Abstract
Carbapenem-resistant Enterobacteriaceae (CRE) are a serious public health threat. Infections due to these organisms are associated with significant morbidity and mortality. Mechanisms of drug resistance in gram-negative bacteria (GNB) are numerous; β-lactamase genes carried on mobile genetic elements are a key mechanism for the rapid spread of antibiotic-resistant GNB worldwide. Transmissible carbapenem-resistance in Enterobacteriaceae has been recognized for the last 2 decades, but global dissemination of carbapenemase-producing Enterobacteriaceae (CPE) is a more recent problem that, once initiated, has been occurring at an alarming pace. In this article, we discuss the evolution of CRE, with a focus on the epidemiology of the CPE pandemic; review risk factors for colonization and infection with the most common transmissible CPE worldwide, Klebsiella pneumoniae carbapenemase–producing K. pneumoniae; and present strategies used to halt the striking spread of these deadly pathogens.
Keywords: epidemiology, gram-negative bacteria, Enterobacteriaceae infections, carbapenemases, drug resistance, antibacterial agents, carbapenems, adult, child, global health
The prevalence of multidrug-resistant organisms (MDROs), a major public health threat, continues to increase on a global level and is associated with significant morbidity and mortality. Historically, MDROs have affected patients in hospital settings, where exposure to antibiotics, frequent and/or long-term hospitalization, use of in-dwelling devices, and host factors provide risks for acquisition [1, 2]. However, the distinction between multidrug-resistant healthcare-acquired and community-onset bacterial infections has become blurred over the last 2 decades, with an explosion in antibiotic resistance genes located on mobile genetic elements (MGEs) capable of efficient spread between bacteria and hosts in and out of hospitals [3].
These trends are highlighted in Enterobacteriaceae, a family of gram-negative bacteria (GNB) responsible for a variety of community and healthcare-acquired infections. In GNB, the major driving force of resistance is the presence of β-lactamases (encoded by bla), a rapidly expanding list of β-lactam–hydrolyzing enzymes for which the number of unique protein sequences as currently cataloged has surpassed 2100 [4]. Many of these organisms carry additional plasmid-borne genes active against other classes of antibiotics, rendering bacteria resistant to multiple drugs [5, 6].
There is a dearth of drugs capable of treating MDR GNB infections [7]. As carbapenem-resistant Enterobacteriaceae (CRE) have become increasingly prevalent worldwide, carbapenems, long a last line of defense, more and more are challenged by MGEs harboring carbapenemases and other drug resistance genes [8]. As the molecular mechanisms of resistance continue to evolve, the epidemiology of CRE is changing, and growing numbers of people worldwide are being affected by these dangerous organisms.
MOLECULAR MECHANISMS OF CARBAPENEM RESISTANCE IN ENTEROBACTERIACEAE
Phenotypic resistance to carbapenems is typically caused by 2 main mechanisms: (1) β-lactamase activity combined with structural mutations and (2) production of carbapenemases, enzymes that hydrolyze carbapenem antibiotics (Table 1) [6, 9]. The former mechanism includes extended-spectrum β-lactamases (ESBLs), which are generally encoded by plasmids, and AmpC cephalosporinases (AmpC), for which expression in Enterobacteriaceae is most often associated with hyperproduction of enzymes from inducible or derepressed chromosomal genes [6]. ESBLs and AmpC are capable of conferring carbapenem resistance when combined with the mutation of porins, a family of proteins of the outer membrane of GNB that, when altered or lost, can retard diffusion of antibiotics across the bacterial membrane to a rate slow enough to facilitate the action of ESBL and AmpC enzymes [9, 17, 18]. Other mechanisms associated with carbapenem-resistance in GNB include drug efflux pumps and alterations in penicillin-binding proteins [8].
Table 1.
Characteristics of Common Acquired Carbapenem-Hydrolyzing β-Lactamases in Enterobacteriaceae
| Ambler Structural Class | Functional Classa | Active Siteb | Inhibitor(s) | Notable Genec | Mobile Genetic Elementsd | Multilocus STse | Retained β-Lactam Susceptibilityf |
|---|---|---|---|---|---|---|---|
| A | 2f | Serine | Commercially available β-lactamase inhibitors | KPC | IncFIIK2, IncF1A, IncI2, multiple types; Tn4401g | CC258 (ST258) dominant,h others | Carbapenems (low-to-high–level hydrolysis) |
| GES | Class I integronsi | … | Carbapenems (low-level hydrolysis) | ||||
| B | 3 | Zinc | Metal-chelating agents (eg, EDTA) | VIM | IncN, IncI1, multiple types; class I integrons | ST147, ST11, others | Monobactams spared |
| IMP | IncL/M, IncA/C, multiple types; class I integrons | … | |||||
| NDM | IncA/C, multiple; ISAba125 | ST101, ST11, several others | |||||
| D | 2d | Serine | NaCl (in vitro) | OXA-48 | IncL/M, Tn1999, IS1999 | ST147, ST11, ST101, ST405, ST395, others | PCN (high-level hydrolysis), carbapenems (low-level hydrolysis), extended-spectrum cephalosporins spared |
| OXA-181 | ColE plasmids, Tn2013, ISEcp1 | … |
Abbreviations: CG, clonal group; EDTA, ethylenediaminetetraacetic acid; GES, Guiana extended spectrum; IMP, active on imipenem; Inc, plasmid incompatibility type; IS, insertion sequence; KPC, Klebsiella pneumoniae carbapenemase; NDM, New Delhi metallo-β-lactamase; OXA, oxacillinase-type carbapenem-hydrolyzing β-lactamase; PCN, penicillin; ST, sequence type; VIM, Verona integron-encoded metallo-β-lactamase.
a Bush-Jacoby-Medeiros functional classification scheme.
b Hydrolytic mechanism. Class B carbapenem-hydrolyzing β-lactamases represent metallo-β-lactamases, and class D represent oxacillinases.
c The most common acquired genes are included and may vary by region. Only certain variants harbor carbapenemase genes (ie, GES-5). Several other genes exist in each class.
d The most common mobile genetic elements are included and may vary by region.
e Only Klebsiella pneumoniae– and Escherichia coli–associated STs are listed.
f The phenotypic profile may vary depending on the genetic variant type and/or if multiple β-lactamase genes are present in an isolate.
g Tn4401 is a Tn3-based transposon and is associated with multiple plasmid types. Tn1999 is associated with IncL/M plasmids.
h CC258 contains 43 STs, including the ST258 pandemic strains (2 major clades exist). Several non-CC258 strains (eg, ST147, ST442, and ST14) harbor blaKPC.
i These are often embedded in conjugative plasmids and/or transposons, facilitating horizontal transfer.
Carbapenemases are classified by their molecular structures and belong to 3 classes of β-lactamases: class A, B, and D of the Ambler classification system [6, 10]. Class A and D carbapenemases require serine at their active site, while class B, the metallo-β-lactamases (MBLs) require zinc for β-lactam hydrolysis [6]. Notable class A carbapenemase genes include Klebsiella pneumoniae carbapenemases (KPCs), Guiana extended spectrum (GES), imipenem resistant (IMI), non–metallo-carbapenemase-A (NMC-A), Serratia marcescens enzyme (SME), and Serratia fonticola carbapenemase (SFC), of which the KPCs are the most common transmissible class A genes circulating in Enterobacteriaceae worldwide [8]. KPCs are capable of hydrolyzing all β-lactams, and strains harboring blaKPC often have acquired resistance to fluoroquinolones, aminoglycosides, and trimethoprim-sulfamethoxazole, creating MDROs [19].
The international spread of KPC-producing Enterobacteriaceae is primarily due to clonal expansion of strains of K. pneumoniae belonging to clonal complex 258 (CC258) and, more specifically, to multilocus sequence type (ST) 258 strains harboring a blaKPC-2 or blaKPC-3 gene located on a Tn3-based transposon, Tn4401 [20, 21]. However, the propagation of blaKPC is much more complex. Circulating ST258 K. pneumoniae strains comprise 2 distinct genetic clades (I and II), and several additional sequence types have been found to carry blaKPC, which is associated with a variety of plasmids [8, 11, 22]. Additionally, KPC-producing strains have low to high level carbapenem resistance with corresponding minimum inhibitory concentrations ranging from susceptible to >16 µg/mL, related to increased blaKPC gene copy number, deletions directly upstream of the blaKPC gene, and/or outer membrane porin losses (OmpK35 and/or OmpK36) [8, 23].
The class D OXA β-lactamases, named somewhat ironically for their oxacillin-hydrolyzing capabilities, are a diverse and heterogeneous group of enzymes found in Acinetobacter species and, increasingly, especially the OXA-48 variants, in Enterobacteriaceae [24, 25]. The backbone most commonly associated with the spread of OXA-48–producing Enterobacteriaceae is an IncL/M-type plasmid with integration of the blaOXA-48 gene through the acquisition of a Tn1999 composite transposon [12, 24–26]. OXA-48 enzymes hydrolyze penicillins at a high level and carbapenems at a low level, while sparing extended-spectrum cephalosporins; however, strains may express multiple ESBLs, rendering them resistant to all β-lactams [12].
The class B MBLs are a complex group of enzymes that hydrolyze all β-lactams, save monobactams, and are not inhibited by commercially available β-lactamase inhibitors [6, 8]. They differ from the serine carbapenemases in the requirement of zinc for β-lactam hydrolysis; thus, their activity is inhibited by metal-chelating agents such as ethylenediaminetetraacetic acid (EDTA) [6, 8]. Notable transmissible MBL genes in Enterobacteriaceae include IMP (active on imipenem), VIM (Verona integron-encoded MBL), and NDM (New Delhi MBL) [6, 8, 27]. There are 3 MBL subclasses (B1–B3), which differ by amino acid sequence homology; almost all clinically important, acquired MBLs belong to subclass B1 [8, 28]. VIM-type and IMP-type MBLs are most commonly embedded in class I integrons and are associated with transposons or plasmids, which facilitate spread.
Although the rapid dissemination of NDM-producing Enterobacteriaceae resembles that of KPC-producing Enterobacteriaceae, the spread of NDM-type MBLs does not appear to be associated with dominate clonal strains and is mediated by several different plasmid incompatibility (Inc) types. The current theory is that the most common circulating NDM MBL gene in Enterobacteriaceae (blaNDM-1) evolved from Acinetobacter baumannii. This view is based on the complete or variant insertion sequence ISAba125 upstream of the blaNDM-1 gene in both blaNDM-harboring A. baumannii and Enterobacteriaceae and the similar coexpression in both genera of blaNDM with bleMBL, a gene responsible for resistance to the cancer drug bleomycin [29, 30]. NDM-type MBL genes have been found in several epidemic clones, including K. pneumoniae ST11 and ST147 and Escherichia coli ST131 and ST101, which are known to harbor other β-lactamase genes and antibiotic resistance determinants [8, 27, 30]. It is thought that the rapid and dramatic spread of NDM MBLs is facilitated by the genetic elements' bacterial promiscuity.
The differentiation between carbapenemase-producing (CP) CRE and non-CP CRE is important epidemiologically and clinically; however, providers need mainly susceptibility patterns and treatment recommendations for patient care. The Centers for Disease Control and Prevention (CDC) has provided updated definitions that can help direct definitions and testing considerations in the diagnosis and management of CRE [31].
THE GLOBAL DISTRIBUTION AND PREVALENCE OF THE MOST COMMON TRANSMISSIBLE CARBAPENEMASE GENES IN ENTEROBACTERIACEAE, BY REGION
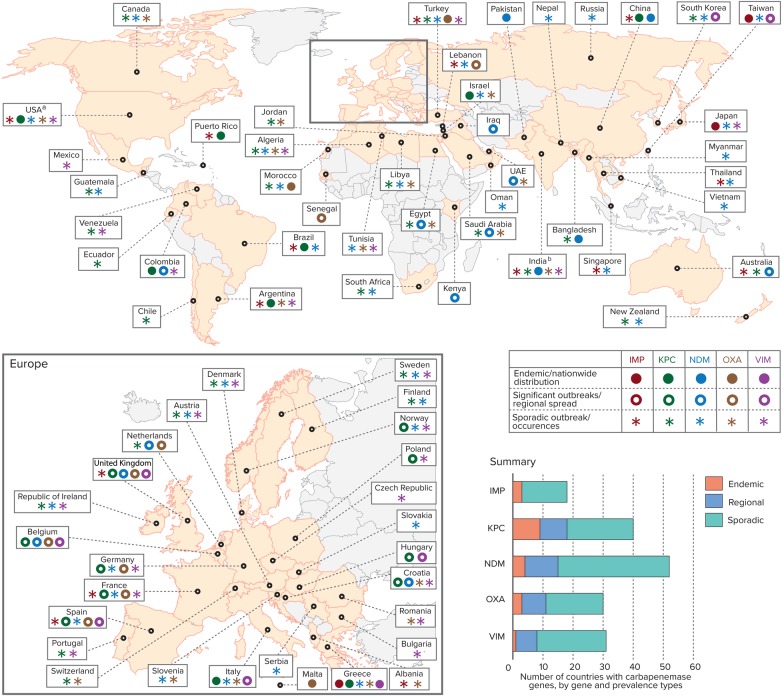
Figure 1 represents a global map that highlights the dramatic worldwide dissemination of carbapenemase genes in Enterobacteriaceae, by country and region. While the first identification of chromosomally based carbapenemase genes was in gram-positive bacilli, by the mid-to-late 1980s, “metalloenzymes,” now referred to as MBLs, were recognized in gram-negative non–lactose-fermenting bacteria [13]. This was followed shortly by description of another set of carbapenem-hydrolyzing enzymes (using serine at their active site) in Enterobacteriaceae [13]. This landscape radically changed in the early 1990s, when plasmid carriage of these originally chromosomally based, species-specific enzymes was recognized in multiple species found in clinical isolates [8, 13]. To date, the most common species of Enterobacteriaceae harboring transmissible carbapenemase genes are K. pneumoniae.
Figure 1.
Global distribution of carbapenemases in Enterobacteriaceae, by country and region. Data are adapted from [8, 12, 13, 15, 25, 32–40]. aKPCs are endemic in some US states; bOXA mainly refers to OXA-48, except in India, where it refers to OXA-181. Abbreviations: IMP, active on imipenem metallo-β-lactamase; KPC, Klebsiella pneumoniae carbapenemase; NDM, New Delhi metallo-β-lactamase; OXA, oxacillinase-type carbapenem-hydrolyzing β-lactamase; VIM, Verona integron-encoded metallo-β-lactamase.
The MBLs
The first major description of a transmissible carbapenemase gene in a clinical Enterobacteriaceae isolate occurred >2 decades ago when a gene, subsequently named IMP-1 MBL, was discovered on an integron in Serratia marcescens in Okazaki, Japan, associated with a plasmid-mediated outbreak in 7 Japanese hospitals. Widespread dissemination of blaIMP-1-harboring Enterobacteriaceae throughout Japan followed [41]. At present, there have been at least 52 variants of IMP genes identified in multiple species with worldwide distribution; however, to date, IMP-type MBL-containing Enterobacteriaceae are endemic only in Japan and Taiwan [32]. For example, a Taiwanese study from a 900-bed hospital in Southern Taiwan in 2002 assessed 9082 clinical Enterobacteriaceae isolates (other than Klebsiella species) for MBL genes and found that 29 of 1261 Enterobacter cloacae isolates (2.9%) harbored blaIMP-8, a variant of blaIMP-2 [42]. Descriptions of IMP MBLs in other countries are mostly of sporadic outbreaks or single reports [8, 9, 32].
The VIM-type MBLs were described in 1996 and 1997 in P. aeruginosa from Verona, Italy (VIM-1), and Marseilles, France (VIM-2) [43, 44]. By the late 1990s to early 2000s, there were several reports of VIM-type MBLs in Enterobacteriaceae [13]. Currently, VIM-2 is the most common VIM-type MBL worldwide, with at least 46 blaVIM variants now cataloged [45]. The epicenter of VIM-type Enterobacteriaceae is Greece, where K. pneumoniae and E. coli containing blaVIM-1 predominate [46]. A study in the intensive care units (ICUs) of 3 teaching hospitals in Athens, Greece, in 2002 recovered 17 K. pneumoniae isolates harboring blaVIM-1 over a 3-month period; at least 12 isolates were clinically relevant [47]. Since that time, several other VIM types have been recovered from gram-negative bacilli in Greece; globally, the majority of regions have reported outbreaks with VIM-producing Enterobacteriaceae [32, 33, 46, 47].
Attention to the epidemic of MBL-producing Enterobacteriaceae increased dramatically in 2008 with the discovery of an ST14 K. pneumoniae with a new MBL gene, blaNDM-1, from a Swedish patient who received healthcare in New Delhi, India [48]. Since then, there has been global dissemination of NDM MBLs with rapid gene transfer between species. In regions of endemicity, such as the Indian subcontinent, NDM-type MBLs predominate over other carbapenemases. In most other regions (except the Middle East and Balkan countries), NDM-type MBLs are described mostly as sporadic occurrences [30]. There are currently 16 cataloged variants of NDM-type MBLs, blaNDM-1 to blaNDM-16 [49].
Increasing colonization rates with blaNDM-producing bacteria have been noted in patients in several Indian and Pakistani hospitals, where reported prevalence rates of carriage of blaNDM-producing bacteria in ICUs range from 2% to 13.5% [50–53]. Additionally, data from the SENTRY Antimicrobial Surveillance Program (SENTRY) suggest that blaNDM may have been circulating in bacteria in India as early as 2006 [54]. In the Study for Monitoring Antimicrobial Resistance Trends 2009 program, of the 235 isolates tested from India, 66 (28%) carried ≥1 carbapenemase gene; the most common gene carried—in 50% of these isolates—was blaNDM-1 [55]. An additional concern particular to the NDM-type MBLs is spread via environmental sources in community settings in lower-income countries. A point-prevalence survey of public tap water and seepage water in India in 2011 found that a striking 4% of drinking water samples and 30% of seepage samples contained blaNDM-1-positive bacteria [56].
The Class D OXA Carbapenem-Hydrolyzing β-Lactamases
The worldwide spread of OXA-type carbapenemases is mainly attributed to the success of OXA-48–producing clones and, to a lesser extent, OXA-181 in certain regions (eg, the Indian subcontinent) [24, 26, 32]. The blaOXA-48 element was discovered in Turkey in 2001 in K. pneumoniae, and since then OXA-producing bacteria have become endemic in that country [57]. Several countries have reported outbreaks with OXA-producing Enterobacteriaceae, but few countries report endemicity. Because of the variable susceptibility profiles of OXA—heterogeneity of hydrolysis of carbapenems, broad-spectrum cephalosporins, and aztreonam and lack of inhibition by EDTA or clavulanic acid—the prevalence of these enzymes may be underestimated [24, 32].
The Class A KPCs
The global rise of KPC-producing Enterobacteriaceae remains one of the most successful MDRO pandemics in the history of GNB. A major focus of the propagation and persistence of KPC-harboring Enterobacteriaceae is the successful ST258 lineage, MDR strains of K. pneumoniae that are endemic in an increasing number of countries and are responsible for many major outbreaks worldwide [8, 34].
KPC-ENDEMIC REGIONS
Greece
Greece has experienced some of the highest carbapenem resistance rates among GNB globally. Prior to 2001, the Greek System for the Surveillance of Antimicrobial Resistance reported carbapenem resistance prevalence of <1%; this increased to 30% in hospital wards and to 60% in ICUs by 2008 [58]. Data from the European Centre for Disease Prevention and Control EARS-Net revealed that, in 2014, of 1088 Greek K. pneumoniae isolates, 678 (62.3%) were resistant to carbapenems [35]. Before 2006, the predominant carbapenemase genes in Enterobacteriaceae recovered in Greece were VIM-1–type MBLs. This changed after the introduction and rapid dissemination of blaKPC-2-producing K. pneumoniae isolates throughout the country in 2007; by the end of 2008, a surveillance study of 21 hospitals found blaKPC-2-producing K. pneumoniae in 18 hospitals across Crete, Thessaloniki, and Athens, with 96% of isolates a single pulsotype and the ST258 lineage [59]. More-recent surveys confirm the ongoing dominance of ST258 K. pneumoniae strains; however, several other ST types harboring blaKPC are circulating among the almost 40% of K. pneumoniae currently harboring blaKPC in Greece [34, 36].
Israel
Israel was the second country (after the United States) to report outbreaks of infection due to KPC-producing K. pneumoniae. A study of carbapenem-resistant K. pneumoniae in Tel-Aviv during 2004–2006 disclosed epidemic blaKPC-2- or blaKPC-3-carrying strains. The peak of the outbreak occurred in 2007, with 55.5 incident nosocomial cases of carbapenem-resistant K. pneumoniae infection per month per 100 000 patient-days [60, 61]. A nationally implemented intervention, employed in 2007–2008, resulted in a decrease in the monthly incidence to 11.7 cases per 100 000 patient-days [61].
Two cross-sectional, point-prevalence national surveys of CP Enterobacteriaceae (CPE) in post-acute-care Israeli hospitals in 2008 and 2013 (before and after the intervention) showed a significant decrease in the overall prevalence of carbapenem resistance among Enterobacteriaceae isolates (184 of 1147 isolates [16%] and 127 of 1287 isolates [9.9%], respectively). Notably, in 2008, all CPE surveyed were KPC-containing K. pneumoniae, while during the 2013 survey, additional carbapenemase genes were found (including blaNDM and blaOXA). However, KPC-carrying K. pneumoniae persisted as the predominant CPE, with an increasing proportion of ST258 K. pneumoniae strains (120 of 184 [65%] in 2008 vs 91 of 113 [80%] in 2013) [37].
Latin America
KPC-producing bacteria disseminated throughout Colombia in the late 2000s after the discovery of K. pneumoniae harboring blaKPC-2 in 2005 in patients with no travel history, followed by an outbreak of infection due to K. pneumoniae carrying blaKPC-3, which was traced to an index patient who had travelled recently to Israel [62–64]. In 2006, Colombia was the first country to report a KPC-producing Pseudomonas aeruginosa [65]. Since that time, other countries, including Argentina, Chile, and Mexico, have reported the introduction of KPC-producing Enterobacteriaceae; the highest prevalence of blaKPC-positive bacteria outside of Colombia is in Brazil, with dissemination throughout the country and reports of KPC-producing isolates in all states [64]. The spread in Brazil has been associated mostly with CC258 K. pneumoniae, including ST258, ST11, and ST437; these strains harbor a blaKPC-2 gene associated with Tn4401b and multiple plasmid (IncFII, IncL/M, and IncN) types [66–68]. Of 70 CRE submitted to SENTRY from Latin hospitals in 2010, 56 strains contained blaKPC-2; 44 (78.6%) were from Brazil, and among 19 Brazilian K. pneumoniae strains tested, 17 (89.5%) were grouped within CC258 [38].
United States
The first KPC-producing K. pneumoniae was discovered in a patient in a North Carolina hospital in 1996 [69]. By 2001, there was an explosion of reports in northeastern United States (with a focus in New York) of KPC-producing bacteria in hospitalized patients [34, 70]. In 2006, the Meropenem Yearly Susceptibility Test Information Collection surveillance program described 57 isolates, with 9.5% of the collection characterized as clonal blaKPC-producing Klebsiella strains, representing a 2-fold increase from the prior year; most isolates were from states in the Mid-Atlantic US Census division, and hospital prevalence rates ranged from 2.4% in Ohio to 50.8% in New York [71, 72].
A nationwide survey by SENTRY in 2007–2009 studied 42 medical centers for CP K. pneumoniae and found an overall blaKPC-positive bacteria prevalence of 5.5%, with significant regional increases in KPC genes detected in K. pneumoniae over the period in the Mid-Atlantic (28.6% overall and 33% in 2009) and East North Central (2.4% overall, 3.8% in 2009) US Census divisions [73]. The expansion of KPC-producing bacteria across the United States is clearly evident; the same surveillance program reported that, by 2010, 28 of 195 Enterobacteriaceae isolates (14.4%) surveyed from 26 US medical centers harbored blaKPC-2 or blaKPC-3; 9 of the 28 were found in Texas [74].
Most recently, a population- and laboratory-based active surveillance study of 7 US metropolitan areas during 2012–2013 found an overall annual CRE incidence of 2.93 cases per 100 000 population; of 188 CRE isolates tested, 90 (47.3%) were identified as CPE, all of which were found to contain blaKPC [75]. As of April 2016, the CDC reported that KPC-producing bacteria have been identified in 48 states, the District of Columbia, and Puerto Rico; however, the endemicity of KPC-producing bacteria within the United States is still focused in regional hot spots [34, 76].
In children, there have been notable increases in CRE prevalence, although few genotypic data are available [14]. A recent study of CRE prevalence in US children, using antimicrobial susceptibility data for Enterobacteriaceae reported to 300 laboratories participating in the Surveillance Network–USA database during January 1999–July 2012, found an overall low pediatric prevalence of CRE (266 of 316 253 isolates [0.08%]); however, there was a significant increase in CRE detected in children over the study period (from 0% in 1999–2000 to 0.47% in 2011–2012) [77]. The highest increases were seen in Enterobacter species, blood culture isolates, and isolates from patients in the ICU (0.0% in 1999–2000 and 5.2%, 4.5%, and 3.2%, respectively, in 2011–2012). While the increases in Enterobacter species may be related to nosocomial ecology and not reflect true carbapenemase production, there were also significant increases in CR E. coli and K. pneumoniae (both 0% in 1999–2000 and 0.14% and 1.7%, respectively, in 2011–2012), which more likely represent CPE [77]. Colonization and infection with KPC and MBL-producing Enterobacteriaceae are being reported increasingly across the United States in pediatric settings [14, 78–82].
CLINICAL EPIDEMIOLOGY OF KPC-PRODUCING ENTEROBACTERIACEAE IN THE UNITED STATES
KPC-producing bacteria in the United States, unlike NDM-type and CTX-M–producing Enterobacteriaceae, generally have not emerged as community pathogens. The majority of CRE infections are mainly a problem for inpatient facilities. CRE infections are associated with mortality rates as high as 40%–50%, and these organisms continue to increase in prevalence in national reports [83, 84]. Because most KPCs (and other carbapenemases) are found in K. pneumoniae, the majority of studies assessing factors associated with CRE have been specific to CR K. pneumoniae or KPC-producing K. pneumoniae. These associations have varied, likely because of differences in study venues and design; risk factors for colonization and/or infection in adults that have been identified in multiple studies include critical illness, comorbid conditions, prolonged hospitalization, multiple invasive medical devices, poor functional status, mechanical ventilation, and receipt of certain antibiotic classes [34, 85–87]. Less is known about the epidemiology of KPC-producing Enterobacteriaceae infections in children; however, similar factors have generally been implicated [14, 82, 88–90].
One important risk for CRE colonization appears to be residence in long-term acute care hospitals (LTACHs), mediated in part by interfacility spread at the time of patient transfers [91]. A point-prevalence study performed in Chicago, Illinois, found that 30.4% of patients (119 of 391) in 7 LTACHs were colonized with KPC-producing Enterobacteriaceae, compared with 3.3% of ICU patients (30 of 910) in 24 short-stay hospitals (prevalence ratio, 9.2; 95% confidence interval, 6.3–13.5) [92]. A recently published case-control study of LTACH patients found that independent factors associated with CRE colonization and infection in this setting included solid organ and stem cell transplantation, mechanical ventilation, fecal incontinence, and exposure in the prior 30 days to carbapenems, vancomycin, and metronidazole [93].
CONTROLLING THE SPREAD OF CRE IN HEALTHCARE SETTINGS
Interventions to curtail the spread of CRE in healthcare facilities most often have involved bundled infection control measures; so, the success of one individual measure cannot simply be compared directly to another. However, successful solutions based on multiple studies include using patient cohorts, contact isolation, and dedicated staffs; daily bathing of all patients with chlorhexidine; educating and training staff; limiting use of invasive devices; shortening the duration of mechanical ventilation; improving hand hygiene rates and antimicrobial stewardship; and, in some studies, enhancing environmental cleaning [84, 85, 94].
In high-prevalence areas, regional surveillance can be extremely useful when paired with the sharing of patient information among facilities; a strategy recommended by the CDC is to “detect and protect” through early identification of patients infected with CRE, followed by prevention of transmission through implementation of infection control precautions [95]. An example of this is the statewide registry of extensively drug-resistant organisms in Illinois, an interactive public health informatics tool that provides a mechanism for standardized reporting of CRE-carrier patients from all healthcare facilities throughout the state. This unique partnership of public health, academia, and non-profit organizations aids in decreasing spread of CRE through communication, which allows for early detection and intervention by receiving facilities [96].
Potential interventions in US facilities where CRE rates are still low include screening high-risk patients for CRE carriage on admission, such as patients transferred from long-term care facilities; while awaiting screening results, hospitals may use preemptive contact precautions for such admissions, especially if rates are high in referring facilities.
LOOMING THREATS
In November 2015, Liu et al reported a new public health threat, transmissible polymyxin resistance in Enterobacteriaceae associated with the plasmid-mediated colistin resistance gene, mcr-1, a member of the phosphoethanolamine transferase enzyme family [97]. E. coli and K. pneumoniae that harbored mcr-1 were found in contaminated retail meat and in colonized food animals and inpatients in 5 Chinese provinces [97]. Shortly after recognition of the threat of plasmid-mediated polymixin resistance, a clinical isolate from a Swiss patient, with no travel history, was discovered to coharbor plasmid-mediated blaMCR-1 and MBL (blaVIM-1) genes [98]. As of March 2016, at least 17 countries had identified mcr-1 in gram-negative organisms in food, animals, and/or humans; several reports have documented isolates coharboring carbapenemase and/or ESBL genes with mcr-1; and studies have suggested a link between this unwelcome emergence and the broad agricultural and veterinary use of polymyxins [98–100].
CONCLUSIONS
CRE continue to evolve, posing an increasing threat to patients of all ages. Mechanisms of carbapenem resistance are variable, and the breadth of MGEs in Enterobacteriaceae—carbapenemase genes and other antibiotic resistance mechanisms and virulence determinants—continues to expand. Early recognition of this global public health threat through molecular characterization, epidemiologic studies, and surveillance may allow for timely approaches in prevention. Bundled infection control measures, education and training, and interventions aimed at healthcare-associated risk factors for colonization and/or infection, as well as proactive assessment of emerging community reservoirs, may help thwart the rapid dissemination of these truly menacing pathogens.
Notes
Acknowledgments. We thank Mr Suraj Pant of the Center for Disease Dynamics, Economics, and Policy, for the creation of the figure; and Drs Sumanth Gandra, Mariam S. Aziz, Robert A. Bonomo, Kenneth M. Boyer, and Mary K. Hayden, for thoughtful comments and guidance.
Disclaimer. This content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health (NIH).
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases, NIH (grant K08AI112506 to L. K. L.), and by the Foglia Foundation (unrestricted grant to R. A. W., via Rush University Medical Center).
Potential conflict of interest. Both authors: No reported conflicts. Both authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Siegel JD, Rhinehart E, Jackson M, Chiarello L, Health Care Infection Control Practices Advisory Committee. 2007 guideline for isolation precautions: preventing transmission of infectious agents in health care settings. Am J Infect Control 2007; 35:S65–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Siegel JD, Rhinehart E, Jackson M, Chiarello L, Healthcare Infection Control Practices Advisory Committee. Management of multidrug-resistant organisms in health care settings, 2006. Am J Infect Control 2007; 35:S165–93. [DOI] [PubMed] [Google Scholar]
- 3. Prabaker K, Weinstein RA. Trends in antimicrobial resistance in intensive care units in the United States. Curr Opin Crit Care 2011; 17:472–9. [DOI] [PubMed] [Google Scholar]
- 4. Bush K. Top 10 Beta-lactamase Papers for 2015. Presented at: ASM Microbe 2016. Boston, MA, 18 June 2016. [Google Scholar]
- 5. Jacoby GA, Munoz-Price L. The new β-lactamases. N Engl J Med 2005; 352:380–91. [DOI] [PubMed] [Google Scholar]
- 6. Bush K, Fisher J. Epidemiological expansion, structural studies, and clinical challenges of new β-lactamases from gram-negative bacteria. Annu Rev Microbiol 2011; 65:455–78. [DOI] [PubMed] [Google Scholar]
- 7. Thomson JM, Bonomo RA. The threat of antibiotic resistance in Gram-negative pathogenic bacteria: β-lactams in peril!. Curr Opin Microbiol 2005; 8:518–24. [DOI] [PubMed] [Google Scholar]
- 8. Patel G, Bonomo RA. “Stormy waters ahead”: global emergence of carbapenemases. Front Microbiol 2013; 4:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bush K, Jacoby GA. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother 2010; 54:969–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ambler RP. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci 1980; 289:321–31. [DOI] [PubMed] [Google Scholar]
- 11. Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother 2009; 53:2227–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Poirel L, Potron A, Nordmann P. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother 2012; 67:1597–606. [DOI] [PubMed] [Google Scholar]
- 13. Queenan AM, Bush K. Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev 2007; 20:440–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Logan LK. Carbapenem-resistant Enterobacteriaceae: an emerging problem in children. Clin Infect Dis 2012; 55:852–9. [DOI] [PubMed] [Google Scholar]
- 15. Pitout JDD. Worldwide spread of carbapenemases: update 2015 and future prospects. Presented at: ICAAC/ICC, San Diego, California, 17–21 September 2015. [Google Scholar]
- 16. Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol 2014; 22:686–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Paterson DL, Bonomo RA. Extended-spectrum β-lactamases: a clinical update. Clin Microbiol Rev 2005; 18:657–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jacoby GA. AmpC beta-lactamases. Clin Microbiol Rev 2009; 22:161–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis 2009; 9:228–36. [DOI] [PubMed] [Google Scholar]
- 20. Kitchel B, Rasheed JK, Patel JB et al. . Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother 2009; 53:3365–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cuzon G, Naas T, Nordmann P. Functional characterization of Tn4401, a Tn3-based transposon involved in blaKPC gene mobilization. Antimicrob Agents Chemother 2011; 55:5370–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen L, Mathema B, Pitout JDD, DeLeo FR, Kreiswirth BN. Epidemic Klebsiella pneumoniae ST258 Is a Hybrid Strain. mBio 2014; 5:e01355–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kitchel B, Rasheed JK, Endimiani A et al. . Genetic factors associated with elevated carbapenem resistance in KPC-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 2010; 54:4201–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Poirel L, Naas T, Nordmann P. Diversity, epidemiology, and genetics of class D β-lactamases. Antimicrob Agents Chemother 2010; 54:24–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Carrer A, Poirel L, Yilmaz M et al. . Spread of OXA-48-encoding plasmid in Turkey and beyond. Antimicrob Agents Chemother 2010; 54:1369–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Poirel L, Bonnin RA, Nordmann P. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob Agents Chemother 2012; 56:559–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Walsh TR. Emerging carbapenemases: a global perspective. Int J Antimicrob Agents 2010; 36:S8–S14. [DOI] [PubMed] [Google Scholar]
- 28. Mojica MF, Bonomo RA, Fast W. B1-metallo-beta-lactamases: where do we stand? Curr Drug Targets 2016; 17:1029–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dortet L, Nordmann P, Poirel L. Association of the emerging carbapenemase NDM-1 with a bleomycin resistance protein in Enterobacteriaceae and Acinetobacter baumannii. Antimicrob Agents Chemother 2012; 56:1693–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dortet L, Poirel L, Nordmann P. Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. Biomed Res Int 2014; 2014:249856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. FAQs About Choosing and Implementing a CRE Definition. https://www.cdc.gov/hai/organisms/cre/definition.html. Accessed 25 June 2016. [Google Scholar]
- 32. Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 2011; 17:1791–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Glasner C, Albiger B, Buist G et al. . Carbapenemase-producing Enterobacteriaceae in Europe: a survey among national experts from 39 countries, February 2013. Euro Surveill 2013; 18:20525. [DOI] [PubMed] [Google Scholar]
- 34. Munoz-Price LS, Poirel L, Bonomo RA et al. . Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 2013; 13:785–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. European Centre for Disease Prevention and Control (EARS-Net). http://ecdc.europa.eu/en/healthtopics/antimicrobial_resistance/database/Pages/table_reports.aspx. Accessed 12 April 2016.
- 36. Giakkoupi P, Papagiannitsis CC, Miriagou V et al. . An update of the evolving epidemic of blaKPC-2-carrying Klebsiella pneumoniae in Greece (2009–10). J Antimicrob Chemother 2011; 66:1510–3. [DOI] [PubMed] [Google Scholar]
- 37. Adler A, Hussein O, Ben-David D et al. . Persistence of Klebsiella pneumoniae ST258 as the predominant clone of carbapenemase-producing Enterobacteriaceae in post-acute-care hospitals in Israel, 2008–13. J Antimicrob Chemother 2015; 70:89–92. [DOI] [PubMed] [Google Scholar]
- 38. Castanheira M, Costello AJ, Deshpande LM, Jones RN. Expansion of clonal complex 258 KPC-2-producing Klebsiella pneumoniae in Latin American hospitals: report of the SENTRY Antimicrobial Surveillance Program. Antimicrob Agents Chemother 2012; 56:1668–9; author reply 1670–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cantón R, Akóva M, Carmeli Y et al. . Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin Microbiol Infect 2012; 18:413–31. [DOI] [PubMed] [Google Scholar]
- 40. Badal R, Kazmierczak K, Hackel M et al. . Geographic distribution of carbapenemases found in Enterobacteriaceae—SMART 2012 [abstract C-799]. Presented at: 54th Interscience Conference on Antimicrobial Agents and Chemotherapy,Washington, DC, 5–9 September 2014. [Google Scholar]
- 41. Ito H, Arakawa Y, Ohsuka S, Wacharotayankun R, Kato N, Ohta M. Plasmid-mediated dissemination of the metallo-beta-lactamase gene blaIMP among clinically isolated strains of Serratia marcescens. Antimicrob Agents Chemother 1995; 39:824–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yan JJ, Ko WC, Chuang CL, Wu JJ. Metallo-beta-lactamase-producing Enterobacteriaceae isolates in a university hospital in Taiwan: prevalence of IMP-8 in Enterobacter cloacae and first identification of VIM-2 in Citrobacter freundii. J Antimicrob Chemother 2002; 50:503–11. [DOI] [PubMed] [Google Scholar]
- 43. Lauretti L, Riccio ML, Mazzariol A et al. . Cloning and characterization of blaVIM, a new integron-borne metallo-beta-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob Agents Chemother 1999; 43:1584–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Poirel L, Naas T, Nicolas D et al. . Characterization of VIM-2, a carbapenem-hydrolyzing metallo-beta-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob Agents Chemother 2000; 44:891–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. OXA-Type β-Lactamases. http://www.lahey.org/studies/other.asp#table1 Accessed 11 May 2016. [Google Scholar]
- 46. Walsh TR, Toleman MA, Poirel L, Nordmann P. Metallo-ß-Lactamases: the Quiet before the Storm? Clin Microbiol Rev 2005; 18:306–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vatopoulos A. High rates of metallo-beta-lactamase-producing Klebsiella pneumoniae in Greece--a review of the current evidence. Euro Surveill 2008; 13:8023. [PubMed] [Google Scholar]
- 48. Yong D, Toleman MA, Giske CG et al. . Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 2009; 53:5046–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. OXA-Type β-Lactamases. http://www.lahey.org/studies/other.asp#table1 Accessed 11 May 2016. [Google Scholar]
- 50. Kumarasamy KK, Toleman MA, Walsh TR et al. . Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 2010; 10:597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bharadwaj R, Joshi S, Dohe V, Gaikwad V, Kulkarni G, Shouche Y. Prevalence of New Delhi metallo-beta-lactamase (NDM-1)-positive bacteria in a tertiary care centre in Pune, India. Int J Antimicrob Agents 2012; 39:265–6. [DOI] [PubMed] [Google Scholar]
- 52. Deshpande P, Shetty A, Kapadia F, Hedge A, Soman R, Rodrigues C. New Delhi metallo 1: have carbapenems met their doom? Clin Infect Dis 2010; 51:1222. [DOI] [PubMed] [Google Scholar]
- 53. Perry JD, Naqvi SH, Mirza IA et al. . Prevalence of faecal carriage of Enterobacteriaceae with NDM-1 carbapenemase at military hospitals in Pakistan, and evaluation of two chromogenic media. J Antimicrob Chemother 2011; 66:2288–94. [DOI] [PubMed] [Google Scholar]
- 54. Castanheira M, Deshpande LM, Mathai D, Bell JM, Jones RN, Mendes RE. Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006–2007. Antimicrob Agents Chemother 2011; 55:1274–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lascols C, Hackel M, Marshall SH et al. . Increasing prevalence and dissemination of NDM-1 metallo-β-lactamase in India: data from the SMART study (2009). J Antimicrob Chemother 2011; 66:1992–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Walsh TR, Weeks J, Livermore DM, Toleman MA. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis 2011; 11:355–62. [DOI] [PubMed] [Google Scholar]
- 57. Poirel L, Heritier C, Tolun V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother 2004; 48:15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Souli M, Galani I, Antoniadou A et al. . An outbreak of infection due to ß-Lactamase Klebsiella pneumoniae carbapenemase 2–producing K. pneumoniae in a Greek university hospital: molecular characterization, epidemiology, and outcomes. Clin Infect Dis 2010; 50:364–73. [DOI] [PubMed] [Google Scholar]
- 59. Giakoupi P, Maltezou H, Polemis M et al. . KPC-2-producing Klebsiella pneumoniae infections in Greek hospitals are mainly due to a hyperepidemic clone. Euro Surveill 2009; 14:19218. [DOI] [PubMed] [Google Scholar]
- 60. Leavitt A, Navon-Venezia S, Chmelnitsky I, Schwaber MJ, Carmeli Y. Emergence of KPC-2 and KPC-3 in carbapenem-resistant Klebsiella pneumoniae strains in an Israeli hospital. Antimicrob Agents Chemother 2007; 51:3026–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schwaber MJ, Lev B, Israeli A et al. . Containment of a country-wide outbreak of carbapenem-resistant Klebsiella pneumoniae in Israeli hospitals via a nationally implemented intervention. Clin Infect Dis 2011; 52:848–55. [DOI] [PubMed] [Google Scholar]
- 62. Mojica MF, Correa A, Vargas DA et al. . Molecular correlates of the spread of KPC-producing Enterobacteriaceae in Colombia. Int J Antimicrob Agents 2012; 40:277–9. [DOI] [PubMed] [Google Scholar]
- 63. Villegas MV, Lolans K, Correa A et al. . First detection of the plasmid-mediated class A carbapenemase KPC-2 in clinical isolates of Klebsiella pneumoniae from South America. Antimicrob Agents Chemother 2006; 50:2880–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Maya JJ, Ruiz SJ, Blanco VM et al. . Current status of carbapenemases in Latin America. Expert Rev Anti Infect Ther 2013; 11:657–67. [DOI] [PubMed] [Google Scholar]
- 65. Villegas MV, Lolans K, Correa A et al. . First identification of Pseudomonas aeruginosa isolates producing a KPC-type carbapenem-hydrolyzing beta-lactamase. Antimicrob Agents Chemother 2007; 51:1553–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Andrade LN, Curiao T, Ferreira JC et al. . Dissemination of blaKPC-2 by the spread of Klebsiella pneumoniae clonal complex 258 clones (ST258, ST11, ST437) and plasmids (IncFII, IncN, IncL/M) among Enterobacteriaceae species in Brazil. Antimicrob Agents Chemother 2011; 55:3579–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Perenguez M, Motoa G, Correa A et al. . Molecular characterization of carbapenemases among Enterobacteriaceae in Latin America (LatAm) [abstract C-801]. Presented at: 54th InterscienceConference on Antimicrobial Agents and Chemotherapy, Washington, DC, 5–9 September 2014. [Google Scholar]
- 68. Pereira PS, de Araujo CFM, Seki LM, Zahner V, Carvalho-Assef APD, Asensi MD. Update of the molecular epidemiology of KPC-2-producing Klebsiella pneumoniae in Brazil: spread of clonal complex 11 (ST11, ST437 and ST340). J Antimicrob Chemother 2013; 68:312–6. [DOI] [PubMed] [Google Scholar]
- 69. Yigit H, Queenan AM, Anderson GJ et al. . Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 2001; 45:1151–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bradford PA, Bratu S, Urban C et al. . Emergence of carbapenem-resistant Klebsiella species possessing the class a carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 β-lactamases in New York City. Clin Infect Dis 2004; 39:55–60. [DOI] [PubMed] [Google Scholar]
- 71. Rhomberg PR, Deshpande LM, Kirby JT, Jones RN. Activity of meropenem as serine carbapenemases evolve in US medical centers: monitoring report from the MYSTIC program (2006). Diagn Microbiol Infect Dis 2007; 59:425–32. [DOI] [PubMed] [Google Scholar]
- 72. Deshpande LM, Rhomberg PR, Sader HS, Jones RN. Emergence of serine carbapenemases (KPC and SME) among clinical strains of Enterobacteriaceae isolated in the United States Medical Centers: report from the MYSTIC Program (1999–2005). Diagn Microbiol Infect Dis 2006; 56:367–72. [DOI] [PubMed] [Google Scholar]
- 73. Kaiser RM, Castanheira M, Jones RN, Tenover F, Lynfield R. Trends in Klebsiella pneumoniae carbapenemase-positive K. pneumoniae in US hospitals: report from the 2007–2009 SENTRY Antimicrobial Surveillance Program. Diagn Microbiol Infect Dis 2013; 76:356–60. [DOI] [PubMed] [Google Scholar]
- 74. Castanheira M, Farrell SE, Deshpande LM, Mendes RE, Jones RN. Prevalence of β-lactamase-encoding genes among Enterobacteriaceae bacteremia isolates collected in 26 U.S. hospitals: report from the SENTRY Antimicrobial Surveillance Program (2010). Antimicrob Agents Chemother 2013; 57:3012–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Guh AY, Bulens SN, Mu Y et al. . Epidemiology of carbapenem-resistant Enterobacteriaceae in 7 US communities, 2012–2013. JAMA 2015; 314:1479–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Centers for Disease Control and Prevention. Tracking CRE. http://www.cdc.gov/hai/organisms/cre/TrackingCRE.html. Accessed 25 June 2016.
- 77. Logan L, Renschler J, Gandra S, Weinstein R, Laxminarayan R. Carbapenem-resistant Enterobacteriaceae in children, United States, 1999–2012. Emerg Infect Dis 2015; 21:2014–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Logan LK, Bonomo RA. Metallo-β-lactamase (MBL)-producing Enterobacteriaceae in United States children. Open Forum Infect Dis 2016; doi:10.1093/ofid/ofw090. [DOI] [PMC free article] [PubMed]
- 79. Suwantarat N, Logan LK, Carroll KC et al. . The prevalence and molecular epidemiology of multidrug-resistant Enterobacteriaceae colonization in a pediatric intensive care unit. Infect Control Hosp Epidemiol 2016; 37:535–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Stillwell T, Green M, Barbadora K et al. . Outbreak of KPC-3 Producing Carbapenem-Resistant Klebsiella pneumoniae in a US Pediatric Hospital. J Pediatric Infect Dis Soc 2015; 4:330–8. [DOI] [PubMed] [Google Scholar]
- 81. Pannaraj P, Bard J, Cerini C, Weissman S. Pediatric carbapenem-resistant Enterobacteriaceae in Los Angeles, California, a high-prevalence region in the United States. Pediatric Infect Dis J 2015; 34:11–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Viau RA, Hujer AM, Marshall SH et al. . “Silent” dissemination of Klebsiella pneumoniae isolates bearing K. pneumoniae carbapenemase in a long-term care facility for children and young adults in Northeast Ohio. Clin Infect Dis 2012; 54:1314–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tumbarello M, Viale P, Viscoli C et al. . Predictors of mortality in bloodstream infections caused by Klebsiella pneumoniae carbapenemase-producing K. pneumoniae: importance of combination therapy. Clin Infect Dis 2012; 55:943–50. [DOI] [PubMed] [Google Scholar]
- 84. Kallen M, Ricks P, Edwards J et al. . Vital signs: carbapenem-resistant Enterobacteriaceae. Morb Mortal Wkly Rep 2013; 62:165–70. [PMC free article] [PubMed] [Google Scholar]
- 85. Temkin E, Adler A, Lerner A, Carmeli Y. Carbapenem-resistant Enterobacteriaceae: biology, epidemiology, and management. Ann N Y Acad Sci 2014; 1323:22–42. [DOI] [PubMed] [Google Scholar]
- 86. Falagas ME, Rafailidis PI, Kofteridis D et al. . Risk factors of carbapenem-resistant Klebsiella pneumoniae infections: a matched case–control study. J Antimicrob Chemother 2007; 60:1124–30. [DOI] [PubMed] [Google Scholar]
- 87. Gupta N, Limbago BM, Patel JB, Kallen AJ. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis 2011; 53:60–7. [DOI] [PubMed] [Google Scholar]
- 88. Chiotos K, Han JH, Tamma PD. Carbapenem-resistant Enterobacteriaceae infections in children. Curr Infect Dis Rep 2016; 18:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Scaggs FA, Charnot-Katsikas A, Bartlett AH et al. . Klebsiella pneumoniae carbapenemase (KPC) producing Enterobacteriaceae infections in children: A two-center study. Presented at: IDWeek 2015, San Diego, California,10October 2015. [Google Scholar]
- 90. Scaggs FA, Charnot-Katsikas A, Bartlett AH et al. . A multi-centered study of factors associated with Klebsiella pneumoniae carbapenemase (KPC)-producing Enterobacteriaceae infections in children. Presented at: Pediatric Academic Societies Annual Meeting, Baltimore, Maryland, 1 May 2016. [Google Scholar]
- 91. Prabaker K, Lin MY, McNally M et al. . Transfer from high-acuity long-term care facilities is associated with carriage of Klebsiella pneumoniae carbapenemase–producing Enterobacteriaceae: a multihospital study. Infect Control Hosp Epidemiol 2012; 33:1193–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lin MY, Lyles-Banks RD, Lolans K et al. . The importance of long-term acute care hospitals in the regional epidemiology of Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae. Clin Infect Dis 2013; 57:1246–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Mills JP, Talati NJ, Alby K, Han JH. The epidemiology of carbapenem-resistant Klebsiella pneumoniae colonization and infection among long-term acute care hospital residents. Infect Control Hosp Epidemiol 2016; 37:55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Munoz-Price LS, Hayden MK, Lolans K et al. . Successful control of an outbreak of Klebsiella pneumoniae carbapenemase—producing K. pneumoniae at a long-term acute care hospital. Infect Control Hosp Epidemiol 2010; 31:341–7. [DOI] [PubMed] [Google Scholar]
- 95. CDC. 2012 CRE toolkit—guidance for control of carbapenem-resistant Enterobacteriaceae (CRE). http://www.cdc.gov/hai/organisms/cre/cre-toolkit/background.html Accessed 1 July 2015.
- 96. Trick WE, Lin MY, Cheng-Leidig R et al. . Electronic public health registry of extensively drug-resistant organisms, Illinois, USA. Emerg Infect Dis 2015; 21:1725–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Liu Y, Wang Y, Walsh TR et al. . Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 2016; 16:161–8. [DOI] [PubMed] [Google Scholar]
- 98. Poirel L, Kieffer N, Liassine N, Thanh D, Nordmann P. Plasmid-mediated carbapenem and colistin resistance in a clinical isolate of Escherichia coli. Lancet Infect Dis 2016; 16:281. [DOI] [PubMed] [Google Scholar]
- 99. Paterson DL, Harris PN. Colistin resistance: a major breach in our last line of defence. Lancet Infect Dis 2016; 16:132–3. [DOI] [PubMed] [Google Scholar]
- 100. Skov R, Monnet D. Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Euro Surveill 2016; 21:30155. [DOI] [PubMed] [Google Scholar]