OsPIN2 is identified as the casual gene responsible for the phenotype of lta1, a rice mutant that displays large root angles and a shallow root system architecture, affecting the polar transport of auxin in the root tip.
Keywords: Auxin, gravitropism, OsPIN2, rice, root growth angle, root system architecture
Abstract
Root system architecture is very important for plant growth and crop yield. It is essential for nutrient and water uptake, anchoring, and mechanical support. Root growth angle (RGA) is a vital constituent of root system architecture and is used as a parameter for variety evaluation in plant breeding. However, little is known about the underlying molecular mechanisms that determine root growth angle in rice (Oryza sativa). In this study, a rice mutant large root angle1 (lra1) was isolated and shown to exhibit a large RGA and reduced sensitivity to gravity. Genome resequencing and complementation assays identified OsPIN2 as the gene responsible for the mutant phenotypes. OsPIN2 was mainly expressed in roots and the base of shoots, and showed polar localization in the plasma membrane of root epidermal and cortex cells. OsPIN2 was shown to play an important role in mediating root gravitropic responses in rice and was essential for plants to produce normal RGAs. Taken together, our findings suggest that OsPIN2 plays an important role in root gravitropic responses and determining the root system architecture in rice by affecting polar auxin transport in the root tip.
Introduction
Roots play a central role in plant growth and development through providing anchorage and taking up nutrients and water from the soil. The root system architecture in the soil determines the scope of resources available to plants and responds to environmental conditions, thus governing the growth and final yields for crop plants (Rogers and Benfey, 2015). Therefore, ideal root system architectures that optimize water and nutrient uptake have been intensively studied in crop breeding and production in recent years. Root growth angle (RGA) is an important parameter of root system architecture in the soil. Large RGAs (shallow root growth) are now being deployed as targets in crop breeding programs for improving nutrient uptake efficiency in stressful soil environments (Lynch, 2013). It has been reported that shallow root systems can promote phosphate uptake from the topsoil in wheat (Manske et al., 2000) and in common bean (Liao et al., 2004; Lynch, 2011). In addition, shallow roots play a vital role in the avoidance of the hypoxic environments and promote the growth of rice (Mano et al., 2005).
Gravitropic response is an important factor that affects RGA (Morita and Tasaka, 2004). Root gravitropism describes the orientation of root growth along the gravity vector. It has been suggested that in Arabidopsis this response requires the coordinated, asymmetric distribution of auxin within the root tip, and depends on the concerted activities of PIN (PIN-FORMED) proteins, AUX1 (AUXIN-INSENSITIVE1), and other members of the auxin transport pathway (Abas et al., 2006). In Arabidopsis, genetic and biochemical studies have revealed the essential roles of the members of PIN, AUX1/LAX (AUXIN-INSENSITIVE1/LIKE AUX1), and ABCB (B subfamily of ABC transporters) families in mediating polar auxin transport and root gravitropism (Geisler et al., 2014). AUX1 is expressed in root apical tissues, and regulates root gravitropism by facilitating auxin transport in Arabidopsis (Marchant et al., 1999). AXR4 is an accessory protein of the endoplasmic reticulum (ER) and regulates AUX1 localization, thus affecting root gravitropism (Hobbie and Estelle, 1995; Dharmasiri et al., 2006).
There are eight PIN proteins in Arabidopsis (Grunewald and Friml, 2010), among which PIN1 is involved in the basipetal movement of auxin and is crucial for shoot vascular development and shoot gravitropic responses (Gälweiler et al., 1998; Vernoux et al., 2000; Friml, 2003). PIN2 and PIN3 are essential for the root gravitropic response. PIN2 polar localizes towards the shoot in the lateral root cap and root epidermis cells, and towards the root in the root cortex cells. Roots of pin2 mutant seedlings are agravitropic. In pin2 mutants, shootward auxin distribution in the lower side of the root is largely repressed during gravity stimulus, thus resulting in an agravitropic phenotype (Chen et al., 1998; Luschnig et al., 1998; Müller et al., 1998; Utsuno et al., 1998). PIN3 localizes in the lower side of the columella cells, and functions in redirecting auxin fluxes to trigger asymmetric growth (Grunewald and Friml, 2010). pin3 seedlings display decreased root gravitropic responses and inhibited hypocotyl and root growth (Friml et al., 2002; Harrison and Masson, 2008; Keuskamp et al., 2010). While PIN4 and PIN7 have partially overlapping functions with other PINs in the root tip, their involvement in root gravitropism is not clear (Grunewald and Friml, 2010). In addition, the B subclass ABC transporters, such as ABCB1 (PGP1), ABCB19 (PGP19/MDR1), and ABCB4 (PGP4), have been shown to affect root gravitropism since their loss-of-function mutants display significantly reduced gravitropic responses (Noh et al., 2001; Santelia et al., 2005; Terasaka et al., 2005; Bouchard et al., 2006; Bailly et al., 2008)
Rice is a model cereal plant that possesses a fibrous root system, which is mainly composed of crown roots emerging post-embryonically from the nodes of the stem (Coudert et al., 2010). The crown root growth angle is an important component for the distribution of rice roots in soil. Recently, several quantitative trait loci (QTLs) have been identified as contributing to the regulation of RGA in rice (Uga et al., 2013a, 2013b; Kitomi et al., 2015). One of these, DEEPER ROOTING 1 (DRO1), is negatively regulated by auxin and is also involved in cell elongation in the root tip (Uga et al., 2013a). Uga et al. (2013b, 2015) have also characterized two other major QTLs controlling RGA: one is located on chromosome 4, and the other, DRO3, is on the long arm of chromosome 7 and might be involved in the DRO1 genetic pathway. Although many genes involved in root gravitropic responses have now been identified in Arabidopsis, only a few have been identified in rice. The molecular mechanisms underlying the regulation of RGA and gravitropic responses in rice remain almost unknown.
In this study, a rice root mutant displaying larger root angles and an agravitropic response was isolated, and was named lra1 (large root angle1) according to the phenotype. Molecular cloning and complementation analysis revealed that a point mutation in OsPIN2 resulted in a premature stop codon and the lra1 phenotype. Further molecular, genetic, and physiological analyses demonstrated that OsPIN2 plays an important role in rice root gravitropism and in determining the RGA via effects on the polar auxin transport in the root tip.
Materials and methods
Plant material and growth conditions
An ethyl-methanesulfonate (EMS)-mutagenized M2 library in the background of Oryza sativa L. cv Hei-Jing2 (HJ2), a Japonica rice variety, was used for screening mutants with altered root structure. lra1, a mutant with a large root growth angle, was isolated and named according to the phenotype. Rice plants were grown in solution culture (Yoshida et al., 1976) with FeCl3 replaced by NaFe(Ⅲ)-EDTA. The pH of the culture solution was adjusted to 5.5 before use and the solution was replaced every 3 d. The solution-cultured mutants and the HJ2 wild-type (WT) were grown in a greenhouse with a 12-h light (30 ℃) / 12-h dark (22 ℃) photoperiod, at approx. 200 μmol m−2 s−1 photon density, and 60% relative humidity. For an agar-gel medium test, rice seeds were sterilized using 75% ethanol for 2 min and 30% bleach for 30 min with gentle shaking, and then washed for 6∼7 times with sterile double-distilled water. Seeds were dried for 3 min, then laid in half Murashige and Skoog (MS) solid medium in tissue-culture flasks (Thermo Fisher Scientific), and then grown vertically for 7 d before taking photographs.
For imaging with X-ray micro-computed tomography (X-ray μCT), plants were grown in polyvinyl chloride columns (80 mm diameter × 180 mm height) containing sieved (<2 mm) sandy clay loam soil (sand 60%, silt 17%, and clay 23%; pH 7.1; organic matter 5%) (‘Sterilised Kettering Loam’, Broughton Ltd., Kettering, UK). The soil was uniformly packed to a bulk density of 1.2 Mg m−3 and saturated overnight from the bottom upwards with deionized water before planting with pre-germinated rice seeds (3 d after germination with emergence of the coleoptile and radicle). Growth conditions were the same as those in the solution culture detailed above.
Gene cloning and complementation tests
To clone the causal gene, the lra1 mutant was backcrossed with the HJ2 wild-type. F2 progeny plants showing large root growth angles were selected for genetic analysis and gene cloning using the MutMap method (Abe et al., 2012). DNA was extracted from 40 F2 individuals and mixed in an equal ratio. Genomic resequencing, single-nucleotide polymorphism (SNP) and InDel analysis, and mutation identification were conducted as previously described (Yang et al., 2016). To confirm that the casual gene OsPIN2 was mutated in the mutant, the coding sequence and genomic DNA of OsPIN2 were amplified (primers OsPIN2-F, OsPIN2-R) from the mutant and HJ2 for sequencing analysis. A cleaved amplified polymorphic sequence (CAPS) marker [primers CAPS-F and CAPS-R (Tsp45I)] was also developed to confirm the mutation of the lra1 mutant. For complementation tests, the sequence of OsPIN2 (6245 bp), including the genomic sequence of the OsPIN2 coding domain (3691 bp) and its native promoter (2554 bp), were PCR-amplified from HJ2 DNA using the primer pairs OsPIN2-infusion-F1 (EcoRI) and OsPIN2-infusion-R1 (HindⅢ) and inserted into the binary vector pCambia1300 between EcoRI and HindⅢ using an Infusion kit (Takara Clontech, Japan). The binary vector was transformed into lra1 mutants by Agrobacterium-mediated transformation (Chen et al., 2003). All the primer sequences are listed in Supplementary Table S1 at JXB online.
β-Glucuronidase (GUS) histochemical analysis
To identify the tissue-specific expression of OsPIN2, pOsPIN2: GUS transgenic plants were generated. The promoter of OsPIN2 was amplified using a pair of specific primers: proOsPIN2-F (which contains the Sal I restriction site) and proOsPIN2-R (which contains the BamHI restriction site). The promoter region was cut using SalI and BamHI and cloned into the vector pBI101.3GUSplus (http://genome-www.stanford.edu/vectordb/vector_descrip/COMPLETE/PBI1013.SEQ.html). The resultant pOsPIN2:GUS construct was transformed into the WT by Agrobacterium-mediated transformation. Histochemical GUS analysis was performed as previously described (Liu et al., 2005). The primers are listed in Supplementary Table S1.
Antibody preparation, immunostaining, and sub-cellular localization
The OsPIN2 antibodies were prepared and purified as described previously (Li et al., 2014)
For immunostaining, the roots of 1-week-old seedlings of WT and lra1 were used as described previously (Murata et al., 2006).
The pOsPIN2:OsPIN2-eGFP expression vector was constructed as described previously by Wu et al. (2015), and the 35S:OsCHL1-mCherry expression vector was constructed as described previously by Lv et al. (2014). The resulting constructs were sequenced to verify the in-frame fusion, and then used for transient transformation in rice protoplasts (Miao and Jiang, 2007). Transformed protoplasts were examined with a confocal microscope (Zeiss LSM 710).
RNA extraction, reverse transcription, and quantitative RT-PCR
Total RNAs were isolated using an RNA extraction kit (NucleoSpin RNA Plant; Macherey-Nagel, Germany). Reverse transcription, RT-PCR, and quantitative RT-PCR were performed as described by Chen et al. (2013). The primers used for RT-PCR and qRT-PCR are listed in Supplementary Table S1.
Measurements of IAA concentration and distribution
For measurement of indole-3-acetic acid (IAA) concentrations, 20 mg samples of roots of 7-d-old WT and lra1 mutants were washed several times with deionized water, and ground into fine powder under liquid nitrogen. The measurement of IAA was performed as described previously (Wang et al., 2014). Rice auxin-inducible reporter DR5-GFP lines were obtained by transforming the DR5-GFP reporter gene into both the WT and the lra1 mutant. The DR5-GFP vector was constructed as described previously (Qi et al., 2012). The auxin distribution revealed by GFP fluorescence in WT and lra1 roots was visualized using a confocal microscope (Zeiss LSM 710).
Root gravistimulation and measurement
Root gravistimulation was performed as previously described (Paciorek et al., 2005). For gravitropic stimulation, 5-d-old vertically grown seedlings were gravistimulated with 90° rotation. Digital images were collected for at least 20 seedlings for each time point and analyzed using the Image J software (https://imagej.nih.gov/ij/).
Identifying the function of OsPIN2 in Arabidopsis
To understand whether OsPIN2 functions similarly to AtPIN2, a construct of pAtPIN2:OsPIN2 was made as follows: the AtPIN2 promoter (2836 bp) was amplified by PCR using the primers proAtPIN2-infusion-F3 (containing the EcoRI restriction site) and proAtPIN2-infusion-R3 (containing the KpnI restriction site), and then inserted into the pCAMIBA1300 vector using In-Fusion™ PCR Cloning Kits (Takara Clontech, Japan). Then the cDNA (2119 bp) of OsPIN2 was amplified by PCR using the primers OsPIN2-cDNA-F (containing the KpnI restriction site) and OsPIN2-cDNA-R (containing the KpnI restriction site) and inserted into the pCAMIBA1300 vector containing the promoter of AtPIN2 at the KpnI site using In-Fusion™ PCR Cloning Kits (Takara Clontech, Japan). The resulting pAtPIN2:OsPIN2 construct was transformed to Atpin2 mutants to get the pAtPIN2:OsPIN2/Atpin2 transgenic lines using the floral dip method (Zhang et al., 2006). All the primers are listed in Supplementary Table S1.
3D reconstruction of root structure based on X-ray µCT scanning
X-ray μCT scans were performed at The Hounsfield Facility (School of Biosciences, The University of Nottingham, UK) using a phoenix v|tome|x m 240kV X-ray CT system (GE Sensing & Inspection Technologies GmbH, Wunstorf, Germany). Two scans were required per plant to obtain the full column height (which were digitally combined following data reconstruction). Each scan acquired 2160 projection images over a 360° rotation of the sample using a detector exposure time of 250 ms, integrated over three averaged images, resulting in a total scan time of 75 min for both scans. Full details of the X-ray CT scanner settings are given in Supplementary Table S2. Following the scans, data were reconstructed using the datos|REC software (GE Sensing & Inspection Technologies GmbH, Wunstorf, Germany). Visualization and quantification of root material in the reconstructed data were preformed using the method of Tracy et al. (2012), employing a combination of the VGStudioMAX v2.2 (Volume Graphics GmbH, Heidelberg, Germany) and RooTrak software (Mairhofer et al., 2012). For the measurement of root angles, a bespoke software tool called RooTh was used (Mairhofer, et al., 2017).
Results
Isolation and phenotypic characterization of the lra1 mutant
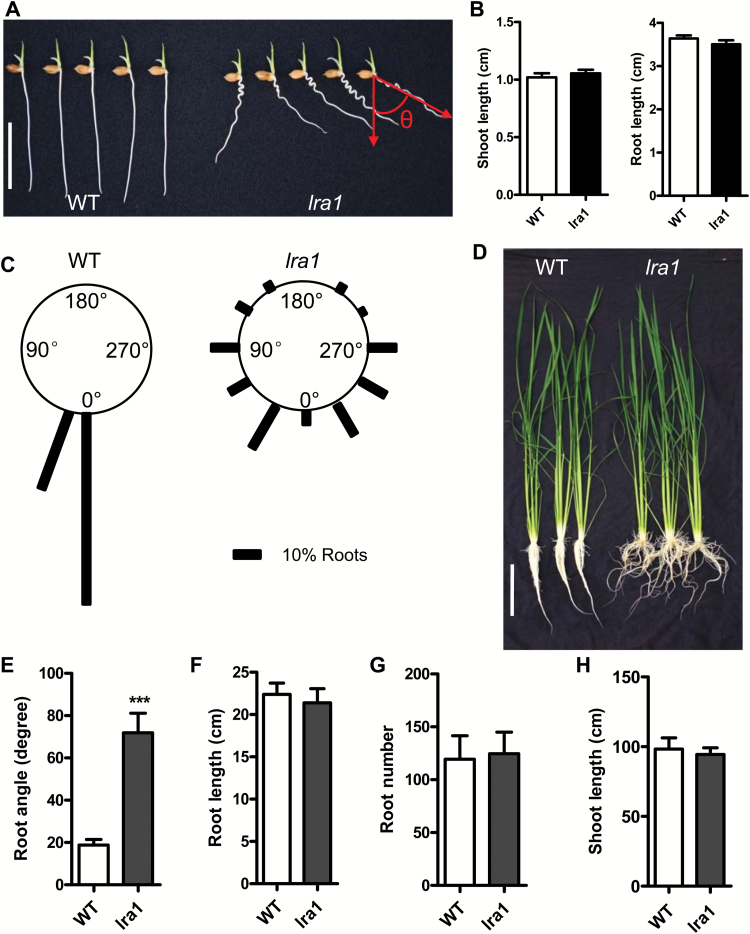
To investigate the molecular mechanisms governing the root system architecture in rice, we obtained a rice root mutant lra1 in a screening of an EMS library. Compared with the WT, 3-d-old lra1 mutant seedlings displayed an agravitropic phenotype with slightly curved roots (Fig. 1A). However, the root and shoot lengths of the seedlings showed no significant differences between lra1 and the WT (Fig. 1B). Root angles of WT seedlings were mainly between 0–20°, but those of lra1 seedlings varied from 0 to 360° (Fig. 1C).
Fig 1.
Phenotype of the lra1 mutant and the wild-type (WT). (A) 3-d-old WT and lra1 seedlings grown in solution culture. Scale bar =2 cm. θ indicates the root angle. (B) Shoot and root lengths of 3-d-old WT and lra1 seedlings as shown in (A). Data are means ±SE (n=20). (C) The root gravitropic response of 3-d-old WT and lra1 (n=100). (D) Phenotype of 4-week-old WT and lra1 seedlings grown in solution culture. Scale bar=15 cm. (E–H) Root angle (E), root length (F), root number (G), and shoot length (H) of 4-week-old WT and lra1 seedlings grown in solution culture. Data are means ±SD (n=10). Asterisks indicate significance differences between WT and lra1 plants as determined by Student’s t-test (***, P<0.005).
The primary root length, shoot length, root number, lateral root number, longest lateral root length, and the total lateral root length were not significantly affected in the lra1 mutant compared with the WT after 7 d in solution culture (Supplementary Fig. S1), but lra1 showed larger root growth angle (Supplementary Fig. S2A) and a reduced gravitropic response (Fig. S2B). After 4 weeks, the average root angle of lra1 mutants (71.9°) was much larger than that of the WT (18.75°) (Fig. 1E, Supplementary Fig. S2C). These results indicated that both the primary root and the later-emerging adventitious roots in lra1 showed reduced gravitropic responses (Supplementary Fig. S2A–C). There were no significant differences in shoot length, root length, and root number in 4-week-old seedlings grown in solution culture between lra1 and WT (Fig.1F–H).
At the reproductive stage in solution culture, the lra1 mutant showed no significant differences in agronomic traits such as plant height, tiller number, and seed-setting compared with the WT (Supplementary Table S3). However, the plant height of lra1 was significant lower than that of the WT, although the other measured traits showed no difference with the WT in soil culture (Supplementary Fig. S3 and Supplementary Table S4).
Cloning and identification of LRA1
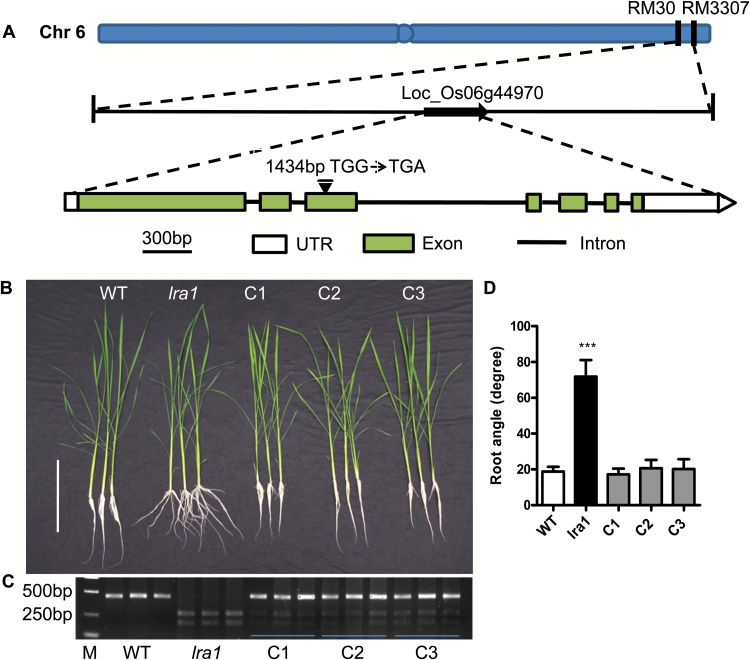
To identify the casual gene of the lra1 mutant, we backcrossed the mutant to the WT. All the plants of the F1 progeny showed the small root angle of the WT. In the F2 progeny, 172 and 59 plants showed the WT and mutant phenotypes, respectively, which fits a ratio of about 3:1 (χ2=9.9, P<0.01). This result indicates that the lra1 mutant is caused by a single recessive gene. After genome resequencing of the bulked mutant-like plants (n=39) and WT plants (n=30) in the F2 progenies, a single nucleotide mutation (G1434A) was found in the third exon of LOC_Os06g44970, which generated a premature stop codon (Fig. 2A). The point mutation in the lra1 mutant was confirmed using a CAPS marker (Fig. 2C). LOC_Os06g44970 is annotated as an auxin efflux transporter OsPIN2 that contains nine transmembrane domains. The mutation in the lra1 mutant would produce a truncated OsPIN2 that has lost the last four transmembrane segments, as predicted by InterPro (http://www.ebi.ac.uk/interpro/search/sequence-search;Supplementary Fig. S4).
Fig 2.
Gene cloning and complementation analysis. (A) Chromosome location and gene structure of OsPIN2. The G-to-A mutation on the third exon is indicated. (B–D) Complementation analysis. (B) Phenotype of 30-d-old seedlings of the wild-type (cv HJ2), lra1 mutant, and three complementation (OsPIN2p:OsPIN2/lra1) lines (C1–C3) grown in hydroponics. Scale bar =20 cm. (C) Molecular characterization of the complementation lines by CAPS maker (primers are listed in Supplementary Table S1, PCR products were digested with Tsp45I). M, molecular marker (DL2000, Takara). (D) The root angle of seedlings shown in (C). Data are means ±SD of ten replicates. Asterisks indicate significance differences as determined by Student’s t-test (***, P<0.005).
In order to confirm the fact that the agravitropic phenotype of lra1 was caused by the point mutation of OsPIN2, the genomic sequence of OsPIN2 driven by its native promoter was transformed to the lra1 mutants. A series of transgenic lines were obtained that showed the same small root angles as the WT. Three representative lines were molecularly characterized using a CAPS marker and showed the bands of lra1 and band of the WT, indicating their transgenes in the lra1 background (Fig. 2). Phenotypic analysis was conducted using the three lines. The root angles of the complementary transgenic lines were comparable to those of WT plants in different growth media (Fig. 2B, D; Supplementary Fig. S2A–C), and the expression levels of OsPIN2 in the complementary lines were comparable with those in the WT (Supplementary Fig. S2D). The results suggest that the phenotypic defect of lra1 is caused by the mutation of OsPIN2.
Expression pattern of OsPIN2
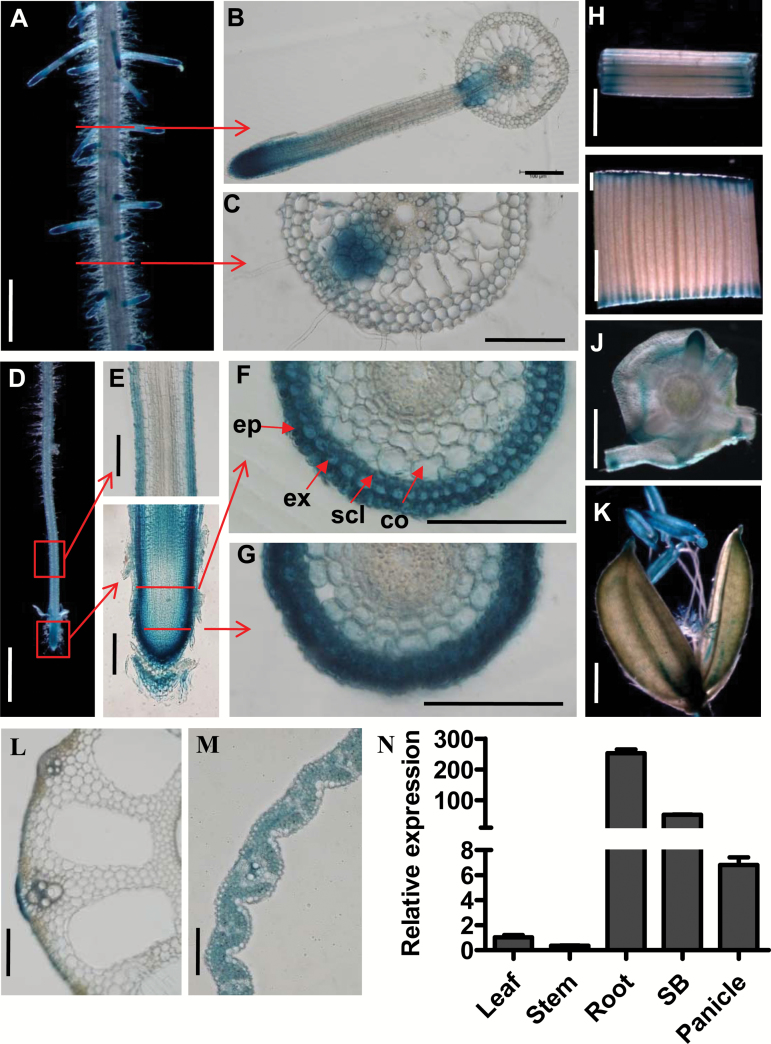
The tissue-specific expression pattern of OsPIN2 was analysed by using transgenic lines in which the GUS reporter gene was driven by the OsPIN2 promoter in WT plants. GUS staining of the representative transgenic lines revealed that OsPIN2 was expressed in lateral root tips (Fig. 3A, B), primary root tips (Fig. 3D), stems (Fig. 3H), leaves (Fig. 3I), the stem base (Fig. 3J), and flowers (Fig. 3K). Cross-sections of different parts of a primary root showed that OsPIN2 was expressed in lateral root primordia and lateral root initiation zones (Fig. 3B, C). Longitudinal and cross-sections of primary root tips showed that OsPIN2 was mainly expressed in the epidermis, exodermis, and sclerenchyma cells of the root meristematic and elongation zones (Fig. 3E–G). Cross-sections of stems and leaves demonstrated that OsPIN2 was also expressed in the epidermal tissues of the stem (Fig. 3L) and in leaf mesophyll cells (Fig. 3M). The results were in agreement with the expression patterns of OsPIN2 that have been described previously (Wang et al., 2009; Chen et al., 2012).
Fig 3.
Tissue-specific expression patterns of OsPIN2. (A–M) GUS staining of transgenic plants harboring the OsPIN2p:GUS reporter gene. (A) Mature root region with lateral roots, (B) cross-section of the region where a lateral root emerges, (C) cross-section of the region where a lateral root primordium is initiated, (D) primary root tip, (E) longitudinal section of primary root tip (lower panel) and the elongation zone (upper panel), (F) cross-section of the elongation zone, (G) cross-sections of a root cap, (H) stem, (I) leaf, (J) stem base, and (K) panicle. Abbreviations: ep, epidermis; ex, exodermis; scl, sclerenchyma; co, cortex. Scale bars indicate 1 mm in (A, D, H–K), 100 µm in (B, C, E–G, M), 200 µm in (L). (N) Quantitative RT-PCR analysis of OsPIN2 mRNA levels in the leaf, stem, root, stem base (SB), and panicle. The primers used are listed in Supplementary Table S1.
Quantitative RT-PCR analysis showed that the expression levels of OsPIN2 were much higher in roots and the stem base (SB) than in leaves, stems, and panicles (Fig. 3N), implying that OsPIN2 plays a role in root development in rice.
Cellular and subcellular localization of OsPIN2
The cellular localization of the OsPIN2 protein was examined by immunostaining using an anti-OsPIN2 polyclonal antibody. In WT plants, signals were observed on the plasma membranes of epidermal and cortex cells in the root tip, and polar localization could be seen on the upper side in epidermal cells and on the lower side in cortex cells (Fig. 4A). In general, the signals in epidermal cells were much stronger than those in cortex cells (Fig. 4A). In contrast, no signals were detected in lra1 roots. These observations confirm that the OsPIN2 protein is truncated in lra1 mutants.
Fig 4.
Tissue-specificity and subcellular localization of OsPIN2. (A) Tissue-specificity in the root tip. Immunostaining was performed with an anti-PIN2 antibody in root tips of the wild-type (WT) and lra1 (longitudinal sections). The boxed areas are magnified in the images to the right. Abbreviations: ep, epidermis; ex, exodermis; scl, sclerenchyma; co, cortex. Arrows indicate polar localization of OsPIN2. Scale bars =20 µm. (B) Subcellular localization of OsPIN2. The OsPIN2-GFP fusion protein driven by the OsPIN2 promoter and CHL1-mCherry (cell membrane marker) driven by the 35S promoter were transiently co-expressed in rice protoplasts. Fluorescent signals as observed by confocal microscopy are shown. Scale bars =5 µm.
The subcellular localization of OsPIN2 was further examined by transient expression in rice protoplasts. The OsPIN2 protein was co-localized with CHL1, a plasma-membrane marker (Lv et al., 2014) (Fig. 4B), indicating that OsPIN2 was indeed localized in the plasma membrane.
OsPIN2 rescues the phenotypic defect of the pin2 mutant in Arabidopsis
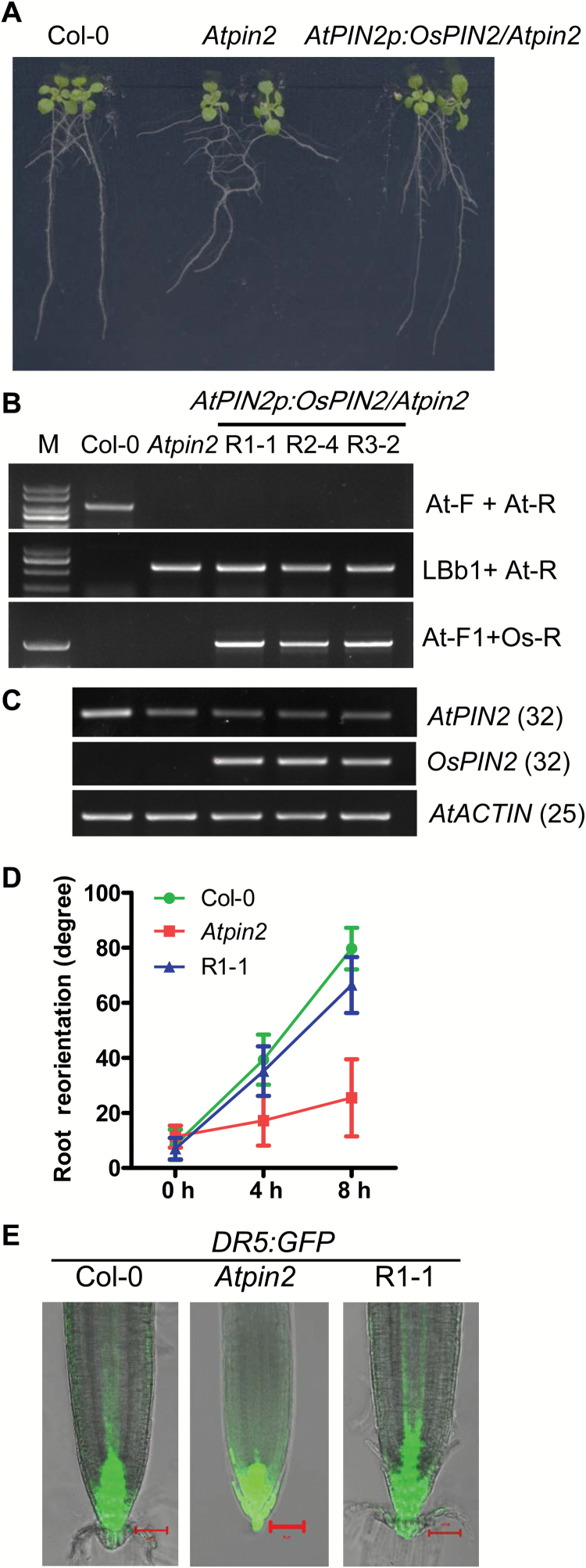
To test whether OsPIN2 functions like AtPIN2 as an auxin efflux carrier (Müller et al., 1998), the OsPIN2 full-length cDNA driven by the AtPIN2 promoter was transformed into the Arabidopsis pin2 mutant (Atpin2). The transgenic lines, which were verified by PCR (Fig. 5B, C), showed normal root growth just like the wild-type plants (Fig. 5A). Gravitropic root reorientation was examined to test whether the response of the transgenic seedlings was rescued. The kinetics of root reorientation of the WT, Atpin2, and the transgenic line (R1-1) were examined when roots were placed horizontally for up to 8 h. The transgenic line showed normal root reorientation similar to that of the WT, while the Atpin2 mutant displayed an agravitropic response (i.e. as seen in Fig. 5A). Furthermore, the root reorientation of R1-1 was comparable to that of the WT, while Atpin2 did not respond to gravistimulation (Fig. 5D). These results indicated that the gravitropic response of Atpin2 was recovered to the level of the WT by expressing OsPIN2 (Fig. 5A). To test whether the auxin distribution in the root tips of R1-1 was rescued, lines of DR5-GFP/Atpin2 and DR5-GFP/R1-1 were constructed. In contrast to Atpin2, the auxin distribution as revealed by the GFP signal in the R1-1 line was the same as that in the WT, and was mostly located in the columella, stele, and epidermis (Fig. 5E). This indicates that OsPIN2 can rescue the defective auxin distribution in Atpin2. These results suggest that OsPIN2 plays a similar role to Arabidopsis PIN2 as an auxin efflux carrier that regulates the root gravitropic response in rice.
Fig 5.
Expression of OsPIN2 rescues the phenotype of the Arabidopsis pin2 mutant. (A) The phenotypes of 10-d-old Col-0, Atpin2, and Atpin2p:OsPIN2/Atpin2 transgenic lines. (B) Molecular characterization of Col-0, Atpin2, and three independent Atpin2p:OsPIN2/Atpin2 transgenic lines using PCR. Primers are listed in Supplementary Table S1. (C) Expression levels of AtPIN2 and OsPIN2 in the transgenic lines using RT-PCR. (D) Kinetics of root reorientation of Col-0, Atpin2, and Atpin2p:OsPIN2/Atpin2 transgenic lines (R1-1): 4-day-old seedlings were placed horizontally and the root angle was measured at time points as indicated. Data are means ±SD (n=20). (E) Fluorescence of DR5:GFP in Col-0, Atpin2, and Atpin2p:OsPIN2/Atpin2 line R1-1; scale bars =50 μm.
The distribution of auxin is altered in root tips of lra1 mutants
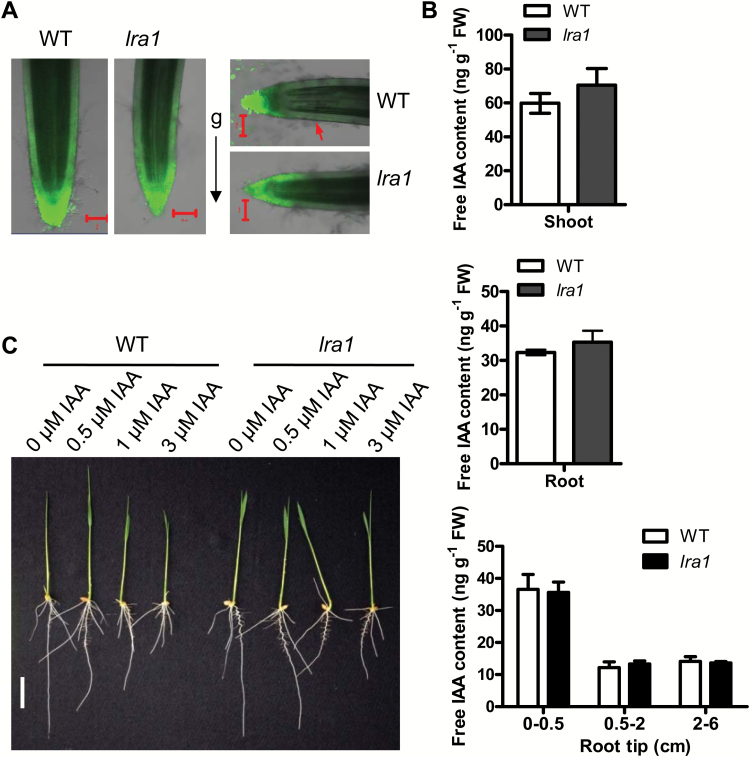
To test whether the distribution of auxin in root tips was different between WT and lra1 seedlings, the DR5-GFP reporter line was developed and crossed with the lra1 mutant. As revealed by GFP fluorescence, the auxin signal in root-cap columella cells of the lra1 mutant was less than that in the WT; no significant differences were observed in the other tissues in the root tip (Fig. 6A). After being placed horizontally for 30 min, GFP fluorescence was higher on the lower side of the root cap in the WT, but not in the lra1 mutant (Fig. 6A). This indicates that OsPIN2 plays an important role in auxin polar distribution in the root tips.
Fig 6.
Mutation of OsPIN2 affects the distribution of auxin in rice root tips. (A) Auxin distribution as revealed by green fluorescence in the roots of 3-d-old DR5:GFP transgenic wild-type (WT) and lra1 plants. The roots shown in the panels to the right were placed horizontally for 30 min before imaging (g indicates direction of gravity). Images are representative of three independent lines. (B) Endogenous free IAA concentrations in different tissues of 7-d-old WT and lra1 mutant plants. Data are means ±SE of three replicates. (C) Phenotypes of WT and lra1 seedlings treated with different concentrations of IAA, as indicated.
The concentrations of endogenous free IAA in the WT and lra1 seedlings were further quantified. As shown in Fig. 6B, there were no differences in concentrations at the whole-shoot and whole-root level between the WT and lra1. Further analysis showed that there were also no differences when individual sections of the roots were examined at various differences from the root tip.
To examine whether the external IAA would affect the phenotype, different concentrations of IAA were applied to WT and lra1 plants. The results showed that lra1 plants were less sensitive to external IAA compared with WT plants, especially at a concentration of 0.5 μM IAA (Fig. 6C, Supplementary Fig S5).
OsPIN2 plays an important role in the root system architecture
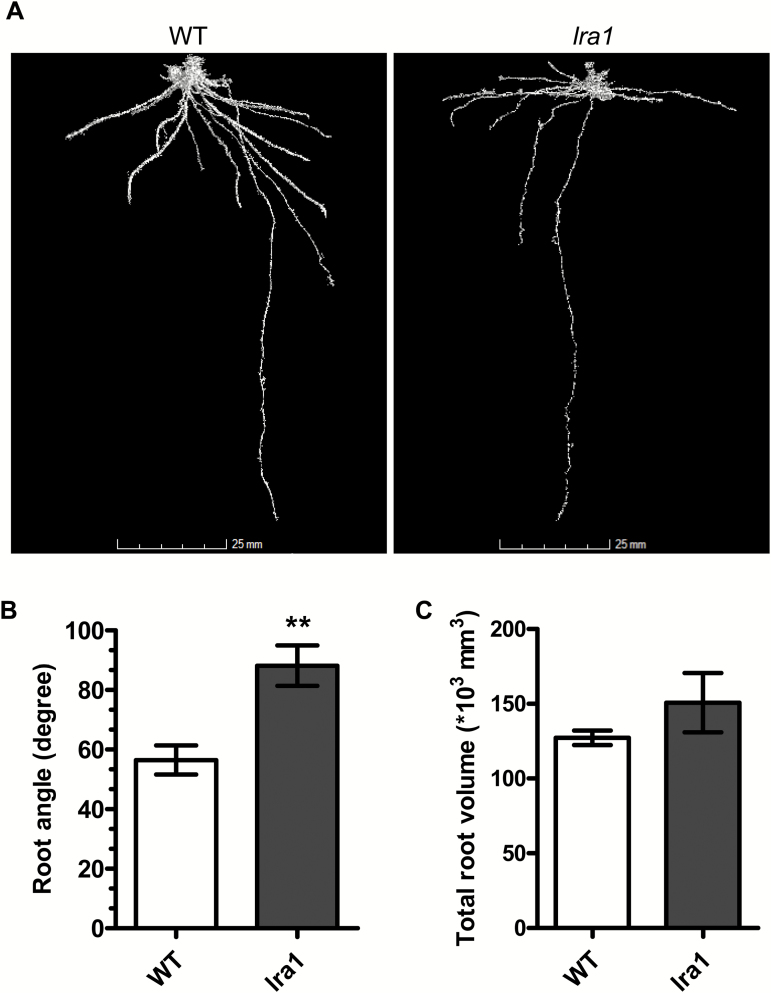
X-ray computed tomography (μCT) is a non-destructive imaging method that permits 3D reconstruction of scanned objects (Tracy et al., 2010). To determine whether lra1 affected the root architecture in of plants grown in soil, 3D root systems of lra1 and WT plants were reconstructed using μCT imaging (representative images are shown in Fig. 7A, and additional images from different angles are also presented in Supplementary Fig. S6). The images revealed that roots of lra1 plants had a larger root angle (88.1°) and were distributed more in the upper surface layer of the soil compared to the WT plants (56.4° and deeper root growth) (Fig. 7A, B), which is consistent with what was observed in solution culture and in MS medium (Supplementary Fig. S2). The total root volume showed no significant difference between the WT and lra1 (Fig 7C). These results suggest that OsPIN2 plays an important role in root system architecture by affecting the root growth angle.
Fig 7.
Root system architecture of wild-type (WT) and lra1 plants as revealed by X-ray computed tomography (CT). (A) Root system architecture of the WT (cv HJ2) and lra1 grown for 21 d in 8-cm diameter pots filled with sterilized Kettering loam. Roots were segmented by a region-growing algorithm. (B) Root angles of the WT and lra1 measured using the software RooTh (provided by University of Nottingham). Asterisks indicate significance differences as determined by Student’s t-test (**P<0.01). (C) Total root volume of the WT and lra1 measured using the software Rootrak_0.3.2 (provided by University of Nottingham).
SNPs within OsPIN2 among different rice varieties were analysed using RiceVarMap (http://ricevarmap.ncpgr.cn/). The results showed that there are 21 SNPs within OsPIN2 among 799 Indica varieties and 497 Japonica varieties. Of the 21 SNPs, four were synonymous, 15 were localized in the intron or in the 3′ untranscribed region (3′UTR) (Supplementary Table S5), and two non-synonymous SNPs (sf0627201163 and sf0627201316) showed differentiation between Indica and Japonica. sf0627201163 displayed A in 98.25% of Indica rice and G in 99.2% of Japonica, while sf0627201316 displayed G in 98.12% of Indica rice and T in 99.2% of Japonica (Supplementary Table S5).
Discussion
Root gravitropism is a complex process during which plant roots grow downwards into the soil. The gravitropic response process contains four steps: sensing the direction of gravity; conversion of a biophysical signal to a biochemical one; transmission of the signal to the responding tissues; and organ bending (Morita and Tasaka, 2004; Philosoph-Hadas et al., 2005). Although the genes involved in root gravitropic responses in Arabidopsis have been well studied, how the response is regulated in rice is still largely unknown. Using forward genetic analysis, this study has identified LRA1 in rice, a gene encoding the auxin efflux transporter OsPIN2, which is required for the root gravitropic response. A single-nucleotide mutation (G1434A) in lra1 produces a truncated OsPIN2 protein without the last four transmembrane segments (Supplementary Fig. S4). The lra1 mutant showed an agravitropic root phenotype with large root growth angles, which could be rescued by the OsPIN2 genomic sequence (Fig. 2B; Supplementary Fig. S2A–D). After roots were placed horizontally, GFP fluorescence in DR5:GFP reporter lines was higher in the lower side of the root tip in the WT, but not in the lra1 mutant (Fig. 6A). Expression of OsPIN2 driven by the promoter of AtPIN2 was able to fully rescue the phenotypic defect in the Atpin2 mutant (Fig. 5). These results suggest that OsPIN2 plays an important role in root gravitropic responses and in the root growth angle in rice.
OsPIN2 affects auxin polar distribution in the root tip
Polar auxin transport and redistribution are essential for root gravitropism (Abas et al., 2006). In Arabidopsis, PIN2 (also named AGR1/EIR1/WAV6) localizes towards the shoot in the lateral root cap and root epidermis cells, and towards the root in the root cortex cells, and is known to be an auxin efflux carrier that facilitates basipetal transport (Chen et al., 1998; Luschnig et al., 1998; Müller et al., 1998; Utsuno et al., 1998). Although PIN2 has been shown to mediate basipetal transport of auxin and gravitropic root bending in Arabidopsis (Feraru and Friml, 2008), it is mostly unknown whether PIN2 orthologs play similar roles in other plants.
Our results showed that OsPIN2 localizes on the plasma membrane of epidermal and cortex cells in the root tip (Fig. 4A). After reorientation of roots to change the direction of the gravity stimulation, auxin redistribution was not changed in the lra1 mutant, as determined by DR5:GFP reporter lines (Fig. 6A). Furthermore, lra1 was less sensitive to external auxin (IAA) treatment compared with WT plants (Fig. 6C, Supplementary Fig. S5). These results suggest that, like PIN2 in Arabidopsis, OsPIN2 plays an important role in polar auxin distribution in root tips.
There are 12 PIN family members in the rice genome (Wang et al., 2009). To investigate whether loss of function of PIN2 affected the expression of other PIN genes, the expression of these genes (OsPIN1a, OsPIN1b, OsPIN1c, OsPIN3a, OsPIN3b, OsPIN4, OsPIN5a, OsPIN5b, OsPIN9, OsPIN10a, and OsPIN10b) were evaluated in lra1 by qRT-PCR. The results indicated that the transcript levels of OsPIN3b and OsPIN10b in shoots were significantly up-regulated in lra1 compared with the WT. In roots, OsPIN3a showed a notable up-regulation in lra1 compared with that in the WT (Supplementary Fig. S7). Phylogenetic analysis has suggested that OsPIN3a and OsPIN3b are much more closely related to AtPIN3 compared with other rice PINs (Miyashita et al., 2010). AtPIN3, mainly located in the columella cell boundaries, is essential for the root gravitropic response (Miyashita et al., 2010). This suggests that loss of function of OsPIN2 up-regulates the expression of OsPIN3, which to some extent compensates for the loss of OsPIN2 function. This is consistent with reports in Arabidopsis that different PINs are ectopically expressed in pin mutants and thus can at least partially compensate for the function of the missing PIN protein (Blilou et al., 2005). It is interesting that the transcript levels of the OsPIN2 were greatly decreased in the lra1 mutant both in the shoot and root compared with the WT (Supplementary Fig. S7). Whether functional PIN2 is required for the stability of its transcript level in rice or the G-to-A replacement causes instability of the PIN2 mRNA remains to be investigated.
PIN2 shows conserved function in root gravitropic responses between monocots and dicots
Phylogenetic analysis indicates that OsPIN2 is the closest rice homolog of Arabidopsis PIN2. The rice PIN2 mutant lra1 showed an agravitropic root phenotype with no significant auxin redistribution to the lower root epidermis cells after roots were reorientated to the horizontal, as determined by DR5:GFP (Fig. 6A). These results are consistent with those found in the Arabidopsis pin2 mutant. AtPIN2 is an auxin efflux transporter (Chen et al., 1998; Luschnig et al., 1998). To understand whether OsPIN2 functions the same as AtPIN2, we transformed a construct of OsPIN2 driven by the promoter of AtPIN2 to the Atpin2 mutant. The transgenic lines showed normal growth and root gravitropic responses just like WT plants (Fig. 5A). The DR5:GFP reporter lines also indicated that the defect of auxin distribution in the root tip was also completely rescued by the expression of OsPIN2 (Fig. 5E). These results suggest that the function of OsPIN2 is conserved in root gravitropic responses between monocots and dicots. They also suggest that OsPIN2 might be an auxin efflux transporter in rice. However, differences exist between rice and Arabidopsis. In rice, the fluorescence in the DR5:GFP lines was weaker in the root cap columella cells of the lra1 mutant than in the WT (Fig. 6A). Although the IAA content in the root tip showed no significant differences between lra1 and the WT (Fig. 6B), this could be because our sampling region encompassed 0.5 cm of the root tip, which not only contained the root cap but also the meristematic region. On the other hand, whether the DR5:GFP lines can indicate the auxin content in rice still needs to be verified. In Arabidopsis, DR5:GFP lines showed comparable GFP florescence in columella cells between the WT and pin2 (Fig. 5E). Furthermore, it has been reported that the Atpin2 mutant responds normally to externally applied auxin (Luschnig et al., 1998), while the lra1 mutant showed less sensitivity to external auxin, especially at a concentration of 0.5 μM IAA (Fig. 6C, Supplementary Fig S5). The results suggest that OsPIN2 and AtPIN2 may affect auxin distribution in different ways, and this needs to be investigated further.
OsPIN2 is a potential candidate for improving root structure
Optimization of root system architecture is an important objective for modern plant breeding, and hence a better understanding of the molecular mechanisms controlling root architecture is essential to improve plant root systems to enhance nutrient uptake efficiency and crop yield. It has been suggested that root systems with large root angles enhance topsoil foraging and that this benefits the acquisition of phosphate in the upper soil layers (Lynch, 2013). Rice is usually grown in lowland conditions and, in most cases, fields are flooded. Fertilizer use efficiency is a very important parameter for sustainable rice production, and the aim is to reduce the amount of fertilizer applied while keeping yields high. As most nutrients are applied in the surface of the soil, a shallow root system should benefit uptake (Pothuluri et al., 1986; Murphy et al., 1998; Lynch and Brown, 2001; Zhu et al., 2005). It has also been reported that shallow root systems play an important role in the avoidance of hypoxic environments and promote the growth of rice (Mano et al., 2005). lra1 showed root growth similar to that observed in shallow-rooted varieties, with normal shoot growth irrespective of whether it was cultured in solution or in soil (Figs 1 and 7), which indicates that OsPIN2 plays an important role in root structure. In addition, among the 21 SNPs that were found in different rice varieties, two non-synonymous ones showed different allele priorities between Indica and Japonica varieties. The sf0627201163 SNP showed A in 98.25% of Indica rice and G in 99.2% Japonica, while sf0627201316 showed G in 98.12% of Indica rice and T in 99.2% of Japonica (Supplementary Table S5). This suggests that these two SNPs could be useful for breeding purposes. Overall, our results suggest that OsPIN2 is a potential candidate gene for improving root system architecture in rice, and possibly other crops.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. The phenotype of the lra1 mutant.
Fig. S2. The root phenotype of the wild-type, lra1 mutant, and complementation lines in different growth media.
Fig. S3. The phenotype of the wild-type and lra1 mutant grown in soil pots.
Fig. S4. Predicted transmembrane topology models of the OsPIN2 protein in the wild-type and lra1 mutant.
Fig. S5. Phenotypic data for wild-type and lra1 seedlings treated with different concentrations of IAA.
Fig. S6. 3D visualization of rice roots grown in soil and viewed from different angles at 21 d after germination.
Fig. S7. qRT-PCR analysis of the expression of PIN family genes in shoots and roots of the wild-type and lra1 mutant.
Table. S1. Primers used in the study.
Table. S2. Sample properties and scanning settings for X-ray micro-computed tomography.
Table. S3. Agronomic traits of the wild-type and lra1 grown in solution culture.
Table. S4. Agronomic traits of the wild-type and lra1 grown in soil pots.
Table. S5. SNPs within OsPIN2 in different rice varieties.
Author contributions
MC, WL, PJ, and LC planned and designed the research; WL, GM. and LY performed the experiments; RW, MX, and WZ analysed the data; WL, MC, YH, CJS, and LC wrote the manuscript.
Acknowledgements
We thank J.W. Pan for kindly providing the Arabidopsis pin2 mutant. This work was supported by the Natural Science Foundation of Zhejiang Province, China (LZ17C020001), the National Key Research and Development Program of China (2016YFD0100700), the National Basic Research and Development Program of China (2015CB942900), BBSRC International Partnering Award (BB/J020443/1), the National Natural Science Foundation of China (31572187,31372120), and the Ministry of Education and Bureau of Foreign Experts of China (B14027).
References
- Abas L, Benjamins R, Malenica N, Paciorek T, Wiśniewska J, Wirniewska J, Moulinier-Anzola JC, Sieberer T, Friml J, Luschnig C. 2006. Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nature Cell Biology 8, 249–256. [DOI] [PubMed] [Google Scholar]
- Abe A, Kosugi S, Yoshida K, et al. . 2012. Genome sequencing reveals agronomically important loci in rice using MutMap. Nature Biotechnology 30, 174–178. [DOI] [PubMed] [Google Scholar]
- Bailly A, Sovero V, Vincenzetti V, Santelia D, Bartnik D, Koenig BW, Mancuso S, Martinoia E, Geisler M. 2008. Modulation of P-glycoproteins by auxin transport inhibitors is mediated by interaction with immunophilins. The Journal of Biological Chemistry 283, 21817–21826. [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. 2005. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433, 39–44. [DOI] [PubMed] [Google Scholar]
- Bouchard R, Bailly A, Blakeslee JJ, et al. . 2006. Immunophilin-like TWISTED DWARF1 modulates auxin efflux activities of Arabidopsis P-glycoproteins. The Journal of Biological Chemistry 281, 30603–30612. [DOI] [PubMed] [Google Scholar]
- Chen R, Hilson P, Sedbrook J, Rosen E, Caspar T, Masson PH. 1998. The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proceedings of the National Academy of Sciences, USA 95, 15112–15117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Jin W, Wang M, Zhang F, Zhou J, Jia Q, Wu Y, Liu F, Wu P. 2003. Distribution and characterization of over 1000 T-DNA tags in rice genome. The Plant Journal 36, 105–113. [DOI] [PubMed] [Google Scholar]
- Chen X, Shi J, Hao X, Liu H, Shi J, Wu Y, Wu Z, Chen M, Wu P, Mao C. 2013. OsORC3 is required for lateral root development in rice. The Plant Journal 74, 339–350. [DOI] [PubMed] [Google Scholar]
- Chen Y, Fan X, Song W, Zhang Y, Xu G. 2012. Over-expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1. Plant Biotechnology Journal 10, 139–149. [DOI] [PubMed] [Google Scholar]
- Coudert Y, Périn C, Courtois B, Khong NG, Gantet P. 2010. Genetic control of root development in rice, the model cereal. Trends in Plant Science 15, 219–226. [DOI] [PubMed] [Google Scholar]
- Dharmasiri S, Swarup R, Mockaitis K, et al. . 2006. AXR4 is required for localization of the auxin influx facilitator AUX1. Science 312, 1218–1220. [DOI] [PubMed] [Google Scholar]
- Feraru E, Friml J. 2008. PIN polar targeting. Plant Physiology 147, 1553–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J. 2003. Auxin transport – shaping the plant. Current Opinion in Plant Biology 6, 7–12. [DOI] [PubMed] [Google Scholar]
- Friml J, Wiśniewska J, Benková E, Mendgen K, Palme K. 2002. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415, 806–809. [DOI] [PubMed] [Google Scholar]
- Gälweiler L, Guan C, Müller A, Wisman E, Mendgen K, Yephremov A, Palme K. 1998. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282, 2226–2230. [DOI] [PubMed] [Google Scholar]
- Geisler M, Wang B, Zhu J. 2014. Auxin transport during root gravitropism: transporters and techniques. Plant Biology 16, 50–57. [DOI] [PubMed] [Google Scholar]
- Grunewald W, Friml J. 2010. The march of the PINs: developmental plasticity by dynamic polar targeting in plant cells. The EMBO Journal 29, 2700–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison BR, Masson PH. 2008. ARL2, ARG1 and PIN3 define a gravity signal transduction pathway in root statocytes. The Plant Journal 53, 380–392. [DOI] [PubMed] [Google Scholar]
- Hobbie L, Estelle M. 1995. The axr4 auxin-resistant mutants of Arabidopsis thaliana define a gene important for root gravitropism and lateral root initiation. The Plant Journal 7, 211–220. [DOI] [PubMed] [Google Scholar]
- Keuskamp DH, Pollmann S, Voesenek LA, Peeters AJ, Pierik R. 2010. Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proceedings of the National Academy of Sciences, USA 107, 22740–22744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitomi Y, Kanno N, Kawai S, Mizubayashi T, Fukuoka S, Uga Y. 2015. QTLs underlying natural variation of root growth angle among rice cultivars with the same functional allele of DEEPER ROOTING 1. Rice 8, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H, Yan X, Rubio G, Beebe SE, Blair MW, Lynch JP. 2004. Genetic mapping of basal root gravitropism and phosphorus acquisition efficiency in common bean. Functional Plant Biology 31, 959–970. [DOI] [PubMed] [Google Scholar]
- Li G, Liang W, Zhang X, Ren H, Hu J, Bennett MJ, Zhang D. 2014. Rice actin-binding protein RMD is a key link in the auxin–actin regulatory loop that controls cell growth. Proceedings of the National Academy of Sciences, USA 111, 10377–10382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang S, Yu X, Yu J, He X, Zhang S, Shou H, Wu P. 2005. ARL1, a LOB-domain protein required for adventitious root formation in rice. The Plant Journal 43, 47–56. [DOI] [PubMed] [Google Scholar]
- Luschnig C, Gaxiola RA, Grisafi P, Fink GR. 1998. EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes & Development 12, 2175–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Q, Zhong Y, Wang Y, et al. . 2014. SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. The Plant Cell 26, 1586–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JP. 2011. Root phenes for enhanced soil exploration and phosphorus acquisition: tools for future crops. Plant Physiology 156, 1041–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JP. 2013. Steep, cheap and deep: an ideotype to optimize water and N acquisition by maize root systems. Annals of Botany 112, 347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JP, Brown KM. 2001. Topsoil foraging – an architectural adaptation of plants to low phosphorus availability. Plant and Soil 237, 225–237. [Google Scholar]
- Mairhofer S, Pridmore T, Johnson J, Wells DM, Bennett MJ, Mooney SJ, Sturrock CJ. 2017. X-ray computed tomography of crop plant root systems grown in soil. Current Protocols in Plant Biology 2, 270–286. [DOI] [PubMed] [Google Scholar]
- Mairhofer S, Zappala S, Tracy SR, Sturrock C, Bennett M, Mooney SJ, Pridmore T. 2012. RooTrak: automated recovery of three-dimensional plant root architecture in soil from x-ray microcomputed tomography images using visual tracking. Plant Physiology 158, 561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano Y, Muraki M, Fujimori M, Takamizo T, Kindiger B. 2005. Identification of QTL controlling adventitious root formation during flooding conditions in teosinte (Zea mays ssp. huehuetenangensis) seedlings. Euphytica 142, 33–42. [Google Scholar]
- Manske GGB, Ortiz-Monasterio JI, Van Ginkel M, González RM, Rajaram S, Molina E, Vlek PLG. 2000. Traits associated with improved P-uptake efficiency in CIMMYT’s semidwarf spring bread wheat grown on an acid Andisol in Mexico. Plant and Soil 221, 189–204. [Google Scholar]
- Marchant A, Kargul J, May ST, Muller P, Delbarre A, Perrot-Rechenmann C, Bennett MJ. 1999. AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. The EMBO Journal 18, 2066–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Jiang L. 2007. Transient expression of fluorescent fusion proteins in protoplasts of suspension cultured cells. Nature Protocols 2, 2348–2353. [DOI] [PubMed] [Google Scholar]
- Miyashita Y, Takasugi T, Ito Y. 2010. Identification and expression analysis of PIN genes in rice. Plant Science 178, 424–428. [Google Scholar]
- Morita MT, Tasaka M. 2004. Gravity sensing and signaling. Current Opinion in Plant Biology 7, 712–718. [DOI] [PubMed] [Google Scholar]
- Müller A, Guan C, Gälweiler L, Tänzler P, Huijser P, Marchant A, Parry G, Bennett M, Wisman E, Palme K. 1998. AtPIN2 defines a locus of Arabidopsis for root gravitropism control. The EMBO Journal 17, 6903–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y, Ma JF, Yamaji N, Ueno D, Nomoto K, Iwashita T. 2006. A specific transporter for iron(III)-phytosiderophore in barley roots. The Plant Journal 46, 563–572. [DOI] [PubMed] [Google Scholar]
- Murphy DV, Sparling GP, Fillery IRP. 1998. Stratification of microbial biomass C and N and gross N mineralisation with soil depth in two contrasting Western Australian agricultural soils. Soil Research 36, 45–56. [Google Scholar]
- Noh B, Murphy AS, Spalding EP. 2001. Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. The Plant Cell 13, 2441–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciorek T, Zazímalová E, Ruthardt N, et al. . 2005. Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435, 1251–1256. [DOI] [PubMed] [Google Scholar]
- Philosoph-Hadas S, Friedman H, Meir S. 2005. Gravitropic bending and plant hormones. Vitamins and Hormones 72, 31–78. [DOI] [PubMed] [Google Scholar]
- Pothuluri JV, Kissel DE, Whitney DA, Thien SJ. 1986. Phosphorus uptake from soil layers having different soil test phosphorus levels1. Agronomy Journal 78, 991–994. [Google Scholar]
- Qi Y, Wang S, Shen C, Zhang S, Chen Y, Xu Y, Liu Y, Wu Y, Jiang D. 2012. OsARF12, a transcription activator on auxin response gene, regulates root elongation and affects iron accumulation in rice (Oryza sativa). New Phytologist 193, 109–120. [DOI] [PubMed] [Google Scholar]
- Rogers ED, Benfey PN. 2015. Regulation of plant root system architecture: implications for crop advancement. Current Opinion in Biotechnology 32, 93–98. [DOI] [PubMed] [Google Scholar]
- Santelia D, Vincenzetti V, Azzarello E, Bovet L, Fukao Y, Düchtig P, Mancuso S, Martinoia E, Geisler M. 2005. MDR-like ABC transporter AtPGP4 is involved in auxin-mediated lateral root and root hair development. FEBS Letters 579, 5399–5406. [DOI] [PubMed] [Google Scholar]
- Terasaka K, Blakeslee JJ, Titapiwatanakun B, et al. . 2005. PGP4, an ATP binding cassette P-glycoprotein, catalyzes auxin transport in Arabidopsis thaliana roots. The Plant Cell 17, 2922–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracy SR, Black CR, Roberts JA, Sturrock C, Mairhofer S, Craigon J, Mooney SJ. 2012. Quantifying the impact of soil compaction on root system architecture in tomato (Solanum lycopersicum) by X-ray micro-computed tomography. Annals of Botany 110, 511–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracy SR, Roberts JA, Black CR, McNeill A, Davidson R, Mooney SJ. 2010. The X-factor: visualizing undisturbed root architecture in soils using X-ray computed tomography. Journal of Experimental Botany 61, 311–313. [DOI] [PubMed] [Google Scholar]
- Uga Y, Kitomi Y, Yamamoto E, Kanno N, Kawai S, Mizubayashi T, Fukuoka S. 2015. A QTL for root growth angle on rice chromosome 7 is involved in the genetic pathway of DEEPER ROOTING 1. Rice 8, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uga Y, Sugimoto K, Ogawa S, et al. . 2013a. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nature Genetics 45, 1097–1102. [DOI] [PubMed] [Google Scholar]
- Uga Y, Yamamoto E, Kanno N, Kawai S, Mizubayashi T, Fukuoka S. 2013b. A major QTL controlling deep rooting on rice chromosome 4. Scientific Reports 3, 3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsuno K, Shikanai T, Yamada Y, Hashimoto T. 1998. AGR, an agravitropic locus of Arabidopsis thaliana, encodes a novel membrane-protein family member. Plant & Cell Physiology 39, 1111–1118. [DOI] [PubMed] [Google Scholar]
- Vernoux T, Kronenberger J, Grandjean O, Laufs P, Traas J. 2000. PIN-FORMED 1 regulates cell fate at the periphery of the shoot apical meristem. Development 127, 5157–5165. [DOI] [PubMed] [Google Scholar]
- Wang JR, Hu H, Wang GH, Li J, Chen JY, Wu P. 2009. Expression of PIN genes in rice (Oryza sativa L.): tissue specificity and regulation by hormones. Molecular Plant 2, 823–831. [DOI] [PubMed] [Google Scholar]
- Wang S, Xu Y, Li Z, Zhang S, Lim JM, Lee KO, Li C, Qian Q, Jiang A, Qi Y. 2014. OsMOGS is required for N-glycan formation and auxin-mediated root development in rice (Oryza sativa L.). The Plant Journal 78, 632–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Xie Y, Zhang J, et al. . 2015. VLN2 regulates plant architecture by affecting microfilament dynamics and polar auxin transport in rice. The Plant Cell 27, 2829–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Gao MX, Hu H, Ding XM, Lin HW, Wang L, Xu JM, Mao CZ, Zhao FJ, Wu ZC. 2016. OsCLT1, a CRT-like transporter 1, is required for glutathione homeostasis and arsenic tolerance in rice. New Phytologist 211, 658–670. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Forno D, Cock J, Gomez K. 1976. Laboratory manual for physiological studies of rice. Manila, Philippines: International Rice Research Institute. [Google Scholar]
- Zhang X, Henriques R, Lin SS, Niu QW, Chua NH. 2006. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nature Protocols 1, 641–646. [DOI] [PubMed] [Google Scholar]
- Zhu J, Kaeppler SM, Lynch JP. 2005. Topsoil foraging and phosphorus acquisition efficiency in maize (Zea mays). Functional Plant Biology 32, 749–762. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.