Abstract
The uneven anatomic distribution of cell subsets that harbor human immunodeficiency virus (HIV) during antiretroviral therapy (ART) complicates investigation of the barriers to HIV cure. Here we propose that while previous studies done largely in blood cells have led to important investigations into HIV latency, other important mechanisms of HIV persistence during ART may not be readily apparent in the bloodstream. We specifically consider as an example the question of ongoing HIV replication during ART. We discuss how growing understanding of key anatomic sanctuaries for the virus can inform future experiments aimed at further clarifying this issue.
Keywords: HIV, lymph nodes, T follicular helper cells, phylogenetics, antiretroviral therapy.
Defining the mechanisms by which human immunodeficiency virus (HIV) persists in vivo despite effective antiretroviral therapy (ART) has been a key focus of HIV cure research for nearly 2 decades. Within this field, the investigation of HIV latency in CD4 T cells has been central. Inferred initially from the detection of HIV proviruses in phenotypically resting blood cells [1] and subsequently from the stability of the circulating, latently infected CD4 T-cell pool defined by virus outgrowth assays (VOAs) [2–6], the prolonged quiescence of replication-competent proviruses within some resting memory CD4 T cells would be expected to prevent HIV eradication even without any cell-to-cell virus transmission under ART. This insight has prompted the development of “shock and kill” strategies [7, 8] that pair HIV latency reversal in vivo [9–14] with one or more means of augmenting infected cell clearance. At the same time, however, interest in alternative or complementary approaches to HIV cure persists. Although this interest is partly due to the difficulty of achieving effective and safe HIV latency reversal in vivo, it also remains possible that HIV persists during ART by multiple mechanisms. In particular, the infection of non-CD4 T-cell types [15–25], the prolonged persistence of infectious virions outside cells [26, 27], and ongoing cellular infections occurring after ART initiation [28] have all been proposed as potential barriers to cure. The incorporation of novel strategies that target such additional mechanisms could increase the chances of clinically meaningful results in future cure trials.
Results from the analysis of HIV infection in tissues further emphasize that the barrier to HIV cure may involve multiple mechanisms. As recently reviewed by Yukl and Wong [29], HIV is detectable in multiple extravascular sites during effective ART, including lymphoid and some nonlymphoid tissues. In certain cases, HIV nucleic acid or in vitro HIV replication has been demonstrated inside terminally differentiated myeloid or parenchymal cell types [15–25]. Such potential reservoirs might not be addressed adequately using latency reversing agents (LRAs) originally targeted to CD4 T cells. Moreover, even if the proviruses that rekindle high-level HIV replication upon ART interruption ultimately prove to be restricted to CD4 T cells, the heterogeneity among these functionally diverse, widely distributed cells still creates major challenges in HIV cure research. These challenges are in part pharmacokinetic, as putative cure agents could fail to reach therapeutic levels in some compartments that contain infected cells. More fundamentally, however, the diversity of CD4 T-cell subsets with distinct genetic programs and anatomic localizations complicates interpretation of basic investigations into how HIV persists in vivo. Although studies in circulating cells led to the identification of the HIV latent reservoir and have since allowed LRAs to be evaluated, circulating cells represent only a tiny fraction of all CD4 T cells in the body at any point in time [30, 31]. Studies performed using these cells may not adequately reflect critical events occurring outside the bloodstream.
In this perspective, we argue that fully understanding the barrier to HIV cure requires detailed consideration of such events and elucidation of the mechanisms that underlie them. To illustrate this, we will specifically examine whether HIV continues replicating during “fully suppressive” ART. This long-debated issue remains controversial in large part because new cellular infections cannot be visualized directly, either in HIV-infected people or in nonhuman primate models. At the same time, indirect experimental approaches to resolving the issue have produced variable results. The analysis of HIV sequence evolution has been especially controversial, with several publications reporting ongoing evolution during ART [28, 32] and others reaching the opposite conclusion [33–38]. Here we propose modifications to the hypothesis underlying this work that we believe can help guide future investigations. We specifically consider mounting evidence on the contributions of lymph node T follicular helper (TFH) cells to HIV replication and persistence in vivo, and on the relationship of HIV within TFH cells to viruses in the bloodstream. We highlight the difficulty of revealing these relationships without a detailed analysis that distinguishes among functional and anatomic subsets of CD4 T cells. We use our own recent findings in individuals with natural control of HIV to help make this point, and conclude by proposing additional experiments that may clarify the role of ongoing HIV replication in lymphoid tissues during ART.
That HIV continually replicates in the secondary lymphoid tissue during untreated, progressive infection is broadly accepted. Supporting evidence includes early studies that showed a higher HIV burden in lymph node biopsy samples than in blood. The quantification of HIV DNA demonstrated higher proportions of infected cells in lymphoid tissue than in blood [39], and the detection of cells containing HIV transcripts suggested virus expression by infected cells in lymphoid tissue [40]. At the same time, abundant extracellular virion RNA has been detected in lymph node during high-level viremia, particularly in association with the follicular dendritic cell (FDC) network [40, 41]; virus proteins and virions are also detectable [41, 42]. More than simply reflecting active virus production by infected cells, these findings have been interpreted to reflect ongoing cycles of virus replication. Although proving ongoing cycles of replication can be difficult, the conditions in lymph nodes, and particularly within B-cell follicles, appear highly favorable for cell-to-cell transmission of the virus. Infectious HIV virions appear to be retained within immune complexes on the surface of FDCs [41, 43]. In close contact with this source of infectious virus are abundant activated CD4 T cells. Furthermore, in vitro studies indicate that FDCs may release soluble factors that promote infection in CD4 T cells [44]. For these and other reasons, lymph nodes appear to be a key site for the high-level ongoing replication that characterizes even the early asymptomatic phase of untreated HIV infection.
In contrast to the findings made during untreated infection, those made under effective ART have led to conflicting conclusions about ongoing virus replication. Multiple direct measures of virus burden in lymphoid tissue decline sharply during the initial months of ART [45–47]. In particular, levels of virion RNA on the FDC network were found to decline >1000-fold over the initial 24 weeks of ART in early studies [45]. Numbers of mononuclear cells containing intracellular virion RNA also decline quickly over this same interval, suggesting a marked reduction in cells actively expressing the virus [45]. Nevertheless, ART does not completely eliminate HIV nucleic acids from lymph nodes. This is true of virion RNA on the FDC network, which remains detectable at very low levels by in situ hybridization even after an initial sharp decline [45]. This is also true of infected cells, as measured by cell-associated HIV DNA [47], and in particular of infected cells expressing the virus genes, as measured by intracellular HIV RNA [45, 48]. Although the persistence of cells expressing HIV genes is consistent with intermittent reactivation of latent cellular infections established before ART initiation, pharmacokinetic analysis has also revealed that lymph node levels of some commonly used antiretrovirals are surprisingly low [49]. In combination with the transient increases in certain nonintegrated forms of cell-associated HIV DNA that have been demonstrated in some ART intensification studies [50, 51], these findings leave open the possibility that new cellular infections occur in lymphoid tissue even when viremia is fully suppressed by ART.
To help resolve the uncertainty surrounding this issue, multiple groups have sought sequence evidence of ongoing virus evolution during effective ART. Either the emergence of drug-resistant virus variants or the progressive divergence of virus sequences from ancestral sequences during ART would represent strong indirect evidence of ongoing replication. Because obtaining sequential lymph node biopsies from a single individual is challenging, however, these studies have typically been done in blood. Using single-genome amplification and sequencing (SGS) protocols to obtain panels of single-copy sequences, virus populations in blood cell–associated DNA or plasma virions have been examined for either the emergence of drug resistance or progressive divergence over time from ancestral sequences. With notable exceptions [28, 32], most such studies performed in blood have found no evidence of HIV evolution during ART [33–38]. An important part of the interpretation of these studies is the premise that new virus sequences created through ongoing replication in lymphoid tissue should eventually disseminate systemically. In support of this, Kearney and colleagues performed a thorough comparison of panels of single-copy virus sequences from blood and multiple tissues in macaques receiving suppressive ART regimens in the setting of infection with an SIV containing an HIV-derived reverse-transcriptase gene (RT-SHIV) [52]. Analyzing between 3 and 42 single-copy virus DNA sequences per tissue, the investigators found that sequences were well mixed both among tissues and between tissues and the bloodstream in each animal. These findings suggest that sequences of the predominant pool of circulating, cell-associated viruses may be an accurate surrogate for sequences inside lymphoid tissue cells. This would in turn suggest that the lack of virus evolution in blood during ART may indicate a similar lack of evolution—and thus of ongoing replication—in lymphoid tissue.
Nevertheless, consideration of the emerging body of work on TFH cells in HIV infection raises questions about this conclusion. Cells of the TFH subset home to lymphoid follicles via the chemokine receptor CXCR5, where they provide help for germinal center B cells to facilitate immunoglobulin affinity maturation [53]. Due perhaps in part to a relative exclusion of virus-specific CD8 T cells from lymphoid follicles [54], the retention of infectious virus on FDCs [27], and the activated state of cells inside follicles, TFH cells carry a particularly high burden of HIV in the absence of ART [55]. Interestingly, they also appear to harbor replication-competent HIV more frequently than do non-TFH cells in the setting of effective ART, with levels that decline over time during virus suppression [56]. Several interpretations of this finding remain possible, including either an accumulation of latent HIV proviruses in TFH cells during ART or intersubset differences in the response to stimulation during VOAs that lead to more reliable recovery of replicating HIV from TFH cells than from other cell subsets in vitro. However, an additional potential interpretation of the relative abundance of replication-competent proviruses within the TFH subset during ART is that they reflect low levels of ongoing replication that is restricted to follicles.
If true, how would the latter interpretation be reconciled with the limited evidence for ongoing HIV replication during ART, as measured by tests for sequence evolution in blood cells and virions? One key consideration is the issue of sampling depth. The TFH subset represents a small proportion of all CD4 T cells in lymph node during ART—generally <10% of all memory cells, which make up a fraction of the total CD4 T-cell pool [56]. Because of this, any viruses in cell-associated DNA that were recently acquired within follicles would be expected to be intermingled with large numbers of the archival and defective proviruses that accumulate in ART-treated individuals (see model in Figure 1). This may be particularly true in the bloodstream: Although the precise surface phenotype of “memory TFH” cells remains unknown, it appears likely that CD4 T cells migrating from lymph node follicles to the systemic circulation are relatively rare [57]. Therefore, without sequencing either very large numbers of blood cell–associated viruses or viruses in narrowly defined subsets enriched for recent follicular emigrants, the virus genetic signatures of recently infected cells that would provide evidence of ongoing replication during ART might easily be missed.
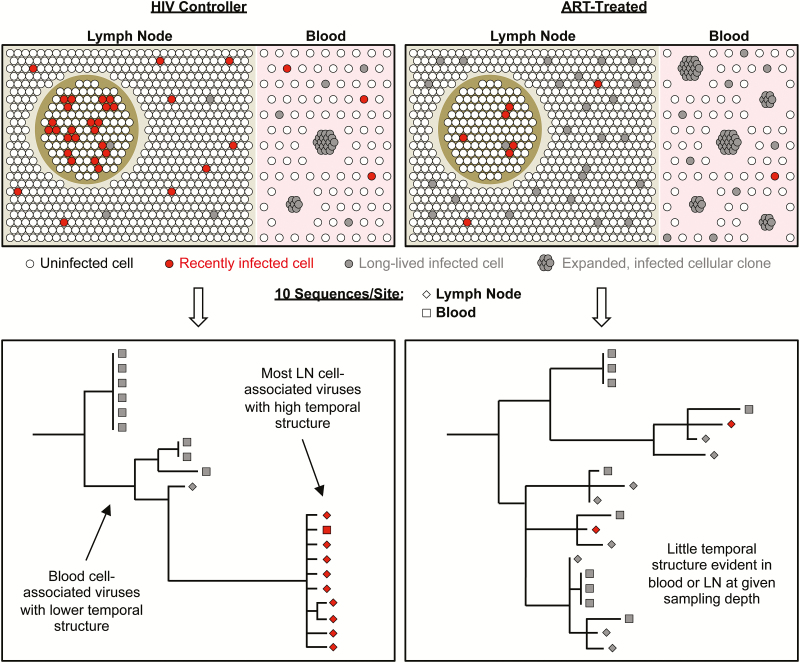
Figure 1.
Proposed model of anatomic and virus genetic relationships among distinct subsets of human immunodeficiency virus (HIV)–infected CD4 T cells during antiretroviral therapy (ART) that could account for the lack of observed evolution of HIV sequences over time in ART-treated individuals despite ongoing replenishment of the infected cell pool by new infections. For comparison, a similar model is proposed for HIV controllers based on our previous findings [58]. In HIV controllers (left), CD4 T cells harboring transcriptionally active viruses are enriched within the T-follicular helper (TFH) subset (illustrated here as within a follicle). Sequences of these viruses are divergent from ancestral sequences and closely related to plasma virion RNA sequences, which were previously shown to evolve over time in HIV controllers [60, 61] and thus likely reflect an actively replicating virus pool. We propose that ongoing cell-to-cell HIV transmissions among TFH cells in HIV controllers are associated with dissemination of related viruses to extrafollicular lymph node (LN) sites, which contain most of the CD4 T cells in lymphoid tissue, and also with hematogenous dissemination of some cells containing closely related viruses. Single-copy sequencing of LN CD4 T cell-associated viruses in HIV controllers reveals genetic markers consistent with recent infection, including high divergence from ancestral and a strong temporal structure (as described in [65]). In the bloodstream in HIV controllers, where cell-to-cell HIV transmissions are likely to be uncommon or absent, most CD4 T-cell–associated HIV DNA sequences are archival. Such sequences are often associated with expanded cellular clones whose proliferation further dilutes the phylogenetic signal from rare, recently infected cells. In ART-treated individuals (right), antiretroviral drugs limit any cell-to-cell HIV transmission that might occur within follicles. At the same time, the numbers of cells harboring archival HIV DNA sequences in LN and blood are likely to be higher in ART-treated individuals than in HIV controllers. As a result, sequencing limited numbers of cell-associated HIV DNA sequences in total LN or blood cells almost exclusively reflects long-lived cells harboring archival viruses. These sequences would not be expected to evolve over time. In addition, new cellular infections within LN in ART-treated individuals may in part result from intermittent reactivations of archival viruses. As a result, HIV DNA sequences from recently infected cells might show neither a temporal structure nor evolution in serial samples.
Our recent study in individuals with natural control of HIV infection illustrates this argument [58]. These individuals, termed HIV controllers, represent a rare group whose antiviral immune responses likely limit HIV replication in vivo in the absence of ART. In previous sequencing studies in these individuals, it was noted that HIV DNA sequences inside blood cells were distinct from plasma virion RNA sequences [59]. Longitudinal sampling further showed evidence of ongoing evolution in plasma virus sequences [60, 61], but not in cell-associated HIV DNA sequences [60, 61]. Our study, which was performed in 14 individuals with plasma viral loads <1000 copies/mL in the absence of ART, made similar findings. By sequencing relatively large total numbers of viruses and by performing this analysis within sorted CD4 T-cell subsets rather than unfractionated cells, we also found that HIV sequences in circulating central, transitional, and effector memory CD4 T cells were ancestral to plasma viruses. Furthermore, HIV DNA sequences in blood cells from HIV controllers frequently included groups of matching single-copy sequences. We found that these groups of matching sequences were enriched in the most differentiated cell subset—the effector memory subset—and using integration site sequencing showed that they originated from expanded cellular clones. Some of these clones had expanded massively, accounting for hundreds of HIV DNA copies/mL of peripheral blood, and many showed lethal genetic defects. In all 3 memory CD4 T-cell subsets in blood from these individuals, cell-associated levels of HIV transcripts were low or undetectable. Overall, these findings suggested a predominance of nonreplicating proviruses in blood cells in HIV controllers, consistent with strong restrictions on ongoing virus replication in vivo.
By contrast, when we examined the lymphoid tissue in these individuals, we found a markedly distinct population of infected cells (see Figure 1). As was demonstrated in SIV-infected macaques with natural virologic control, infected cells in lymph node from HIV controllers harbored viruses that appeared to be actively replicating. We inferred this based on their closer genetic similarity to plasma HIV sequences than to most blood cell–associated HIV DNA sequences, and because they were associated with high levels of spliced and unspliced virus transcripts. As in the macaque studies, infected cells harboring these viruses were enriched in the TFH subset. Interestingly, by stimulating large numbers of circulating cells in vitro and then sequencing genomic RNA from virions released in culture, we found rare inducible viruses that were genetically similar both to viruses in lymph node cells and to plasma viruses and were also highly divergent from ancestral sequences. Thus, by excluding the noninducible proviruses in cell-associated DNA from blood cells, we uncovered the small minority of cells that appeared to have been recently infected. It is critical to emphasize that these cells, which we propose had recently trafficked from lymphoid tissue, represented on the order of 1% of all circulating cells containing HIV DNA. For this reason, standard SGS approaches in which tens of single-copy virus sequences are obtained would not be expected to detect these cells reliably.
Thus, one can propose the hypothesis that evidence of low-level, ongoing HIV replication may be more readily detected by either (1) very deep sampling of the total infected circulating cell pool, as has been reported [62], or (2) targeted analysis of small cell populations that are closely related to the extravascular foci where replication could occur. Such an approach could prove useful in the setting of ART. As in HIV controllers, blood cell–associated HIV DNA sequences from ART-treated individuals are likely enriched for nonreplicating proviruses [63, 64] that could obscure important, genetically distinct virus subpopulations. The task of uncovering these subpopulations may be quantitatively more difficult during ART, as cycles of ongoing replication may be fewer and infected cells harboring archival proviruses more abundant in ART-treated individuals than in HIV controllers. Furthermore, new cellular infections could replenish the infected CD4 T-cell pool intermittently rather than in sustained fashion, and could in some cases be initiated by reactivation of latently infected cells, thus failing to produce sequence evolution over time. Nevertheless, direct sampling of TFH cells from lymph node or closely related subsets in blood could yet reveal groups of temporally clustered virus sequences that would indicate recent infection. Pairing this approach with in vitro stimulation and virion RNA recovery to filter out large numbers of archival, defective proviruses may lead to increased sensitivity. Thus, the extent to which low levels of ongoing replication in lymph node contribute to the persistence of inducible viruses in the circulation during ART may be quantified. Such experiments could help understand the extent to which new antiretrovirals with better tissue penetration might advance emerging cure strategies.
Overall, these arguments illustrate the need to investigate the mechanisms of HIV persistence during ART with reference to mounting knowledge of the cellular and anatomic organization of the immune system. Due to the very large number of replicative cycles that occur in the course of HIV infection in any individual, even the infrequent occurrence of nonproductive cellular infections can lead to considerable accumulated evidence of replication-incompetent viruses. The challenge for cure approaches is to determine the ratios of defective—and thus likely harmless—proviruses to intact proviruses within different tissues. Layered upon this are the probability that distinct gene expression patterns among infected cells in these tissues confer different degrees of latency to intact cell-associated proviruses, and the apparent ability of some infected cells to change their phenotypes and genetic programs without expressing the viruses they harbor. Thus, the total body CD4 T-cell reservoir for HIV likely consists of multiple distinct reservoirs that may have different requirements for purging.
Notes
Financial support. D. C. D. and E. A. B. are funded by the National Institutes of Health (NIH) Intramural Research Program. D. C. D. is also funded by the National Institute of Allergy and Infectious Diseases Division of AIDS and the NIH Office of AIDS Research.
Supplement sponsorship. This supplement was supported by grants from Merck & Co., Inc. and Gilead Sciences, Inc..
Potential conflicts of interest. Both authors: No reported conflicts. Both authors have submitted the ICMJE Form for Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med 1995; 1:1284–90. [DOI] [PubMed] [Google Scholar]
- 2. Chun TW, Carruth L, Finzi D, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 1997; 387:183–8. [DOI] [PubMed] [Google Scholar]
- 3. Chun TW, Stuyver L, Mizell SB, et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A 1997; 94:13193–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Finzi D, Hermankova M, Pierson T, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 1997; 278:1295–300. [DOI] [PubMed] [Google Scholar]
- 5. Wong JK, Hezareh M, Günthard HF, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 1997; 278:1291–5. [DOI] [PubMed] [Google Scholar]
- 6. Finzi D, Blankson J, Siliciano JD, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 1999; 5:512–7. [DOI] [PubMed] [Google Scholar]
- 7. Deeks SG. HIV: Shock and kill. Nature 2012; 487:439–40. [DOI] [PubMed] [Google Scholar]
- 8. Lehrman G, Hogue IB, Palmer S, et al. Depletion of latent HIV-1 infection in vivo: a proof-of-concept study. Lancet 2005; 366:549–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Routy JP, Tremblay CL, Angel JB, et al. Valproic acid in association with highly active antiretroviral therapy for reducing systemic HIV-1 reservoirs: results from a multicentre randomized clinical study. HIV Med 2012; 13:291–6. [DOI] [PubMed] [Google Scholar]
- 10. Rasmussen TA, Tolstrup M, Brinkmann CR, et al. Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial. Lancet HIV 2014; 1:e13–21. [DOI] [PubMed] [Google Scholar]
- 11. Spivak AM, Andrade A, Eisele E, et al. A pilot study assessing the safety and latency-reversing activity of disulfiram in HIV-1-infected adults on antiretroviral therapy. Clin Infect Dis 2014; 58:883–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Søgaard OS, Graversen ME, Leth S, et al. The depsipeptide romidepsin reverses HIV-1 latency in vivo. PLoS Pathog 2015; 11:e1005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Archin NM, Liberty AL, Kashuba AD, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 2012; 487:482–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Elliott JH, McMahon JH, Chang CC, et al. Short-term administration of disulfiram for reversal of latent HIV infection: a phase 2 dose-escalation study. Lancet HIV 2015; 2:e520–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carter CC, McNamara LA, Onafuwa-Nuga A, et al. HIV-1 utilizes the CXCR4 chemokine receptor to infect multipotent hematopoietic stem and progenitor cells. Cell Host Microbe 2011; 9:223–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Christofinis G, Papadaki L, Sattentau Q, Ferns RB, Tedder R. HIV replicates in cultured human brain cells. AIDS 1987; 1:229–34. [PubMed] [Google Scholar]
- 17. Harouse JM, Kunsch C, Hartle HT, et al. CD4-independent infection of human neural cells by human immunodeficiency virus type 1. J Virol 1989; 63:2527–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marras D, Bruggeman LA, Gao F, et al. Replication and compartmentalization of HIV-1 in kidney epithelium of patients with HIV-associated nephropathy. Nat Med 2002; 8:522–6. [DOI] [PubMed] [Google Scholar]
- 19. Moses AV, Bloom FE, Pauza CD, Nelson JA. Human immunodeficiency virus infection of human brain capillary endothelial cells occurs via a CD4/galactosylceramide-independent mechanism. Proc Natl Acad Sci U S A 1993; 90:10474–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rytik PG, Eremin VF, Kvacheva ZB, et al. Susceptibility of primary human glial fibrillary acidic protein-positive brain cells to human immunodeficiency virus infection in vitro: anti-HIV activity of memantine. AIDS Res Hum Retroviruses 1991; 7:89–95. [DOI] [PubMed] [Google Scholar]
- 21. Sattentau QJ, Stevenson M. Macrophages and HIV-1: an unhealthy constellation. Cell Host Microbe 2016; 19:304–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Semenzato G, Agostini C, Ometto L, et al. CD8+ T lymphocytes in the lung of acquired immunodeficiency syndrome patients harbor human immunodeficiency virus type 1. Blood 1995; 85:2308–14. [PubMed] [Google Scholar]
- 23. Sundstrom JB, Ellis JE, Hair GA, et al. Human tissue mast cells are an inducible reservoir of persistent HIV infection. Blood 2007; 109:5293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tornatore C, Nath A, Amemiya K, Major EO. Persistent human immunodeficiency virus type 1 infection in human fetal glial cells reactivated by T-cell factor(s) or by the cytokines tumor necrosis factor alpha and interleukin-1 beta. J Virol 1991; 65:6094–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Watkins BA, Dorn HH, Kelly WB, et al. Specific tropism of HIV-1 for microglial cells in primary human brain cultures. Science 1990; 249:549–53. [DOI] [PubMed] [Google Scholar]
- 26. Smith BA, Gartner S, Liu Y, et al. Persistence of infectious HIV on follicular dendritic cells. J Immunol 2001; 166:690–6. [DOI] [PubMed] [Google Scholar]
- 27. Keele BF, Tazi L, Gartner S, et al. Characterization of the follicular dendritic cell reservoir of human immunodeficiency virus type 1. J Virol 2008; 82:5548–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lorenzo-Redondo R, Fryer HR, Bedford T, et al. Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature 2016; 530:51–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wong JK, Yukl SA. Tissue reservoirs of HIV. Curr Opin HIV AIDS 2016; 11:362–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Di Mascio M, Paik CH, Carrasquillo JA, et al. Noninvasive in vivo imaging of CD4 cells in simian-human immunodeficiency virus (SHIV)-infected nonhuman primates. Blood 2009; 114:328–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Westermann J, Pabst R. Distribution of lymphocyte subsets and natural killer cells in the human body. Clin Investig 1992; 70:539–44. [DOI] [PubMed] [Google Scholar]
- 32. Günthard HF, Frost SD, Leigh-Brown AJ, et al. Evolution of envelope sequences of human immunodeficiency virus type 1 in cellular reservoirs in the setting of potent antiviral therapy. J Virol 1999; 73:9404–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Josefsson L, von Stockenstrom S, Faria NR, et al. The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc Natl Acad Sci U S A 2013; 110:E4987–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kearney MF, Spindler J, Shao W, et al. Lack of detectable HIV-1 molecular evolution during suppressive antiretroviral therapy. PLoS Pathog 2014; 10: e1004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kieffer TL, Finucane MM, Nettles RE, et al. Genotypic analysis of HIV-1 drug resistance at the limit of detection: virus production without evolution in treated adults with undetectable HIV loads. J Infect Dis 2004; 189:1452–65. [DOI] [PubMed] [Google Scholar]
- 36. Mens H, Pedersen AG, Jørgensen LB, et al. Investigating signs of recent evolution in the pool of proviral HIV type 1 DNA during years of successful HAART. AIDS Res Hum Retroviruses 2007; 23:107–15. [DOI] [PubMed] [Google Scholar]
- 37. Persaud D, Siberry GK, Ahonkhai A, et al. Continued production of drug-sensitive human immunodeficiency virus type 1 in children on combination antiretroviral therapy who have undetectable viral loads. J Virol 2004; 78:968–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ruff CT, Ray SC, Kwon P, et al. Persistence of wild-type virus and lack of temporal structure in the latent reservoir for human immunodeficiency virus type 1 in pediatric patients with extensive antiretroviral exposure. J Virol 2002; 76:9481–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pantaleo G, Graziosi C, Butini L, et al. Lymphoid organs function as major reservoirs for human immunodeficiency virus. Proc Natl Acad Sci U S A 1991; 88:9838–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Haase AT, Henry K, Zupancic M, et al. Quantitative image analysis of HIV-1 infection in lymphoid tissue. Science 1996; 274:985–9. [DOI] [PubMed] [Google Scholar]
- 41. Schuurman HJ, Krone WJ, Broekhuizen R, Goudsmit J. Expression of RNA and antigens of human immunodeficiency virus type-1 (HIV-1) in lymph nodes from HIV-1 infected individuals. Am J Pathol 1988; 133:516–24. [PMC free article] [PubMed] [Google Scholar]
- 42. O’Hara CJ, Groopman JE, Federman M. The ultrastructural and immunohistochemical demonstration of viral particles in lymph nodes from human immunodeficiency virus-related and non-human immunodeficiency virus-related lymphadenopathy syndromes. Hum Pathol 1988; 19:545–9. [DOI] [PubMed] [Google Scholar]
- 43. Joling P, Bakker LJ, Van Strijp JA, et al. Binding of human immunodeficiency virus type-1 to follicular dendritic cells in vitro is complement dependent. J Immunol 1993; 150:1065–73. [PubMed] [Google Scholar]
- 44. Thacker TC, Zhou X, Estes JD, et al. Follicular dendritic cells and human immunodeficiency virus type 1 transcription in CD4+ T cells. J Virol 2009; 83:150–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cavert W, Notermans DW, Staskus K, et al. Kinetics of response in lymphoid tissues to antiretroviral therapy of HIV-1 infection. Science 1997; 276:960–4. [DOI] [PubMed] [Google Scholar]
- 46. Tamalet C, Lafeuillade A, Fantini J, Poggi C, Yahi N. Quantification of HIV-1 viral load in lymphoid and blood cells: assessment during four-drug combination therapy. AIDS 1997; 11:895–901. [DOI] [PubMed] [Google Scholar]
- 47. Wong JK, Günthard HF, Havlir DV, et al. Reduction of HIV-1 in blood and lymph nodes following potent antiretroviral therapy and the virologic correlates of treatment failure. Proc Natl Acad Sci U S A 1997; 94:12574–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lafeuillade A, Chollet L, Hittinger G, Profizi N, Costes O, Poggi C. Residual human immunodeficiency virus type 1 RNA in lymphoid tissue of patients with sustained plasma RNA of <200 copies/mL. J Infect Dis 1998; 177:235–8. [DOI] [PubMed] [Google Scholar]
- 49. Fletcher CV, Staskus K, Wietgrefe SW, et al. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc Natl Acad Sci U S A 2014; 111:2307–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Buzón MJ, Massanella M, Llibre JM, et al. HIV-1 replication and immune dynamics are affected by raltegravir intensification of HAART-suppressed subjects. Nat Med 2010; 16:460–5. [DOI] [PubMed] [Google Scholar]
- 51. Hatano H, Strain MC, Scherzer R, et al. Increase in 2-long terminal repeat circles and decrease in D-dimer after raltegravir intensification in patients with treated HIV infection: a randomized, placebo-controlled trial. J Infect Dis 2013; 208:1436–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kearney MF, Anderson EM, Coomer C, et al. Well-mixed plasma and tissue viral populations in RT-SHIV-infected macaques implies a lack of viral replication in the tissues during antiretroviral therapy. Retrovirology 2015; 12:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Crotty S. Follicular helper CD4 T cells (TFH). Annu Rev Immunol 2011; 29:621–63. [DOI] [PubMed] [Google Scholar]
- 54. Connick E, Mattila T, Folkvord JM, et al. CTL fail to accumulate at sites of HIV-1 replication in lymphoid tissue. J Immunol 2007; 178:6975–83. [DOI] [PubMed] [Google Scholar]
- 55. Perreau M, Savoye AL, De Crignis E, et al. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med 2013; 210:143–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Banga R, Procopio FA, Noto A, et al. PD-1(+) and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nat Med 2016; 22:754–61. [DOI] [PubMed] [Google Scholar]
- 57. Ueno H. Human circulating T follicular helper cell subsets in health and disease. J Clin Immunol 2016; 36suppl 1:34–9. [DOI] [PubMed] [Google Scholar]
- 58. Boritz EA, Darko S, Swaszek L, et al. Multiple origins of virus persistence during natural control of HIV infection. Cell 2016; 166:1004–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bailey JR, Williams TM, Siliciano RF, Blankson JN. Maintenance of viral suppression in HIV-1-infected HLA-B*57+ elite suppressors despite CTL escape mutations. J Exp Med 2006; 203:1357–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mens H, Kearney M, Wiegand A, et al. HIV-1 continues to replicate and evolve in patients with natural control of HIV infection. J Virol 2010; 84:12971–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. O’Connell KA, Brennan TP, Bailey JR, Ray SC, Siliciano RF, Blankson JN. Control of HIV-1 in elite suppressors despite ongoing replication and evolution in plasma virus. J Virol 2010; 84:7018–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Buzón MJ, Codoñer FM, Frost SD, et al. Deep molecular characterization of HIV-1 dynamics under suppressive HAART. PLoS Pathog 2011; 7:e1002314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bruner KM, Murray AJ, Pollack RA, et al. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med 2016; 22:1043–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ho YC, Shan L, Hosmane NN, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 2013; 155:540–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nickle DC, Jensen MA, Shriner D, et al. Evolutionary indicators of human immunodeficiency virus type 1 reservoirs and compartments. J Virol 2003; 77:5540–6. [DOI] [PMC free article] [PubMed] [Google Scholar]