Aluminum tolerance of aluminum-sensitive rice was enhanced under darkness by multiple changes in membrane sterols: decreased stigmasterol, increased precursor partitioning for sterols biosynthesis, and increased expression of HMG genes.
Keywords: Al tolerance, carotenoid, dark conditions, HMG gene, rice, sterol, stigmasterol
Abstract
Aluminum-sensitive rice (Oryza sativa L.) cultivars showed increased Al tolerance under dark conditions, because less Al accumulated in the root tips (1 cm) under dark than under light conditions. Under dark conditions, the root tip concentration of total sterols, which generally reduce plasma membrane permeabilization, was higher in the most Al-sensitive japonica cultivar, Koshihikari (Ko), than in the most Al-tolerant cultivar, Rikuu-132 (R132), but the phospholipid content did not differ between the two. The Al treatment increased the proportion of stigmasterol (which has no ability to reduce membrane permeabilization) out of total sterols similarly in both cultivars under light conditions, but it decreased more in Ko under dark conditions. The carotenoid content in the root tip of Al-treated Ko was significantly lower under dark than under light conditions, indicating that isopentenyl diphosphate transport from the cytosol to plastids was decreased under dark conditions. HMG2 and HMG3 (encoding the key sterol biosynthetic enzyme 3-hydroxy-3-methylglutaryl CoA reductase) transcript levels in the root tips were enhanced under dark conditions. We suggest that the following mechanisms contribute to the increase in Al tolerance under dark conditions: inhibition of stigmasterol formation to retain membrane integrity; greater partitioning of isopentenyl diphosphate for sterol biosynthesis; and enhanced expression of HMGs to increase sterol biosynthesis.
Introduction
To adapt to acidic soil environments, plants have developed various strategies to protect root tips from aluminum (Al). The molecular basis and the role of Al sensing and signaling in Al tolerance have been reviewed (Liu et al., 2014).
A few studies have revealed some of the mechanisms by which the composition of plasma membrane (PM) lipids affects Al tolerance. Dysfunction of phosphatidyl phosphohydrolase in an Arabidopsis mutant led to the accumulation of phospholipids in the PM and a decrease in Al tolerance (Kobayashi et al., 2013). The expression level of PsCYP51 (encoding OBT 14DM in pea, Pisum sativum L.) was correlated with Al tolerance, and knocked-down CYP51A expression suppressed Al tolerance in Arabidopsis. The decreased expression level of CYP51 lowered the sterol content and increased the PM permeability in Al-treated root tips (Wagatsuma et al., 2015). Sterols are essential for the maintenance of membrane fluidity and permeability in living cells, and the ion leakage from roots (as an index of membrane integrity) was markedly increased in cyp51 mutants (Kim et al., 2005). All sterol species in plants affect membrane fluidity, but each one has different effects depending on its structure (Hartmann, 1998). Among all the sterols, sitosterol most strongly restricts the mobility of the surrounding phospholipid acyl chains. The trans-oriented double bond at C22 in the side chain of stigmasterol significantly reduces its ordering ability, with an efficiency that can be measured by the reduction in water permeability through the membrane; the order of efficiency is sitosterol>campesterol>>stigmasterol (Schuler et al., 1991). Therefore, higher proportions of stigmasterol lead to greater membrane permeability. To date, there are no reports on the contribution of different sterol species to differences in Al tolerance.
The synthesis of lipids in the PM is under complex regulation, and responds to various environmental conditions other than Al. Seedlings grown under −P pretreatment showed enhanced Al tolerance, which was mainly attributed to a decrease in phospholipids and increase in galactolipids in the PM of root cells (Maejima et al., 2014). Enzymes in sterol biosynthesis pathways show different activities under light and dark conditions. For example, 3-hydroxy-3-methyglutaryl CoA reductase (HMGR, encoded by HMG), the key limiting enzyme in phytosterol biosynthesis via the mevalonate (MVA) pathway (Schaller et al., 1995), was induced under dark conditions in Arabidopsis seedlings, except in the roots (Enjuto et al., 1994; Learned, 1996). In rice roots, expression of HMG2, which is also involved in sterol biosynthesis, was slightly increased in the dark (Ha et al., 2001). The overexpression of HMG in tobacco (Nicotiana tabacum L.) increased sterol contents (Schaller et al., 1995). On the basis of those results, we speculated that a dark treatment could confer Al tolerance via increased expression of HMGs.
Sterols are isoprenoid-derived molecules that are found in a wide range of organisms including plants (Schaller, 2004). All isoprenoids are biosynthesized from a common precursor, i.e. the five-carbon (C5) molecule isopentenyl diphosphate (IPP; see Fig. 7). In plants, IPP is produced via both the MVA and the methylerythritol 4-phosphate (MEP) pathways, although they are localized in different cell compartments, i.e. the cytosol/ER and plastids, respectively. The MEP pathway was discovered only relatively recently (Lichtenthaler et al., 1997); consequently, little is known about the crosstalk between the MEP and MVA pathways, and especially their roles in producing IPP in the roots under dark conditions.
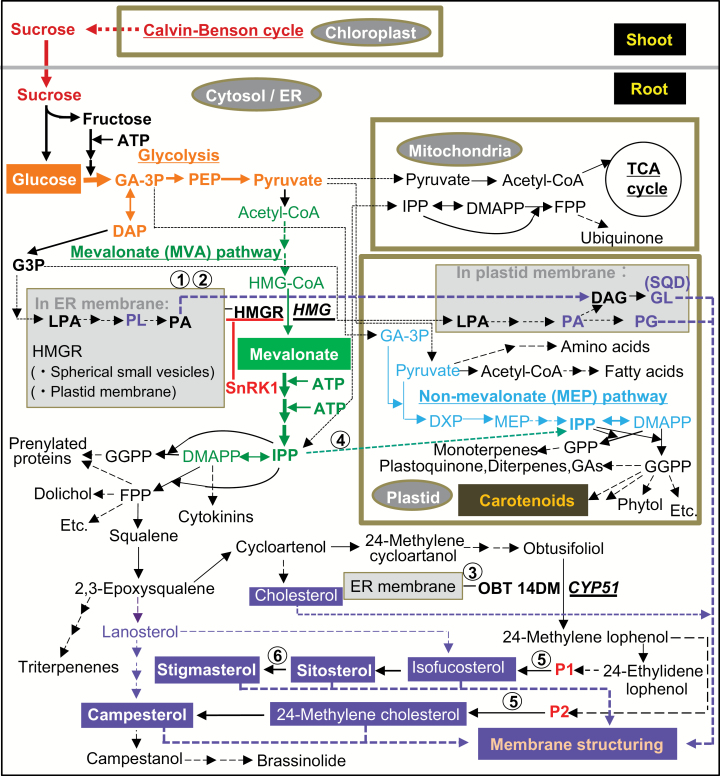
Fig. 7.
Model for biosynthesis of sterols and other isoprenoids in rice roots. Two independent pathways produce IPP, the precursor of all isoprenoids: the cytosolic acetate/mevalonate (MVA) pathway and the plastidial non-MVA methylerythritol 4-phosphate (MEP) pathway. Figure was compiled by reference to Fig. 1 of Fujioka et al. (1997), Fig. 1 of Rodríguez-Concepción et al. (2004), and Fig. 1 of Wang et al. (2012). Thicker arrows or bold letters in cytosol indicate presumed high concentrations of metabolites under dark conditions with mevalonate and glucose supplied (this study; see Discussion). Glycolysis, MVA, and MEP pathways are shown in orange, green, and blue letters, respectively. Purple dashed arrow shows biosynthetic pathway to phytosterol via lanosterol in dicots. ①, localization of HMGR at endoplasmic reticulum (ER) membrane and partially at spherical small vesicles (Leivar et al. 2005); ②, localization of HMGR at plastid membrane (Wong et al. 1982); ③, localization site of OBT 14DM at ER membrane (Bak et al. 1997); ④, greater inhibition of cytosolic IPP partitioning from the cytosol to the plastid by Al treatment in Al-sensitive Ko under DMG than under L; ⑤, Δ5,7-sterol Δ7-reductase encoded by DWARF5; ⑥, sterol-Δ22-desaturase encoded by CYP710A1; DAP, dihydroxyacetone phosphate; DMAPP, dimethylallyl diphosphate; DXP, 1-deoxy-D-xylulose-5-phosphate; FPP, farnesyl diphosphate; GA-3P, glyceraldehyde-3-phosphate; GAs, gibberellins; GGPP, geranylgeranyl diphosphate; G3P, glycerol 3-phosphate; GL, galactolipids; GPP, geranyl diphosphate; HMG, 3-hydroxy-3-methylglutaryl-CoA reductase encoding gene; HMGR, HMG reductase; IPP, isopentenyl diphosphate; LPA, lysophosphatidate; MEP, methylerythritol-4-phosphate; P1, stigmasta-5,7,Z-24(241)-trien-3β-ol; P2, ergosta-5,7,24(241)-trien-3β-ol; PEP, phosphoenol pyruvate; PG, phosphatidylglycerol; PL, phospholipids; SnRK1, sucrose non-fermenting 1-related kinase 1; SQD, sulfoquinovosyldiacylglycerol.
In this study, Al-sensitive rice cultivars showed increased Al tolerance under dark conditions. Therefore, we conducted molecular and physiological analyses to explore the reasons for the difference in Al tolerance between dark-grown and light-grown plants. Changes in the sterol status in the dark led to enhanced Al tolerance of temperate japonica rice cultivars. These changes were related to IPP crosstalk between the cytosol and the plastid, and the increased expression of HMG genes in the root tip in the dark.
Materials and methods
Plant material and growth conditions
We used six cultivars of rice: five temperate japonica subpopulations (Rikuu-132, Hidekomochi, Norin-21, Rikuu-20, and Koshihikari) and one Indica aus subpopulation (Kasalath) (Xu et al., 2009). Seeds were soaked in tap water with aeration for 24 h at 27 °C in a growth room and germinated under fluorescent white light (80 µmol m−2 s−1). The germinated seeds were spread on a nylon screen, placed on a container filled with 9 l tap water containing (in mg l−1) Ca 8.0, Mg 2.92, K 1.95, and minor quantities of other minerals (P, Fe, Mn, Zn, and Cu) (Khan et al., 2009), and grown for 5 d.
Analysis for Al tolerance under different illumination conditions
Twelve seedlings of each cultivar were used as a set of seedlings for root growth experiments. Sets of seedlings with similar root length (ca 4 cm) were pre-incubated in control solution (pH 4.9; 0.2 mM CaCl2) for 6 h. Then, each set was transferred to the Al-toxic solution containing 10 µM AlCl3 (+Al) or control solution (−Al). Root elongation was measured after 24 h of incubation and then relative root elongation (+Al/−Al, expressed as a percentage) was calculated as an index of Al tolerance (Khan et al., 2009). Seedlings were kept in the light (L; continuous light; 80 µmol m−2 s−1), in the dark (D) or in the dark with MVA (1 mM; Sigma-Aldrich, St Louis, MO, USA) and glucose (1 mM) (DMG). The pH of all solutions was maintained at 4.9 by adjusting at 2 h and 18 h after the start of the treatment. The root growth experiment was replicated independently using five sets for L, four sets for D, and two sets for DMG. The same trends were observed in each replicated experiment, and so the results from all experiments were pooled and used to calculate average values and standard errors.
Visualization of Al accumulation in roots
After treatment with or without Al, whole roots were stained with hematoxylin (0.2% hematoxylin in 0.02% sodium iodide, w/w, pH 4.8) for 15 min as described by Khan et al. (2009), and Al accumulation in the root tip was observed under a stereoscope (SMZ-10, Nikon, Tokyo, Japan).
Real-time qRT-PCR
Total RNA was extracted from 1-cm root tips of cv. Rikuu-132 (R132) and cv. Koshihikari (Ko) after 24 h treatment with 0.2 mM CaCl2 with or without 10 µM AlCl3 (pH 4.9) under L or DMG. Extraction and purification of the total RNA was performed using an RNAqueous column with Plant RNA Isolation Aid (Ambion, Austin, TX, USA). cDNA was synthesized from 1 µg total RNA with a QuantiTech reverse transcription kit (Qiagen, Hilden, Germany). Real-time qRT-PCR using SYBR Green I was carried out with a TP800 thermal cycler (Takara Bio, Shiga, Japan) as described previously (Wagatsuma et al., 2015) using the following gene-specific primers; 5′-GGACGTGGAAAGTCTGTGGT-3′ (sense) and 5′-AACAGCTGAACCAGCAAGGT-3′ (antisense) for OsHMG2, and 5′-AAGGCCTTCTTGGATTC-3′ (sense) and 5′-GCAGCAGCTGAATCTCATGT-3′ (anti-sense) for OsHMG3. The gene transcript levels were quantified by the standard curve method using a complementary DNA dilution series as described by Bustin et al. (2009). The transcript levels of each gene were normalized to that of 18S rRNA.
Extraction and quantification of sterols and phospholipids in the roots
Sterols were extracted and quantified as described by Suzuki et al. (2004). Briefly, the freeze-dried 1-cm root tips of 5-day-old seedlings of three rice cultivars [Kasalath (Ka), Ko, and R132] after 24 h treatment with 0.2 mM CaCl2 with or without 10 µM AlCl3 (pH 4.9) under L or DMG were extracted with CHCl3–methanol (1:1), and [25,26,26,26,27,27,27-2H7]cholesterol was added to the extract as an internal standard. The extract was dried and chromatographed on a silica gel column with hexane–ethyl acetate (2:1) and CHCl3–methanol (1:1). The hexane-ethyl acetate eluent was dried, saponified with methanol and 20% KOH, and its eluent with CHCl3–methanol was dried. The residue and the debris from extraction were combined and hydrolysed with MeOH and 4 M HCl. These mixtures were extracted with hexane, and the combined hexane layer was dried. The residue was trimethylsilylated and analysed by GC-MS (GC: 6890A, Agilent Technologies, Wilmington, DE, USA; MS: JMS-AM SUN200, JEOL, Tokyo).
Phospholipids were extracted and quantified as described by Khan et al. (2009). Briefly, phospholipids were extracted by the modified Bligh and Dyer method [isopropanol: chloroform: H2O (1:1:1, v/v/v)]. After purification and dehydration of the extract, phosphorus was quantified by the molybdenum blue spectrophotometric method.
Extraction and quantification of carotenoids in the root
Carotenoids were extracted and quantified as described by Şükran et al. (1998). Briefly, fresh 1-cm root tips of 5-day-old seedlings of two rice cultivars (Ko, R132) after 24 h treatment with 0.2 mM CaCl2 with or without 10 μM AlCl3 (pH 4.9) under L or DMG were extracted with 96% MeOH. The absorbance of the extract was determined at 470, 653, and 666 nm with a spectrophotometer. The carotenoid concentration was calculated as follows: carotenoids (μg ml−1)=(1000A470−2.86chloropyll a−129.2chlorophyll b)/245.
Statistical analysis
All data were analysed using Fisher’s least significant difference (LSD) test (Fisher, 1958).
Results
Aluminum tolerance of rice cultivars and Al accumulation in root tip under different illumination conditions
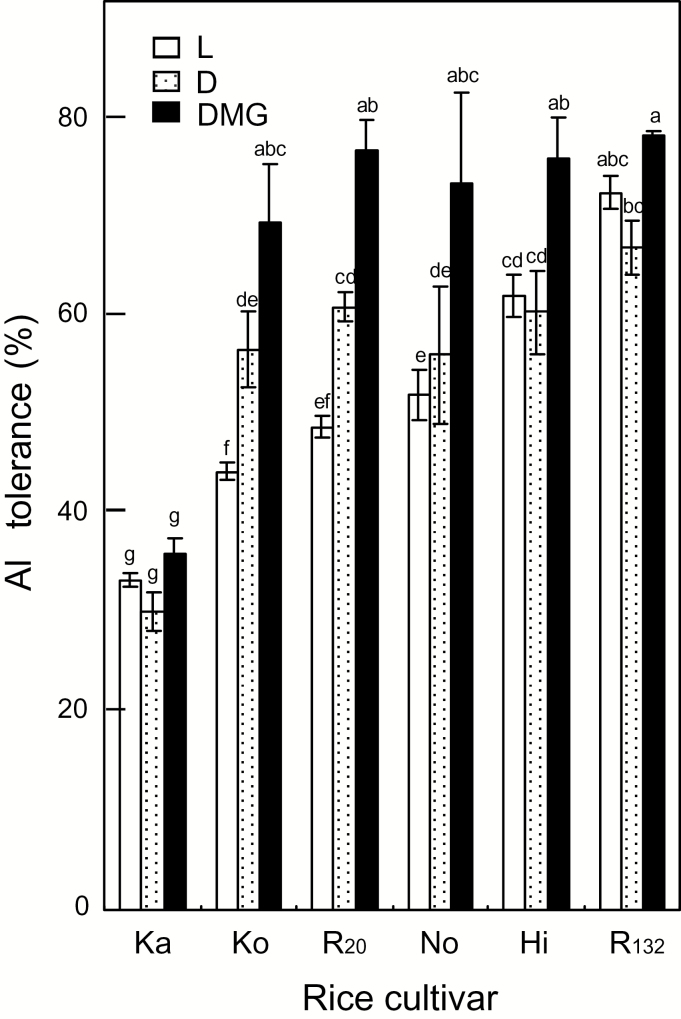
Under L, the order of Al tolerance among the rice cultivars was as follows: R132≥Hidekomochi (Hi)>Norin-21 (No)≥Rikuu-20 (R20)≥Ko>Ka (Fig. 1). Under D, the Al tolerance became similar among R132, Hi, No, R20, and Ko, because of an increase in the Al tolerance of the Al-sensitive cultivars. The Al tolerance of the most Al-sensitive indica-type Ka and the most Al-tolerant japonica-type R132 was unchanged by the different illumination conditions. Addition of MVA and glucose (sterol precursors) to the Al toxic solution further enhanced the Al tolerance of all Al-sensitive japonica-type cultivars (Fig. 1). All japonica-type cultivars showed similar growth in the Al toxic solution. The most Al-sensitive japonica-type cultivar under L, Ko, grew comparably to the most Al-tolerant one, R132, under D.
Fig. 1.
Aluminum tolerance of six rice cultivars under different light conditions. Five-day-old rice seedlings were treated for 24 h with 10 µM AlCl3 in the presence of 0.2 mM CaCl2 (pH 4.9), and Al tolerance was calculated as the ratio of net elongation of the longest root in the Al treatment to that in the control (0.2 mM CaCl2 without Al). Twelve seedlings with similar root length (ca 4 cm) were used for one set in Al tolerance analysis. Analysis was carried out independently for five sets for L, four sets for D, and two sets for DMG. L, light; D, dark; DMG, dark in the presence of 1 mM mevalonate and 1 mM glucose. Rice cultivars: Ka, Kasalath; Ko, Koshihikari; Hi, Hidekomochi; No, Norin-21; R20, Rikuu-20; R132, Rikuu-132. Values are means of independent replicates±standard error. Different letters above bars indicate significant differences (P<0.05; Fisher’s LSD).
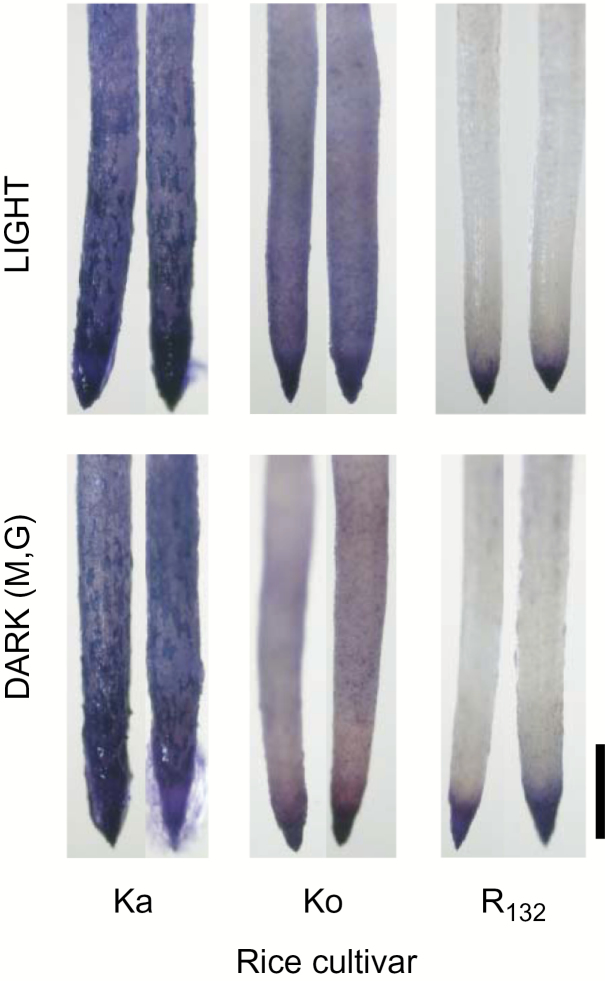
We compared Al accumulation in the root tip of the three cultivars with significant differences in Al tolerance (Fig. 2). Under L, more Al accumulated in the Al-sensitive cultivars than in R132, as judged by the blue color intensity of hematoxylin staining. The blue color in the Ko root tip was lighter under DMG, suggesting that the DMG treatment improved the Al tolerance of Ko by decreasing Al accumulation in the root tip.
Fig. 2.
Aluminum accumulation in root tip. Roots of 5-day-old seedlings of three rice cultivars, i.e. Ka (Kasalath), Ko (Koshihikari), and R132 (Rikuu-132), were treated for 24 h with 10 μM AlCl3 + 0.2 mM CaCl2 (pH 4.9) under light (LIGHT) or dark conditions in the presence of 1 mM mevalonate and 1 mM glucose (DARK[M,G]). More than six root tips were used for each cultivar, and two representative tips are shown. Aluminum accumulation was observed by hematoxylin staining; denser purple staining indicates greater Al accumulation. Scale bar: 1 mm.
Sterol profile and phospholipid content in the root tip under different illumination conditions
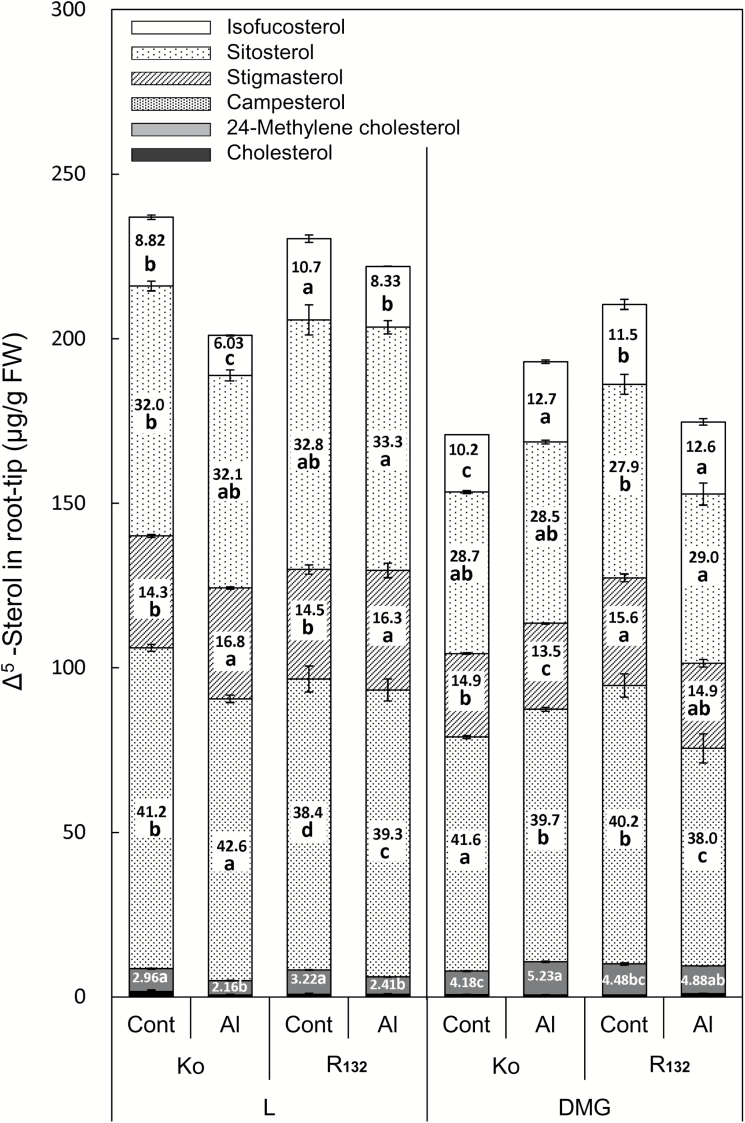
Irrespective of the genotype, Al treatment, or illumination conditions, the major sterol species were sitosterol, stigmasterol, and campesterol (together accounting for 81–92% of total sterols) and the minor sterol species were isofucosterol and 24-methylene cholesterol (together accounting for 8.2–18.2% of total sterols) (Fig. 3). Other sterol species accounted for only small proportions of total sterols: i.e. cholesterol, 0.33–0.69%; cycloartenol, 0.12–0.87%; unidentified sterols, 0.44–0.81% (data not shown).
Fig. 3.
Sterol composition in 1-cm root tip of two rice cultivars (Ko, Koshihikari; R132, Rikuu-132) under different illumination conditions. Length of each bar indicates concentration of Δ5-sterols (µg g−1 FW of root tips); number within each bar indicates relative proportion of each sterol to total Δ5-sterols in that treatment (%). As the composition of cholesterol was too low (0.33–0.69%), it is concealed below that of 24-methylene cholesterol. Five-day-old rice seedlings were treated for 24 h with 0.2 mM CaCl2 in the presence or absence of 10 µM AlCl3 (pH 4.9) under light conditions (L) or dark conditions in the presence of 1 mM mevalonate and 1 mM glucose (DMG). Values are means of three independent replicates±standard error. Different letters in bars indicate significant differences (P<0.05; Fisher’s LSD) among the same sterol species in all treatments under the same illumination conditions.
Under L, the Al treatment caused a greater decrease in the total sterol content in the Al-sensitive Ko than in the Al-tolerant R132, while the sterol content was similar in the two cultivars in the absence of Al under L (Fig. 3). The Al treatment also affected the sterol profile. Under L, the proportion of the two major sterol species (stigmasterol and campesterol) out of total sterols was significantly higher in the Al treatment (stigmasterol, 16.3 ± 0.40% in R132 and 16.8 ± 0.13% in Ko; campesterol, 39.3 ± 0.10% in R132 and 42.6 ± 0.17% in Ko) than in the control (stigmasterol, 14.5 ± 0.12% in R132 and 14.3 ± 0.07% in Ko; campesterol, 38.4 ± 0.09% in R132 and 41.2 ± 0.28% in Ko) (P<0.05; LSD test). The Al-induced increase in the proportion of the major sterol species was slightly greater in the Al-sensitive Ko than in Al-tolerant R132 (Fig. 3 and Table 1). Under L, the proportion of sitosterol was unchanged by the Al treatment (32.8 ± 0.44% [control] and 33.3 ± 0.30% [Al treatment] in R132 and 32.0 ± 0.12% [control] and 32.1 ± 0.38% [Al treatment] in Ko).
Table 1.
Proportional ratios of major sterol species under different conditions (calculated from data shown in Fig. 3)
Al/Cont: proportion of each sterol species to total sterols in Al/that in control. AlDMG/AlL: proportion of each sterol species to total sterols in Al under DMG/that in Al under L.
| Sitosterol | Stigmasterol | Campesterol | |||
|---|---|---|---|---|---|
| Al/Cont | L | R132 | 1.02 | 1.12 | 1.02 |
| Ko | 1.00 | 1.17 | 1.04 | ||
| DMG | R132 | 1.04 | 0.96 | 0.95 | |
| Ko | 0.99 | 0.91 | 0.95 | ||
| AlDMG/AlL | R132 | 0.87 | 0.91 | 0.97 | |
| Ko | 0.89 | 0.80 | 0.94 |
Under L, the proportion of the two minor sterol species (isofucosterol and 24-methylene cholesterol) out of total sterols was significantly lower in the Al treatment (isofucosterol, 8.33 ± 0.27% in R132 and 6.03 ± 0.16% in Ko; 24-methylene cholesterol, 2.41 ± 0.07% in R132 and 2.16 ± 0.09b in Ko) than in the control (isofucosterol, 10.7 ± 0.06% in R132 and 8.82 ± 0.14% in Ko; 24-methylene cholesterol, 3.22 ± 0.12% in R132 and 2.96 ± 0.03% in Ko). The decrease in the proportion of the minor sterol species by the Al treatment was larger in the Al-sensitive Ko than in the Al-tolerant R132 (Fig. 3 and Supplementary Table S1 at JXB online).
Under DMG, the total sterol content was decreased by the Al treatment in the Al-tolerant R132, as under L. However, the total sterol content in the Al-sensitive Ko was increased by the Al treatment (Fig. 3). The effect of the Al treatment on the sterol profile under DMG was the reverse of that observed under L, that is, the proportion of the two major sterol species (stigmasterol or campesterol) out of total sterols was decreased significantly by the Al treatment in both cultivars except for stigmasterol in R132 (stigmasterol, 15.6 ± 0.15% in the control and 14.9 ± 0.35% in the Al treatment for R132, and 14.9 ± 0.09% in the control and 13.5 ± 0.09% in the Al treatment for Ko; campesterol, 40.2 ± 0.15% in the control and 38.0 ± 0.31% in the Al treatment for R132, and 41.6 ± 0.09% in the control and 39.7 ± 0.23% in the Al treatment for Ko). In contrast, the Al treatment increased significantly the proportion of the minor two sterol species except for 24-methylene cholesterol (isofucosterol, 11.5 ± 0.23% in the control and 12.6 ± 0.24% in the Al treatment for R132, and 10.2 ± 0.07% in the control and 12.7 ± 0.18% in the Al treatment for Ko; 24-methylene cholesterol, 4.48 ± 0.05b% in the control and 4.88 ± 0.27% in the Al treatment for R132, and 4.18 ± 0.08% in the control and 5.23 ± 0.07% in the Al treatment for Ko). The decrease in one of the major sterol species (stigmasterol) (Fig. 3 and Table 1) and the increase in the minor sterol species (Fig. 3 and Supplementary Table S1) by the Al treatment were larger in the Al-sensitive Ko. The proportion of sitosterol out of the total sterol content was slightly increased or similar for the Al treatment in both cultivars (27.9 ± 0.33% in the control and 29.0 ± 0.47% in the Al treatment for R132, and 28.7 ± 0.12% in the control and 28.5 ± 0.06% in the Al treatment for Ko).
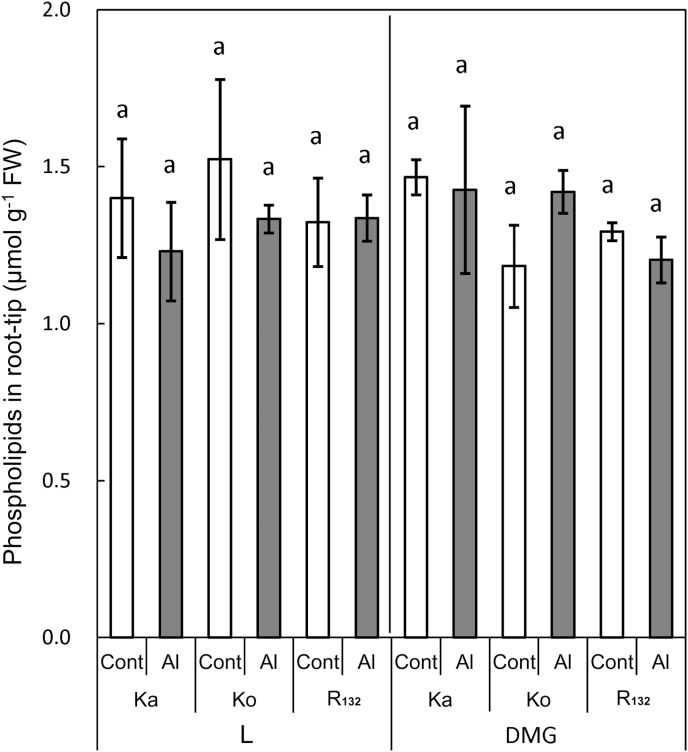
The phospholipid content in the root tip did not differ significantly among the three rice cultivars, i.e. Ka, Ko, and R132, irrespective of the Al treatment and illumination conditions (Fig. 4).
Fig. 4.
Phospholipids content in 1-cm root tip of three rice cultivars with or without Al treatment under different illumination conditions (μmol g−1 FW). Five-day-old rice seedlings were treated for 24 h with 0.2 mM CaCl2 in the presence or absence of 10 μM AlCl3 (pH 4.9) under light (L) or dark conditions in the presence of 1 mM mevalonate and 1 mM glucose (DMG). Ka, the most Al-sensitive indica aus cv. Kasalath; Ko, Al-sensitive japonica cv. Koshihikari; R132, Al-tolerant japonica cv. Rikuu-132. Values are means of independent replicates±standard error. The same letters above bars indicate non-significant differences (P<0.05; Fisher’s LSD).
Carotenoid content in root tip under different illumination conditions
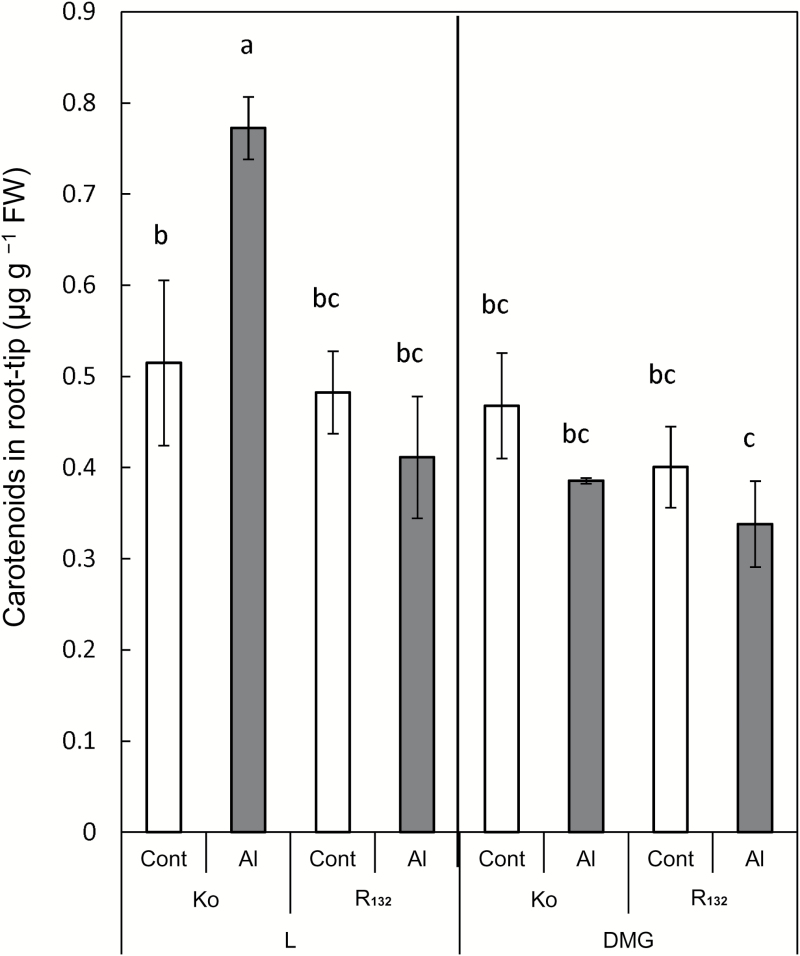
To explore the crosstalk between the MVA and MEP pathways under different illumination conditions or Al treatment, we measured the carotenoid content as a measure of IPP transport from the cytosol to the plastids, although plastids contain not only carotenoids but also plastoquinone, monoterpenes, diterpenes, GAs, phytols, and other products (see Fig. 7). Under L, the Al treatment slightly decreased the carotenoid content in the root tip of the Al-tolerant R132, but markedly increased that in the root tip of Al-sensitive Ko (Fig. 5). Under D, the carotenoid content in the root tip was slightly decreased by the Al treatment in both R132 and Ko.
Fig. 5.
Carotenoid content in 1-cm root tip of two rice cultivars under different illumination conditions. Ko, Koshihikari; R132, Rikuu-132; L, light conditions; DMG, dark conditions in the presence of 1 mM mevalonate and 1 mM glucose. Values are means of independent replicates±standard error. Letters (a–c) above error bars indicate significant differences (P<0.05; Fisher’s LSD).
Transcript levels of HMG2 and HMG3 in root tip under different illumination conditions
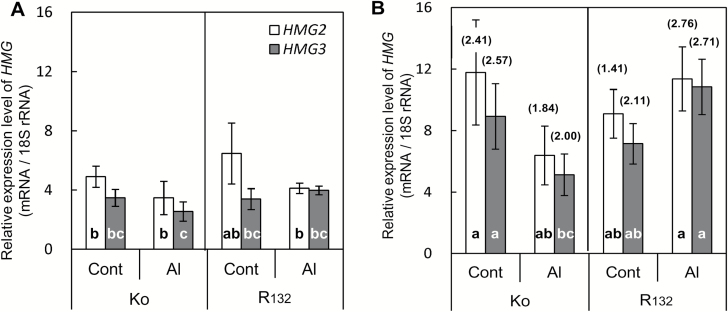
Among the HMG genes, both HMG2 and HMG3 are thought to be related to sterol biosynthesis although their functions are different (Ha et al., 2001; Ohyama et al., 2007). We measured the mRNA levels of these genes. Under L, both HMGs generally showed slightly higher transcript levels in the Al-tolerant R132 root tips than in the Al-sensitive Ko root tips, irrespective of the Al treatment. Their transcript levels were generally decreased by the Al treatment (Fig. 6A). The transcript levels of both HMGs were higher under DMG than under L (1.41–2.76 times higher) (Fig. 6B). The Al treatment decreased HMG transcript levels in the Al-sensitive Ko, but increased them in the Al-tolerant R132. Irrespective of the illumination conditions, the transcript level of HMG2 was always higher than that of HMG3.
Fig. 6.
Relative transcript levels of HMG2 and HMG3 in 1-cm root tip of two rice cultivars under different light conditions. Five-day-old rice seedlings were treated for 24 h with 0.2 mM CaCl2 under L (light conditions) (A) or DMG (dark conditions with 1 mM mevalonate and 1 mM glucose) (B). Total RNA was extracted from frozen 1-cm root tips and used for real-time qRT-PCR. Relative transcript levels of HMGs were normalized to that of 18S rRNA (internal control). Numbers in parentheses indicate relative transcript level of each HMG under DMG to that under L. Rice cultivars: Ko, Koshihikari; R132, Rikuu-132. Values are means of three independent replicates±standard error. Different letters in bars indicate significant differences (P<0.05; Fisher’s LSD) among all treatments.
Discussion
New finding that enhanced Al tolerance of rice cultivars in the dark is not related to changes in phospholipids in the root tip
Several studies have focused on the effects of different ions in the rhizosphere (OH-, H+, Ca2+, Mg2+, phosphate, SO42-) on Al tolerance (Tanaka et al., 1987). However, none has focused on the effects of aboveground conditions on Al tolerance. Here, we have shown for the first time that the Al tolerance of Al-sensitive temperate japonica cultivars was enhanced under darkness, especially in the presence of the precursors MVA and glucose (under DMG; Fig. 1). Aside from genetic engineering techniques, no other methods have successfully produced rice lines with enhanced Al tolerance. Dark or DMG conditions did not retard net root elongation in the absence of Al, and the net root elongation in the Al treatment under D or DMG was considerably increased in the most Al-sensitive temperate japonica cultivar Ko, compared with that under L (ca 60% increase under DMG) (Supplementary Table S2). These results indicate that the increase in Al tolerance under DMG is not an artefact of the calculation of the relative value for Al tolerance, but reflects an actual increase in root elongation under DMG.
The phospholipid content in the root tip did not differ significantly among the three rice cultivars, irrespective of the Al treatment and the illumination conditions (Fig. 4). The weaker negative charge associated with lower phospholipid content in the PM has been reported as an effective Al-tolerance mechanism in rice, Arabidopsis, and tobacco (Khan et al., 2009; Kobayashi et al., 2013, Maejima et al., 2014; Wagatsuma et al., 2015; Zhang et al., 2016). However, we found that the phospholipid content did not contribute to the differences in Al tolerance among cultivars in this study. Greater permeation of Al through the PM into the cytoplasm is suggested to result not only from increased phospholipid content in the PM, but also from increased contents of other lipid species without a negative charge, such as sterols, which affects the permeability of the PM.
Enhanced Al tolerance of Al-sensitive Ko in the dark is due to quantitative and qualitative changes in sterols and regulated partitioning of IPP to plastids
The lower Al accumulation in the root tip of the Al-tolerant cultivar under L (Fig. 2) is consistent with the results of a previous study (Khan et al., 2009). Under DMG, the Al accumulation in the root tip was remarkably decreased only in the most Al-sensitive temperate japonica cultivar Ko. There are no previous reports of a decrease in Al accumulation in the root tip of rice under DMG.
Several studies have isolated and characterized Al tolerance genes from rice (Liu et al., 2014). However, fewer studies have focused on Al tolerance mechanisms or genes related to lipid metabolism.
Under L, the total sterol content in the Al treatment was higher in the Al-tolerant R132 than in the Al-sensitive Ko (Fig. 3). Under DMG, the increase in Al tolerance in Al-sensitive Ko was accompanied by an increase in the total sterol content and the relatively greater decrease in Al accumulation in the root tip, compared with that in the root tip of the Al-tolerant R132 in the Al treatment (Figs 1–3). Therefore, our results indicate that higher Al tolerance is related to higher sterol content and lower Al accumulation in the root tip. These points agreed with the results of other studies on pea (P. sativum), triticale (×Triticosecale Wittmark cv. Currency), maize (Zea mays), wheat (Triticum aestivum), sorghum (Sorghum bicolor), rice, and Arabidopsis (Khan et al., 2009; Wagatsuma et al., 2015).
Campestanol, P1 (stigmasta-5,7,Z-24(241)-trien-3β-ol) and P2 (ergosta-5,7,24(241) -trien-3β-ol) (Fig. 7) were far lower than those of the three major sterols and the two minor sterol species detected in this study (Fujioka et al., 1997; Kim et al., 2005). Therefore, the changes in the sterol profile can be discussed based on the fate of these five sterol species. In normal light conditions, the proportional ratio (Al/Cont) of the two major sterols (sitosterol and campesterol) in the Al-tolerant R132 was almost 1.0. The proportional ratio of stigmasterol was increased to 1.12 in the Al-tolerant R132 and 1.17 in the Al-sensitive Ko (Table 1). Under DMG, however, the proportional ratio of stigmasterol was decreased in both cultivars (0.96 in Al-tolerant R132 and 0.91 in Al-sensitive Ko), and the decrease was greater in the Al-sensitive Ko. There was a greater decrease in the ratio of the proportion of stigmasterol to total sterols in the Al treatment under DMG to that under L (AlDMG/AlL) in the Al-sensitive Ko than in the Al-tolerant R132 (0.91 in Al-tolerant R132 and 0.80 in Al-sensitive Ko). Stigmasterol has no ability to reduce membrane permeabilization because of the trans-oriented double bond at C22 in its side chain (Schuler et al., 1991; Hartmann, 1998).
The initial Δ5-sterols, i.e. isofucosterol and 24-methylene cholesterol, are formed from P1 and P2, respectively (Fig. 7), in a reaction catalysed by Δ5,7-sterol Δ7-reductase encoded by DWARF5 (Choe et al., 2000; Benveniste, 2002) (⑤ in Fig. 7). Formation of stigmasterol from sitosterol is catalysed by sterol-Δ22-desaturase encoded by CYP710A1 (Morikawa et al., 2006). We suggest that the reduced proportion of stigmasterol in the Al-sensitive Ko under DMG may be due to inhibition of sterol-Δ22-desaturase (⑥ in Fig. 7) by Al that accumulated to moderately high level in the Al-sensitive Ko (Fig. 2).
The proposed effects of Al on sterol synthesis, based on the data obtained in this study, can be summarized as follows. Under L, Al inhibited the activity of Δ5,7-sterol Δ7-reductase, especially in the Al-sensitive Ko, as shown in the sterol profiles in Figs 3 and 7 and Table 1 (decreased isofucosterol and increased stigmasterol in the Al treatment). Under D, Al treatment inhibited the activity of sterol-Δ22-desaturase, especially in the Al-sensitive Ko as shown in Figs 3 and 7 and Table 1 (increased isofucosterol and decreased stigmasterol in the Al treatment). The higher Al accumulation in the Al-sensitive Ko (Fig. 2) supports these results. The Al-sensitive Ko may have greater potential to change the Al-targeted enzyme from Δ5,7-sterol Δ7-reductase to sterol-Δ22-desaturase to enhance Al tolerance under D. This is promising information for a new strategy to generate Al-tolerant rice lines, although its detailed mechanisms are unknown. The most Al-sensitive indica aus, Ka, showed the largest increase in the proportion of stigmasterol out of total sterols in response to Al, irrespective of the illumination conditions: 20.5 ± 0.26% in the Al treatment and 16.2 ± 0.27% in the control (P<0.01; t-test) under L, and 20.5 ± 0.73 in the Al treatment and 14.0 ± 0.18% in the control under DMG (P<0.01; t-test) (Supplementary Fig. S1). This result suggests that the same mechanisms operate in the most Al-sensitive indica aus, Ka, as in the two temperate japonica cultivars.
Under dark conditions, instant activation of the MVA pathway provides increased amounts of precursors for the biosynthesis of sterols, which are required for growth in search of light (Rodríguez-Concepción, 2006). The pivotal MVA intermediate IPP is transported to the plastid for the production of carotenoids under dark conditions (Park et al., 2002). An active MEP pathway has been found in some non-photosynthetic tissues such as roots (Walter et al., 2002; Hampel et al., 2005; Seemann et al., 2006; Goldwasser et al., 2008; López-Ráez et al., 2008; Kohlen et al., 2011; Abe et al., 2014). Although active crosstalk of cytosolic IPP from the MVA pathway has been reported between the cytosol and the plastid, there is no information on IPP crosstalk in the roots under different illumination conditions. In the Al-sensitive Ko, carotenoid production was inhibited by Al treatment under DMG (Fig. 5), although greater carotenoid accumulation occurred in the Al treatment under L. Carotenoids are not the only plastidial compounds biosynthesized in the MEP pathway, but their fate is a measure of the crosstalk of cytosolic IPP between the cytosol and the plastids (④ in Fig. 6). These results suggested that the Al-sensitive Ko was able to shut down the cytosolic IPP transporter system under Al treatment in DMG to supply more IPP for increased sterol biosynthesis.
Enhanced expression of HMGs and its relationship with sterol biosynthesis in the rice root tip under DMG
We detected HMG1 (data not shown), but this gene is probably a pseudogene and is not a functional gene in temperate japonica rice (Nelson et al., 1994).
The transcript levels of HMG genes, especially HMG2, were slightly higher in the whole roots of dark-grown seedlings than in those of light-grown seedlings of temperate japonica rice (Ha et al., 2001). The HMG transcription patterns and levels detected in rice root tips in this study were consistent with the results of Ha et al. (2001). The transcript levels of HMG genes under DMG were 1.41–2.76 times higher than those under L irrespective of the rice genotype or the presence of Al (Fig. 6). The transcript level of HMG2 was always higher than that of HMG3. Rice HMG2 is homologous to Arabidopsis HMG1, which is a sterol biosynthesis housekeeping gene (Ha et al., 2001). There is no direct evidence for the function of HMG3 in rice, but its encoded product is thought to be involved in both sterol biosynthesis and triterpenoids biosynthesis. Previous reports have shown that MVA in the medium is incorporated rapidly into sterols (Adler and Kasprzyk, 1975; Atallar et al., 1975). In the present experiment, therefore, we added 1 mM MVA to the medium under D to supply supplementary substrate for sterol biosynthesis (Fig. 7).
Under L, the total sterol content in the root tip was generally parallel to the transcript levels of HMGs (Figs 3 and 6A). In the Al treatment, the transcript levels of HMGs and the total sterol content were lower in the Al-sensitive Ko than in the Al-tolerant R132. The lower total sterol content in the Al-sensitive Ko than in the Al-tolerant R132 in the Al treatment (Fig. 3) is proposed to result from the greater inhibition of HMGR activity resulting from the higher concentration of root tip Al (Fig. 2) and the slightly lower transcript levels of HMGs (Fig. 6). Under DMG, however, the sterol content in the root tip was not parallel to the transcript levels of HMGs (Figs 3 and 6B). One of the reasons for this discrepancy may be the post-translational control of HMGR by sucrose non-fermenting (SNF)-1 related protein kinase-1 (SnRK1), which inactivates HMGR by phosphorylation of Ser-577 (Halford et al., 2003). Further studies are required to clarify the roles of such post-translational regulation. Another possible reason for this discrepancy may be the unknown lipid homeostatic mechanisms observed in several previous studies (Wang et al., 2014; Wagatsuma et al., 2015; Zhang et al., 2016). That is, under normal growth conditions, there was no increase in monogalactosyldiacylglycerol synthase (MGD) in the leaves of tobacco overexpressing MGD (Wang et al., 2014; Zhang et al., 2016), nor was there a decrease in sterol content in Arabidopsis plants with knocked-down expression of CYP51 (which encodes obtusifoliol 14α-demethylase, the enzyme converting obtusifoliol to 24-methylene lophenol, the precursor of Δ5-sterols) (Kushiro et al., 2001; Wagatsuma et al., 2015).
Although the expression levels of HMGs under DMG were almost double those under L (Fig. 6B), the total sterol content in all root tips was lower under DMG than under L (72.2–96.1% of that under L) (Fig. 3). The light conditions affect many aspects of metabolism, especially sugar-related metabolism. Therefore, the differences in sterol contents under the different illumination conditions reflected natural changes in metabolism. The greater decrease in the total sterol content in Ko under DMG (72.2% of that under L, Fig. 3) than in R132 under DMG (90.9% of that under L, Fig. 3) in the control may be due to a larger decrease in sucrose translocation from the shoot to the root tip under dark conditions.
In this study, we found that the Al-sensitive temperate japonica rice cultivars showed enhanced Al tolerance under dark conditions. Changes in phospholipids were not the determinant of increased Al permeation into the cytoplasm or differences in Al tolerance among rice cultivars under Al treatment. The results of the study suggest that the Al-sensitive temperate japonica rice cultivar had the following control mechanisms in the Al treatment under dark conditions: (i) inhibition of sterol-Δ22-desaturase, which decreased the production of stigmasterol; (ii) inhibition of cytosolic IPP transport to the plastid, which increased the supply of the precursor for sterol biosynthesis; and (iii) enhanced expression of HMGs, which increased sterols biosynthesis. These findings have identified new targets for the generation of new Al-tolerant plants.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Proportion of minor sterol species under different conditions.
Table S2. Net root elongation of 5-day-old rice cultivars under different illumination conditions.
Fig. S1. Sterol contents in 1-cm root tip of rice cv. Ka under different illumination conditions.
Acknowledgements
This study was supported by a Grant-in-Aid for Scientific Research (Nos. 18208008, 23380041) to TW from the Japan Society for the Promotion of Science (JSPS).
Glossary
Abbreviations:
- CYP51
cytochrome P450 encoding gene
- HMG
3-hydroxy-3- methylglutaryl CoA reductase (HMGR) encoding gene
- IPP
isopentenyl diphosphate
- MEP
methylerythritol-4-phosphate
- MVA
mevalonate
- OBT 14DM
obtusifoliol 14α-demethylase.
References
- Abe S, Sado A, Tanaka K, et al. . 2014. Carlactone is converted to carlactoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proceedings of the National Academy of Sciences, USA 111, 18084–18089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler G, Kasprzyk Z. 1975. The incorporation of mevalonate-[2-14C] into the sterol fractions of Calendula officinalis. Phytochemistry 14, 723–726. [Google Scholar]
- Atallar AM, Aexel RT, Ramsey RB, Nicholas HJ. 1975. Biosynthesis of sterols and triterpenes in Pelargonium hortorum. Phytochemistry 14, 1529–1533. [Google Scholar]
- Bak S, Kahn RA, Olsen CE, Halkier BA. 1997. Cloning and expression in Escherichia coli of the obtusifoliol 14α-demethylase of Sorghum bicolor (L.) Moench, a cytochrome P450 orthologous to the sterol 14α-demethylases (CYP51) from fungi and mammals. The Plant Journal 11, 191–201. [DOI] [PubMed] [Google Scholar]
- Benveniste P. 2002. Sterol metabolism. The Arabidopsis Book 1, e0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, et al. . 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry 55, 611–622. [DOI] [PubMed] [Google Scholar]
- Choe S, Tanaka A, Noguchi T, Fujioka S, Takatsuto S, Ross AS, Tax FE, Yoshida S, Feldmann KA. 2000. Lesions in the sterol Δ7 reductase gene of Arabidopsis cause dwarfism due to a block in brassinosteroid biosynthesis. The Plant Journal 21, 431–443. [DOI] [PubMed] [Google Scholar]
- Enjuto M, Balcells L, Campos N, Caelles C, Arró M, Boronat A. 1994. Arabidopsis thaliana contains two differentially expressed 3-hydroxy-3-methylglutaryl-CoA reductase genes, which encode microsomal forms of the enzyme. Proceedings of the National Academy of Sciences, USA 91, 927–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. 1958. Statistical methods for research workers, 13th edn. Edinburgh: Oliver & Boyd. [Google Scholar]
- Fujioka S, Li J, Choi YH, et al. . 1997. The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. The Plant Cell 9, 1951–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldwasser Y, Yoneyama K, Xie X, Yoneyama K. 2008. Production of strigolactones by Arabidopsis thaliana responsible for Orobanche aegyptiaca seed germination. Plant Growth Regulation 55, 21–28. [Google Scholar]
- Ha SH, Lee SW, Kim YM, Hwang YS. 2001. Molecular characterization of Hmg2 gene encoding a 3-hydroxy-methylglutaryl-CoA reductase in rice. Molecules and Cells 11, 295–302. [PubMed] [Google Scholar]
- Halford NG, Hey S, Jhurreea D, Laurie S, McKibbin RS, Paul M, Zhang Y. 2003. Metabolic signalling and carbon partitioning: role of Snf1-related (SnRK1) protein kinase. Journal of Experimental Botany 54, 467–475. [DOI] [PubMed] [Google Scholar]
- Hampel D, Mosandl A, Wüst M. 2005. Biosynthesis of mono- and sesquiterpenes in carrot roots and leaves (Daucus carota L.): metabolic cross talk of cytosolic mevalonate and plastidial methylerythritol phosphate pathways. Phytochemistry 66, 305–311. [DOI] [PubMed] [Google Scholar]
- Hartmann M-A. 1998. Plant sterols and the membrane environment. Trends in Plant Science 3, 170–174. [Google Scholar]
- Khan MS, Tawaraya K, Sekimoto H, et al. . 2009. Relative abundance of Δ5-sterols in plasma membrane lipids of root-tip cells correlates with aluminum tolerance of rice. Physiologia Plantarum 135, 73–83. [DOI] [PubMed] [Google Scholar]
- Kim HB, Schaller H, Goh CH, Kwon M, Choe S, An CS, Durst F, Feldmann KA, Feyereisen R. 2005. Arabidopsis cyp51 mutant shows postembryonic seedling lethality associated with lack of membrane integrity. Plant Physiology 138, 2033–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Kobayashi Y, Watanabe T, Shaff JE, Ohta H, Kochian LV, Wagatsuma T, Kinraide TB, Koyama H. 2013. Molecular and physiological analysis of Al³⁺ and H⁺ rhizotoxicities at moderately acidic conditions. Plant Physiology 163, 180–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlen W, Charnikhova T, Liu Q, Bours R, Domagalska MA, Beguerie S, Verstappen F, Leyser O, Bouwmeester H, Ruyter-Spira C. 2011. Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis. Plant Physiology 155, 974–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushiro M, Nakano T, Sato K, Yamagishi K, Asami T, Nakano A, Takatsuto S, Fujioka S, Ebizuka Y, Yoshida S. 2001. Obtusifoliol 14α-demethylase (CYP51) antisense Arabidopsis shows slow growth and long life. Biochemical and Biophysical Research Communications 285, 98–104. [DOI] [PubMed] [Google Scholar]
- Learned RM. 1996. Light suppresses 3-hydroxy-3-methylglutaryl coenzyme A reductase gene expression in Arabidopsis thaliana. Plant Physiology 110, 645–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, González VM, Castel S, Trelease RN, López-Iglesias C, Arró M, Boronat A, Campos N, Ferrer A, Fernàndez-Busquets X. 2005. Subcellular localization of Arabidopsis 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Plant Physiology 137, 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK, Schwender J, Disch A, Rohmer M. 1997. Biosynthesis of isoprenoids in higher plant chloroplasts proceeds via a mevalonate-independent pathway. FEBS Letters 400, 271–274. [DOI] [PubMed] [Google Scholar]
- Liu J, Piñeros MA, Kochian LV. 2014. The role of aluminum sensing and signaling in plant aluminum resistance. Journal of Integrative Plant Biology 56, 221–230. [DOI] [PubMed] [Google Scholar]
- López-Ráez JA, Charnikhova T, Gómez-Roldán V, et al. . 2008. Tomato strigolactones are derived from carotenoids and their biosynthesis is promoted by phosphate starvation. New Phytologist 178, 863–874. [DOI] [PubMed] [Google Scholar]
- Maejima E, Watanabe T, Osaki M, Wagatsuma T. 2014. Phosphorus deficiency enhances aluminum tolerance of rice (Oryza sativa) by changing the physicochemical characteristics of root plasma membranes and cell walls. Journal of Plant Physiology 171, 9–15. [DOI] [PubMed] [Google Scholar]
- Morikawa T, Mizutani M, Aoki N, et al. . 2006. Cytochrome P450 CYP710A encodes the sterol C-22 desaturase in Arabidopsis and tomato. The Plant Cell 18, 1008–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson AJ, Doerner PW, Zhu Q, Lamb CJ. 1994. Isolation of a monocot 3-hydroxy-3-methylglutaryl coenzyme A reductase gene that is elicitor-inducible. Plant Molecular Biology 25, 401–412. [DOI] [PubMed] [Google Scholar]
- Ohyama K, Suzuki M, Masuda K, Yoshida S, Muranaka T. 2007. Chemical phenotypes of the hmg1 and hmg2 mutants of Arabidopsis demonstrate the in-planta role of HMG-CoA reductase in triterpene biosynthesis. Chemical & Pharmaceutical Bulletin 55, 1518–1521. [DOI] [PubMed] [Google Scholar]
- Park H, Kreunen SS, Cuttriss AJ, DellaPenna D, Pogson BJ. 2002. Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. The Plant Cell 14, 321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Concepción M. 2006. Early steps in isoprenoid biosynthesis: multilevel regulation of the supply of common precursors in plant cells. Phytochemistry Reviews 5, 1–15. [Google Scholar]
- Rodríguez-Concepción M, Forés O, Martinez-García JF, González V, Phillips MA, Ferrer A, Boronat A. 2004. Distinct light-mediated pathways regulate the biosynthesis and exchange of isoprenoid precursors during Arabidopsis seedling development. The Plant Cell 16, 144–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller H. 2004. New aspects of sterol biosynthesis in growth and development of higher plants. Plant Physiology and Biochemistry 42, 465–476. [DOI] [PubMed] [Google Scholar]
- Schaller H, Grausem B, Benveniste P, Chye ML, Tan CT, Song YH, Chua NH. 1995. Expression of the Hevea brasiliensis (H.B.K.) Mull. Arg. 3-hydroxy-3-methylglutaryl-coenzyme A reductase 1 in tobacco results in sterol overproduction. Plant Physiology 109, 761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler I, Milon A, Nakatani Y, Ourisson G, Albrecht A-M, Benveniste P, Hartmann M-A. 1991. Differential effects of plant sterols on water permeability and on acyl chain ordering of soybean phosphatidylcholine bilayers. Proceedings of the National Academy of Sciences, USA 88, 6926–6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann M, Tse Sum Bui B, Wolff M, Miginiac-Maslow M, Rohmer M. 2006. Isoprenoid biosynthesis in plant chloroplasts via the MEP pathway: direct thylakoid/ferredoxin-dependent photoreduction of GcpE/IspG. FEBS Letters 580, 1547–1552. [DOI] [PubMed] [Google Scholar]
- Şükran D, Güneş T, Sivaci R. 1998. Spectrophotometric determination of chlorophyll-A, B and total carotenoid contents of some algae species using different solvents. Turkish Journal of Botany 22, 13–18. [Google Scholar]
- Suzuki M, Kamide Y, Nagata N, et al. . 2004. Loss of function of 3-hydroxy-3-methylglutaryl coenzyme A reductase 1 (HMG1) in Arabidopsis leads to dwarfing, early senescence and male sterility, and reduced sterol levels. The Plant Journal 37, 750–761. [DOI] [PubMed] [Google Scholar]
- Tanaka A, Tadano T, Yamamoto K, Kanamura N. 1987. Comparison of toxicity to plants among Al3+, AlSO4+, and Al-F complex ions. Soil Science and Plant Nutrition 33, 43–55. [Google Scholar]
- Wagatsuma T, Khan MS, Watanabe T, et al. . 2015. Higher sterol content regulated by CYP51 with concomitant lower phospholipid content in membranes is a common strategy for aluminium tolerance in several plant species. Journal of Experimental Botany 66, 907–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MH, Hans J, Strack D. 2002. Two distantly related genes encoding 1-deoxy-D-xylulose 5-phosphate synthases: differential regulation in shoots and apocarotenoid-accumulating mycorrhizal roots. The Plant Journal 31, 243–254. [DOI] [PubMed] [Google Scholar]
- Wang H, Nagegowda DA, Rawat R, Bouvier-Navé P, Guo D, Bach TJ, Chye ML. 2012. Overexpression of Brassica juncea wild-type and mutant HMG-CoA synthase 1 in Arabidopsis up-regulates genes in sterol biosynthesis and enhances sterol production and stress tolerance. Plant Biotechnology Journal 10, 31–42. [DOI] [PubMed] [Google Scholar]
- Wang S, Uddin MI, Tanaka K, et al. . 2014. Maintenance of chloroplast structure and function by overexpression of the rice monogalactosyldiacylglycerol synthase gene leads to enhanced salt tolerance in tobacco. Plant Physiology 165, 1144–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong RJ, McCormack DK, Russell DW. 1982. Plastid 3-hydroxy-3-methylglutaryl coenzyme A reductase has distinctive kinetic and regulatory features: properties of the enzyme and positive phytochrome control of activity in pea seedlings. Archives of Biochemistry and Biophysics 216, 631–638. [DOI] [PubMed] [Google Scholar]
- Xu Y, This D, Pausch RC, Vonhof WM, Coburn JR, Comstock JP, McCouch SR. 2009. Leaf-level water use efficiency determined by carbon isotope discrimination in rice seedlings: genetic variation associated with population structure and QTL mapping. Theoretical and Applied Genetics 118, 1065–1081. [DOI] [PubMed] [Google Scholar]
- Zhang M, Deng X, Yin L, Qi L, Wang X, Wang S, Li H. 2016. Regulation of galactolipid biosynthesis by overexpression of the rice MGD gene contributes to enhanced aluminum tolerance in tobacco. Frontiers in Plant Science 7, 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.