Summary
Transgenic parasites harboring a naturally-arising polymorphism in PfRh2b reveal no invasion pathway difference but are differentially inhibited by IgG from three endemic regions. Transgenic parasites provide a useful tool to assess specificity of natural or vaccine-induced inhibitory antibodies.
Keywords: growth inhibition, invasion, PfRh2b, transgenic, vaccine
Abstract
Background
Plasmodium falciparum reticulocyte-binding protein homologue 2b (PfRh2b) is an invasion ligand that is a potential blood-stage vaccine candidate antigen; however, a naturally occurring deletion within an immunogenic domain is present at high frequencies in Africa and has been associated with alternative invasion pathway usage. Standardized tools that provide antigenic specificity in in vitro assays are needed to functionally assess the neutralizing potential of humoral responses against malaria vaccine candidate antigens.
Methods
Transgenic parasite lines were generated to express the PfRh2b deletion. Total immunoglobulin G (IgG) from individuals residing in malaria-endemic regions in Tanzania, Senegal, and Mali were used in growth inhibition assays with transgenic parasite lines.
Results
While the PfRh2b deletion transgenic line showed no change in invasion pathway utilization compared to the wild-type in the absence of specific antibodies, it outgrew wild-type controls in competitive growth experiments. Inhibition differences with total IgG were observed in the different endemic sites, ranging from allele-specific inhibition to allele-independent inhibitory immune responses.
Conclusions
The PfRh2b deletion may allow the parasite to escape neutralizing antibody responses in some regions. This difference in geographical inhibition was revealed using transgenic methodologies, which provide valuable tools for functionally assessing neutralizing antibodies against vaccine-candidate antigens in regions with varying malaria endemicity.
Malaria caused by Plasmodium falciparum represents a major public health challenge with at least 400000 deaths annually, occurring primarily in young children and pregnant women [1]. The process of merozoite invasion is a critical step in the Plasmodium life cycle, and the multiple ligand–receptor interactions involved in invasion influence parasite virulence and disease severity [2]. To establish infection within the human host, the merozoite faces 2 selective pressures: erythrocyte receptor polymorphism and the humoral immune response. Such selective pressures are manifested both in variant expression of parasite ligands and in sequence polymorphisms in the genome.
With the rise in antimalarial drug resistance, an effective vaccine is ever more critical. While most vaccines currently under development target the sporozoite stage of the life cycle, there has been a recent resurgence in interest in blood-stage vaccines targeting merozoite invasion ligands [3–6]. In assessing vaccine candidates, it is important to identify polymorphisms under selection and determine whether such polymorphisms contribute to altered receptor binding (eg, erythrocyte-binding antigen [EBA]–181]) [7] or immune evasion (eg, apical membrane antigen 1 [AMA-1]) [8].
The P. falciparum reticulocyte-binding protein homologue 2b (PfRh2b) is an important sialic acid–independent invasion ligand [9] and is a member of a multigene family of reticulocyte binding–like (RBL) proteins involved in the commitment step of invasion. PfRh2a and PfRh2b are identical in sequence for their first N-terminal 2700 amino acids, yet diverge in their c-terminal 400–500 amino acids—a region that includes variable heptad repeats, a unique ectodomain that differs between PfRh2a and PfRh2b, a transmembrane domain, and a cytoplasmic tail [10, 11]. The PfRh2a/2b erythrocyte-binding domain has been mapped to the N-terminus of the protein; however the c-terminal unique region of PfRh2a has been shown to have some erythrocyte-binding activity as well [12]. Variation in PfRh2b expression is associated with alternative invasion pathway use in both laboratory and field isolates [13, 14]. A large polymorphism in the c-terminal unique ectodomain region of PfRh2b was described in the P. falciparum line T996 [15] and was identified in a large percentage of field isolates in Senegal [16]. In 2 studies in Senegal, the PfRh2b deletion was associated with invasion pathway utilization [16, 17]. The deletion allele is highly prevalent within Africa and is proposed to be under strong immune selection [18].
Population genetic analyses can infer the type of selection acting at these loci [19, 20], but genetic methodologies can be employed to precisely determine the role of a given polymorphism. By using transgenic methodologies, a specific polymorphism can be studied in an isogenic background, controlling for unrelated polymorphisms in many antigens. In this study, we use genetic replacement to test the role of the PfRh2b deletion polymorphism in both invasion pathway utilization (receptor binding) and immune evasion in 3 regions with dramatically different levels of malaria endemicity.
METHODS
Study Subjects and Samples
Patients were recruited in Thiès, a region in Senegal with low P. falciparum endemicity and seasonal malaria transmission (entomological inoculation rate [EIR] = 1–5 infectious bites/person/year) [21] in 2008–2009, when parasite positivity rate in children between 2–10 years of age (PfPr2–10) has been modeled to be 6% [22]; and in Mlandizi, a region of Tanzania of high malaria endemicity and perennial transmission (EIR = 200) [23] in 2003–2004, when PfPr2–10 has been modeled to be 23%–26% [22]. In both Thiès and Mlandizi, patients diagnosed with uncomplicated malaria were enrolled in the study after informed consent was obtained. In Kenieroba, Mali, an area of high malaria endemicity and perennial transmission (EIR > 100) (personal communication, Adama Dao and Nafomon Sogoba, International Center for Excellence in Research (ICER)/FMOS/USTTB, Bamako, Mali), adults known to be highly malaria-exposed were recruited from May 2013 to May 2014, when PfPr2–10 has been modeled to be 42%–43% [22], and donated blood for plasma collection after informed consent was obtained. For the purposes of population transmission comparisons, here we consider low transmission to be EIR < 10; medium transmission, EIR = 10–100; and high transmission, EIR > 100 [24].
Transgenic Parasite Lines
The PfRh2b deletion single crossover plasmid was generated by amplifying the c-terminal 1.3 kilo base pairs from a sequenced Senegalese field strain genomic DNA containing the PfRh2b deletion (and all other sequence identical to 3D7). The PfRh2b wild-type replacement parasite line (1A) was generated and characterized previously [25]. PfRh2b knockout parasites (3D7∆2b, C2; W2Mef∆2b, C23) were also described previously [9, 25]. Details of transgenic parasite line characterization (Western blots, immunofluorescence microscopy, and quantitative polymerase chain reaction [qPCR]) are included in Supplementary Methods. Here, we refer to PfRh2b wild-type replacement parasite line (1A) as “WT,” the PfRh2b deletion single crossover parasites as “DEL,” and 3D7∆2b, C2 and W2Mef∆2b, C23 parasites as “KO.” Two independent transgenic deletion clones were characterized in each genetic background: in 3D7, DEL A (clone A12), and DEL B (clone H11); and in W2Mef, DEL C (clone A10) and DEL D (clone C10).
Invasion Assay
Invasion assays were performed as previously described [26, 27]. Additional details can be found in Supplementary Methods.
Invasion Phenotype Measurements
Synchronized ring-stage parasites were plated at 1% parasitemia and monitored by microscopy at the ring (T = 0 hours), trophozoite/early schizont (T = 40 hours), late schizont/reinvaded ring (T = 48 hours), and ring/trophozoite (T = 56 hours) stages. Parasite multiplication rate was calculated as follows: (final parasitemia)/(initial parasitemia). The selectivity index was counted as previously described [28, 29]. Additional details can be found in Supplementary Methods. Average merozoite numbers (Av Mero) were counted for late-stage segmented schizonts only (T = 48), and 100 schizonts were counted per experiment to determine the average merozoite number. Invasion efficiency was calculated as described [30]: Invasion efficiency = (final parasitemia)/[(average merozoite number) × (initial parasitemia)].
Invasion Inhibition Assays
Invasion inhibition assays were performed similarly to invasion assays with minor modifications. Wild-type 3D7 parasites and DEL A (3D7-PfRh2b deletion) were used and invasion inhibition was measured into untreated erythrocytes (Roswell Park Memorial Institute medium) as well as neuraminidase-treated erythrocytes (Nm). Parasites were plated at 0.25% parasitemia in 2% hematocrit in 25 μL final volume. Protein A–purified antibodies from rabbits immunized with the unique domain of PfRh2b [31, 32] were added to synchronized ring-stage parasites at a final concentration of 250 μg/mL and incubated for 1 cycle of reinvasion.
Growth Inhibition Assays
Growth inhibition assays were performed as previously described [33], using protein G-purified total immunoglobulin G (IgG) from Thiès, Senegal (n = 39), Mlandizi, Tanzania (n = 33), and Kenieroba, Mali (n = 27), at a final concentration of 10 mg/mL and transgenic parasites: WT, DEL A, and KO. Transgenic parasites were continuously grown under WR-99210 pressure.
RESULTS
Generation of Transgenic Parasites to Test the Role of the PfRh2b Deletion Allele
To determine the specific role of the PfRh2b deletion polymorphism on invasion pathway, efficiency, and immune evasion, we generated transgenic parasite lines that harbored the PfRh2b deletion (DEL3D7), or the PfRh2b full-length allele (WT) [25], in a genetically identical 3D7 parental background (Supplementary Figure 1). We introduced the PfRh2b deletion into the PfRh2b locus by homologous recombination and single crossover (Supplementary Figure 1A). We confirmed disruption of the endogenous locus by Southern blot (Supplementary Figure 1B) and sequencing (Supplementary Figure 1D). We also created transgenic PfRh2b deletion (DELW2Mef) parasites in the W2Mef background (Supplementary Figure 1C).
The PfRh2b Deletion Does Not Alter Gene Expression, Protein Expression, Proteolytic Cleavage, or Localization of Other Invasion Ligands
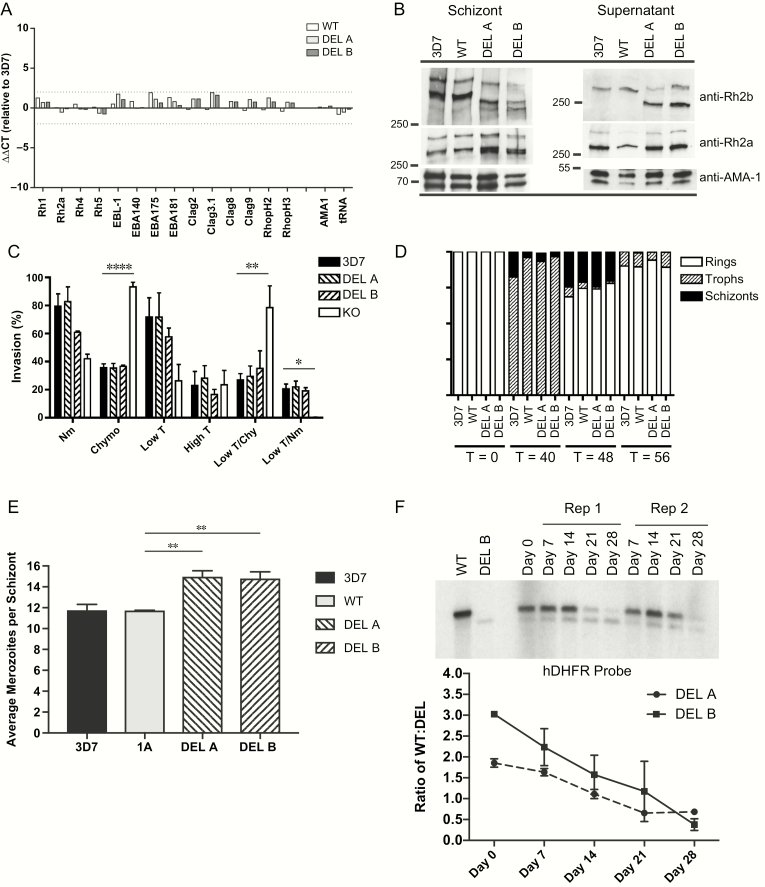
To determine whether the deletion influences gene expression, we performed qPCR on complementary DNA prepared from schizont-stage ribonucleic acid for 3D7, WT, DEL A, and DEL B. The ∆∆CT values ranged from –0.72 to 1.98, and revealed no significant fold change for all transgenic lines relative to 3D7 (Figure 1A). To determine whether the deletion might affect protein expression and cleavage of other important invasion ligands, we performed Western blots of both schizont and invasion supernatant extracts (Figure 1B) for 3D7, WT, and DEL A and DEL B clones, and probed with anti-PfRh2a and anti-PfRh2b antibodies. We found no difference in expression levels of PfRh2a and PfRh2b (other than the size difference in the PfRh2b protein due to the deletion). Additionally, we found no change in apical merozoite localization of PfRh2b, adjacent to the rhoptry bulb, as indicated by partial colocalization with RhopH3 (Supplementary Figure 1D).
Figure 1.
The PfRh2b deletion does not affect alternative invasion pathway utilization but shows a competitive growth advantage over time. A, Quantitative PCR was employed to determine differences in invasion gene expression between DEL clones and WT control. CT values were compared relative to AMA-1 (∆CT). Data are expressed as fold change (∆∆CT) values relative to 3D7. Clag 3.2 is not shown as it is not expressed in the 3D7 clone used. B, Western blots of synchronous schizont material and invasion supernatants were performed. Samples were adjusted relative to the schizont stage–specific proteins PfAMA-1 for both schizont blots and supernatant blots. In schizonts, expected size of PfRh2b full-length protein (3D7, WT) is 382 kDa, which after proteolytic processing results in a 297 kDa cleavage product [45]; whereas the Deletion (DEL) results in a shorter protein which runs approximately 360 kDa with the cleavage product at 275 kDa. For PfRh2a, the unprocessed form runs at 370 kDa with a 285-kDa processed form. In supernatants, further processing results in an additional processed form of 290 kDa for PfRh2b (3D7, WT), 268 kDa (DEL), and 279 kDa for PfRh2a [45]. C, Invasion assays were performed to determine the erythrocyte receptor utilization for DEL clones compared to wild-type 3D7 and KO (3D7ΔPfRh2b). Percent invasion into enzyme-treated erythrocytes was calculated relative to invasion of the same parasite line into RPMI-treated erythrocytes. Assays were repeated 5 times, in triplicate, except for KO and DEL B (which were repeated twice, in triplicate). Values shown are the mean of all experiments; error bars represent SD. Overall significance is assessed by 1-way ANOVA; differences between columns are assessed with Dunnett test for multiple comparisons. Asterisks indicate significant differences (*P < .05; **P < .01; ***P < .001; ****P < .0001). KO invasion into lowT/Nm was tested, but is undetectable. D, Detailed measurements of invasion and growth were performed for PfRh2b deletion and control strains for 3 independent experiments, plated in duplicate. Erythrocytic cycle duration was measured for the 2 PfRh2b deletion (DEL A and DEL B) and control strains (3D7, WT) over 1 cycle. Stages were quantified morphologically at T = 0, T = 40, T = 48, and T = 56 hours postinvasion. A total of 300 parasitized cells were counted per time point; the average of the 3 experiments, plated in duplicate, is shown. E, Average merozoite number per schizont was calculated for transgenic parasites and controls. Data shown are from 3 independent experiments. Means and 95% CIs are shown. Overall significance is assessed by 1-way ANOVA; differences between columns are assessed with Dunnett test for multiple comparisons. Asterisks indicate significant differences (*P < .05; **P < .01; ***P < .001). F, Competitive growth assays were conducted in which DEL and WT were mixed in equal proportions and kept in in vitro culture for 1 month, with weekly gDNA harvests. Relative proportions of DEL and WT were assessed by Southern blot hybridization using a probe to human DHFR. A representative experiment is shown with DEL B and WT. Relative ratios of WT compared to DEL over time were calculated for both DEL A and DEL B clones, with 2 independent experiments per clone; error bars represent the range of the 2 experiments.
Abbreviations: ANOVA, analysis of variance; CIs, confidence intervals; CT, cycle threshold; DEL, PfRh2b deletion single crossover parasites; DHFR, dihydrofolate reductase; gDNA, genomic DNA; kDa, kilodalton; KO, 3D7∆2b, C2 parasites; lowT, low trypsin-treated erythrocytes; Nm, neuraminidase-treated erythrocytes; PCR, polymerase chain reaction; PfRh2b, Plasmodium falciparum reticulocyte-binding protein homologue 2b; RPMI, Roswell Park Memorial Institute; T, trypsin-treated erythrocytes; WT, PfRh2b wild-type replacement parasite line (1A).
The PfRh2b Deletion Does Not Alter Invasion Pathway or Measurements of Invasion Efficiency
While past reports from Senegal have shown that the PfRh2b deletion was mildly associated with sialic acid–dependent invasion [16], high trypsin-resistant invasion, and marginally, low trypsin/chymotrypsin-resistant invasion [17], we observed no difference in invasion pathway utilization between the 3D7 and DEL parasite lines for all enzyme treatments tested, in contrast to a striking difference in invasion pathway compared to the PfRh2b knockout (KO) (Figure 1C). The most significant differences were observed into chymotrypsin treated (3D7, P = .0001; DEL A, P = .0001; DEL B, P = .0001; relative to KO) and low trypsin/chymotrypsin-treated cells (3D7, P = .0028; DEL A, P = .0058; DEL B, P = .0346; relative to KO), the characteristic enzyme treatments defining the PfRh2b receptor. Thus, the deletion does not mimic a PfRh2b “loss of function” phenotype, as is observed with KO compared to 3D7. The same result was found in the W2Mef genetic background (Supplementary Figure 2).
To address whether the PfRh2b deletion may confer a fitness advantage in the absence of in vivo selective pressures, we compared multiple measures of invasion efficiency for the 3D7, WT, and DEL transgenic lines. In 3 independent experiments, erythrocytic cycle length (Figure 1D), parasite multiplication rate, selectivity index, and invasion efficiency of the deletion strains were not found to differ significantly from 3D7 and WT (Table 1). There was a small difference in the average merozoite number per schizont (Av Mero) for the 2 deletion clones (DEL A P = .01 and DEL B P = .02 relative to 3D7; DEL A P = .005 and DEL B P = .008 relative to WT) (Figure 1E); however, this difference was not manifested in a difference in invasion efficiency, which takes merozoite number into account (Table 1).
Table 1.
Measures of Invasion Efficiency for PfRh2b Allele Transgenic Strains
| PMR | SI | IE | |
|---|---|---|---|
| Strain (mean, CI) | |||
| 3D7 | 5.72 (4.25, 7.17) | 8.93 (4.48, 13.39) | 49.17 (26.25, 72.08) |
| WT | 4.37 (3.02, 5.72) | 12.70 (7.20, 18.20) | 37.30 (25.50, 49.10) |
| DEL A | 7.21 (3.02, 11.40) | 9.10 (6.32, 11.88) | 48.32 (19.95, 76.69) |
| DEL B | 5.80 (2.45, 9.16) | 12.24 (5.52, 18.97) | 39.69 (12.40, 66.98) |
| 1-way ANOVA | P = .0927 | P = .1023 | P = .3679 |
PMR, SI, and IE are calculated for transgenic parasites and controls. Data shown is from 3 independent experiments. Means and 95% CIs are shown. Overall significance is assessed by 1-way ANOVA.
Abbreviations: ANOVA, analysis of variance; CI, confidence interval; DEL, PfRh2b deletion single crossover parasites; IE, invasion efficiency; PfRh2b, Plasmodium falciparum reticulocyte-binding protein homologue 2b; PMR, parasite multiplication rate; SI, selectivity index; WT, PfRh2b wild-type replacement parasite line (1A).
The PfRh2b Deletion Influences Parasite Growth Rate Over Time
While subtle differences in invasion efficiency may not be significant in a single round of parasite replication, they can amplify over multiple rounds of growth. We performed in vitro competition experiments, similar to those described by others [34], between the WT and DEL clones (DEL A or DEL B) over 4 weeks, corresponding to 14 cycles of parasite replication. We observed that the DEL A and DEL B clones outgrew the WT controls at a rate of 8.4% per cycle for DEL A and 18.9% per cycle for DEL B, with a combined 13% per cycle (Figure 1E), implying that the deletion influences invasion over time.
The PfRh2b Deletion Allows Parasites to Escape the Effect of PfRh2b-Specific Inhibitory Antibodies
The 2 main selective pressures driving polymorphism in vivo are erythrocyte-receptor polymorphism and humoral immunity. We tested the hypothesis that parasites with the deletion can escape inhibitory antibodies by performing antibody inhibition assays in the presence of antibodies against the PfRh2b unique region, which were previously shown to inhibit invasion of parasite isolates invading by a trypsin-resistant pathway [32]. IgG from rabbits immunized with the PfRh2b unique domain did inhibit wild-type (3D7) parasites modestly, yet significantly more than DEL parasites (P = .0419), but only when the parasites were limited to a sialic acid–independent pathway through treatment with neuraminidase (Supplementary Figure 3).
Population-Specific Development of PfRh2b Allele-Transcendent Immune Responses
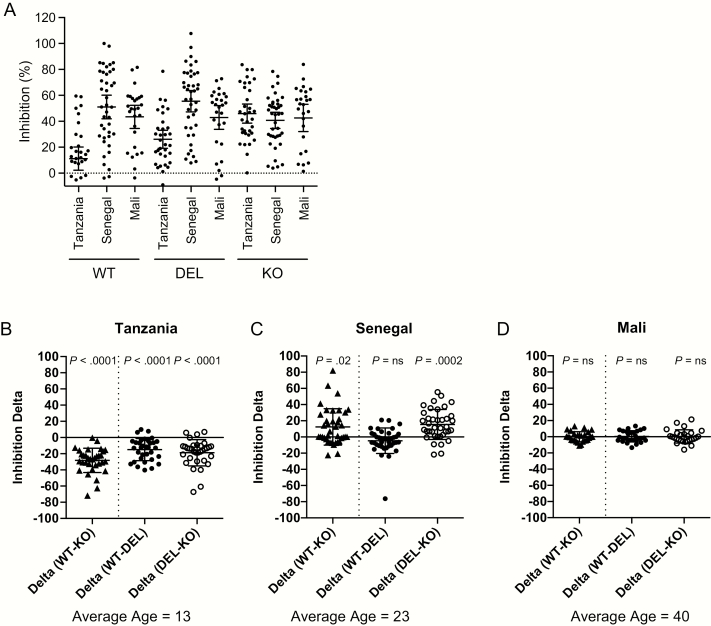
To determine the inhibitory potential of naturally acquired IgG from malaria-endemic populations, we employed the highly standardized growth inhibition assay [33] using IgG from children in a highly endemic region of Tanzania, from adolescents and adults in a low-endemicity region of Senegal, and adults in a highly endemic region in Mali. We observed strain-specific differences in growth inhibition for WT, DEL A, and KO strains (Figure 2A). When stratified by endemic region, in Tanzania, inhibition was significantly different for all strains, with WT inhibited less than DEL A, and DEL A inhibited less than KO (WT < DEL < KO) (Figure 2B). In contrast, in Senegal, the KO is inhibited less than WT and DEL A (WT = DEL > KO) (Figure 2C). In Mali, for adults with significant malaria exposure, while overall inhibition was high, there were no differences between alleles (WT = DEL = KO), suggesting that the PfRh2b allelic differences do not account for overall differences in inhibition, and a more strain-independent immunity has been achieved (Figure 2D). Because age distributions of the 3 geographic populations were not similar, we conducted a multivariate analysis with both age and site as covariates. For inhibition of WT parasites, the age effect was significant at P = .0083; the site effect was significant at P < .0001 (Senegal > Mali, P = .0432; Senegal > Tanzania, P < .0001). For inhibition of DEL parasites, the age effect was marginally significant at P = .0478; the site effect was significant at P < .0001 (Senegal > Mali, P = .0102; Senegal > Tanzania, P = .0002). For inhibition of KO parasites, inhibition was high across all ages and sites, and no significant age effect (P = .0983) or site effect (P = .3728) was observed.
Figure 2.
PfRh2b deletion reveals region-specific differences in immune-mediated inhibition of invasion. A, Purified IgG from Tanzania, Senegal, and Mali were used in highly standardized GIAs with transgenic parasite lines (WT, DEL A, and KO). Inhibition means and 95% CIs are shown. B–D, Inhibitory profiles in each endemic region show differences by strain. Inhibition is displayed as “Inhibition ∆” showing the difference between strains. On the left of each graph is the Inhibition ∆ for WT-KO, with any differences representing differences in inhibition of the PfRh2b (WT) and non-PfRh2b (KO) invasion pathways; in the middle, the Inhibition ∆ are shown for specific allelic inhibition of either PfRh2b full-length (WT) compared to PfRh2b deletion (DEL); and on the right, Inhibition ∆ are shown for specific allelic inhibition of either PfRh2b deletion (DEL) compared to KO.
Significant P values from comparing inhibition of WT to DEL A, and DEL A to KO, using nonparametric-paired t tests (Wilcoxon matched pairs) are indicated.
Abbreviations: CIs, confidence intervals; DEL, PfRh2b deletion single crossover parasites; GIAs, growth inhibition assays; IgG, immunoglobulin G; KO, 3D7∆2b, C2 parasites; ns, not significant; PfRh2b, Plasmodium falciparum reticulocyte-binding protein homologue 2b; WT, PfRh2b wild-type replacement parasite line (1A).
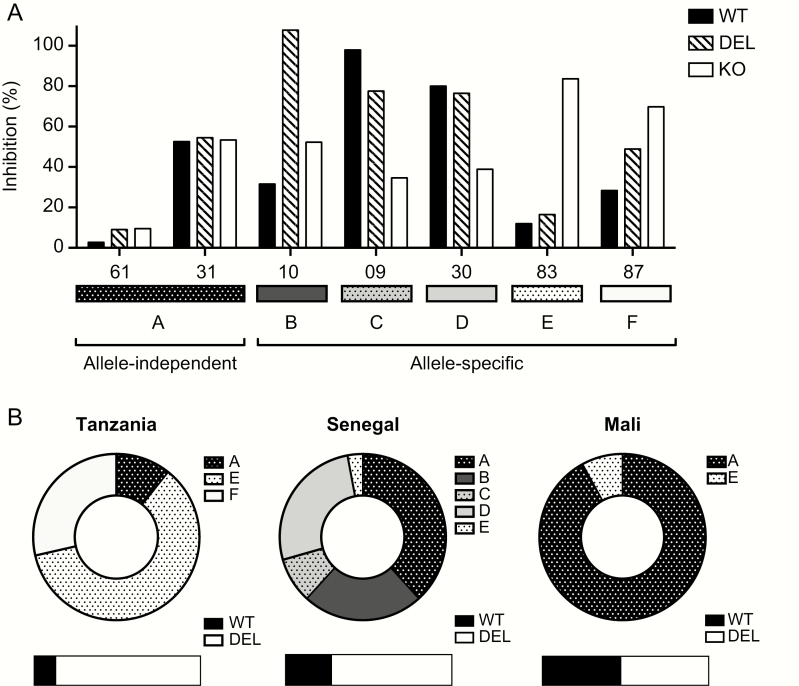
A Model of PfRh2b-Dependent and -Independent Inhibitory Immune Responses
Transgenic parasites expressing allelic variants of a key invasion ligand in an isogenic background allow for (1) a specific determination of the role of a given allele on the increase or decrease in immune neutralization, and (2) the comparison of the role of a given allele on the overall level of inhibitory immunity. These 2 situations are observed using selected individual samples (Figure 3A) and total samples from each African population (Figure 3B). Both allele-specific and allele-independent inhibitory patterns are observed, at different frequencies in each population. In situation A, there is no significant difference in immune inhibition between the 3 transgenic strains, implying that all alleles of PfRh2b (WT, DEL A, and KO) are recognized equivalently. This can be seen in sample 61 where there is very little inhibition, as well as in sample 31 where inhibition is high but is not due to PfRh2b (Figure 3A, situation A). This inhibitory situation is rarely observed in Tanzania, found more frequently in Senegal, and represents the majority situation in Malian samples (Figure 3B). In situations B, C, D, E, and F, different kinds of allele-specific inhibitory profiles are revealed (Figure 3A, situations B–F). Situation C, which is only observed in Senegal, represents high inhibition of PfRh2b WT, partial escape from inhibition with the deletion, and further escape when PfRh2b is absent. Situations E and F represent significantly greater inhibition of the KO than the WT and DEL, implying inhibition of a non-PfRh2b pathway. By assessing the invasion inhibition using allele-specific tools, the inhibitory potential of naturally acquired IgG can be classified as allele-specific or allele-independent, having important implications for vaccine design and candidate prioritization in diverse geographical regions with varying malaria endemicity.
Figure 3.
Model of PfRh2b-dependent and -independent inhibitory immune responses. Examples of immune neutralization that is either PfRh2b allele-specific or allele transcendent is illustrated using individual samples from Senegal (A) and all samples from Tanzania, Senegal, and Mali (B). A significant difference is considered greater than 15% difference in inhibition, the degree of difference tolerated in replicates of the same strain in the GIA. A, Situation A shows no significant difference in immune inhibition between the 3 transgenic strains, whether low inhibition or high inhibition, termed allele-independent immunity. Situations B–F demonstrate differences in inhibition specifically due to the allelic form of PfRh2b (allele-specific immunity). Situation B shows increased inhibition of DEL relative to the WT and KO. Such a result could indicate that the deletion in the protein is revealing an alternative epitope of PfRh2b. In situation C, there is an “immune escape” phenotype observed for both the PfRh2b deletion as well as the KO, implying that the differences in inhibition observed are specifically due to the allele of PfRh2b present. Situation D shows “immune escape” for the PfRh2b KO only. Situation E shows enhanced inhibition of both the WT and the deletion allele, and situation F shows incremental increases in inhibition from WT, to PfRh2b deletion, to PfRh2b KO. B, The fraction of the total for each type of inhibitory situation is shown for each population. The population genetic prevalence of the PfRh2b full-length and PfRh2b deletion alleles are shown below as bars.
Abbreviations: DEL, PfRh2b deletion single crossover parasites; GIA, growth inhibition assay; KO, 3D7∆2b, C2 parasites; PfRh2b, Plasmodium falciparum reticulocyte-binding protein homologue 2b; WT, PfRh2b wild-type replacement parasite line (1A).
DISCUSSION
In the evaluation of novel blood-stage vaccine-candidate antigens, it is critical to consider not only the impact of variant expression of invasion ligands but also the degree to which genetic diversity in amino acid sequence and structure influences immune inhibition. Associative studies implicated the PfRh2b deletion in alternative invasion pathway utilization [16, 17], and population genetic analyses suggest that it may be selected for by strong directional immune pressure [18]. Further, the PfRh2b locus has been implicated in continental-level natural selection, with the coding region deletion present in Africa and a larger 5.9 kilo base pair deletion (spanning the c-terminus and into the neighboring PfRh6 pseudogene) identified in South East Asia [35]. Generating the PfRh2b deletion in an isogenic parasite background made it possible to determine the role of this polymorphism in either invasion pathway utilization or immune evasion, independently from other polymorphisms. Using these transgenic parasites, we found that the deletion does not influence invasion pathway as previously proposed. It is possible that this result is due to the deletion being linked with other polymorphisms in field-isolated parasites that influence the invasion pathway. Alternatively, it is possible that the deletion does not alter the invasion pathway in the 3D7 genetic background used in generating the transgenic lines. However, we tested PfRh2b deletion transgenic clones generated in the W2Mef genetic background and similarly found no influence of the deletion on invasion pathway (Supplementary Figure 2). Short-term measures of cycle length and invasion efficiency were not significantly different for the deletion parasites compared to controls; however, an increased growth rate over time was observed for deletion lines. This enhanced growth rate over multiple cycles of invasion could provide the parasites with a selective advantage in vivo in either the presence or absence of neutralizing antibodies.
We observed that the PfRh2b deletion allows parasites to escape the inhibitory effect of PfRh2b c-terminal unique domain-specific antibodies. When total IgG from malaria-endemic regions is tested, various inhibitory scenarios are observed, including PfRh2b-independent inhibition (situation A), enhanced inhibition of the PfRh2b deletion (situation B), immune escape for parasites with the PfRh2b deletion (situation C), immune escape of the KO (situation D), and enhanced inhibition of the KO, implying inhibition of a non-PfRh2b pathway (situations E and F) (Figure 3). The overall pattern of allele-specific versus allele-independent inhibitory immune responses varies by population (Figure 3B), likely due to differences in invasion pathways used in each population [13, 16, 17], differences in exposure to particular allelic variants, as well as the cumulative frequency of malaria exposure in the individual. Having the ability to distinguish these different inhibitory situations sheds light on the mechanisms of anti-PfRh2b immunity generated in natural populations. While both site- and age- specific differences were observed for WT and DEL parasites, KO parasites showed consistently high levels of inhibition in all sites. The PfRh2b knockout (KO) parasite is more dependent on a neuraminidase-sensitive, chymotrypsin-resistant invasion pathway. Equivalent inhibition of KO across age groups and sites, and with high inhibition (even in younger children), might imply that antibodies against sialic acid–dependent, chymotrypsin-independent pathways (such as those used by EBA-175) might be generated first, as has been observed using serum samples from Kenya [36], and has been suggested by others who found an inverse correlation between EBA-175 expression and age in The Gambia, presumably due to immune selection [37]. An alternative explanation is that a KO of any invasion ligand results in a decrease in the overall repertoire of inhibitable ligands—making the strain more susceptible to inhibition in general [36, 38]. This could be possible in the case of the PfRh2b KO, as there is no upregulation of alternative ligands, just a revealing of alternative ligands without a compensatory change [39].
Besides invasion pathway, another factor influencing geographic differences in PfRh2b allele-specific immune responses is the population prevalence of the wild-type or deletion alleles. While the population prevalence of the deletion is relatively high within African countries, it varies dramatically in Latin America and Southeast Asia [18]. It is possible that in regions where more inhibitory responses exist against PfRh2b, the deletion allele may be selected to higher frequencies. Studies in Papua New Guinea have implicated antibodies against PfRh2b in protection from malaria [40] and, intriguingly, there is a high frequency of the deletion in not-too-distant Malaysia (Sabah). Alternatively, inhibitory antibodies against the deletion could simply reflect greater prevalence, thus greater immune exposure, to the deletion allele. In the populations studied here, while Tanzania and Senegal both have high prevalence of the deletion (87% and 72%, respectively; Figure 3B [18],), Mali has a deletion prevalence of 52.5% (Figure 3B), meaning infected individuals in Mali are equally likely to be infected with the full length compared to the deletion. Interestingly, Mali shows few allele-specific differences in inhibition, whereas dramatic differences are observed in both Tanzania and Senegal, regions with more variation in allelic frequency; and in the case of Tanzania, high variant expression and alternative invasion pathway usage [13]. While the data here might tend to favor the argument that dramatic differences in the prevalence of the allele in the population will result in allele-specific inhibitory antibodies directed against that allele, further studies from diverse geographic locations with varying endemicities are needed to determine the association between PfRh2b deletion frequency and immune pressure at this locus.
Interest in pursuing blood-stage vaccines for P. falciparum is growing, and the PfRh and PfEBA families of invasion ligands are potential candidates, whether singly or in combination [38, 41]. However, dynamics such as variant expression and polymorphism will be very important to consider before moving forward with such vaccines. Interestingly, the malaria field has not rapidly adapted transgenic approaches to assess immune inhibition and invasion [8, 36, 42, 43]. To date, only 1 study has used allelic replacement to study the impact of naturally acquired polymorphisms in the context of immune inhibition [8]. While some have used chimeric parasites or knockouts to assess immune inhibition from naturally acquired antibodies from a single-endemic country [42, 44], none have done so comparatively in different malaria-endemic countries. Our study combines transgenic parasites expressing naturally occurring PfRh2b alleles with naturally acquired IgG to assess the PfRh2b allelic contribution to invasion inhibition comparatively from 3 malaria-endemic countries. From this data, we can conclude that rather than having a role in alternative invasion pathway utilization, the deletion could have been selected through population-specific immune-mediated selection. The genetic methodologies described here provide a method for assessing inhibitory immune responses to allelic variants in vitro and facilitate comparison of the specific contribution of allele-specific antibodies to the overall inhibitory effect—a critical aspect of prioritizing vaccine candidates in the face of genetic diversity.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Acknowledgments. We are grateful to Dr Johanna Daily, Younous Diedhiou, Amadou Makhtar Mbaye, Omar Ly, Lamine Ndiaye, and Dior Diop for the collection and contribution of patient samples in Thiès, Senegal. We are also grateful to Dr Saibou Doumbia, Dr Drissa Konate, and Mr Abdoul Salam Keita for their assistance with Kenieroba, Mali, sample collection. We thank Christopher Membi, Billy Ngasala, and Marcelina Mubi for assistance with the collection of patient samples in Mlandizi, Tanzania. We also wish to acknowledge all the patients who participated in the studies. Growth inhibition assay establishment in Senegal was performed with the assistance of Iguosadolo Nosamiefan, and we are grateful to Samuel E. Moretz (National Institute of Allergy and Infectious Diseases [NIAID]) for assistance with IgG purification. Immunofluorescent images were acquired at the Mental Retardation Developmental Disability Research Center (MRDDRC) Imaging Core, supported by grant no. NIH-P30-HD-18655, with assistance from Lihong Bu and Anthony Hill.
Financial support. This work was supported by the National Institutes of Health (NIH) grants R01AI057919 and 1R03TW008053 (to M. T. D.), and partially supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health. A. K. B. is supported by a US Department of State Fulbright Fellowship, a Harvard Institute for Global Health fellowship, a Centers for Disease Control and Prevention grant (R36 CK000119-01), an Epidemiology of Infectious Disease and Biodefense Training grant (2T32 AI007535-12), and an International Research Scientist Development Award (1K01TW010496). A. D. A. is a Fogarty trainee supported by an NIH grant (5D43TW001503-09) to Dr Dyann Wirth. J. D. D. is a Pediatric Scientist Development Program Fellow supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (K12-HD000850) and the NIAID (K08-AI087874 and R01-AI102907). M. T. D. is a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Diseases.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. World Malaria Report Geneva, Switzerland: World Health Organization, 2015. [Google Scholar]
- 2. Chotivanich K, Udomsangpetch R, Simpson JA et al. Parasite multiplication potential and the severity of falciparum malaria. J Infect Dis 2000; 181:1206–9. [DOI] [PubMed] [Google Scholar]
- 3. Richards JS, Beeson JG. The future for blood-stage vaccines against malaria. Immunol Cell Biol 2009; 87:377–90. [DOI] [PubMed] [Google Scholar]
- 4. Osier FH, Mackinnon MJ, Crosnier C et al. New antigens for a multicomponent blood-stage malaria vaccine. Sci Transl Med 2014; 6:247ra102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goodman AL, Draper SJ. Blood-stage malaria vaccines—recent progress and future challenges. Ann Trop Med Parasitol 2010; 104:189–211. [DOI] [PubMed] [Google Scholar]
- 6. Tham WH, Beeson JG, Rayner JC. Plasmodium vivax vaccine research—we’ve only just begun. Int J Parasitol 2017; 47:111–8. [DOI] [PubMed] [Google Scholar]
- 7. Mayer DC, Mu JB, Kaneko O, Duan J, Su XZ, Miller LH. Polymorphism in the Plasmodium falciparum erythrocyte-binding ligand JESEBL/EBA-181 alters its receptor specificity. Proc Natl Acad Sci USA 2004; 101:2518–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Healer J, Murphy V, Hodder AN et al. Allelic polymorphisms in apical membrane antigen-1 are responsible for evasion of antibody-mediated inhibition in Plasmodium falciparum. Mol Microbiol 2004; 52:159–68. [DOI] [PubMed] [Google Scholar]
- 9. Duraisingh MT, Triglia T, Ralph SA et al. Phenotypic variation of Plasmodium falciparum merozoite proteins directs receptor targeting for invasion of human erythrocytes. EMBO J 2003; 22:1047–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rayner JC, Galinski MR, Ingravallo P, Barnwell JW. Two Plasmodium falciparum genes express merozoite proteins that are related to Plasmodium vivax and Plasmodium yoelii adhesive proteins involved in host cell selection and invasion. Proc Natl Acad Sci USA 2000; 97:9648–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tham WH, Healer J, Cowman AF. Erythrocyte and reticulocyte binding-like proteins of Plasmodium falciparum. Trends Parasitol 2012; 28:23–30. [DOI] [PubMed] [Google Scholar]
- 12. Gunalan K, Gao X, Liew KJ, Preiser PR. Differences in erythrocyte receptor specificity of different parts of the Plasmodium falciparum reticulocyte binding protein homologue 2a. Infect Immun 2011; 79:3421–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bei AK, Membi CD, Rayner JC et al. Variant merozoite protein expression is associated with erythrocyte invasion phenotypes in Plasmodium falciparum isolates from Tanzania. Mol Biochem Parasitol 2007; 153:66–71. [DOI] [PubMed] [Google Scholar]
- 14. Triglia T, Duraisingh MT, Good RT, Cowman AF. Reticulocyte-binding protein homologue 1 is required for sialic acid–dependent invasion into human erythrocytes by Plasmodium falciparum. Mol Microbiol 2005; 55:162–74. [DOI] [PubMed] [Google Scholar]
- 15. Taylor HM, Grainger M, Holder AA. Variation in the expression of a Plasmodium falciparum protein family implicated in erythrocyte invasion. Infect Immun 2002; 70:5779–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jennings CV, Ahouidi AD, Zilversmit M et al. Molecular analysis of erythrocyte invasion in Plasmodium falciparum isolates from Senegal. Infect Immun 2007; 75:3531–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lantos PM, Ahouidi AD, Bei AK et al. Erythrocyte invasion profiles are associated with a common invasion ligand polymorphism in Senegalese isolates of Plasmodium falciparum. Parasitology 2009; 136:1–9. [DOI] [PubMed] [Google Scholar]
- 18. Ahouidi AD, Bei AK, Neafsey DE et al. Population genetic analysis of large sequence polymorphisms in Plasmodium falciparum blood-stage antigens. Infect Genet Evol 2010; 10:200–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Binks RH, Baum J, Oduola AM et al. Population genetic analysis of the Plasmodium falciparum erythrocyte binding antigen-175 (eba-175) gene. Mol Biochem Parasitol 2001; 114:63–70. [DOI] [PubMed] [Google Scholar]
- 20. Conway DJ, Cavanagh DR, Tanabe K et al. A principal target of human immunity to malaria identified by molecular population genetic and immunological analyses. Nat Med 2000; 6:689–92. [DOI] [PubMed] [Google Scholar]
- 21. Trape JF, Lefebvre-Zante E, Legros F et al. Vector density gradients and the epidemiology of urban malaria in Dakar, Senegal. Am J Trop Med Hyg 1992; 47:181–9. [DOI] [PubMed] [Google Scholar]
- 22. Bhatt S, Weiss DJ, Cameron E et al. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature 2015; 526:207–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Premji Z, Ndayanga P, Shiff C, Minjas J, Lubega P, MacLeod J. Community based studies on childhood mortality in a malaria holoendemic area on the Tanzanian coast. Acta Trop 1997; 63:101–9. [DOI] [PubMed] [Google Scholar]
- 24. Carneiro I, Smith L, Ross A et al. Intermittent preventive treatment for malaria in infants: a decision-support tool for sub–Saharan Africa. Bull World Health Organ 2010; 88:807–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. DeSimone TM, Bei AK, Jennings CV, Duraisingh MT. Genetic analysis of the cytoplasmic domain of the PfRh2b merozoite invasion protein of Plasmodium falciparum. Int J Parasitol 2009; 39:399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Duraisingh MT, Maier AG, Triglia T, Cowman AF. Erythrocyte-binding antigen 175 mediates invasion in Plasmodium falciparum utilizing sialic acid-dependent and -independent pathways. Proc Natl Acad Sci USA 2003; 100:4796–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reed MB, Caruana SR, Batchelor AH, Thompson JK, Crabb BS, Cowman AF. Targeted disruption of an erythrocyte binding antigen in Plasmodium falciparum is associated with a switch toward a sialic acid–independent pathway of invasion. Proc Natl Acad Sci USA 2000; 97:7509–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Simpson JA, Silamut K, Chotivanich K, Pukrittayakamee S, White NJ. Red cell selectivity in malaria: a study of multiple-infected erythrocytes. Trans R Soc Trop Med Hyg 1999; 93:165–8. [DOI] [PubMed] [Google Scholar]
- 29. Chotivanich KT, Pukrittayakamee S, Simpson JA, White NJ, Udomsangpetch R. Characteristics of Plasmodium vivax–infected erythrocyte rosettes. Am J Trop Med Hyg 1998; 59:73–6. [DOI] [PubMed] [Google Scholar]
- 30. Reilly HB, Wang H, Steuter JA, Marx AM, Ferdig MT. Quantitative dissection of clone-specific growth rates in cultured malaria parasites. Int J Parasitol 2007; 37:1599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Triglia T, Thompson J, Caruana SR, Delorenzi M, Speed T, Cowman AF. Identification of proteins from Plasmodium falciparum that are homologous to reticulocyte binding proteins in Plasmodium vivax. Infect Immun 2001; 69:1084–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rayner JC, Vargas-Serrato E, Huber CS, Galinski MR, Barnwell JW. A Plasmodium falciparum homologue of Plasmodium vivax reticulocyte binding protein (PvRBP1) defines a trypsin-resistant erythrocyte invasion pathway. J Exp Med 2001; 194:1571–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Miura K, Zhou H, Moretz SE et al. Comparison of biological activity of human anti-apical membrane antigen-1 antibodies induced by natural infection and vaccination. J Immunol 2008; 181:8776–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Van Tyne D, Uboldi AD, Healer J, Cowman AF, Wirth DF. Modulation of PF10_0355 (MSPDBL2) alters Plasmodium falciparum response to antimalarial drugs. Antimicrob Agents Chemother 2013; 57:2937–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cheeseman IH, Miller B, Tan JC et al. Population structure shapes copy number variation in malaria parasites. Mol Biol Evol 2016; 33:603–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Persson KE, McCallum FJ, Reiling L et al. Variation in use of erythrocyte invasion pathways by Plasmodium falciparum mediates evasion of human inhibitory antibodies. J Clin Invest 2008; 118:342–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gomez-Escobar N, Amambua-Ngwa A, Walther M, Okebe J, Ebonyi A, Conway DJ. Erythrocyte invasion and merozoite ligand gene expression in severe and mild Plasmodium falciparum malaria. J Infect Dis 2010; 201:444–52. [DOI] [PubMed] [Google Scholar]
- 38. Lopaticki S, Maier AG, Thompson J et al. Reticulocyte and erythrocyte binding–like proteins function cooperatively in invasion of human erythrocytes by malaria parasites. Infect Immun 2011; 79:1107–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baum J, Maier AG, Good RT, Simpson KM, Cowman AF. Invasion by P. falciparum merozoites suggests a hierarchy of molecular interactions. PLOS Pathog 2005; 1:e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reiling L, Richards JS, Fowkes FJ et al. Evidence that the erythrocyte invasion ligand PfRh2 is a target of protective immunity against Plasmodium falciparum malaria. J Immunol 2010; 185:6157–67. [DOI] [PubMed] [Google Scholar]
- 41. Richards JS, Beeson JG. The future for blood-stage vaccines against malaria. Immunol Cell Biol 2009; 87:377–90. [DOI] [PubMed] [Google Scholar]
- 42. O’Donnell RA, de Koning-Ward TF, Burt RA et al. Antibodies against merozoite surface protein (MSP)-1(19) are a major component of the invasion-inhibitory response in individuals immune to malaria. J Exp Med 2001; 193:1403–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. O’Donnell RA, Saul A, Cowman AF, Crabb BS. Functional conservation of the malaria vaccine antigen MSP-119across distantly related Plasmodium species. Nat Med 2000; 6:91–5. [DOI] [PubMed] [Google Scholar]
- 44. Persson KE, Lee CT, Marsh K, Beeson JG. Development and optimization of high-throughput methods to measure Plasmodium falciparum–specific growth inhibitory antibodies. J Clin Microbiol 2006; 44:1665–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Triglia T, Chen L, Lopaticki S et al. Plasmodium falciparum merozoite invasion is inhibited by antibodies that target the PfRh2a and b binding domains. PLOS Pathog 2011; 7:e1002075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.