Summary
To meet the need for new pathways to develop antibacterial agents to treat drug-resistant infections, we present feasible trial designs that could enable conduct of narrow-spectrum clinical trials and ultimately approval of critically needed new antibacterial agents.
Keywords: antimicrobial resistance, antibiotic development
Abstract
Despite progress in antimicrobial drug development, a critical need persists for new, feasible pathways to develop antibacterial agents to treat people infected with drug-resistant bacteria. Infections due to resistant gram-negative bacilli continue to cause unacceptable morbidity and mortality rates. Antibacterial agents have been historically studied in noninferiority clinical trials that focus on a single site of infection (eg, complicated urinary tract infections, intra-abdominal infections), yet these designs may not be optimal, and often are not feasible, for study of infections caused by drug-resistant bacteria. Over the past several years, multiple stakeholders have worked to develop consensus regarding paths forward with a goal of facilitating timely conduct of antimicrobial development. Here we advocate for a novel and pragmatic approach and, toward this end, present feasible trial designs for antibacterial agents that could enable conduct of narrow-spectrum, organism-specific clinical trials and ultimately approval of critically needed new antibacterial agents.
(See the editorial commentary by Drusano, on pages 150–2.)
Rising rates of antimicrobial resistance increasingly threaten physicians’ ability to provide care that relies on effective antimicrobial drugs. Globally, it is conservatively estimated that >700000 persons die annually due to infections caused by multidrug-resistant pathogens, with millions more suffering serious infectious complications. For these patients, the threat of a postantibiotic era is already a devastating reality. Left unchecked, annual deaths attributable to antimicrobial resistance are estimated to reach 10 million by 2050, outpacing cancer, diabetes, diarrheal diseases, and automobile accidents [1]. Physicians are often forced to treat these infections using drugs with limited clinical data and/or significant toxic effects. For example, colistin and polymixin B are now routinely administered via inhaled and parenteral routes to treat multidrug-resistant infections, including pneumonia and bloodstream and abdominal infections, despite a paucity of data supporting dosing, efficacy, or safety for these indications [2]. In outpatients, fosfomycin is frequently administered to treat infections caused by extended-spectrum β-lactamase–producing gram-negative organisms, despite the lack of data supporting its use for indications other than uncomplicated cystitis [1].
Further accelerating global anxiety, a transferable colistin resistance gene, mcr-1, was discovered in China in November 2015; a second transferrable colisitin resistance gene, mcr-2, was reported shortly thereafter [3]. This development is particularly worrisome because colistin is a last-resort antibiotic for treatment of infection caused by many multidrug-resistant bacteria [4]. The mcr-1 gene is thought to have emerged in relation to high levels of agricultural colistin use in China. It was soon recognized in other countries in Asia, Europe, and North America—including the United States—reminding us that antimicrobial resistance does not respect national borders [5]. This gene has been found in carbapenem-resistant organisms (blaKPC and blaNDM organisms), demonstrating that true pan-resistant pathogens are possible [6].
The global response to the continued escalation of antimicrobial resistance requires a unified, multidimensional approach of increased epidemiological surveillance, appropriate antimicrobial use in humans and animals, enhanced infection prevention, and development of new therapies by reinvigorating the antimicrobial research and development pipeline. Efforts include the Combating Antibiotic-resistant Bacteria Action Plan and Presidential Advisory Council on Combating Antibiotic-Resistant Bacteria (PACCARB) in the United States, the Innovative Medicines Initiative New Drugs for Bad Bugs program, CARB-X, the World Health Organization, and the Margolis Center for Health Policy at Duke University, as well as the recent high-level United Nations meeting [6–8]. Stewardship and prevention efforts, though essential, can only slow or limit the development of resistance, not stop it, because certain infections still require antimicrobial therapy. Thus, patients will continue to need new antimicrobial treatment options.
Focusing on development of new antimicrobial drugs, pathways are needed to ensure the availability of a diverse, renewable and robust pipeline of antimicrobial agents. The most pressing needs include drugs to treat the most resistant gram-negative pathogens, including narrow-spectrum agents targeting resistant Pseudomonas aeruginosa and Acinetobacter and Klebsiella species. Fortunately, infections caused by such multidrug-resistant or extensively drug-resistant pathogens are uncommon overall and are encountered almost exclusively in critically ill patients, who are difficult to include in clinical trials. However, in this complex patient population, these infections are being encountered more frequently, underscoring the urgent need for new therapeutic options. This white paper aims to provide options for developing drugs to treat infections caused by a single pathogen and/or in the population of patients with the greatest unmet medical need.
Over the past several years, stakeholder groups, including the US Food and Drug Administration (FDA), the Infectious Diseases Society of America, and the National Institutes of Health, have met to discuss potential means of developing antimicrobial drugs for unmet needs, specifically, for drugs targeting a single pathogen. Developing narrow-spectrum drugs has been difficult outside a few specific examples, including certain infections caused by Staphylococcus aureus (acute bacterial skin and skin structure infection), Neisseria gonorrheae (sexually transmitted infection), Clostridium difficile infection, and P. aeruginosa infections in patients with cystic fibrosis. A path is desperately needed for the development of narrow-spectrum agents for other indications [9, 10]. Major gaps also remain for treating infections caused by P. aeruginosa and Acinetobacter and Klebsiella species; however, plausible candidate drugs and pathways are emerging. For the study of these life-threatening infections, patients are few, and neither noninferiority nor superiority designs are routinely feasible. Development of rapid diagnostic tests may help but alone cannot solve the problem, because they serve only to identify these patients—the patients are (fortunately) rare, and large screening efforts would still be needed.
STAKEHOLDER INPUT: FDA WORKSHOP
In July 2016, the FDA convened a 2-day workshop to address challenges and emphasize the need for new options in developing narrow-spectrum drugs [11]. The discussion focused on a very specific example of a hypothetical drug candidate to treat P. aeruginosa infection [11]. This hypothetical drug candidate was of a novel class and had activity limited to P. aeruginosa. In vitro resistance was uncommon and seemed to develop only rarely. The hypothetical parenteral drug had a straightforward pharmacological mechanism and pharmacokinetic-pharmacodynamic (PK/PD) information that identified a well-justified target exposure, and a dose regimen was found that produced this exposure. The drug was found to penetrate into the human lung in a phase 1 epithelial lining fluid study and was shown in a small phase 2 study to have an effect on bacterial burden in adults with bronchiectasis not associated with cystic fibrosis. In short, the hypothetical drug candidate was an entirely plausible candidate for development.
The problem then posed at the workshop was that the clinical development program would be difficult owing to the relatively low frequency of this pathogen at any given body site (data from a single body site, rather than data pooled across multiple body sites, is typically currently required for FDA approval) and the need for initial treatment with a second agent to ensure that the empirical spectrum was adequate. Hypothetical preclinical, phase 1, and phase 2 data were provided, and the workshop participants were tasked with thinking about options for a clinical program. Several phase 3 scenarios were presented and showed increasingly difficult situations built around the theme of a narrow-spectrum agent that targets a low-frequency pathogen. The ability to enroll patients with the target pathogen was progressively reduced in each scenario. Time was devoted to debating the merits of the scenarios, as well as a number of other options. After 2 days of deliberation, all agreed that no single option was perfect and that several seemed less desirable. Consensus did emerge, however, regarding key elements of both preclinical and clinical development of drugs for such pathogens, and although no single path was identified as most optimal, we submit that these key elements can be applied to each candidate as a means toward successful development.
DEVELOPMENT PROGRAM SCENARIOS
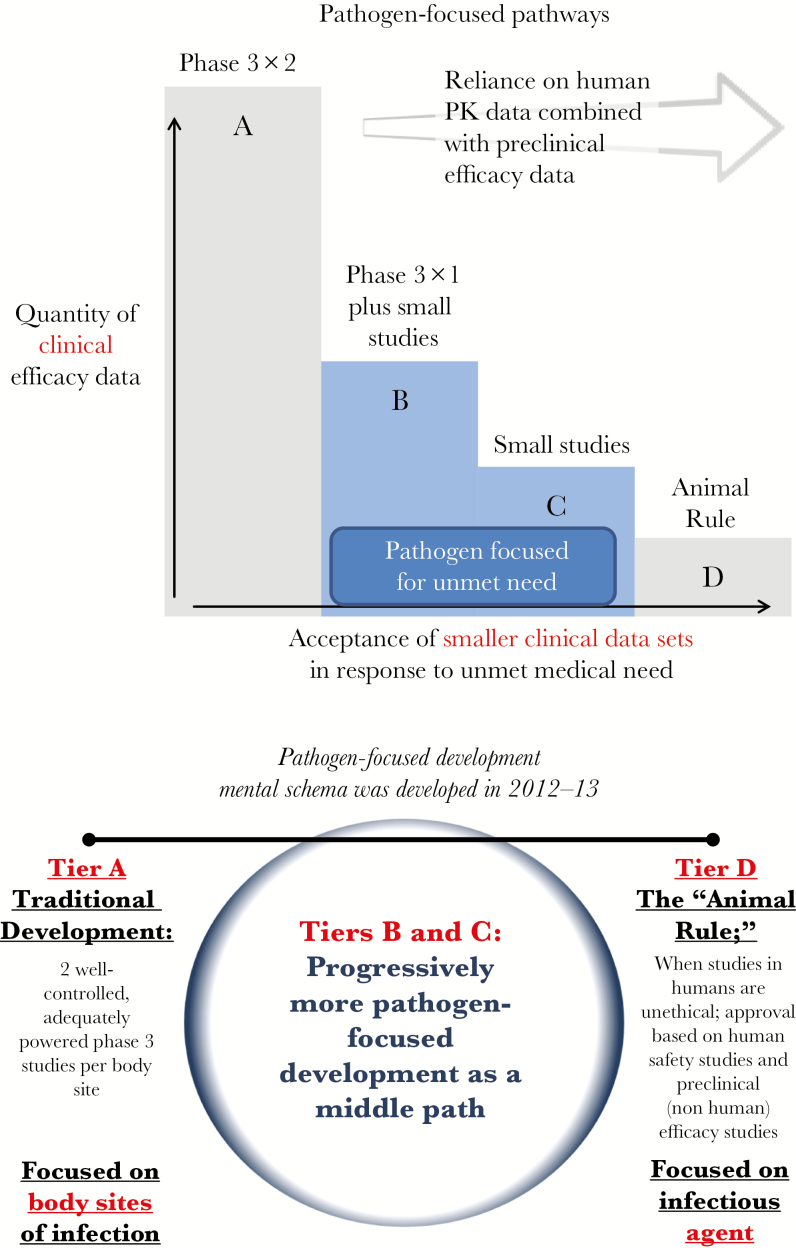
In presenting the development scenarios, a published framework was reviewed for tiers of approval from standard programs with plentiful clinical data to very small programs with limited data based on the quantity of clinical efficacy data that can be generated [12]. Within this framework, moving from tier A to tier D requires increased reliance on human pharmacokinetic data combined with preclinical efficacy data, and ultimately on the Animal Rule in certain circumstances (Figure 1 and Table 1; Animal Rule discussed in detail below). Accordingly, wording in the approved drug labeling reflects “areas of residual uncertainty” [12].
Figure 1.
Pathogen-focused pathways and development [12].
Table 1.
Tiered Labeling Framework for Development of Antibacterial Agentsa
| Tier | Typical Efficacy Database | Typical Label Wording |
|---|---|---|
| A | Two phase 3 trialsb in infection Y; additional indications can be added after single phase 3 studies | Drug X is indicated for treatment of infection Y when proved or strongly suspected to be caused by drug X–susceptible strains of [list of pathogens] |
| B | One phase 3 trialb in infection Y, plus small prospective studies and descriptive data focused on the tier C pathogen(s) in a range of standard infections | Drug X is indicated for treatment of infection Y and [list of studied infections from tier C database] when proved or strongly suspected to be caused by drug X–susceptible strains of [list of pathogens]; because data for drug X in these infections are limited, drug X should be used only if other alternatives are known or suspected to be less suitable |
| C | Small prospective studies and descriptive data focused on the tier C pathogen(s) in a range of standard infections | Drug X is indicated for treatment of [list of studied infections from tier C database] when proved or strongly suspected to be caused by drug X–susceptible strains of [list of pathogens]; because data for drug X in these infections are limited, drug X should be used only if other alternatives are known or suspected to be less suitable |
| D | Animal studies | Drug X is indicated for the emergency treatment of infection Y caused by susceptible strains of organism Z; drug X should not be used for infection Y unless other options are unavailable |
aSource: Rex et al [12].
bThese trials should meet standard requirements for disease definitions, end points, and statistical design elements.
Tier A development programs based on traditional large designs in which replicate noninferiority studies are used to provide empiric verification of causality. As noted above, this pathway is impossible for developing narrow-spectrum drugs.
Several tier B development programs are underway and supported by FDA and European Medicines Agency guidance, but these programs have been criticized as potentially providing less-than-optimal data. Previous efforts to pursue direct study of multidrug-resistant pathogens and/or to demonstrate superiority have failed. In a notable example, the team at Achaogen reported screening 659 patients in order to enroll 14 (screen-enrollment rate of 2.1%) in their planned Combating Antibiotic Resistant Enterobacteriaceae (CARE) study designed to evaluate patients with carbapenem-resistant Enterobacteriaceae [11]. The most common reasons for screening failure included identifying a pathogen other than carbapenem-resistant Enterobacteriaceae and prior receipt of empirical antibiotic therapy for >72 hours [11]. Experience with tier C–type designs has been even more challenging in practice. The workshop subsequently addressed the potential use of improved diagnostic tests and merits of superiority versus noninferiority study designs.
ROLE OF RAPID DIAGNOSTIC TESTS
Diagnostic tests that expedite the identification of the infecting pathogen are desperately needed and should constitute a vital component of antimicrobial stewardship and drug development [13, 14]. Diagnostic tests do not change the rate of infection due to a particular pathogen, but they can identify patients earlier than culture-based tests and can reduce the time to enrollment and treatment with the study drug. It is also important to realize that if a diagnostic test has a false-negative rate of >0%, the number of screened patients needed to enroll the required number of infected patients becomes even larger.
SUPERIORITY STUDIES NOT OPTIMAL, FEASIBLE, OR TRACTABLE
Superiority studies were explored in the workshop and have been explored in practice; they were not deemed a viable or reliable path forward. During the workshop, a superiority trial design of a new drug against extensively drug-resistant P. aeruginosa was considered. This design required emergence of extensively drug-resistant strains, thus creating a situation where there was no best available standard-of-care comparator. Such an occurrence is both unpredictable and undesirable from a public health perspective. An alternative design of comparing a new drug combined with standard of care to standard of care in a superiority design was similarly vetted. This design is analogous to the optimized background therapy trials used in human immunodeficiency virus and antituberculosis drug development trials. Challenges in this situation include the ethical need to design the study to ensure that the standard of care is reliably active, in which case it is unlikely that the new drug combined with the standard of care would outperform standard of care alone [15].
As previously discussed, it is unethical to expose patients with serious and life-threatening infections to drugs with known inferior activity or tolerability when efficacious and safe alternatives exist. In the infrequent instance when a superiority trial might be possible, the window of opportunity may close in the midst of the trial should another drug be approved, one that changes the standard of care that ethically must then be provided to patients [9]. Moreover, it is undesirable at a population level to be able to show superiority of a new antibacterial drug when that depends on studying appreciable numbers of patients infected with a pathogen resistant to the current standard of care. We never want the current public health crisis of antimicrobial resistance to expand such that we lack effective treatment options in such large numbers of patients that we can readily conduct such clinical trials. Although we as a community must be vigilant to contain resistance, we must also recognize that it is unlikely that superiority over a fully dosed modern comparator will be observed when the pathogen is susceptible to both the comparator and test antibiotic [9, 12].
RECOMMENDATIONS ON DEVELOPING DRUGS FOR NARROW-SPECTRUM INDICATIONS: ADDRESSING UNCERTAINTY
Randomized clinical trials are conducted to reduce bias when evaluating a potential new treatment [16, 17]. Typically, after phase 1 and 2 studies are completed, 2 randomized clinical trials are conducted to enhance certainty about the safety and efficacy of a potential new treatment. This approach is reasonable, because better certainty is desirable given that science is built on the assumption of experimental reproducibility [18]. Unfortunately, as mentioned earlier, the conduct of a single randomized clinical trial involving new antimicrobial drugs for rare pathogens, let alone 2 such trials, is not practicable.
Consequently, we must consider other mechanisms to provide the requisite evidence of antimicrobial drug effect that forms the scientific foundation for regulatory drug approval and clinical use. We support the inclusion of the following elements, each providing a measure of certainty, to support antimicrobial drug approval and clinical use: (1) PK/PD-based dosing regimen selection; (2) confirmation that targeted drug exposures are attainable in relevant patient populations, including children; (3) regimen efficacy confirmation using a range of animal models; (4) validated external controls; and (5) very small clinical data sets, possibly with pooling of data from multiple body sites. Each element will be discussed in greater detail below. Ideally, at least some data will be generated from all key body sites in infected patients. These ideas follow the concept of using causal evidence combined with clinical data as the basis for substantial evidence of efficacy [19].
The example of meropenem therapy for hospital-acquired or ventilator-associated bacterial pneumonia caused by P. aeruginosa illustrates the success of this strategy. In vitro potency of meropenem against target pathogens, including P. aeruginosa, was demonstrated using a panel of clinical isolates from a wide geographic region [20]. Murine models demonstrated the efficacy of meropenem combined with tobramycin in treating P. aeruginosa pneumonia [21]. Penetration of meropenem into epithelial lining fluid of patients with ventilator-associated pneumonia was demonstrated, and data from clinical trials were used in a population pharmacokinetic model that demonstrated adequate target attainment of meropenem in critically ill patients with this life-threatening infection [22, 23].
PK/PD-BASED DOSE REGIMEN SELECTION
The first layer of certainty supporting the adequacy of an antimicrobial drug dosing regimen can be obtained from preclinical animal and in vitro PK/PD infection models [24, 25]. Here, at least 3 critical parameters can forecast the clinical efficacy and durability of an antimicrobial regimen. These include the determination of the PK/PD measure(s) associated with efficacy (ie, ratio of the area under the curve to the minimum inhibitory concentration [MIC], ratio of the maximum concentration to the MIC, and/or the percentage of time above the MIC), the magnitude of the PK/PD measure necessary for efficacy, and the relationship between antimicrobial drug exposure and time to in vitro emergence of bacterial drug resistance [24–29]. Exposure-response relationships derived from preclinical infection models have been used successfully to forecast effective dosing regimens in several patient populations and to explain clinical trial program failures [30–35].
The PK/PD preclinical infection model should be appropriate to support the scientific question posed. For instance, the PK/PD measure(s) associated with efficacy can be discriminated with a number of short-duration preclinical in vitro (eg, chemostat, hollow-fiber) and in vivo (eg, nonneutropenic and neutropenic murine thigh and lung) PK/PD infection models, whereas characterizing the relationship between drug exposure and time to emergence of bacterial resistance can be evaluated using long-duration preclinical in vitro (eg, hollow-fiber) models [28, 29, 36–38].
The challenge isolates used in preclinical PK/PD infection models should reflect those groups or species expected to be treated clinically and should be genetically well characterized. The numbers of challenge isolates should be large enough to capture variance between isolates in their response to the antimicrobial drug studied. Moreover, the challenge panel optimally should contain a wide range of isolates, both with and without resistance determinants to the study drug. Data from such studies are critical to demonstrating any impact of resistance determinants on the drug exposure magnitude necessary for efficacy [39]. Such information is essential for effective dose regimen forecasting.
It should be recognized that the pharmacokinetic profile in plasma and at the infection site varies greatly from site to site and between experimental animal models and humans [40]. Thus, in addition to the evaluation of plasma pharmacokinetics, effect site pharmacokinetics should be determined during the conduct of preclinical animal PK/PD infection models. Although generating high-quality clinical data on kinetics at specific sites is often challenging, any available information could be integrated with preclinical data used to inform dose regimen selection decisions as well as selection of in vitro susceptibility test interpretive criteria.
PATIENT POPULATION PHARMACOKINETICS
Dose regimen justification should be viewed as an iterative process. The first dose justification iteration typically involves the integration of healthy volunteer (phase 1) pharmacokinetic and preclinical efficacy data through Monte Carlo simulation in the context of relevant pathogen MIC distributions [31, 32]. Typically, these analyses account for between-species differences in pharmacokinetics (eg, drug clearance, protein binding, and effect site penetration). The pharmacokinetic data should at a minimum include data from male and female subjects who received single and multiple doses and data from those with clearing organ dysfunction. Effect site pharmacokinetic studies are advisable for some target patient populations (eg, patients with meningitis or pneumonia).
However, it is critical to recognize that there are often drug disposition differences between healthy volunteers and infected patient populations [35]. Therefore, the second dose justification iteration involves obtaining target patient population pharmacokinetic data and dose forecasting refinement before the conduct of clinical trials. These studies may involve single-dose or multiple-dose data obtained from infected patients being treated with other active agents for their infection.
The final dose justification iteration involves obtaining pharmacokinetic data from treated patients. Pharmacokinetic data should be obtained from all treated patients enrolled in clinical trials. Such data enable pharmacokinetic model refinement, exposure-response analyses and, ultimately, final dose justification and any needed dose adjustments for clearing organ dysfunction.
ANIMAL MODELS, INCLUDING ANIMAL RULE–COMPLIANT VALIDATED MODELS
In certain circumstances, most notably settings where human efficacy studies are not ethical or feasible, US regulations allow for drug approval based largely on animal studies via what is known as the Animal Rule (21 CFR §314.600 for drugs and 21 CFR §601.90 for biological products) [41]. The following 4 criteria must be met for evidence from animal studies to provide substantial evidence of effectiveness: (1) There is a reasonably well-understood pathophysiological mechanism of toxicity of the substance and its prevention or substantial reduction; (2) the effect is demonstrated in >1 animal species expected to react with a response predictive for humans, unless it is demonstrated in a single species that represents a sufficiently well-characterized animal model for predicting the response in humans; (3) the animal study end point is clearly related to the desired end point in humans, generally the enhancement of survival or prevention of major morbid effects; and (4) the PK/PD data or information the product, or other relevant data or information, in humans and animals, allows selection of an effective dose in humans.
Adequate and well-controlled animal efficacy studies are required for approval under the Animal Rule. In this circumstance, animal efficacy studies substitute for human efficacy trials. Therefore, the assessment of efficacy in animals should use end points that demonstrate an important clinical benefit, generally the enhancement of survival or prevention of major morbid effects. Studies should be designed to mimic the ultimate clinical use of the investigational drug and to achieve meaningful outcomes similar to the effectiveness desired in humans. Data from in vitro studies, other types of animal studies, and human studies may be supportive.
The animal species selected for the adequate and well-controlled efficacy studies must exhibit key characteristics of the human disease when the animal is exposed to the challenge organism, and the drug’s effect in the animal species should be expected to be predictive of the effect in humans. This allows extrapolation from the animal data to an effective dose and regimen for humans. Generally, the efficacy of the drug should be demonstrated in >1 animal species expected to have a response predictive for humans.
There are 3 additional requirements for a drug approved under the Animal Rule: (1) postmarketing studies (eg, field studies) to provide evaluation of safety and clinical benefit if circumstances arise in which a study would be feasible and ethical (ie, in the event an emergency arises and the drug is used); (2) restrictions to ensure safe use, if needed (eg, restricting distribution to facilities or health care practitioners with special training, requiring specified types of follow-up, or imposing record-keeping requirements); and (3) information to be provided in the labeling, explaining, that for ethical or feasibility reasons, the drug’s approval was based on efficacy studies conducted in animals alone [41]. We believe that these principles represent an important starting point for an approach to the approval of narrow-spectrum agents, but we also recognize that the requisite validated animal models are not available today. It is expected that development of these animal models may take 3–5 years, perhaps longer. Moreover, it is uncertain that these models can be successfully developed.
Given our current lack of these models and the uncertainly surrounding their feasibility and time of availability, we think that the principles underpinning this approach can be combined with limited clinical development programs to achieve rational product approval. In particular, we would suggest that a combination of a variety of animal models could reasonably be implemented in support of an investigational agent. The collection of models should include infections produced in >1 animal species, at >1 body site, and using >1 strain of the relevant bacteria. End points might initially focus on surrogate measures, such as colony-forming units per gram of infected tissue, but a reasonable range of models should be conducted using survival end points. Humanized exposures of drugs might be used. In addition, control agents having little activity (negative controls), intermediate activity, and potent activity (positive controls, when possible) might usefully be employed to demonstrate that the individual models are able both to detect strong drugs effects and to discriminate between agents possessing intermediate effects at the target exposures. As mentioned earlier, these types of studies have served well in forecasting effective regimens and explaining regimen failures.
CLINICAL TRIALS WITH VERY SMALL CLINICAL DATA SETS: SMALL RANDOMIZED CONTROLLED TRIALS, SMALL TIER C–TYPE STUDIES, VALIDATED EXTERNAL-CONTROL STUDIES, AND SMALL NONCOMPARATIVE STUDIES
Given these limitations, it is likely that clinical development of narrow-spectrum drugs (or broad-spectrum drugs with activity against targeted organisms) will be possible only in very small numbers of patients. As discussed above, the study of even a small number of patients permits the clinical program to have a strength that cannot be attained with a purely Animal Rule–based approval. In effect, the postmarketing study requirement can be addressed before marketing and at a time when controlled observations can be made without the pressure of a public demand for rapid access to an incompletely tested agent.
Several options for development should be considered. These programs may include small randomized controlled trials (RCTs), small tier C–type studies with external controls, or small clinical data sets based on noncomparative studies.
Although large RCTs are likely not feasible, use of a RCT that allows for greater statistical uncertainty should be pursued if feasible. For example, a study could employ uneven randomization and use the full M1 (effect of active control assumed to be present in the study) for a larger noninferiority margin, especially in cases where clinical outcomes are highly objective (eg, death) [42]. This might make a randomized trial feasible and seems tractable for infections associated with high mortality rates in the absence of therapy, such as or ventilator-associated bacterial pneumonia or certain bloodstream infections.
Studies using validated external controls may be considered, especially if the treatment effect is large, though a number of limitations should be addressed. Both historical and reasonably contemporaneous controls, representing patients who actually could have been enrolled in a randomized trial, may be considered. An earlier white paper reviewed the regulatory basis for considering externally controlled studies [43]. The guidance emphasizes the importance of selecting controls with similar characteristics to the treatment group, selecting objective end points, assuring that covariates influencing outcome are well characterized, and using this design in diseases for which treatment is associated with a dramatic effect and the outcome without therapy highly predictable [44].
A criticism of external controls in general is that retrospective identification of cases will identify both patients who could and those who could not have enrolled in a given trial. The subset not enrolled (eg, patients with disease diagnosed at autopsy) would tend to reduce response rates and thus bias in favor of seeing a new therapy as being better than the external controls. As an approach to resolving this issue, a database of patients with a given infection could be compiled prospectively. If patients were screened for likely eligibility criteria for a given trial, only those eligible to be part of a control group could be used for estimation of response rates. This approach is the intellectual inverse of the idea of sequential parallel comparison design, in which an initial screening step is used to remove placebo responders [45]. Once created, such a database could be shared widely across developers and would facilitate future drug development. A clinical trials network could provide the foundation that allows study of less frequently encountered pathogens and could also accelerate development of improved external control data.
Comparative studies producing very small clinical data sets might also serve as part of the basis for approval. One example involves use of a standard drug plus the test drug versus the standard drug plus an often, though not always, effective agent. Workshop participants deliberated an example of test drug with meropenem versus aminoglycoside plus meropenem to treat P. aeruginosa infection. With an expectation of an approximately 20% resistance rate of P. aeruginosa to meropenem, such a study would possibly include a subset, perhaps 20% of the population, effectively comparing the test drug alone with the aminoglycoside alone. A focused analysis would be possible in this group, though noninferiority testing would not be feasible unless the sample size were increased 5-fold. An alternative strategy involves treating patients with infections at a variety of body sites (eg, complicated urinary tract infection, bloodstream infection, or hospital-acquired or ventilator-associated bacterial pneumonia) with a new agent and analyzing pooled data.
In any of these scenarios, careful attention must be paid to developing strong case definitions, to including patients with severe infections when possible, to minimizing prior effective therapy, and to generating high-quality clinical data. Clinical trial networks may facilitate patient enrollment, and several groups, including The Wellcome Trust and the National Institutes of Health Antibacterial Resistance Leadership Group, are currently developing plans for such networks [46, 47]. Other means to augment the clinical development program include increasing the safety data set via expanded phase 1 data packages and/or phase 2 or 3 studies of a more routine indication. No matter what combination of studies is selected, patients with well-characterized, unremitting infections and likely poor outcome without effective therapy are a key group. In this population, responses in even a small number of patients can be compelling.
ENSURING LIMITED AND APPROPRIATE USE
As discussed, it will generally not be possible to study large numbers of patients treated with drugs for these narrow-spectrum indications, and traditional development programs guided by existing FDA guidance will not be feasible (see Introduction). In addition, effective treatment of multidrug-resistant pathogens is a critical unmet need that can be sustained only through ensuring stewardship and optimal use of such newly approved drugs. It is also likely that the market for these drugs will be substantially smaller than for other drugs, even among anti-infective products.
In this context, a new regulatory paradigm of the Limited Population Antimicrobial Drug (LPAD) has emerged that will allow for conduct of smaller, feasible clinical trials yet meet existing statutory regulatory standards [10]. The House of Representatives approved LPAD as part of the 21st Century Cures Act, HR 34, on 30 November 2016, the Senate followed suit on 6 December 2016, and President Obama signed the legislation 13 December 2016. The LPAD pathway will allow smaller, more efficient, and potentially feasible clinical trials and includes safeguards to guide appropriate use of the drugs that gain approval under this pathway.
In addition to the assurance of limited labeling via the LPAD mechanism, appropriate restriction of use via robust antibiotic stewardship programs led by infectious diseases (ID) physicians will enable expert management of all patients in whom these medicines are used [6, 14, 48]. The Joint Commission issued a new antimicrobial stewardship standard, effective 1 January 2017, requiring hospitals to establish antibiotic stewardship programs that are aligned with the Centers for Disease Control and Prevention’s Core Elements of Hospital Antibiotic Stewardship Programs [49, 50]. The Joint Commission’s leadership will contribute significantly toward ensuring that ID physician–led stewardship programs are a reality throughout the country. The Centers for Medicare & Medicaid Services issued a proposed rule mandating stewardship in 2016 [49, 50]. The antibiotic use module of the Centers for Disease Control and Prevention’s National Healthcare Safety Network reporting system provides the vehicle for reporting antibiotic use and has been cited as the preferred means of performing surveillance of usage [7].
DRUG LABELING/MULTIPLE BODY SITES
Drug labeling of new narrow-spectrum drugs to treat resistant bacterial infections would clearly state the limited population and expected use. In addition, we support inclusion of information about penetration and pharmacology of the drug at all studied body sites. Where available, effectiveness data at less common sites of infection should also be provided.
CONCLUSIONS
Developing narrow-spectrum drugs for treatment of patients with infections caused by drug-resistant pathogens is problematic, and all approaches to development of these drugs pose major scientific challenges. Investigators and regulators must work together to use a collection of the limited tools that are available as the basis for registration.
The patients needed for such studies are uncommon, and we want this to remain true. Neither noninferiority- nor superiority-based approaches are routinely tractable. Improved diagnostics and early detection will not solve the problem in that they do not create patients with the target pathogen—they only help identify them and may facilitate enrollment. That said, screening for such rare patients would be facilitated by layering the study of such drugs on top of a clinical trial network that is actively running more traditional registrational studies [47]. Excellent preclinical PK/PD programs are now possible and need to be better leveraged.
In the face of the global crisis of antimicrobial resistance, inaction is not acceptable [6]. A reliable path seems possible by combining robust PK/PD studies, validated animal models, validated external controls, especially if the treatment effect is large, and very small clinical data sets. Adequate, well-controlled data from small RCTs with wide noninferiority margins and uneven randomization or small tier C–type studies with external controls can both serve as a basis of a strong clinical development program. In all clinical studies, strong case definitions and maximizing inclusion of patients with severe infections will increase the likelihood of generating high-quality clinical data. Including multiple body sites and infection types provides useful clinical data to clinicians treating these patients.
A guidance document is needed but should not delay progress. Along the way, public consensus should continue to be built via PACCARB, FDA public meetings or advisory committees, workshops, public meetings, and/or other means. The LPAD mechanism ensures use in limited population with needed safeguards, and ID physician-led stewardship facilitates expert involvement in the management of all patients in whom these medicines are used.
Finally, to succeed in a sustained fashion and return to a robust and vibrant, diverse antimicrobial development pipeline, we must engage the larger group of stakeholders and make push-and-pull incentives work in order to address economic barriers to antimicrobial drug development. This will require a path forward that is unique and pragmatic. Not finding such a pathway would force future patients and physicians to accept even more uncertainty as antibiotic resistance continues to emerge and threaten public health. Such a course is unacceptable to our patients and future generations.
Notes
Potential conflicts of interest. H. W. B. is a consultant to Actelion (Data Safety Monitoring Committee) and the National Institute of Allergy and Infectious Disease (Adjudication Committee). P. G. A. is president of the Institute for Clinical Pharmacodynamics, a special government employee for the Department of Health and Human Services, and scientific advisor and consultant for Achaogen, Actelion, AiCuris, Arsanis, Basilea, Cellceutix, Cempra, Cidara, Contrafect, Debiopharm, Geom, GlaxoSmithKline, Insmed, Kalya, Medicines Company, Meiji, Melinta, Merck, Nabriva, Nexcida, Northern Antibiotics, Novartis, Paratek, PolyPhor, Roche, Spero, Takeda, Theravance, Tetraphase, VenatoRx, Wockhardt, and Zavante. H. F. C. receives grant support from Genentech and Allergan. R. H. E. is a patentee on antibacterial drug development, a consultant to Genomatica, and a biomedical experts panel member for the, Skolkovo Foundation. A. J. is an employee of the Infectious Diseases Society of America. B. E. M. receives grant support from Merck, Theravance, and Actavis and is a consultant for Cempra and Paratek. J. G. N. is a consultant for RPS Diagnostics. J. H. R. is chief strategy officer for CARB-X, chief medical officer and director for F2G, nonexecutive director and consultant for Adenium Biotech, operating partner and consultant for Advent Life Sciences, and expert-in-residence for Wellcome Trust; is on the scientific advisory boards of Macrolide Pharmaceuticals, Spero Therapeutics, and Bugworks Research; is a shareholder in AstraZeneca Pharmaceuticals, F2G, Adenium Biotech, Advent Life Sciences, Macrolide Pharmaceuticals, and Bugworks Research; and has received consulting fees from Phico Therapeutics, ABAC Therapeutics, Polyphor, Heptares Therapeutics, and Ganagen. B. O. reports no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. O’Neill, J; The Review on Antimicrobial Resistance. Tackling drug-resistant infections globally: final report and recommendations 2016. http://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf Accessed 25 August 2016.
- 2. Pogue JM, Ortwine JK, Kaye KS. Optimal usage of colistin: are we any closer? Clin Infect Dis 2015; 61:1778–80. [DOI] [PubMed] [Google Scholar]
- 3. Xavier BB, Lammens C, Ruhal R, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill 2016; 21:1–6. doi:10.2807/1560,7917.ES.2016.21.27.30280. [DOI] [PubMed] [Google Scholar]
- 4. McGann P, Snesrud E, Maybank R, et al. Escherichia coli harboring mcr-1 and blaCTX-M on a novel IncF plasmid: first report of mcr-1 in the United States. Antimicrob Agents Chemother 2016; 60:4420–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Abbasi J. Infectious disease expert sees threat from colistin-resistant superbug. JAMA 2016; 316:806–7. [DOI] [PubMed] [Google Scholar]
- 6. Boucher HW, Bakken JS, Murray BE. The United Nations and the urgent need for coordinated global action in the fight against antimicrobial resistance. Ann Intern Med 2016; 165:812–3. [DOI] [PubMed] [Google Scholar]
- 7. Presidential Advisory Council on Combating Antibiotic Resistant Bacteria. Report 1: initial assessments of the national action plan for combating antibiotic- resistant bacteria 2016. https://www.hhs.gov/sites/default/files/paccarb-final- report-03312016.pdf Accessed 25 August 2016
- 8. World Health Organization. Antimicrobial resistance: global report on surveillance 2014. http://apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf?ua=1 Accessed 25 August 2016.
- 9. Nambiar S, Laessig K, Toerner J, Farley J, Cox E. Antibacterial drug development: challenges, recent developments, and future considerations. Clin Pharmacol Ther 2014; 96:147–9. [DOI] [PubMed] [Google Scholar]
- 10. Spellberg B, Brass EP, Bradley JS, et al. White paper: recommendations on the conduct of superiority and organism-specific clinical trials of antibacterial agents for the treatment of infections caused by drug-resistant bacterial pathogens. Clin Infect Dis 2012; 55:1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Workshop Participants. Facilitating antibacterial drug development for patients with unmet need and developing antibacterial drugs that target a single species 2016. hhttp://www.fda.gov/Drugs/NewsEvents/ucm497650.htm Accessed 15 November 2016.
- 12. Rex JH, Eisenstein BI, Alder J, et al. A comprehensive regulatory framework to address the unmet need for new antibacterial treatments. Lancet Infect Dis 2013; 13:269–75. [DOI] [PubMed] [Google Scholar]
- 13. Caliendo AM, Gilbert DN, Ginocchio CC, et al. ; Infectious Diseases Society of America (IDSA) Better tests, better care: improved diagnostics for infectious diseases. Clin Infect Dis 2013; 57(suppl 3):S139–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barlam TF, Cosgrove SE, Abbo LM, et al. Implementing an Antibiotic Stewardship Program: Guidelines by the Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America. Clin Infect Dis 2016; 62:e51–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tamma PD, Cosgrove SE, Maragakis LL. Combination therapy for treatment of infections with gram-negative bacteria. Clin Microbiol Rev 2012; 25:450–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Streptomycin in Tuberculosis Trials Committee. Streptomycin treatment of pulmonary tuberculosis: a medical research council investigation. Br Med J 1948; 2:769.18890300 [Google Scholar]
- 17. Stolberg HO, Norman G, Trop I. Randomized controlled trials. AJR Am J Roentgenol 2004; 183:1539–44. [DOI] [PubMed] [Google Scholar]
- 18. Casadevall A, Fang FC. Reproducible science. Infect Immun 2010; 78:4972–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peck CC, Rubin DB, Sheiner LB. Hypothesis: a single clinical trial plus causal evidence of effectiveness is sufficient for drug approval. Clin Pharmacol Ther 2003; 73:481–90. [DOI] [PubMed] [Google Scholar]
- 20. EUCAST. MIC distributions and ECOFFs 2016. www.eucast.org/mic_distributions_and_ecoffs/ Accessed 25 July 2016.
- 21. Louie A, Liu W, Fikes S, Brown D, Drusano GL. Impact of meropenem in combination with tobramycin in a murine model of Pseudomonas aeruginosa pneumonia. Antimicrob Agents Chemother 2013; 57:2788–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mattioli F, Fucile C, Del Bono V, et al. Population pharmacokinetics and probability of target attainment of meropenem in critically ill patients. Eur J Clin Pharmacol 2016; 72:839–48. [DOI] [PubMed] [Google Scholar]
- 23. Lodise TP, Sorgel F, Melnick D, Mason B, Kinzig M, Drusano GL. Penetration of meropenem into epithelial lining fluid of patients with ventilator-associated pneumonia. Antimicrob Agents Chemother 2011; 55:1606–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Craig WA. Pharmacokinetic/pharmacodynamic parameters: rationale for antibacterial dosing of mice and men. Clin Infect Dis 1998; 26:1–10; quiz 11–2. [DOI] [PubMed] [Google Scholar]
- 25. Ambrose PG, Bhavnani SM, Rubino CM, et al. Pharmacokinetics-pharmacodynamics of antimicrobial therapy: it’s not just for mice anymore. Clin Infect Dis 2007; 44:79–86. [DOI] [PubMed] [Google Scholar]
- 26. Jumbe N, Louie A, Leary R, et al. Application of a mathematical model to prevent in vivo amplification of antibiotic-resistant bacterial populations during therapy. J Clin Invest 2003; 112:275–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Drusano GL. Antimicrobial pharmacodynamics: critical interactions of ‘bug and drug’. Nat Rev Microbiol 2004; 2:289–300. [DOI] [PubMed] [Google Scholar]
- 28. Tam VH, Louie A, Deziel MR, Liu W, Leary R, Drusano GL. Bacterial-population responses to drug-selective pressure: examination of garenoxacin’s effect on Pseudomonas aeruginosa. J Infect Dis 2005; 192:420–8. [DOI] [PubMed] [Google Scholar]
- 29. Vanscoy B, Mendes RE, Castanheira M, et al. Relationship between ceftolozane-tazobactam exposure and drug resistance amplification in a hollow-fiber infection model. Antimicrob Agents Chemother 2013; 57:4134–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Andes D, Craig WA. In vivo activities of amoxicillin and amoxicillin-clavulanate against Streptococcus pneumoniae: application to breakpoint determinations. Antimicrob Agents Chemother 1998; 42:2375–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Drusano GL, Preston SL, Hardalo C, et al. Use of preclinical data for selection of a phase II/III dose for evernimicin and identification of a preclinical MIC breakpoint. Antimicrob Agents Chemother 2001; 45:13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bhavnani SM, Hammel JP, Cirincione BB, Wikler MA, Ambrose PG. Use of pharmacokinetic-pharmacodynamic target attainment analyses to support phase 2 and 3 dosing strategies for doripenem. Antimicrob Agents Chemother 2005; 49:3944–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ambrose PG, Forrest A, Craig WA, et al. Pharmacokinetics-pharmacodynamics of gatifloxacin in a lethal murine Bacillus anthracis inhalation infection model. Antimicrob Agents Chemother 2007; 51:4351–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ambrose PG. Use of pharmacokinetics and pharmacodynamics in a failure analysis of community-acquired pneumonia: implications for future clinical trial study design. Clin Infect Dis 2008; 47(suppl 3):S225–31. [DOI] [PubMed] [Google Scholar]
- 35. Ambrose PG, Bhavnani SM, Ellis-Grosse EJ, Drusano GL. Pharmacokinetic-pharmacodynamic considerations in the design of hospital-acquired or ventilator-associated bacterial pneumonia studies: look before you leap! Clin Infect Dis 2010; 51(suppl 1):S103–10. [DOI] [PubMed] [Google Scholar]
- 36. Craig WA. Interrelationship between pharmacokinetics and pharmacodynamics in determining dosage regimens for broad-spectrum cephalosporins. Diagn Microbiol Infect Dis 1995; 22:89–96. [DOI] [PubMed] [Google Scholar]
- 37. Andes D, Craig WA. Pharmacodynamics of the new fluoroquinolone gatifloxacin in murine thigh and lung infection models. Antimicrob Agents Chemother 2002; 46:1665–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Srivastava S, Modongo C, Siyambalapitiyage Dona CW, Pasipanodya JG, Deshpande D, Gumbo T. Amikacin optimal exposure targets in the hollow-fiber system model of tuberculosis. Antimicrob Agents Chemother 2016; 60:5922–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Andes D, Craig WA. Treatment of infections with ESBL-producing organisms: pharmacokinetic and pharmacodynamic considerations. Clin Microbiol Infect 2005; 11(suppl 6):10–7. [DOI] [PubMed] [Google Scholar]
- 40. Rodvold KA, Nicolau DP, Lodise TP, et al. Identifying exposure targets for treatment of staphylococcal pneumonia with ceftobiprole. Antimicrob Agents Chemother 2009; 53:3294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. U.S. Department of Health and Human Services Food and Drug Administration Center for Drug Evaluation and Research (CDER), Center for Biologics Evaluation and Research (CBER). Guidance for industry: product development under the animal rule 2015. http://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm399217.pdf Accessed 17 November 2016.
- 42. U.S. Department of Health and Human Services. Non-inferiority clinical trials to establish effectiveness; guidance for industry 2016. https://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm202140.pdf Accessed 28 February 2017.
- 43. Infectious Diseases Society of America. White paper: recommendations on the conduct of superiority and organism-specific clinical trials of antibacterial agents for the treatment of infections caused by drug-resistant bacterial pathogens. Clin Infect Dis 2012; 55:1031–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use (ICH). 2001. Guidance for industry E 10: choice of control group and related issues in clinical trials http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2009/09/WC500002925.pdf Accessed 16 November 2016.
- 45. Fava M, Evins AE, Dorer DJ, Schoenfeld DA. The problem of the placebo response in clinical trials for psychiatric disorders: culprits, possible remedies, and a novel study design approach. Psychother Psychosom 2003; 72:115–27. [DOI] [PubMed] [Google Scholar]
- 46. Wellcome Trust. Clinical trial networks for antibiotic development: why they’re important and how they should be developed 2016. https://wellcome.ac.uk/sites/default/files/clinical-trial-networks-for-antibiotic-development-wellcome-oct16.pdf Accessed 15 November 2015.
- 47. McDonnell A, Rex JH, Goossens H, Bonten M, Fowler VG, Jr, Dane A. Efficient delivery of investigational antibacterial agents via Sustainable Clinical Trial Networks. Clin Infect Dis 2016; 63(suppl 2):S57–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cosgrove SE, Hermsen ED, Rybak MJ, et al. Guidance for the knowledge and skills required for antimicrobial stewardship leaders. Infect Control Hosp Epidemiol 2014; 35:1444–51. [DOI] [PubMed] [Google Scholar]
- 49. The Joint Commission. New antimicrobial stewardship standard 2016. https://www.jointcommission.org/assets/1/6/HAP-CAH_Antimicrobial_Prepub.pdf Accessed 5 December 2016.
- 50. Centers for Disease Control and Prevention. Core elements of hospital antibiotic stewardship programs 2016. http://www.cdc.gov/getsmart/healthcare/implementation/core-elements.html Accessed 5 December 2016.