Yersiniabactin has been implicated in promotion of respiratory tract colonization and infection through evasion of lipocalin 2, an innate immune protein, owing to structural differences from other siderophores that limit lipocalin 2 binding [14]. Enterobactin appears to be important for growth under iron-limited conditions [13], whereas aerobactin is expressed by HvKP strains and has been shown to potentiate invasive infection in in vitro and mouse models [80]. Given that siderophores are highly linked to HvKP strains, they may play a particularly important role in CA invasive disease, raising the possibility that the contribution of capsule type to K. pneumoniae hypervirulence may be overestimated [20]. However, yersiniabactin has also been detected in several multidrug-resistant (MDR) clones, including KPC-Kp ST258 [14, 20], although its effect on clinical CRKP infections has not been determined.
Additional genes participating in iron metabolism and other metabolic pathways have been implicated in K. pneumoniae virulence. kfu is involved in iron acquisition and is associated with invasive infection and increased virulence in mice [15]. Clinical isolates causing PLA, including NTUH-K2044, were also found to have a unique chromosomal region containing genes involved in allantoin metabolism, which enables nitrogen fixation in K. pneumoniae, and to highly express the allS gene, which is required for growth in allantoin minimal medium [81]. The LD50 significantly decreased following intragastric inoculation of mice by an allS mutant, compared with WT K2044, but not intraperitoneal infection, suggesting genetic specialization for this virulence phenotype.
Contribution of Plasmids and Genomic Islands
Although ST258 has emerged as a globally dominant MDR clone associated with high mortality, it lacks capsule types and other factors associated with virulence [7]. Murine intratracheal and septicemia models have demonstrated that ST258 is nonlethal, including in high-inoculum infections [25, 82]. Given that ST258 is well known to primarily affect patients with immunocompromise and previous antibiotic exposure [3, 5], this supports its role as a nosocomial MDR pathogen producing opportunistic infections. ST258 isolates lacking KPC are remarkably uncommon [83], suggesting that the success of this clone depends to some degree on blaKPC, which it likely acquired prior to dissemination [44], and its resistance phenotype. Moreover, KPC-negative ST258 isolates showed limited persistence and lack of association with invasive infection, suggesting low epidemic potential. However, blaKPC carriage is unlikely to fully explain the selective emergence of ST258, including its apparent ability to disseminate rapidly in nosocomial settings.
The success of HvKP may also be in part due to plasmid-encoded virulence factors. Despite differences in clonal background, hypervirulent K1 and K2 strains contain homologues of a large virulence plasmid [84], which was detected in NTUH-K2044 (K1/ST23) [53] and closely resembles the pLVPK plasmid of a hypervirulent K2 isolate [85]. It harbors multiple virulence genes, including 2 siderophores and rmpA/2, and has been strongly associated with infection in humans [20]. This plasmid was detected in CC23 isolates and other diverse K2 isolates. Comparative genomics suggests that it was initially acquired once by each clone, with subsequent clonal expansion and divergence [84]. The possibility of transfer of plasmid-encoded virulence genes into CRKP isolates thus represents an alternative mechanism for intersection of hypervirulence and AMR.
Genomic islands may further harbor virulence determinants. In isolate Kp52.145, 4 islands were identified and included genes encoding the synthesis of a putative outer membrane cytotoxin, secretion systems, nucleases, and a phospholipase D family protein (PLD1) [86]. PLD1 likely plays a role in cardiolipin metabolism and was associated with K. pneumoniae isolates from severe infections. In a mouse pneumonia model, high-inoculum infection (108 CFU) with a pld1 mutant was nonlethal in mice, while infection with a WT and complemented strain produced 100% lethality.
HOST RESPONSE AND IMMUNE EVASIVE MECHANISMS
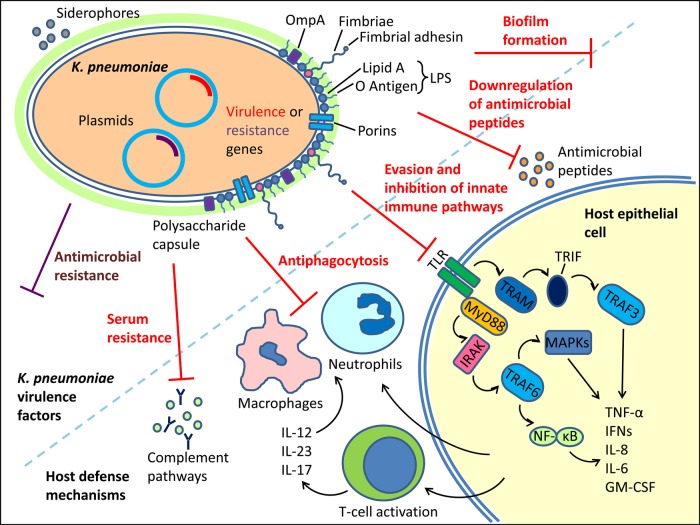
In addition to virulence mechanisms, K. pneumoniae uses a range of strategies to overcome or evade host immune clearance mechanisms (Figure 1). Broadly, these mechanisms address various components of the immune response, from innate immune signaling to phagocytosis, and they vary by strain and inoculation site. Most studies addressing K. pneumoniae–specific immune responses also use antibiotic-susceptible laboratory strains in murine pneumonia or other models. Whether CRKP and other nosocomial strains use specific immune evasive mechanisms to disseminate within hospital settings has yet to be determined.
Figure 1.
Schematic summary of Klebsiella pneumoniae, host cell response, and intracellular signaling. Capsular polysaccharide plays a critical role in infection, antiphagocytosis, and evading the host innate immune pathways. Additional virulence factors include other outer membrane proteins and siderophores. Antimicrobial resistance contributes to worse outcomes but has limited overlap with markers of virulence. Plasmid-encoded antimicrobial resistance or virulence harbors the potential for novel hypervirulent carbapenem-resistant K. pneumoniae clones. Abbreviations: GM-CSF, granulocyte-macrophage colony-stimulating factor; IFNs, interferons; IRAK, IL-1 receptor-associated kinase; IL-12, interleukin 12; IL-17, interleukin 17, IL-23, interleukin 23; LPS, lipopolysaccharide; MAPK, mitogen-activated protein kinase; NF-κB, nuclear factor κB; TLR, toll-like receptor.
Immune Signaling
Once K. pneumoniae organisms penetrate mechanical host defenses, they encounter pattern recognition receptors expressed by epithelial and immune cells, which are responsible for initiating proinflammatory signaling cascades and recruiting phagocytes (Figure 1). These receptors, of which Toll-like receptors (TLRs), including TLR2 and TLR4, are particularly important, respond primarily to cell wall components such as LPS and fimbriae, as well as to bacterial DNA [87]. In the respiratory tract, ligand binding triggers host signaling pathways via MyD88 and TRIF that lead to activation of NF-κB and mitogen-activated protein kinases (MAPKs) and, subsequently, to production of proinflammatory cytokines and chemokines [88]. MyD88-dependent signaling dominated over TRIF-dependent pathways in a mouse pneumonia model, where MyD88-deficient (MyD88−/−) mice showed more-pronounced reductions than TRIF−/− mice in bacterial clearance, cytokine expression levels, and neutrophil recruitment, as well as survival, compared with WT mice [88].
Well-documented study of the antibiotic-susceptible ATCC 43816 strains has revealed the importance of the early inflammatory response to the eradication of K. pneumoniae infections from the lung. Activation of interleukin 12 and interleukin 23p40 via these innate signaling pathways elicits downstream activation of IFN-γ, interleukin 17 (IL-17), TNF-α, and interleukin 1β (IL-1β) and leads to recruitment of neutrophils and macrophages and induction of T-cell–dependent humoral and cell-mediated immune responses [89]. In addition to MyD88, mice deficient in proteins that mediate signaling, such as the TNF receptor 1 (TNFR1) and STAT4, developed more-severe or lethal disease following infection with ATCC 43816 [90, 91]. IL-17A production in the setting of TNF-induced ILC3 lymphocyte recruitment in early infection was also critical in monocyte-mediated eradication of infection with K. pneumoniae ST258, in which neutrophil clearance plays a less important role [92]. IL-1 receptor–associated kinase M (IRAK-M) is a proximal inhibitor of TLR signaling expressed by epithelial cells and macrophages in the lung and is upregulated in lungs of WT mice following K. pneumoniae infection. IRAK-M−/− murine alveolar macrophages were more responsive to K. pneumoniae and K. pneumoniae LPS in vitro, and the extent of lung inflammation was increased in IRAK-M−/− mice shortly after intranasal K. pneumoniae inoculation [93]. However, IRAK-M−/− mice did not show increased cytokine or chemokine levels in their lungs after infection. The absence of IRAK-M resulted in a strongly improved host defense as reflected by reduced bacterial growth in the lungs, diminished dissemination to distant body sites, less peripheral tissue injury, and better survival rates.
Cellular Clearance
On the cellular level, neutrophils serve a critical role in the phagocytosis and intracellular killing of K. pneumoniae. Neutrophils are recruited to the site of infection in a T-helper type 17 cell signaling–dependent manner [89]. Other innate immune cells contributing to clearance of K. pneumoniae include natural killer cells, macrophages, and antigen-presenting cells, such as dendritic cells. Monocyte populations also play an important role in the effective host response to K. pneumoniae infection [92, 94]. Mice lacking resident lung macrophages were more likely to die when exposed to K. pneumoniae, owing to insufficient neutrophil recruitment. Macrophages themselves have been shown to be recruited by expression of CXCL10, which serves as a chemokine for a number of cell types. However, it appears that K. pneumoniae does not directly mediate strong activation of macrophages [95].
The expression of proinflammatory cytokines is also regulated by the receptor for advanced glycation end products (RAGE) [96]. This multiligand receptor of the immunoglobulin superfamily is expressed on different cell types and binds various endogenous innate danger signals (DAMPs or alarmins) and triggers several proinflammatory signaling cascades. RAGE was shown to play an important role in the effective antibacterial host response during K. pneumoniae infection, in part mediated by a reduced ability of phagocytosis in RAGE−/− neutrophils in vitro [96].
Immune Evasion Mechanisms
K. pneumoniae has evolved a number of mechanisms to evade host immune defenses, including recognition and killing by mediators of the innate and cellular immune systems. CPS and other bacterial membrane components play an important role in shielding K. pneumoniae against innate immune mechanisms, including resisting and downregulating antimicrobial β-defensins in the lungs, evading complement deposition and opsonization, and reducing recognition and adhesion by epithelial cells and phagocytes [97]. CPS has also been strongly implicated in resistance of K. pneumoniae to phagocytosis, in particular for some strain variants. More recently, sialic acid has been described as a non-immunogenic component of CPS because it is widely present in eukaryotic cells [98]. As discussed above, diversity in lipid A modifications induced in vivo has been suggested to counteract the innate immune response [68]. An ompA mutant was found to be important for K. pneumoniae immune evasion in vitro and in vivo by inducing the secretion of inflammatory mediators via activation of NF-KB and MAPKs [12]. Additional bacterial factors that inhibit opsonization are Omp, wzm (LPS O-antigen transporter), and the clps protease subunit [12, 99]. It has also been suggested that K. pneumoniae can survive within macrophages in a vacuolar compartment, triggering apoptotic pathways and avoiding delivery to lysosomes [100].
On the molecular level, a number of studies have dissected the antiinflammatory effects of K. pneumoniae infection in the mouse pneumonia model. One pathway involves the interference with the NF-κB canonical pathway by a K2 isolate (52145) [16, 101]. K. pneumoniae inhibited Rac1 activation, and inhibition of Rac1 activity triggered CYLD and MKP-1 expression via NOD1, an intracellular pattern recognition receptor that recognizes a peptidoglycan motif of gram-negative bacteria. This in turn attenuated IL-1β–induced interleukin 8 (IL-8) secretion, which was not seen in a cps mutant. However, purified CPS neither reduced IL-8 secretion nor induced the expression of CYLD and MKP-1, thereby indicating that CPS is necessary but not sufficient to attenuate inflammation. In contrast, CPS appears critical in the activation of the epidermal growth factor receptor signaling pathway. This in turn induced the expression of CYLD, which diminished the cytokine-dependent nuclear translocation of NF-κB [101]. Other putative triggers for activation of NF-κB include LPS and the pullulanase type II secretion system [17].
As highly successful CRKP clones appear to lack strong virulence factors, much of the observed disease burden is likely a reflection of an impaired host immune response, consistent with the clinical picture of these organisms causing infections in hospitalized, immunocompromised individuals [82]. However, evidence has been accumulating that CRKP may have acquired a unique ability to evade and inhibit host innate immune clearance from lung tissue. This may involve a relative lack of immunogenicity, enabling persistence in the airway and avoiding phagocytosis. It currently remains unclear whether this is due to a targeted antiinflammatory process, inhibition of host cell signaling, or structural differences in pathogen-associated molecular patterns [102]. The decreased virulence of CRKP might also reflect adaptation to inhibit the proinflammatory signaling necessary for clearance by phagocytes. As such, this would enhance the colonization potential and long-term persistence, even during antibiotic pressure. Other studies suggest isolate-specific differences between K. pneumoniae and CRKP isolates in the cellular immune response. When comparing the ATCC isolate to 4 clinical samples, including one CRKP ST258 isolate [103], there was isolate-dependent variation in the contribution of neutrophils and CCR2+ monocytes to the clearance of K. pneumoniae pulmonary infection. Monocyte depletion worsened infection with all 5 isolates, whereas neutrophils contributed to the clearance of all but the CRKP isolates [103].
SUMMARY AND FUTURE DIRECTIONS
Much progress has been made on our understanding of the molecular and cellular factors governing the recent emergence of CRKP infections. WGS has emerged as a powerful tool to more comprehensively identify novel virulence or resistance factors in successful clones and has painted a picture of a species with high genome plasticity. These studies largely benefit from considering the entire K. pneumoniae population structure, rather than focusing on select resistant or virulent clones alone. Comparison of CRKP with coemerging HvKP strains has facilitated identification of virulence factors and demonstrated minimal overlap in AMR and virulence gene content between CRKP and HvKP clonal lineages. By more clearly mapping the location of AMR and virulence genes, they support 2 mechanisms for potential widespread Hv-CRKP: transfer of blaKPC-plasmids into HvKP or transfer of plasmids harboring virulence genes into CRKP isolates. Last, many virulence and host-pathogen investigations have relied on laboratory-adapted, nonresistant K. pneumoniae. However, preliminary evidence suggests a high diversity in infection potential of single isolates. Using clinically diverse isolates in genomic and functional studies may provide important insights into the clinical course of infection, including how K. pneumoniae thwarts the host immune response, and the identification of targets for adjunct immunotherapies.
Notes
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (R01AI116939 and R01AI116939-S01 to A.-C. U.) and the Columbia University Irving Institute (to A. G.-S. and A.-C. U.).
Potential conflict of interest. A.-C. U. has received research grants from Merck. A. G.-S. certifies no potential conflicts of interest. Both authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States, 2013. http://www.cdc.gov/drugresistance/threat-report-2013/. Accessed 15 March 2016.
- 2. Siu LK, Yeh KM, Lin JC, Fung CP, Chang FY. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect Dis 2012; 12:881–7. [DOI] [PubMed] [Google Scholar]
- 3. Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis 2009; 9:228–36. [DOI] [PubMed] [Google Scholar]
- 4. Gomez-Simmonds A, Greenman M, Sullivan SB et al. Population structure of Klebsiella pneumoniae causing bloodstream infections at a New York City tertiary care hospital: Diversification of multidrug-resistant isolates. J Clin Microbiol 2015; 53:2060–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Munoz-Price LS, Poirel L, Bonomo RA et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 2013; 13:785–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dortet L, Poirel L, Nordmann P. Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. Biomed Res Int 2014; 2014:249856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bialek-Davenet S, Criscuolo A, Ailloud F et al. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis 2014; 20:1812–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brisse S, Fevre C, Passet V et al. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS One 2009; 4:e4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lawlor MS, Hsu J, Rick PD, Miller VL. Identification of Klebsiella pneumoniae virulence determinants using an intranasal infection model. Mol Microbiol 2005; 58:1054–73. [DOI] [PubMed] [Google Scholar]
- 10. Shankar-Sinha S, Valencia GA, Janes BK et al. The Klebsiella pneumoniae O antigen contributes to bacteremia and lethality during murine pneumonia. Infect Immun 2004; 72:1423–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lugo JZ, Price S, Miller JE et al. Lipopolysaccharide O-antigen promotes persistent murine bacteremia. Shock 2007; 27:186–91. [DOI] [PubMed] [Google Scholar]
- 12. March C, Moranta D, Regueiro V et al. Klebsiella pneumoniae outer membrane protein A is required to prevent the activation of airway epithelial cells. J Biol Chem 2011; 286:9956–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lawlor MS, O'Connor C, Miller VL. Yersiniabactin is a virulence factor for Klebsiella pneumoniae during pulmonary infection. Infect Immun 2007; 75:1463–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bachman MA, Oyler JE, Burns SH et al. Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipocalin 2. Infect Immun 2011; 79:3309–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ma LC, Fang CT, Lee CZ, Shun CT, Wang JT. Genomic heterogeneity in Klebsiella pneumoniae strains is associated with primary pyogenic liver abscess and metastatic infection. J Infect Dis 2005; 192:117–28. [DOI] [PubMed] [Google Scholar]
- 16. Regueiro V, Moranta D, Frank CG et al. Klebsiella pneumoniae subverts the activation of inflammatory responses in a NOD1-dependent manner. Cell Microbiol 2011; 13:135–53. [DOI] [PubMed] [Google Scholar]
- 17. Tomas A, Lery L, Regueiro V et al. Functional Genomic Screen Identifies Klebsiella pneumoniae Factors Implicated in Blocking Nuclear Factor kappaB (NF-kappaB) Signaling. J Biol Chem 2015; 290:16678–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen JH, Siu LK, Fung CP et al. Contribution of outer membrane protein K36 to antimicrobial resistance and virulence in Klebsiella pneumoniae. J Antimicrob Chemother 2010; 65:986–90. [DOI] [PubMed] [Google Scholar]
- 19. Tsai YK, Fung CP, Lin JC et al. Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence. Antimicrob Agents Chemother 2011; 55:1485–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Holt KE, Wertheim H, Zadoks RN et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A 2015; 112:E3574–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Deleo FR, Chen L, Porcella SF et al. Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc Natl Acad Sci U S A 2014; 111:4988–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen L, Mathema B, Pitout JD, DeLeo FR, Kreiswirth BN. Epidemic Klebsiella pneumoniae ST258 is a hybrid strain. MBio 2014; 5:e01355–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 2005; 43:4178–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cryz SJ Jr, Mortimer PM, Mansfield V, Germanier R. Seroepidemiology of Klebsiella bacteremic isolates and implications for vaccine development. J Clin Microbiol 1986; 23:687–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Diago-Navarro E, Chen L, Passet V et al. Carbapenem-resistant Klebsiella pneumoniae exhibit variability in capsular polysaccharide and capsule associated virulence traits. J Infect Dis 2014; 210:803–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pan YJ, Lin TL, Chen CT et al. Genetic analysis of capsular polysaccharide synthesis gene clusters in 79 capsular types of Klebsiella spp. Sci Rep 2015; 5:15573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pan YJ, Lin TL, Lin YT et al. Identification of capsular types in carbapenem-resistant Klebsiella pneumoniae strains by wzc sequencing and implications for capsule depolymerase treatment. Antimicrob Agents Chemother 2015; 59:1038–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brisse S, Passet V, Haugaard AB et al. wzi Gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J Clin Microbiol 2013; 51:4073–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brisse S, Issenhuth-Jeanjean S, Grimont PA. Molecular serotyping of Klebsiella species isolates by restriction of the amplified capsular antigen gene cluster. J Clin Microbiol 2004; 42:3388–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wyres KL, Gorrie C, Edwards DJ et al. Extensive capsule locus variation and large-scale genomic recombination within the Klebsiella pneumoniae clonal group 258. Genome Biol Evol 2015; 7:1267–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen L, Mathema B, Chavda KD et al. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol 2014; 22:686–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cuzon G, Naas T, Nordmann P. Functional characterization of Tn4401, a Tn3-based transposon involved in blaKPC gene mobilization. Antimicrob Agents Chemother 2011; 55:5370–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Poirel L, Potron A, Nordmann P. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother 2012; 67:1597–606. [DOI] [PubMed] [Google Scholar]
- 34. Peirano G, Lascols C, Hackel M, Hoban DJ, Pitout JD. Molecular epidemiology of Enterobacteriaceae that produce VIMs and IMPs from the SMART surveillance program. Diagn Microbiol Infect Dis 2014; 78:277–81. [DOI] [PubMed] [Google Scholar]
- 35. Mathers AJ, Stoesser N, Sheppard AE et al. Klebsiella pneumoniae carbapenemase (KPC)-producing K.: pneumoniae at a single institution: insights into endemicity from whole-genome sequencing. Antimicrob Agents Chemother 2015; 59:1656–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bonura C, Giuffre M, Aleo A et al. An Update of the Evolving Epidemic of blaKPC Carrying Klebsiella pneumoniae in Sicily, Italy 2014: Emergence of Multiple Non-ST258 Clones. PLoS One 2015; 10:e0132936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ocampo AM, Chen L, Cienfuegos AV et al. A two-year surveillance in five colombian tertiary care hospitals reveals high frequency of non-CG258 clones of carbapenem-resistant Klebsiella pneumoniae with distinct clinical characteristics. Antimicrob Agents Chemother 2015; 60:332–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ruiz-Garbajosa P, Curiao T, Tato M et al. Multiclonal dispersal of KPC genes following the emergence of non-ST258 KPC-producing Klebsiella pneumoniae clones in Madrid, Spain. J Antimicrob Chemother 2013; 68:2487–92. [DOI] [PubMed] [Google Scholar]
- 39. Gaiarsa S, Comandatore F, Gaibani P et al. Genomic epidemiology of Klebsiella pneumoniae in Italy and novel insights into the origin and global evolution of its resistance to carbapenem antibiotics. Antimicrob Agents Chemother 2015; 59:389–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Turton JF, Englender H, Gabriel SN et al. Genetically similar isolates of Klebsiella pneumoniae serotype K1 causing liver abscesses in three continents. J Med Microbiol 2007; 56(Pt 5):593–7. [DOI] [PubMed] [Google Scholar]
- 41. Luo Y, Wang Y, Ye L, Yang J. Molecular epidemiology and virulence factors of pyogenic liver abscess causing Klebsiella pneumoniae in China. Clin Microbiol Infect 2014; 20:O818–24. [DOI] [PubMed] [Google Scholar]
- 42. Lin JC, Koh TH, Lee N et al. Genotypes and virulence in serotype K2 Klebsiella pneumoniae from liver abscess and non-infectious carriers in Hong Kong, Singapore and Taiwan. Gut Pathog 2014; 6:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Snitkin ES, Zelazny AM, Thomas PJ et al. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med 2012; 4:148ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bowers JR, Kitchel B, Driebe EM et al. Genomic analysis of the emergence and rapid global dissemination of the clonal group 258 Klebsiella pneumoniae pandemic. PLoS One 2015; 10:e0133727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang Y, Zeng J, Liu W et al. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J Infect 2015; 71:553–60. [DOI] [PubMed] [Google Scholar]
- 46. Andrade LN, Vitali L, Gaspar GG et al. Expansion and evolution of a virulent, extensively drug-resistant (polymyxin B-resistant), QnrS1-, CTX-M-2-, and KPC-2-producing Klebsiella pneumoniae ST11 international high-risk clone. J Clin Microbiol 2014; 52:2530–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang Y, Zhao C, Wang Q et al. High prevalence of hypervirulent Klebsiella pneumoniae infection in China: geographic distribution, clinical characteristics and antimicrobial resistance. Antimicrob Agents Chemother 2016:pii:AAC.01127-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Carattoli A, Bertini A, Villa L et al. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 2005; 63:219–28. [DOI] [PubMed] [Google Scholar]
- 49. Wright MS, Perez F, Brinkac L et al. Population structure of KPC-producing Klebsiella pneumoniae isolates from midwestern U.S.: hospitals. Antimicrob Agents Chemother 2014; 58:4961–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Turton JF, Doumith M, Hopkins KL et al. Clonal expansion of Escherichia coli ST38 carrying chromosomally-integrated OXA-48 carbapenemase gene. J Med Microbiol 2016; 65:538–46. [DOI] [PubMed] [Google Scholar]
- 51. Conlan S, Thomas PJ, Deming C et al. Single-molecule sequencing to track plasmid diversity of hospital-associated carbapenemase-producing Enterobacteriaceae. Sci Transl Med 2014; 6:254ra126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Marsh JW, Krauland MG, Nelson JS et al. Genomic epidemiology of an endoscope-associated outbreak of Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae. PLoS One 2015; 10:e0144310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wu KM, Li LH, Yan JJ et al. Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J Bacteriol 2009; 191:4492–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fodah RA, Scott JB, Tam HH et al. Correlation of Klebsiella pneumoniae comparative genetic analyses with virulence profiles in a murine respiratory disease model. PLoS One 2014; 9:e107394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ogawa W, Li DW, Yu P et al. Multidrug resistance in Klebsiella pneumoniae MGH78578 and cloning of genes responsible for the resistance. Biol Pharm Bull 2005; 28:1505–8. [DOI] [PubMed] [Google Scholar]
- 56. Domenico P, Salo RJ, Cross AS, Cunha BA. Polysaccharide capsule-mediated resistance to opsonophagocytosis in Klebsiella pneumoniae. Infect Immun 1994; 62:4495–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Simoons-Smit AM, Verweij-van Vught AM, MacLaren DM. The role of K antigens as virulence factors in Klebsiella. J Med Microbiol 1986; 21:133–7. [DOI] [PubMed] [Google Scholar]
- 58. Campos MA, Vargas MA, Regueiro V et al. Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect Immun 2004; 72:7107–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Balestrino D, Ghigo JM, Charbonnel N, Haagensen JA, Forestier C. The characterization of functions involved in the establishment and maturation of Klebsiella pneumoniae in vitro biofilm reveals dual roles for surface exopolysaccharides. Environ Microbiol 2008; 10:685–701. [DOI] [PubMed] [Google Scholar]
- 60. Yu WL, Ko WC, Cheng KC et al. Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin Infect Dis 2006; 42:1351–8. [DOI] [PubMed] [Google Scholar]
- 61. Cheng HY, Chen YS, Wu CY et al. RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J Bacteriol 2010; 192:3144–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lin TL, Yang FL, Yang AS et al. Amino acid substitutions of MagA in Klebsiella pneumoniae affect the biosynthesis of the capsular polysaccharide. PLoS One 2012; 7:e46783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kabha K, Nissimov L, Athamna A et al. Relationships among capsular structure, phagocytosis, and mouse virulence in Klebsiella pneumoniae. Infect Immun 1995; 63:847–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lawlor MS, Handley SA, Miller VL. Comparison of the host responses to wild-type and cpsB mutant Klebsiella pneumoniae infections. Infect Immun 2006; 74:5402–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Favre-Bonte S, Licht TR, Forestier C, Krogfelt KA. Klebsiella pneumoniae capsule expression is necessary for colonization of large intestines of streptomycin-treated mice. Infect Immun 1999; 67:6152–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Struve C, Krogfelt KA. Role of capsule in Klebsiella pneumoniae virulence: lack of correlation between in vitro and in vivo studies. FEMS Microbiol Lett 2003; 218:149–54. [DOI] [PubMed] [Google Scholar]
- 67. Hsieh PF, Lin TL, Yang FL et al. Lipopolysaccharide O1 antigen contributes to the virulence in Klebsiella pneumoniae causing pyogenic liver abscess. PLoS One 2012; 7:e33155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Llobet E, Martinez-Moliner V, Moranta D et al. Deciphering tissue-induced Klebsiella pneumoniae lipid A structure. Proc Natl Acad Sci U S A 2015; 112:E6369–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sebghati TA, Korhonen TK, Hornick DB, Clegg S. Characterization of the type 3 fimbrial adhesins of Klebsiella strains. Infect Immun 1998; 66:2887–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Langstraat J, Bohse M, Clegg S. Type 3 fimbrial shaft (MrkA) of Klebsiella pneumoniae, but not the fimbrial adhesin (MrkD), facilitates biofilm formation. Infect Immun 2001; 69:5805–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Struve C, Bojer M, Krogfelt KA. Characterization of Klebsiella pneumoniae type 1 fimbriae by detection of phase variation during colonization and infection and impact on virulence. Infect Immun 2008; 76:4055–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Struve C, Bojer M, Krogfelt KA. Positive autoregulation of mrkHI by the cyclic di-GMP-dependent MrkH protein in the biofilm regulatory circuit of Klebsiella pneumoniae. J Bacteriol 2015; 197:1659–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Stahlhut SG, Struve C, Krogfelt KA, Reisner A. Biofilm formation of Klebsiella pneumoniae on urethral catheters requires either type 1 or type 3 fimbriae. FEMS Immunol Med Microbiol 2012; 65:350–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Doumith M, Ellington MJ, Livermore DM, Woodford N. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp.: clinical isolates from the UK. J Antimicrob Chemother 2009; 63:659–67. [DOI] [PubMed] [Google Scholar]
- 75. De Majumdar S, Yu J, Fookes M et al. Elucidation of the RamA regulon in Klebsiella pneumoniae reveals a role in LPS regulation. PLoS Pathog 2015; 11:e1004627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ruzin A, Visalli MA, Keeney D, Bradford PA. Influence of transcriptional activator RamA on expression of multidrug efflux pump AcrAB and tigecycline susceptibility in Klebsiella pneumoniae. Antimicrob Agents Chemother 2005; 49:1017–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hentschke M, Wolters M, Sobottka I, Rohde H, Aepfelbacher M. ramR mutations in clinical isolates of Klebsiella pneumoniae with reduced susceptibility to tigecycline. Antimicrob Agents Chemother 2010; 54:2720–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Choi MJ, Ko KS. Loss of hypermucoviscosity and increased fitness cost in colistin-resistant Klebsiella pneumoniae sequence type 23 strains. Antimicrob Agents Chemother 2015; 59:6763–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kim SY, Choi HJ, Ko KS. Differential expression of two-component systems, pmrAB and phoPQ, with different growth phases of Klebsiella pneumoniae in the presence or absence of colistin. Curr Microbiol 2014; 69:37–41. [DOI] [PubMed] [Google Scholar]
- 80. Russo TA, Olson R, MacDonald U, Beanan J, Davidson BA. Aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniae ex vivo and in vivo. Infect Immun 2015; 83:3325–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chou HC, Lee CZ, Ma LC et al. Isolation of a chromosomal region of Klebsiella pneumoniae associated with allantoin metabolism and liver infection. Infect Immun 2004; 72:3783–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tzouvelekis LS, Miriagou V, Kotsakis SD et al. KPC-producing, multidrug-resistant Klebsiella pneumoniae sequence type 258 as a typical opportunistic pathogen. Antimicrob Agents Chemother 2013; 57:5144–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Adler A, Paikin S, Sterlin Y et al. A swordless knight: epidemiology and molecular characteristics of the blaKPC-negative sequence type 258 Klebsiella pneumoniae clone. J Clin Microbiol 2012; 50:3180–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Struve C, Roe CC, Stegger M et al. Mapping the Evolution of Hypervirulent Klebsiella pneumoniae. MBio 2015; 6:e00630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Chen YT, Chang HY, Lai YC et al. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene 2004; 337:189–98. [DOI] [PubMed] [Google Scholar]
- 86. Lery LM, Frangeul L, Tomas A et al. Comparative analysis of Klebsiella pneumoniae genomes identifies a phospholipase D family protein as a novel virulence factor. BMC Biol 2014; 12:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Takeda K, Akira S. Toll receptors and pathogen resistance. Cell Microbiol 2003; 5:143–53. [DOI] [PubMed] [Google Scholar]
- 88. Cai S, Batra S, Shen L, Wakamatsu N, Jeyaseelan S. Both TRIF- and MyD88-dependent signaling contribute to host defense against pulmonary Klebsiella infection. J Immunol 2009; 183:6629–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Happel KI, Dubin PJ, Zheng M et al. Divergent roles of IL-23 and IL-12 in host defense against Klebsiella pneumoniae. J Exp Med 2005; 202:761–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Moore TA, Perry ML, Getsoian AG et al. Increased mortality and dysregulated cytokine production in tumor necrosis factor receptor 1-deficient mice following systemic Klebsiella pneumoniae infection. Infect Immun 2003; 71:4891–00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Deng JC, Zeng X, Newstead M et al. STAT4 is a critical mediator of early innate immune responses against pulmonary Klebsiella infection. J Immunol 2004; 173:4075–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Xiong H, Keith JW, Samilo DW et al. Innate Lymphocyte/Ly6C(hi) Monocyte Crosstalk Promotes Klebsiella Pneumoniae Clearance. Cell 2016; 165:679–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hoogerwerf JJ, van der Windt GJ, Blok DC et al. Interleukin-1 receptor-associated kinase M-deficient mice demonstrate an improved host defense during Gram-negative pneumonia. Mol Med 2012; 18:1067–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Herold S, Tabar TS, Janssen H et al. Exudate macrophages attenuate lung injury by the release of IL-1 receptor antagonist in gram-negative pneumonia. Am J Respir Crit Care Med 2011; 183:1380–90. [DOI] [PubMed] [Google Scholar]
- 95. Zeng X, Moore TA, Newstead MW et al. Interferon-inducible protein 10, but not monokine induced by gamma interferon, promotes protective type 1 immunity in murine klebsiella pneumoniae pneumonia. infect immun 2005; 73:8226–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Achouiti A, de Vos AF, van 't Veer C et al. receptor for advanced glycation end products (RAGE) serves a protective role during Klebsiella pneumoniae - induced pneumonia. PLoS One 2016; 11:e0141000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Moranta D, Regueiro V, March C et al. Klebsiella pneumoniae capsule polysaccharide impedes the expression of beta-defensins by airway epithelial cells. Infect Immun 2010; 78:1135–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Lee CH, Chang CC, Liu JW, Chen RF, Yang KD. Sialic acid involved in hypermucoviscosity phenotype of Klebsiella pneumoniae and associated with resistance to neutrophil phagocytosis. Virulence 2014; 5:673–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Pan PC, Chen HW, Wu PK et al. Mutation in fucose synthesis gene of Klebsiella pneumoniae affects capsule composition and virulence in mice. Exp Biol Med (Maywood) 2011; 236:219–26. [DOI] [PubMed] [Google Scholar]
- 100. Cano V, March C, Insua JL et al. Klebsiella pneumoniae survives within macrophages by avoiding delivery to lysosomes. Cell Microbiol 2015; 17:1537–60. [DOI] [PubMed] [Google Scholar]
- 101. Frank CG, Reguerio V, Rother M et al. Klebsiella pneumoniae targets an EGF receptor-dependent pathway to subvert inflammation. Cell Microbiol 2013; 15:1212–33. [DOI] [PubMed] [Google Scholar]
- 102. Parker D, Ahn D, Cohen T, Prince A. Innate immune signaling activated by MDR bacteria in the airway. Physiol Rev 2016; 96:19–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Xiong H, Carter RA, Leiner IM et al. Distinct contributions of neutrophils and CCR2+ monocytes to pulmonary clearance of different Klebsiella pneumoniae strains. Infect Immun 2015; 83:3418–27. [DOI] [PMC free article] [PubMed] [Google Scholar]