A model based solely on mass-balance constraints refines our understanding of the trade-offs, energy requirements, leaf-level fluxes, and plasticity mechanisms in different biochemical types of assimilation.
Keywords: Assimilation, bioengineering, carbon-concentrating mechanism, constraint, dark reactions, flux balance, flux-balance analysis, NAD-ME, NADP-ME, PEPCK
Abstract
The goal of suppressing photorespiration in crops to maximize assimilation and yield is stimulating considerable interest among researchers looking to bioengineer carbon-concentrating mechanisms into C3 plants. However, detailed quantification of the biochemical activities in the bundle sheath is lacking. This work presents a general stoichiometric model for C3, C2, C2+C4, and C4 assimilation (SMA) in which energetics, metabolite traffic, and the different decarboxylating enzymes (NAD-dependent malic enzyme, NADP-dependent malic enzyme, or phosphoenolpyruvate carboxykinase) are explicitly included. The SMA can be used to refine experimental data analysis or formulate hypothetical scenarios, and is coded in a freely available Microsoft Excel workbook. The theoretical underpinnings and general model behaviour are analysed with a range of simulations, including (i) an analysis of C3, C2, C2+C4, and C4 in operational conditions; (ii) manipulating photorespiration in a C3 plant; (iii) progressively upregulating a C2 shuttle in C3 photosynthesis; (iv) progressively upregulating a C4 cycle in C2 photosynthesis; and (v) manipulating processes that are hypothesized to respond to transient environmental inputs. Results quantify the functional trade-offs, such as the electron transport needed to meet ATP/NADPH demand, as well as metabolite traffic, inherent to different subtypes. The SMA refines our understanding of the stoichiometry of photosynthesis, which is of paramount importance for basic and applied research.
Introduction
Interest in engineering a biochemical carbon-concentrating mechanism (CCM, abbreviations listed in Table 1) to suppress photorespiration in crops is increasing (Furbank et al., 2015; Long et al., 2015). The metabolic activities of a CCM are shared between mesophyll (M) and bundle sheath (BS) cells. Structurally, the leaf parenchyma is organized in concentric layers of cells, with an outer mesophyll encircling one or two layers of BS cells. In some species, the BS cells are isolated from the surroundings by a gas-tight suberized cell wall (Lundgren et al., 2014). Biochemically, the compartmentalization of glycine decarboxylase (GDC) activity in the BS allows plants to take advantage of photorespiratory CO2 release (Sage et al., 2012; Mallmann et al., 2014), giving rise to a mechanism that delivers CO2 around the Rubisco in the BS – the so-called C2 shuttle (Schulze et al., 2013; Keerberg et al., 2014). The ‘C4 cycle’ is a further adaptation involving an energy-dependent carboxylation‒decarboxylation cycle. CO2 is initially fixed into four-carbon (C4) organic (amino) acids in the M by phosphoenolpyruvate (PEP) carboxylase (PEPC). These then diffuse to the BS where they are decarboxylated. C4 plants have traditionally been grouped into three subtypes (Table 2) according to the main decarboxylating enzyme, NAD-dependent malic enzyme (NAD-ME), NADP-dependent malic enzyme (NADP-ME), or phosphoenolpyruvate carboxykinase (PEPCK) (Hatch, 1987), but there is considerable diversity in the degree of engagement of the biochemical activities of the CCM between subtypes. For instance, maize (Zea mays) has been shown to operate two BS decarboxylation enzymes (NADP-ME and PEPCK) and two CO2 delivery pathways (via malate, MAL, or aspartate, ASP, respectively) (Furumoto et al., 1999, 2000; Wingler et al., 1999). There is also overlap between BS and M functions. In maize, sucrose is synthesized mainly in the M, while starch is generally accumulated in the BS, although both possess enzymes to synthesize starch (Rascio et al., 1980; Spilatro and Preiss, 1987). Both the BS and M reduce 3-phosphoglyceric acid (PGA). Further, pyruvate phosphate dikinase (PPDK) has traditionally thought to be confined to the M; however, PPDK was shown to be present and active in the BS (Aoyagi and Nakamoto, 1985; Majeran et al., 2010), although the role of PPDK in the BS remains elusive.
Table 1.
Acronyms, definitions, and variables. Quantities with dimensions are consistent with assimilation (μmol m−2 s−1).
| A | Net assimilation |
| ALA, ALA | Alanine, ALA diffusion rate |
| ASP, ASP | Aspartate, ASP diffusion rate |
| ATP, ATPTOT, ATPBS, ATPM | Adenosine triphosphate, rate of ATP demand: total, in the BS, or in the M respectively |
| BS | Bundle sheath |
| CCM | Carbon-concentrating mechanism |
| CEF | Cyclic electron flow |
| CS, CSTOT, CSBS, CSM | Carbohydrate synthesis, rate of carbohydrate synthesis: total, in the BS, or in the M, respectively |
| DHAP, DHAP | Dihydroxyacetone phosphate, DHAP diffusion rate, respectively |
| DHAP RPP, DHAPRPPM, DHAPRPPBS | Rate of DHAP entering the RPP, in the M, or in the BS respectively |
| F | Rate of photorespiratory CO2 release |
| f C, fO, fRLIGHT, fPR, fCS, fPPDK | Input parameters defining, relative to total, the BS fraction of: Rubisco rate of carboxylation, Rubisco rate of oxygenation, Respiration in the light, PGA reduction, carbohydrate synthesis, and PPDK activity, respectively |
| GA | Gross assimilation (A + RLIGHT) |
| GDC, GDCTOT, GDCBS, GDCM | Glycine decarboxylase, GDC reaction rate: total, in the BS, or in the M, respectively |
| GLA | Glycolic acid |
| GLY, GLY | Glycine, GLY diffusion rate, respectively |
| L | Leak rate, i.e. magnitude of CO2 flux diffusing out of the BS, Eqn S19 |
| LEF | Linear electron flow (flow of electrons derived from the photo-oxidation of water) |
| M | Mesophyll |
| MAL, MAL | Malate, MAL diffusion rate, respectively |
| MDH, MDHBS , MDHM | Malate dehydrogenase, MDH reaction rate in the BS or M, respectively |
| ME, ME | Malic enzyme, ME reaction rate, respectively |
| NADPH TOT , NADPH BS | NADPH demand: total or in the BS, respectively |
| OAA, OAA | Oxaloacetate, OAA diffusion rate, respectively |
| PCO | Photosynthetic carbon oxygenation (cycle), also known as photorespiratory cycle |
| PEP | Phosphoenolpyruvate |
| PEP, PEP | Phosphoenolpyruvate, PEP diffusion rate, respectively |
| PEPC | Phosphoenolpyruvate carboxylase |
| PEPCK, PEPCK | Phosphoenolpyruvate carboxykinase, PEPCK reaction rate, respectively |
| PGA, PGA | 3-phosphoglyceric acid, PGA diffusion rate, respectively |
| PGLA | 2-phosphoglycolic acid |
| PPDK, PPDK | Pyruvate phosphate dikinase, PPDK reaction rate, respectively |
| PR, PRTOT, PRBS, PRM | PGA reduction, reduction rate: total, in the BS, or in the M, respectively |
| PYR, PYR | Pyruvate, PYR diffusion rate, respectively |
| R | Rate of CO2 and NH3 release in the BS associated with the operation of the C2 shuttle, Eqn S16 |
| R LIGHT , R LIGHT BS , R LIGHT M | Respiration in the light: total, in the BS, or in the M, respectively |
| r O/C | Input defining leaf-level Rubisco rate of oxygenation relative to carboxylation, also referred to as φ or VO/VC |
| r PEPCK | input parameter defining the activity of PECK relative to VP |
| RPP | Reductive pentose phosphate (cycle); also known as Calvin‒Benson‒Bassham cycle or photosynthetic carbon reduction cycle |
| Rubisco | RuBP carboxylase oxygenase |
| RuBP | Ribulose–1,5–bisphosphate |
| RuP | Ribulose–5–phosphate |
| RuP phosp, RuPphosp M, RuPphosp BS | Rate of RuP phosphorylation: total, in the M, or in the BS, respectively |
| SER, SER | Serine, SER diffusion rate, respectively |
| SMA | Stoichiometric model of assimilation |
| T, T | Transamination, Transamination rate |
| V C, VCM, VCBS | Rubisco rate of carboxylation: total, in the M, or in the BS, respectively |
| V O, VOM, VOBS | Rubisco rate of oxygenation: total, in the M, or in the BS, respectively |
| V P | PEPC rate of carboxylation |
| αKG | alpha–Ketoglutarate |
Table 2.
Functional classification of photosynthetic types and example species.
| Type | C3 | Proto-Kranz | C2 | C2+C4 | C4 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Subtype | - | - | - | NADP-ME | NAD-ME | PCK | NADP-ME | NADP-ME (+PCK) | NAD-ME | PEPCK (NADP-ME) | PEPCK (NAD-ME) |
| Example | Triticum aestivum | Heliotropium procumbens | Mollugo verticillata | Flaveria pubescens | Alternanthera tenella | Alloteropsis semialata | Sorghum bicolor | Zea mays | Panicum sp. (sensu stricto) | Alloteropsis semialata ssp. semialata | Spartina sp. |
| GDC compartmentalization | no | partial | full | full | full | full | full | full | full | full | full |
| Rubisco compartmentalization | no | partial | partial | partial | partial | partial | full | full | full | full | full |
| PEPC engagement | no | no | no | partial | partial | partial | full | full | full | full | full |
| PEPCK engagement | no | no | no | no | no | partial | no | partial | no | full | full |
| MDH engagement in the M | no | no | no | partial | no | partial | full | full | no | potentially active | no |
Detailed quantification of ATP and NADPH supply and demand in the BS and M is critical for understanding the physiology and regulation of photosynthesis. In terms of supply, the partitioning of ATP and NADPH production varies between cell types depending on the light available locally in the BS or M (Bellasio and Griffiths, 2014c). The dependence of ATP generation upon anatomical traits in the evolutionary continuum from C3 to C4 has recently been studied (Bellasio and Lundgren, 2016), and will not be addressed here. In this work, I shall concentrate on ATP and NADPH demand.
Quantifying ATP and NADPH demand in the M and BS requires detailed mechanistic understanding of assimilatory biochemistry. Mathematical modelling offers valid support for integrating knowledge at the systems level (Morandini, 2013; Singh et al., 2014). Classical photosynthetic models have allowed the simulation of leaf-level assimilation in C3, C2, C2+C4, and C4 plants using a mechanistic description based on Rubisco PEPC kinetics (von Caemmerer, 1989, 2000, 2013). These models are based on several simplifications, which limit their applicability. First, they do not account for spatial segregation of biochemical processes, offering only limited support when the separate requirements of the BS and M are being studied. Second, classical model(s) do not distinguish between biochemical subtypes, making it difficult to evaluate the particular requirements of each subtype. Finally, the models were primarily developed to predict leaf-level CO2 exchange, while the stoichiometry, energetics, and fluxes between the BS and M, which represent a critical bottleneck for C4 photosynthesis (Pick et al., 2011), are not treated sufficiently, and a dedicated model is consequently needed.
The aim of this work was to develop a stoichiometric model of assimilation (hereafter SMA) which (i) generalizes all pathways of assimilation (C3, C2, C2+C4, and C4 including C4 photosynthetic subtypes); (ii) is based only on stoichiometry and therefore does not rely on kinetic measurements; and (iii) includes all main photosynthetic reactions, but is user-friendly for non-specialists. The theoretical underpinnings of the SMA are described in detail and a range of simulations to exemplify the model rationale and behaviour are provided. Numerous topics are covered, including (i) an analysis of C3 and C2 photosynthesis and all subtypes of C2+C4 and C4 (NADP-ME, NAD-ME, and PEPCK, in different combinations) in operational conditions; (ii) the energetics involved in manipulating photorespiration in a C3 plant; (iii) the consequences of progressively upregulating a C2 shuttle in a background of C3 photosynthesis; and (iv) the consequences of progressively upregulating a C4 cycle in a background of C2 photosynthesis. Results quantify ATP and NADPH demand, which link dark and light reactions; refine our understanding of the stoichiometry of photosynthesis and the trade-offs inherent to different photosynthetic subtypes; and represent a useful framework for the integration of existing biochemical models.
SMA development
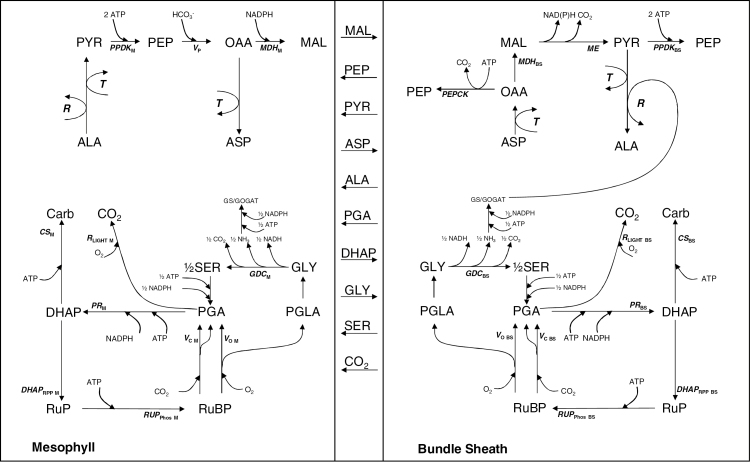
The SMA was developed on the basis of a stoichiometric model of NADP-ME C4 photosynthesis (Bellasio and Griffiths, 2014c; McQualter et al., 2016) to augment all the pathways of carbon assimilation in a single tool. The SMA calculates key reaction rates, and ATP and NADPH requirements, in the M and BS as well as fluxes between the BS and M when the following parameters are known: the locality of GDC and Rubisco, leaf-level Rubisco rates of carboxylation and oxygenation (VO, VC), and PEP carboxylation rate (VP). The ATP and NADPH requirements are SMA outputs and they are not related to light reactions at this stage. Reactions are typically grouped by the biochemical function of the pathways, of which only the entry point is calculated. The SMA is based only on well-established reaction stoichiometry. The SMA accounts for the interactions between C2 and C4 cycles, including the fluxes associated with amino group rebalancing, and for the NADPH and ATP demand of assimilatory processes. The theory underpinning the SMA can be followed in Fig. 1. Owing to space limitations, the full description of the SMA is reported in Supplementary file 1.
Fig. 1.
SMA schematic. The C4 CCM appears at the top, while C3 metabolism is at the bottom, partitioned between M and BS contributions. Metabolites for which fluxes are calculated are listed at the M–BS interface. The Excel workbook provided in Supplementary file 2 renders outputs according to this scheme.
Parameterization
The SMA has 12 input quantities (Table 3): three define Rubisco activity and assimilation (net assimilation, A; respiration in the light, RLIGHT; and Rubisco rate of oxygenation relative to carboxylation, rO/C), two define the activity of the CCM (PEPC engagement, as VP; and PEPCK activity relative to VP, rPEPCK), and seven partition key processes between the BS and M (fC, for Rubisco carboxylation rate; fO, for Rubisco oxygenation rate; fGDC, for glycine decarboxylase; fRLIGHT, for respiration in the light; fPR, for PGA reduction; fCS, for carbohydrate synthesis; fPPDK, for pyruvate phosphate dikinase). In addition, for NAD-ME subtypes it is possible to constrain malate dehydrogenase (MDH) activity to zero. Inputs can be constrained in different ways depending on the research questions. When the goal is to refine the analysis of a particular metabolic state of the leaf, input quantities may represent realistic biochemistry, otherwise inputs can be freely manipulated to simulate bioengineering or explore hypothetical scenarios. A, RLIGHT, and can be measured with accuracy (Bellasio et al., 2014; Bellasio et al., 2016a, b). Quantification of VP is less straightforward, and can be achieved through an in vitro assay of PEPC activity (Pfeffer and Peisker, 1998). This is complicated by PEPC-sensitive regulation and feedback inhibition, and the consequent necessity of reproducing physiological metabolite and ion concentrations in the reaction mixture. In alternative A, and VP can be predicted with biochemical models. Two types of formulations exist, either based on enzyme kinetics (and referred to as enzyme-limited), or based on the rate of ATP and NADPH made available by light reactions (and referred to as light-limited). Integrating enzyme-limited formulations in the SMA is straightforward, because the A, , and VP output by the biochemical model can be directly input into the SMA (see references below). Integrating light-limited formulations is more complicated and will be addressed in a dedicated paper. Physiological values are available for rPEPCK, gained through extensive biochemical work (Kanai and Edwards, 1999; Koteyeva et al., 2015). The traits underpinning fO, fC, fGDC, and fRLIGHT may require evolution or long acclimation periods to change (Sage et al., 2012; Christin and Osborne, 2014) and are therefore considered constant during gas exchange experiments. In the SMA, fO, fC, and fGDC represent the fraction of enzyme activity in the BS, rather than the physical distribution of the enzyme, but when enzyme compartmentalization is complete (Table 2), these become equal. The distribution of Rubisco and GDC can be quantified through proteomics, biochemical assays, or immunolocalization [e.g. Aoyagi and Nakamoto (1985), Majeran et al. (2005), and Keerberg et al. (2014)] and are generally known for model species (Edwards and Ku, 1987). Cases of intermediate GDC distribution are rare (Sage et al., 2014), and, even in these cases, the enzyme distribution may be confidently taken as fGDC because substrate concentrations and regulation may be similar in the M and BS. For Rubisco, when compartmentalization is incomplete (as in C2+C4 species), predicting fC and fO requires that the increased CO2 concentration in the BS be modelled, for instance using the validated models for C4, C3, C2, and C2+C4 photosynthetic subtypes (Farquhar et al., 1980; von Caemmerer, 1989, 2000, 2013; von Caemmerer and Furbank, 1999). fRLIGHT is often assumed to be 0.5 in C4 plants and 0.2 in C2+C4 plants (von Caemmerer, 1989, 2000), and may be determined from the relative BS/M mitochondrial abundance, or simply from the relative BS/M volume (Edwards and Ku, 1987). Circumstantial evidence suggests that fCS, fPR, rPEPCK, and fPPDK may rapidly change and this has likely contributed to the difficulty of distinguishing them experimentally. Exact empirical parameterization may therefore be of limited interest, and it may be more informative to define physiological maxima and minima (Bellasio and Griffiths, 2014c). Within these limits, the adjustment of fCS, fPR, rPEPCK and fPPDK may help the plant maximize assimilation under transient environmental inputs, such as changes in light quality, which may unbalance the partitioning of ATP and NADPH supply (Bellasio and Griffiths, 2014c). The effect of varying these inputs will be simulated in the subsection below dedicated to C4 photosynthesis to show how these mechanisms can re-balance ATP and NADPH demand.
Table 3.
Input quantities used in dynamic computer simulations shown in Supplementary Figure S1 and Figures 2–4
| Simulation | 1.2 C3 photorespiration | 2.2 Proto-Kranz and C2 | 3.2 C2 + C4 | 4.2.1 Partitioning PGA reduction | 4.2.2 Manipulating PEPCK activity | 4.2.3 Partitioning carbohydrate synthesis | 4.2.4 PPDK engagement in the BS | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Subtype | - | - | - | NADP-ME | NAD-ME | NADP-ME | NAD-ME | NADP-ME | NAD-ME | NADP-ME | NAD-ME |
| Figure | S1 | 2 | 3 | 4A | 4B | 4C | 4D | 4E | 4F | 4G | 4H |
| Basic quantities | |||||||||||
| R LIGHT (μmol m−2 s−1) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| A (μmol m−2 s−1) | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 |
| r O/C | variable | 0.45 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | |
| CCM | |||||||||||
| V P (μmol m−2 s−1) | 0 | 0 | fitted for L = 0 | 10.85 | 10.85 | 10.85 | 10.85 | 10.85 | 10.85 | 10.85 | 10.85 |
| r PEPCK | 0 | 0 | 0 | 0 | variable | variable | 0 | 0 | 0 | 0 | |
| MDH M | irrelevant | Eqn S19 | Eqn S19 | Eqn S19 | MDH M = 0 | Eqn S19 | MDH M = 0 | Eqn S19 | MDH M = 0 | Eqn S19 | MDH M = 0 |
| BS contribution | |||||||||||
| Slow response | |||||||||||
| f C , f O | 0 | fitted for L = 0 | variable | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| f GDC | 0 | variable | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| f RLIGHT | 0 | 0.2 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| Fast response | |||||||||||
| f PR | 0 | 0 | 0 | variable | variable | 0.25 | 0.25 | 0 | 0 | 0 | 0 |
| f CS | 0 | 0 | 0 | 0 | 0 | 0 | 0 | variable | variable | 0 | 0 |
| f PPDK | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | variable | variable |
SMA simulations
The following simulations were selected to illustrate the capabilities, rationale, and behaviour of the SMA, while at the same time making some considerations of interest for the theme of this special issue. Simulations are grouped for photosynthetic types (C3, proto-Kranz and C2, C2+C4, and C4). For each type, two sets of simulations are presented: static scenarios, where the SMA is calculated for one set of inputs, representing realistic operational conditions (Supplementary Table S1), and dynamic simulations where inputs are varied (Table 3).
Simulation 1. C3 photosynthesis
Simulation 1.1. Operational conditions
Outputs for C3 photosynthesis in operational conditions are shown in Supplementary Fig. S1. The C4 CCM is not operational. Carbon fixation and the reductive pentose phosphate (RPP) cycle operate in the M, while no carbon fixation is occurring in the BS. Although photorespiration and GDC activity are high, no glycine (GLY) is exported in the BS, and there is no net metabolite flux at the M–BS interface. All ATP and NADPH demand is in the M.
Simulation 1.2. Dynamic simulations
A dynamic scenario for a C3 photosynthetic type was simulated by varying rO/C between 0 and 1 while keeping other quantities at 0 (Table 3). The resultant ATP and NADPH demand as a function of rO/C is plotted in Supplementary Fig. S2A while the ratio between ATP and NADPH demand is plotted in Supplementary Fig. S2B.
Simulation 2. C2 and proto-Kranz
Simulation 2.1. Operational conditions
Outputs for C2 photosynthesis are shown in Supplementary Fig. S3. To operate a C2 shuttle, GDC is absent in the M and the photorespiratory GLY produced in the M is decarboxylated in the BS and fixed by a small fraction of Rubisco located in the BS. Serine (SER) diffuses back to the M, exporting half of the GLY amino groups. The excess ammonia produced by GDC in the BS is fixed by glutamine synthetase/glutamine oxoglutarate aminotransferase (GS/GOGAT), transaminated to alanine (ALA), and diffuses back to the M so as to re-balance the amino groups. For the C2 shuttle to operate, an import flux of pyruvate (PYR) equimolar to R is required in the M. The C4 CCM is not operational. In this example, PGA is reduced mainly in the M and a substantial BS↔M triose phosphate exchange occurs. Because the CCM is not operational, there is no net export of NADPH from M to BS and the NADPH demand in BS of 1.1 μmol m−2 s−1 must be met by linear electron flow (LEF); however, cyclic electron flow (CEF) may be preponderant as the ratio of ATPBS to NADPHBS was ~8.
Simulation 2.2. Dynamic simulations
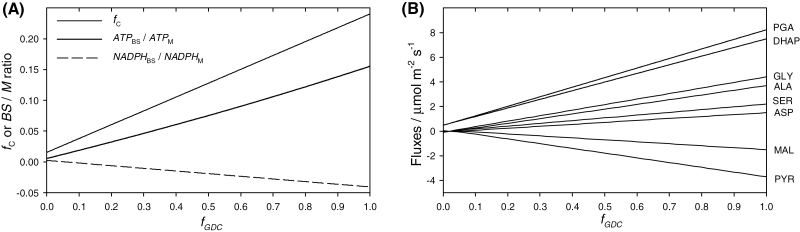
f GDC was varied from 0 to 1 in an idealized C3 plant, thus simulating the transition to C2 photosynthesis through intermediate states of GDC compartmentalization, which are generally referred to as proto-Kranz (Sage et al., 2014). At each fGDC level, fC was fitted so that CO2 leakage was zero (Supplementary Equation S34). The resultant fC as a function of fGDC is plotted in Fig. 2A. Values of fC above the curve will result in a net influx of CO2 into the BS driven by Rubisco fixation, and in a CO2 concentration in the BS lower than that in the M. Values of fC below the curve will result in a net CO2 efflux out of the BS, and in a CO2-concentrating effect of the C2 shuttle. Moving Rubisco and the photosynthetic carbon oxygenation (PCO) cycle to the BS results in changing the locality of ATP and NADPH demand (Fig. 2A) and requires a substantial traffic of metabolites (Fig. 2B). The flux of PGA out of the BS and the opposite flux of dihydroxyacetone phosphate (DHAP; which is lower than that of PGA by a value equal to carbohydrate synthesis, CS) results from setting fPR = 0 in this simulation. This constraint also determines an excess of reducing power in the BS because the NADH produced by GDC in the BS is not used by PR. In these conditions the SMA predicts MAL to diffuse from the BS to M to shuttle the excess reducing power in the BS. MAL is oxidized to oxaloacetate (OAA) in the M and transaminated to ASP, which diffuses back to the BS and is transaminated back to OAA to supply MDH in the BS. The pair PYR/ALA balances the amino groups resulting from OAA/ASP transamination, and the amino groups resulting from the flux of GLY and SER are directly dependent on the operation of the C2 shuttle. Alternative scenarios may involve NADH resulting from GDC activity sustaining a minimal level of PR in the BS (shown in Supplementary Fig. S3).
Fig. 2.
Simulation 2.2. From C3 to C2 photosynthesis. The BS partitioning of GDC activity (fGDC) was varied between 0 and 1. Panel (A) shows the partitioning of Rubisco activity that resulted in no net CO2 flux across the BS–M interface, together with the resultant ATP and NADPH partitioning. Panel (B) shows the corresponding metabolite fluxes. Inputs are shown in Table 3.
Simulation 3. C2+C4
C2+C4 photosynthesis has complete GDC compartmentalization to the BS and intermediate states of CCM engagement (Table 2), corresponding to moderate VP and incomplete Rubisco compartmentalization. The CCM subtype is defined by the engagement of MDH in the M and PEPCK in the BS.
Simulation 3.1. Operational conditions
An example of NADP-ME C2+C4 is shown in Supplementary Fig. S4A. The C4 CCM is operational and CCM activity is sufficient to exceed Rubisco carbon fixation in the BS, and there is a net CO2 efflux from BS (L > 0). PR is mainly located in the M, and triose phosphate trafficking is higher than for the C2 photosynthetic type. Because a large part of Rubisco is mainly located with GDC in the BS, and photorespiration is reduced by the activity of the CCM, the effectiveness of the C2 shuttle (as R) is reduced relative to C2 photosynthesis. PR in the BS consumes all NADPH available through MDHM, therefore T ≈ 0; however, there is no residual NADPH deficit, and LEF is not required in the BS.
In a NAD-ME C2+C4 photosynthetic subtype (Supplementary Fig. S4B), the C4 CCM results in the same export of CO2 to the BS as in the NADP-ME C2+C4 photosynthetic subtype; however, MDH activity in the M is zero, all OAA is transaminated, and the CCM does not export reducing power to the BS. PR and glycolate recycling in the BS consumes NADPH at a rate of 2.32 μmol m−2 s−1, which must be generated through LEF.
In a PEPCK C2+C4 photosynthetic subtype (Supplementary Fig. S4C), the C4 CCM results in the same export of CO2 to the BS as in the NADP-ME and NAD-ME C2+C4 photosynthetic subtypes. However, although MDH is present in the M, PEPCK activity in the BS diverts OAA produced by PEPC in the M, making OAA unavailable for MDH activity in the M, which is consequently zero. Furthermore, because MDH activity is zero, the CCM does not export reducing power to the BS, and the NADPH must be supplied through LEF in the BS.
Simulation 3.2. Dynamic simulations
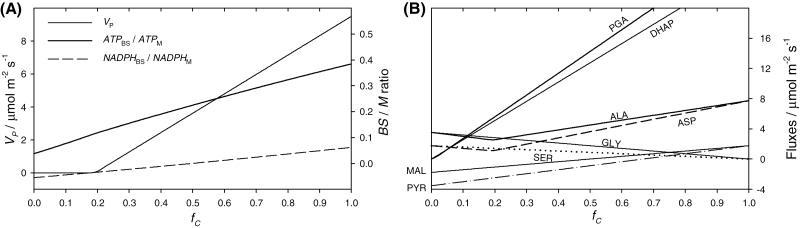
Intermediate states of ‘C4ness’, represented by intermediate degrees of fC and VP, were explored to simulate the transition from C2 photosynthesis to C4 photosynthesis. fC was incrementally varied, and, at each fC level, VP was iteratively fitted so that the CO2 leak rate (L) remained zero (Supplementary Equation S34). The resultant VP as a function of fC is plotted in Fig. 3A. Values of VP above the curve will result in an effective CCM. Values of VP below the curve are insufficient to sustain Rubisco fixation, which will drive a net influx of CO2 into the BS. For fC < 0.2, Rubisco fixation in the BS is supplied by the C2 shuttle, meaning the predicted VP is zero. Moving Rubisco to the BS causes the locality of ATP and NADPH demand to change (Fig. 3A). Reducing power demand in the BS is low and is only used by the PCO cycle by setting fPR at zero. This requires PGA to diffuse out of the BS and in an opposite flux of DHAP, which is lower than that of PGA by a value equal to CS. In addition, ALA and ASP are used by the CCM to bypass MDH in the M. The concave trend of ALA and ASP fluxes reflects a decreasing recruitment for the C2 shuttle at low fC and an increasing recruitment for the CCM at high fC. GLY and SER are recruited only by the C2 shuttle and decrease to zero with fC. MAL and PYR are used by the C4 cycle and their fluxes increase linearly with fC.
Fig. 3.
Simulation 3.2. From C2 to C4 photosynthesis. The BS partitioning of Rubisco activity (fC) was varied between 0 and 1. Panel (A) shows the rate of PEPC activity (VP) that resulted in no net CO2 flux across the BS–M interface, and the resultant ATP and NADPH partitioning. Panel (B) shows the corresponding metabolite fluxes. Inputs are shown in Table 3.
Simulation 4. C4 photosynthetic subtypes
Here, Rubisco and GDC are completely compartmentalized to the BS and VP exceeds VC.
Simulation 4.1. Operational conditions
Simulation 4.1.1.
Outputs for a typical NADP-ME subtype are shown in Supplementary Fig. S5A. The activity of the CCM is strong enough to exceed Rubisco carbon fixation in the BS and a positive CO2 leakage out of the BS. PR is mainly located in the M and metabolite traffic is up to five-times that of gross assimilation (GA). The net effect of the C2 shuttle (as R), which depends on Rubisco oxygenating activity in the M, is zero. The CCM supplies all NADPHBS (6.54 μmol m−2 s−1), no LEF is required in the BS, and there is excess OAA that is not reacted with by MDH and is subsequently transaminated (T).
Simulation 4.1.2.
Supplementary Fig. S5B shows a typical NADP-ME subtype with engagement of PEPCK. PEPCK activity regenerates part of the PEP required by PEPC, driving a positive PEP flux out of the BS, which reduces the activity of PPDK in the M. PEPCK consumes half the ATP of PPDK, resulting in a 4% lower ATP/GA than for the NADP-ME subtype. However, PEPCK activity is generally low, all NADPHBS is supplied by the CCM, and no LEF is required in the BS.
Simulation 4.1.3.
Supplementary Fig. S5C shows a typical NAD-ME C4 photosynthetic subtype with no engagement of PEPCK. The CCM exports CO2 at the same rate as the NADP-ME subtype; however, MDH activity in M is zero, all OAA is transaminated, and the NADPH demand in the BS has to be generated in the BS through LEF.
Simulation 4.1.4.
Supplementary Fig. S5D shows a background NADP-ME metabolism using PEPCK as a sole decarboxylase (rPEPCK = 1). The CCM exports CO2 at the same rate as the other subtypes. Because MDH is present in the M, the CCM would export reducing power to the BS if rPEPCK < 1, but, here, PEPCK activity in the BS diverts OAA produced by PEPC in the M, making OAA unavailable for MDH activity in the M, which is consequently zero. Furthermore, because MDH activity is zero, the CCM does not export reducing power to the BS, and the NADPH must be supplied through LEF in the BS.
Simulation 4.1.5.
Supplementary Fig. S5E shows SMA output for a background NAP-ME metabolism using PEPCK as a sole decarboxylase (rPEPCK = 1). The CCM exports CO2 at the same rate as the other subtypes. Here, MDH in M is not operational, and the CCM does not have the potential to export reducing power to the BS.
Simulation 4.2. Dynamic simulations (C4 plasticity mechanisms)
For the next four simulations, rO/C was set at a typical C4 value of 0.05 (Bellasio et al., 2014); VP was set at 10.85 estimated after von Caemmerer (2000); and gradual transitions of fCS, fPR, rPEPCK, and fPPDK were simulated by calculating the SMA for 21 discrete values of fCS, fPR, rPEPCK , and fPPDK between 0 and 1. Other model inputs are listed in Table 3.
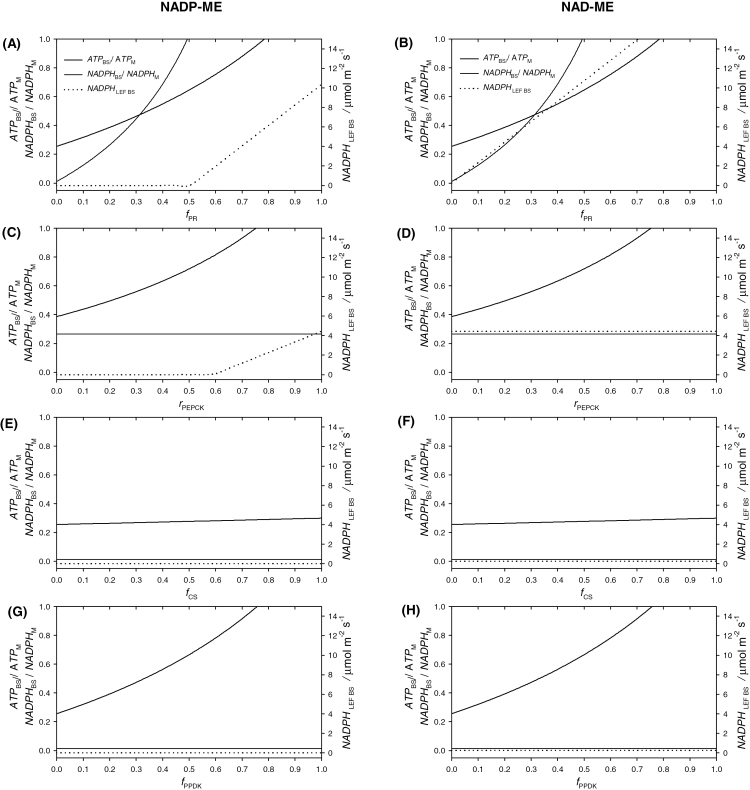
Simulation 4.2.1. Partitioning PR.
The fraction of PGA reduced in the BS was manipulated through the input parameter fPR, while rPEPCK, fPPDK, and fCS were kept at zero. An increase in fPR caused an increase in the ATP and NADPH demand in the BS, which occurred in both NADP-ME and NAD-ME subtypes (Fig. 4A, B). The NADPH demand to be supplied by LEF in the BS followed different trends. In the NADP-ME subtype, with an fPR of up to 0.5, the NADPH demand for PR was met by the CCM through the MAL shuttle, and the resultant NADPH demand through LEF was zero. Additional levels of PR (fPR > 0.5) required the engagement of LEF in the BS. In the NAD-ME subtype, the demand for LEF in the BS increased linearly for fPR > 0.
Fig. 4.
Simulation 4.2. SMA simulations showing the effect of varying the BS engagement in PR (A, B), PEPCK activity (C, D), the BS engagement in CS (E, F), the BS fraction of PPDK activity (G, H), on the partitioning of ATP demand (thick lines), the partitioning of NADPH demand (thin lines), and on the rate of NADPH produced by LEF in the BS (dotted lines, right axes) in a background NADP-ME (left) or NAD-ME (right) subtype. Inputs are shown in Table 3.
Simulation 4.2.2. Manipulating PEPCK activity.
The engagement of PEPCK was manipulated through the input parameter rPEPCK. fPR was set at 0.25 to highlight differences between NADP-ME and NAD-ME subtypes, while fPPDK and fCS were kept at zero (Table 3). An increase in rPEPCK increased the ATP demand in the BS in both the NADP-ME and NAD-ME subtypes (Fig. 4C, D), but the partitioning of NADPH demand was unaffected. The NADPH demand to be supplied by LEF in the BS, however, followed different trends. In the NADP-ME subtype, with an rPEPCK of up to 0.6, the NADPH demand through LEF was zero, and increased linearly up to 4.5 μmol m−2 s−1 for rPEPCK > 0.6. In the NAD-ME subtype, the NADPH demand to be supplied by LEF was constant at 4.5 μmol m−2 s−1, regardless of rPEPCK.
Simulation 4.2.3. Partitioning CS.
In the simulation, the BS fraction of CS was manipulated to increase through the input parameter fCS, while rPEPCK, fPPDK, and fPR were kept at zero. Increasing fCS determined a marginal increase in ATPBS/ATPM from 0.26 to 0.29 in both the NADP-ME and NAD-ME subtypes, while the NADPH demand was unaffected (Fig. 4E, F).
Simulation 4.2.4. Effect of PPDK engagement in the BS.
The effect of PPDK engagement in the BS was studied by manipulating the input parameter fPPDK, while rPEPCK, fCS, and fPR were kept at zero. The ATP demand in the BS increased substantially, while the NADPH demand was unaffected in both the NADP-ME and NAD-ME subtypes (Fig. 4G, H).
Discussion
Flux-balance analysis models are constraint-based models in which steady state fluxes are predicted in a metabolic network by applying mass-balance constraints based on reaction stoichiometry (Sweetlove and Ratcliffe, 2011). Lately the complexity of models has grown to embrace a suite of photosynthetic processes (Laisk et al., 2009; Wang et al., 2014a; Wang et al., 2014b) and reconstruct genome-wide metabolism (Dal’Molin et al., 2010; Saha et al., 2011). Given this complexity, these models are only available for a few well-studied species and modifying them requires considerable coding effort, meaning they are not ideal for studying bio-manipulation or testing hypotheses. The metabolic model developed by Bellasio and Griffiths (2014c) facilitated straightforward modifications and changes to metabolic pathways. For instance, the model computed the effects of partitioning biochemical work between the BS and M on the locality of ATP and NADPH demand in photosynthesizing C4 leaves, and was modified to support the interpretation of biochemical, gas exchang,e and transcriptomic data in engineered sugarcane (McQualter et al., 2016). Based on these theoretical underpinnings, the SMA was developed to account for the interactions between C2 and C4 cycles, including the fluxes associated with amino group rebalancing, recently described in Mallmann et al. (2014), and their effect on NADPH and ATP availability. The SMA integrates assimilatory metabolism and energetics as well as calculating key reaction rates, metabolite traffic fluxes, and ATP and NADPH requirements in the M and BS when the locality of GDC (as fGDC) and Rubisco (as fO, and fC) activity, leaf-level Rubisco rates of carboxylation and oxygenation (as rO/C), and PEP engagement (as VP) are known. While previously published models are tailored for a particular species or a discrete photosynthetic type that is fixed a priori, the SMA allows rates to vary continuously between boundaries defined by what is biochemically feasible. The SMA is designed for integration with existing biochemical models. However, these models rely on assumptions that are specific to a particular photosynthetic type and, as such, they were not included in the SMA at this stage. The SMA is based upon logical constraints and well-established reaction stoichiometry, and is, consequently, of general use. Compared with other recent flux-balance models [e.g. Dal’Molin et al. (2010)], the SMA has several distinctive features. It is focused on assimilation; it generalizes all pathways of carbon assimilation in a single tool, except CAM, which requires an explicit temporal dynamic (Owen and Griffiths, 2013); it allows straightforward modification by the user; and it explicitly accounts for NADPH and ATP demand, allowing the study of the effect of biochemical regulation on energetics.
The following discussion refers to the simulations described in simulations 4.2.1–4.2.4 (C4 plasticity mechanisms, Fig. 4), which are of particular interest for this special issue. Possible mechanisms that C4 plants can exploit to stabilize their energy and redox state were recently reviewed (Stitt and Zhu, 2014). Reversible reactions linking PGA and triose phosphates, the reversibility of the PPDK reaction, the inter-conversion of PEP and PGA, and the transient build-up and utilization of PGA and triose phosphate require explicit temporal dynamics to be modelled and cannot be addressed by the SMA. Nevertheless, the futile cycles involved probably lower the biochemical efficiency of assimilation and are likely to be downregulated under steady state conditions. The other processes reviewed by Stitt and Zhu (2014) are shown in the simulation results. Specifically, the rate of ASP/MAL decarboxylation was mechanistically linked to reducing power requirement in the BS through Supplementary Equation S21, and can be followed in Supplementary file 2 (workbook cell O14); CO2 leakage and leakiness are calculated by balancing all CO2 fluxes in and out of the BS (Supplementary Equation S34) and can be followed in Supplementary file 2 (workbook cells O19 and O6, respectively); the exchange of triose phosphate and PGA and the effect of switching decarboxylase (ME versus PEPCK) are covered in simulations 4.2.1 and 4.2.2. In addition to the list of Stitt and Zhu (2014), here I investigated a possible role of CS by manipulating fPR (simulation 4.2.3), and I suggest a possible role for PPDK in the BS, which was investigated by manipulating fPPDK (simulation 4.2.4). The simulations were carried out as a single factor analysis in which only one input was varied at a time (Table 3). This approach has the benefit of allowing a comparison of the effectiveness of different plasticity mechanisms. Combining effects, however, is also straightforward: when multiple inputs are manipulated, the total effect is the sum of single effects. The four simulations are discussed separately.
Partitioning PR and regulation of NADPH demand
When BS engagement in PR was manipulated to increase, ATP and NADPH demand in the BS also increased. In particular, in NADP–ME subtypes above a fPR of 0.5, there was a substantial demand for NADPH through LEF in the BS. While ATP is produced by BS photophosphorylation in both NADP-ME and NAD-ME plants, BS chloroplasts in NADP-ME plants are generally not able to oxidize water and produce substantial amounts of NADPH. In NADP-ME plants, high levels of fPR are therefore unlikely, consistent with the observation that, in maize, PGA is mainly reduced in the M (Friso et al., 2010). Confining O2 production to M chloroplasts, away from Rubisco carboxylating sites, contributes to photorespiration suppression (Majeran and van Wijk, 2009), but imposes functional trade-offs. The inability to sustain high levels of PR in the BS requires PGA to diffuse to M cells, and DHAP to return to BS cells in order to complete the RPP cycle (Fig. 1). This imposes substantial traffic across the M–BS interface that is five-times that of GA (Supplementary Fig. S5A–E). For this traffic, high BS conductance would be beneficial, but imposes high levels of CO2 retrodiffusion, and limits the effectiveness of the CCM (Bellasio and Griffiths, 2014a). Because of this trade-off, BS conductance is thought to be tightly regulated (Kromdijk et al., 2014), and was observed to scale with assimilation to optimize the efficiency of the CCM (Ubierna et al., 2013; Bellasio and Griffiths, 2014b; von Caemmerer and Furbank, 2016). The available evidence, however, is indirect and further investigation is required.
Manipulating PEPCK activity
While ATP is directly used in PEPCK reactions, PEPCK activity does not directly require NADPH, as shown by the plot of NADPH demand in Fig. 4C. The effect on NADPH demand through LEF is therefore indirect and depends on the subtype (NADP-ME or NAD-ME). NADP-ME subtypes have the potential to export reducing power to the BS through MDH activity in the M: when PEPCK activity increases, increasing levels of OAA are required, which are not available for MDH in the M and cannot be used to shuttle reducing power from the M to the BS through MAL. In other words, the NADPH potentially available through the CCM decreases. Ultimately, when PEPCK is fully engaged, there is no surplus of OAA available for exporting NAPDH to the BS and all NADPH demand in the BS must be met by LEF. In NAD-ME subtypes, which do not have the capacity to export reducing power, all NADPH demand has to be met by LEF. When PEPCK is the only decarboxylating enzyme, irrespective of the presence of MDH in the M (that is, in both NAD-ME and NADP-ME subtypes), no reducing power can be exported to the BS (Fig. 2C, D). Wang et al. (2014a) noted considerable variability in PEPCK engagement. For instance, in the NADP-ME subtypes there is a gradient from Sorghum bicolor with virtually no PEPCK engagement, through low engagement in Flaveria species, intermediate engagement in maize and sugarcane (Saccharum species), to virtually complete engagement in the atypical C4Alloteropsis semialata (Gutierrez et al., 1974; Ueno and Sentoku, 2006; Wang et al., 2014a; Lundgren et al., 2016; Dunning et al., in review). Similarly, in NAD-ME subtypes the gradient spans Panicum species with virtually no PEPCK engagement, Cleome C4 species with intermediate PEPCK activity, and Spartina maritima where PEPCK engagement is virtually complete (Gowik and Westhoff, 2011; Bellasio and Griffiths, 2014c; Lundgren et al., 2016). Regardless of how this variability is classified (Furbank, 2011; Wang et al., 2014a; Koteyeva et al., 2015), the SMA allows rates to vary continuously between boundaries set by what is biochemically realistic.
Given that PEPCK regenerates PEP with half the ATP required by PPDK, moderate levels of PEPCK engagement could potentially increase the biochemical efficiency of assimilation (compare ATPTOT/GA in Supplementary Fig. S5A with Fig. S5D). The rapid regulation of PPDK (Leegood and Walker, 1999) could contribute to the flexibility and efficiency of the CCM, supported by the observation that redundant decarboxylating pathways appeared multiple times at late evolutionary stages (Christin and Osborne, 2014). At high levels of PEPCK engagement (Supplementary Fig. S5D, E), the ATP/GA predicted is substantially lower than for the other photosynthetic subtypes. However, biochemical efficiency may be reduced by the need to hydrolyse part of the newly synthetized PEP to drive the PEPCK reaction, which is close to thermodynamic equilibrium and may be too slow to support physiological decarboxylation rates (Huber and Edwards, 1975). Clarification is still required, and there may be interspecific variability (Smith and Woolhouse, 1983); however, circumstantial evidence gained in comparative experiments have shown lower quantum efficiency of PEPCK plants [Furbank (2011) and references therein].
PEPCK is required for the CCM to work, and when PEPCK is the only decarboxylating enzyme it is likely to be modulated solely by the requirements of the CCM. As a consequence, in PEPCK plants, PEPCK activity cannot contribute to fine-tuning ATP demand, and hence the biochemical plasticity of the CCM is lower. The additional ATP used by PEPCK can therefore be considered part of the minimum ATP demand in the BS, ATPBSMIN = RuPphospBS + PEPCK + VOBS + R, corresponding to a minimum ATP demand partitioning ratio of which is approximately three times that of other C4 subtypes (see below). Although PEPCK plants have numerous chloroplasts in the BS (Dengler and Nelson, 1999; Bellasio and Lundgren, 2016), limiting environmental conditions (e.g. dim diffuse sky light) may prevent the BS from supplying ATPBSMIN. In these conditions ATP could be synthetized by mitochondria, thus countering the lower biochemical plasticity of the CCM with additional flexibility in ATP generation. The isolated BS of some PEPCK plants can, under ATP starvation, convert MAL-derived NADH into ATP through mitochondrial oxidation of NADH (Carnal et al., 1993). This process is less energy efficient than photophosphorylation (Buckley and Adams, 2011; Kramer and Evans, 2011), and may contribute to the generation of ATPBSMIN only when illumination of the BS chloroplast is insufficient.
Partitioning CS
The main photosynthetic products in C4 species are starch and sucrose. Both can be synthesized in the M and BS (Majeran and van Wijk, 2009; Friso et al., 2010), but sucrose is preferentially synthesized in the M, while starch disproportionately accumulates in the BS (Furbank et al., 1985; Lunn and Furbank, 1997). It has been observed that accumulation of sucrose in the leaf does not directly influence the partitioning between sucrose and starch (Lunn and Hatch, 1995), apparently discounting a role for starch synthesis as a sink for carbon overspill when sucrose synthesis is inhibited by sucrose build-up (Lunn and Hatch, 1995). Experiments conducted with Panicum species examined the effect of illumination on the distribution of sucrose–phosphate synthase activity between the M and BS, showing contrasting activation patterns between subtypes (Ohsugi and Huber, 1987). Here, a role of energetics is unlikely given the overall negligible ATP cost of CS. Alternative explanations involve an effect of the ratio of PGA to inorganic phosphate on the activity of ADP-glucose pyrophosphorylase [Stitt et al., 1987; Lunn and Furbank, 1997; for further considerations see Ap Rees (1987), Furbank (1992), Leegood and Walker (1999), Zeeman et al. (2007), Kotting et al. (2010), and Weise et al. (2011)].
Effect of PPDK engagement in the BS and fast regulation of ATP demand
PPDK was shown to be present and active in the BS (Aoyagi and Nakamoto, 1985; Friso et al., 2010), although its elusive role is generally not included in textbook descriptions of C4 photosynthesis. The engagement of PPDK increases ATP demand in the BS in the same way as PR, as both processes require two ATP per catalytic turnover. However, PPDK does not require NADPH, whereas PR does. I propose that the tight regulation of PR and PPDK activities (and, in some cases, PEPCK, see above) contribute to fine-tuning ATP and NADPH demand in the BS in response to illumination of the BS chloroplast. Light availability in the BS is determined by anatomy and changes dynamically according to light intensity and quality. The locality of ATP production is therefore largely independent of metabolic control [Bellasio and Lundgren (2016) and references therein]. Local ATP imbalances cannot be rebalanced by ATP diffusion because maintaining ATP concentration and a high ATP to ADP ratio in each cell compartment is critical. The only way to balance supply and demand in each compartment is to regulate the locality of ATP demand (Evans et al., 2007; Bellasio and Griffiths, 2014c). For instance, blue-rich diffuse sky radiation is strongly absorbed in the superficial M and results in preferential ATP production in the M (Evans et al., 2007). In these conditions BS activity can be downregulated, and countered by a proportional increase in M activity, to maintain assimilation and biochemical efficiency. However, operating the RPP, the C2, and PCO cycles in the BS impose a limit, and a threshold of ATP demand in the BS (ATPBSMIN = RuPphospBS + VOBS + R), corresponding to an ATP partitioning ratio of If ATP supply in the BS is lower than the minimum demand, C4 photosynthesis ceases, leading to stunted phenotypes (Bellasio and Griffiths, 2014c; McQualter et al., 2016). More penetrating light (e.g. red light or direct sunlight) may drive higher ATP synthesis in the BS. In this case, PR and PEPCK may be upregulated, but are limited by the amount of NADPH available in the BS through the MAL shuttle or through LEF, as discussed above. PPDK may be engaged to take advantage of additional ATP, which may be made available under transient exposure to even more penetrating light qualities (e.g. green-enriched canopy light).
Conclusion
A SMA has been developed as a modelling framework based solely on mass-balance constraints, which generalizes all pathways of assimilation (except CAM) in a single tool. A range of examples detailed the energetics and metabolite fluxes involved in the gradual activation of the C2 shuttle and the CCM along a spectrum of photosynthetic subtypes from C3 to C4 photosynthesis. This knowledge is important for basic and applied research, to support advanced breeding techniques, or to study natural variability of biochemical traits. For instance, by providing quantitative estimates for fluxes and energy requirements in the BS and M, it is possible to set realistic targets for bioengineering projects. Future work will integrate biochemical models to allow the SMA to respond directly to environmental variables.
Availability
SMA is coded in an Excel workbook that is freely available to download. Macros are avoided.
Supplementary data
Supplementary data are available at JXB online.
File 1. SMA development and equations S1–S34; simulation of static scenarios under physiological operational conditions: Table S1 and Figures S1, S3–S5E; simulation 1.2, a dynamic scenario for C3 photosynthesis: Figure S2.
File 2. Excel workbook coding the SMA.
Supplementary Material
Acknowledgments
I am grateful to Joe Quirk for help with the manuscript; to Pascal-Antoine Christin and Marjorie Lundgren for critical review and discussion; and to Richard Leegood, Rowan Sage, and Colin Osborne for useful suggestions. I acknowledge funding through an ERC advanced grant (CDREG, 322998) awarded to David J Beerling.
References
- Aoyagi K, Nakamoto H. 1985. Pyruvate, Pi dikinase in bundle sheath strands as well as in mesophyll cells in maize leaves. Plant Physiology 78, 661–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ap Rees T. 1987. Compartmentation of plant metabolism. In: Davies D, ed. The biochemistry of plants, Vol. 12 San Diego: Academic Press, 87–113. [Google Scholar]
- Bellasio C, Beerling DJ, Griffiths H. 2016. a. Deriving C4 photosynthetic parameters from combined gas exchange and chlorophyll fluorescence using an Excel tool: theory and practice. Plant, Cell & Environment 39, 1164–1179. [DOI] [PubMed] [Google Scholar]
- Bellasio C, Beerling DJ, Griffiths H. 2016. b. An Excel tool for deriving key photosynthetic parameters from combined gas exchange and chlorophyll fluorescence: theory and practice. Plant, Cell & Environment 39, 1180–1197. [DOI] [PubMed] [Google Scholar]
- Bellasio C, Burgess SJ, Griffiths H, Hibberd JM. 2014. A high throughput gas exchange screen for determining rates of photorespiration or regulation of C4 activity. Journal of Experimental Botany 65, 3769–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellasio C, Griffiths H. 2014. a. Acclimation of C4 metabolism to low light in mature maize leaves could limit energetic losses during progressive shading in a crop canopy. Journal of Experimental Botany 65, 3725–3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellasio C, Griffiths H. 2014. b. Acclimation to low light by C4 maize: implications for bundle sheath leakiness. Plant, Cell & Environment 37, 1046–1058. [DOI] [PubMed] [Google Scholar]
- Bellasio C, Griffiths H. 2014. c. The operation of two decarboxylases (NADPME and PEPCK), transamination and partitioning of C4 metabolic processes between mesophyll and bundle sheath cells allows light capture to be balanced for the maize C4 pathway. Plant Physiology 164, 466–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellasio C, Lundgren MR. 2016. Anatomical constraints to C4 evolution: light harvesting capacity in the bundle sheath. New Phytologist DOI: 10.1111/nph.14063. [DOI] [PubMed] [Google Scholar]
- Buckley TN, Adams MA. 2011. An analytical model of non-photorespiratory CO2 release in the light and dark in leaves of C3 species based on stoichiometric flux balance. Plant, Cell & Environment 34, 89–112. [DOI] [PubMed] [Google Scholar]
- Carnal NW, Agostino A, Hatch MD. 1993. Photosynthesis in phosphoenolpyruvate carboxykinase-type C4 plants: mechanism and regulation of C4 acid decarboxylation in bundle sheath cells. Archives of Biochemistry and Biophysics 306, 360–367. [DOI] [PubMed] [Google Scholar]
- Christin PA, Osborne CP. 2014. The evolutionary ecology of C4 plants. New Phytologist 204, 765–781. [DOI] [PubMed] [Google Scholar]
- Dal’Molin CGD, Quek LE, Palfreyman RW, Brumbley SM, Nielsen LK. 2010. C4GEM, a genome-scale metabolic model to study C4 plant metabolism. Plant Physiology 154, 1871–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengler NG, Nelson T. 1999. Leaf structure and development in C4 plants. In: Sage RF, Monson RK, eds. C4 plant biology. San Diego, CA: Academic Press, 133–172. [Google Scholar]
- Edwards GE, Ku MSB. 1987. Biochemistry of C3-C4 intermediates. In: Stumpf PK, Conn EE, eds. The biochemistry of plants. San Diego, CA: Academic Press, 275–326. [Google Scholar]
- Evans JR, von Caemmerer S, Vogelmann TC. 2007. Balancing light capture with distributed metabolic demand during C4 photosynthesis. In: Sheehy JE, Mitchell PL, Hardy B, eds. Charting new pathways to C4 rice. Los Baños, Philippines: IRRI International Rice Research Institute. [Google Scholar]
- Farquhar GD, von Caemmerer S, Berry JA. 1980. A biochemical-model of photosynthetic CO2 assimilation in leaves of C3 species. Planta 149, 78–90. [DOI] [PubMed] [Google Scholar]
- Friso G, Majeran W, Huang MS, Sun Q, van Wijk KJ. 2010. Reconstruction of metabolic pathways, protein expression, and homeostasis machineries across maize bundle sheath and mesophyll chloroplasts: large-scale quantitative proteomics using the first maize genome assembly. Plant Physiology 152, 1219–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furbank R. 1992. Metabolic regulation and genetic engineering of sucrose and starch synthesis in C4 leaves. Improvement of yield in sugarcane through increased sucrose accumulation - workshop report. Bardon, Australia: CSIRO, 51. [Google Scholar]
- Furbank RT. 2011. Evolution of the C4 photosynthetic mechanism: are there really three C4 acid decarboxylation types?Journal of Experimental Botany 62, 3103–3108. [DOI] [PubMed] [Google Scholar]
- Furbank RT, Quick WP, Sirault XRR. 2015. Improving photosynthesis and yield potential in cereal crops by targeted genetic manipulation: prospects, progress and challenges. Field Crops Research 182, 19–29. [Google Scholar]
- Furbank RT, Stitt M, Foyer CH. 1985. Intercellular compartmentation of sucrose synthesis in leaves of Zea mays L. Planta 164, 172–178. [DOI] [PubMed] [Google Scholar]
- Furumoto T, Hata S, Izui K. 1999. cDNA cloning and characterization of maize phosphoenolpyruvate carboxykinase, a bundle sheath cell-specific enzyme. Plant Molecular Biology 41, 301–311. [DOI] [PubMed] [Google Scholar]
- Furumoto T, Hata S, Izui K. 2000. Isolation and characterization of cDNAs for differentially accumulated transcripts between mesophyll cells and bundle sheath strands of maize leaves. Plant and Cell Physiology 41, 1200–1209. [DOI] [PubMed] [Google Scholar]
- Gowik U, Westhoff P. 2011. The path from C3 to C4 photosynthesis. Plant Physiology 155, 56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez M, Gracen VE, Edwards GE. 1974. Biochemical and cytological relationships in C4 plants. Planta 119, 279–300. [DOI] [PubMed] [Google Scholar]
- Hatch MD. 1987. C4 photosynthesis - a unique blend of modified biochemistry, anatomy and ultrastructure. Biochimica et Biophysica Acta 895, 81–106. [Google Scholar]
- Huber SC, Edwards GE. 1975. Regulation of oxaloacetate, aspartate, and malate formation in mesophyll protoplast extracts of 3 types of C4 plants. Plant Physiology 56, 324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai R, Edwards GE. 1999. The biochemistry of C4 photosynthesis. In: Sage RF, Monson RK, eds. C4 plant biology. San Diego, CA: Academic Press, 49–87. [Google Scholar]
- Keerberg O, Pärnik T, Ivanova H, Bassüner B, Bauwe H. 2014. C2 photosynthesis generates about 3-fold elevated leaf CO2 levels in the C3–C4 intermediate species Flaveria pubescens. Journal of Experimental Botany 65, 3649–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koteyeva NK, Voznesenskaya EV, Edwards GE. 2015. An assessment of the capacity for phosphoenolpyruvate carboxykinase to contribute to C4 photosynthesis. Plant Science 235, 70–80. [DOI] [PubMed] [Google Scholar]
- Kotting O, Kossmann J, Zeeman SC, Lloyd JR. 2010. Regulation of starch metabolism: the age of enlightenment?Current Opinion in Plant Biology 13, 321–329. [DOI] [PubMed] [Google Scholar]
- Kramer DM, Evans JR. 2011. The importance of energy balance in improving photosynthetic productivity. Plant Physiology 155, 70–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kromdijk J, Ubierna N, Cousins AB, Griffiths H. 2014. Bundle-sheath leakiness in C4 photosynthesis: a careful balancing act between CO2 concentration and assimilation. Journal of Experimental Botany 65, 3443–3457. [DOI] [PubMed] [Google Scholar]
- Laisk A, Eichelmann H, Oja V. 2009. Leaf C3 photosynthesis in silico: integrated carbon/nitrogen metabolism. In: Laisk A, Nedbal L, Govindjee, eds. Photosynthesis in silico, Vol. 29 Dordrecht: Springer Netherlands, 295–322. [Google Scholar]
- Leegood RC, Walker AP. 1999. Regulation of the C4 pathway. In: Sage RF, Monson RK, eds. C4 plant biology. San Diego, CA: Academic Press, 89–121. [Google Scholar]
- Long SP, Marshall-Colon A, Zhu X-G. 2015. Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell 161, 56–66. [DOI] [PubMed] [Google Scholar]
- Lundgren MR, Christin P-A, Gonzalez Escobar E, Ripley BS, Besnard G, Long CM, Hattersley PW, Ellis RP, Leegood RC, Osborne CP. 2016. Evolutionary implications of C3-C4 intermediates in the grass Alloteropsis semialata. Plant, Cell & Environment DOI: 10.1111/pce.12665. [DOI] [PubMed] [Google Scholar]
- Lundgren MR, Osborne CP, Christin P-A. 2014. Deconstructing Kranz anatomy to understand C4 evolution. Journal of Experimental Botany 65, 3357–3369. [DOI] [PubMed] [Google Scholar]
- Lunn JE, Furbank RT. 1997. Localisation of sucrose-phosphate synthase and starch in leaves of C-4 plants. Planta 202, 106–111. [DOI] [PubMed] [Google Scholar]
- Lunn JE, Hatch MD. 1995. Primary partitioning and storage of photosynthate in sucrose and starch in leaves of C4 plants. Planta 197, 385–391. [Google Scholar]
- Majeran W, Cai Y, Sun Q, van Wijk KJ. 2005. Functional differentiation of bundle sheath and mesophyll maize chloroplasts determined by comparative proteomics. The Plant Cell 17, 3111–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W, Friso G, Ponnala L, et al. 2010. Structural and tetabolic transitions of C4 leaf development and differentiation defined by microscopy and quantitative proteomics in maize. The Plant Cell 22, 3509–3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W, van Wijk KJ. 2009. Cell-type-specific differentiation of chloroplasts in C4 plants. Trends in Plant Science 14, 100–109. [DOI] [PubMed] [Google Scholar]
- Mallmann J, Heckmann D, Brautigam A, Lercher MJ, Weber APM, Westhoff P, Gowik U. 2014. The role of photorespiration during the evolution of C4 photosynthesis in the genus Flaveria. Elife 3, 02478 DOI: 10.7554/eLife.02478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQualter RB, Bellasio C, Gebbie L, Petrasovits LA, Palfreyman R, Hodson M, Plan M, Blackman D, Brumbley S, Nielsen L. 2016. Systems biology and metabolic modelling unveils limitations to polyhydroxybutyrate accumulation in sugarcane leaves; lessons for C4 engineering. Plant Biotechnology Journal 14, 567–580. [DOI] [PubMed] [Google Scholar]
- Morandini P. 2013. Control limits for accumulation of plant metabolites: brute force is no substitute for understanding. Plant Biotechnology Journal 11, 253–267. [DOI] [PubMed] [Google Scholar]
- Ohsugi R, Huber SC. 1987. Light-modulation and localization of sucrose phosphate synthase activity between mesophyll cells and bundle sheath cells in C4 species. Plant Physiology 84, 1096–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen NA, Griffiths H. 2013. A system dynamics model integrating physiology and biochemical regulation predicts extent of crassulacean acid metabolism (CAM) phases. New Phytologist 200, 1116–1131. [DOI] [PubMed] [Google Scholar]
- Pfeffer M, Peisker M. 1998. CO2 gas exchange and phosphoenolpyruvate carboxylase activity in leaves of Zea mays L. Photosynthesis Research 58, 281–291. [Google Scholar]
- Pick TR, Brautigam A, Schluter U, et al. 2011. Systems analysis of a maize leaf developmental gradient redefines the current C4 model and provides candidates for regulation. The Plant Cell 23, 4208–4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rascio N, Colombo PM, Orsenigo M. 1980. The ultrastructural development of plastids in leaves of maize plants exposed to continuous illumination. Protoplasma 102, 131–139. [Google Scholar]
- Sage RF, Khoshravesh R, Sage TL. 2014. From proto-Kranz to C4 Kranz: building the bridge to C4 photosynthesis. Journal of Experimental Botany 65, 3341–3356. [DOI] [PubMed] [Google Scholar]
- Sage RF, Sage TL, Kocacinar F. 2012. Photorespiration and the evolution of C4 photosynthesis. Annual Review of Plant Biology 63, 19–47. [DOI] [PubMed] [Google Scholar]
- Saha R, Suthers PF, Maranas CD. 2011. Zea mays iRS1563: a comprehensive genome-scale metabolic reconstruction of maize metabolism. PloS One 6, e21784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze S, Mallmann J, Burscheidt J, Koczor M, Streubel M, Bauwe H, Gowik U, Westhoff P. 2013. Evolution of C4 photosynthesis in the genus Flaveria: establishment of a photorespiratory CO2 pump. The Plant Cell 25, 2522–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Pandey P, James D, Chandrasekhar K, Achary VMM, Kaul T, Tripathy BC, Reddy MK. 2014. Enhancing C3 photosynthesis: an outlook on feasible interventions for crop improvement. Plant Biotechnology Journal 12, 1217–1230. [DOI] [PubMed] [Google Scholar]
- Smith AM, Woolhouse HW. 1983. Metabolism of phosphoenolpyruvate in the C4 cycle during photosynthesis in the phosphoenolpyruvate-carboxykinase C4 grass Spartina Anglica Hubb. Planta 159, 570–578. [DOI] [PubMed] [Google Scholar]
- Spilatro SR, Preiss J. 1987. Regulation of starch synthesis in the bundle sheath and mesophyll of Zea mays L. Intercellular compartmentalization of enzymes of starch metabolism and the properties of the ADP glucose pyrophosphorylases. Plant Physiology 83, 621–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M, Huber S, Kerr P. 1987. Control of photosynthetic sucrose formation. In: Hatch MD, Boardman NK, eds. The biochemistry of plants: a comprehensive treatise (USA), Vol. 10 San Diego, CA: Academic Press, 327–409. [Google Scholar]
- Stitt M, Zhu X-G. 2014. The large pools of metabolites involved in intercellular metabolite shuttles in C4 photosynthesis provide enormous flexibility and robustness in a fluctuating light environment. Plant, Cell & Environment 37, 1985–1988. [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Ratcliffe RG. 2011. Flux-balance modeling of plant metabolism. Frontiers in Plant Science 2, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubierna N, Sun W, Kramer DM, Cousins AB. 2013. The efficiency of C4 photosynthesis under low light conditions in Zea mays, Miscanthus X giganteus and Flaveria bidentis. Plant, Cell & Environment 36, 365–381. [DOI] [PubMed] [Google Scholar]
- Ueno O, Sentoku N. 2006. Comparison of leaf structure and photosynthetic characteristics of C3 and C4 Alloteropsis semialata subspecies. Plant, Cell & Environment 29, 257–268. [DOI] [PubMed] [Google Scholar]
- von Caemmerer S. 1989. A model of photosynthetic CO2 assimilation and carbon-isotope discrimination in leaves of certain C3−C4 intermediates. Planta 178, 463–474. [DOI] [PubMed] [Google Scholar]
- von Caemmerer S. 2000. Biochemical models of leaf photosynthesis. Collingwood: CSIRO Publishing. [Google Scholar]
- von Caemmerer S. 2013. Steady-state models of photosynthesis. Plant, Cell & Environment 36, 1617–1630. [DOI] [PubMed] [Google Scholar]
- von Caemmerer S, Furbank RT. 1999. Modelling C4 photosynthesis. In: Sage RF, Monson RK, eds. The biology of C4 photosynthesis. London: Academic Press, 173–211. [Google Scholar]
- von Caemmerer S, Furbank RT. 2016. Strategies for improving C4 photosynthesis. Current Opinion in Plant Biology 31, 125–134. [DOI] [PubMed] [Google Scholar]
- Wang Y, Bräutigam A, Weber APM, Zhu X-G. 2014. a. Three distinct biochemical subtypes of C4 photosynthesis? A modelling analysis. Journal of Experimental Botany 65, 3567–3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Long SP, Zhu X-G. 2014. b. Elements required for an efficient NADP-malic enzyme type C4 photosynthesis. Plant Physiology 164, 2231–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise SE, van Wijk KJ, Sharkey TD. 2011. The role of transitory starch in C(3), CAM, and C(4) metabolism and opportunities for engineering leaf starch accumulation. Journal of Experimental Botany 62, 3109–3118. [DOI] [PubMed] [Google Scholar]
- Wingler A, Walker RP, Chen ZH, Leegood RC. 1999. Phosphoenolpyruvate carboxykinase is involved in the decarboxylation of aspartate in the bundle sheath of maize. Plant Physiology 120, 539–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman SC, Smith SM, Smith AM. 2007. The diurnal metabolism of leaf starch. Biochemical Journal 401, 13–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.