The formation of the haustorium in holoparasitic plants as demonstrated in Phelipanche ramosa is initiated through perception of a host-exudated cytokinin and a cytokinin signaling response.
Keywords: Brassica napus, broomrape, cytokinins, haustorium, microarray, parasitic plant, Phelipanche ramosa, root development, root exudates
Abstract
The heterotrophic lifestyle of parasitic plants relies on the development of the haustorium, a specific infectious organ required for attachment to host roots. While haustorium development is initiated upon chemodetection of host-derived molecules in hemiparasitic plants, the induction of haustorium formation remains largely unknown in holoparasitic species such as Phelipanche ramosa. This work demonstrates that the root exudates of the host plant Brassica napus contain allelochemicals displaying haustorium-inducing activity on P. ramosa germinating seeds, which increases the parasite aggressiveness. A de novo assembled transcriptome and microarray approach with P. ramosa during early haustorium formation upon treatment with B. napus root exudates allowed the identification of differentially expressed genes involved in hormone signaling. Bioassays using exogenous cytokinins and the specific cytokinin receptor inhibitor PI-55 showed that cytokinins induced haustorium formation and increased parasite aggressiveness. Root exudates triggered the expression of cytokinin-responsive genes during early haustorium development in germinated seeds, and bio-guided UPLC-ESI(+)-/MS/MS analysis showed that these exudates contain a cytokinin with dihydrozeatin characteristics. These results suggest that cytokinins constitutively exudated from host roots play a major role in haustorium formation and aggressiveness in P. ramosa.
Introduction
Some terrestrial plant species have developed unique features in the course of evolution, leading them to adopt a heterotrophic lifestyle. Plant parasitism is thought to have evolved independently 12–13 times in angiosperms, allowing the occurrence of a diversity of parasitic behavior ranging from opportunistic facultative parasitism to obligatory parasitism (Westwood et al., 2010). The transition from autotrophy to parasitism lies in the development of a multicellular specialized infectious organ, the haustorium, which allows adhesion to and penetration of host tissues by the parasitic plant and the establishment of vascular connections between the host and the parasite (Yoshida et al., 2016).
Most plants in the Orobanchaceae family are root parasitic organisms, including the facultative parasites Triphysaria versicolor and Phtheirospermum japonicum, and the obligatory parasites Striga sp. (witchweeds), and Orobanche and Phelipanche sp. (broomrapes). Facultative parasites can grow to maturity without host connections, although in the vicinity of a host root, they can develop lateral haustoria to connect to the host vascular system (Albrecht et al., 1999). Lateral haustoria develop at the root elongation zone without interfering with the meristematic activity of the root apex, allowing the root growth to resume after the initiation of haustorium formation (Matvienko et al., 2001a). Obligatory parasites develop terminal haustoria at the apex of the radicle of germinated seeds which cause a complete root growth arrest and a redirection of cellular extension from longitudinal to radial (Joel and Losner-Goshen, 1994; Hood et al., 1998). In chlorophyllous species (hemiparasites) including witchweeds and facultative parasites, the development of early haustorial structures (EHSs) starts with the swelling of the root tip due to the redirection of cell division. Concomitantly, a polar outgrowth of epidermal cells is observed which give rise to haustorial hairs that serve as structures for adhesion to the host (Baird and Riopel, 1983; Hood et al., 1998; Bandaranayake and Yoder, 2013). In achlorophyllous species (holoparasites) such as Orobanche cumana and Phelipanche aegyptiaca, the early haustorium development is also revealed by a swelling of the root tip, caused by the extension of the apical cells and a polar outgrowth of epidermal cells, but it gives rise to short cell extensions termed papillae (Joel and Losner-Goshen, 1994; Fernández-Aparicio et al., 2016). Both haustorial hairs and papillae produce adhesive molecules meant to anchor the haustorium to the host root (Baird and Riopel, 1985; Joel and Losner-Goshen, 1994).
EHS formation is triggered after a host-derived chemical stimulation by so-called haustorium-inducing factors (HIFs), which are mainly phenolic acids (Albrecht et al., 1999). Among them, 2,6-dimethoxy-1,4-benzoquinone (DMBQ) was isolated from abraded Sorghum roots and shown to trigger haustorium formation in T. versicolor and Striga hermonthica (Lynn et al., 1981; Chang and Lynn, 1986). DMBQ is released by oxidation of host cell wall compounds upon host peroxidase activity triggered by hydrogen peroxide production at the tip of the Striga radicle (Keyes et al., 2000). Such a model explains how the parasite extracts HIFs and develops an haustorium only upon close contact with the host root (Keyes et al., 2007). DMBQ recognition involves a modification of redox potential, in which semiquinone intermediates are formed to trigger a redox-sensitive signal transduction pathway (Keyes et al., 2000). This redox cycling involves two quinone oxidoreductase-encoding genes (TvQR1 and TvQR2) which are transcriptionally up-regulated in T. versicolor roots in response to DMBQ (Bandaranayake et al., 2010; Ishida et al., 2017). Treatments with phytohormones also trigger EHS formation (i.e. haustorial hairs) (O’Malley and Lynn, 2000), but with differences in the degree and size of the swelling and in the precise and synchronous location of the haustorial hairs with respect to the root apex (Tomilov et al., 2005). Nevertheless, indole acetic acid (IAA) treatment enhances haustorial hair formation in T. versicolor when low DMBQ concentrations are applied. Ishida et al. (2016) showed that the auxin biosynthesis gene YUC3 encoding a flavin monooxygenase is up-regulated in haustorial cells of P. japonicum. Overexpression of YUC3 in P. japonicum roots triggers haustorium formation, and IAA levels increase at the haustorium formation site after DMBQ treatment. These results suggest that the auxin biosynthetic and regulatory pathways may be recruited in these facultative parasites during EHS formation.
Although no known host-derived HIF is active on holoparasites, recent findings showed that the mycotoxins sphaeropsidones may trigger EHS formation in O. cumana and O. crenata (Fernández-Aparicio et al., 2016). These molecules are not related in structure to DMBQ, which suggests that other receptors and signaling pathways leading to EHS formation in broomrapes may exist. Although recent transcriptomic studies have unraveled the existence of core parasitism genes expressed in response to DMBQ in S. hermonthica, T. versicolor, and P. aegyptiaca (Yang et al., 2015), and strengthened the involvement of hormone signaling in EHS development in P. japonicum and Santalum album (Zhang et al., 2015; Ishida et al., 2016), the mechanisms leading to haustorium formation in the holoparasite P. aegyptiaca remain largely unknown.
Here, using transcriptomic and biochemical approaches, we examined the induction of haustorium formation in the holoparasitic species P. ramosa in response to host plant root exudates (Brassica napus), and demonstrated that this event is triggered by a cytokinin (CK) signal.
Materials and methods
Plant material and growth conditions
Phelipanche ramosa (L.) Pomel seeds were collected in 2014 from mature broomrape flowering spikes in an oilseed rape field at Saint Martin de Fraigneau (France) and stored at 25 °C in the dark before use. The highly susceptible B. napus cultivar ES Aliénor (EURALIS, Lescar, France) was chosen for co-cultivation experiments in Petri dishes and exudation assays. Seeds were surface-sterilized using 2.4% sodium hypochlorite (Lechat et al., 2012). Phelipanche ramosa seeds were then suspended (10 mg ml–1) in 1 mM HEPES, pH 7.5, 0.1% PPM (incubation medium) and placed at 21 °C in the dark for 7 d (conditioning period; Lechat et al., 2015).
Brassica napus seeds were placed on a plate between two moistened filter papers and incubated at 21 °C in a growth chamber (16 h light, 8 h dark) for 7 d. Two hundred plantlets were then distributed into 20 drained plastic pots [700 ml, 9 × 11 × 12 cm (W×L×H), Nicoplast] nested in undrained pots and filled with glass beads (Ø 3 mm). Plantlets were first watered with 100 ml of 50% Coïc medium pH 6.8 (Coic and Lesaint, 1975) once a week and then every 2 d from week 3 on. Culture medium (root exudates) from each drained pot was recovered and pooled at the end of weeks 3 and 4 (weeks with exudates displaying the highest EHS induction activity).
Bioassays for haustorial induction
After the conditioning period, the incubation medium was removed and P. ramosa seeds were rinsed three times with sterile distilled water and suspended (2.5 mg ml–1) in germination medium [0.5 mM HEPES, pH 7.5, 0.05% PPM, rac-GR24 10–7 M (synthetic germination stimulant, kindly provided by Dr B. Zwanenburg)]. A 100 µl aliquot of GR24-treated P. ramosa seeds (~25 seeds) was then distributed in each well in 96-well plates (Cell Culture Multiwell Plate Cellstar; Greiner Bio-One) and placed to germinate at 21 °C in the dark for 4 d.
The germination solution was then removed from the wells and seeds were suspended in 100 µl of test solutions. Test solutions for EHS induction assays contain 0.5 mM HEPES, pH 7.5, 0.05% PPM (final concentration) and various concentrations of commercial standards and organic or aqueous root exudates from B. napus (Supplementary Table S1 at JXB online). Induction of EHS was monitored 3 d after treatments.
Confocal laser and light microscopy
Observations of haustorium formation at the apex of P. ramosa germinated seeds were conducted using a confocal laser scanning microscope (MCBL Nikon A1). Germinated seeds were treated or not with B. napus root exudates for 72 h and then stained in acridine orange (0.02%) for 5 min and rinsed three times with water. The excitation and emission wavelengths were set at 488 nm and 500–530 nm, respectively.
Germinated seeds were fixed and embedded in LRW resin (London Resin White acrylic, Sigma Aldrich) (Chateigner-Boutin et al., 2014). Sections of 1.5 µm thickness were prepared using a Leica EM UC7 ultramicrotome, stained using toluidine blue (Toluidine blue 1%, Na2CO3 2.5%) for 5 min, and rinsed with distilled water, then observed using a Nikon AZ100 macroscope.
Aggressiveness bioassay
For co-cultivation experiments, 7-day-old B. napus plantlets were transferred onto filter paper each covered with 20 mg of germinated P. ramosa seeds previously treated or not for 2 d with root exudates and placed in cut square plates (120 × 120 × 17 mm, Greiner, France) containing a uniform layer of rockwool moisturized with 50 ml of Coïc medium. Five plantlets per plate and five plates per condition were used. Plates were sealed and incubated vertically at 21 °C in a growth chamber (16 h light, 8 h dark, 70% humidity) for 17 d and supplied every 2 d with 10 ml of Coïc medium. The development of a functional haustorium was monitored daily using a phloem-mobile fluorescent dye (6-Carboxyfluorescein, Sigma Aldrich, 10 mM, pH 6.3). Droplets of carboxyfluorescein were applied on abraded host leaves, and fluorescent tubercles were observed using a fluorescence binocular equipped with a GFP2 filter: 480/40 nm excitation; 510 nm emission (Leica), showing functional phloem connections between the host and parasite within the haustorium (Péron et al., 2017).
Total RNA extraction
RNA was extracted using a Nucleospin RNA Plant kit (Macherey Nagel) according to the manufacturer’s instruction. Genomic DNA was removed from RNA samples by treatment with 6.8 U of RNase-free DNase I (Qiagen, Cat. No 79254) followed by a clean-up step with an RNA clean up XS spin column (Macherey Nagel) according to the manufacturer’s instruction. RNA was quantified using a NanoDrop ND-1000 (NanoDrop Technologies), and RNA quality was assessed with an Agilent 2100 Bioanalyzer.
cDNA sequencing and de novo transcriptome assembly
Total RNAs were extracted from P. ramosa seeds imbibed for 24 h, seeds conditioned for 7 d, germinated seeds (72 h after GR24 treatment), young tubercles, ‘spider’ stage tubercles, young flowering stems, and mature flowering stems. A Roche 454 normalized library was constructed with mRNAs isolated from each sample and mixed in equal proportion, and single read sequences were obtained with Roche GS-FLX+ 454 pyrosequencing systems equipped with on-instrument Data Analysis Software Modules v.2.3 (Life Science). cDNA library construction and sequencing were performed by Eurofins Genomics Company (Ebersberg, Germany).
For de novo transcriptome assembly, sequences were treated using the Mira sequence assembler Version 3.4 by Eurofins Genomics Company. Blast annotation of the translated sequences (BlastX) was performed using the BioEdit software v7.2.5 against Arabidopsis thaliana (TAIR 10) and Solanum lycopersicum (SlGDB, http://www.plantgdb.org) libraries. The contigs were then assigned to the PlantTribes 2.0 orthogroups (Wall et al., 2008). Gene Ontology (GO) terms were assigned for each unigene based on GO slim terms using the PANTHER algorithm (http://www.pantherdb.org) (Mi et al., 2016).
Microarray analysis
Contigs obtained from de novo transcriptome assembly were used to design the Agilent-069670 Phelipanche_v1 chip. It was in situ synthesized by Agilent (see the manufacturer’s web site at http://www.agilent.com/) and contained 105 086 oligoprobes of 60-mers as follows: 52 543 predicted sense and 52 543 complementary antisense sequences corresponding to the predicted coding genes. All probes were designed within the CDS and with a positive bias toward the 3′ end of the genes, and fixed on Agilent 4 X 180K microarray slides (Gene Expression Omnibus under the accession number GPL23512).
Total RNA was extracted from 200 mg of germinated seeds treated or not with crude B. napus root exudates for 12 h and 24 h then deep frozen in liquid nitrogen. mRNA was amplified, labeled as described in Celton et al. (2014), and co-hybridized according to the Agilent two-color microarray-based gene expression analysis protocol.
For each comparison, two biological replicates were analyzed in dye-switch design as described in Depuydt et al. (2009). All statistical analyses were performed using the R language (R Development Core Team, 2009) as described in Celton et al. (2014). Transcriptome data have been submitted to the Gene Expression Omnibus as Series GSE99377. GO term enrichment of differentially expressed genes (DEGs) was carried out using the AGRIGO algorithm (http://bioinfo.cau.edu.cn/agriGO) (Du et al., 2010) against the GO assignment of P. ramosa de novo transcriptome assembly using Fisher’s exact test corrected as described in Benjamini and Hochberg (1995) [false discovery rate (FDR) <0.05].
Quantitative real-time PCR (RT-qPCR) analysis
cDNA was synthesized from 0.5 μg of total RNA using SuperMIX qScript cDNA (Quanta) according to the manufacturer’s instructions. cDNA was diluted 20 times and used in 25 µl RT-PCR experiments with 12.5 µl of PerfeCTa SYBR Green SuperMix (Quanta) and 0.3 mM of each primer, carried out on an Applied Biosystems 7300 real-time PCR system (Applied Biosystems) as described in Lechat et al. (2015) using the amplicon of the constitutive 18S, which showed low cycle threshold variation (SD <0.5 cycle threshold), as an internal control to normalize all the data. The gene-specific primers used for each amplification are presented in Supplementary Table S2. A control experiment without cDNA was included for each PCR mix.
Bioassay-guided reversed phase (RP)-HPLC purification of HIFs
Two liters of crude B. napus root exudates were loaded on a Strata-X 33µm Polymeric Solid Phase Extraction column (SPE, Phenomenex) and then washed with 4 ml of distilled water. Flow-through and washing volumes were pooled (FT). Compounds eluted with 4 ml of acetonitrile (ACN) were further evaporated using an R II Rotavapor (Buchi) and suspended in 200 µl of 50% ACN (SPE extract).
The SPE extract was further fractionated by using a LaChrom Elite HPLC System (Hitachi) equipped with an L-2100 pump, L-2200 Autosampler, L-2400 UV Detector, and column thermostat Jetstream 2 Plus. A 90 µl aliquot of SPE extract was loaded on a Kromasil C18 guard column (5 µm, 250 × 4.6 mm, Sigma-Aldrich). Chromatographic conditions were: flow rate of 0.5 ml min−1; column temperature of 30 °C; and the following binary gradient: 0 min, 95:5; 7 min, 95:5; 28 min, 0:100; 39 min, 0:100 (time, water–0.1% acetic acid:ACN–0.1% acetic acid). Absorbance was monitored at 260 nm. Fractions 0.5 ml to 2.5 ml of the purified exudate were collected from 0 to 39 min and tested in an haustorium induction assay.
Detection and quantification of CKns by UPLC-ESI (+)-MS/MS in root exudates
CK identification in the fractionated exudates was carried out by UPLC-ESI(+)-MS/MS according to Svačinová et al. (2012) with some modifications. An Acquity UPLC System (Waters) linked to a triple quadrupole mass spectrometer Xevo™ TQ-S (Waters MS Technologies) equipped with an electrospray interface (ESI) and the unique performance of a collision cell (ScanWave) was used. MS data were processed by MassLynx™ software with the TargetLynx™ program (version 4.2, Waters).
A 2 µl aliquot of samples (SPE extract or fractions) was injected onto a reversed-phase column (Acquity UPLC BEH C18, 1.7 μm, 2.1 × 100 mm, Waters). The flow rate was fixed at 0.4 ml min−1; column temperature was 40 °C; sample temperature was 4 °C; and the following binary gradient (A, water–0.5% formic acid; B, methanol–0.5% formic acid) was applied: 0 min, 95:5 (A:B); 4.0 min, 95:5; 9.0 min, 50:50; 10.0 min, 0:100; 1.0 min, 0:100; 13.0 min, 95:5; 15.0 min, 95:0. Selective reaction monitoring mode using mass-to-charge (m/z) transitions of precursor and product ions was optimized: capillary voltage 3.0 kV; source/desolvation gas temperature 150/400 °C; cone/desolvation gas flow rates 150/650 l h−1.
Results
Brassica napus root exudates induce EHSs at the apex of the P. ramosa radicle
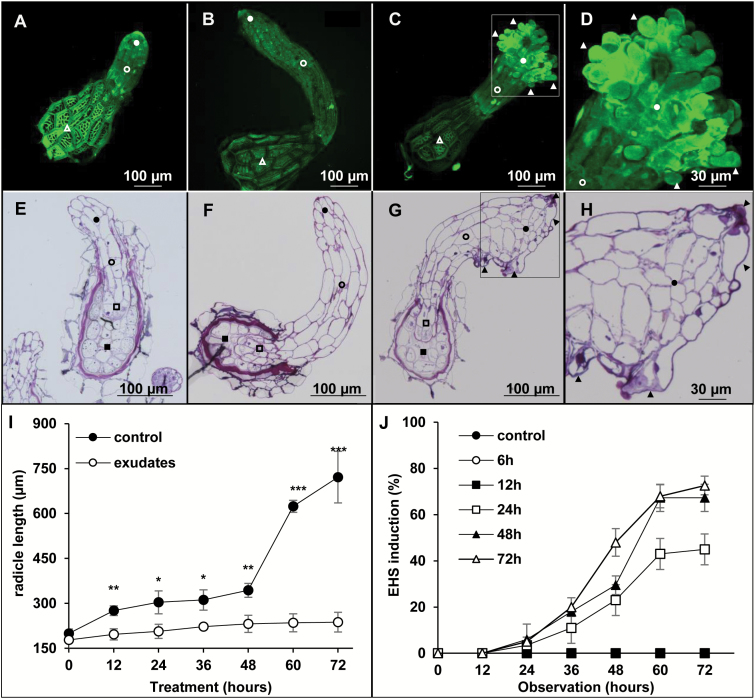
By choosing B. napus as a major host for P. ramosa, we tested if the signal allowing the induction of haustorium formation is of allelochemical nature and present in the root exudates. Germinated seeds (96 h post 10−7 M GR24 treatment, T0 seeds) were treated or not with B. napus root exudates during 72 h. While T0 seeds and untreated seeds displayed radicles with a thin and regular conical apex (Fig. 1A, B, E, F), treated seeds displayed a larger apex with irregular lobes and larger cells (Fig. 1C, G). Moreover, a complete root growth arrest was observed in treated germinated seeds in comparison with the untreated radicles (Fig. 1I). This suggests that root exudates trigger a change in cell expansion and root elongation. In addition, papillae were observed in the crown at the apex of treated radicles, each arising from single epidermis cells (Fig. 1C, D, G, H). EHSs are characterized by the swelling of the root apex, shorter roots, and outgrowth of epidermal cells (formation of papillae).
Fig. 1.
Brassica napus root exudates can trigger the formation of early haustorial structures in P. ramosa. (A–D) Confocal laser scanning microscope observation of P. ramosa germinated seeds stained with acridine orange. (E–H) Light microscope observation of thin sections of P. ramosa germinated seeds stained with toluidine blue. Open squares, embryo; filled squares, albumen; open circles, radicle; filled circles, apex; open triangles, testa; filled triangles, papillae. (A, E) Seeds before treatment. (B, F) Seeds incubated for 72 h in control buffer. (C, G) Seeds incubated for 72 h in half-diluted B. napus root exudates. Sale bars=100 µm. (D, H) Close-up view of the root apex of seeds incubated for 72 h in B. napus root exudates. Scale bars=30 µm. (I) Assessment of radicle length of P. ramosa germinated seeds upon treatment with B. napus root exudates during 72 h. Exudates were half diluted in buffer solution (0.5 mM HEPES, 0.05% PPM). The control condition corresponds to a treatment with the buffer solution alone. Means are values ±SE (n=6). Means annotated with asterisks are significantly different from the control points (t-test, *P<0.05, **P<0.01, and ***P<0.001). (J) Time course assessment of EHS development upon continuous treatment with B. napus root exudates for 6, 12, 24, 48, and 72 h, respectively. At the end of each treatment time, germinated seeds were rinsed, and exudates were discarded and replaced by buffer solution. Means are values ±SE (n=6).
A time course treatment was used to determine the kinetics of EHS formation as well as the minimum treatment time required. Untreated seeds and seeds treated for 6 h and 12 h did not show any EHS formation in the course of the experiment, whereas seeds treated for 24, 48, and 72 h all displayed a significant EHS formation at 36 h and reached up to 72% of radicles displaying EHSs at 72 h (Fig. 1J). A dose–response pattern was observed using diluted extracts (Supplementary Fig. S1).
Taken together, these results (root growth arrest and EHS formation) suggest that B. napus root exudates contain molecules acting as HIFs.
Induction of EHSs enhances aggressiveness of P. ramosa germinated seeds
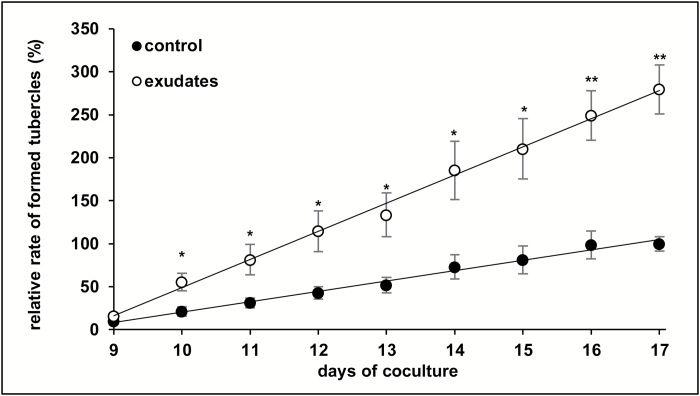
Co-cultivation assays were used in order to demonstrate the importance of the observed EHSs for the parasite aggressiveness. Germinated P. ramosa seeds were treated or not for 48 h with B. napus root exudates before being put in contact with roots of B. napus plantlets. The number of tubercles with vascular connections reflecting the development of functional haustoria was assessed using the phloem tracer, 6-carboxyfluorescein. The results were expressed as a percentage of the maximum number of tubercles observed in control conditions (no treatment with B. napus root exudates). The number of tubercles in treated conditions was already significantly higher (55 ± 10%) than in untreated conditions (21 ± 6%) from the 10th day on and reached 280 ± 28% at the end of the experiment (Fig. 2), indicating that pre-inducing the development of EHSs clearly increased the aggressiveness of the parasite.
Fig. 2.
Induction of EHSs at the apex of P. ramosa germinated seeds enhances parasite aggressiveness. A 20 mg aliquot of P. ramosa germinated seeds was incubated for 48 h in B. napus root exudates or control solution before placing them in contact with B. napus roots. Five plantlets per plate and five plates per condition were used. The number of developing tubercles was assessed using the phloem-mobile dye 6-carboxyfluorescein and the results were expressed as a percentage of the maximum number of tubercles observed in control conditions. Exudates were half diluted in buffer solution (0.5 mM HEPES, 0.05% PPM). The control condition corresponds to a treatment with the buffer solution alone. Means are values ±SE (n=5 plates). Means annotated with asterisks are significantly different from the control points (t-test, *P<0.05 and **P<0.01).
High-throughput sequencing and de novo assembly of the P. ramosa transcriptome
To obtain de novo assembly of the P. ramosa transcriptome, a GS-FLX+ 454 pyrosequencing approach (Life Science) was used. Total RNAs were extracted from various developmental stages from seeds to mature flowering shoots. A normalized cDNA library was constructed from mRNAs isolated from each sample and mixed in equal proportion. A total of 1 151 288 reads with an average length of 437 bp and a total length of 503 Mbp were generated. The assembly yielded 58 Mbp distributed in 53 511 unigenes (non-redundant sequences; Supplementary Table S3). The unigenes had an average size of 952 bp and an N50 contig length of 1071 (L50=20 080).
The coverage rate of the P. ramosa transcriptome was assessed by examining the proportion of conserved orthologous genes in the assembly data set. Two sets of conserved genes were used, namely the UCOs containing 357 single-copy genes highly conserved in A. thaliana, humans, mice, yeast, fruit flies, and Caenorhabditis elegans, and APVO sequences containing 959 single-copy genes from A. thaliana, Populus trichocarpa, Vitis vinifera, and Oryza sativa plant genomes (Wall et al., 2008; Duarte et al., 2010): 98% of UCOs (with a threshold blast e-value of 1 × 1010) and 82% of APVO sequences were detected in P. ramosa transcriptome assembly, indicating the high coverage rates of P. ramosa transcripts (Supplementary Table S3).
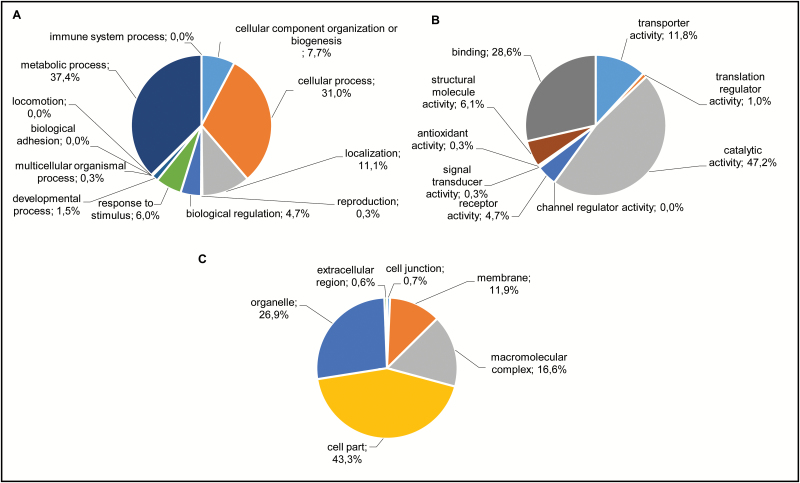
Local alignment using the BLASTx algorithm (cut-off e-value 1 × 1010) showed that 51% and 55% of P. ramosa contigs/unigenes have homologous sequences in the A. thaliana genome (TAIR 10) and in the S. lycopersicum (GeneBank 160) genome, respectively (Supplementary Table S3). Phelipanche ramosa contigs/unigenes were then classified into 6227 slim GO terms which were regrouped in 30 main terms (Fig. 3A–C, Supplementary Data set S1). Among the terms represented, the biological process category was mainly classified into metabolic process and cellular process, accounting for ~34% and 31% of the transcripts, respectively. The molecular function category was dominated by catalytic activity and binding. Of these, hydrolase activity (31%), transferase activity (29%), oxidoreductase activity (15%), nucleic acid binding (57%), and protein binding (30%) were the most represented. In the cellular component category, 43% of the contigs were classified in cell part, of which 82% account for intracellular and 14% account for plasma membrane localization (Fig. 3A–C; Supplementary Data set S1).
Fig. 3.
Gene Ontology analyses of all contigs. Pram contigs were assigned to GO slim terms for biological processes (A), molecular functions (B), and cellular components (C) using the PANTHER algorithm (http://www.pantherdb.org) (Mi et al., 2016). Numbers indicate percentages of each GO slim term within main ontologies.
Identification and characterization of P. ramosa genes induced during haustorium formation
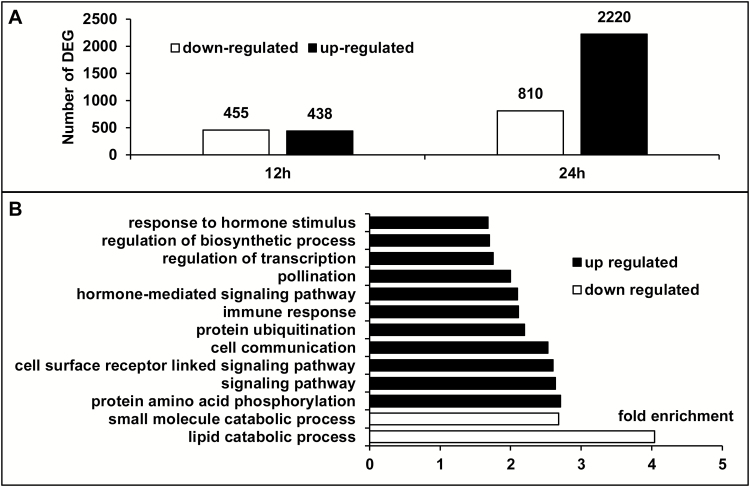
To identify genes involved in haustorium formation and especially genes regulated before the visible onset of EHSs, a microarray based on the P. ramosa transcriptome assembly was designed. Germinated seeds were treated with B. napus root exudates and gene transcriptional levels were analyzed by microarray hybridization. According to the time course experiment (Fig. 1J), two time points (12 h and 24 h) were chosen for the microarray analysis in order to focus on early transcriptional changes. In these conditions, 893 DEGs displayed a 1.5-fold higher or 0.75-fold lower expression 12 h after treatment, and 3030 were detected after 24 h of treatment (Fig. 4A). RT-qPCR analyses were performed on a set of 21 randomly selected DEGs to validate the microarray results. Expression profiles showed patterns similar to those obtained from microarray analysis, thus demonstrating the reproducibility of microarray data (Supplementary Fig. S2).
Fig. 4.
Analysis of differentially expressed genes (DEGs) during haustorial development. (A) Number of DEGs with log2 (FC) (fold change between B. napus exudate-treated/control P. ramosa germinated seeds) ≥1.5 or ≥ –0.75 at 12 h and 24 h. (B) Selected significant GO terms for pooled annotated DEGs. GO term enrichment analysis was carried out on the pooled DEGs using the AGRIGO algorithm. Fisher’s exact test (Hochberg FDR <0.05).
DEGs were divided into two pools according to their up- or down-regulation, and the average gene expression values (fold changes against the control sample) was calculated for each pool and displayed in Table 1.
Table 1.
Average expression values of annotated DEGs as log2 (FC)
DEGs at 12 h or 24 h were pooled in two clusters according to their over- or under expression status.
| Cluster | Sequence number with hit blast A. thaliana | Average log 2 (FC) |
|---|---|---|
| Pool up-regulated | 974 | 1.4 ± 0.7 |
| Pool down-regulated | 720 | –0.8 ± 0.3 |
Means are values ±SD.
To investigate a functional shift in the gene population, GO term enrichment analyses were carried out on the two pools of DEGs. Among the sequences assigned to the GO term biological process, repressed genes showed an over-representation of GO terms corresponding to lipid catabolic process and small molecule catabolic process. Up-regulated genes were notably enriched for terms corresponding to protein amino acid phosphorylation, signaling pathway, regulation of transcription, and hormone response, which suggests an involvement of hormone perception and signaling in EHS induction (Fig. 4B; Supplementary Data set S2).
Multiple hormone responses
Among DEGs, 79 genes were related to a broad hormone response (Supplementary Data set S2). Considering this result, the impact of plant hormones on EHS formation was assessed using a set of hormones including auxin [1-naphthaleneacetic acid (NAA)], CK [c/t-zeatin (c/tZ)], gibberellic acid (GA3), abscisic acid (ABA), ethylene [1-aminocyclopropane-1-carboxylic acid (ACC)], jasmonic acid (JA), brassinosteroids (castosterone), and strigolactones (rac-GR24) at physiological concentrations ranging from 10–4 M to 10–9 M. c/tZ was the only molecule to induce EHS formation, with ~80% of the radicles harboring these structures at a concentration range of 10–8–10−7 M (Table 2A). As expected, no induction of EHSs was observed when germinated seeds were treated using molecules previously identified as HIFs in hemiparasitic plants, including DMBQ, syringic acid, vanilic acid, and coniferyl alcohol (Supplementary Table S4A).
Table 2.
Effect of treatments with phytohormones on EHS induction.
(A) P. ramosa germinated seeds were treated with an array of phytohormones at concentrations ranging from 10–4 M to 10−10 M. (B) Inhibiting activities were determined using a co-treatment with either c/tZ at optimum concentration (10–7 M) and NAA or ABA at concentrations ranging from 10–4 M to 10−10 M, or root exudates (Ex) and NAA or ABA at concentrations ranging from 10–4 M to 10−10 M. Results for treatments with c/tZ or exudates alone are displayed in (A). Exudates were half-diluted in buffer solution (0.5 mM HEPES, 0.05% PPM).
| Compound | EHS induction (%) | |||||||
|---|---|---|---|---|---|---|---|---|
| 10–4M | 10–5M | 10–6M | 10–7M | 10–8M | 10–9M | / | ||
| A | GR24 | NI | NI | NI | NI | NI | NI | |
| Cstosterone | NI | NI | NI | NI | NI | NI | ||
| GA3 | NI | NI | NI | NI | NI | NI | ||
| ABA | NI | NI | NI | NI | NI | NI | ||
| NAA | NI | NI | NI | NI | NI | NI | ||
| ACC | NI | NI | NI | NI | NI | NI | ||
| JA | NI | NI | NI | NI | NI | NI | ||
| c/tZ | NI | NI | NI | 82 ± 4 | 81 ± 1 | NI | ||
| Exudates | – | – | – | / | / | / | 88 ± 2 | |
| B | c/tZ (10–7 M)+NAA | NI | NI | 2 ± 2*** | 87 ± 2 | 86 ± 2 | 87 ± 3 | |
| c/tZ (10–7 M)+ABA | NI | 19 ± 3*** | 40 ± 2*** | 85 ± 1 | 86 ± 1 | 85 ± 2 | ||
| Ex+NAA | NI | NI | 87 ± 2 | 84 ± 4 | 88 ± 2 | 86 ± 3 | ||
| Ex+ BA | 36 ± 8*** | 50 ± 6*** | 92 ± 2 | 89 ± 1 | 95 ± 3 | 91 ± 2 | ||
NI, non-induced.
Means are values ±SE (n=6 wells).
Means annotated with asterisks are significantly different from the highest induction value (t-test, ***P<0.001).
These results show that c/tZ triggers EHS formation in P. ramosa whereas NAA and ABA tended to have an antagonistic action, thus suggesting the involvement of the CK response in this process.
Direct CK treatments induce EHS at the apex of the P. ramosa radicle
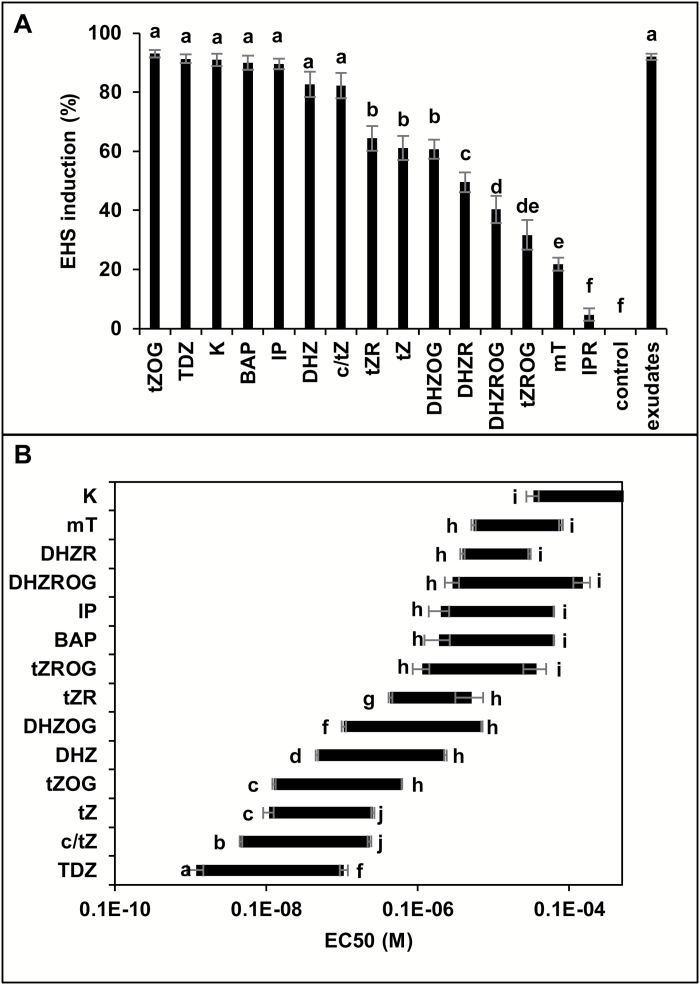
To confirm the role of CKs in EHS development, germinated seeds were treated independently with a set of 15 adenine-type CKs and phenylurea-type CKs known for their different structure–activity relationship (SAR) (Holub et al., 1998; Spíchal, 2012). Adenine-type CKs include biologically active CKs [isopentenyladenine (IP), zeatin (Z), and dehydrozeatin (DHZ)], glycosylated and ribosylated CKs known to have reduced biological activities [zeatin O-glucoside (ZOG), DHZ O-glucoside (DHZOG), IP riboside (IPR), Z riboside (ZR), DHZ riboside (DHZR), DHZ riboside-O-glucoside (DHZROG), and tZ riboside-O-glucoside (tZROG)], and 6-benzyl, 6-furfuryl, and 6-hydrobenzyl CKs [6-benzylaminopurine (BAP), kinetin (K), and meta-tomalin (mT)]. The highly biologically active phenylurea-type CK thidiazuron (TDZ) was also included (Supplementary Table S1). For each CK, the maximum haustorium induction activity (Fig. 5A) and the dose–response activity (Fig. 5B) were determined for concentrations ranging between 10−10 M and 10–3 M. Interestingly, as shown in Fig. 5B, each CK displayed two EC50 values, meaning that the efficiency of the compound can be determined with the concentration value but also with the active concentration range. In these conditions, TDZ was the most efficient compound with a maximum activity of 91 ± 1% (Fig. 5A) and active concentrations ranging from 1.2 × 10−9±2.8 × 10−10 M to 10–7±1.3 × 10−8 M (Fig. 5B). The most active group of CKs contains the isoprenoid CKs tZOG, c/tZ, DHZ, IP, and the aromatic CKs K and BAP, inducing >80% of EHSs. A second group was formed with tZ, DHZOG, and tZR, with an induction activity found between 60% and 70%. DHZR induced 50 ± 8% of EHSs and, below this value, a last group with the least active CKs (DHZROG and tZROG) was found (Fig. 5A). Taken together, these results suggest a strong role for CKs in EHS development.
Fig. 5.
Effect of treatments with cytokinins on EHS induction. (A) Maximum EHS induction at the apex of P. ramosa germinated seeds after 72 h of treatment with an optimal concentration of tZOG (10–7 M), TDZ (10–8 M), K (10–4 M), BAP (10–5 M), c/tZ (10–7 M), DHZ (10–7 M), IP (10–5 M), tZ (10–7 M), DHZOG (10–6 M), tZR (10–6 M), DHZR (10–5 M), DHZROG (10–5 M), mT (10–5 M), IPR (10–5 M), and tZROG (10–6 M). Means are values ±SE (n=6). Values with the same letter are not significantly different from the control points (ANOVA P<0.01). (B) Concentration-dependent stimulation by tZOG, TDZ, K, BAP, c/tZ, DHZ, IP, tZ, DHZOG, tZR DHZR DHZROG, mT, and tZROG with concentrations ranging from 10–4 M to 10−10 M. EC50=half-maximal effective concentration. Means are values ±SE (n=6). Values with the same letter are not significantly different from the control points (ANOVA P<0.01).
However, CKs with glycosylated or aromatic substituents display reduced efficiency. Indeed, it was found to be reduced 2–5 times when a glucose substituent was added (tZ>tZOG; DHZ>DHZOG; DHZR>DHZROG) and 20–70 times when a ribose substituent was added (tZ>tZR; DHZ>DHZR; DHZOG>DHZROG). The addition of furfuryl or hydroxybenzyl substituents also decreased the overall efficiency (Fig. 5B).
Cytokinin receptor inhibitor decreases EHS formation and parasite aggressiveness
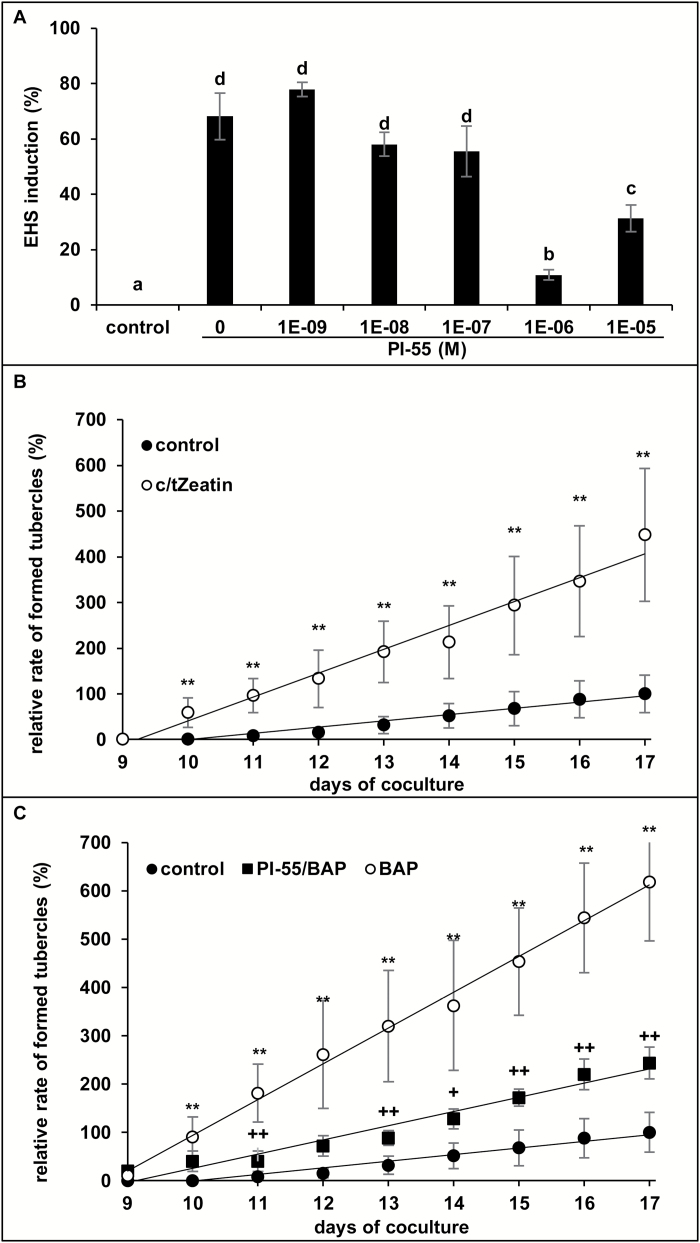
To determine if CK perception is crucial for EHS formation in P. ramosa, PI-55, a specific inhibitor of BAP perception, was used. PI-55 is structurally related to BAP and was shown to inhibit competitively BAP binding on Arabidopsis-specific receptors CRE1/AHK4 and AHK3 (Spíchal et al., 2009). Since the P. ramosa transcriptome also contains CRE1/AHK4 homolog sequence (Pram_00096), germinated seeds were treated with 10–5 M BAP alone or concomitantly with PI-55 (concentrations ranging from 10–5 M to 10–9 M) for 72 h, and EHS induction was monitored. BAP alone induced 68 ± 8% of EHSs. A maximum reduction of 83.8% was observed at 10–6 M PI-55 in the percentage of EHS induction (Fig. 6A).
Fig. 6.
Induction of EHSs and parasite aggressiveness are controlled by cytokinins. (A) P. ramosa germinated seeds were co-treated with BAP (10−5 M) and PI-55 at concentrations ranging from 10–6 M to 10–9 M. The control condition corresponds to a treatment with BAP (10–5 M) alone. Means are values ±SE (n=6). Means annotated with different letters are significantly different from the control point (t-test, **P<0.001). (B) A 20 mg aliquot of P. ramosa germinated seeds was incubated for 48 h in c/tZ (10–7 M) or control solution before placing them in contact with B. napus roots. Five plantlets per plate and five plates per condition were used. The number of developing tubercles was assessed using the phloem-mobile dye 6-carboxyfluorescein and the results were expressed as a percentage of the maximum number of tubercles observed in control conditions. The control condition corresponds to a treatment with the buffer solution alone (0.5 mM HEPES, 0.05% PPM). Means are values ±SE (n=5). Means annotated with asterisks are significantly different from the control points (t-test, *P<0.05 and **P<0.01). (C) A 20 mg aliquot of P. ramosa germinated seeds was incubated for 48 h in BAP (10–5 M), BAP (10–5 M)/PI-55 (10–6 M), or control solution before placing the seeds in contact with B. napus roots. Five plantlets per plate and five plates per condition were used. The control condition corresponds to a treatment with the buffer solution alone (0.5 mM HEPES, 0.05% PPM). Means are values ±SE (n=5). Means annotated with astericks are significantly different from the control points (t-test, **P<0.01). Means annotated with plus signs are significantly different from the BAP alone points (t-test, +P<0.05 and ++P<0.01, respectively).
In order to determine the involvement of CKs in the aggressiveness of P. ramosa, a co-cultivation assay was used. Germinated seeds were treated or not with either 10–7 M c/tZ or 10–5 M BAP or with 10–5 M BAP together with 10–6 M PI-55 for 48 h before placing them in contact with roots of B. napus plantlets. The number of tubercles in c/tZ- and BAP-treated conditions was already significantly higher (59 ± 32% and 91 ± 42%, respectively) than in untreated conditions (0%) from the 10th day on, and reached 448 ± 145% and 619 ± 123%, respectively, at the end of the experiment (Fig. 6B, C). During co-treatment with BAP/PI-55, a 56% reduction in the number of tubercles was observed at 10 d and a 60.5% reduction was observed at 17 d (Fig. 6C).
These results demonstrate that pre-inducing the development of EHS with CKs clearly increased the aggressiveness of the parasite in a similar way to that observed with B. napus root exudates, and that the inhibition of CK perception decreased both the number of EHSs and the subsequent aggressiveness of the parasite.
Root exudates induce a cytokinin response pattern
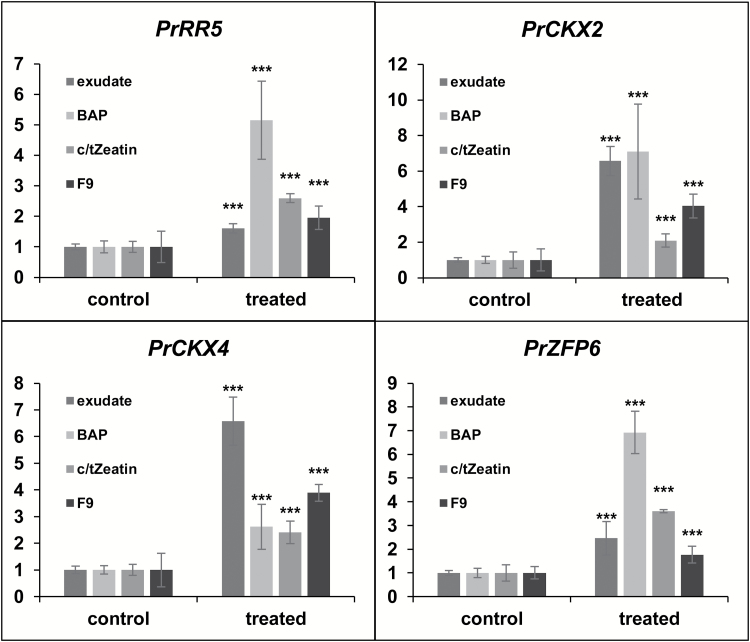
To determine if the formation of EHSs induced by B. napus root exudates was accompanied by a similar gene response to that observed in response to CKs, P. ramosa germinated seeds were treated with exudates, 10–7 M c/tZ, 10–5 M BAP, or control buffer for 24 h. Total RNA was extracted and qRT-PCR experiments were carried out on a set of four genes (PrRR5, PrCKX2, PrCKX4, and PrZFP6), which were found to be differentially expressed in the present microarray analysis (Supplementary Data set S2) and which are described as CK marker genes (Bhargava et al., 2013). As observed in Fig. 7, an accumulation of transcripts in response to either exudates or CK treatments was observed in the case of all marker genes, whereas no induction was observed in control conditions, which suggests that B. napus root exudates trigger a CK-like transcriptional response. Moreover, concomitant treatments with root exudates or c/tZ at optimum concentration (10–7 M) together with known antagonists of CK (NAA or ABA) decreased the number of EHSs at the apex of P. ramosa radicles (Table 2B). Indeed, induction triggered by c/tZ at 10–7 M or root exudates was completely abolished at 10–6 M and 10–5 M NAA, respectively. An ABA co-treatment at 10–6 M and 10–5 M decreased the induction triggered by c/tZ at 10–7 M or root exudates of ~50% of ABA.
Fig. 7.
Brassica napus root exudates trigger the expression of cytokinin-responsive genes. (A–C) Expression levels of cytokinin-responsive genes (PrRR5, PrCKX2, PrCKX4, and PrZFP6) in P. ramosa germinated seeds treated for 24 h with either B. napus root exudates, BAP (10–5 M), c/tZ (10–7 M), or purified fraction 9 measured by RT-qPCR. The values correspond to the fold change compared with the control treated germinated seeds. The control condition corresponds to a treatment with buffer solution (0.5 mM HEPES, 0.05% PPM) supplemented with the corresponding solvent. Exudates and fraction 9 were half-diluted and 130-fold diluted in buffer solution (0.5 mM HEPES, 0.05% PPM), respectively. Two biological replicates were performed, each in three technical replicates. Means are values ±SD (n=6). Means annotated with asterisks are significantly different from the control points (t-test, ***P<0.001).
Characterization of HIFs released by Brassica napus roots
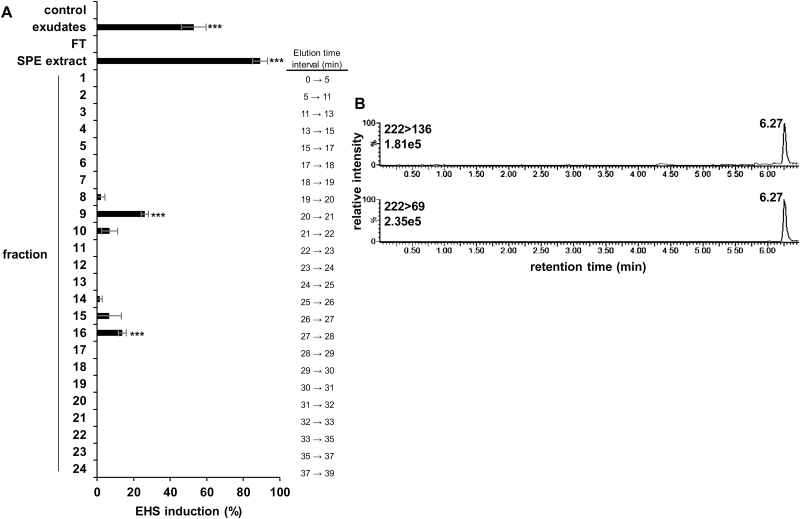
Brassica napus plants were grown hydroponically for 5 weeks and root exudates from weeks 3, 4, and 5 were collected and assayed for EHS induction. Active crude exudates were pooled, extracted by SPE, and separated by a RP-HPLC in 24 fractions. EHS induction was assessed throughout the purification process. The activity of B. napus root exudates was efficiently concentrated in the SPE extract (Fig. 8A). Regarding the different RP-HPLC fractions, EHS induction activity was significantly associated with only fractions 9 and 16 which eluted at 20–21 min and 27–28 min, respectively (Fig. 8A). The retention times of each fraction were compared with those of known CKs and with compounds previously tested for haustorium induction activity in hemiparasites (Kim et al., 1998; Albrecht et al., 1999). The results showed that fraction 16 (27–28 min) co-eluted with isorhamnetin which had no haustorium induction activity on P. ramosa (Supplementary Tables S4A, S5B), and interestingly fraction 9 co-eluted with different CK standards displaying EHS induction, including c/tZ, tZ, tZR, tZROG, DHZ, DHZR, and DHZROG (Supplementary Table S5A; Fig. 5A, B).
Fig. 8.
Fractionation of B. napus root exudates. (A) Distribution of EHS induction activity after RP-HPLC fractionation of SPE concentrated B. napus root exudates. Half-diluted exudates and SPE extracts (1000-fold diluted) were used as positive controls, Coïc medium was used as negative control. HPLC-purified fractions were 100-fold diluted. All dilutions were performed in buffer solution (0.5 mM HEPES, 0.05% PPM). Means are values ±SE (n=3). Means annotated with asterisks are significantly different from the negative control (t-test, ***P<0.001). (B). UPLC-ESI(+)-MS/MS analysis of fraction 9 detected in the MRN channel of DHZ.
All fractions were further examined for the presence of 12 known CKs by using UPLC-ESI(+)-/MS/MS. No tested CK standard was detected in any fractions. Nevertheless, two transitions of m/z 222>136 and 222>69 normally associated with DHZ were detected specifically in fraction 9 at a retention time of 6.27 min (Fig. 8B; Supplementary Table S6). These results show that the active fraction 9 contains a CK which shares a DHZ structure. Moreover, as observed in response to crude root exudates, fraction 9 also showed a CK-inducing potential at the gene expression level since all four tested marker genes were induced in response to this fraction (Fig. 7).
Discussion
The molecular determinism of the key development step of haustorium formation is largely unknown in root holoparasitic plants (Joel and Losner-Goshen, 1994). In the present study, we demonstrated that radicles of P. ramosa germinated seeds treated with uninfected B. napus root exudates displayed a growth arrest, a swollen apex, and epidermal cell outgrowth corresponding to EHS development previously described in either hemiparasitic or holoparasitic plants (Baird and Riopel, 1983; Joel and Losner-Goshen, 1994; Bandaranayake and Yoder, 2013; Cui et al., 2016). These results show that B. napus roots constitutively produce active molecules which trigger durable changes in cell expansion, cell division, and cell differentiation at the apex of P. ramosa radicles.
Haustorium formation has been studied for decades in either holoparasites or facultative and obligate hemiparasitic plants and, whereas EHS formation has been shown to be an inducible process in the latter, the role of these structures in the overall aggressiveness of the parasite has long been uncertain and linked to the presence of attachment substances at the surface of haustorial hairs or papillae (Baird and Riopel, 1983; Joel and Losner-Goshen, 1994). The present study highlights the role of EHS development in P. ramosa aggressiveness. Indeed, a strong increase in the number of broomrape attachments (tubercles) was observed when germinated seeds were pre-treated with B. napus root exudates to induce EHSs before placing them in contact with B. napus roots (Fig. 2). Similarly, a recent study in P. japonicum showed that plants displaying the haustorial hair defective (hhd) mutation fail to form haustorial hairs upon DMBQ treatment and form fewer functional haustoria when put in contact with host roots (Cui et al., 2016).
The transcriptome assembly obtained in this work yielded 53 511 contigs, 51% and 55% of which have a BLASTx hit with the Arabidopsis TAIR 10 database and the S. lycopersicum (GeneBank 160) genome, respectively. Although this value is weaker than the one found in a recent study in P. japonicum (60.23%) with 57 939 unigenes (Ishida et al., 2016), our de novo assembled transcriptome still provides a reasonable coverage of P. ramosa genes. The P. ramosa microarray analysis focused on time points surrounding EHS formation in response to B. napus root exudates. GO term enrichment analysis of the overall DEGs did not reveal any over- representation of GO terms linked to reactive oxygen species (ROS) generation or response but an under-representation of the GO term GO:0016491 oxidoreductase activity. This finding differs from the over-representation of this GO term observed in previous transcriptomic analyses focusing on haustorium development in hemiparasitic species in response to DMBQ treatment (Yang et al., 2015; Ishida et al., 2016). Indeed, although ROS-related genes have been previously proposed to play an important role in the signaling leading to haustorium development in these species (Matvienko et al., 2001b; Bandaranayake et al., 2010; Ngo et al., 2013; Yang et al., 2015; Ishida et al., 2016), ROS-related compounds such as DMBQ have no effect on haustorium formation in P. ramosa. The GO term pollination (GO:0009856) was over-represented in P. ramosa germinated seeds in response to root exudates, which might be surprising but it has previously been proposed that haustorial growth may be related to pollen tube growth (Thorogood and Hiscock, 2010; Pérez-de-Luque, 2013), and genes linked to this process were also detected during haustorium formation in hemiparasitic plants (Yang et al., 2015). The GO analysis also revealed an increase inthe expression of genes related to signaling pathway, hormone-mediated signaling pathway, or regulation of transcription. This shift in the GO terms underlines the involvement of hormone signaling, which was previously described during haustorium development (Tomilov et al., 2005; Ishida et al., 2016). Indeed, auxin induces the formation of lateral haustoria in T. versicolor and P. japonicum. In contrast, it inhibits haustorium formation in the obligate hemiparasite S. asiatica which, like P. ramosa, forms terminal haustoria (Keyes et al., 2000; O’Malley and Lynn, 2000; Tomilov et al., 2005; Ishida et al., 2016). The present study confirms those results since auxin did not show any EHS induction on P. ramosa germinated seeds and had an inhibitory effect when applied together with root exudates.
Among the different plant hormones tested in this study, only c/tZ triggered EHS formation with a maximum activity similar to B. napus root exudate treatment. Although the synthetic CK kinetin has already been shown to act in a similar way to DMBQ by inducing EHS formation in S. asiatica (Keyes et al., 2000; O’Malley and Lynn, 2000), the swollen apex induced by this hormone was described as smaller than the one induced by DMBQ treatment whereas haustorial hair growth was exaggerated, giving rise to a somewhat distorted haustorium with no demonstrated functionality. The results presented here show that in contrast to DMBQ which has no effect, CKs trigger EHS formation in P. ramosa and increase the parasite aggressiveness. Moreover, when an array of CKs was tested, the SAR observed during EHS induction matched the one previously described in other development processes such as callus growth (Matsubara, 1980). The CK signal is mediated by the histidine kinase receptors CRE1/AHK4, AHK3, and AHK2 in A. thaliana (Inoue et al., 2001; Suzuki et al., 2001; Ueguchi et al., 2001; Yamada et al., 2001; Nishimura et al., 2004), which act as a two-component signaling pathway (Hwang et al., 2002). PI-55 is a purine derivative which specifically competitively inhibits the binding of BAP to CRE1/AHK4 in A. thaliana and thus disrupts this two-component signaling pathway. However, this molecule also displays a partial agonist activity on CK perception (Spíchal et al., 2009). In the present study, the co-treatment of P. ramosa germinated seeds with PI-55 together with BAP yielded a massive reduction in the number of the observed EHSs but without completely abolishing their formation. Pharmacological approaches using PI-55 usually fail to be 100% successful. PI-55 effectively blocks the activity of CRE1/AHK4 CK receptor in Arabidopsis, but at high concentrations it has its own weak CK activity, most probably through another receptor (AHK3) (Spíchal et al., 2009). This could explain the observed ‘mixed’ behavior of PI-55 at high concentrations (10−6 M and 10−5 M), which can cause an incomplete blocking of EHS development, especially when CK activity promotes it. PI-55 treatment also reduced the overall aggressiveness of P. ramosa when applied with BAP in comparison with BAP alone, suggesting that the signaling pathway leading to EHS formation involves histidine kinase receptors homologous to CRE1/AHK4 and AHK3. A transcript accumulation analysis of a set of known CK-responsive genes conducted during EHS induction in P. ramosa with either B. napus root exudates or CKs (c/tZ and BAP) revealed similar gene expression patterns. Interestingly, bio-guided RP-HPLC fractionation of B. napus root exudates revealed that two fractions (9 and 16) displayed high EHS induction activities and fraction 9 co-eluted with c/tZ, tZ, tZR, tZROG, DHZ, DHZR, and DHZROG standards. A subsequent UPLC-ESI(+)-/MS/MS analysis revealed that only the active fraction 9 contained a molecule displaying the transition m/z 222>136 and 222>69, which are normally associated with DHZ. This finding is also concordant with the CK-inducing potential of this fraction here demonstrated at the gene expression level. Although a further determination of the exact structure of this CK is required as well as the identification of the active compound contained in fraction 16, these results suggest that B. napus root exudates contain CKs. Such a finding is in accordance with previous results since the presence of plant-derived CKs has already been mentioned in the rhizosphere of tomato and rice (Davey and van Staden, 1976; Soejima et al., 1992; Yang et al., 2002).
CKs are known to control A. thaliana root growth and shape through multiple nodes of interaction with auxin (Schaller et al., 2015). In this complex interplay, CKs tend to inhibit root growth by reducing the size of the root apical meristem and to promote cell differentiation (Dello Ioio et al., 2007) through the involvement of the Aux/IAA protein SHY2 (Dello Ioio et al., 2008; Moubayidin et al., 2010). This demonstrated role of CKs is consistent with the phenotypical changes such as root growth arrest and differentiation of papillae from epidermal cells that occur upon B. napus root exudate or direct CK treatments at the root apex of P. ramosa germinated seeds. Ishida et al. (2016) have shown that auxin levels increased during haustorium induction and that the overexpression of YUC3, an auxin biosynthesis gene, was sufficient to induce EHS formation in the hemiparasitic species P. japonicum. A direct auxin treatment does not induce EHS formation in obligate root parasitic plants such as P. ramosa and S. hermontica that form terminal haustoria (Keyes et al., 2000; O’Malley and Lynn, 2000; this study). However, it is effective in facultative hemiparasitic plants that form lateral haustoria (Tomilov et al., 2005; Ishida et al., 2016). Interestingly, CKs have been shown to be effective in both obligate and facultative parasitic plants. Although this suggests that CKs and auxin may interact to shape lateral haustorium formation in facultative parasitic plants and CKs alone may govern the establishment of the terminal haustorium in obligate root parasitic plants, more detailed studies are required on the involvement of such an interplay in this specific root development.
The results presented here demonstrate that EHS formation in P. ramosa is an inducible process under the control of host root exudate compounds, notably of an as yet unidentified CK contained in B. napus root exudates. This sheds new light on the establishment of the haustorium in parasitic plants by putting an emphasis on the involvement of host-exudated CKs as HIFs.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Stock solution used.
Table S2. List of primers used for RT-qPCR analysis.
Table S3. Summary of the de novo assembly of the P. ramosa transcriptome.
Table S4. Effect of treatments with known HIFs and solvent effect on haustorium formation in P. ramosa.
Table S5. Chromatographic behavior of standard compounds during RP-HPLC fractionation.
Table S6. Chromatographic behavior of cytokinin standard compounds during UPLC-ESI(+)-MS/MS analysis.
Fig. S1. Dose–response effect of exudate dilution.
Fig. S2. RT-qPCR validation of the expression profiles of selected genes.
Data set S1. GO term enrichment analysis on de novo assembly of the P. ramosa transcriptome.
Data set S2. GO term enrichment analysis on the DEG set.
Data deposition. Transcriptome sequence data have been deposited in the Parasitic Plant Genome Project (Westwood et al., 2012; Yang et al., 2015; http://ppgp.huck.psu.edu/). Microarray data have been deposited in the Gene Expression Omnibus under the accession numbers GPL23512 and GSE99377.
Supplementary Material
Acknowledgements
VG and EB were financially supported by PhD grants from the French Ministry of Education and Research and from the Region Pays de Loire (RFI Objectif Végétal project), respectively. We thank the ANAN platform of the SFR Quasav, the IBiSA platform ‘Biopolymers-Structural Biology’ (INRA Research Unit 1268 BIA), and the LABERCA laboratory of the CORSAIRE facility for access to microarray facilities, microscopy analyses, and UPLC-MS analyses, respectively. LS was supported by the Ministry of Education, Youth and Sports of the Czech Republic (grant LO1204 from the National Program of Sustainability).
Glossary
Abbreviation:
- CK
cytokinin
- DEG
differentially expressed gene
- EHS
early haustorial structure
- GO
Gene Ontology
- HIF
haustorium-inducing factor.
References
- Albrecht H, Yoder JI, Phillips DA. 1999. Flavonoids promote haustoria formation in the root parasite Triphysaria versicolor. Plant Physiology 119, 585–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird WV, Riopel JL. 1983. Experimental studies of the attachment of the parasitic angiosperm Agalinis purpurea to a host. Protoplasma 118, 206–218. [Google Scholar]
- Baird WV, Riopel JL. 1985. Surface characteristics of root and haustorial hairs of parasitic Scrophulariaceae. Botanical Gazette 146, 63–69. [Google Scholar]
- Bandaranayake PC, Filappova T, Tomilov A, Tomilova NB, Jamison-McClung D, Ngo Q, Inoue K, Yoder JI. 2010. A single-electron reducing quinone oxidoreductase is necessary to induce haustorium development in the root parasitic plant Triphysaria. The Plant Cell 22, 1404–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandaranayake PCG, Yoder JI. 2013. Haustorium initiation and early development. In: Joel DM, Gressel J, Musselman LJ, eds. Parasitic Orobanchaceae. Berlin: Springer, 61–74. [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B: Methodological 57, 289–300. [Google Scholar]
- Bhargava A, Clabaugh I, To JP, Maxwell BB, Chiang YH, Schaller GE, Loraine A, Kieber JJ. 2013. Identification of cytokinin-responsive genes using microarray meta-analysis and RNA-Seq in Arabidopsis. Plant Physiology 16, 279–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celton JM, Dheilly E, Guillou MC, Simonneau F, Juchaux M, Costes E, Laurens F, Renou JP. 2014. Additional amphivasal bundles in pedicel pith exacerbate central fruit dominance and induce self-thinning of lateral fruitlets in apple. Plant Physiology 164, 1930–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Lynn DG. 1986. The haustorium and the chemistry of host recognition in parasitic angiosperms. Journal of Chemical Ecology 12, 561–579. [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, Bouchet B, Alvarado C, Bakan B, Guillon F. 2014. The wheat grain contains pectic domains exhibiting specific spatial and development-associated distribution. PLoS One 9, e89620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coic Y, Lesaint C. 1975. La nutrition minérale et en eau des plantes en horticulture avancée. Doc Tech SCPA. [Google Scholar]
- Cui S, Wakatake T, Hashimoto K, Saucet SB, Toyooka K, Yoshida S, Shirasu K. 2016. Haustorial hairs are specialized root hairs that support parasitism in the facultative parasitic plant Phtheirospermum japonicum. Plant Physiology 170, 1492–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey JE, van Staden J. 1976. Cytokinin translocation: changes in zeatin and zeatin-riboside levels in the root exudate of tomato plants during their development. Planta 130, 69–72. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S. 2007. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Current Biology 17, 678–682. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S. 2008. A genetic framework for the control of cell division and differentiation in the root meristem. Science 322, 1380–1384. [DOI] [PubMed] [Google Scholar]
- Depuydt S, Trenkamp S, Fernie AR, Elftieh S, Renou JP, Vuylsteke M, Holsters M, Vereecke D. 2009. An integrated genomics approach to define niche establishment by Rhodococcus fascians. Plant Physiology 149, 1366–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. 2010. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Research 38, W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte JM, Wall PK, Edger PP, Landherr LL, Ma H, Pires JC, Leebens-Mack J, dePamphilis CW. 2010. Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels. BMC Evolutionary Biology 10, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Aparicio M, Masi M, Maddau L, Cimmino A, Evidente M, Rubiales D, Evidente A. 2016. Induction of haustorium development by sphaeropsidones in radicles of the parasitic weeds striga and orobanche. A structure–activity relationship study. Journal of Agricultural and Food Chemistry 64, 5188–5196. [DOI] [PubMed] [Google Scholar]
- Holub J, Hanuš J, Hanke DE, Strnad M. 1998. Biological activity of cytokinins derived from ortho- and meta-hydroxybenzyladenine. Plant Growth Regulation 26, 109–115. [Google Scholar]
- Hood ME, Condon JM, Timko MP, Riopel JL. 1998. Primary haustorial development of Striga asiatica on host and nonhost species. Phytopathology 88, 70–75. [DOI] [PubMed] [Google Scholar]
- Hwang I, Chen HC, Sheen J. 2002. Two-component signal transduction pathways in Arabidopsis. Plant Physiology 129, 500–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T. 2001. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409, 1060–1063. [DOI] [PubMed] [Google Scholar]
- Ishida JK, Wakatake T, Yoshida S, Takebayashi Y, Kasahara H, Wafula E, dePamphilis CW, Namba S, Shirasu K. 2016. Local auxin biosynthesis mediated by a YUCCA flavin monooxygenase regulates haustorium development in the parasitic plant Phtheirospermum japonicum. The Plant Cell 28, 1795–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida JK, Yoshida S, Shirasu K. 2017. Quinone oxidoreductase 2 is involved in haustorium development of the parasitic plant Phtheirospermum japonicum. Plant Signaling and Behavior 12, e1319029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joel DM, Losner-Goshen D. 1994. The attachment organ of the parasitic angiosperms Orobanche cumana and O. aegyptiaca and its development. Canadian Journal of Botany 72, 564–574. [Google Scholar]
- Keyes WJ, O’Malley RC, Kim D, Lynn DG. 2000. Signaling organogenesis in parasitic angiosperms: xenognosin generation, perception, and response. Journal of Plant Growth Regulation 19, 217–231. [DOI] [PubMed] [Google Scholar]
- Keyes WJ, Palmer AG, Erbil WK, Taylor JV, Apkarian RP, Weeks ER, Lynn DG. 2007. Semagenesis and the parasitic angiosperm Striga asiatica. The Plant Journal 51, 707–716. [DOI] [PubMed] [Google Scholar]
- Kim D, Kocz R, Boone L, Keyes WJ, Lynn DG. 1998. On becoming a parasite: evaluating the role of wall oxidases in parasitic plant development. Chemistry and Biology 5, 103–117. [DOI] [PubMed] [Google Scholar]
- Lechat MM, Brun G, Montiel G, Véronési C, Simier P, Thoiron S, Pouvreau JB, Delavault P. 2015. Seed response to strigolactone is controlled by abscisic acid-independent DNA methylation in the obligate root parasitic plant, Phelipanche ramosa L. Pomel. Journal of Experimental Botany 66, 3129–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechat MM, Pouvreau JB, Péron T et al. . 2012. PrCYP707A1, an ABA catabolic gene, is a key component of Phelipanche ramosa seed germination in response to the strigolactone analogue GR24. Journal of Experimental Botany 63, 5311–5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn DG, Steffens JC, Kamut VS, Graden DW, Shabanowitz J, Riopel JL. 1981. Isolation and characterization of the first host recognition substance for parasitic angiosperms. Journal of the American Chemical Society 103, 1868–1870. [Google Scholar]
- Matsubara S. 1980. Structure–activity relationships of cytokinins. Phytochemistry 19, 2239–2253. [Google Scholar]
- Matvienko M, Torres MJ, Yoder JI. 2001a. Transcriptional responses in the hemiparasitic plant Triphysaria versicolor to host plant signals. Plant Physiology 127, 272–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matvienko M, Wojtowicz A, Wrobel R, Jamison D, Goldwasser Y, Yoder JI. 2001b. Quinone oxidoreductase message levels are differentially regulated in parasitic and non-parasitic plants exposed to allelopathic quinones. The Plant Journal 25, 375–387. [DOI] [PubMed] [Google Scholar]
- Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. 2016. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Research 44, D336–D342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moubayidin L, Perilli S, Dello Ioio R, Di Mambro R, Costantino P, Sabatini S. 2010. The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Current Biology 20, 1138–1143. [DOI] [PubMed] [Google Scholar]
- Ngo QA, Albrecht H, Tsuchimatsu T, Grossniklaus U. 2013. The differentially regulated genes TvQR1 and TvPirin of the parasitic plant Triphysaria exhibit distinctive natural allelic diversity. BMC Plant Biology 13, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C. 2004. Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. The Plant Cell 16, 1365–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley RC, Lynn DG. 2000. Expansin message regulation in parasitic angiosperms: marking time in development. The Plant Cell 12, 1455–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-de-Luque A. 2013. Haustorium invasion into host tissues. In: Joel DM, Gressel J, Musselman LJ, eds. Parasitic Orobanchaceae. Berlin: Springer, 75–86. [Google Scholar]
- Péron T, Candat A, Montiel G, Veronesi C, Macherel D, Delavault P, Simier P. 2016. New insights into phloem unloading and expression of sucrose transporters in vegetative sinks of the parasitic plant Phelipanche ramosa L. (Pomel). Frontiers in Plant Science 7, 2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team 2009. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Schaller GE, Bishopp A, Kieber JJ. 2015. The yin–yang of hormones: cytokinin and auxin interactions in plant development. The Plant Cell 27, 44–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soejima H, Sugiyama T, Ishihara K. 1992. Changes in cytokinin activities and mass spectrometric analysis of cytokinins in root exudates of rice plant (Oryza sativa L.): comparison between cultivars Nipponbare and Akenohoshi. Plant Physiology 100, 1724–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spíchal L. 2012. Cytokinins—recent news and views of evolutionally old molecules. Functional Plant Biology 39, 267–284. [DOI] [PubMed] [Google Scholar]
- Spíchal L, Werner T, Popa I, Riefler M, Schmülling T, Strnad M. 2009. The purine derivative PI-55 blocks cytokinin action via receptor inhibition. FEBS Journal 276, 244–253. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Miwa K, Ishikawa K, Yamada H, Aiba H, Mizuno T. 2001. The Arabidopsis sensor His-kinase, AHk4, can respond to cytokinins. Plant and Cell Physiology 42, 107–113. [DOI] [PubMed] [Google Scholar]
- Svačinová J, Novák O, Plačková L, Lenobel R, Holík J, Strnad M, Doležal K. 2012. A new approach for cytokinin isolation from Arabidopsis tissues using miniaturized purification: pipette tip solid-phase extraction. Plant Methods 8, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorogood CJ, Hiscock SJ. 2010. Compatibility interactions at the cellular level provide the basis for host specificity in the parasitic plant Orobanche. New Phytologist 186, 571–575. [DOI] [PubMed] [Google Scholar]
- Tomilov AA, Tomilova NB, Abdallah I, Yoder JI. 2005. Localized hormone fluxes and early haustorium development in the hemiparasitic plant Triphysaria versicolor. Plant Physiology 138, 1469–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi C, Koizumi H, Suzuki T, Mizuno T. 2001. Novel family of sensor histidine kinase genes in Arabidopsis thaliana. Plant and Cell Physiology 42, 231–235. [DOI] [PubMed] [Google Scholar]
- Wall PK, Leebens-Mack J, Müller KF, Field D, Altman NS, dePamphilis CW. 2008. PlantTribes: a gene and gene family resource for comparative genomics in plants. Nucleic Acids Research 36, D970–D976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westwood JH, Yoder JI, Timko MP, dePamphilis CW. 2010. The evolution of parasitism in plants. Trends in Plant Science 15, 227–235. [DOI] [PubMed] [Google Scholar]
- Westwood JH, dePamphilis CW, Das M, Fernández-Aparicio M, Honaas LA, Timko MP, Wafula EK, Wickett NJ, Yoder JI. 2012. The Parasitic Plant Genome Project: new tools for understanding the Biology of Orobanche and Striga. Weed Science 60, 295–306. [Google Scholar]
- Yamada H, Suzuki T, Terada K, Takei K, Ishikawa K, Miwa K, Yamashino T, Mizuno T. 2001. The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant and Cell Physiology 42, 1017–1023. [DOI] [PubMed] [Google Scholar]
- Yang J, Zhang J, Wang Z, Zhu Q, Liu L. 2002. Abscisic acid and cytokinins in the root exudates and leaves and their relationship to senescence and remobilization of carbon reserves in rice subjected to water stress during grain filling. Planta 215, 645–652. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wafula EK, Honaas LA et al. . 2015. Comparative transcriptome analyses reveal core parasitism genes and suggest gene duplication and repurposing as sources of structural novelty. Molecular Biology and Evolution 32, 767–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Cui S, Ichihashi Y, Shirasu K. 2016. The haustorium, a specialized invasive organ in parasitic plants. Annual Review of Plant Biology 67, 643–667. [DOI] [PubMed] [Google Scholar]
- Zhang X, Berkowitz O, Teixeira da Silva JA, Zhang M, Ma G, Whelan J, Duan J. 2015. RNA-Seq analysis identifies key genes associated with haustorial development in the root hemiparasite Santalum album. Frontiers in Plant Science 6, 661. doi:10.3389/fpls.2015.00661 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.