Backbone-cyclized peptides, which have applications in the pharmaceutical and agricultural industries, can be made efficiently in plants by co-expressing them with a cyclizing enzyme.
Keywords: Asparaginyl endopeptidase, cyclic peptide, cyclotide, kalata B1, Nicotiana benthamiana, plant-made pharmaceutical, transient expression, SFTI
Abstract
Cyclotides are ultra-stable, backbone-cyclized plant defence peptides that have attracted considerable interest in the pharmaceutical industry. This is due to their range of native bioactivities as well as their ability to stabilize other bioactive peptides within their framework. However, a hindrance to their widespread application is the lack of scalable, cost-effective production strategies. Plant-based production is an attractive, benign option since all biosynthetic steps are performed in planta. Nonetheless, cyclization in non-cyclotide-producing plants is poor. Here, we show that cyclic peptides can be produced efficiently in Nicotiana benthamiana, one of the leading plant-based protein production platforms, by co-expressing cyclotide precursors with asparaginyl endopeptidases that catalyse peptide backbone cyclization. This approach was successful in a range of other plants (tobacco, bush bean, lettuce, and canola), either transiently or stably expressed, and was applicable to both native and engineered cyclic peptides. We also describe the use of the transgenic system to rapidly identify new asparaginyl endopeptidase cyclases and interrogate their substrate sequence requirements. Our results pave the way for exploiting cyclotides for pest protection in transgenic crops as well as large-scale production of cyclic peptide pharmaceuticals in plants.
Introduction
Cyclotides are plant defence peptides that are exceptionally stable. They have a head-to-tail cyclic peptide backbone of around 30 amino acid residues and together with three interlocking disulfide bonds form a cyclic cystine knot structure. Cyclotides are attractive candidates for application within the pharmaceutical and agricultural industries both directly, by harnessing their intrinsic bioactivities, and indirectly by presenting foreign bioactive peptides within their stable framework (Craik et al., 1999; Göransson et al., 2012; Poth et al., 2013; Weidmann and Craik, 2016).
Despite their complex structure, cyclotides can be produced synthetically, and numerous studies describe the synthesis of a range of native as well as engineered cyclic peptides (Daly et al., 1999; Gunasekera et al., 2006). These studies are generally based on solid-phase peptide synthesis and involve the use of expensive reagents and the generation of large amounts of waste. On an industrial scale, these factors could have a significant impact on the cost and environmental impact of production. Alternatively, there are various biochemical approaches, both in vitro and in vivo, for cyclizing peptides generated by standard recombinant techniques (Aboye and Camarero, 2012). These are promising for the development of cyclic peptide libraries that can be screened for biological activity and for the rapid elucidation of structure–function relationships.
Plant-based production of therapeutic proteins, although still in its infancy, has developed rapidly over the past two decades. This evolution has been driven by advantages of production speed, cost, and safety over traditional microbial and mammalian cell culture systems (Yao et al., 2015). Given that cyclotides can accumulate to high levels in their native plant hosts (as high as 2 mg g−1 of plant material; Gran, 1970), plant-based production could be the most efficient approach for large-scale manufacture of cyclic peptides for therapeutic and other uses. Furthermore, heterologous production in plants is crucial for exploiting the potential pest control applications of cyclotides in transgenic crops. The plant species for which cyclotides have been most studied, such as Oldenlandia affinis, Clitoria ternatea, and Viola species, are seldom used in plant biotechnology and their amenability to gene transfer methods is not established. Regardless, large-scale cyclic peptide production would be best suited to plants that lack a background of native cyclotides and this is the case for species already established as protein production platforms such as tobacco, potato, lettuce, duckweed, and maize. However, when cyclotide genes are transferred to plants that are not native cyclotide-producers, cyclization efficiency is poor and acyclic misprocessed versions of the cyclotides predominate (Gillon et al., 2008; Conlan et al., 2012).
The maturation of cyclotide precursors involves successive enzymatic removal of the ER signal sequence and N-terminal propeptide (NTPP) followed by a transpeptidation reaction in which a C-terminal propeptide (CTPP) is released and the N- and C-termini of the cyclotide domain are joined (Gillon et al., 2008; Harris et al., 2015; Shafee et al., 2015; Fig. 1A). This final reaction is catalysed by members of the asparaginyl endopeptidase (AEP) family of enzymes (EC 3.4.22.34; Nguyen et al., 2014; Harris et al., 2015). Although AEPs more commonly act as proteases, a subset have evolved a ligase function in cyclotide-producing plants (Shafee et al., 2015). Plants that lack the appropriate AEP isoform required for backbone cyclization are therefore intrinsically ill-suited for recombinant cyclotide production (Gillon et al., 2008).
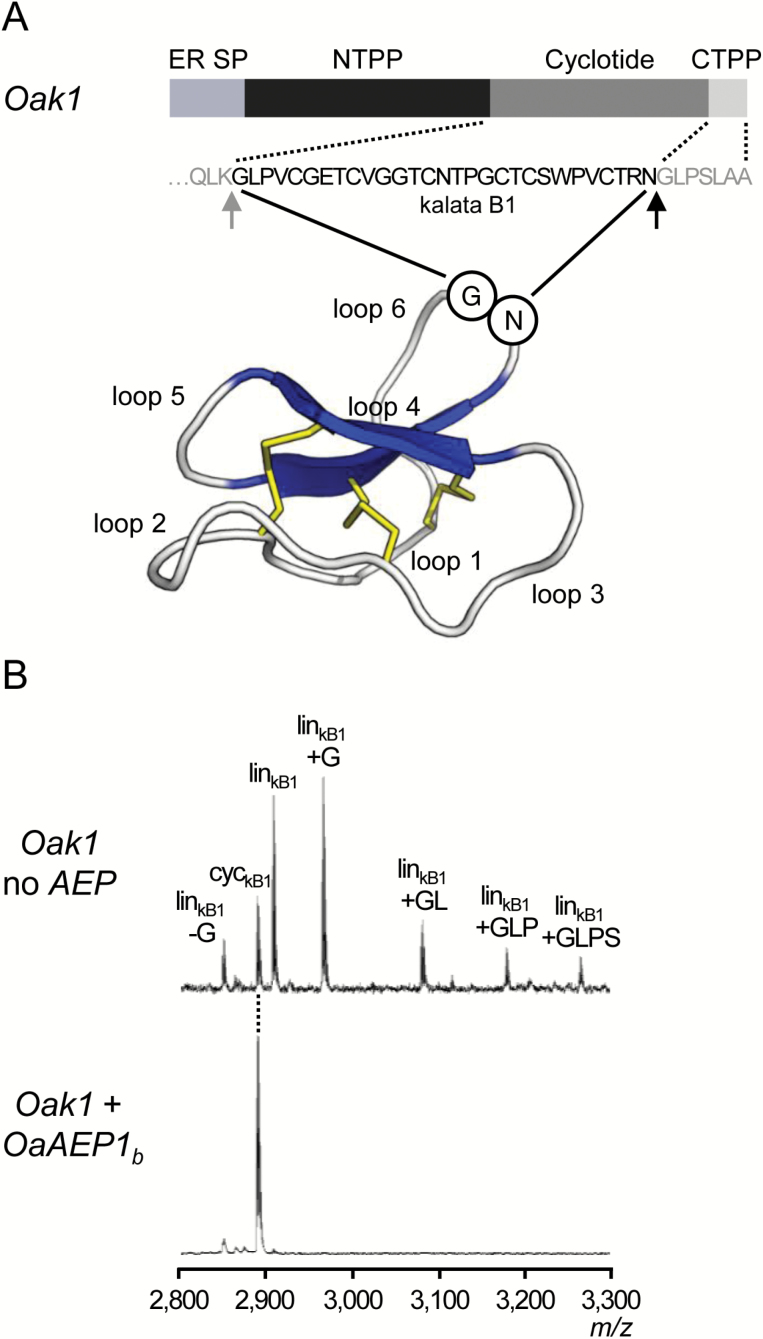
Fig. 1.
Cyclotide structure and production in N. benthamiana. (A) The prototypic cyclotide, kalata B1, is encoded by Oak1, which is composed of an ER signal peptide (ER SP), an N-terminal propeptide (NTPP), the mature cyclotide domain and a C-terminal propeptide (CTPP). The precursor protein is proteolytically processed to mature kalata B1 with the final steps involving cleavage at the NTPP (grey arrow) followed by cleavage at the CTPP (black arrow) and subsequent cyclization by peptide bond formation between the first glycine residue and the last asparagine residue of the mature cyclotide domain. The cyclic protein backbone and the knotted arrangement of three disulfide bonds (yellow) impart exceptional stability whilst the loop regions contribute native bioactivities or can be replaced with other bioactive peptides. The ribbon structure is based on PDB code 1NB1. (B) Representative MALDI-TOF mass spectra of peptides produced by transient expression of Oak1 alone or co-expressed with OaAEP1b in N. benthamiana. The production of predominantly linear peptides when Oak1 is expressed alone in N. benthamiana has been reported previously (Gillon et al., 2008; Conlan et al., 2012). The position of cyclic kB1 is indicated with a dotted line; cyc, cyclic product; lin, linear product.
Here we show that co-expression of a cyclizing AEP with a cyclotide gene significantly improves cyclization efficiency in non-cyclotide producing plants, resulting in a much higher proportion of cyclic product than with the cyclotide gene alone. This approach was successful in a range of food and non-food plants, and is applicable to both native and engineered cyclotides as well as the trypsin inhibitor class of cyclic peptides.
Materials and methods
Production of gene constructs
DNA encoding either AEPs or substrates was cloned into a vector containing the cauliflower mosaic virus 35S promoter and terminator sequences (Tabe et al., 1995) before transfer into the binary vector pBIN19 (Bevan, 1984). The pBIN19 expression vectors were then transformed into Agrobacterium tumefaciens (strain LBA4404) by electroporation. All gene constructs were made using a variety of standard molecular biology techniques such as cDNA cloning, PCR, restriction site cloning and Gibson assembly; some sequences were also generated by gene synthesis (GenScript). The following sequences used in this study have GenBank accession numbers: Oak1 (AF393825), OaAEP1 (KR259377 with substitutions 9A>G and 1112A>T to produce OaAEP1b), OaAEP2 (KR259378 with a single substitution 14C>T), OaAEP3 (KR259379), CtAEP1 (KF918345), CtAEP2 (KR912009), CtAEP6 (KY640209), Oak4 (AF393828), and Cter M (JF501210). Other sequences are listed in Supplementary Figs S8A and S10F at JXB online. For co-expression of the p19 suppressor, the pEAQexpress-GFP-HT construct was used whilst Oak1 was also expressed with pEAQ-HT-DEST1 (Sainsbury et al., 2009).
Transient expression in plants
Agrobacterium cells were spread as a bacterial lawn on agar plates containing yeast mannitol medium supplemented with kanamycin (50 μg ml−1) and streptomycin (100 μg ml−1). The cells were grown in the dark at 30 °C for 3 d. The lawn of bacteria was harvested and resuspended to an OD600 of 1.0 in infiltration buffer (10 mM MgCl2 and 10 μM acetosyringone). The resuspended bacteria were incubated at room temperature for 2–4 h. Nicotiana benthamiana plants were grown from seed in either a glasshouse or a growth cabinet at 25 °C for 4–6 weeks. In the latter, there was a 16 h/8 h day/night cycle. Leaves were infiltrated with the resuspended Agrobacterium (or 1:1 mixtures of the relevant Agrobacterium suspensions for co-expression) by gently pressing a 1 ml syringe (without a needle) against the underside of the leaf. The infiltrated area of the leaf was outlined with a permanent marker. Plants were grown for a further 4 d before the infiltrated areas were excised. Other plant tissues used were bush bean cotyledons (Phaseolus vulgaris cv. Royal Burgundy), grown for 10 d, and lettuce leaves (Lactuca sativa cv. Green Cos), grown for approximately 4 weeks. Infiltrated leaf segments were weighed, placed in microfuge tubes with a ball bearing and ground to a fine powder in liquid nitrogen using a mixer mill (30 s−1, 2 × 15 s). Proteins were extracted in aqueous 50% (v/v) acetonitrile–0.1% (v/v) trifluoroacetic acid (TFA) using 1 μl mg−1 wet weight together with 5 mg of insoluble poly(vinylpolypyrrolidone). Samples were centrifuged and the supernatants were then analysed by matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS). Alternatively, whole N. benthamiana plants were vacuum infiltrated by submerging them in 2 litres Agrobacterium suspensions in a desiccation chamber. The Agrobacterium had been grown in liquid LB medium supplemented with antibiotics for 2 d (200 rpm, 30 °C), centrifuged and resuspended in infiltration buffer to an OD600 of 0.5. A vacuum was applied to 100 mbar and then slowly released; this process was repeated. The plants were then grown for a further 6 d. The total leaf tissue from each plant was weighed, lyophilized, weighed again and then ground to a fine powder using ball bearings in a tissue homogenizer (Geno/Grinder 2010) (1500 rpm, 3 min). Proteins were extracted with 50% acetonitrile–0.1% TFA using 20 ml g−1 dry weight.
Stable expression in plants
N. tabacum and N. benthamiana leaf discs were used for Agrobacterium-mediated plant transformations (Horsch et al., 1985). A single copy, homozygous line of N. benthamiana transformed with OaAEP1b was generated by measuring copy number/zygosity of T1 plants using digital droplet PCR (Głowacka et al., 2016) and the single copy NbSo gene (GenBank accession no. AEV40816) as the reference. Stably transformed canola (Brassica napus cv. RI64) plants were produced essentially as described (Cardoza and Stewart, 2006); for these transformations, the NptII transcription unit in the pBIN19 binary vector was replaced by a bialaphos resistance (bar) selectable marker gene from the cloning vector pSAT1A-ocsAocsP-bar-ocsT (Chung et al., 2005) and transgenic canola shoots were selected on phosphinothricin (glufosinate ammonium).
Peptide purification
Kalata B1 was purified from leaf tissue harvested from 17 whole N. benthamiana plants vacuum infiltrated with an equal mix of Agrobacterium suspensions harbouring the double stack expression construct Oak1//OaAEP1b and the p19 suppressor gene construct (pEAQexpress-GFP-HT). Peptides were extracted in aqueous 50% acetonitrile–1% formic acid using 20 ml g−1 dry weight and filtered to remove insoluble material. The extract was then lyophilized and the crude solid redissolved in aqueous 10% acetonitrile–1% formic acid for initial solid phase extraction using a gravity fed C18 Sep-pak column (Waters). The kB1-containing fraction was detected using MALDI-TOF MS and further purified by successive RP-HPLC using Zorbax C18 columns (21.2 × 250 mm, 7 μm, 300 Å and 9.4 × 250 mm, 5 μm, 300 Å). Purity and yield of kB1-containing fractions were determined by RP-UHPLC and gravimetrically.
Nuclear magnetic resonance spectroscopy
Nuclear magnetic resonance (NMR) spectroscopy was used to confirm the correct fold of the isolated in planta-produced kB1. NMR experiments were performed on a Bruker Avance III 600 spectrometer (Bruker AXS GmbH, Karlsruhe, Germany) equipped with a cryogenically cooled probe. Approximately 1 mg of peptide (99% pure by RP-UHPLC) was dissolved in 550 μl of H2O/D2O (9:1) containing 20 µg 4,4-dimethyl-4-silapentane-1-sulfonic acid as internal standard. NMR spectra, including 1D 1H, as well as 2D total correlation spectroscopy (TOCSY), nuclear Overhauser effect spectroscopy (NOESY) and 1H-13C heteronuclear single quantum correlation (HSQC) spectra were recorded at 298 K and used for sequential assignment of the peaks from individual amino acids. The resultant Hα-chemical shifts provide a sensitive probe of peptide secondary and tertiary structure and were identical to literature values for kB1 isolated from Oldenlandia affinis (Rosengren et al., 2003) (see Supplementary Fig. S3).
Peptide analysis by MALDI-TOF MS
Leaf extracts were desalted and concentrated using C18 ZipTips (Merck Millipore) and then mixed 1:1 with α-cyano-4-hydroxycinnamic acid (5 mg ml−1 in 50% acetonitrile–0.1% TFA–5 mM (NH4)H2PO4) before they were spotted onto a ground steel sample plate and air dried. Mass analysis was performed in positive ion reflector mode on a Bruker Ultraflex III MALDI TOF/TOF mass spectrometer. Two hundred spectra at each of 10 randomly selected positions were accumulated per spot between 1000 and 4000 Da. Calibration was conducted using a mixture of peptide standards (Bruker Daltonics). Data were acquired and processed using Bruker flexAnalysis software; peaks were detected using the software’s ‘Snap’ algorithm. For relative quantification of cyclic and linear peptides, the sum of the integrated peak areas of the first three isotopic peaks corresponding to each assigned peptide was taken as 100% of the expressed peptides. The percentage of cyclic and linear peptides within the sample could then be calculated (Conlan et al., 2012). Peptide assignments and relative quantification of cyclic and linear products from all transiently expressed constructs are shown in Supplementary Figs S2, S4, S5 and S8–S10.
Absolute quantification of cyclic kB1
A MALDI-TOF MS-based method to quantify the cyclic kB1 present in N. benthamiana extracts was based on a previous study (Saska et al., 2008) with the difference being the use of a 15N-labelled cyclic kB1 (Mylne and Craik, 2008) as the internal standard. Synthetic oxidized kB1 (0.078–1.25 μg ml−1 final concentration) was mixed with 15N-kB1 (0.53 μg ml−1 final concentration). Samples were then mixed 1:1 with α-cyano-4-hydroxycinnamic acid and analysed by MALDI-TOF MS as above. The solvent was 50% acetonitrile–0.1% TFA and all data were collected in a background of endogenous N. benthamiana protein extract at a final concentration of 0.45 mg ml−1. Peaks were detected and peak areas integrated using the ‘Centroid’ algorithm of the flexAnalysis software. The relative peak area of unlabelled to labelled kB1 was plotted against unlabelled kB1 concentration to generate a standard curve (see Supplementary Fig. S7A). For quantification of the kB1 content in unknown samples, the total protein concentration was determined by BCA assay (Pierce) and adjusted to 0.9 mg ml−1 using 50% acetonitrile–0.1% TFA. This was then mixed 1:1 with 15N-kB1 (0.53 μg ml−1 final concentration) and analysed in triplicate by MALDI-TOF MS as above.
Results
Co-expression of a cyclizing AEP with a cyclotide precursor produces a high proportion of cyclic product
We used transient gene expression in Nicotiana benthamiana, a non-cyclotide producing plant and a common platform for producing heterologous proteins, to deliver both Oak1 (encoding the precursor to the prototypic cyclotide kalata B1 [kB1]) and a cyclizing AEP (OaAEP1b from Oldenlandia affinis or CtAEP1 (butelase 1) from Clitoria ternatea) by syringe agroinfiltration (Sparkes et al., 2006). In the first example, Oak1 and the cyclizing AEP were placed under the control of the constitutive cauliflower mosaic virus 35S promoter and terminator, and transiently expressed alone or in combination. Oak1 alone produced a suite of peptides whose masses correspond to linear oxidized kB1 along with other linear forms that lack the N-terminal Gly (−G) and/or retain successive residues from the CTPP (+G, +GL, +GLP) (Fig. 1B). Some cyclic kB1 was observed, but this constituted only a minor proportion of the total kB1 products detected. None of these peptides was detected in leaves infiltrated with empty vector or OaAEP1b alone (see Supplementary Fig. S1). However, when OaAEP1b was co-infiltrated with the cyclotide precursor, the relative proportion of cyclic kB1 increased substantially (Figs 1B and 2B and Supplementary Fig. S2). This increase also occurred when CtAEP1 was co-infiltrated with Oak1-HV, which is Oak1 with its CTPP changed from GLPSLAA to HV to reflect the reported specificity of CtAEP1 (Nguyen et al., 2014) (Fig. 2C, D and Supplementary Fig. S2). Cyclic kB1 produced transiently in N. benthamiana was isolated by HPLC and the integrity of its 3D structure was confirmed by NMR (see Supplementary Fig. S3).
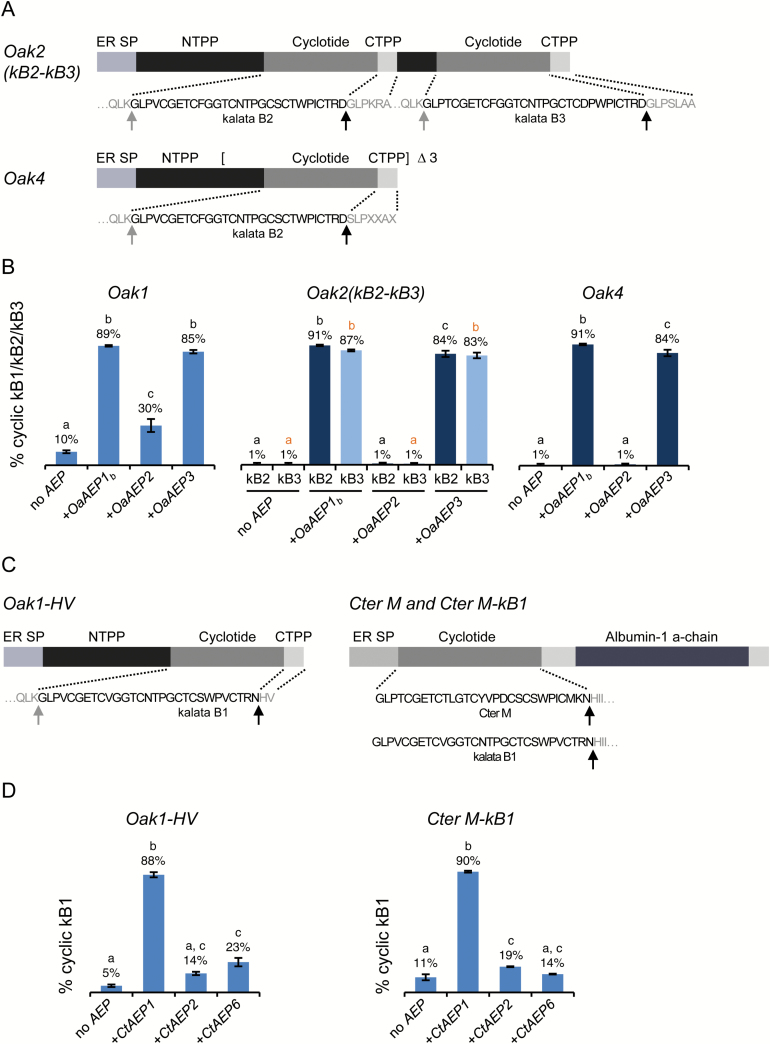
Fig. 2.
Production of kalata B1, kalata B2 and kalata B3 by transient co-expression of the cyclotide precursors with a cyclizing AEP in N. benthamiana. (A) Oak2(kB2-kB3) and Oak4 are multidomain cyclotide sequences that present different residues from Oak1 at the P1-P1′ positions of the C-terminal AEP processing site (Oak1: NG; Oak2: DG; Oak4: DS). See Fig. 1 for a description of domains and processing steps. (B) Mean percentage of cyclic kB1 (blue bars), cyclic kB2 (dark blue bars), and cyclic kB3 (light blue bars) relative to all assigned peptides±SEM based on mass spectra peak areas for Oak1, Oak2(kB2-kB3), and Oak4 expressed alone or co-expressed with OaAEP1b, OaAEP2, or OaAEP3. (C) Schematic representation of substrates co-infiltrated with AEPs from Clitoria ternatea. In Oak1-HV the native CTPP sequence of Oak1 (GLPSLAA) was replaced with HV. Cter M is a native cyclotide precursor from C. ternatea comprising the cyclotide domain embedded within an albumin precursor protein (Poth et al., 2011). In Cter M-kB1, the native cyclotide domain was replaced with kB1 within the Cter M precursor. (D) Mean relative percentage of cyclic kB1 relative to all assigned peptides±SEM based on mass spectra peak areas for Oak1-HV and Cter M-kB1 expressed alone or co-expressed with CtAEP1, CtAEP2, or CtAEP6. All data were derived from a minimum of three independent replicates. Different letters indicate significant differences found by Tukey’s ANOVA (P<0.05). Representative MALDI-TOF mass spectra, number of replicates, observed monoisotopic masses and mean relative percentages of all assigned peptides are shown in Supplementary Fig. S2.
Kalata B2 and kalata B3, which contain different C-terminal precursor processing sites (D29–G30/S30 and D30–G31, respectively) from the kB1 precursor (N29–G30), were also efficiently produced when constructs containing multiple cyclotide domains were co-infiltrated with OaAEP1b demonstrating that the inter-plant transfer of processing capability is not limited to the prototypic cyclotide, kB1 (Fig. 2A, B and Supplementary Fig. S2). Furthermore, a C. ternatea cyclotide, Cter M, was cyclized by CtAEP1, though not by OaAEP1b, when presented within its native Cter M precursor gene (see Supplementary Fig. S4A, B). The Cter M precursor is atypical since the cyclotide domain is embedded within an albumin precursor that lacks the canonical NTPP sequence; in this case the ER signal sequence is directly followed by the cyclotide domain, eliminating the requirement for enzymatic removal of the NTPP (Poth et al., 2011; Fig. 2C). Importantly, kB1 could also be presented within the Cter M albumin precursor and was efficiently cyclized in planta by both CtAEP1 (Fig. 2D) and OaAEP1b (Supplementary Fig. S4C). These data highlight the robustness of this approach: it is applicable to multiple cyclotides presented in single and multi-domain formats as well as within typical or atypical cyclotide precursors.
The high efficiency of cyclotide maturation was independent of whether the substrate and cyclizing AEP were in separate constructs that were co-infiltrated, or in a single, double-stack construct (see Supplementary Fig. S5). In the latter configuration, the substrate and AEP remain under control of their own promoter and terminator, but are on a single T-DNA, streamlining transformations. We also produced N. benthamiana transgenic plants that constitutively express either OaAEP1b or OaAEP3. Transient expression of Oak1 alone within these AEP-expressing plants also resulted in an increased proportion of cyclic kB1 across multiple plant lines (Supplementary Fig. S6). Thus, a plant line optimally expressing a cyclizing AEP could be used for producing cyclic peptides by transient expression of the appropriate substrate.
The yield of cyclic kB1 was determined using MALDI-TOF MS (Saska et al., 2008) to be 75 ± 7 μg g−1 dry weight (DW) of leaf tissue from N. benthamiana plants transiently expressing the Oak1//OaAEP1b double stack construct. This represents at least an 8-fold increase in absolute cyclic kB1 production compared with expressing Oak1 without OaAEP1b, where the cyclic kB1 yield was below the quantification limit (<8.5 μg g−1 DW) (see Supplementary Fig. S7B). A further increase in yield was achieved when the Oak1//OaAEP1b double stack construct was co-expressed with the p19 suppressor of gene silencing from tomato bushy stunt virus (Scholthof, 2007) (139 ± 13 μg g−1 DW). As an alternative strategy for increased yield, we also expressed Oak1 using a pEAQ vector (Sainsbury et al., 2009) in a stable transgenic N. benthamiana line expressing OaAEP1b, obtaining 199 ± 21 μg g−1 DW compared with <10.8 μg g−1 DW when this construct was expressed in wild-type N. benthamiana (Supplementary Fig. S7B).
Evaluation of substrate sequence requirements for cyclization in planta
Oak1 sequence requirements for the production of mature, cyclic kB1 were earlier investigated in N. benthamiana without co-expression of an AEP (Gillon et al., 2008; Conlan et al., 2012). However, this approach was limited by the low proportion of cyclic kB1 produced, suggesting that the endogenous N. benthamiana AEPs do not act preferentially as cyclases. Having demonstrated that co-expression of a cyclizing enzyme in N. benthamiana resulted in efficient cyclization of kB1, we re-examined Oak1 sequence requirements by co-expressing a series of Oak1 variants with OaAEP1b (see Supplementary Fig. S8). The minimal CTPP for efficient cyclization was Gly30Leu31, and strict requirement for Asn29 or Asp29 as the C-terminal residue was observed. Gly30Ala and Leu31Ala substitutions were also acceptable, although the proportion of cyclic product from the latter decreased from 89% to 69%.
Screening AEPs for cyclization ability
AEPs are typically thought of as proteases and thus far only two have been identified as preferential cyclases (Shafee et al., 2015). Therefore, another application of our expression system is the rapid screening of uncharacterized AEPs for cyclizing ability. To investigate their preferred function, previously uncharacterized AEPs from O. affinis (Harris et al., 2015) and C. ternatea (Gilding et al., 2016) were co-expressed with several different substrates (Fig. 2B, D). OaAEP3 had similar cyclization efficiency to OaAEP1b, producing a high proportion of cyclic kB1, kB2, and kB3, identifying it as a preferential cyclase. In contrast, OaAEP2 increased the proportion of cyclic kB1 from Oak1 only slightly (30% cyclic product compared with 10% without AEP co-expression). Furthermore, the reduction in the proportion of linear species retaining residues from the CTPP suggests that this AEP acts predominantly as a protease, removing the CTPP without subsequent cyclization (see Supplementary Fig. S7A). OaAEP2 had no effect on the processing of kB2 or kB3 from Oak2(kB2-kB3) or Oak4, generating profiles very similar to those obtained without AEP co-expression (Supplementary Fig. S7B, C). This suggests that OaAEP2 may have a substrate preference for cleaving N–G rather than D–G or D–S.
CtAEP2 and CtAEP6 were also poor cyclases, increasing the proportion of cyclic product from Oak1-HV only slightly (14% and 23% cyclic product, respectively, compared with 5% without AEP co-expression and 88% for CtAEP1). Furthermore, they generated only 19% and 14% cyclic product, respectively, from the Cter M-kB1 substrate compared with 11% without AEP co-expression and 90% for CtAEP1. Like OaAEP2, CtAEP6 eliminated the accumulation of linear processing variants carrying residues from the CTPP, suggesting efficient CTPP cleavage and a role as a protease (see Supplementary Fig. S7D, E).
Successful cyclotide production is transferrable to crop species
Two double-stack constructs (Oak1//OaAEP1b and Oak1-HV//CtAEP1) were transiently expressed in bush bean and lettuce and compared with constructs expressing the respective substrates alone. In each case, the presence of the cyclizing enzymes significantly increased the proportion of cyclic kB1, showing that this technology is transferrable to other plants, including edible species (see Supplementary Fig. S9). Efficient cyclic kB1 production observed in transient expression assays also extended to the stable transformation of two tobacco species (N. tabacum and N. benthamiana) and canola (Supplementary Fig. S6).
Grafted versions of kalata B1 can be produced in planta
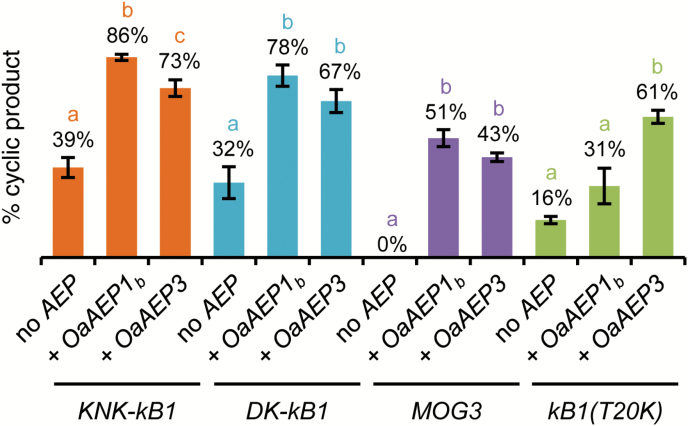
We first tested whether two kB1 variants ([W19K,P20N,V21K]kB1 (KNK-kB1) and [P20D,V21K]kB1 (DK-kB1)) engineered to explore the structural plasticity of the cyclotide framework (Clark et al., 2006) could be made in N. benthamiana. Although the proportion of cyclic product for both molecules was higher than obtained with native kB1 when they were expressed without a cyclizing AEP (KNK-kB1, 39%; DK-kB1, 32%; native kB1, 10%), the proportion of cyclic product was significantly increased when an AEP was co-expressed (OaAEP1b: KNK-kB1 86%, DK-kB1 78%; OaAEP3: KNK-kB1 73%, DK-kB1 67%) (Fig. 3). Next, we tested the in planta production of two kB1 variants (MOG3 and kB1(T20K)) that have shown promising activity in a mouse model of multiple sclerosis (Wang et al., 2014; Thell et al., 2016). Cyclic MOG3 was not detected when the precursor was expressed without a cyclizing AEP but comprised 51% and 43% of total MOG3 peptides when co-expressed with OaAEP1b or OaAEP3, respectively. More cyclic kB1(T20K) was also produced in N. benthamiana when co-expressed with a cyclizing AEP. For this molecule, OaAEP3 produced a higher proportion of cyclic kB1(T20K) than OaAEP1b with 61% cyclic peptide compared with 31% (Fig. 3).
Fig. 3.
Production of grafted cyclic peptides in N. benthamiana. Mean relative percentage of cyclic peptide relative to all assigned peptides±SEM based on mass spectra peak areas for Oak1-KNK-kB1 (orange bars), Oak1-DK-kB1 (aqua bars), Oak1-MOG3 (purple bars), and Oak1-kB1(T20K) (green bars) expressed alone or co-expressed with OaAEP1b or OaAEP3. All data were derived from a minimum of three independent replicates. Different letters indicate significant differences found by Tukey’s ANOVA (P<0.05). Representative MALDI-TOF mass spectra, number of replicates, observed monoisotopic masses and mean relative percentages of all assigned peptides are shown in Fig. S10.
The cyclic peptide SFTI-1 can be produced in planta
To test whether this approach would also be successful for a different class of cyclic peptide, the kB1 sequence within the Oak1 precursor was replaced with the sunflower trypsin inhibitor, SFTI-1 (Mylne et al., 2011). The peptide was not detected when the precursor was expressed alone in N. benthamiana, but upon co-expression with OaAEP1b, cyclic SFTI-1 was efficiently produced (see Supplementary Fig. S10E). Successful in planta production was also extended to an engineered SFTI-1 variant (SFTI-FCQR) that inhibits human kallikrein-related peptidase 4, a target for prostate cancer treatment (Supplementary Fig. S10E) (Swedberg et al., 2009).
Discussion
Understanding the mechanism of cyclotide biosynthesis advanced significantly when it was shown that members of the asparaginyl endopeptidase (AEP) family of enzymes catalyse the backbone cyclization of cyclotides (Nguyen et al., 2014; Harris et al., 2015). When cyclotide genes are expressed without a cyclizing AEP in plants that do not natively produce cyclotides, the efficiency of cyclization is poor. Instead, endogenous enzymes, which may include AEPs that are preferential proteases with limited cyclase activity and/or carboxypeptidases that trim successive residues from the CTPP, produce predominantly linear, misprocessed versions of the cyclotide. In each of the plants tested in our study (N. benthamiana, N. tabacum, bush bean, and lettuce), none of which natively produce cyclotides, co-expression of a cyclizing AEP with a cyclotide precursor outcompeted endogenous processes that produce linear forms. Thus, the low cyclizing efficiency of plants that do not naturally produce cyclotides can be overcome.
By co-expressing previously uncharacterized AEPs with Oak1, we have identified OaAEP3 as another cyclizing AEP. OaAEP3 shares 79% identity with OaAEP1b and both enzymes cyclized kalata B1, B2, and B3 with similar efficiency in planta. However, OaAEP3 was significantly better at cyclizing kB1(T20K), suggesting subtle differences in substrate specificities. It should be noted that while an AEP that produces a high proportion of cyclic product can be categorized as a cyclizing AEP, low apparent cyclization may result from other factors, such as sub-optimal substrates, low expression levels or slow kinetics such that cyclization is outcompeted by degradation by endogenous enzymes.
Previous investigations into Oak1 sequence requirements for cyclization involved the expression of Oak1 variants in N. benthamiana, N. tabacum, or A. thaliana (Gillon et al., 2008; Conlan et al., 2012). The low cyclization efficiency for wild-type Oak1 in these plants, however, hampered detailed substrate structure–cyclization efficiency studies. Moreover, results may also be influenced by the nature of the endogenous AEPs whose primary roles are unlikely to involve cyclization. The highly efficient cyclization of Oak1 by OaAEP1b makes our in planta system ideal for investigating substrate sequence requirements for cyclization. For example, in previous studies without AEP co-expression, no cyclic product was observed from the Leu31Ala variant (Conlan et al., 2012) or Asn29Asp variant (Gillon et al., 2008) whereas with OaAEP1b co-expression, we observed 69% and 87% cyclic kB1, respectively. The reduction in the relative proportion of cyclic kB1 in the former is consistent with the slower in vitro processing of this precursor with recombinant purified enzyme (Harris et al., 2015). The minimal CTPP for efficient cyclization (Gly30Leu31) and strict requirement for Asn29 or Asp29 as the C-terminal residue were also consistent with experiments with recombinant purified OaAEP1b (Harris et al., 2015). Our system therefore facilitates rapid, accurate screening of optimal precursor substrate sequences for a given AEP.
The quantification of cyclic kB1 produced in N. benthamiana shows that the increase in the relative proportion of cyclic kB1 produced from Oak1 alone or co-expressed with OaAEP1b (from 10% to 89%) also applies in absolute terms where the Oak1//OaAEP1b double stack produced around 9-fold more cyclic kB1 than Oak1 alone (see Supplementary Fig. S7B). Although the highest levels of kB1 reported here (199 μg g−1 DW) are around 10-fold less than present in the leaves of the native plant, O. affinis, (1.82 mg g−1 DW for plants grown in a greenhouse) (Seydel and Dörnenburg, 2006), large amounts of N. benthamiana leaf biomass can be grown rapidly for recombinant protein production by agroinfiltration (Chen et al., 2013; Holtz et al., 2015), offering a feasible option for large-scale cyclic peptide production. The reported yields of other plant-produced therapeutic proteins, such as monoclonal antibodies and vaccines (Gleba et al., 2014), are generally much higher on a mass basis than what we have demonstrated with cyclic kB1. However the much smaller size of cyclotides means that on a molar basis, our current best yield is broadly comparable. Moreover, other strategies, such as the use of self-replicating plant virus-based expression vectors (Gleba et al., 2014), might further increase cyclotide yield.
Cyclic peptides have also been produced in vivo using yeast and bacterial expression systems. In this approach, backbone cyclization is achieved by intein-mediated splicing and cyclic peptide yields of up to 180 µg l−1 have been reported (Jagadish et al., 2013, 2015; Li et al., 2016). Whilst the focus of these studies has been the production of cyclotide libraries that could be rapidly screened for a desired biological activity, this approach may also be applicable to large-scale cyclotide production. Further work could compare the feasibility of the plant and microbe systems for this purpose.
The exceptional stability of cyclotides and their reported orally delivered bioactivity from plant material, even after boiling (Gran, 1970), provides tantalising evidence that bioactive cyclotides could be introduced into edible plants and directly administered as medicines. Successful transient cyclic peptide production in N. benthamiana was transferrable to both bush bean and lettuce as well as stably introduced into canola. Although we have focused on production in leaf, it will be necessary to investigate other plant tissues such as seeds, fruits, and tubers to determine how broadly cyclic peptides can be produced in edible plant tissues.
The intrinsic insecticidal and nematicidal activity of some cyclotides (Jennings et al., 2001; Gilding et al., 2016) means that their production in transgenic crops could confer pest protection. Our evidence that cyclotides can be efficiently produced in bush bean, lettuce, tobacco and canola by co-expressing a cyclizing AEP suggests that this approach could be applied to protect agriculturally important crops. Plants that naturally produce cyclotides typically contain dozens of different cyclotides that accumulate in different tissues, suggesting that they are combinatorial mediators of defence (Gilding et al., 2016). While cyclotide-producing plants are likely to express multiple AEPs, our evidence that OaAEP1b cyclizes multiple cyclotides with high efficiency suggests an approach where a transgenic crop constitutively expressing a single cyclizing AEP but multiple cyclotide precursors could be protected against a broad range of pests. Alternatively, such a strategy could be used to pyramid cyclotide-based pest-protective traits, analogous to the stacking of Bt genes (Zhao et al., 2003), to delay the onset of resistance.
To be broadly applicable for agricultural or pharmaceutical applications, it is important that non-native cyclotides with other, engineered activities can also be produced in planta. Our data confirm that the cyclization efficiency of non-native cyclotides in planta can be improved when a cyclizing AEP is co-expressed. The relative percentages of cyclic product for these grafted kB1s upon AEP co-expression were lower than those for native kB1 and there was a significant difference in the cyclization efficiency for kB1(T20K) between OaAEP1b and OaAEP3. These grafted kB1s all have the same sequences around the AEP recognition/cleavage site as native kB1. Therefore we speculate that the grafted peptides are creating subtle conformational changes that affect the efficiency of AEP processing. Alternatively, these changes could be favourable for processing by endogenous, non-cyclizing enzymes. Determining optimal substrate–cyclizing enzyme combinations will be crucial for maximizing the proportion of cyclic product.
The successful production of a different class of cyclic peptide, the trypsin inhibitor SFTI-1 and a grafted version of this molecule, suggests that co-expression of a cyclizing AEP may find broad application for the production of a range of cyclic peptides. The absence of linear SFTI-1 products, in both the presence and the absence of the cyclizing AEP, suggests that misprocessed molecules may be rapidly degraded by endogenous enzymes, highlighting the stabilizing effect of cyclization.
In conclusion, plants can be used for the production of natural cyclotides and other cyclic peptides engineered for therapeutic use, by co-expressing target peptides with a cyclizing AEP. The establishment of agroinfiltration for heterologous protein production in N. benthamiana on an industrial scale (Chen et al., 2013; Holtz et al., 2015) means that large-scale, affordable production of cyclic peptides may be possible. Furthermore, the applicability of this technology to food crops establishes the feasibility of supplying therapeutic cyclotides directly from plant sources or exploiting the potential of cyclotides to enhance pest resistance in transgenic crops. Finally, this approach can also be used to identify and characterize AEPs with cyclizing ability and to probe substrate sequence requirements for cyclization in planta.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Representative MALDI-TOF MS spectra of Oak1 transiently expressed in N. benthamiana compared with transient expression of OaAEP1b or an empty vector.
Fig. S2. Production of kalata B1, kalata B2 and kalata B3 by transient co-expression of the cyclotide precursors with a cyclizing AEP in N. benthamiana.
Fig. S3. NMR analysis of kB1 isolated from N. benthamiana transiently expressing Oak1//OaAEP1b.
Fig. S4. Transient expression of cyclotides in the Cter M precursor.
Fig. S5. Transient expression of double stack constructs.
Fig. S6. Cyclotide production in stable transformants.
Fig. S7. MALDI-TOF MS quantification of cyclic kalata B1 produced in N. benthamiana.
Fig. S8. Transient co-expression of Oak1 variants with OaAEP1b in N. benthamiana.
Fig. S9. Cyclotide production in bush bean and lettuce.
Fig. S10. Production of grafted cyclic peptides in N. benthamiana.
Acknowledgements
We thank Dr Vijay Kaul and Ms Ekaterina Mouradova for stable transgenic plant generation and Dr Gianna Kalc, Mr Bruce McGinness, and Ms Hannah Noorda for growing plants. We also thank Dr Amanda Gillon and Dr Brendon Conlan for gene constructs that were used in their earlier studies. The pEAQ vectors were kindly provided by Prof. George Lomonossoff at the John Innes Centre and Plant Bioscience Ltd. We acknowledge funding from the Australian Research Council (ARC Laureate Fellowship (FL150100146) to DJC, ARC grant DP0984390 to DJC and MAA, and ARC grant DP150100443 to DJC, EKG, and TD). This research was also supported by the 2015 Ramaciotti Biomedical Research Award to DJC and MAA, and by Hexima Ltd.
Glossary
Abbreviations:
- AEP
asparaginyl endopeptidase
- CTPP
C-terminal propeptide
- kB1
kalata B1
- NTPP
N-terminal propeptide
- SFTI
sunflower trypsin inhibitor.
References
- Aboye TL, Camarero JA. 2012. Biological synthesis of circular polypeptides. The Journal of Biological Chemistry 287, 27026–27032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M. 1984. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Research 12, 8711–8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoza V, Stewart CN. 2006. Canola (Brassica napus L.). Methods in Molecular Biology 343, 257–266. [DOI] [PubMed] [Google Scholar]
- Chen Q, Lai H, Hurtado J, Stahnke J, Leuzinger K, Dent M. 2013. Agroinfiltration as an effective and scalable strategy of gene delivery for production of pharmaceutical proteins. Advanced Techniques in Biology & Medicine 1, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SM, Frankman EL, Tzfira T. 2005. A versatile vector system for multiple gene expression in plants. Trends in Plant Science 10, 357–361. [DOI] [PubMed] [Google Scholar]
- Clark RJ, Daly NL, Craik DJ. 2006. Structural plasticity of the cyclic-cystine-knot framework: implications for biological activity and drug design. Biochemical Journal 394, 85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlan BF, Colgrave ML, Gillon AD, Guarino R, Craik DJ, Anderson MA. 2012. Insights into processing and cyclization events associated with biosynthesis of the cyclic peptide kalata B1. The Journal of Biological Chemistry 287, 28037–28046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craik DJ, Daly NL, Bond T, Waine C. 1999. Plant cyclotides: A unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. Journal of Molecular Biology 294, 1327–1336. [DOI] [PubMed] [Google Scholar]
- Daly NL, Love S, Alewood PF, Craik DJ. 1999. Chemical synthesis and folding pathways of large cyclic polypeptides: studies of the cystine knot polypeptide kalata B1. Biochemistry 38, 10606–10614. [DOI] [PubMed] [Google Scholar]
- Gilding EK, Jackson MA, Poth AG, Henriques ST, Prentis PJ, Mahatmanto T, Craik DJ. 2016. Gene coevolution and regulation lock cyclic plant defence peptides to their targets. New Phytologist 210, 717–730. [DOI] [PubMed] [Google Scholar]
- Gillon AD, Saska I, Jennings CV, Guarino RF, Craik DJ, Anderson MA. 2008. Biosynthesis of circular proteins in plants. The Plant Journal 53, 505–515. [DOI] [PubMed] [Google Scholar]
- Gleba YY, Tusé D, Giritch A. 2014. Plant viral vectors for delivery by Agrobacterium. In: Palmer K, Gleba Y, eds. Plant viral vectors. Berlin, Heidelberg: Springer, 155–192. [DOI] [PubMed] [Google Scholar]
- Głowacka K, Kromdijk J, Leonelli L, Niyogi KK, Clemente TE, Long SP. 2016. An evaluation of new and established methods to determine T-DNA copy number and homozygosity in transgenic plants. Plant, Cell & Environment 39, 908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göransson U, Burman R, Gunasekera S, Strömstedt AA, Rosengren KJ. 2012. Circular proteins from plants and fungi. The Journal of Biological Chemistry 287, 27001–27006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gran L. 1970. An oxytocic principle found in Oldenlandia affinis DC. An indigenous, Congolese drug “Kalata-Kalata” used to accelerate delivery. Meddelelser Fra Norsk Farmaceutisk Selskap 32, 173–180. [Google Scholar]
- Gunasekera S, Daly NL, Anderson MA, Craik DJ. 2006. Chemical synthesis and biosynthesis of the cyclotide family of circular proteins. IUBMB Life 58, 515–524. [DOI] [PubMed] [Google Scholar]
- Harris KS, Durek T, Kaas Q, et al. 2015. Efficient backbone cyclization of linear peptides by a recombinant asparaginyl endopeptidase. Nature Communications 6, 10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtz BR, Berquist BR, Bennett LD, et al. 2015. Commercial-scale biotherapeutics manufacturing facility for plant-made pharmaceuticals. Plant Biotechnology Journal 13, 1180–1190. [DOI] [PubMed] [Google Scholar]
- Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT. 1985. A simple and general method for transferring genes into plants. Science 227, 1229–1231. [DOI] [PubMed] [Google Scholar]
- Jagadish K, Borra R, Lacey V, Majumder S, Shekhtman A, Wang L, Camarero JA. 2013. Expression of fluorescent cyclotides using protein trans-splicing for easy monitoring of cyclotide–protein interactions. Angewandte Chemie International Edition 52, 3126–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadish K, Gould A, Borra R, Majumder S, Mushtaq Z, Shekhtman A, Camarero JA. 2015. Recombinant expression and phenotypic screening of a bioactive cyclotide against α-synuclein-induced cytotoxicity in baker’s yeast. Angewandte Chemie International Edition 54, 8390–8394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings C, West J, Waine C, Craik D, Anderson M. 2001. Biosynthesis and insecticidal properties of plant cyclotides: The cyclic knotted proteins from Oldenlandia affinis. Proceedings of the National Academy of Sciences, USA 98, 10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Aboye T, Breindel L, Shekhtman A, Camarero JA. 2016. Efficient recombinant expression of SFTI-1 in bacterial cells using intein-mediated protein trans-splicing. Biopolymers 106, 818–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylne JS, Colgrave ML, Daly NL, Chanson AH, Elliott AG, McCallum EJ, Jones A, Craik DJ. 2011. Albumins and their processing machinery are hijacked for cyclic peptides in sunflower. Nature Chemical Biology 7, 257–259. [DOI] [PubMed] [Google Scholar]
- Mylne JS, Craik DJ. 2008. 15N cyclotides by whole plant labeling. Peptide Science 90, 575–580. [DOI] [PubMed] [Google Scholar]
- Nguyen GK, Wang S, Qiu Y, Hemu X, Lian Y, Tam JP. 2014. Butelase 1 is an Asx-specific ligase enabling peptide macrocyclization and synthesis. Nature Chemical Biology 10, 732–738. [DOI] [PubMed] [Google Scholar]
- Poth AG, Chan LY, Craik DJ. 2013. Cyclotides as grafting frameworks for protein engineering and drug design applications. Biopolymers 100, 480–491. [DOI] [PubMed] [Google Scholar]
- Poth AG, Colgrave ML, Lyons RE, Daly NL, Craik DJ. 2011. Discovery of an unusual biosynthetic origin for circular proteins in legumes. Proceedings of the National Academy of Sciences, USA 108, 10127–10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosengren KJ, Daly NL, Plan MR, Waine C, Craik DJ. 2003. Twists, knots, and rings in proteins. Structural definition of the cyclotide framework. The Journal of Biological Chemistry 278, 8606–8616. [DOI] [PubMed] [Google Scholar]
- Sainsbury F, Thuenemann EC, Lomonossoff GP. 2009. pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnology Journal 7, 682–693. [DOI] [PubMed] [Google Scholar]
- Saska I, Colgrave ML, Jones A, Anderson MA, Craik DJ. 2008. Quantitative analysis of backbone-cyclised peptides in plants. Journal of Chromatography B 872, 107–114. [DOI] [PubMed] [Google Scholar]
- Scholthof HB. 2007. Heterologous expression of viral RNA interference suppressors: RISC management. Plant Physiology 145, 1110–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydel P, Dörnenburg H. 2006. Establishment of in vitro plants, cell and tissue cultures from Oldenlandia affinis for the production of cyclic peptides. Plant Cell, Tissue and Organ Culture 85, 247–255. [Google Scholar]
- Shafee T, Harris K, Anderson M. 2015. Biosynthesis of cyclotides. Advances in Botanical Research 76, 227–269. [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C. 2006. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nature Protocols 1, 2019–2025. [DOI] [PubMed] [Google Scholar]
- Swedberg JE, Nigon LV, Reid JC, et al. 2009. Substrate-guided design of a potent and selective kallikrein-related peptidase inhibitor for kallikrein 4. Chemistry & Biology 16, 633–643. [DOI] [PubMed] [Google Scholar]
- Tabe LM, Wardley-Richardson T, Ceriotti A, Aryan A, McNabb W, Moore A, Higgins TJ. 1995. A biotechnological approach to improving the nutritive value of alfalfa. Journal of Animal Science 73, 2752–2759. [DOI] [PubMed] [Google Scholar]
- Thell K, Hellinger R, Sahin E, et al. 2016. Oral activity of a nature-derived cyclic peptide for the treatment of multiple sclerosis. Proceedings of the National Academy of Sciences, USA 113, 3960–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CK, Gruber CW, Cemazar M, et al. 2014. Molecular grafting onto a stable framework yields novel cyclic peptides for the treatment of multiple sclerosis. ACS Chemical Biology 9, 156–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidmann J, Craik DJ. 2016. Discovery, structure, function, and applications of cyclotides: circular proteins from plants. Journal of Experimental Botany 67, 4801–4812. [DOI] [PubMed] [Google Scholar]
- Yao J, Weng Y, Dickey A, Wang KY. 2015. Plants as factories for human pharmaceuticals: applications and challenges. International Journal of Molecular Sciences 16, 28549–28565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JZ, Cao J, Li YX, Collins HL, Roush RT, Earle ED, Shelton AM. 2003. Transgenic plants expressing two Bacillus thuringiensis toxins delay insect resistance evolution. Nature Biotechnology 21, 1493–1497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.