A novel transcription factor, EjNAC3, is involved in chilling-induced lignification in loquat via direct interaction with a CAD-like gene, which differs from other ‘master-switch’ NAC genes.
Keywords: CAD-like, chilling injury, lignin biosynthesis, loquat, NAC, transcriptional regulations
Abstract
Lignin is an important component of many plant secondary cell walls. In the fruit of loquat (Eriobotrya japonica), lignification of cell walls in the fleshy tissue occurs when fruit are subjected to low-temperature storage, which is commonly used to avoid the rapid senescence that occurs at room temperature. In this study, two NAC domain genes, EjNAC3 and EjNAC4, were isolated and shown to be significantly induced at 0 °C, which was concomitant with an increase in the fruit lignification index. Lignification and expression of both EjNAC3 and EjNAC4 were inhibited by low-temperature conditioning and by heat treatment. In addition, EjNAC3 trans-activated the lignin biosynthesis-related EjCAD-like promoter, which was measured using a dual-luciferase assay. Further analysis with yeast one-hybrid and electrophoretic mobility shift assays indicated that EjNAC3 could physically bind to the promoter of the EjCAD-like gene. Thus, EjNAC3 is a direct regulator of loquat chilling-induced lignification, via regulations of EjCAD-like.
Introduction
Low-temperature storage is routinely applied to extend the post-harvest life of fruit; however, various chilling injury symptoms commonly occur in fruit subjected to inappropriately low temperatures or long-term storage. For example, the skin of pomegranate becomes brown and pitted if stored below 5 °C (Sayyari et al., 2009), while peach fruit stored at 5 °C exhibits more severe symptoms of flesh browning than fruit stored at 0 or 8 °C (Zhang et al., 2011). Unlike other fruit, loquat (Eriobotrya japonica), a species of Rosaceae, not only undergoes flesh browning in response to low-temperature storage (0 °C), but also exhibits typical lignification symptoms, such as an increase in lignin content and firmness (Zheng et al., 1999; Cai et al., 2006d). Due to the significant impact of lignification on fruit quality, numerous treatments have been developed to manipulate loquat fruit lignification, including low-temperature conditioning (LTC) (Cai et al., 2006d), heat treatment (HT) (Xu et al., 2014), salicylic acid treatment (Ding et al., 2002; Cai et al., 2006b), methyl jasmonate (MeJA) treatment (Cao et al., 2009), and 1-methylcyclopropene (1-MCP) treatment (Cai et al., 2006a). Thus, loquat fruit represents well-researched material on which to investigate the underlying mechanisms controlling fruit-flesh lignification.
Based on research from model plants and woody trees, the biochemical and molecular basis for loquat fruit lignification has been investigated, and it has been shown to be correlated with the activities of enzymes involved in the phenylpropanoid pathway, including L-phenylalanine ammonia lyase (PAL), 4-coumarate:coenzyme A ligase (4CL), and cinnamyl alcohol dehydrogenase (CAD): these enzymes are significantly repressed by treatments with LTC, 1-MCP, acetylsalicylic acid (ASA) (Cai et al., 2006a, b, c, d), and MeJA (Cao et al., 2010), which also reduce lignification.
In model plants and woody trees, transcription factors, especially NAC and MYB, have been widely reported to be involved in lignin biosynthesis and secondary cell wall construction, and a complicated network is formed through interactions at the transcriptional and/or post-translational levels (Kubo et al., 2005; Mitsuda et al., 2005; Zhong et al., 2008; Ohashi-Ito et al., 2010; Yamaguchi et al., 2011). Many MYB-type transcription factors, for example AtMYB58, AtMYB63 (Zhou et al., 2009), AtMYB85 (Zhong et al., 2008), and AtBLH6 (Cassan-Wang et al., 2013), are activators of lignin biosynthesis while AtMYB4, AtMYB7, and AtMYB32 act as repressors (Preston et al., 2004; Wang and Dixon, 2012), and in some cases the gene promoters they interact with have been identified. For instance, 4CL genes, which function in the upstream part of the phenylpropanoid pathway, were confirmed to be a direct target of both AtMYB58 and AtMYB6 (Zhou et al., 2009). In contrast, most NAC-domain proteins operate at the top level of the regulatory network, which means that evidence that these NAC genes physically interact with structural genes has not been found. They are designated as ‘master switches’ of secondary cell wall formation, and include AtNST1 and AtNST2, AtVND1 to AtVND7, and AtSND1 (Kubo et al., 2005; Mitsuda et al., 2005; Zhong et al., 2006; Bennett et al., 2010), although AtSND1 also directly activates expression of MtF5H, which encodes ferulate-5-hydroxylase in Medicago truncatula by binding to its promoter (Zhao et al., 2010), and all have crucial roles during secondary cell wall formation.
The first transcription factors shown to be involved in loquat fruit-flesh lignification were EjMYB1 and EjMYB2 (Xu et al., 2014). EjMYB1 was found to be capable of transcriptional activation of a number of lignin biosynthesis-related genes, such as EjPAL1, Ej4CL1, and Ej4CL5, and transient over-expression of EjMYB1 in tobacco leaves resulted in ectopic accumulation of lignin. EjMYB2 was characterized as a repressor of loquat fruit lignification, and an AP2/ERF family gene, EjAP2-1, indirectly regulated lignification via protein–protein interactions with either EjMYB1 or EjMYB2 (Zeng et al., 2015). In addition to MYB transcription factors, EjNAC1 was also shown to enhance expression during loquat fruit chilling-induced lignification; however, it could not bind directly to promoters of lignin biosynthesis genes (Xu et al., 2015), indicating an indirect mechanism of activation. These reports identified several components of the transcriptional regulatory mechanisms underlying loquat fruit-flesh lignification; however, the relationships between them and the overall control of the pathway are not clear. For instance, all of the known transcription factors, EjMYB1, EjMYB2, and EjAP2-1, have significant regulatory effects on EjPAL1 and Ej4CL1 promoters (upstream genes of the phenylpropanoid pathway) but have limited effects on downstream pathway genes.
In the present study, two NAC-domain genes were isolated from loquat fruit. Their transcript levels in response to low-temperature conditioning and heat treatment were characterized, together with their trans-activation functions. In addition, the relationship between these NAC genes and lignification was investigated and potential targets of the NAC genes were screened by dual luciferase assays, and their physical interactions were confirmed using a yeast one-hybrid system and electrophoretic mobility shift assays.
Materials and methods
Plant materials and treatments
Fruits of loquat (Eriobotrya japonica Lindl., red-fleshed variety ‘Luoyangqing’) were collected in 2011 at Luqiao, Zhejiang province, China. The fruit material and treatments have been described in detail previously by Xu et al., (2014). In brief, uniform fruit were selected and divided into three batches with about 150 fruit in each. One batch was stored at 40 °C for 4 h as the heat treatment (HT) and was then transferred to 0 °C for storage. For the low-temperature conditioning (LTC), fruit were kept at 5 °C for 6 d followed by storage at 0 °C. The third batch of fruit was immediately stored at 0 °C, as a control. Three biological replicates were used for all treatments and samplings.
Gene isolation and phylogenetic analysis
Differentially expressed unigenes, within the NAC domain, were obtained from previously generated RNA-seq results (Xu et al., 2014). The full coding DNA sequences (CDS) of two genes were amplified with a SMART RACE cDNA amplification kit (Clontech), using the primers listed in Supplementary Table S1 at JXB online. These genes were aligned with Arabidopsis NAC genes obtained from the TAIR database (https://www.arabidopsis.org/), using the neighbor-joining (NJ) method with ClustalX (v.1.81). The results of the alignments were visualized using FigTree (v. 1.3.1).
RNA extraction and cDNA synthesis
Total RNA used to synthesize cDNA was extracted from the fruit flesh according to the protocol described by Shan et al., (2008). Potential genomic DNA contamination was eliminated using a TURBO DNA-free kit (Ambion). The quality and quantity of RNA extracted from the fruit flesh was determined by gel electrophoresis and spectrophotometry (Implen). To synthesize first-strand cDNA, 1 µg DNA-free total RNA was added to a mixture of cDNA synthesis reagents (BioRad).
Real-time PCR analysis
Real-time PCR was carried out using the CFX96 Real-Time System (Biorad), with SsoFast EvaGreen Supermix (Biorad). The primers were designed using Primer3 (version 4.0.0; http://bioinfo.ut.ee/primer3/) and are listed in Supplementary Table S2. All of the primers were tested to ensure their specificity for unique genes (Yin et al., 2010).
Dual luciferase assay
Dual luciferase assays (described previously by Yin et al., 2010; Min et al., 2014) were performed to analyse the potential relationships between NAC transcription factors and loquat lignin biosynthesis genes and previously characterized transcription factors involved in loquat lignification. Full-length NAC genes were amplified with primers (listed in Supplementary Table S3) and integrated into the pGreen II 0029 62-SK vector (SK). Promoters of lignin synthesis-related genes from both loquat and Arabidopsis were inserted into the pGreen II 0800-LUC vector (LUC), as described by Xu et al., (2014). Promoters regions of the EjMYB1, EjMYB2, and EjAP2-1 transcription factors were constructed into the pGreen II 0800-LUC vector.
All of the recombinant SK and LUC vectors were transfected into Agrobacterium tumefaciens GV3101. The glycerol stocks with transfected Agrobacterium were grown in lysogeny broth (LB) plates with agar and 50 µg ml–1 kanamycin and 25 µg ml–1 gentamycin for 2 d and then restreaked onto new LB plates for 1 d. Agrobacterium were suspended in infiltration buffer (10 mM MES, 10 mM MgCl2, 150 mM acetosyringone, pH 5.6) to an optimal density (OD600=0.75), then 1 ml of Agrobacterium cultures containing transcription factors were mixed with 100 µl of Agrobacterium containing promoters. The mixtures were then injected into tobacco (Nicotiana tabacum) leaves with needleless syringes, and 3 d after infiltration the LUC and REN fluorescence intensities were measured using dual luciferase assay reagents (Promega). Five replicates were conducted for each transcription factor and promoter combination.
Yeast one-hybrid assay
Yeast one-hybrid assays were conducted to test the physical interactions between transcription factors and promoters identified from the dual luciferase assay. Yeast one-hybrid assays were performed using the Matchmaker Gold Yeast One-Hybrid Library Screening System (Clontech, USA). The promoter of EjCAD-like was amplified and inserted into the pAbAi vector. Similarly, the pGADT7 vector with full-length EjNAC3 was constructed (primers are listed in Supplementary Table S4). The recombinant EjCAD-like-pAbAi vector was linearized and transformed into the Y1HGold yeast strain. After testing for autoactivation, the Y1HGold[EjCAD-like/AbAi] yeast strain was transfected with the EjNAC3-pGADT7 plasmid and grown on SD/–Leu medium containing 0–500 ng ml– of AbA at 30 °C for 3 d. The empty pGADT7 plasmid was transformed as a negative control.
Electrophoretic mobility shift assay (EMSA)
The intact open reading frame of EjNAC3 was inserted into pET-32a (Clontech) to generate an EjNAC3-His fusion protein. The reconstructed plasmid was transformed into Escherichia coli strain BL21. The transformed cells were treated with 1mM isopropyl β-D-1-thiogalactopyranoside (IPTG) followed by incubation at 16 °C for 20 h. After extraction of the recombined protein, a HisTALON™ Gravity Column (Clontech) was used for protein purification, following the steps described in the official user manual.
A Lightshift Chemiluminescent EMSA kit (Thermo) was used to perform EMSA experiments according to the manufacturer’s instructions. Single-strand oligonucleotides were synthesized and biotinylated by the JINBAIAO company (Shanghai, China). After mixing forward and reverse oligonucleotides together, the mixture was incubated at 95 °C for 5 min and the temperature gradually decreased to 25 °C at the rate of 0.1 °C s–1, allowing the formation of double-strand probes. The binding buffer provided with the kit was used together with 5 mM MgCl2, 2.5% glycerol, 0.05% NP-40, 50 ng of purified protein 2 nM probe, and 50 ng µl–1 of poly(dI-dC) to set reactions. For competition reactions, the cold probe was incubated with the protein for 20 min followed by addition of the biotinylated probe. The solution was analysed using 6% native polyacrylamide gel electrophoresis for 30 min after incubation for 20 min at room temperature. The voltage was set to 120 (constant voltage mode) and 0.5× Tris-borate/EDTA buffer was pre-chilled. The fractionated products were transferred onto nitrocellulose membranes with 0.5× Tris-borate/EDTA at 280 mA (constant electric current mode) for 30 min with cold 0.5× Tris-borate/EDTA buffer (4 °C). Cross-linking was performed under a UV-light for 30 min. The membrane was then incubated in blocking buffer with shaking for 15 min, transferred to conjugate/blocking buffer (16.75 µl stabilized streptavidin-horseradish peroxidase conjugate with 5 ml blocking buffer) and washed four times, each for 5 min with 1× washing buffer. Detection of biotinylated DNA probes was achieved using the ChemiDoc™ MP Imaging System (Bio-Rad). The probes used for EMSAs are listed in Supplementary Table S5.
Statistical analysis
The statistical significance of differences was determined using Student’s t-test. Least-significant differences (LSD) at the 5% level were calculated using DPS7.05 (Zhejiang University, Hangzhou, China). Figures were drawn using Origin 8.0 (Microcal Software Inc., Northampton, MA, USA).
Results
Isolation of loquat NAC genes and phylogenetic analysis
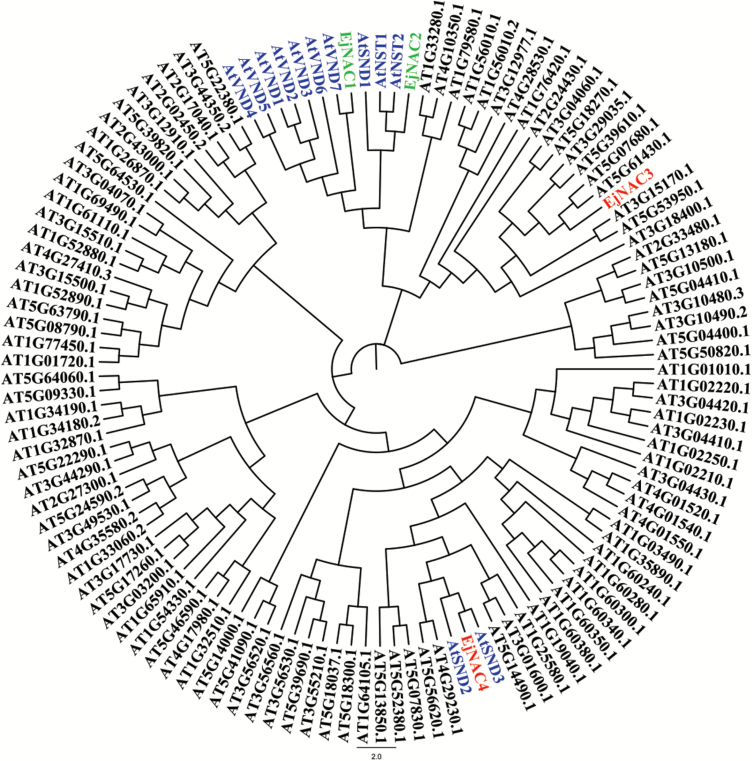
Two loquat NAC genes, EjNAC3 and EjNAC4, were isolated based on unigenes from the loquat RNA-seq database (data not shown). Phylogenetic tree analysis indicated that EjNAC4 was clustered with lignin-related AtSND2/3, while EjNAC3 was not clustered with any secondary cell wall-related NAC genes (Fig. 1).
Fig. 1.
Phylogenetic analyses of loquat NAC genes. Amino acid sequences of Arabidopsis NAC genes were downloaded from TAIR (https://www.arabidopsis.org/). Secondary cell wall-related genes from Arabidopsis are highlighted in blue, and previously reported EjNAC1 and EjNAC2 are highlighted in green.
Expression levels of NAC genes in fruit flesh after HT and LTC treatments
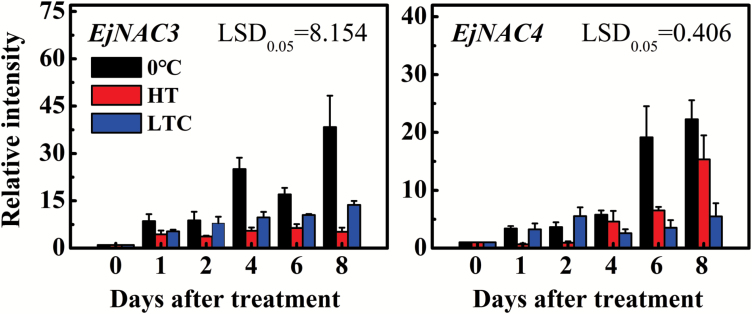
HT and LTC treatments both alleviate chilling-induced loquat fruit lignification to some extent (Xu et al., 2014; Zeng et al., 2015). Thus, the relationship between NAC genes and fruit lignification was investigated using fruit subject to HT and LTC (Xu et al., 2014). mRNAs for EjNAC3 and EjNAC4 accumulated in response to 0 °C, and at 8 d showed 38-fold and 22-fold increases, respectively (Fig. 2). The transcript levels for each gene were significantly depressed by prior HT or LTC. For example, the relative level of EjNAC3 at 4 d was 5.5 and 9.7 in HT and LTC fruit, compared with 25.1 for control fruit (0 °C), and this difference was even more marked at day 8 (Fig. 2). EjNAC4 transcripts also increased, but this was initiated two days later, and although prior HT and LTC reduced transcript levels, HT was less effective than LTC (Fig. 2).
Fig. 2.
Expression of loquat NAC genes in response to cold (0 °C), heat treatment (HT), and low-temperature conditioning (LTC). Values are presented as relative expression, with the value at 0 d set to 1. The materials were collected and treatments conducted as described by Xu et al. (2014). Error bars indicate SE from three biological replicates. LSD indicates least-significant difference at P=0.05.
Trans-activation by NAC genes of promoters of genes involved in lignin biosynthesis
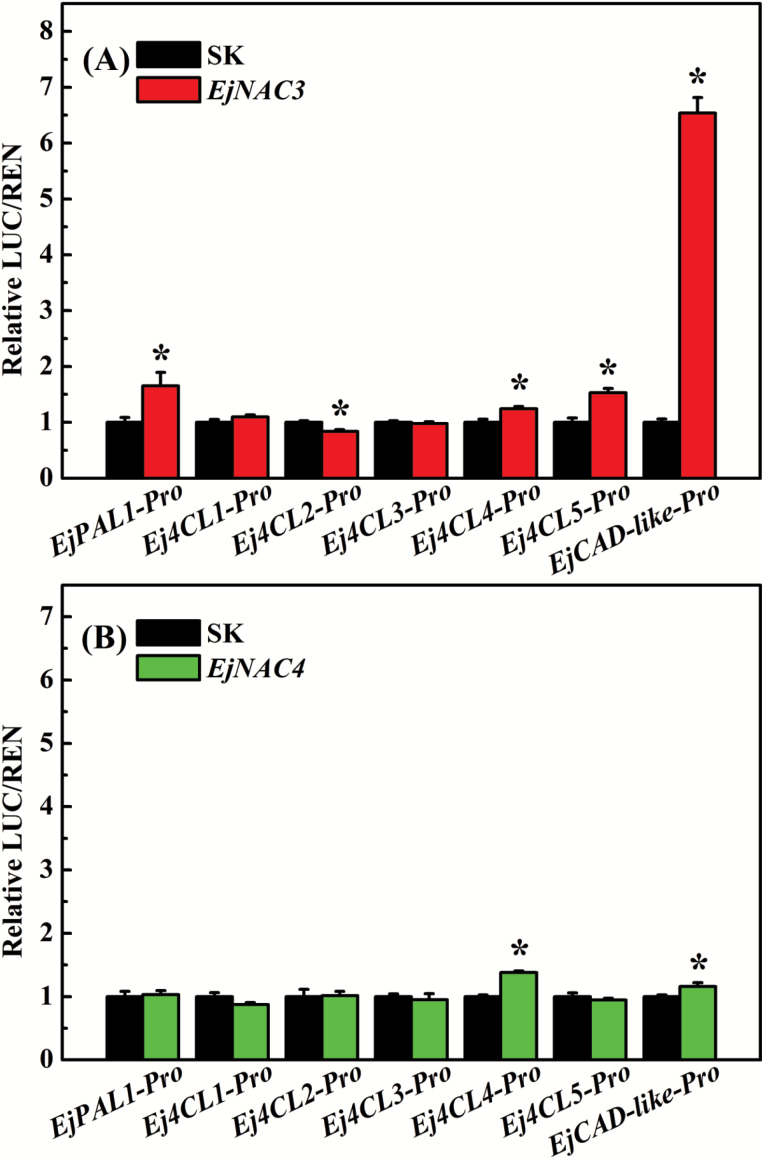
According to data derived from real-time PCR, EjNAC3 and EjNAC4 transcript levels are highly correlated with lignification of fruit flesh (Xu et al., 2014), which implies that these NAC genes might act as transcriptional regulators of lignin biosynthesis genes. To verify this conjecture, previously isolated promoters of seven genes from the phenylpropanoid pathway, namely EjPAL1, Ej4CL1, Ej4CL2, Ej4CL3, Ej4CL4, Ej4CL5, and EjCAD1, were selected for testing. Of these, Ej4CL1 and EjCAD1 were considered to be particularly important because they had been previously implicated in the control of lignification (Shan et al., 2008; Xu et al., 2014). Isolation of the full-length EjCAD1 indicated that it is an atypical CAD member, with only the NAD(P)-binding Rossmann-fold domain, and lacking the conserved zinc-binding signature GHEXXGXXXXXGXXV (Kim et al., 2004). Therefore, it was renamed as EjCAD-like (Genbank no. EF685346). Subcellular analysis indicated that EjNAC3 was localized in the nucleus (see Supplementary Fig. S1), which is similar to most transcription factors. The dual-luciferase assay indicated that promoter activity of EjCAD-like was increased 6.54-fold higher than the control after transient overexpression of EjNAC3 in tobacco leaves, but EjNAC3 had little effect on the other gene promoters tested (Fig. 3A). Despite the correlation of EjNAC4 with fruit lignification (Fig. 2), it had very limited effects on the promoters of lignin biosynthesis genes (Fig. 3B).
Fig. 3.
Regulatory effects of (A) EjNAC3 and (B) EjNAC4 on the promoters of loquat lignin biosynthesis genes as determined using dual-luciferase assays. The ratio of LUC/REN fluorescence obtained with the empty vector (SK) plus the promoter was used as a calibrator (set as 1). Error bars indicate SE from five replicates. Asterisks indicate that the difference between the control and treatment is significant at P<0.05.
Interaction between EjNAC3 and the EjCAD-like promoter
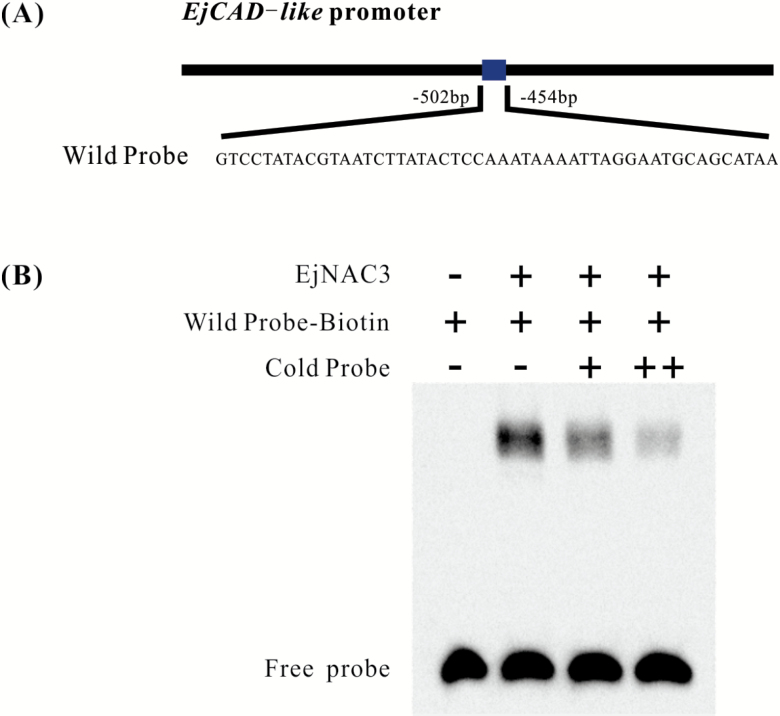
In order to clarify the interaction between EjNAC3 and the EjCAD-like promoter, yeast one-hybrid assays were performed. The promoter sequence of EjCAD-like was inserted into the pAbAi vector and transformed into the Y1H strain. The results indicated that the EjCAD-like promoter was not activated without protein binding (Fig. 4A), and the interaction between EjNAC3 and the EjCAD-like promoter was therefore analysed. Yeast containing EjNAC3 exhibited normal growth under 100 ng ml–1 AbA, while growth of the negative control with the empty vector was inhibited (Fig. 4B). EMSAs were performed aimed at locating more precisely the cis-element involved in binding. It has been reported that some NAC genes bind to TTGCGT or TTACGT core motifs (Lindemose et al., 2014), and after searching the promoter of EjCAD-like we found there are five TTRCGT sites within an 870-bp region upstream of the start codon (Fig. 5A). Sequences from the region –502 bp to –454 bp were used as the native promoter. The biotinylated probe was able to bind EjNAC3 protein, and the addition of a high concentration of cold probe significantly reduced the binding affinity of the biotinylated probe. (Fig. 5B).
Fig. 4.
Yeast one-hybrid analysis of the ability of EjNAC3 to bind the promoter of EjCAD-like. (A) Autoactivation was tested on SD medium lacking Ura with 100 ng ml–1 Aureobasidin A (AbA). (B) Interactions were determined on SD medium lacking Leu in the presence of AbA (SD/–Leu +100 ng ml–1 AbA). The empty pGADT7 vector was used as a negative control.
Fig. 5.
Electrophoretic mobility shift assay (EMSA) of EjNAC3 binding to the EjCAD-like promoter. (A) The probe sequence used for EMSA. (B) Purified EjNAC3 protein and biotin-labeled DNA probe were mixed and analysed on 6% native polyacrylamide gels. The presence (+) or absence (–) of specific probes is indicated. The concentration of the cold probe was 100 nM (+) or 300 nM (++), while that of the biotinylated probe was 1 nM.
Discussion
Lignin participates in various physiological processes, including the development of morphological characteristics, and the development of xylem strongly influences the conversion efficiency of lignocellulosic biomass to ethanol and the digestibility of forage crops (Jung, 1989; Ragauskas et al., 2006; Fu et al., 2011). Despite extensive studies on energy plants and model plants (e.g. Populus, Eucalyptus, switchgrass, sugarcane, and Arabidopsis), the regulatory mechanisms of fruit lignification are not well understood, and no transcription factors related to lignification have been reported in other fruit apart from loquat. Lignification in fruit is widely associated with stresses, such as chilling (Cai et al., 2006d) and wounding (Kamdee et al., 2014), and significantly influences fruit quality, consumer preference, and marketability. Thus, understanding the regulation of lignin biosynthesis is important not only in energy plants and model plant systems, but also for the fruit industry. Loquat fruit undergoes lignification after harvest, and LTC, HT, and application of MeJA have been developed to alleviate this (Cai et al., 2006d;Cao et al., 2009; Rui et al., 2010). Moreover, the molecular basis for fruit lignification has also been investigated, and the role of some structural genes of the phenylpropanoid pathway has been reported (Shan et al., 2008). In addition, four transcription factors, EjMYB1, EjMYB2, EjAP2-1, and EjNAC1, have been reported to be involved in lignification in loquat fruit. Further studies showed that the regulatory mechanisms of EjMYB1 and EjMYB2 were similar to their homologs from model plants (Xu et al., 2014), while EjAP2-1 was a novel regulator of lignin biosynthesis (Zeng et al., 2015), and EjNAC1 is an indirect regulator, although the underlying mechanisms have remained unclear (Xu et al., 2015). Compared to the extensively reported transcription factors from energy plants and model species, and in view of its economic importance, the transcription factors involved in fruit lignification require more detailed investigation.
Association between EjNAC expression and lignification of loquat fruit
In Arabidopsis, the NAC genes AtNST1 and AtNST2, AtVND1 to AtVND7, and AtSND1 are top-layer regulators and have a strong influence on the synthesis of constituents of the secondary cell wall (Mitsuda et al., 2005; Zhong et al., 2006; Zhou et al., 2014). Among these NAC genes, AtVNDs are expressed specifically in vessels while AtSND1, AtNST1, and AtNST2 are expressed in xylem fibers and extraxylary fibers or the anther endothecium. Mutation of these important regulatory genes results in abnormal secondary cell wall formation (Mitsuda et al., 2005; Zhong et al., 2006). Loquat EjNAC1 is associated with lignin accumulation, and is capable of trans-activation of the promoters of EjPAL1 and Ej4CL1. However, EjNAC1 lacks a physical interaction with the target promoters (Xu et al., 2015). In the current study, two new NAC genes were isolated, in addition to the previously reported EjNAC1 and EjNAC2. The expression analysis indicated that EjNAC3 and EjNAC4 transcripts were positively correlated with lignin accumulation in fruit flesh in response to storage at 0 °C, and expression was inhibited by LTC and HT, with a corresponding alleviation of lignification (Xu et al., 2014).
EjNAC3, unlike most other lignin-related NAC genes, is a direct regulator of the lignin biosynthesis pathway
In Arabidopsis, NAC genes have mainly been considered as switches for secondary cell wall metabolisms, rather than being the direct regulators of structural genes (Zhong et al., 2006), although there are rare exceptions such as AtVNT7 and AtSND1 (Zhao et al., 2010; Yamaguchi et al., 2011). However, enzymes of the phenylpropanoid pathway are not targets of AtVNT7, while AtSND1 binds Medicago truncatula MtF5H but not the Arabidopsis endogenous homolog. The present findings using dual-luciferase assays, yeast one-hybrid assays, and EMSAs indicated that EjNAC3 is a direct regulator of an EjCAD-like gene, via physical binding to TTRCGT motifs in the promoter. However, EjNAC4 had very limited effects on lignin biosynthesis genes. Further investigation of the relationship between EjNAC3 and known loquat fruit lignification-related transcription factors showed that both EjNAC3 and EjNAC4 had little effect on the promoters of EjMYB1, EjMYB2, and EjAP2-1 (data not shown). In addition, EjNAC3 is not only structurally different from most master-switch NAC members, but also NACs such as AtVND7 and AtSND1 that are capable of binding promoters of AtCesA4 and MtF5H, respectively (Zhao et al., 2010; Yamaguchi et al., 2011). Taken together, this evidence indicates that EjNAC3 is a new NAC transcription factor with a distinct role in regulating lignin biosynthesis.
The second candidate gene, EjNAC4, with a similar expression pattern to EjNAC3, was not, however, able to influence the activities of the target-gene promoters tested. It is possible that EjNAC4 might play a similar role to its homolog AtSND2, whose target is AtMYB103 (Hussey et al., 2011), and this is something that should be tested.
Role of EjNAC3 in fruit-flesh lignification
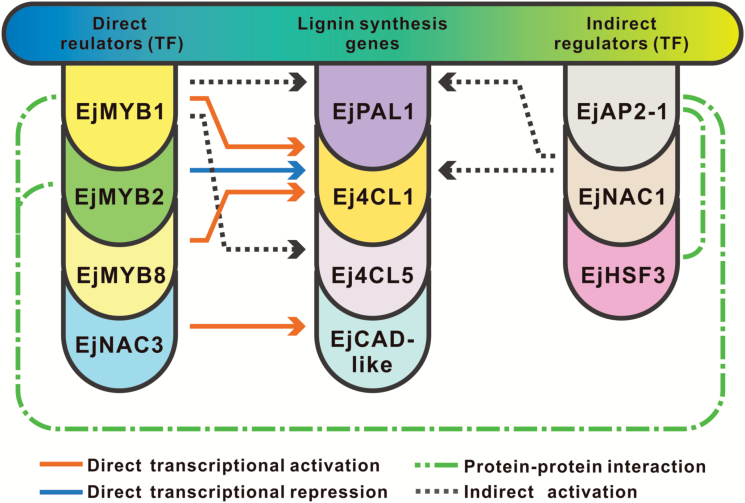
According to earlier studies (Xu et al., 2014; Wang et al., 2016), in loquat, Ej4CL1 was the only enzyme to be affected by previously characterized transcription factors, such as EjMYB1, EjMYB2, and EjMYB8 (Fig. 6). The lignin biosynthesis pathway is composed of 11 enzymes in addition to 4CL (Boerjan et al., 2003). Theoretically, each step in the pathway could be accessible to regulators. The discovery of EjNAC3 identifies a new regulatory step in the pathway, in addition to the control of 4CL. CADs catalyse the last step of monolignol synthesis, and the interaction between EjNAC3 and EjCAD-like broadens the regulatory range of lignin-related transcription factors. So far, two NAC family genes have been confirmed as being involved in fruit lignification, EjNAC1 (Xu et al., 2015), which regulates EjPAL1 and Ej4CL1, and EjNAC3. Interestingly, they seemed to function in parallel, since their targets are different. These clues indicate the complicated roles that NAC genes play in the hierarchy of regulatory interactions in the overall network (Fig. 6).
Fig. 6.
The regulatory network of lignin biosynthesis in loquat fruit. Although the network that regulates key enzymes in the phenylpropanoid pathway is far from complete, some transcription factors together with their targets have been identified, including NAC-domain proteins, MYB-domain proteins, a HSF protein, and an AP2-domain protein. Generally, these transcription factors can be classified into two groups: direct regulators that have physical interactions with identified target promoters, and indirect regulators that activate or repress expression of specific genes via pathways yet to be established.
In the present study, a chilling-inducible NAC transcription factor (EjNAC3) was characterized from loquat fruit. Phylogenetic analysis of its amino acid sequence indicated that it has a distinct structure compared with SND, NST, and VND genes. Dual-luciferase assays, Y1H assays, and EMSAs suggested that EjNAC3 could recognize and regulate the promoter of an EjCAD-like gene, indicating that EjNAC3 has the ability to directly regulate structural genes, which only few NAC genes possess. In loquat, chilling, as an environmental stimulus, can increase lignin production in the fruit flesh by inducing transcripts of EjNAC3 that act on a CAD-like gene.
Supplementary Data
Supplementary data are available at JXB online.
Table S1. Primers for used for RACE.
Table S2. Primers used for real time PCR.
Table S3. Primers used for full-length sequences of EjNAC gene isolation.
Table S4. Primers used for pGADT-7 vector construction.
Table S5. Probes used for EMSAs.
Fig. S1. Subcellular localization of EjNAC3.
Supplementary Material
Acknowledgements
This research was supported by the National Key Research and Development Program (2016YFD0400102), the National Natural Science Foundation of China (31630067, 31372041), the Natural Science Foundation of Zhejiang Province, China (LR16C150001), the Project of the Science and Technology Department of Zhejiang Province (2016C04001), and the 111 project (B17039). The authors have no conflict of interest.
Glossary
Abbreviations:
- 1-MCP
1-methylcyclopropene
- 4CL
4-coumarate-CoA ligase
- AbA
aureobasidin A
- ASA
acetylsalicylic acid
- CAD
cinnamyl alcohol dehydrogenase
- F5H
ferulate-5-hydroxylase
- HT
heat treatment
- LTC
low-temperature conditioning
- PAL
phenylalanine ammonia lyase
References
- Bennett T, van den Toorn A, Sanchez-Perez GF, Campilho A, Willemsen V, Snel B, Scheres B. 2010. SOMBRERO, BEARSKIN1, and BEARSKIN2 regulate root cap maturation in Arabidopsis. The Plant Cell 22, 640–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M. 2003. Lignin biosynthesis. Annual Review of Plant Biology 54, 519–546. [DOI] [PubMed] [Google Scholar]
- Cai C, Chen KS, Xu WP, Zhang WS, Li M, Ferguson I. 2006a. Effect of 1-MCP on postharvest quality of loquat fruit. Postharvest Biology and Technology 40, 155–162. [Google Scholar]
- Cai C, Li X, Chen KS. 2006b. Acetylsalicylic acid alleviates chilling injury of postharvest loquat (Eriobotrya japonica Lindl.) fruit. European Food Research and Technology 223, 533–539. [Google Scholar]
- Cai C, Xu CJ, Li X, Ferguson I, Chen KS. 2006c. Accumulation of lignin in relation to change in activities of lignification enzymes in loquat fruit flesh after harvest. Postharvest Biology and Technology 40, 163–169. [Google Scholar]
- Cai C, Xu CJ, Shan LL, Li X, Zhou CH, Zhang WS, Ferguson I, Chen KS. 2006d. Low temperature conditioning reduces postharvest chilling injury in loquat fruit. Postharvest Biology and Technology 41, 252–259. [Google Scholar]
- Cao SF, Zheng YH, Wang KT, Jin P, Rui HJ. 2009. Methyl jasmonate reduces chilling injury and enhances antioxidant enzyme activity in postharvest loquat fruit. Food Chemistry 115, 1458–1463. [Google Scholar]
- Cao SF, Zheng YH, Wang KT, Rui HJ, Tang SS. 2010. Effect of methyl jasmonate on cell wall modification of loquat fruit in relation to chilling injury after harvest. Food Chemistry 118, 641–647. [Google Scholar]
- Cassan-Wang H, Goué N, Saidi MN, Legay S, Sivadon P, Goffner D, Grima-Pettenati J. 2013. Identification of novel transcription factors regulating secondary cell wall formation in Arabidopsis. Frontiers in Plant Science 4, 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding CK, Wang CY, Gross KC, Smith DL. 2002. Jasmonate and salicylate induce the expression of pathogenesis-related-protein genes and increase resistance to chilling injury in tomato fruit. Planta 214, 895–901. [DOI] [PubMed] [Google Scholar]
- Fu C, Mielenz JR, Xiao X et al. . 2011. Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proceedings of the National Academy of Sciences, USA 108, 3803–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey SG, Mizrachi E, Spokevicius AV, Bossinger G, Berger DK, Myburg AA. 2011. SND2, a NAC transcription factor gene, regulates genes involved in secondary cell wall development in Arabidopsis fibres and increases fibre cell area in Eucalyptus. BMC Plant Biology 11, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HG. 1989. Forage lignins and their effects on fiber digestibility. Agronomy Journal 81, 33–38. [Google Scholar]
- Kamdee C, Imsabai W, Kirk R, Allan AC, Ferguson IB, Ketsa S. 2014. Regulation of lignin biosynthesis in fruit pericarp hardening of mangosteen (Garcinia mangostana L.) after impact. Postharvest Biology and Technology 97, 68–76. [Google Scholar]
- Kim SJ, Kim MR, Bedgar DL, Moinuddin SG, Cardenas CL, Davin LB, Kang C, Lewis NG. 2004. Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. Proceedings of the National Academy of Sciences, USA 101, 1455–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T. 2005. Transcription switches for protoxylem and metaxylem vessel formation. Genes & Development 19, 1855–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemose S, Jensen MK, Van de Velde J, O’Shea C, Heyndrickx KS, Workman CT, Vandepoele K, Skriver K, De Masi F. 2014. A DNA-binding-site landscape and regulatory network analysis for NAC transcription factors in Arabidopsis thaliana. Nucleic Acids Research 42, 7681–7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min T, Fang F, Ge H, Shi YN, Luo ZR, Yao YC, Grierson D, Yin XR, Chen KS. 2014. Two novel anoxia-induced ethylene response factors that interact with promoters of deastringency-related genes from persimmon. PLoS ONE 9, e97043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. 2005. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. The Plant Cell 17, 2993–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K, Oda Y, Fukuda H. 2010. Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. The Plant Cell 22, 3461–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston J, Wheeler J, Heazlewood J, Li SF, Parish RW. 2004. AtMYB32 is required for normal pollen development in Arabidopsis thaliana. The Plant Journal 40, 979–995. [DOI] [PubMed] [Google Scholar]
- Ragauskas AJ, Williams CK, Davison BH et al. . 2006. The path forward for biofuels and biomaterials. Science 311, 484–489. [DOI] [PubMed] [Google Scholar]
- Rui H, Cao S, Shang H, Jin P, Wang K, Zheng Y. 2010. Effects of heat treatment on internal browning and membrane fatty acid in loquat fruit in response to chilling stress. Journal of the Science of Food and Agriculture 90, 1557–1561. [DOI] [PubMed] [Google Scholar]
- Sayyari M, Babalar M, Kalantari S, Serrano M, Valero D. 2009. Effect of salicylic acid treatment on reducing chilling injury in stored pomegranates. Postharvest Biology and Technology 53, 152–154. [Google Scholar]
- Shan LL, Li X, Wang P et al. . 2008. Characterization of cDNAs associated with lignification and their expression profiles in loquat fruit with different lignin accumulation. Planta 227, 1243–1254. [DOI] [PubMed] [Google Scholar]
- Wang HZ, Dixon RA. 2012. On–off switches for secondary cell wall biosynthesis. Molecular Plant 5, 297–303. [DOI] [PubMed] [Google Scholar]
- Wang WQ, Zhang J, Ge H, Li SJ, Li X, Yin XR, Grierson D, Chen KS. 2016. EjMYB8 transcriptionally regulates flesh lignification in loquat fruit. PLoS ONE 11, e0154399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Wang WQ, Zeng JK, Zhang J, Grierson D, Li X, Yin XR, Chen KS. 2015. A NAC transcription factor, EjNAC1, affects lignification of loquat fruit by regulating lignin. Postharvest Biology and Technology 102, 25–31. [Google Scholar]
- Xu Q, Yin XR, Zeng JK, Ge H, Song M, Xu CJ, Li X, Ferguson IB, Chen KS. 2014. Activator- and repressor-type MYB transcription factors are involved in chilling injury induced flesh lignification in loquat via their interactions with the phenylpropanoid pathway. Journal of Experimental Botany 65, 4349–4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Mitsuda N, Ohtani M, Ohme-Takagi M, Kato K, Demura T. 2011. VASCULAR-RELATED NAC-DOMAIN 7 directly regulates the expression of a broad range of genes for xylem vessel formation. The Plant Journal 66, 579–590. [DOI] [PubMed] [Google Scholar]
- Yin XR, Allan AC, Chen KS, Ferguson IB. 2010. Kiwifruit EIL and ERF genes involved in regulating fruit ripening. Plant Physiology 153, 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng JK, Li X, Xu Q, Chen JY, Yin XR, Ferguson IB, Chen KS. 2015. EjAP2-1, an AP2/ERF gene, is a novel regulator of fruit lignification induced by chilling injury, via interaction with EjMYB transcription factors. Plant Biotechnology Journal 13, 1325–1334. [DOI] [PubMed] [Google Scholar]
- Zhang B, Xi WP, Wei WW, Shen JY, Ferguson I, Chen KS. 2011. Changes in aroma-related volatiles and gene expression during low temperature storage and subsequent shelf-life of peach fruit. Postharvest Biology and Technology 60, 7–16. [Google Scholar]
- Zhao Q, Wang H, Yin Y, Xu Y, Chen F, Dixon RA. 2010. Syringyl lignin biosynthesis is directly regulated by a secondary cell wall master switch. Proceedings of the National Academy of Sciences, USA 107, 14496–14501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Li S, Xi Y, Su X, Yi Y. 1999. Polyamine changes and chilling injury in cold-stored loquat fruits. Acta Botanica Sinica 42, 824–827. [Google Scholar]
- Zhong R, Demura T, Ye ZH. 2006. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. The Plant Cell 18, 3158–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH. 2008. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. The Plant Cell 20, 2763–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lee C, Zhong R, Ye ZH. 2009. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. The Plant Cell 21, 248–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Zhong R, Ye ZH. 2014. Arabidopsis NAC domain proteins, VND1 to VND5, are transcriptional regulators of secondary wall biosynthesis in vessels. PLoS ONE 9, e105726. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.