To facilitate bioinformatics analysis of high-throughput sequencing (HTS) data for the detection of both known and novel viruses, we have developed a new reference viral database (RVDB) that provides a broad representation of different virus species from eukaryotes by including all viral, virus-like, and virus-related sequences (excluding bacteriophages), regardless of their size. In particular, RVDB contains endogenous nonretroviral elements, endogenous retroviruses, and retrotransposons. Sequences were clustered to reduce redundancy while retaining high viral sequence diversity. A particularly useful feature of RVDB is the reduction of cellular sequences, which can enhance the run efficiency of large transcriptomic and genomic data analysis and increase the specificity of virus detection.
KEYWORDS: RVDB, adventitious viruses, bioinformatics analysis, high-throughput sequencing, reference virus database, viral sequences, virus detection
ABSTRACT
Detection of distantly related viruses by high-throughput sequencing (HTS) is bioinformatically challenging because of the lack of a public database containing all viral sequences, without abundant nonviral sequences, which can extend runtime and obscure viral hits. Our reference viral database (RVDB) includes all viral, virus-related, and virus-like nucleotide sequences (excluding bacterial viruses), regardless of length, and with overall reduced cellular sequences. Semantic selection criteria (SEM-I) were used to select viral sequences from GenBank, resulting in a first-generation viral database (VDB). This database was manually and computationally reviewed, resulting in refined, semantic selection criteria (SEM-R), which were applied to a new download of updated GenBank sequences to create a second-generation VDB. Viral entries in the latter were clustered at 98% by CD-HIT-EST to reduce redundancy while retaining high viral sequence diversity. The viral identity of the clustered representative sequences (creps) was confirmed by BLAST searches in NCBI databases and HMMER searches in PFAM and DFAM databases. The resulting RVDB contained a broad representation of viral families, sequence diversity, and a reduced cellular content; it includes full-length and partial sequences and endogenous nonretroviral elements, endogenous retroviruses, and retrotransposons. Testing of RVDBv10.2, with an in-house HTS transcriptomic data set indicated a significantly faster run for virus detection than interrogating the entirety of the NCBI nonredundant nucleotide database, which contains all viral sequences but also nonviral sequences. RVDB is publically available for facilitating HTS analysis, particularly for novel virus detection. It is meant to be updated on a regular basis to include new viral sequences added to GenBank.
IMPORTANCE To facilitate bioinformatics analysis of high-throughput sequencing (HTS) data for the detection of both known and novel viruses, we have developed a new reference viral database (RVDB) that provides a broad representation of different virus species from eukaryotes by including all viral, virus-like, and virus-related sequences (excluding bacteriophages), regardless of their size. In particular, RVDB contains endogenous nonretroviral elements, endogenous retroviruses, and retrotransposons. Sequences were clustered to reduce redundancy while retaining high viral sequence diversity. A particularly useful feature of RVDB is the reduction of cellular sequences, which can enhance the run efficiency of large transcriptomic and genomic data analysis and increase the specificity of virus detection.
INTRODUCTION
Of the various advanced nucleic acid-based technologies that have recently been developed for broad virus detection (1), high-throughput sequencing (HTS) has demonstrated broad capabilities for the detection of known and novel viruses in a variety of different sample types, including environmental, clinical, and biological samples such as cell lines and biological products (2, 3). Methods that can detect known and novel viruses can be useful for demonstrating the absence of adventitious (“unwanted”) viruses, particularly when new cell lines are used to manufacture biologics (4). An ongoing challenge is the analysis of large amounts (often gigabytes or even terabytes) of nucleotide sequences that are generated from HTS, which can often result in a “bioinformatics bottleneck” because of limits of computational capacity, data storage, or data transfer (5). Additionally, although there are several public databases available for sequence analysis, they have some limitations for the analysis of large HTS data sets. In particular, the detection of distantly related sequences in novel viruses can be difficult because of incomplete representation of all viral sequences in a single database (2). GenBank currently serves as the largest publicly available nucleic acid sequences data bank (6) and is maintained by the National Center for Biotechnology Information (NCBI), National Library of Medicine, National Institutes of Health (Bethesda, MD). NCBI further maintains the (partially) nonredundant nucleotide (nr/nt) database, which contains entries from traditional divisions of GenBank, as well as from the European Molecular Biology Laboratory-European Bioinformatics Institute Nucleotide Sequence Database (EMBL-EBI) (7) maintained by the European Nucleotide Archive (ENA; Cambridge, United Kingdom), and the DNA Data Bank of Japan (DDBJ; Mishima, Japan) (8), and also includes the Reference Sequence (RefSeq) collection (9) and the Protein Data Bank (PDB) (10) sequences.
The NCBI nr/nt database is widely used by researchers for sequence analysis; however, detection of viral sequences can be missed for several reasons, such as the lack of a closely related sequence, misannotation of sequences (especially those archived in GenBank, since these can only be corrected by the original submitter), or the presence of a large number of cellular sequences that may overshadow a positive viral hit. In such cases, follow-up analysis of hits becomes more complex. One approach to overcome this challenge of HTS data analysis for virus detection could be to limit the BLAST search to viruses; however, this would miss the detection of viral elements such as endogenous retroviruses, which in many cases are associated with flanking host genomic sequences and therefore have been assigned the host taxonomy. The viral RefSeq data set provides manually curated reference genome records for each viral species (11). Typically, each viral species includes one RefSeq reference, and when possible, these references include high-quality annotation provided by NCBI staff or members of the scientific community. The viral RefSeq data set is intended to be nonredundant and is derived from full-length or nearly full-length viral genomes within the viral and phage divisions (VRL and PHG) in GenBank. This means that the viral RefSeq data set does not include viral sequences classified into other GenBank divisions, viral sequences that are part of host genomes, partial sequences, or transposable elements, and it lacks the intraspecies sequence diversity found in the NCBI nr/nt database. The NCBI Viral Genomes data set includes RefSeq sequences and so-called genome neighbors, which are validated complete or nearly complete viral genomes (11), and is available as a downloadable set of approximately 105,000 complete eukaryotic viral genomes and nearly 4,000 bacterial viral genomes (bacteriophages) (as of April 2017). While this second data set better represents the sequence diversity found within each viral species and can be searched by using BLAST, it does not contain the full-spectrum of viral, virus-related, and virus-like sequences that may be available as complete or partial genomes.
Numerous other resources for viral sequences exist, but these generally tend to be virus family specific, with a particular focus on pathogenic viruses responsible for high-impact infectious diseases, such as hepatitis C virus, HIV-1, hemorrhagic fever virus, influenza A virus, dengue virus, and West Nile virus (12). The International Committee on Taxonomy of Viruses (ICTV), which is responsible for classifying the ~3,000 species of viruses at its ICTV Taxonomy website (https://talk.ictvonline.org/taxonomy/) (13), has recently recognized the need to update the virus taxonomy with considerations for virus sequence discovery by using HTS (14, 15). With the flood of viral sequences in recent years as a result of next-generation sequencing technologies, there has been an explosion in the number of novel bioinformatics tools that focus on fundamental aspects of sequence and taxonomic analysis, such as genome annotation, prediction of open reading frames (ORFs), and genotyping, and each is usually limited to a small group of viruses (12). However, since these resources generally extend previous repositories of family-specific viral sequence information, there is a need for a more comprehensive reference virus sequence database. At the other end of the spectrum, the NCBI Sequence Read Archive of HTS data (16) may have sequences for endogenous retroviruses and retrotransposons in addition to those in GenBank but without sequence annotation, and at ~10,000 TB and ~1.1 × 1016 bases (as of April 2017), searching this resource is a major bioinformatics endeavor on its own.
More recently, other databases have been created containing only endogenous viral elements including endogenous retrovirus and retrotransposon sequences. Sequences in gEVE (17) have been selected by using bioinformatics tools and include over 700,000 endogenous retroviral ORFs/motifs mined from 20 genomes obtained from 19 different mammalian species. However, it should be noted that the majority of gEVE-predicted sequences have yet to be experimentally validated and the retroviral sequences have yet to be characterized. Additionally, there also exist human endogenous retroviral sequence databases, HERVd (18) and the new HERVgDB4 (19), and a mouse endogenous retroviral sequence database, ERE (20). Gypsy Database (GyDB) release 2.0 (21) provides an extensive collection of retroelements, which includes complete genomes, long terminal repeats (LTRs), and core sequences, along with the organization of the elements into families and lineages. GyDB is also extensively linked to external sources of information by its Wiki framework and provides hidden Markov model profiles for over 300 mobile elements. GyDB can serve as a useful hub for the organization and integration of information regarding retrotransposons; however, it is not ideally suited as a repository of information for interrogation, since in contains only 2,579 sequences and has not been updated since 2010. Another database that contains repetitive elements in eukaryotic genomes is RepBase, which is updated regularly and contains over 30,000 sequences (22, 23); however, only a fraction of the sequences are LTR retroelements (endogenous retroviruses and retrotransposons), and these are consensus sequences. DFAM (24), which relies heavily on RepBase as a reference, contains 2,656 families of LTR retrotransposons. Therefore, like GyDB, RepBase and DFAM are useful primarily as reference databases for retroelements. In addition to these databases, several other databases of retrotransposons exist that are focused on specific organisms, including soyTMdb for soybean elements (25), BmTEdb for silkworm elements (26), MnTEdb for mulberry elements (27), and DPTEdb for dioecious plant elements (28).
The majority of the resources described above are specific to a taxonomic group or type of viral element, and some may also contain a high degree of redundancy. The goal of our study was to create a nonredundant, well-characterized reference viral database (RVDB) that includes all viral, virus-related, and virus-like entries, mainly from eukaryotes, representing complete viral genomes or partial viral sequences. Additionally, there is an overall reduction of host cell sequence content, which is expected to enhance HTS data analysis for known and novel virus detection.
RESULTS
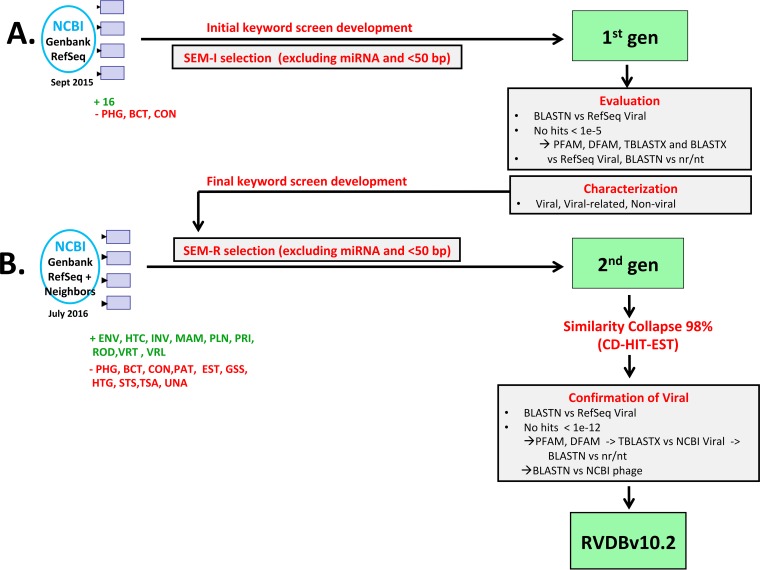
RVDB was created by using a semantic data mining approach, which broadened the selection of viral entries from GenBank to include virus-related and virus-like sequences such as LTR retrotransposons, regardless of sequence length (>50 bp). Figure 1 shows schematically the different GenBank divisions used to develop an initial viral database (designated first-generation VDB) and the generation of a second-generation VDB, which resulted in the development of the final RVDB. Briefly, the first-generation VDB was generated by using positive keywords to capture all viruses and retroelements and negative keywords to exclude nonviral sequences, as well as size and microRNA (miRNA) exclusion criteria and negative rules (for details, see Materials and Methods). These initial selection criteria were designated semantic initial or SEM-I. The resulting first-generation VDB contained a total of 3,724,251 sequences. The sequences selected by SEM-I were further queried by using an array of sequence homology tools to confirm their viral identity (described in Materials and Methods). Further review and characterization of the sequences in the first-generation VDB led to the development of refined semantic selection criteria (designated semantic refined or SEM-R), which consisted of a final extended set of positive and negative keywords, rules, and regular expressions that was used to develop a second-generation VDB. SEM-R contained fewer positive words than the original SEM-I but had an increased number of negative words, rules, and regular expressions aimed at removing nonviral sequences from the database. The final RVDB was clustered at 98% nucleotide sequence identity to reduced redundancy and retain diversity. The following sections present details of the development and characterization of RVDBv10.2, which was the first public version of the database.
FIG 1 .
Workflow for the development of RVDB. (A) Development and characterization of the first-generation (gen) VDB by using an initial semantic screening, SEM-I. Review of sequences obtained from SEM-I, including BLASTX, TBLASTX, PFAM, and DFAM analyses, led to the development of a refined semantic screening, SEM-R. (B) Redevelopment of the second-generation VDB by using SEM-R and generation of the final RVDBv10.2. The enhanced screening SEM-R was used to select sequences from the TPA and GenBank divisions again in July 2016 (excluding PHG, BCT, CON, and an additional seven divisions), and NCBI Viral Genomes (RefSeq+Neighbors) were added directly without any screening. Furthermore, the nonphage sequences in VRL were also directly added without any screening. The sequences were then clustered by using CD-HIT-EST at 98% nucleotide sequence identity. Viral identity was confirmed by using various bioinformatics searches and a BLASTN search of the nr/nt database identified bacteriophage sequence and sequences with no viral or virus-related hits in the nr/nt database for removal from RVDB.
Characterization of unclustered and clustered sequences in RVDBv10.2.
RVDBv10.2 contained 2,073,535 sequences in its unclustered form (designated U-RVDBv10.2), of which 1,944,656 (93.8%) were from the VRL division of GenBank, while of the remaining 128,879 sequences, 91,968 were from NCBI Viral Genomes (including 5,058 RefSeq and 86,910 neighbors), and 36,911 were from the ENV, HTC, INV, MAM, PLN, PRI, ROD, TPA, and VRT divisions of GenBank (Table 1). The clustered form of RVDBv10.2 (designated C-RVDBv10.2), which was generated to retain viral diversity and reduce redundancy by clustering at 98% sequence identity, contained 561,676 clustered representative sequences (creps), of which 511,731 (91.1%) were from the VRL division of GenBank, while of the remaining 49,945 sequences, 28,021 were from NCBI Viral Genomes (consisting of 5,035 RefSeq and 22,986 neighbor sequences), and 21,924 sequences were from the ENV, HTC, INV, MAM, PLN, PRI, ROD, TPA (third-party annotation), and VRT divisions of GenBank (Table 1).
TABLE 1 .
Number of entries in major categories of sequences in RVDBv10.2 by GenBank divisions or NCBI collectionsa
| Division(s) | No. of sequences in: |
|||||
|---|---|---|---|---|---|---|
| Exogenous viral |
Endogenous nonretroviral |
Endogenous retroviral |
LTR retrotransposon |
Unassigned viral genes/fragments |
All of RVDB |
|
| U-RVDB | ||||||
| VRL | 1,943,041 | 0 | 1,530 | 85 | 0 | 1,944,656 |
| ENV | 6,368 | 0 | 0 | 12 | 311 | 6,691 |
| HTC | 105 | 0 | 13 | 31 | 104 | 253 |
| INV | 40 | 117 | 39 | 2,550 | 561 | 3,307 |
| MAM | 26 | 15 | 2,028 | 16 | 106 | 2,191 |
| PLN | 205 | 356 | 228 | 17,250 | 637 | 18,676 |
| PRI | 88 | 130 | 3,560 | 89 | 94 | 3,961 |
| ROD | 148 | 1 | 553 | 86 | 73 | 861 |
| TPA | 20 | 0 | 0 | 51 | 0 | 71 |
| VRT | 64 | 63 | 443 | 237 | 93 | 900 |
| RefSeq Viral |
5,051 | 1 | 6 | 0 | 0 | 5,058 |
| NCBI Viral Genomesb |
86,893 | 0 | 17 | 0 | 0 | 86,910 |
| All | 2,042,049 | 683 | 8,417 | 20,407 | 1,979 | 2,073,535 |
| C-RVDB | ||||||
| VRL | 511,415 | 0 | 290 | 26 | 0 | 511,731 |
| ENV | 2,434 | 0 | 0 | 11 | 208 | 2,653 |
| HTC | 73 | 0 | 9 | 27 | 97 | 206 |
| INV | 25 | 112 | 39 | 1,496 | 262 | 1,934 |
| MAM | 10 | 13 | 859 | 14 | 32 | 928 |
| PLN | 131 | 156 | 139 | 12,549 | 414 | 13,389 |
| PRI | 55 | 2 | 1,517 | 54 | 56 | 1,684 |
| ROD | 64 | 1 | 325 | 63 | 59 | 512 |
| TPA | 20 | 0 | 0 | 51 | 0 | 71 |
| VRT | 17 | 46 | 227 | 192 | 65 | 547 |
| RefSeq Viral |
5,028 | 1 | 6 | 0 | 0 | 5,035 |
| NCBI Viral Genomesb |
22,971 | 0 | 15 | 0 | 0 | 22,986 |
| All | 542,243 | 331 | 3,426 | 14,483 | 1,193 | 561,676 |
NCBI RefSeq Viral and Viral Genomes minus RefSeq sequences are shown.
Minus RefSeq Viral.
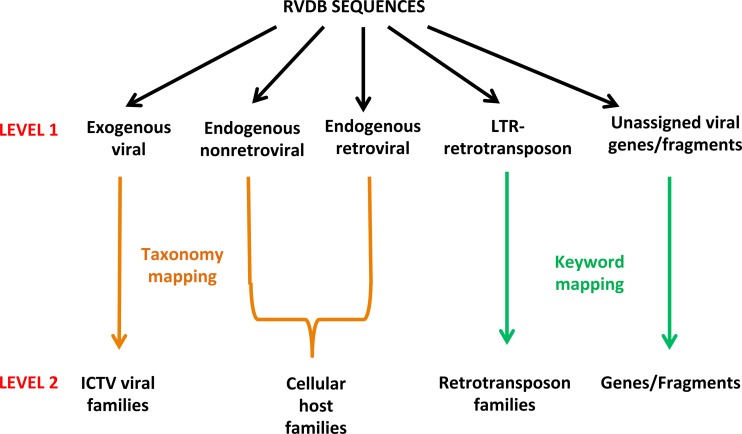
RVDB sequences were further characterized at two progressive levels (Fig. 2). At level 1, all of the viral, virus-related, and virus-like sequences were categorized as exogenous viral, endogenous nonretroviral, endogenous retroviral, and LTR retrotransposon. This characterization was done by using SEM-R after dividing it into the four corresponding categories and sequences that remained unallocated to any of the four categories after this process were placed in an unassigned viral genes/fragments category. The scripts used for level 1 characterization were designated RVDB_characterization.py; their description and instructions for use are provided in Materials and Methods (see also the characterization script in Text S2 in the supplemental material). At level 2, families were created by using different approaches based on the level 1 category. For the exogenous viral category, sequences were broken down by viral family taxonomic identifier; sequences in the endogenous nonretroviral and endogenous retroviral categories were further characterized by host family taxonomic identifier; sequences in the LTR retrotransposon category were further characterized by keywords into major families of retroviral elements; and sequences in the unassigned viral genes/fragments category were characterized by specific keywords related to gene/fragment names, e.g., pol, gag. Some manual review was required to properly group all of the endogenous retroviral sequences into their host families, since some sequences had been labeled as family Retroviridae rather than their host family. To evaluate the distribution of sequences in the different databases, the number of sequences assigned to the different categories and families in RVDBv10.2 were compared with the similar characterization of NCBI Viral Genomes (July 2016 version) in both the unclustered and clustered forms of each database. Additionally, the exogenous viral sequences were also compared with the May 2017 version of NCBI Viral Genomes (unclustered) to compare the addition of new sequences to this database since July 2016. The results are presented in Table S1A to E, characterization (Table S1A to E correspond to exogenous viral [ExViral], endogenous nonretroviral [ENRV], endogenous retroviral [ERV], LTR retrotransposons, and unassigned viral genes/fragments, respectively). The analysis indicated viral sequence families that were underrepresented in RVDBv10.2 and in NCBI Viral Genomes or not represented in the latter at all. Furthermore, sequences were identified that could not be taxonomically mapped in current virus families or had host taxonomy (indicated in the boxed region in Table S1A and C). Details about the taxonomic grouping are described in Materials and Methods.
FIG 2 .
Characterization of RVDB sequences. Sequences in RVDB were sorted into viral categories by using specific criteria derived from SEM-R (level 1) and then further grouped into families on the basis of taxonomy or group-specific keywords (level 2). An overview of the two-tier approach used is shown.
Sequences in RVDB and NCBI Viral Genomes were characterized by using the python script designated RVDB_characterization.py into the level 1 categories exogenous viral (ExViral), endogenous nonretroviral (ENRV), endogenous retroviral (ERV), and LTR retrotransposon, and the remaining viral sequences were categorized as unassigned genes/fragments. The number of entries based on family taxonomic identifiers or names is indicated. Entries that could not be mapped on the basis of a taxonomic identifier or were outdated are boxed. The characterization script used is available at https://github.com/ArifaKhanLab/RVDB, along with the instructions for its use (the latter is also provided in Text S2). Download TABLE S1, XLSX file, 0.1 MB (29.6KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The majority of the sequences were from the exogenous viral category and were primarily from the GenBank VRL division, (1,943,041 in U-RVDB and 511,415 in C-RVDB; Table 1) and from NCBI Viral Genomes (shown separately from RefSeq Viral in Table 1; the total including RefSeq Viral was 91,944 in U-RVDB and 27,999 in C-RVDB). However, a number of exogenous viral sequences were also from non-VRL GenBank divisions, primarily from the GenBank ENV division, with 7,064 in U-RVDB and 2,829 in C-RVDB (Table 1). There were only a small number of endogenous nonretroviral sequences compared to other viral categories, with 683 in U-RVDB and 331 in C-RVDB (Table 1); these were predominantly from the INV, PLN, and PRI divisions of GenBank. Nearly half of all endogenous retroviral sequences were from the PRI division of GenBank, with the remainder predominantly from the VRL, MAM, ROD, and VRT divisions. In the case of endogenous retroviruses, there were 8,417 entries in U-RVDB and 3,426 in C-RVDB (Table 1). LTR retrotransposon sequences were largely selected from the INV, PLN, and VRT divisions of GenBank, and small numbers were selected from all of the other divisions (there were a total of 20,407 sequences in U-RVDB and 14,483 in C-RVDB; Table 1). However, there were 0 LTR retrotransposon sequences in NCBI RefSeq Viral and NCBI Viral Genomes. Viral genes that could not be assigned to any of the previous categories (1,979 in U-RVDB and 1,193 in C-RVDB) were fairly evenly distributed across the GenBank divisions (Table 1).
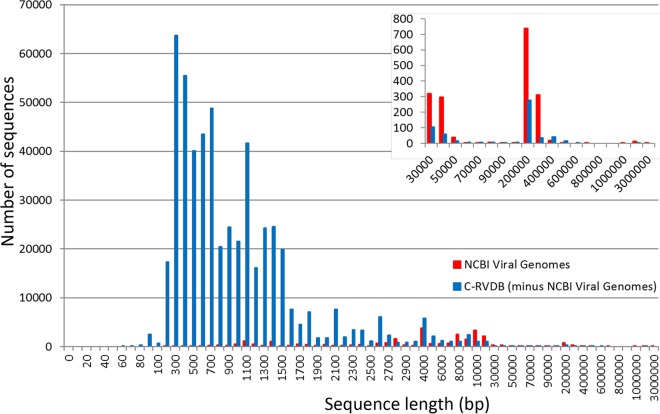
The distribution of sequences on the basis of their size/length was analyzed in C-RVDBv10.2 and in clustered NCBI Viral Genomes to compare the differences; C-RVDB was analyzed without Viral Genomes. The results indicated that the size distribution in the creps in NCBI Viral Genomes was distinct from that in the creps in the rest of C-RVDB (Fig. 3, red and blue, respectively). The majority of the creps in NCBI Viral Genomes were in the size range of approximately 4 to 10 kb, whereas the sizes of the majority of the sequences in C-RVDB ranged from approximately 300 bp to 1.5 kb. The large number of fragments in the small size range in C-RVDB suggested a preponderance of subgenomic viral sequences or small viral genomes (29). Interestingly, the difference in the number of sequences in these two size ranges was 2 orders of magnitude between NCBI Viral Genomes and RVDB; there were no sequences of <200 bp in NCBI Viral Genomes. Additionally, at >10 kb, NCBI Viral Genomes had more sequences than RVDB (Fig. 3, inset). This was most likely due to large viruses (e.g., herpesvirus) and giant viruses (e.g., megaviruses and mimiviruses). However, for these families of viruses, RVDB had additional sequences not included in NCBI Viral Genomes, even for large and rare genomes >100 kb in length (Fig. 3, inset). This analysis highlights the importance of including NCBI Viral Genomes in its entirety in RVDB, which extended the range of sequence lengths from short viral fragments and genes to full-length virus genomes.
FIG 3 .
Comparison of sequence length distribution in RVDB and NCBI Viral Genomes. For this analysis, sequences in NCBI Viral Genomes were also clustered and excluded from C-RVDBv10.2. Sequences in both were binned by length and plotted by frequency. The distribution of sequence length was different in the two databases; most of the sequences in C-RVDB (blue) were between 200 and 1,500 bp long, whereas most of the sequence in C-NCBI Viral Genomes were 4 to 10 kb long.
Characterization of unique sequences in C-RVDBv10.2
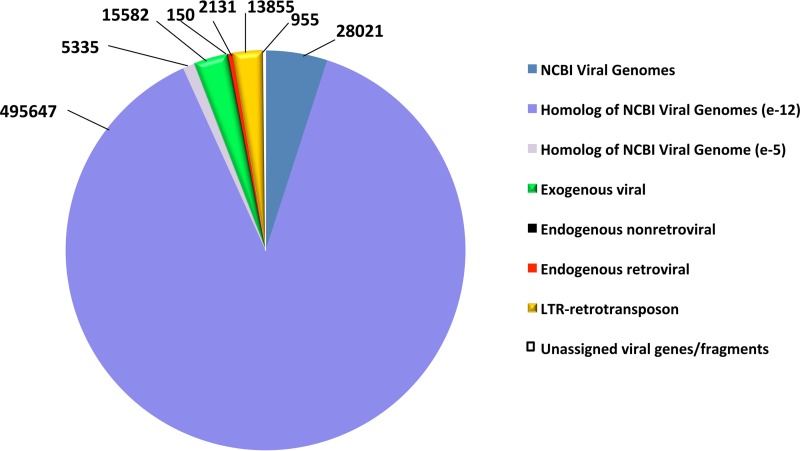
All of the sequences in RVDB were expected to be viral since they were selected by using SEM-R. However, the creps in RVDB that had an E value of greater than (i.e., less significant than) 1e-12 by BLASTN search of NCBI Viral Genomes were further evaluated to verify these sequences as viral by using various bioinformatics tools such as BLASTX, TBLASTX, PFAM, and DFAM. There were 529,003 sequences in C-RVDB that were either part of NCBI Viral Genomes or found to have a hit by using BLASTN with an E value of less than (i.e., more significant than) 1e-5 to at least one sequence in NCBI Viral Genomes; the latter were termed homologs of NCBI Viral Genomes. Therefore, RVDB included 28,021 creps directly from NCBI Viral Genomes (Fig. 4, dark blue) and 500,982 creps that were homologs of NCBI Viral Genomes, which consisted of 495,647 sequences with an E value of <1e-12 (Fig. 4, medium blue) and 5,335 sequences with an E value of <1e-5 (Fig. 4, light blue). Sequences with a hit that had an E value of greater than (i.e., less significant than) 1e-5 were termed unique sequences. The distribution of these 32,673 unique sequences on the basis of their different viral categories (exogenous viral, endogenous nonretroviral, endogenous retroviral, LTR retrotransposon, and unassigned viral genes/fragments) is shown in Fig. 4, and their selection on the basis of the different GenBank divisions is shown in Table 2. It was noted that that the total number of exogenous viral creps in RVDBv10.2 (542,243 out of 561,676 in Table 1) were greatly reduced compared to the total number of unique creps (15,582 out of 32,673 in Table 2) (i.e., 96.6% of all creps versus 47.7% of the unique sequences). However, the 47.7% of unique sequences still constituted a large number of sequences that were clearly viral (14,071 from VRL) and not present in NCBI Viral Genomes. Conversely, other level 1 categories of sequences were only slightly reduced in number in the unique sequences; for example, there were 14,483 LTR retrotransposons among the creps and 13,855 among the unique sequences (Tables 1 and 2, respectively). Thus, 95.7% of LTR retrotransposons in the RVDBv10.2 were identified as unique with respect to NCBI Viral Genomes.
FIG 4 .
Sequences in RVDB. C-RVDBv10.2 contained NCBI Viral Genomes and sequences that were homologous to NCBI Viral Genomes, designated homologs of NCBI Viral Genomes, which were distinguished on the basis of their E values (<e-12 and <e-5). The remaining unique sequences were grouped into the five viral categories indicated.
TABLE 2 .
Categorization of C-RVDBv10.2 uniquea sequences in GenBank divisions
| GenBank division(s) |
No. of sequences in: |
|||||
|---|---|---|---|---|---|---|
| Exogenous viral |
Endogenous nonretroviral |
Endogenous retroviral |
LTR retrotransposon |
Unassigned viral genes/fragments |
All of RVDB |
|
| VRL | 14,071 | 0 | 102 | 0 | 0 | 14,173 |
| ENV | 1,283 | 0 | 0 | 11 | 31 | 1,325 |
| HTC | 51 | 0 | 6 | 26 | 86 | 169 |
| INV | 8 | 110 | 32 | 1,458 | 255 | 1,863 |
| MAM | 7 | 13 | 445 | 11 | 28 | 504 |
| PLN | 61 | 4 | 77 | 12,037 | 408 | 12,587 |
| PRI | 38 | 0 | 1,156 | 35 | 43 | 1,272 |
| ROD | 32 | 0 | 178 | 44 | 47 | 301 |
| TPA | 20 | 0 | 0 | 51 | 0 | 71 |
| VRT | 11 | 23 | 135 | 182 | 57 | 408 |
| All | 15,582 | 150 | 2,131 | 13,855 | 955 | 32,673 |
Unique sequences in RVDB are defined by results of BLASTN searching of NCBI Viral.
Unique sequences in C-RVDBv10.2 were confirmed as viral by using an array of different sequence homology detection tools. For this analysis, the LTR retrotransposon sequences were not included because of the absence of a relevant reference (see Materials and Methods). Furthermore, 41 sequences were removed from the database on the basis of a lack of viral or virus-related hits by any of the confirmation analyses described as follows and therefore not shown in Table 3. Thus, analyses were done with 18,800 unique sequences that did not include LTR retrotransposons. Furthermore, 4,821 sequences that were homologs of NCBI Viral Genomes, with a best E value between 1e-12 and 1e-5, were included in the analyses. These 4,821 also did not include 514 LTR retrotransposons in the database (shown collectively as 5,335 in Fig. 4).
TABLE 3 .
Confirmation of unique sequences in RVDBa
| Unique sequences | No. of sequences |
||||
|---|---|---|---|---|---|
| PFAM/DFAM | TBLASTX vs NCBI Viral Genomes |
BLASTN vs RVDB |
TBLASTX vs RVDB |
No hits | |
| GenBank divisions | |||||
| VRL | 7,099 | 2,502 | 3,222 | 671 | 679 |
| ENV | 339 | 164 | 150 | 175 | 486 |
| HTC | 108 | 4 | 6 | 15 | 10 |
| INV | 293 | 45 | 30 | 13 | 24 |
| MAM | 402 | 45 | 23 | 0 | 5 |
| PLN | 307 | 25 | 105 | 40 | 73 |
| PRI | 1,177 | 6 | 36 | 2 | 16 |
| ROD | 234 | 1 | 9 | 2 | 11 |
| TPA | 20 | 0 | 0 | 0 | 0 |
| VRT | 167 | 22 | 19 | 1 | 17 |
| All | 10,146 | 2,814 | 3,600 | 919 | 1,321 |
| Viral categories | |||||
| Exogenous viral | 7,688 | 2,695 | 3,397 | 854 | 1,230 |
| Endogenous nonretroviral | 85 | 43 | 18 | 2 | 2 |
| Endogenous retroviral | 1,678 | 60 | 76 | 3 | 14 |
| Unassigned viral genes/fragments | 695 | 16 | 109 | 60 | 75 |
| All | 10,146 | 2,814 | 3,600 | 919 | 1,321 |
Unique sequences in RVDBv10.2 were confirmed as viral by using various tools and categorized based on GenBank divisions and viral categories. Results are shown based on the tools used to get hits (or no hits).
A total of 23,621 sequences from RVDB (consisting of 18,800 unique sequences and 4,821 homologs with intermediate E values) were subjected to confirmation analyses for viral identity by using BLAST and HMMER searches of different databases. The results for the 18,800 unique sequences in C-RVDBv10.2 assigned in the different GenBank divisions and viral categories are shown in Table 3. The confirmed unique sequences were predominantly exogenous viral (82.8%); a majority (10,146 out of 18,800) had hits to either viral domains from the PFAM database or hits to DFAM (Table 3, column PFAM/DFAM). Of the remaining 8,654 sequences, 2,814 had hits by TBLASTX against NCBI Viral Genomes, 95.7% of which were exogenous viral (including 5.8% from the ENV division) (Table 3, column TBLASTX versus NCBI Viral Genomes). Of the remaining 5,840 sequences, 3,600 (Table 3, column BLASTN versus RVDB) had hits by BLASTN against C-RVDB, the majority of which were from VRL, with the next highest numbers from ENV and then from PLN. Of the remaining 2,240 unique sequences, 919 had hits by TBLASTX against C-RVDB (Table 3, column TBLASTX versus RVDB) and 1,321 sequences remained unconfirmed by these analyses. The latter were almost all from the exogenous viral category and were further queried against the nr/nt database by using BLASTN to verify viral identity. More sequences in the ENV division were confirmed as viral by manual review of virus names (n = 486; 37.0%) than by any single one of the preceding bioinformatics analyses. The PFAM/DFAM analysis also confirmed that the majority of sequences in the endogenous nonretrovirus, endogenous retrovirus, and unassigned viral genes/fragments categories (2,720 of the total of 3,236) had a viral or virus-related identity.
Since NCBI Viral Genomes contained only a small number endogenous nonretroviral, endogenous retroviral, and LTR retrotransposon sequences, we further determined the extent to which RVDB contained unique sequences compared to other, specialized databases that had greater numbers of virus-related and virus-like sequence than RefSeq Viral and NCBI Viral Genomes. A BLASTN analysis of C-RVDBv10.2 with gEVE resulted in 147,420 of 561,698 creps in RVDB with significant hits (E values of <1e-12) to 67,051 of the 736,771 sequences in gEVE (unpublished data). The 147,420 RVDBv10.2 creps with hits contained 3,610 unique sequences (11%). The BLASTN comparison of C-RVDBv10.2 and Repbase resulted in 111,477 of 561,698 creps with significant hits (E values of <1e-12) to 11,185 of the 46,062 sequences in Repbase. The 111,477 RVDBv10.2 creps with hits contained 14,361 unique sequences (44%). These results indicate that although there are additional sequences in the other databases specific for endogenous viral elements and retrotransposons, RVDB contains endogenous nonretroviral, endogenous retroviral, and LTR retrotransposon sequences that are lacking in the other databases.
Performance evaluation of RVDBv10.2
The NCBI nr/nt database is the largest public database of viral sequences and a useful resource for analyzing HTS data for virus investigations. Therefore, the run efficiency of RVDBv10.2 was compared with the NCBI nr/nt database (downloaded to the local computing cluster in March 2015) by using an in-house HTS data set (designated K-10 [described in Materials and Methods], which was a 2.2-GB data set containing 7,124,833 paired-end Illumina reads). The results indicated the equivalent of 9,464 h, 56 min of runtime on an x86_64, 2,667-MHz central processing unit (CPU) was required for analysis against the nr/nt database, compared to 35 h, 48 min of runtime against RVDBv10.2. This was expected on the basis of the differences in the size of the nr/nt database compared with RVDB (135 and 1.0 GB, respectively), which can be attributed largely to the abundant nonviral sequences in the nr/nt database compared to the reduced content of such sequences in RVDB (discussed below). The extended runtime against the nr/nt database was most likely due to hits with cellular sequences present along with viral sequences in the query data set. The runtime of RVDB with the K-10 data set was also compared with NCBI Viral Genomes (version July 2016), which was found to be only 4 h, 22 min. This was expected because of the smaller size of the Viral Genomes database (0.74 GB), which specifically contains complete or nearly complete virus genomes. However, detection of some viruses may be missed since NCBI Viral Genomes does not include viruses for which only short or partial viral sequences are available, as well as many endogenous viruses (including nonretroviral and retroviral) and retrotransposons (30, 31).
We also evaluated the specificity of virus detection by performing BLASTN searches in RVDBv10.2, the nr/nt database, and Viral Genomes by individually interrogating with nine distinct fragments containing insect endogenous retrovirus sequences (errantiviruses) isolated from Spodoptera frugiperda (30; unpublished data). Although similar BLASTN results were seen with RVDBv10.2 and the nr/nt database, in the latter case, there were a large number of cellular hits reflecting the presence of uncharacterized viral sequences in these entries; additionally, some of the viral hits had a greater E value (less significant) than in RVDB. The latter hits were below the detection threshold of 1e-5, and therefore virus detection would be missed by the analysis. For example, the Sf-17 viral fragment had a hit with an E value of 4e-6 to the Drosophila melanogaster Zam element in RVDBv10.2, but its E value in the nr/nt database was 7e-4. Similarly, in our previous study, BLASTN analysis of the Sf9 transcriptome by using the nr/nt database initially failed to detect a novel rhabdovirus in Sf9 cells because of abundant cellular hits (2, 32). These results indicated that databases with a large number of cellular hits with E values more significant than (less than) or similar to those of the viral hits could displace the viral hits in the BLAST output, thus missing the detection of viral hits. Furthermore, BLASTN searches by using the nine insect endogenous retroviral fragments produced no hits in NCBI Viral Genomes, whereas a search of RVDBv10.2 produced the expected self-hits and hits to reverse transcriptase (RT) genes from other endogenous retroviruses of various insect species (Trichoplusia ni, Ephestia kuehniella, Lymantria dispar, D. melanogaster, Cotesia sesamiae) and plants (Helianthus petiolaris, Hypochaeris chillensis) (data not shown). It was also noted in our earlier analysis that Sf9 rhabdovirus was not detected in the Sf9 transcriptome by BLAST searches of NCBI RefSeq Viral and Viral Genomes because of a lack of the partial Taastrup virus sequence in these databases (2). However, since our original study, several insect viruses have been added to NCBI Viral Genomes, which can now facilitate the detection of Sf9 rhabdovirus. The analyses with the different databases demonstrate the robustness of RVDB for virus detection by HTS.
RVDB update.
While this report was being written, RVDB was updated by using the May 2017 GenBank release. The same SEM-R screening was used to pull in sequences from GenBank and TPA. The unclustered form of v11.5 has 2,282,754 sequences, compared to 2,073,535 for v10.2. Most of the additional sequences in v11.5 (198,849) are from the GenBank VRL division, although there were an additional 508 from other divisions and 9,860 from NCBI Viral Genomes. As a result of a change in the GenBank format in September 2016 in which GI numbers were removed, the RVDB headers in v11.5 were modified accordingly. Sequence provenance has also been modified to include GenBank, TPA, REFSEQ, or NEIGHBOR values. Furthermore, GenBank sequences corresponding to RefSeq entries were removed in v11.5 to remove duplicate sequence entries. Clustered and unclustered forms of v11.5 are available publicly along with the previous version.
The proteic versions of RVDB were kindly generated by Marc Eloit and Thomas Bigot (Institut Pasteur, Paris, France) since v10.2 and are also available along with notes about their development on the same website as the nucleotidic databases (details will be published elsewhere).
Development of a pipeline for updating RVDB.
To facilitate the update process, which is intended to be performed on a regular basis, concurrent with official GenBank releases, the scripts and procedures involved in updating were assembled into a pipeline that can be called with only a small number of command lines or blocks executed in the Windows cmd.exe command shell. In addition to the nine GenBank divisions, RefSeq Viral sequences and NCBI Viral Genomes were downloaded and parsed in an automated manner, which allowed for efficient removal of RefSeq duplicates in GenBank and indication of sequence provenance (GenBank, TPA, REFSEQ, or NEIGHBOR). The keyword screening script for updating RVDB, designated SEM-R_PIPE.py, contains the screens for positive keyword, size/miRNA, and negative keyword. The scripts for the pipeline are freely available online at https://github.com/ArifaKhanLab/RVDB, and a link is provided on the same database URL as the RVDB (for a detailed description and instructions for the generation of both the U-RVDB and C-RVDB, see Text S1). An update can now be run in less than a day with minimal manual review. Any further refinement of the update pipeline will be incorporated in GitHub.
Instructions and scripts for updating RVDB and generating the unclustered and clustered fasta files. These are also available at https://github.com/ArifaKhanLab/RVDB. Download TEXT S1, DOCX file, 0.03 MB (37.5KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
DISCUSSION
RVDBv10.2 was created to address the limitations in the detection of novel and distantly related viruses in publicly available databases. The challenges of bioinformatics analysis for virus detection by using large HTS data sets, particularly those containing large amounts of cellular sequences such as transcriptomes, were recognized by HTS technology users (33) and in our own laboratory studies using HTS, which resulted in the identification of a novel rhabdovirus in the Sf9 insect cell line (2, 32). On the basis of discussions in a subgroup of the Advanced Virus Detection Technologies Interest Group (AVDTIG) (34) that focused on identifying the strengths and weaknesses in databases, we undertook the development of a new, comprehensive RVDB that would facilitate HTS bioinformatics analysis for novel virus detection. Our strategy was to include all eukaryotic viral, virus-related, and virus-like sequences of all species, regardless of size (>50 bp). Furthermore, to increase the efficiency of HTS transcriptomic and genomic data analyses, efforts were made to also reduce the cellular content. These unique features distinguish RVDB from other custom databases such as NCBI Viral Genomes, which generally contains full-length or nearly complete genomes, including those of bacteriophages; gEVE, which contains only the protein domains and homologous coding regions of endogenous viruses, including retroviruses; and Gypsy, which is composed of only retrotransposons. The RVDB described in this paper is version 10.2; however, it was updated to v11.5 while this report was being prepared. It should be noted that internal testing of the RVDB (data not shown) indicated value in using both the unclustered and clustered versions and therefore both are publicly available to offer flexibility in bioinformatics analyses. For example, a BLASTN search of U-RVDB may be useful to detect viruses with a high nucleotide sequence identity level, while TBLASTX analysis of C-RVDB may aid in the detection of distantly related viruses on the basis of translated amino acid identity. Additionally, the proteic versions of RVDB generated by Marc Eloit and Thomas Bigot (Institut Pasteur) are currently available along with nucleotidic RVDB (details will be described elsewhere) and provide additional bioinformatics resources to identify novel viruses on the basis of identity at the amino acid level.
RVDB contains sequences of retroelements such as endogenous retroviruses and LTR retrotransposons, as well as other endogenous virus sequences from different species. Although there have been many studies to investigate the structure and function of human endogenous retroviruses, genomes of other species, for example, insects and plants, have been studied less. The identification and characterization of active endogenous viral sequences in a host species can help assess if they pose a potential safety concern in cells used to manufacture biologics (4). For example, genomic and biological characterization of endogenous retroviral particles that were chemically induced from an African green monkey cell line (35, 36) or constitutively expressed from chicken embryo fibroblast cells demonstrated that they were not infectious (37–39), whereas endogenous retrovirus from a porcine xenograft was shown to infect human cells in vitro (40, 41). Moreover, although a majority of retrotransposons are noninfectious, some can encode an env-like protein and are infectious; for example, the Ty3/gypsy LTR retrotransposon can infect D. melanogaster (42, 43). Furthermore, the giant retrotransposon Ogre in the plant, although not infectious, was found to be constitutively expressed at high levels. The Ogre retrotransposon has complete Ty3/gypsy anatomy, possessing all of the genes required for infection (44). Further understanding of the structure and function of endogenous retroelements other endogenous virus sequences can help assess their importance and relevance in HTS analysis for virus detection.
RVDB was designed to include all virus-related sequences, regardless of length, to include some virus families that are represented only by a short sequence. For example, the Sf9 errantivirus sequences seem to represent distinct endogenous retroviruses based on sequence analysis but these are only available currently as short sequence fragments (about 200 to 900 bp) (30). These are the only representatives of errantiviruses from S. frugiperda and will be replaced in future updates of RVDB as longer or full-length sequences become available. It should be noted that the clustering step in the generation of the database will remove shorter sequences that have 98% identity with a longer one that has been deposited in GenBank.
Some entries in RVDBv10.2 contain cellular or vector sequences adjacent to the viral sequences (such as endogenous retroviruses and retrotransposons, which are integrated in the host genome); however, in the majority of cases, the cellular portion has been annotated in the GenBank features. Future efforts directed toward comprehensive annotation of sequences in RVDB will help distinguish viral hits from nonviral hits, thus determining the need for additional follow-up studies to evaluate HTS results. A variety of tools and approaches can be used to identify viral sequences and annotate entries that contain unannotated cellular flanking sequences. Some approaches can include an all-versus-all BLAST search of RVDBv10.2 against itself, which would align viral sequences on the basis of homology and identification of coding regions. Additional tools such as DFAM (24) could help locate LTRs of retroelements.
The characterization of virus-related sequences in RVDB highlighted the need for revision of the existing viral taxonomy. Although the ICTV is the most complete resource of virus taxa (13), the system currently classifies only exogenous viruses. The situation for nonexogenous, endogenous virus-related sequences is still poorly defined since many endogenous viruses, including endogenous retroviruses and retrotransposons, are tagged with their hosts’ taxonomic identifiers, which makes them difficult to recognize as viruses. Therefore, annotation efforts to identify and characterize such viruses can aid in their taxonomic classification and facilitate efforts to develop a complete viral database. The assignment of retrotransposons in current existing discrete groups would be more appropriate than classification based on host taxonomy. For instance, retrotransposons in Ty1/Copia and Ty3/Gypsy have more in common among their group members than they do with their species. Focused, in-depth studies of retrotransposons are currently limited because of mislabeling on the basis of host taxonomy. Additionally, the nomenclature for retrotransposons is poorly organized; for Ty1/Copia and Ty3/Gypsy groups of retrotransposons, over two dozen individual, sometimes cell line-specific, names for elements were given, requiring manual review of the publication itself for discovery of group membership. As there is no standardization of names, there is no standardization of lineage, making evolutionary studies difficult to pursue without prior expert knowledge of the elements, and global studies may become cumbersome, even for experts. Finally, while dedicated databases of retrotransposons do exist, notably the Gypsy database (21), these databases are often incomplete or poorly maintained. Our RVDB provides a uniquely complete collection of endogenous retroviruses and retrotransposons. More in-depth characterization of the sequences in our retrotransposon “other” category, which makes up 24.4% of all retrotransposon cluster representatives in RVDBv10.2, may lead not only to enhanced formalism in the taxonomic classification of retrotransposons but potentially to the discovery of novel families of retroelements.
There are several advantages of the RVDB over other available references databases. First, the viral sequence space coverage exceeds that of NCBI Viral Genomes, which is a broadly used public resource (11). While the NCBI Viral Genomes resource is large and well organized, it is a genome-centric model that is based on the availability of complete or nearly complete genomes; therefore, viruses for which only short or partial sequences are available remain underrepresented. The inclusion of all viral sequences, regardless of their size, resulted in a number of unique sequences in RVDB. The presence of unique sequences is a second advantage of RVDBv10.2. The unique sequences are of particular importance to the RVDB because they represent certain viral taxonomic groups that are underrepresented in current publicly available resources and in some cases entirely not represented at all; these include endogenous nonretroviruses, endogenous retroviruses, and LTR retrotransposons. Another major advantage is that RVDBv10.2 is a comprehensive eukaryotic, virus-specific database containing viral, virus-related, and virus-like sequences. Other comprehensive viral databases, such as the NCBI nr/nt database, contain large numbers of cellular, bacterial, and bacterial virus (bacteriophage) sequences, as well as non-LTR retrotransposons. Overall, the distinct features of RVDBv10.2 are expected to provide a public resource to enhance novel virus detection in research investigations and the evaluation of potential adventitious viruses in biologics. The increased viral sequence diversity in RVDB should aid in novel virus discovery and characterization. We expect continued refinement of RVDB with improvement in sequences deposited in GenBank (annotation and full genome sequences) and real-time user feedback.
MATERIALS AND METHODS
Development of first-generation VDB.
Efforts to develop a new viral database were initiated by downloading sequence files (September 2015) from 17 of the 20 divisions of the GenBank ftp site (ftp://ftp.ncbi.nih.gov/genbank). Initially, only the BCT (bacterial), CON (constructed sequences), and PHG (phage) divisions were excluded. In the development of the first-generation VDB, the GenBank VRL (viral) division, which contained 1,831,042 sequences as of September 2015, was used in its entirety. Additional sequences (1,893,209) were pulled in by semantic mining of 16 other GenBank divisions for viral and LTR retrotransposon sequences by using a list of knowledge-based keywords that were developed to broadly capture all viral, virus-related, and virus-like sequences in GenBank, with less emphasis on the exclusion of nonviral entries. Therefore, initial positive keywords included general terms such as virus and viral. The keywords were also designed to pull in all retrotransposons and therefore included both generic terms such as “retro,” “transpos,” and “repetitive element,” as well as names of major families of retrotransposons such as “copia” and “gypsy element.” After further analysis including manual review, additional keywords were added for specific names of retroelements, which would not be selected by using the general keywords. An initial set of negative keywords and rules was used to remove obvious nonviral entries that contained virus-related keywords (e.g., virus receptor). In this paper, we refer to these initial selection criteria as semantic initial or SEM-I. SEM-I consisted of 60 positive keywords, a size and miRNA screen, 34 negative keywords, and eight negative rules (sequences of <50 bp and miRNAs were removed to reduce the likelihood of spurious hits). The resulting first-generation VDB contained a total of 3,724,251 sequences.
(i) Evaluation of first-generation VDB with sequence homology-based tools.
The sequences selected by SEM-I were further queried by using an array of sequence homology tools to confirm their viral identity. Initially, all of the startup nucleotide sequences were run against the RefSeq Viral nucleotide sequences (October 2015 release, version 72, minus the phage sequences) by using BLASTN (45) with the equivalent of the NCBI online server (http://blast.ncbi.nlm.nih.gov/Blast.cgi) “somewhat similar” parameters. At this stage, only RefSeq Viral was used. The parameters were word_size 11, reward 2, penalty −3, gapopen 5, gapextend 2, and a maximum number of hits per query of 250. To count a hit as significant, an aligned query sequence needed to have an E value of <1e-12. Under these criteria, just over half of the sequences (1,895,743 including 1,778,161 or 97.1% of VRL) had significant hits with at least one RefSeq Viral sequence. Nucleotide sequences without significant BLASTN hits were then examined further by using additional tools from BLAST+ (46) (BLASTX to search a protein database by using translated queries and TBLASTX to search a translated database by using translated queries) against RefSeq Viral (E value of <1e-5). HMMERv3.1b2 was also used to align translated query sequences against virus-related domain profiles in the Protein Families (PFAM) database v28, which had been initially determined by using translated RefSeq viral genomic sequences (47). To confirm the identity of retrotransposons, NHHMER was used to align nucleotide query sequences against profiles in the Repetitive DNA Element (DFAM) database v1.4 (24). For BLASTX and PFAM analyses, DNA query sequences were translated to protein sequences in all six reading frames. Finally, nucleotide sequences without significant BLASTN hits against RefSeq Viral were run against the nr/nt database by using BLASTN.
The results generated from the additional BLAST, PFAM, and DFAM analyses were initially grouped into viral, virus-related, and nonviral groups and manually reviewed in conjunction with the entry headers. This resulted in the development of refined semantic selection criteria (designated semantic refined or SEM-R), consisting of a final extended set of positive and negative keywords, rules, and regular expressions that was used to develop a second-generation RVDB as described below. SEM-R contained 50 positive keywords, four positive rules, three positive regular expressions, a size and miRNA screen, 252 negative keywords, 119 negative rules, and seven negative regular expressions. SEM-R contained fewer positive words than the original SEM-I but had a greater number of negative words, rules, and regular expressions since the goal was to remove all nonviral sequences from the database. For example, the keywords env and envelope were often the only indication that a particular entry contained a viral env gene; however, we needed to exclude entries containing nuclear envelope, membrane envelope, and outer envelope. Also, coat protein always contained the positive keyword viral or virus when referring to viral coat proteins. Some of the nonspecific positive keywords in SEM-I, such as repetitive element, insert, and insertion sequence, were removed since they were found to select mostly nonviral sequences, and a broad positive keyword such as retro was replaced with the more specific retrotranspos and Retrovir root strings. This resulted in the elimination of false-positive nonviral entries. The overall strategy used to develop the first-generation VDB and the refined semantic selection criteria (SEM-R) used for the final keyword screening are summarized in Fig. 1A.
It should be noted that early draft versions were distributed to volunteers of the AVDTIG (34) for testing and further database refinement. The generation of the final RVDBv10.2, which was initially released for beta testing and is now publically available (https://hive.biochemistry.gwu.edu/rvdb), is described below.
Generation of RVDBv10.2.
A second-generation VDB was developed by using SEM-R to select sequences from eight GenBank divisions (ENV [environmental], HTC [high-throughput cDNA], INV [invertebrate sequences], MAM [other mammalian sequences], PLN [plant and fungal sequences], PRI [primate sequences], ROD [rodent sequences], and VRT [other vertebrate sequences]), as well as TPA from GenBank (July 2016). Nonphage sequences from the VRL division of GenBank were added without semantic screening. Additionally, NCBI Viral sequences were added directly without semantic screening, after the removal of phage sequences (July 2016), comprising about 90,000 eukaryotic viral sequences. TPA sequences were added because, although not technically part of the central GenBank database, they were found to contain certain unique viral and virus-related sequences. NCBI Viral was added to provide the maximal compression of sequences against the existing standard during clustering and also to reveal more distantly related groups of sequences. The selected GenBank divisions and number of starting sequences in each are indicated in Table 4.
TABLE 4 .
GenBank divisions and entries included in second-generation RVDBa
| GenBank division | Description | No. of entries |
|---|---|---|
| VRL | Viral | 2,030,643 |
| ENV | Environmental sampling | 7,904,590 |
| HTC | High-throughput cDNA sequencing | 608,888 |
| INV | Invertebrate | 6,319,850 |
| MAM | Other mammalian | 468,842 |
| PLN | Plant (including fungi and algae) | 4,137,915 |
| PRI | Primate | 828,230 |
| ROD | Rodent sequence entries | 524,368 |
| TPA | Third party annotated sequences | 278,453 |
| VRT | Other vertebrate | 2,490,585 |
| RefSeq Viral | Representative genomes | 5,119 |
| NCBI Viral Genomes | Representative genomes and neighbor genomes | 91,968 |
Prior to SEM-R screening (July 2016).
As in the case of the first-generation VDB, sequences from BCT, CON, and PHG were excluded. Additionally, the following divisions were not included: expressed sequence tags (EST), genomic survey sequences (GSS), high-throughput genome sequencing (HTG), patented sequences (PAT), sequence-tagged sites (STS), synthetic constructs (SYN), transcriptome shotgun assembly sequences (TSA), and unannotated sequences (UNA). EST, GSS, HTG, STS, and TSA sequences were excluded because they were found to be largely redundant with existing genomic sequences in the other divisions. PAT sequences were excluded because they contained a disproportionate amount of short sequences and/or flanking regions for inserts. SYN sequences were discarded because they contained a large proportion of modified DNA sequences. UNA sequences were excluded because they often lacked sufficient annotation for the semantic selection to be performed with confidence. GenBank sequences were formatted to make headers similar to NCBI entries.
(i) Clustering.
The clustering tool CD-HIT-EST (48) was used to reduce sequence redundancy in the second-generation VDB. Although primarily intended for shorter sequences, CD-HIT-EST was chosen because of its efficiency at clustering at high sequence identity. PSI-CD-HIT is more efficient for the clustering of larger sequences (e.g., whole genomes); however, it is optimized for clustering at low sequence identity. By using 40 CPUs (x86_64, 2,667 MHz each), clustering of the entirety of the second-generation VDB was performed in just under 12 h. Clustering was performed at 98% sequence identity to reduce redundancy but retain viral diversity, by using a k-mer length of 11 (maximum) since greater k-mer lengths are preferred for clustering at higher sequence identities. Cluster representatives (creps) were the longest sequences in their clusters, except when a RefSeq Viral or neighbor sequence was present, in which case the RefSeq Viral sequence became the crep or if no RefSeq Viral sequence was present, the neighbor sequence became the crep.
(ii) Confirmation of viral identity.
Clustered sequences were confirmed as viral with various computational tools. Initially, sequences were confirmed on the basis of significant E values (<1e-12) by a BLASTN search of NCBI Viral. Retrotransposons were excluded from the confirmation analysis (i.e., accepted without confirmation) since they were not expected to be present in NCBI Viral and therefore would be inadvertently removed because of the lack of a hit. The sequences that did not have hits were further analyzed by BLASTN searching by using a customized NCBI phage data set to remove the residual phage sequences (1,135) that had a significant hit (E value of <1e-12). The sequences were further confirmed in a sequential stepwise manner by HMMER search of viral PFAM (v27) and DFAM (v2.0) families. Queries with hits were confirmed, while queries still without hits were run by a TBLASTX search of NCBI Viral by using an E value of <1e-5 as the threshold of significant TBLASTX homology, and again, those with hits were confirmed. Entries that still remained with no hits were then run against the C-RVDB (with self-hits masked) by using BLASTN (E value of <1e-12), and subsequently against self, by using TBLASTX (E value of <1e-5) and finally against the nr/nt database by using BLASTN (E value of <1e-12), each time by using the same reductive approach. For the final step, BLASTN hits in the nr/nt database were considered viral if they passed the SEM-R keyword screening. Queries with viral hits were retained, while those without any viral hits were manually reviewed. Forty-one sequences were found to be nonviral and excluded from the resulting final version of VDB (RVDBv10.2). This final screening also resulted in some further refinement of the second-generation VDB by retroactively removing the same sequences that were excluded from RVDBv10.2, resulting in the final U-RVDB.
Clustered sequences were also run by BLASTN searches of both the recent gEVE database of endogenous viral elements and the Repbase database of repetitive elements. However, the results were used only for corroboration of viral identity as established by the techniques described above.
Characterization of sequences in RVDBv10.2.
Sequences in RVDB were characterized at two progressive levels. Level 1 categories included exogenous viral, endogenous nonretroviral, endogenous retroviral, LTR retrotransposon, and unassigned viral genes/fragments. Level 2 assignments were based on ICTV viral families, cellular host families, retrotransposon families, and names of genes/fragments. The python script for characterization at level 1, designated RVDB_characterization.py, is available at https://github.com/ArifaKhanLab/RVDB along with the instructions for its use (the latter are also provided in Text S2).
Instructions and scripts for characterization of entries into level 1 categories. These are also available at https://github.com/ArifaKhanLab/RVDB. Download TEXT S2, DOCX file, 0.01 MB (16KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Details of the taxonomic groupings are described below.
(i) Exogenous viruses.
Sequences were mapped to the exogenous viral category by using exogenous-virus-specific positive keywords from SEM-R. For the second level of characterization, sequences were mapped by using the NCBI taxonomy database (49). First, GenBank identifiers were mapped to NCBI taxonomic identifiers (mapping file available at ftp://ftp.ncbi.nih.gov/pub/taxonomy/gi_taxid_nucl.dmp.gz) (49). These taxonomic identifiers were then used as starting points to “climb” the taxonomic tree by using NCBI parent-child taxonomic identifier definitions (taxonomic definition file available at ftp://ftp.ncbi.nih.gov/pub/taxonomy/taxcat.zip; nodes.dmp file) to the family level (Fig. 2, left). Family taxonomic identifiers were mapped to their organism names (taxonomic name file available at ftp://ftp.ncbi.nih.gov/pub/taxonomy/taxcat.zip; names.dmp file). These NCBI family names were matched to their counterparts in ICTV by using fuzzy string matching to ensure that the family existed in ICTV. Viral sequences were also characterized as being either full or partial length. For the number of sequences belonging to each exogenous viral family (exviral), both complete genomes and partial, see Table S1A.
(ii) Endogenous nonretroviruses and retroviruses.
Sequences were mapped to the endogenous nonretroviral and endogenous retroviral categories by using virus-specific positive keywords from SEM-R. After the application of these keywords, the endogenous viral sequences were manually reviewed to transfer any remaining endogenous retroviral and retrotransposon sequences into relevant categories. For the second level of characterization, endogenous nonretroviruses and retroviruses were mapped down to the family phylogenetic level, as with exogenous viruses. However, in this case, no formal viral/virus-related taxonomy such as the ICTV classification was available as a reference, since no endogenous viral or endogenous retroviral taxa are formally described by ICTV. Therefore, the NCBI taxonomy was used as a default, which led to mapping up to the host family level. Mapping to the family phylogenetic level and mapping from identifier to name was performed in the same manner as it was for viruses (by using nodes.dmp and names.dmp files from ftp://ftp.ncbi.nih.gov/pub/taxonomy/taxcat.zip) (Fig. 2). Generally, the endogenous retroviral sequences were associated with their host species, with only 294 out of 2,447 classified as viruses in the family Retroviridae. These 294 retroviruses were grouped along with other endogenous retroviruses on the basis of their host families. For the number of sequences belonging to each endogenous nonretroviral host family (ENRV), see Table S1B, and for the number of sequences belonging to each endogenous retroviral host family (ERV), see Table S1C.
(iii) LTR retrotransposons.
LTR-containing retrotransposons were further characterized into specific families. For this second level of characterization, because of the lack of a formal taxonomic representation for retrotransposons, taxonomic classification for LTR retrotransposons was performed by using keywords based on specific family and subfamily names for LTR retrotransposons; four main families exist, Ty1/Copia, Ty3/Gypsy, BEL/Pao, and DIRS. Mapping to each of these four categories was done by using keywords including the canonical name (e.g., ty1), as well as more specific names of individual elements (Fig. 2, right middle). Ty1/Copia, Ty3/Gypsy, BEL/Pao, and DIRS elements were mapped by using 28, 38, 5, and 13 element-specific names, respectively. Any retrotransposons lacking names of the four main families or specific elements were counted in a fifth category, unassigned viral genes. The LTR retrotransposon groups Morgane and TRIM were found in such low numbers, possibly because of incomplete annotation, that they were added to unassigned viral genes as well. For the number of sequences belonging to each retrotransposon family, see Table S1D.
(iv) Unassigned viral genes/fragments.
Sequences were placed in the unassigned viral gene/fragment category if they failed to be placed in any of the other four categories (Fig. 2, right). This category was manually reviewed and found to contain some sequences that were identified on the basis of the names of specific viruses, which were not included in the positive keywords in SEM-R; such sequences were accordingly placed in their respective level 1 viral categories. The remaining unassigned genes contained only gene names (e.g., gag, pol, and env) as evidence of viral or retrotransposon identity and were placed into 13 categories based on these gene or genome fragments. These categories are LTR, env, capsid, gag, pol, RT, polyprotein, integrase, intracisternal A particle, endogenous, dUTPase, polycomb response element, and replicase. For the number of sequences belonging to each unassigned gene/fragment, see Table S1E.
Analysis of sequence length distribution in C-RVDBv10.2.
Clustered sequences were divided into two groups, NCBI Viral Genomes and RVDB (minus NCBI Viral Genomes), where the latter consisted of all remaining cluster representatives after the removal of NCBI Viral Genomes from RVDB. A size (sequence length) distribution was generated for each of these two groups, and the two distributions were compared. The clustered sequences were then organized into the five level 1 viral groups (described above), as well as by GenBank divisions. Size distributions were generated for each taxonomic groups/GenBank division. For each size distribution generated, the redundancy or the ratio of the number of unclustered to clustered sequences was also calculated.
Performance evaluation.
We evaluated performance efficiency as the runtime on a single CPU. All analyses were performed on the FDA CDRH White Oak supercomputing grid, Betsy cluster, many in a parallelized format. Run files from all of the CPUs involved in an analysis were used to add up the total run time of a single CPU. These analyses were performed with processors with the following specifications: eight x86_64 CPUs with a processor speed of 2,667 MHz each. An in-house data set of HTS reads that was obtained from supernatant of an insect cell line was used to test the runtimes of C-RVDBv10, the NCBI nr/nt database, and NCBI Viral Genomes. This data set, designated K-10 here, composed of 2.2 GB and contained 14.2 million paired-end Illumina MiSeq reads containing viral and cellular sequences.
Nine fragments from S. frugiperda (sf-17, sf-18, sf-19, sf-31, sf-37, sf-58, sf-67, sf-70, and sf-311) containing distinct endogenous retroviral sequences from the pol gene (30) were queried against RVDBv10.2, the NCBI nr/nt database, and NCBI Viral Genomes by using BLASTN. Since RVDBv10.2 contains all nine of the Sf errantiviral pol genes, the query sequences were masked from the “self” hits during the collection of the BLAST search results. Because the actual queries themselves were masked in the hits, the performance of the analysis was assessed primarily in terms of robustness, which we define here as the ability to get hits to sequences that were similar to the query, both taxonomically and by sequence identity.
Database URL.
The RVDB URL is https://hive.biochemistry.gwu.edu/rvdb. This link is also available at https://precision.fda.gov/.
ACKNOWLEDGMENTS
We thank J. Rodney Brister for initial discussions related to databases and critical review of the manuscript. We are grateful to Raja Mazumder, Naila Gulzar, and Robel Kahsay for making the RVDB available to the public through the George Washington University HIVE and to Debbie Sieff and her staff at the FDA for providing large data storage and transfer space. We appreciate discussions from members of the AVDTIG related to RVDB content, particularly Kavitha Bekkari, John Thompson, Paul Duncan, Robert Charlesbois, Christophe Lambert, and Fabio La Neve, who performed testing and provided comments for database refinement. We thank Marc Eloit and Thomas Bigot for generating and publicly sharing the proteic databases for RVDB (http://rvdb-prot.pasteur.fr/).
This project was funded, in part, by the FDA Medical Countermeasures Initiative.
Footnotes
This paper was submitted via the mSphereDirect™ pathway.
Contributor Information
Michael J. Imperiale, University of Michigan—Ann Arbor.
Eric Delwart, Blood Systems Research Institute.
David Wang, Washington University in St. Louis School of Medicine.
REFERENCES
- 1.Khan AS, King KE, Brack K, Cassart J-P, Chiu C, Dehghani H, Duncan P, Jaing C, Kolman J, Munroe D, Palermo A, Plavsic M, Sampath R, Slezak T, Takle G, Taliaferro LP, Toso E, Vacante D, Willkommen H. 2015. Emerging methods for virus detection. Parenteral Drug Association, Bethesda, MD. [Google Scholar]
- 2.Ma H, Galvin TA, Glasner DR, Shaheduzzaman S, Khan AS. 2014. Identification of a novel rhabdovirus in Spodoptera frugiperda cell lines. J Virol 88:6576–6585. doi: 10.1128/JVI.00780-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Victoria JG, Wang C, Jones MS, Jaing C, McLoughlin K, Gardner S, Delwart EL. 2010. Viral nucleic acids in live-attenuated vaccines: detection of minority variants and an adventitious virus. J Virol 84:6033–6040. doi: 10.1128/JVI.02690-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.U.S. Food and Drug Administration 2010. Characterization and qualification of cell substrates and other biological materials used in the production of viral vaccines for infectious disease indications. U.S. Food and Drug Administration, Bethesda, MD: http://www.fda.gov/downloads/BiologicsBloodVaccines/GuidanceComplianceRegulatoryInformation/Guidances/Vaccines/UCM202439.pdf. Accessed 15 November 2017. [Google Scholar]
- 5.Scholz MB, Lo CC, Chain PS. 2012. Next generation sequencing and bioinformatic bottlenecks: the current state of metagenomic data analysis. Curr Opin Biotechnol 23:9–15. doi: 10.1016/j.copbio.2011.11.013. [DOI] [PubMed] [Google Scholar]
- 6.Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2014. GenBank. Nucleic Acids Res 42:D32–D37. doi: 10.1093/nar/gkt1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanz C, Aldebert P, Althorpe N, Baker W, Baldwin A, Bates K, Browne P, van den Broek A, Castro M, Cochrane G, Duggan K, Eberhardt R, Faruque N, Gamble J, Diez FG, Harte N, Kulikova T, Lin Q, Lombard V, Lopez R, Mancuso R, McHale M, Nardone F, Silventoinen V, Sobhany S, Stoehr P, Tuli MA, Tzouvara K, Vaughan R, Wu D, Zhu W, Apweiler R. 2005. The EMBL Nucleotide Sequence Database. Nucleic Acids Res 33:D29–D33. doi: 10.1093/nar/gki098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tateno Y, Imanishi T, Miyazaki S, Fukami-Kobayashi K, Saitou N, Sugawara H, Gojobori T. 2002. DNA Data Bank of Japan (DDBJ) for genome scale research in life science. Nucleic Acids Res 30:27–30. doi: 10.1093/nar/30.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O’Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rose PW, Bi C, Bluhm WF, Christie CH, Dimitropoulos D, Dutta S, Green RK, Goodsell DS, Prlic A, Quesada M, Quinn GB, Ramos AG, Westbrook JD, Young J, Zardecki C, Berman HM, Bourne PE. 2013. The RCSB Protein Data Bank: new resources for research and education. Nucleic Acids Res 41:D475–D482. doi: 10.1093/nar/gks1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brister JR, Ako-Adjei D, Bao Y, Blinkova O. 2015. NCBI viral genomes resource. Nucleic Acids Res 43:D571–D577. doi: 10.1093/nar/gku1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma D, Priyadarshini P, Vrati S. 2015. Unraveling the web of viroinformatics: computational tools and databases in virus research. J Virol 89:1489–1501. doi: 10.1128/JVI.02027-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lefkowitz EJ, Dempsey DM, Hendrickson RC, Orton RJ, Siddell SG, Smith DB. 2018. Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res 46:D708–D717. doi: 10.1093/nar/gkx932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams MJ, Lefkowitz EJ, King AM, Harrach B, Harrison RL, Knowles NJ, Kropinski AM, Krupovic M, Kuhn JH, Mushegian AR, Nibert ML, Sabanadzovic S, Sanfaçon H, Siddell SG, Simmonds P, Varsani A, Zerbini FM, Orton RJ, Smith DB, Gorbalenya AE, Davison AJ. 2017. 50 years of the International Committee on Taxonomy of Viruses: progress and prospects. Arch Virol 162:1441–1446. doi: 10.1007/s00705-016-3215-y. [DOI] [PubMed] [Google Scholar]
- 15.Simmonds P, Adams MJ, Benkő M, Breitbart M, Brister JR, Carstens EB, Davison AJ, Delwart E, Gorbalenya AE, Harrach B, Hull R, King AM, Koonin EV, Krupovic M, Kuhn JH, Lefkowitz EJ, Nibert ML, Orton R, Roossinck MJ, Sabanadzovic S, Sullivan MB, Suttle CA, Tesh RB, van der Vlugt RA, Varsani A, Zerbini FM. 2017. Consensus statement: virus taxonomy in the age of metagenomics. Nat Rev Microbiol 15:161–168. doi: 10.1038/nrmicro.2016.177. [DOI] [PubMed] [Google Scholar]
- 16.Kodama Y, Shumway M, Leinonen R, International Nucleotide Sequence Database Collaboration . 2012. The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res 40:D54–D56. doi: 10.1093/nar/gkr854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakagawa S, Takahashi MU. 2016. gEVE: a genome-based endogenous viral element database provides comprehensive viral protein-coding sequences in mammalian genomes. Database 2016:baw087. doi: 10.1093/database/baw087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paces J, Pavlícek A, Zika R, Kapitonov VV, Jurka J, Paces V. 2004. HERVd: the Human Endogenous RetroViruses Database: update. Nucleic Acids Res 32:D50. doi: 10.1093/nar/gkh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becker J, Pérot P, Cheynet V, Oriol G, Mugnier N, Mommert M, Tabone O, Textoris J, Veyrieras JB, Mallet F. 2017. A comprehensive hybridization model allows whole HERV transcriptome profiling using high density microarray. BMC Genomics 18:286. doi: 10.1186/s12864-017-3669-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kao D, Hsu K, Chiu S, Tu V, Chew A, Lee KH, Lee YK, Kwon DN, Greenhalgh DG, Cho K. 2012. ERE database: a database of genomic maps and biological properties of endogenous retroviral elements in the C57BL/6J mouse genome. Genomics 100:157–161. doi: 10.1016/j.ygeno.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Llorens C, Futami R, Covelli L, Domínguez-Escribá L, Viu JM, Tamarit D, Aguilar-Rodríguez J, Vicente-Ripolles M, Fuster G, Bernet GP, Maumus F, Munoz-Pomer A, Sempere JM, Latorre A, Moya A. 2011. The Gypsy Database (GyDB) of mobile genetic elements: release 2.0. Nucleic Acids Res 39:D70–D74. doi: 10.1093/nar/gkq1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bao W, Kojima KK, Kohany O. 2015. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA 6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kapitonov VV, Jurka J. 2008. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat Rev Genet 9:411–412; author reply 414. doi: 10.1038/nrg2165-c1. [DOI] [PubMed] [Google Scholar]
- 24.Hubley R, Finn RD, Clements J, Eddy SR, Jones TA, Bao W, Smit AF, Wheeler TJ. 2016. The Dfam database of repetitive DNA families. Nucleic Acids Res 44:D81–D89. doi: 10.1093/nar/gkv1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du J, Grant D, Tian Z, Nelson RT, Zhu L, Shoemaker RC, Ma J. 2010. SoyTEdb: a comprehensive database of transposable elements in the soybean genome. BMC Genomics 11:113. doi: 10.1186/1471-2164-11-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu HE, Zhang HH, Xia T, Han MJ, Shen YH, Zhang Z. 2013. BmTEdb: a collective database of transposable elements in the silkworm genome. Database (Oxford) 2013:bat055. doi: 10.1093/database/bat055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma B, Li T, Xiang Z, He N. 2015. MnTEdb, a collective resource for mulberry transposable elements. Database 2015:bav004. doi: 10.1093/database/bav004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li SF, Zhang GJ, Zhang XJ, Yuan JH, Deng CL, Gu LF, Gao WJ. 2016. DPTEdb, an integrative database of transposable elements in dioecious plants. Database 2016:baw078. doi: 10.1093/database/baw078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campillo-Balderas JA, Lazcano A, Becerra A. 2015. Viral genome size distribution does not correlate with the antiquity of the host lineages. Front Ecol Evol 3:143. doi: 10.3389/fevo.2015.00143. [DOI] [Google Scholar]
- 30.Menzel T, Rohrmann GF. 2008. Diversity of Errantivirus (retrovirus) sequences in two cell lines used for baculovirus expression, Spodoptera frugiperda and Trichoplusia ni. Virus Genes 36:583–586. doi: 10.1007/s11262-008-0221-5. [DOI] [PubMed] [Google Scholar]
- 31.Geisler C, Jarvis DL. 2016. Rhabdovirus-like endogenous viral elements in the genome of Spodoptera frugiperda insect cells are actively transcribed: implications for adventitious virus detection. Biologicals 44:219–225. doi: 10.1016/j.biologicals.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan AS, Ma H, Taliaferro LP, Galvin TA, Shaheduzzaman S. 2014. New technologies and challenges of novel virus detection. PDA J Pharm Sci Technol 68:661–666. doi: 10.5731/pdajpst.2014.01029. [DOI] [PubMed] [Google Scholar]
- 33.Khan AS, Vacante DA. 2014. Introduction and workshop summary: advanced technologies for virus detection in the evaluation of biologicals—applications and challenges. PDA J Pharm Sci Technol 68:546–547. doi: 10.5731/pdajpst.2014.01028. [DOI] [PubMed] [Google Scholar]
- 34.Khan AS, Vacante DA, Cassart JP, Ng SH, Lambert C, Charlebois RL, King KE. 2016. Advanced Virus Detection Technologies Interest Group (AVDTIG): efforts on high throughput sequencing (HTS) for virus detection. PDA J Pharm Sci Technol 70:591–595. doi: 10.5731/pdajpst.2016.007161. [DOI] [PubMed] [Google Scholar]
- 35.Ma H, Ma Y, Ma W, Williams DK, Galvin TA, Khan AS. 2011. Chemical induction of endogenous retrovirus particles from the Vero cell line of African green monkeys. J Virol 85:6579–6588. doi: 10.1128/JVI.00147-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Onions D, Côté C, Love B, Toms B, Koduri S, Armstrong A, Chang A, Kolman J. 2011. Ensuring the safety of vaccine cell substrates by massively parallel sequencing of the transcriptome. Vaccine 29:7117–7121. doi: 10.1016/j.vaccine.2011.05.071. [DOI] [PubMed] [Google Scholar]
- 37.Shahabuddin M, Sears JF, Khan AS. 2001. No evidence of infectious retroviruses in measles virus vaccines produced in chicken embryo cell cultures. J Clin Microbiol 39:675–684. doi: 10.1128/JCM.39.2.675-684.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson JA, Heneine W. 2001. Characterization of endogenous avian leukosis viruses in chicken embryonic fibroblast substrates used in production of measles and mumps vaccines. J Virol 75:3605–3612. doi: 10.1128/JVI.75.8.3605-3612.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weissmahr RN, Schüpbach J, Böni J. 1997. Reverse transcriptase activity in chicken embryo fibroblast culture supernatants is associated with particles containing endogenous avian retrovirus EAV-0 RNA. J Virol 71:3005–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bartosch B, Stefanidis D, Myers R, Weiss R, Patience C, Takeuchi Y. 2004. Evidence and consequence of porcine endogenous retrovirus recombination. J Virol 78:13880–13890. doi: 10.1128/JVI.78.24.13880-13890.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patience C, Scobie L, Quinn G. 2002. Porcine endogenous retrovirus—advances, issues and solutions. Xenotransplantation 9:373–375. doi: 10.1034/j.1399-3089.2002.02056_3.x. [DOI] [PubMed] [Google Scholar]
- 42.Kim A, Terzian C, Santamaria P, Pélisson A, Purd’homme N, Bucheton A. 1994. Retroviruses in invertebrates: the gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc Natl Acad Sci U S A 91:1285–1289. doi: 10.1073/pnas.91.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim AI, Lyubomirskaya NV, Belyaeva ES, Shostack NG, Ilyin YV. 1994. The introduction of a transpositionally active copy of retrotransposon GYPSY into the Stable Strain of Drosophila melanogaster causes genetic instability. Mol Gen Genet 242:472–477. doi: 10.1007/BF00281799. [DOI] [PubMed] [Google Scholar]
- 44.Neumann P, Pozárková D, Macas J. 2003. Highly abundant pea LTR retrotransposon Ogre is constitutively transcribed and partially spliced. Plant Mol Biol 53:399–410. doi: 10.1023/B:PLAN.0000006945.77043.ce. [DOI] [PubMed] [Google Scholar]
- 45.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 46.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. Blast+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. 2014. Pfam: the protein families database. Nucleic Acids Res 42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 49.Federhen S. 2012. The NCBI Taxonomy database. Nucleic Acids Res 40:D136–D143. doi: 10.1093/nar/gkr1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequences in RVDB and NCBI Viral Genomes were characterized by using the python script designated RVDB_characterization.py into the level 1 categories exogenous viral (ExViral), endogenous nonretroviral (ENRV), endogenous retroviral (ERV), and LTR retrotransposon, and the remaining viral sequences were categorized as unassigned genes/fragments. The number of entries based on family taxonomic identifiers or names is indicated. Entries that could not be mapped on the basis of a taxonomic identifier or were outdated are boxed. The characterization script used is available at https://github.com/ArifaKhanLab/RVDB, along with the instructions for its use (the latter is also provided in Text S2). Download TABLE S1, XLSX file, 0.1 MB (29.6KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Instructions and scripts for updating RVDB and generating the unclustered and clustered fasta files. These are also available at https://github.com/ArifaKhanLab/RVDB. Download TEXT S1, DOCX file, 0.03 MB (37.5KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Instructions and scripts for characterization of entries into level 1 categories. These are also available at https://github.com/ArifaKhanLab/RVDB. Download TEXT S2, DOCX file, 0.01 MB (16KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.