CD4 T-cell transcriptomic results reveal significant changes associated with disease severity in primary respiratory syncytial virus infection, with evidence of Th2 skewing, and provide insight into early T-cell responses associated with disease severity.
Keywords: RNA sequencing, respiratory syncytial virus, T cell, disease severity, gene espression
Abstract
Background
Nearly all children are infected with respiratory syncytial virus (RSV) within the first 2 years of life, with a minority developing severe disease (1%–3% hospitalized). We hypothesized that an assessment of the adaptive immune system, using CD4+ T-lymphocyte transcriptomics, would identify gene expression correlates of disease severity.
Methods
Infants infected with RSV representing extremes of clinical severity were studied. Mild illness (n = 23) was defined as a respiratory rate (RR) < 55 and room air oxygen saturation (SaO2) ≥ 97%, and severe illness (n = 23) was defined as RR ≥ 65 and SaO2 ≤ 92%. RNA from fresh, sort-purified CD4+ T cells was assessed by RNA sequencing.
Results
Gestational age, age at illness onset, exposure to environmental tobacco smoke, bacterial colonization, and breastfeeding were associated (adjusted P < .05) with disease severity. RNA sequencing analysis reliably measured approximately 60% of the genome. Severity of RSV illness had the greatest effect size upon CD4 T-cell gene expression. Pathway analysis identified correlates of severity, including JAK/STAT, prolactin, and interleukin 9 signaling. We also identified genes and pathways associated with timing of symptoms and RSV group (A/B).
Conclusions
These data suggest fundamental changes in adaptive immune cell phenotypes may be associated with RSV clinical severity.
Respiratory syncytial virus (RSV), the most important cause of respiratory tract illness in infants and young children, infects 50%–70% of infants during the first year of life [1]. Although most infections are relatively mild, 1%–3% of infected infants require hospitalization, accounting for 74000–126000 admissions of infants aged < 1 year annually in the United States [2, 3]. Additionally, RSV-related emergency department visits for infants aged ≤ 1 year of age range from 39 to 69 per 1000, and RSV-related office visits are 3 times as many [4]. Although many risk factors for severe disease are recognized, such as prematurity, congenital heart disease, pulmonary disease, and neurologic and immunosuppressive conditions, the majority of infants brought to medical attention are healthy full-term infants. Host and environmental factors also associated with more severe disease, although less overt, are male sex, lack of breastfeeding, tobacco smoke exposure, and low levels of maternally derived protective antibody [5].
Host immune responses are also thought to influence disease manifestations, including severity [6–9]. Experimental animal models of RSV infection suggest that Th2 CD4 immune-dominant responses, as well as diminished or impaired anti-inflammatory T-regulatory (Treg) cell function and increased Th17 responses, contribute to increased lung pathology [6, 7, 10]. Data from infant studies are less compelling, with some supporting and others refuting that a T2-dominant response is responsible for severe disease [9, 11–15].
Recently gene expression analysis of whole blood collected during infection has been used to assess the immune response to RSV [16–18]. One study noted increased expression of interferon signaling and neutrophil gene pathways and diminished T- and B-cell gene pathways [17]. Because CD4 T cells are critical in the development of adaptive immunity following infection and also influence the degree of inflammation, we sought to investigate gene expression patterns using high-throughput RNA sequencing (RNA-seq) of isolated CD4 T cells in healthy full-term infants aged <10 months at the time of primary RSV infection by comparing infants with mild and severe clinical disease. We identified unique gene expression patterns, implicating biological pathways associated with disease severity, which provide novel insight into pathogenesis of RSV infection in this population.
METHODS
Subjects
The Research Subject Review Boards of the University of Rochester Medical Center (URMC) and Rochester General Hospital approved the study, and a parent provided written informed consent. Respiratory syncytial virus–infected infants were selected for this analysis from 3 cohorts as part of the AsPIRES study of RSV pathogenesis. A birth cohort, enrolled between August and December of both 2012 and 2013, were followed by a combination of passive and active surveillance for development of RSV infection during the subsequent winter. Infants with respiratory symptoms were evaluated at home for RSV infection using an RSV group–specific reverse transcriptase-polymerase chain reaction (RT-PCR) assay [19]. A second cohort was enrolled when seen for acute respiratory symptoms in pediatric offices or emergency rooms and tested for RSV by antigen detection (Quidel, San Diego, CA) and/or RT-PCR. The third cohort consisted of infants hospitalized with RSV at URMC’s Golisano Children’s Hospital. Eligible subjects were full-term (>36 wk gestation) healthy infants born after the previous May 1 and aged <10 months at RSV infection to ensure primary infection. The hospitalized infants were seen daily until discharged, and charts were reviewed for signs of respiratory illness and lowest room air oxygen saturation (SaO2).
Specimen Collection
Infants were evaluated by 2 members of the study team (a physician and a project nurse). Demographic data, illness symptoms, findings on physical examination, and results of standard laboratory and chest radiograph results, when available, were recorded and defined as visit 1 (acute). Illness onset was determined by physician interview of parent(s) during evaluation. Following evaluation, a nasal swab using an infant-sized flocked swab (FLOQSwabs catalog no. 525CS01, Copan, Murrieta, CA) was placed in 2 mL of sterile ultraviolet-inactivated water and 2–3 mL of heparinized blood collected. A second blood sample was collected during a second visit (convalescent) 12–16 days after illness onset.
Detection of Pathogenic Virus and Bacteria
The nasal swab was used for detection of other respiratory virus coinfection, Streptococcus pneumoniae, and Haemophilus influenzae using a TaqMan multiplex assay and separately for Moraxella catarrhalis according to published methods [20].
Cell Purification and RNA Isolation
Heparinized blood was maintained at room temperature for up to 2 hours, and peripheral blood mononuclear cells were isolated by Ficoll-hypaque gradient, flow-sorted into subsets including CD3+CD4+CD8− lymphocytes, and stored in RNA lysis buffer at −20°C [21].
Library Construction and Sequencing
RNA sequencing was performed as previously described, starting with 1 ng of RNA and using the SMARter Ultra Low amplification kit (Clontech, Mountain View, CA) [20]. Libraries were constructed using the NexteraXT library kit (Illumina, San Diego, CA) and sequenced on the Illumina HiSeq2500 to generate approximately 20 million 100-bp single end-reads per sample. Preanalysis data processing was as previously described [20].
Univariate and Multivariate Gene Selection
For categorical variables, we used SAMseq to identify genes with significant differences in mean expression (q < 0.05). Both Pearson and Spearman correlation tests, with Benjamini-Hochberg correction to control false discovery rate at a 0.05 level, were used to select genes with significant correlation with continuous variables. We also conducted a multivariate linear mixed-effects regression analysis to study the linear association between the expression of individual genes (response variables) and various important demographic, clinical, and environment variables (see Supplementary Methods).
Pathway Analysis and Transcription Factor Target Identification
Differentially expressed genes were used for canonical pathway identification and upstream regulator analysis using Ingenuity Pathway Analysis (Qiagen, Redwood City, CA). To assess T-cell transcription factor activity, we interpreted CD4 gene expression patterns associated with severity using Bayesian estimation of transcription factor activity [22, 23].
Quantitative Polymerase Chain Reaction
Quantitative polymerase chain reaction was performed on selected genes for confirmation of the gene expression as described [20].
Statistical Analysis of Clinical Data
A P value < .05 was considered statistically significant. For continuous clinical variables, we performed 2-sample Welch t tests to check the equality of mean values between 2 patient groups defined by disease severity. For binary variables, Fisher’s exact test was used. Breastfeeding was modeled as a categorical variable (None < Some < Exclusive) and tested using Spearman’s rank correlation.
RESULTS
Subjects and Clinical Characteristics
Of 86 RSV-infected infants enrolled in the study, we selected 46 representing the extreme ends of the severity spectrum: 23 with mild disease (n = 10 birth cohort, n = 13 second cohort), defined as maximum respiratory rate (RRmax) < 55 per minute and SaO2 ≥ 97%; and 23 with severe disease (all from the hospitalized cohort), defined as RRmax ≥ 65 and SaO2 ≤ 92%. Subjects with more severe illness were significantly younger (2.2 vs 4.0 months; P < .01), more likely to be exposed to environmental tobacco smoke (23% vs 4%; P = .01), and less likely to be breastfed (65% vs 87%; P = .05) (Table 1). Viral coinfection was similar between groups. The groups were equally colonized with Moraxella catarrhalis, but the more severely ill group was significantly more likely to be colonized with S. pneumoniae and/or H. influenzae (65% vs 30%; P = .04). Although all infants were considered full term, those with severe clinical symptoms were born at slightly lower gestational age.
Table 1.
Demographic, Clinical, and Microbiological Data for 46 Respiratory Syncytial Virus–Infected Infants
| Clinical variable | Mild disease (n = 23) | Severe disease (n = 23) | P value |
|---|---|---|---|
| Gestational age, wk; mean ± SE | 39.6 ± 1.6 | 38.4 ± 1.4 | .05 |
| Birth weight, kg; mean ± SE | 3.4 ± .05 | 3.4 ± .08 | .92 |
| Gender, male; no. (%) | 11 (48) | 11 (48) | 1.00 |
| Race; no. (%) | |||
| Caucasian only | 13 (57) | 14 (61) | |
| African American only | 7 (30) | 5 (22) | .84 |
| Other | 3 (13) | 4 (17) | |
| Age, mo.; mean ± SE | 4.0 ± 2.2 | 2.0 ± 1.4 | <.01 |
| Days of illness; mean ± SE | 5.0 ± 1.9 | 5.1 ± 2.2 | .83 |
| Breast feedinga; no. (%) | |||
| Exclusive | 16 (70) | 10 (43) | |
| Some | 4 (17) | 5 (22) | .05 |
| Never | 3 (13) | 4 (17) | |
| Exposure to tobacco smoke; no. (%) | 1 (4) | 9 (23) | .01 |
| Hospitalized; no. (%) | 1 (4) | 23 (100) | <.0001 |
| RSV group A, B; no. each group | 13, 10 | 10, 12 | .56 |
| Viral coinfectionb; no. (%) | 7 (30) | 4 (17) | .49 |
| Bacterial colonizationc; no. (%) | |||
| Streptococcus pneumoniae and/or Haemophilus influenza | 7 (30) | 15 (65) | .04 |
| Moraxella catarrhalis | 12 (52) | 9 (39) | .55 |
| Wheezing; no. (%) | 2 (9) | 22 (96) | <.0001 |
| Rales/rhonchi; no. (%) | 1 (4) | 21 (91) | <.0001 |
| Retractions; no. (%) | 6 (26) | 23 (100) | <.0001 |
| Cyanosis; no. (%) | 0 (0) | 3 (13) | .23 |
| Apnea; no. (%) | 0 (0) | 1 (4) | 1.00 |
| Poor air movement; no. (%) | 0 (0) | 9 (39) | .0004 |
| Lethargy; no. (%) | 0 (0) | 13 (57) | <.0001 |
| Max RR; mean ± SD; no. (%) | 43 ± 6 | 79 ± 12 | <.0001 |
| Worst SaO2; mean ± SD | 98 ± 1 | 83 ± 5 | <.0001 |
Abbreviations: RR, respiratory rate; SaO2, room air oxygen saturation.
a Defined as any breastfeeding after discharge from birth hospitalization.
b Mild disease group: human metapneumovirus (1), rhinovirus (1), parechovirus (1), coronavirus (3), coronavirus plus rhinovirus (1). Severe disease group: rhinovirus (1), coronavirus (2), adenovirus (1).
c Polymerase chain reaction– positive for Streptoccoccus pneumoniae, Haemophilus influenzae, or Moraxella catarrhalis in nasal swab specimen.
Global Adaptive Immune Cell Gene Expression Patterns
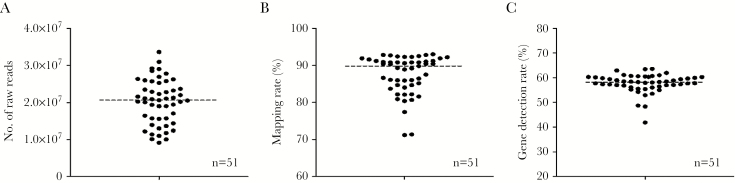
The CD3+/CD4+/CD8− (CD4+) T cells from this group of 46 infants were sorted and subjected to RNA-seq analysis. A total of 51 samples from 38 subjects (n = 19 in each group) passed quality control (QC), and the remaining samples were removed from analysis. The raw reads, mapping rate, and gene detection rates are shown in Figure 1. Samples averaged approximately 20M reads with >90% mapping rate, and our filtered analytical data set included expression values for 10446 genes. Interestingly CD4+ T cells appeared to express approximately 60% of the genome, consistent with prior data on sorted lymphocytes [21]. An assessment of cell type−specific markers (eg, CD3, CD4, CD8, CD19, MPO) confirmed the purity of the sorted cells (Supplementary Figure 1).
Figure 1.
Sequencing statistics of respiratory syncytial virus–infected CD4 samples. A, Number of total raw reads. B, Genome mapping rate of overall samples. C, Gene detection rate of 51 samples.
Univariate analysis was used to identify gene expression patterns associated with each clinical or demographic variable (Table 2). Our analysis demonstrated that clinical severity was associated with the greatest effect size upon gene expression (n = 551 genes). The effect size for clinical severity was much greater than that for sex (n = 74 genes), RSV group (A/B; n = 53 genes), or days since onset of clinical symptoms (n = 35 genes). Interestingly, we found that bacterial cocolonization seems to have a greater effect (n = 12 genes) than viral coinfection (n = 3 genes).
Table 2.
Univariate and Multivariate Regression Modeling to Identify Gene Expression Changes Associated With Intrinsic (Age, Days Since Onset Of Illness) and Extrinsic (Tobacco Smoke Exposure and Bacterial Colonization or Viral Infection) Factors
| Clinical variable | No. of genes univariate model |
No. of genes multivariate model |
|---|---|---|
| Illness severity | 551 | 140 |
| sex | 74 | 113 |
| Tobacco smoke exposure | 1 | 7 |
| Infecting respiratory syncytial virus group | 53 | 68 |
| Viral coinfection and/or bacterial colonization | 3 | 67 |
| Bacterial colonization with Streptococcus pneumonia and/or Haemophilus influenza | 12 | 44 |
| Gestational age | 1 | 41 |
| Visit age | 2 | 18 |
| Time since onset of illness; acute vs convalescent | 35 | 63 |
| Time since onset of illness; day of illness | 41 | 6 |
Shown are the numbers of genes identified as significant for each variable and model.
Appreciating the potential of confounding variables to influence identification of significant gene expression patterns, we constructed a multivariate linear mixed-effects regression model that included the variables with greatest marginal effects. Numbers of significant genes identified by this more conservative analysis are summarized in Table 2. Details for genes identified, including their relationships to individual variables, are provided in Supplementary Table 1. In this multivariate model, severity of illness continued to have the strongest association with gene expression (n = 140 genes), albeit now only slightly greater than sex (n = 113 genes). The multivariate model again showed significant gene expression associated with days since onset of clinical symptoms (n = 63 genes), RSV group (n = 68 genes), and gestationl age (n = 41 genes). In addition, the multivariate model also identified a substantial number of genes significantly associated with viral coinfection/bacterial colonization status (n = 67 genes) and found that pathogenic bacteria colonization alone is significantly associated with gene expression (n = 44 genes).
Gene Expression Correlates of Disease Severity
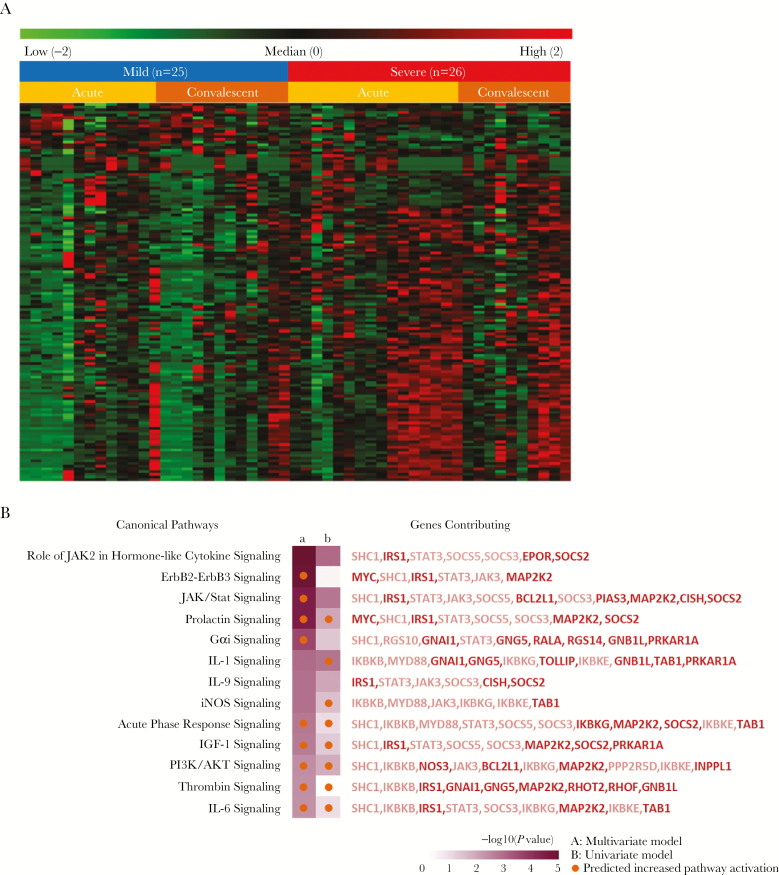
A heat map of the 140 genes associated with disease severity identified by multivariate analysis and stratified by time since onset of clinical symptoms is shown in Figure 2A. These data demonstrated population-level heterogeneity (particularly in the severely affected subjects) and suggested greater differences in gene expression in the acute phase than in the convalescent phase.
Figure 2.
Heat map and pathway analysis of CD4 T cell expression. A, Gene expression associated with clinical severity. Shown are normalized expression levels for the 140 genes selected by multivariate analysis; rows represent genes, and columns represent samples. Red indicates higher expression; green indicates low/no expression. Samples are grouped by phenotype (mild, severe) and by time when sample was obtained (acute, convalescent). B, Pathways associated with severe phenotype. Ingenuity pathway analysis (IPA) was used to identify canonical pathways represented by genes associated with severity in CD4 lymphocytes from respiratory syncytial virus–infected subjects. The variables used to generate gene sets for IPA were multivariate severity phenotype (a; n = 140) and univariate severity phenotype (b; n = 551). Thirteen pathways are shown where Fisher’s exact test P values were <.05 for at least 1 variable. Orange and blue circles indicate predicted increased or decreased pathway activation (activation z score), respectively. Genes included in each pathway are listed and are colored red if increased in severe subjects or green if decreased in severe subjects. Genes with >1.5-fold increases are bold.
Gene-set analysis was used to interpret the expression pattern of genes associated with severity, in both the more liberal univariate analysis (n = 551 genes) and the more conservative multivariate analysis (n = 140 genes), identifying a number of signaling pathways and intercellular signaling molecules that may be associated with clinical responses in RSV-infected infants (Figure 2B and Supplementary Figure 2). Although there was limited confirmation of significance at the individual gene level, with only 64 of 140 multivariate genes also identified in univariate analysis, there was a high degree of confirmation for canonical pathway discovery (Figure 2B). These observations are consistent with the robustness of a pathway-based approach to analysis of transcriptomics data.
Genes associated with severity predicted activation of JAK/STAT, prolactin, Gα1, interleukin 1, iNOS, IGF1, and phosphatidylinositol 3-kinase (PI3K)/AKT signaling pathways were noted in severely affected subjects. Interleukin 9 (IL-9) pathway signaling was also predicted to be altered, but it was unclear whether this pathway was activated or inhibited. Identification of these pathways was predominantly driven by genes demonstrating significant increases in expression, including IRS1, CISH, and SOCS2, among others in severe subjects. Next, we performed a survey for upstream regulatory molecules capable of explaining global changes in gene expression associated with disease severity (Supplementary Figure 2). Interestingly, prostaglandin E synthase (PTGES)– and cyclooxygenase-2 (PTGS2)–mediated regulation was associated with CD4+ T-cell gene expression in severely ill infants, implicating shifts in arachidonate metabolism. Activation of NRF2 (NFE2L2), the major antioxidant pathway, was also associated with CD4+ T-cell gene expression in severely ill infants. This analysis also suggested activation of a number of regulators of mitogenesis, including MYC, KRAS, KIP1 (CDKN1B), and p53 (TP53).
Together these data support the conclusion that broad changes in CD4 T-cell activity and survival are associated with clinical severity in RSV-infected infants. In fact, some observed gene expression changes specifically implicated responses associated with CD4 T-cell subtype, such as regulation of IL-9 signaling (Figure 2B). Also noted were increased activation of JAK2 and increased expression of suppressor of cytokine signaling (SOCS) genes in CD4 T cells from severe subjects, particularly SOCS2, which antagonizes the other SOCS proteins, and together with JAK2 regulates early Th1/Th2 differentiation [24–26]. In an effort to directly test whether severity-associated changes in gene expression were consistent with alterations in T-cell subtype, we implemented Bayesian estimation of transcription factor activity to predict transcription factor activity based upon CD4 gene expression patterns associated with severity [23]. We found changes in GATA3 activity were predicted to be significantly associated with severity as defined by the univariate model (P = 1.62E-05), whereas RORA was marginally significant (P = .04) and STAT1/2/4 and SMADs were not significant (P > .05). GATA3 is a transcriptional activator that is required for T-helper 2 (Th2) differentiation. It also plays a role in differentiation of Th9 cells. These data support the interpretation that changes in CD4 T-cell subtype differentiation are associated with severity in RSV infection.
Gene Expression Correlates of Clinical Progression and Respiratory Syncytial Virus Group
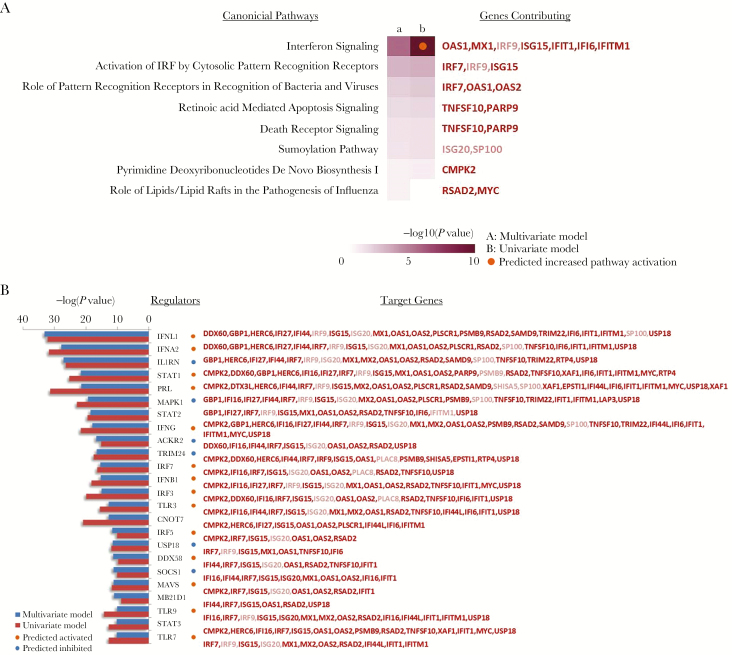
We explored biological interpretations of genes associated with the time since onset of clinical symptoms (Figure 3). For this analysis, we focused on the set of genes significantly associated with the stage of visit—acute versus convalescent. As anticipated, activation of interferon signaling was noted early in the clinical course, based on the assessment of canonical pathway gene expression (Figure 3A) and upstream regulator prediction (Figure 3B). Interestingly, both type I (IFNA/B), type II (IFNG), and type III (IFNL) interferon responses were implicated, which was confirmed by significant enrichment of STAT1-STAT2 and IRF1 target genes (P = 9.033E-05 and .0004, respectively). This was associated with downstream changes in inflammasome activation, as implicated by predictions of MAVS and IL1RN as upstream regulators (Figure 3B). Changes in viral pattern recognition signaling, potentially driven predominantly by increases in TLR3/7/9–associated genes, were also identified. Interestingly, decreases in proliferative activity in CD4 T cells, as indicated by reductions in CNOT7 and MAPK1, were also observed.
Figure 3.
Canonical pathways and regulators associated with time since onset of RSV infection. A, Pathways associated with stage of infection. Ingenuity pathway analysis (IPA) was used to identify canonical pathways represented by genes associated with multivariate modeling of time since onset of clinical symptoms (a; acute vs convalescent; n = 63) or univariate analysis of time since onset of clinical symptoms (b; acute vs convalescent; n = 35). Eight pathways are shown where Fisher’s exact test P values were <.05 for at least 1 analysis. Orange and blue circles indicate predicted increased or decreased pathway activation (activation z score), respectively. Genes included in each pathway are listed and are colored red if increased in acute stage or green if decreased in acute stage and in bold for genes with >1.5-fold changes in expression. B, Regulators associated with stage of infection. Ingenuity pathway analysis was used to identify putative regulators represented by genes associated with multivariate modeling of time since onset of clinical symptoms (a; acute vs convalescent; blue bars) or univariate analysis of time since onset of illness (b; acute vs convalescent; red bars). Twenty-four upstream regulators are shown where P values were <.05 for at least 1 gene list. Orange and blue circles indicate a predicted activation/inhibition state based upon the expression of targets. Upstream regulator targets are listed in red if upregulated and green if downregulated in acute stage, and in bold for genes with >1.5-fold changes in expression.
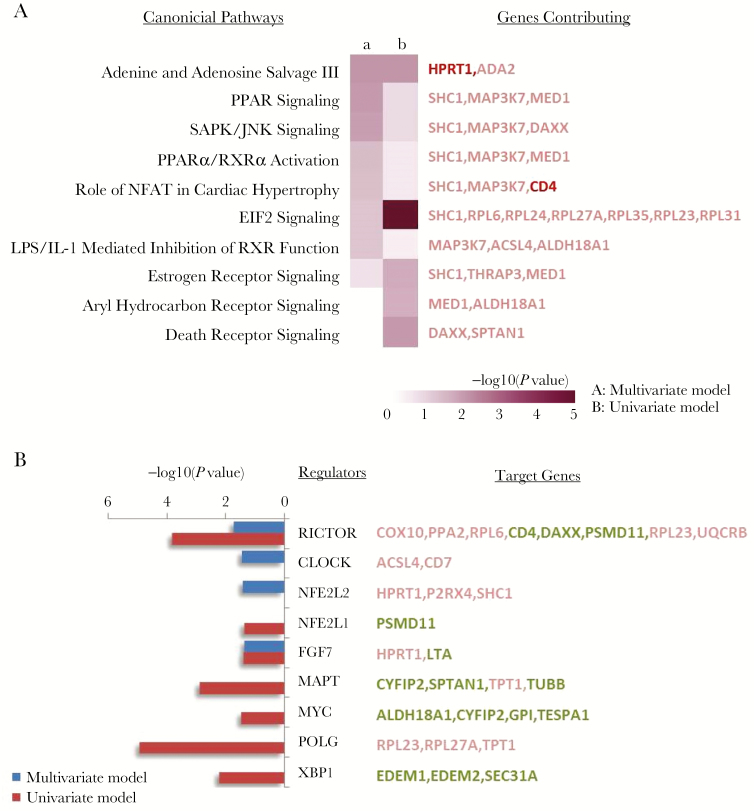
The RSV group (A/B) was not associated with clinical disease severity independently but was associated with significant differences in CD4 T-cell transcriptome status (Table 2). Pathway-based interpretation of these gene expression responses identified signaling pathways that may be differentially affected by infection with these 2 RSV groups (Figure 4A). Based upon canonical pathway analysis, adenine/adenosine salvage, PPAR, and SAPK/JNK signaling were all predicted to be differentially regulated by RSV subgroup infection. Interestingly, upstream regulator analysis (Figure 4B) suggested RICTOR (mTOR2 related), CLOCK, and NFE2L2 (NRF2) may play a significant role in these responses.
Figure 4.
Pathways and regulators associated with RSV group A and B. A, Pathways associated with respiratory syncytial virus (RSV) group infection. Ingenuity pathway analysis (IPA) was used to identify canonical pathways represented by genes associated with infection by RSV group (A/B) in CD4 lymphocytes. The variables used to generate gene sets for IPA were derived from multivariate analyses (a; n = 68) or univariate analysis (b; n = 53). Ten pathways are shown where Fisher’s exact test P values were <.05 for at least 1 gene list. Genes included in each pathway are listed and are colored red if increased in RSV strain A or green if decreased in strain A and in bold for genes with >1.5-fold changes in expression. B, Regulators associated with RSV group infection. Ingenuity pathway analysis was used to identify upstream regulators associated with infection by RSV group (A/B) using gene sets identified with multivariate (blue; n = 68 genes) or univariate (red; n = 53 genes) analysis. Nine upstream regulators are shown where P values were <.05 for at least 1 gene list. Upstraem regulator targets are listed in red if upregulated and green if downregulated in RSV A strain–infected subjects.
Quantitative Polymerase Chain Reaction Validation
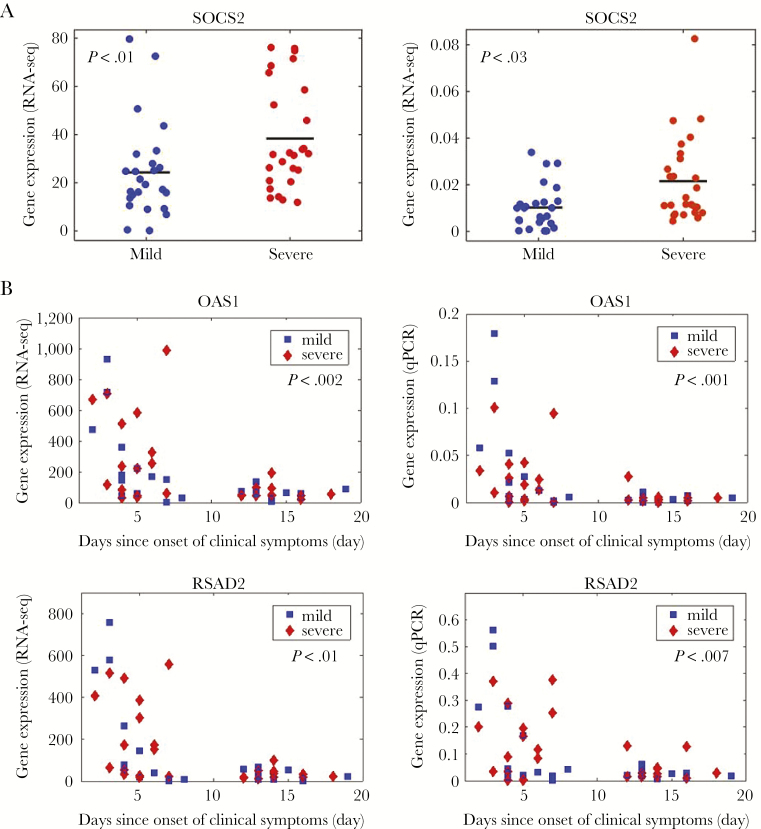
We attempted to validate expression estimates for 6 genes (CD4, CD7, FKBP5, OAS1, RSAD2, SOCS2) selected based upon RNA-seq detection levels and biological interest (Figure 5 and Supplementary Table 2). Expression of 5 of these genes (CD4, FKBP5, OAS1, RSAD2, SOCS2) demonstrated significant concordance with RNA-seq data (all P < .001). CD7 expression levels were low, which is likely the source of failure to validate expression estimates. Among these 5 genes, quantitative polymerase chain reaction expression estimates confirmed SOCS2 expression was significantly associated with severity (Figure 5A and Supplementary Table 2), whereas CD4, OAS1, and RSAD2 were all significantly associated with time since onset of clinical symptoms (Figure 5B and Supplementary Table 2).
Figure 5.
Quantitative polymerase chain reaction validation (qPCR). A, We validated expression of SOCS2, which is associated with illness severity. Left, Gene expression by RNA sequencing (RNA-seq). Right, Gene expression by qPCR. Gene expression is plotted relative to severity. B, We validated expression of OAS1 and RSAD2, which are associated with stage of infection. Left, Gene expression by RNA-seq. Right, Gene expression by qPCR. Gene expression is plotted relative to the day of onset of clinical symptoms, separately for subjects with mild or severe phenotypes. P value indicates significance level for association with stage. Abbreviations: qPCR, quantiative polymerase chain reaction; RNA-seq, RNA sequencing.
DISCUSSION
A full understanding of the cellular immune mechanisms underlying disease severity during RSV infection in infants has been elusive, especially in normal, full-term, healthy infants, the population that comprises the majority of infants brought to medical attention. Most studies investigating RSV disease pathogenesis have measured the presence and levels of various inflammatory proteins, such as cytokines and chemokines in blood or respiratory secretions, or their in vitro production by peripheral blood mononuclear cells, often with disparate results [9, 11–15].
Recently, analysis of gene expression in whole-blood samples using microarray from RSV-infected infants was reported from 2 centers [16–18]. A study of 21 RSV-infected infants showed extensive activation of innate immune responses, specifically of the interferon signaling network, but could not identify differences according to disease severity [16]. In a much larger study involving 90 RSV-infected infants with mild and severe RSV disease, Mejias and colleagues reported overexpression of neutrophil, inflammation, and interferon genes and suppression of T- and B-cell genes [17]. In this study, severe illness was associated with greater expression of neutrophil and inflammation genes than in mildly ill subjects, whereas mildly ill subjects showed overexpression of innate immunity genes.
Because CD4 T cells are important in the early adaptive immune response to RSV and are associated with the degree of inflammation during infection, we chose to investigate gene expression patterns in isolated CD4 T cells from infants with primary RSV infection of differing severity during their first year of life. To optimize identification of differentially expressed genes, we selected infants at the extremes of disease severity. Interrogation of CD4 T-cell gene expression in these subjects identified a number of pathways of interest, including prolactin signaling, JAK/STAT signaling, PI3K/AKT signaling, and IL-9 signaling.
Our data indicated severely ill RSV-infected infants displayed greater activation of prolactin signaling in CD4 T cells. Prolactin, a polypeptide hormone originating from the pituitary gland, has been shown to bind to the prolactin receptor expressed on CD4 T cells and to have a variety of effects on CD4 T cells [27, 28]. Prolactin induction of T-bet transcription through phosphorylation of STAT2 and STAT5 is inversely dose dependent, and it has been suggested that exposure of CD4 T cells to high levels of prolactin might reduce Th1 function [27]. Another study found that Treg cells express prolactin receptor and exposure to prolactin inhibited their suppressive effect on Th1 cells in vitro [28]. Interestingly, it has been reported that infants with severe RSV disease admitted to the intensive care unit had significantly higher serum prolactin levels than moderately ill infants [29].
The JAK2 cytokine and JAK/STAT signaling pathways are integral to the production of the innate type I interferons IFNα and IFNβ, and thus it is not surprising that these pathways are differentially expressed according to disease severity. Confirming that such an effect is detectable in circulating, purified CD4 T cells is novel. It is known that the RSV NS1 and NS2 proteins block type I effectively via degradation of the transcription activator STAT2 [30]. The SOCS genes are included in the JAK/STAT pathways, and the SOCS2, SOCS3, and SOCS5 genes were all overexpressed in the severely ill subjects. It has been reported that SOCS3 is expressed during Th2 immune responses and in vitro experiments after exposure of murine respiratory epithelial cells to RSV [31]. Both STATs and SOCS are key regulators of T-cell differentiation, maturation, and function [32]. Similar to STATs, SOCS may cross-regulate one another, and the proteins are differentially expressed in Th cell lineage [33]. SOCS1 and SOCS3 favor Th2 and Th17 differentiation, and SOCS2 favors the differentiation of Th17 by the compensation of Th2 differentiation [34–36]. It also was not surprising to find that the PI3K/AKT pathway activation was increased in severe RSV disease. PI3K signaling is thought to have an important role in CD4 T-cell differentiation and function, including Tregs [37]. Respiratory syncytial virus has been shown to rapidly activate this pathway, which is associated with inhibition of cellular apoptosis as well as increased inflammatory cytokine production [38].
Interleukin 9, a Th2 cytokine produced by CD4 T cells, eosinophils, and neutrophils, has been found in the upper and lower airway secretions of infants with RSV bronchiolitis at relatively high levels but has not been previously correlated with disease severity [39, 40]. In a murine model of RSV infection, depletion of IL-9 enhanced clearance of virus from the lungs, and the authors concluded that IL-9 promoted a Th2-type inflammatory response [41]. Together, our data are supportive of a model where shifts in CD4 T-cell subtype toward a Th2 phenotype are associated with severity of illness in RSV-infected infants. Further studies, particularly in larger and more heterogeneous populations, will be required to confirm this observation and to determine whether these changes are mechanistic or are purely a result of the illness itself.
The exclusion of infants with well-known risk factors for severe disease allowed us to remove these influences from our analysis. Although we did not use a formal severity scoring system to define disease severity in our population, we believe that the clinical criteria selected provided a valid separation of mild and severe disease. The use of sorted CD4 T cells, rather than whole blood, and RNA-seq are unique aspects of this study and add to the existing gene expression literature. A potential limitation of this method is that some of the findings might have been affected by manipulation of the CD4 T cells during purification and sorting. However, and critically, identical procedures were used for processing samples from all subjects, regardless of disease severity. All blood samples were collected between 8 and 10 am, kept at room temperature, and processed immediately. Therefore, any impact of sample processing should be applicable to all subjects and result mostly in limitations of sensitivity.
In summary, we were able to identify a number of differentially expressed genes and gene pathways involved in primary RSV infection in full-term healthy infants that are associated with increased disease severity. The results may help to ultimately identify potential biomarkers of severe disease and provide putative intermediate, molecular phenotypes of disease that can be assayed using experimental in vitro or in vivo infection models of RSV.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Acknowledgments. We would like to thank the following individuals whose efforts were instrumental in facilitating the research described: Brenda Tesini, Cynthia MacDonald, Doreen Francis, Gerry Lofthaus, Christopher Slaunwhite, Soumyaroop Bhattacharya, Jami McGrath, Mary Criddle, Lisa Denmark, Lynne Shelley, Amy Murphy, Melissa Bowman, Mary Ann Formica, Ken Schnabel, Jennifer Carnahan, Steven Gill, Alex Grier, John Ashton, and the URMC Genomics Research Center. We also express our sincere gratitude to all of our subjects and their families.
Funding source. This project has been funded in whole or in part with federal funds from the National Institute of Allergy and Infectious Diseases, Division of Microbiology and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract no. HHSN272201200005C.
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: International RSV Meetings, San Bariloche, Argentina, September 2016; and Society of Academic Pediatricians, Baltimore, MD, April 2016.
References
- 1. Hall CB. Respiratory syncytial virus and parainfluenza virus. N Engl J Med 2001; 344:1917–28. [DOI] [PubMed] [Google Scholar]
- 2. Hall CB, Weinberg GA, Blumkin AK et al. . Respiratory syncytial virus–associated hospitalizations among children less than 24 months of age. Pediatrics 2013; 132:e341–8. [DOI] [PubMed] [Google Scholar]
- 3. Zhou H, Thompson WW, Viboud CG et al. . Hospitalizations associated with influenza and respiratory syncytial virus in the United States, 1993–2008. Clin Infect Dis 2012; 54:1427–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hall CB, Weinberg GA, Iwane MK et al. . The burden of respiratory syncytial virus infection in young children. N Engl J Med 2009; 360:588–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Meissner HC. Viral bronchiolitis in children. N Engl J Med 2016; 374:62–72. [DOI] [PubMed] [Google Scholar]
- 6. Collins PL, Fearns R, Graham BS. Respiratory syncytial virus: virology, reverse genetics, and pathogenesis of disease. Curr Top Microbiol Immunol 2013; 372:3–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Openshaw PJ, Chiu C. Protective and dysregulated T cell immunity in RSV infection. Curr Opin Virol 2013; 3:468–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mella C, Suarez-Arrabal MC, Lopez S et al. . Innate immune dysfunction is associated with enhanced disease severity in infants with severe respiratory syncytial virus bronchiolitis. J Infect Dis 2013; 207:564–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Russell CD, Unger SA, Walton M, Schwarze J. The human immune response to respiratory syncytial virus infection. Clin Microbiol Rev 2017; 30:481–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Christiaansen AF, Knudson CJ, Weiss KA, Varga SM. The CD4 T cell response to respiratory syncytial virus infection. Immunol Res 2014; 59:109–17. [DOI] [PubMed] [Google Scholar]
- 11. Legg JP, Hussain IR, Warner JA, Johnston SL, Warner JO. Type 1 and type 2 cytokine imbalance in acute respiratory syncytial virus bronchiolitis. Am J Respir Crit Care Med 2003; 168:633–9. [DOI] [PubMed] [Google Scholar]
- 12. Bendelja K, Gagro A, Bace A et al. . Predominant type-2 response in infants with respiratory syncytial virus (RSV) infection demonstrated by cytokine flow cytometry. Clin Exp Immunol 2000; 121:332–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Román M, Calhoun WJ, Hinton KL et al. . Respiratory syncytial virus infection in infants is associated with predominant Th-2-like response. Am J Respir Crit Care Med 1997; 156:190–5. [DOI] [PubMed] [Google Scholar]
- 14. Bont L, Heijnen CJ, Kavelaars A et al. . Peripheral blood cytokine responses and disease severity in respiratory syncytial virus bronchiolitis. Eur Respir J 1999; 14:144–9. [DOI] [PubMed] [Google Scholar]
- 15. Garofalo RP, Patti J, Hintz KA, Hill V, Ogra PL, Welliver RC. Macrophage inflammatory protein-1alpha (not T helper type 2 cytokines) is associated with severe forms of respiratory syncytial virus bronchiolitis. J Infect Dis 2001; 184:393–9. [DOI] [PubMed] [Google Scholar]
- 16. Bucasas KL, Mian AI, Demmler-Harrison GJ et al. . Global gene expression profiling in infants with acute respiratory syncytial virus broncholitis demonstrates systemic activation of interferon signaling networks. Pediatr Infect Dis J 2013; 32:e68–76. [DOI] [PubMed] [Google Scholar]
- 17. Mejias A, Dimo B, Suarez NM et al. . Whole blood gene expression profiles to assess pathogenesis and disease severity in infants with respiratory syncytial virus infection. PLoS Med 2013; 10:e1001549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de Steenhuijsen Piters WA, Heinonen S, Hasrat R et al. . Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am J Respir Crit Care Med 2016; 194:1104–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Walsh EE, Peterson DR, Kalkanoglu AE, Lee FE, Falsey AR. Viral shedding and immune responses to respiratory syncytial virus infection in older adults. J Infect Dis 2013; 207:1424–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chu CY, Qiu X, Wang L et al. . The healthy infant nasal transcriptome: a benchmark study. Sci Rep 2016; 6:33994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Misra RS, Bhattacharya S, Huyck HL et al. . Flow-based sorting of neonatal lymphocyte populations for transcriptomics analysis. J Immunol Methods 2016; 437:13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Van Twisk D, Murphy SP, Thakar J. Optimized logic rules reveal interferon-γ-induced modes regulated by histone deacetylases and protein tyrosine phosphatases. Immunology 2017; 151:71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thakar J, Hartmann BM, Marjanovic N, Sealfon SC, Kleinstein SH. Comparative analysis of anti-viral transcriptomics reveals novel effects of influenza immune antagonism. BMC Immunol 2015; 16:46,015-0107-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol 2007; 7:454–65. [DOI] [PubMed] [Google Scholar]
- 25. Knosp CA, Johnston JA. Regulation of CD4+ T-cell polarization by suppressor of cytokine signalling proteins. Immunology 2012; 135:101–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Akhtar LN, Benveniste EN. Viral exploitation of host SOCS protein functions. J Virol 2011; 85:1912–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tomio A, Schust DJ, Kawana K et al. . Prolactin can modulate CD4+ T-cell response through receptor-mediated alterations in the expression of T-bet. Immunol Cell Biol 2008; 86:616–21. [DOI] [PubMed] [Google Scholar]
- 28. Legorreta-Haquet MV, Flores-Fernández R, Blanco-Favela F et al. . Prolactin levels correlate with abnormal B cell maturation in MRL and MRL/lpr mouse models of systemic lupus erythematosus-like disease. Clin Dev Immunol 2013; 2013:287469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tasker RC, Roe MF, Bloxham DM, White DK, Ross-Russell RI, O’Donnell DR. The neuroendocrine stress response and severity of acute respiratory syncytial virus bronchiolitis in infancy. Intensive Care Med 2004; 30:2257–62. [DOI] [PubMed] [Google Scholar]
- 30. Schlender J, Bossert B, Buchholz U, Conzelmann KK. Bovine respiratory syncytial virus nonstructural proteins NS1 and NS2 cooperatively antagonize alpha/beta interferon-induced antiviral response. J Virol 2000; 74:8234–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moore EC, Barber J, Tripp RA. Respiratory syncytial virus (RSV) attachment and nonstructural proteins modify the type I interferon response associated with suppressor of cytokine signaling (SOCS) proteins and IFN-stimulated gene-15 (ISG15). Virol J 2008; 5:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Palmer DC, Restifo NP. Suppressors of cytokine signaling (SOCS) in T cell differentiation, maturation, and function. Trends Immunol 2009; 30:592–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Egwuagu CE, Yu CR, Zhang M, Mahdi RM, Kim SJ, Gery I. Suppressors of cytokine signaling proteins are differentially expressed in Th1 and Th2 cells: implications for Th cell lineage commitment and maintenance. J Immunol 2002; 168:3181–7. [DOI] [PubMed] [Google Scholar]
- 34. Tanaka K, Ichiyama K, Hashimoto M et al. . Loss of suppressor of cytokine signaling 1 in helper T cells leads to defective Th17 differentiation by enhancing antagonistic effects of IFN-gamma on STAT3 and Smads. J Immunol 2008; 180:3746–56. [DOI] [PubMed] [Google Scholar]
- 35. Piessevaux J, Lavens D, Montoye T et al. . Functional cross-modulation between SOCS proteins can stimulate cytokine signaling. J Biol Chem 2006; 281:32953–66. [DOI] [PubMed] [Google Scholar]
- 36. Seki Y, Inoue H, Nagata N et al. . SOCS-3 regulates onset and maintenance of T(H)2-mediated allergic responses. Nat Med 2003; 9:1047–54. [DOI] [PubMed] [Google Scholar]
- 37. Han JM, Patterson SJ, Levings MK. The role of the PI3K signaling pathway in CD4(+) T cell differentiation and function. Front Immunol 2012; 3:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Thomas KW, Monick MM, Staber JM, Yarovinsky T, Carter AB, Hunninghake GW. Respiratory syncytial virus inhibits apoptosis and induces NF-kappa B activity through a phosphatidylinositol 3-kinase-dependent pathway. J Biol Chem 2002; 277:492–501. [DOI] [PubMed] [Google Scholar]
- 39. McNamara PS, Flanagan BF, Baldwin LM, Newland P, Hart CA, Smyth RL. Interleukin 9 production in the lungs of infants with severe respiratory syncytial virus bronchiolitis. Lancet 2004; 363:1031–7. [DOI] [PubMed] [Google Scholar]
- 40. Semple MG, Dankert HM, Ebrahimi B et al. . Severe respiratory syncytial virus bronchiolitis in infants is associated with reduced airway interferon gamma and substance P. PLoS One 2007; 2:e1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dodd JS, Lum E, Goulding J, Muir R, Van Snick J, Openshaw PJ. IL-9 regulates pathology during primary and memory responses to respiratory syncytial virus infection. J Immunol 2009; 183:7006–13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.