Vaccine effectiveness estimates for 2015–2016 seasonal influenza vaccine are reported from Canada. Findings suggest that agent-host and immuno-epidemiologic factors beyond antigenic match—including viral genomic variation, birth (immunological) cohort effects, repeat vaccination, and potential within-season waning immunity—may influence vaccine performance.
Keywords: Influenza, influenza vaccine, vaccine effectiveness, influenza A subtype, influenza B lineage, sequencing, hemagglutination inhibition, birth cohort effects, original antigenic sin, repeat vaccination
Abstract
Background
Vaccine effectiveness (VE) estimates for 2015–2016 seasonal influenza vaccine are reported from Canada’s Sentinel Practitioner Surveillance Network (SPSN). This season was characterized by a delayed 2009 pandemic influenza A(H1N1) virus (A[H1N1]pdm09) epidemic and concurrent influenza B(Victoria) virus activity. Potential influences on VE beyond antigenic match are explored, including viral genomic variation, birth cohort effects, prior vaccination, and epidemic period.
Methods
VE was estimated by a test-negative design comparing the adjusted odds ratio for influenza test positivity among vaccinated compared to unvaccinated participants. Vaccine-virus relatedness was assessed by gene sequencing and hemagglutination inhibition assay.
Results
Analyses included 596 influenza A(H1N1)pdm09 and 305 B(Victoria) cases and 926 test-negative controls. A(H1N1)pdm09 viruses were considered antigenically related to vaccine (unchanged since 2009), despite phylogenetic clustering within emerging clade 6B.1. The adjusted VE against A(H1N1)pdm09 was 43% (95% confidence interval [CI], 25%–57%). Compared to other age groups, VE against A(H1N1)pdm09 was lower for adults born during 1957–1976 (25%; 95% CI, −16%–51%). The VE against A(H1N1)pdm09 was also lower for participants consecutively vaccinated during both the current and prior seasons (41%; 95% CI, 18%–57%) than for those vaccinated during the current season only (75%; 95% CI, 45%–88%), and the VE among participants presenting in March–April 2016 (19%; 95% CI, −15%–44%) was lower than that among those presenting during January–February 2016 (62%; 95% CI, 44%–74%). The adjusted VE for B(Victoria) viruses was 54% (95% CI, 32%–68%), despite lineage-level mismatch to B(Yamagata) vaccine. The further variation in VE as observed for A(H1N1)pdm09 was not observed for B(Victoria).
Conclusions
Influenza VE findings may require consideration of other agent-host and immuno-epidemiologic influences on vaccine performance beyond antigenic match, including viral genomic variation, repeat vaccination, birth (immunological) cohort effects, and potential within-season waning of vaccine protection.
(See the editorial commentary by Belongia on pages 1477–80.)
The 2015–2016 influenza epidemic in Canada and the United States was characterized by a delayed peak in March (week 10)—one of the latest on recent record—due predominately to 2009 pandemic influenza A(H1N1) viruses (A[H1N1]pdm09) but with cocirculation of influenza B viruses mostly belonging to the B(Victoria) lineage [1, 2]. The last prior season during which A(H1N1)pdm09 dominated was 2013–2014, with an earlier epidemic peak in December–January (week 52–week 4) [3–5].
The test-negative design (TND) is an efficient method for annual influenza vaccine effectiveness (VE) estimation [6]. In Canada, the TND is superimposed on a community-based Sentinel Practitioner Surveillance Network (SPSN) through which VE against medically attended laboratory-confirmed influenza is directly correlated with the genetic and antigenic profile of contributing viruses [7]. The Canadian SPSN estimated that the VE against A(H1N1)pdm09 was approximately 90% for the AS03-adjuvanted monovalent A(H1N1)pdm09 vaccine [8] and approximately 60%–80% for nonadjuvanted seasonal vaccines used thereafter between 2010–2011 and 2013–2014 [4, 9–12]. Throughout that period, the same A/California/7/2009-like A(H1N1)pdm09 vaccine strain was used, and it was retained for the seventh consecutive year in 2015–2016 [13].
The antigenic-distance hypothesis predicts negative interference (ie, reduced VE) with serial vaccination when the prior season’s vaccine (v1) and current season’s vaccine (v2) antigens are similar (ie, v1≈v2) but v1 is antigenically distinct from the current season’s epidemic strain (ie, v1≠e) [14]. However, positive interference (ie, enhanced VE) may occur when v1 is antigenically closer to e (ie, v1≈e). Pronounced negative interference is anticipated when v1 and v2 are identical and distinct from e (ie, v1≡v2≠e) [14]. In Canada, a pattern of positive interference was suggested for A(H1N1)pdm09 during the initial postpandemic seasons, when v1≡v2≈e [9, 10, 15].
In 2012–2013, a new genetic variant of A(H1N1)pdm09, called clade 6B, emerged, distinguished by a K163Q substitution (H1 numbering) in antigenic site Sa atop the hemagglutinin (HA) surface protein [4]. Q163-bearing clade 6B viruses that dominated during the subsequent 2013–2014 epidemic were still considered antigenically similar to the K163-bearing A/California/07/2009 component of the vaccine, based on results of the conventional hemagglutination inhibition (HI) assay [4]. However, HI assays that use antisera collected from previously influenza-naive ferrets may not reflect the diverse exposure histories and immunological landscape of humans, especially adults, that also potentially influence vaccine performance [16–22].
Past exposures may create immunological cohort effects such as those invoked by Linderman et al to explain differential age-specific A(H1N1)pdm09 susceptibility [18]. According to the Linderman et al hypothesis, detailed in our Supplement 1, original childhood priming exposures may have induced preferential K163-specific memory responses among middle-aged adults in 2013–2014, especially for those born during 1940–1984 (notably those born during 1965–1979, among whom K163-specific responses were reported to be highest) [18]. Through the phenomenon of original antigenic sin [23], hierarchical antibody refocusing toward early childhood priming specificity is reinforced with each infection or vaccination involving viruses sharing the immunodominant K163 epitope. Linderman et al propose that younger adults (ie, those born after 1985) were spared such targeted specificity owing to their childhood priming to A(H1N1) viruses that had acquired glycosylation sites masking the K163 epitope [18]. Impaired immunity, including lower VE in 2013–2014 against clade 6B viruses bearing Q163, might be anticipated for those with K163-specific prime-boost experiences. Despite these potential effects, moderate VE against A(H1N1)pdm09 was reported among adults in both Canada (approximately 75%) and the United States (approximately 55%–65%) in 2013–2014 [4, 24, 25]. However, marginally lower VE against A(H1N1)pdm09 was observed that season among v2+v1 recipients, compared with v2 only recipients, in both Canada (approximately 70% vs approximately 80%) and the United States (approximately 50% vs approximately 60%) [4, 15, 24].
Beginning in October 2015, a further subclade of A(H1N1)pdm09, called 6B.1, emerged, defined by additional HA mutations, including S162N, adjacent to Q163, conferring a potential gain of glycosylation [13]. The 2015–2016 epidemic thus provided opportunity to explore VE for the unchanged A(H1N1)pdm09 vaccine with further evolved epidemic viruses. Here we examine the VE of 2015–2016 seasonal influenza vaccine, incorporating agent-host considerations such as viral genomic evolution, repeat vaccination, immunological cohort effects, and potential waning of immunity across the season’s delayed epidemic.
METHODS
Canadian SPSN
Nasal/nasopharyngeal specimens were collected from patients presenting to sentinel sites in provinces participating in the Canadian SPSN (Alberta, British Columbia, Ontario, and Quebec) within 7 days of influenza-like illness (ILI) onset, defined as fever, cough, and ≥1 of the following: sore throat, myalgia, arthralgia, or prostration. Fever was not required for elderly adults ≥65 years old. Epidemiological information was based on self-report documented by sentinel practitioners using a standard questionnaire at the time of specimen collection. All participants or their parent/guardian provided verbal consent. Ethics approval was obtained in each participating province.
Influenza Diagnosis and Characterization
Influenza was diagnosed on the basis of reverse-transcription polymerase chain reaction assay. Sequencing of HA in original specimens obtained from patients included in the VE analysis was attempted to establish clade designation and identify whether amino acids at antigenic sites differed from those in the egg-adapted high-growth reassortant vaccine strain. Antigenic characterization of virus isolates was performed using an HI assay [16]. Details about laboratory methods are provided in Supplement 2. Relevant GenBank accession numbers are as follows: MF195302–MF196091; MF348042–MF348066, except MF348058, MF348062, and MF348065; KU821061–KU821078; and KU821080–KU821100.
Influenza Vaccination and VE Estimation
For the 2015–2016 trivalent influenza vaccine, the World Health Organization recommended the same A(H1N1)pdm09 antigen as used since 2009 but a clade-level change from 2014–2015 for the A(H3N2) and B(Yamagata) components. Quadrivalent vaccine included the same B(Victoria) strain as recommended since 2009–2010 (Supplement 3).
Owing to the delayed 2015–2016 epidemic (Supplement 4), VE analyses were limited to specimens collected from 3 January to 30 April 2016 (weeks 1–17). The VE was derived by the TND: cases tested positive for influenza virus, and controls tested negative. Patients who self-reported receipt of at least 1 2015–2016 influenza vaccine dose ≥2 weeks before ILI onset were considered vaccinated. Logistic regression models were used to derive odds ratios (ORs) for medically attended, laboratory-confirmed influenza in vaccinated participants compared to unvaccinated participants and adjusting for recognized potential confounders. The VE was derived as [1 − OR] × 100%. Serial/repeat vaccination effects were assessed among participants ≥9 years old through indicator-variable analyses based on vaccination history during the current and up to 2 prior seasons. The effect of prior 2009 monovalent AS03-adjuvanted A(H1N1)pdm09 vaccination was also explored.
Stratified analyses were primarily conducted using an interaction term for vaccination status and the stratification variable (eg, age group, month, and epidemic period). Age regrouping was undertaken for the A(H1N1)pdm09 VE analysis to explore potential cohort effects based on priming epoch, as defined in Supplement 1; this was modeled using the interaction term age group*vaccination status and was also assessed by subset analysis. To further explore these effects, VE for each birth year (based on age at time of specimen collection) was also modeled for A(H1N1)pdm09, with age smoothed as a restricted cubic spline function, using 5 knots based on percentiles, and an interaction term for single year of age and vaccination status, as described elsewhere [26]. Owing to sparse data for very old participants, additional age-based evaluations were limited to participants 1–76 years old in 2016 (ie, birth cohorts 1940–2015). As explained in Supplement 1, a maximum delay of 9 years from birth to first influenza A(H1N1) exposure was incorporated in delineating and interpreting cohort effects, recognizing, however, that immunological priming likely occurs before 6 years of age [27–29].
Details related to influenza vaccines and VE estimation are provided in Supplement 3.
RESULTS
Virologic Profile
Overall
Overall, 2008 specimens contributed to VE analyses, of which about half tested positive for influenza virus: approximately 60% were positive for influenza A virus, and approximately 40% were positive for influenza B virus (Table 1). Among the detected influenza A and B viruses with known subtype and lineage information, >90% were A(H1N1)pdm09 and >75% were B(Victoria), respectively. Genetic characterization was successful for approximately 80%–90% of viruses contributing to VE analyses. HI characterization was available for about two thirds of A(H1N1)pdm09 and 80% of influenza B viruses (Table 1).
Table 1.
Virologic Profile of Influenza Viruses Collected From Participants in the 2015–2016 Influenza Vaccine Effectiveness Evaluation of the Canadian Sentinel Practitioner Surveillance Network
| Variable | Specimens, No. (%) |
|---|---|
| Influenza diagnosis | 2008 (100) |
| Influenza virus negative | 926 (46) |
| Influenza virus positive | 1082 (54) |
| Influenza virus type,a subtype/lineage | |
| Influenza A | 664 (61) |
| A(H1N1)pdm09 | 596 (90) |
| A(H3N2) | 55 (8) |
| A (subtype unknown) | 13 (2) |
| Influenza B | 423 (39) |
| B(Victoria) | 305 (72) |
| B(Yamagata) | 85 (20) |
| B (lineage unknown) | 33 (8) |
| Genetic characterization | |
| A(H1N1)pdm09b | 467 (78) |
| Clade 6B | 10 (2) |
| Clade 6B.1 | 452 (97) |
| Clade 6B.2 | 5 (1) |
| A(H3N2)c | 43 (78) |
| Clade 3C.2a | 38 (88) |
| Clade 3C.3a | 5 (12) |
| B(Victoria)d | 277 (91) |
| Clade 1A | 277 (100) |
| B(Yamagata)e | 73 (86) |
| Clade 3 | 73 (100) |
| Antigenic characterizationf | |
| A/California/07/2009(H1N1)pdm09-like (turkey; allantoic); trivalent vaccine strain | 383g,h (64) |
| <4-fold | 364 (95) |
| ≥4-fold | 19 (5) |
| ≥8-fold | 1i (0) |
| A/Switzerland/9715293/2013(H3N2)-like (guinea pig; allantoic or cell); clade 3C.3a trivalent vaccine strainj | 11g (20) |
| <4-fold | 9 (82) |
| ≥4-fold | 2 (18) |
| ≥8-fold | 1 (9) |
| B/Brisbane/60/2008(Victoria)-like (turkey; allantoic); clade 1A quadrivalent vaccine strain | 255g (84) |
| <4-fold | 30 (12) |
| ≥4-fold | 225 (88) |
| ≥8-fold | 149 (58) |
| B/Brisbane/60/2008(Victoria)-like (turkey; cell)k | 149 (49) |
| <4-fold | 137 (92) |
| ≥4-fold | 12 (8) |
| ≥8-fold | 1 (1) |
| B/Phuket/3073/2013(Yamagata)-like (turkey; cell); clade 3 trivalent vaccine strain | 69 (81) |
| <4-fold | 69 (100) |
aFive participants were coinfected with influenza A(H1N1)pdm09 and influenza B. The sum of the subtotals of individuals infected with influenza A and B is greater than the total number of influenza virus–positive individuals.
bSequencing details are provided in Supplement 5 and Supplement 6.
cSequencing details are provided in Supplement 7.
dSequencing details are provided in Supplement 8.
eSequencing details are provided in Supplement 9.
fDefined as the fold reduction in HI titer of study viruses relative to vaccine reference virus (erythrocytes; allantoic or cell culture isolate). Antigenic distinction was defined by a ≥8-fold reduction in HI titer.
gAdditional viruses had insufficient titers for HI characterization, including 7 A(H1N1)pdm09 isolates (2%), 30 A(H3N2) isolates (73%), and 1 B(Victoria) isolate.
hIncludes 325 clade 6B.1, 8 clade 6B, and 5 clade 6B.2 viruses based on sequencing of original patient specimens. Sequencing findings after culture isolation were available for 27 of 325 clade 6B.1 viruses (8%) that were characterized by an HI assay and revealed that all retained the N162 glycosylation site.
iA clade 6B.1 virus.
jCell-cultured reference virus used until 29 April 2016 (in 4 of 11 cases), after which allantoic reference virus was used (in 7 of 11), including the single virus found to be antigenically distinct. Antigenic characterization of influenza A(H3N2) clade 3C.2a viruses was compromised by previously described variability in the agglutination of erythrocytes.
kOnly B(Victoria) viruses showing a ≥8-fold reduction to an allantoic B/Brisbane/60/2008(Victoria)-like virus (n = 149) were antigenically characterized against a cell-culture B/Brisbane/60/2008(Victoria)-like virus.
Abbreviations: A(H1N1)pdm09, 2009 pandemic influenza A(H1N1) virus; HI, hemagglutination inhibition.
A(H1N1)pdm09
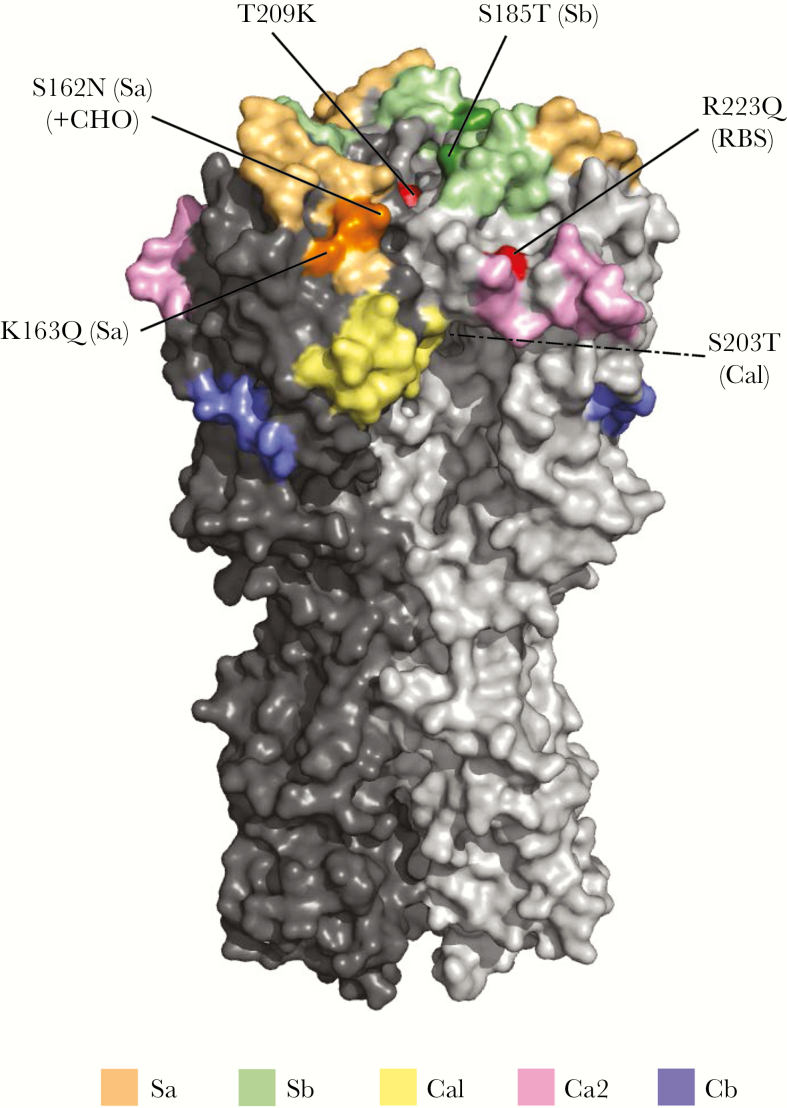
Virtually all (452 of 467 [97%]) sequenced A(H1N1)pdm09 viruses belonged to the emergent clade 6B.1, of which almost all bore 4 antigenic site substitutions relative to vaccine (Figure 1 and Supplement 5). This includes S203T[Ca1] acquired early (since 2009) among circulating A(H1N1)pdm09 viruses and S185T[Sb], located near the RBS—both are common to all clade 6 (and 7) A(H1N1)pdm09 viruses dominant in Canada since at least 2011 (Supplement 6) [10, 11]. It also includes K163Q[Sa], which has been common to all clade 6B viruses since 2012–2013, and the adjacent S162N[Sa] substitution, which confers a potential gain of N-linked glycosylation newly acquired among clade 6B.1 viruses since 2015–2016 [4, 12, 13]. All sentinel A(H1N1)pdm09 viruses but 1 were considered antigenically similar to the egg-adapted vaccine strain.
Figure 1.
Crystal structure (trimer) of hemagglutinin (HA) of dominant clade 6B.1 2009 pandemic influenza A(H1N1) viruses (A[H1N1]pdm09) detected by the Canadian Sentinel Practitioner Surveillance Network, during the 2015–2016 season, with key substitutions relative to the egg-adapted X-179A high-growth reassortant (HGR) vaccine strain. Three-dimensional structural model shows key substitutions in the HA1 of dominant sentinel A(H1N1)pdm09 clade 6B.1 viruses as compared to the 2015–2016 egg-adapted A/California/07/2009-like HGR vaccine strain (namely X-179A most used in Canada). The trimeric HA protein of A(H1N1)pdm09 was constructed using the crystal structure of the A/Washington/05/2011 HA (Protein Data Bank accession number 4LXV) with substitutions relevant to clade 6B.1 viruses added in the PyMOL Molecular Graphics System (version 1.6; Schrödinger), using the Mutagenesis Wizard. Antigenic sites Sa, Sb, Ca1, Ca2, and Cb are shown in pastel colors. Substitutions are labelled and the involved antigenic site is indicated in parentheses and darker shading. Nonantigenic site substitutions arising from egg passage and/or in the HGR (both X-179A and X-181) are shown in red; a third such substitution (N129D) in relation to X-181 alone is not displayed. The dotted line from S203T (Ca1) indicates that the highlighted mutation is not visible from the point of view shown in the crystal structure. Note that amino acid numbering is based on the H1 scheme and begins with the signal peptide removed. RBS, receptor-binding site; +CHO, potential gain of glycosylation site.
B(Victoria)
Predominant B(Victoria) viruses were lineage mismatched to the B(Yamagata) trivalent influenza vaccine strain. All sequenced B(Victoria) viruses belonged to clade 1A (Table 1). The majority bore 3–4 antigenic-site substitutions relative to the egg-adapted clade 1A B(Victoria) quadrivalent vaccine component, which notably had lost the potential N197 glycosylation site present in the cell-passaged consensus sequence and all sentinel viruses (Supplement 8). The majority of B(Victoria) sentinel viruses were considered antigenically distinct from the egg-adapted B(Victoria) quadrivalent vaccine strain, whereas all but 1 of these same viruses were considered antigenically similar to the cell-passaged B(Victoria) referent.
Participant Profile
Nonelderly adults 20–64 years old contributed most to the VE analysis (1284 of 2008 participants [64%]; Table 2). The profile among test-negative controls was similar to that during 2013–2014 and included the same median age (37 years) and proportion vaccinated (33%) [4]. Cases were younger in 2015–2016 versus 2013–2014 (median age, 33 vs 39 years; P < .01)—a difference driven by influenza B virus (23 vs 43.5 years; P < .01) not A(H1N1)pdm09 (38 years during both seasons; Supplement 10) [4]. Most participants ≥9 years old vaccinated in 2015–2016 were previously vaccinated in 2014–2015 (387 of 449 [86%]) and in both 2014–2015 and 2013–2014 (353 of 432 [82%]).
Table 2.
Epidemiological Profile of Participants Included in the 2015–2016 Influenza Vaccine Effectiveness Evaluation, Canadian Sentinel Practitioner Surveillance Network
| Characteristic | All Participants, No. (column %) | Vaccinated Participants, No. (row %) | ||||||
|---|---|---|---|---|---|---|---|---|
| Total | Cases | Controls | P a | Total | Cases | Controls | P a | |
| Overall | 2008/2008 (100) | 1082/2008 (54) | 926/2008 (46) | 510/2008 (25) | 204/1082 (19) | 306/926 (33) | ||
| Age, y | <.01 | <.01 | ||||||
| 1–8 | 281/2008 (14) | 169/1082 (16) | 112/926 (12) | 39/281 (14) | 16/169 (9) | 23/112 (21) | ||
| 9–19 | 260/2008 (13) | 145/1082 (13) | 115/926 (12) | 40/260 (15) | 16/145 (11) | 24/115 (21) | ||
| 20–49 | 902/2008 (45) | 502/1082 (46) | 400/926 (43) | 174/902 (19) | 73/502 (15) | 101/400 (25) | ||
| 50–64 | 382/2008 (19) | 194/1082 (18) | 188/926 (20) | 138/382 (36) | 57/194 (29) | 81/188 (43) | ||
| ≥65 | 183/2008 (9) | 72/1082 (7) | 111/926 (12) | 119/183 (65) | 42/72 (58) | 77/111 (69) | ||
| Median (range)b | 34.5 (1–96) | 33 (1–96) | 37 (1–92) | <.01 | 50 (1–96) | 48.5 (1–96) | 50 (1–92) | <.01 |
| Sex | <.01 | .09 | ||||||
| Female | 1239/2008 (62) | 637/1082 (59) | 602/926 (65) | 331/1239 (27) | 123/637 (19) | 208/602 (35) | ||
| Male | 769/2008 (38) | 445/1082 (41) | 324/926 (35) | 179/769 (23) | 81/445 (18) | 98/324 (30) | ||
| Comorbidityc | <.01 | <.01 | ||||||
| No | 1650/2008 (82) | 927/1082 (86) | 723/926 (78) | 328/1650 (20) | 137/927 (15) | 191/723 (26) | ||
| Yes | 358/2008 (18) | 155/1082 (14) | 203/926 (22) | 182/358 (51) | 67/155 (43) | 115/203 (57) | ||
| Province | <.01 | <.01 | ||||||
| Alberta | 466/2008 (23) | 230/1082 (21) | 236/926 (25) | 139/466 (30) | 49/230 (21) | 90/236 (38) | ||
| British Columbia | 491/2008 (24) | 253/1082 (23) | 238/926 (26) | 126/491 (26) | 47/253 (19) | 79/238 (33) | ||
| Ontario | 690/2008 (34) | 370/1082 (34) | 320/926 (35) | 181/690 (26) | 72/370 (19) | 109/320 (34) | ||
| Quebec | 361/2008 (18) | 229/1082 (21) | 132/926 (14) | 64/361 (18) | 36/229 (16) | 28/132 (21) | ||
| Collection interval, d | <.01 | .34 | ||||||
| ≤4 | 1520/2008 (76) | 863/1082 (80) | 657/926 (71) | 378/1520 (25) | 165/863 (19) | 213/657 (32) | ||
| 5–7 | 488/2008 (24) | 219/1082 (20) | 269/926 (29) | 132/488 (27) | 39/219 (18) | 93/269 (35) | ||
| Median (range)b | 3 (0–7) | 3 (0–7) | 3 (0–7) | <.01 | 3 (0–7) | 3 (0–7) | 3 (0–7) | .03 |
| Specimen collection mod | <.01 | .36 | ||||||
| January | 382/2008 (19) | 147/1082 (14) | 235/926 (25) | 91/382 (24) | 22/147 (15) | 69/235 (29) | ||
| February | 685/2008 (34) | 396/1082 (37) | 289/926 (31) | 164/685 (24) | 61/396 (15) | 103/289 (36) | ||
| March | 705/2008 (35) | 430/1082 (40) | 275/926 (30) | 187/705 (27) | 100/430 (23) | 87/275 (32) | ||
| April | 236/2008 (12) | 109/1082 (10) | 127/926 (14) | 68/236 (29) | 21/109 (19) | 47/127 (37) | ||
| 2015–2016 seasonal influenza vaccination status | ||||||||
| Anye | 533/2031 (26) | 211/1089 (19) | 322/942 (34) | <.01 | … | … | … | |
| ≥2 wk before ILI onset | 510/2008 (25) | 204/1082 (19) | 306/926 (33) | <.01 | … | … | … | |
| LAIVf | 22/302 (7) | 8/122 (7) | 14/180 (8) | .69 | … | … | … | |
| QIVg | 62/287 (22) | 20/107 (19) | 42/180 (23) | .36 | … | … | … | |
| Adjuvantedh | 29/61 (48) | 10/20 (50) | 19/41 (46) | .79 | … | … | … | |
| Influenza vaccination historyi | ||||||||
| 2014–2015 seasonal | 561/1638 (34) | 248/863 (29) | 313/775 (40) | <.01 | 387/561 (69) | 159/248 (64) | 228/313 (73) | <.01 |
| 2013–2014 seasonal | 552/1589 (35) | 240/834 (29) | 312/755 (41) | <.01 | 372/552 (67) | 150/240 (63) | 222/312 (71) | <.01 |
| 2009 monovalent | 580/1432 (41) | 290/768 (38) | 290/664 (44) | .02 | 287/580 (49) | 116/290 (40) | 171/290 (59) | <.01 |
aDifferences between cases and controls and between vaccinated and unvaccinated participants were compared using χ2 or Wilcoxon rank sum tests.
bAmong all participants, data are for 2008 total participants, 1082 cases, and 926 controls. Among vaccinated participants, data are for 510 total participants, 204 cases, and 306 controls.
cIncludes chronic comorbidities that place individuals at higher risk of serious complications from influenza, as defined by the NACI, including heart, pulmonary (including asthma), renal, metabolic (such as diabetes), blood, cancer, or immunocompromising conditions; conditions that compromise management of respiratory secretions and increase risk of aspiration; or morbid obesity (defined as a body mass index of ≥40) [27].
dMissing collection dates were imputed as the laboratory accession date minus 2 days.
eParticipants who received seasonal 2015–2016 seasonal influenza vaccine <2 weeks before ILI onset or for whom vaccination timing was unknown were excluded from primary analysis. They were included for assessing any vaccination, regardless of timing, for comparison with other sources of vaccination coverage.
fData are for participants 2–59 years old who received 2015–2016 influenza vaccine ≥2 weeks before ILI onset and had known information for the type of vaccine received. Among participants 2–17 years old for whom LAIV was recommended by NACI [27], 38% (21 of 55, including 7 cases) with known information had received LAIV. Among participants 2–5 years old for whom LAIV was preferentially recommended by the NACI [27], 32% (7 of 22, including 4 cases) with known information had received LAIV.
gData are for participants who received 2015–2016 influenza vaccine ≥2 weeks before ILI onset and had known information for TIV versus QIV. QIV includes both inactivated influenza vaccine and LAIV products.
hData are for participants ≥65 years old who received 2015–2016 influenza vaccine ≥2 weeks before ILI onset and had known information for MF59-adjuvanted vaccine receipt.
iData are for participants ≥9 years old who had known information for prior season(s)' influenza vaccine receipt.
Abbreviations: ILI, influenza-like illness; LAIV, live attenuated influenza vaccine; NACI, Canadian National Advisory Committee on Immunization; QIV, quadrivalent influenza vaccine; TIV, trivalent influenza vaccine.
Participant details are provided in Table 2 and Supplement 10.
VE Estimates
Overall
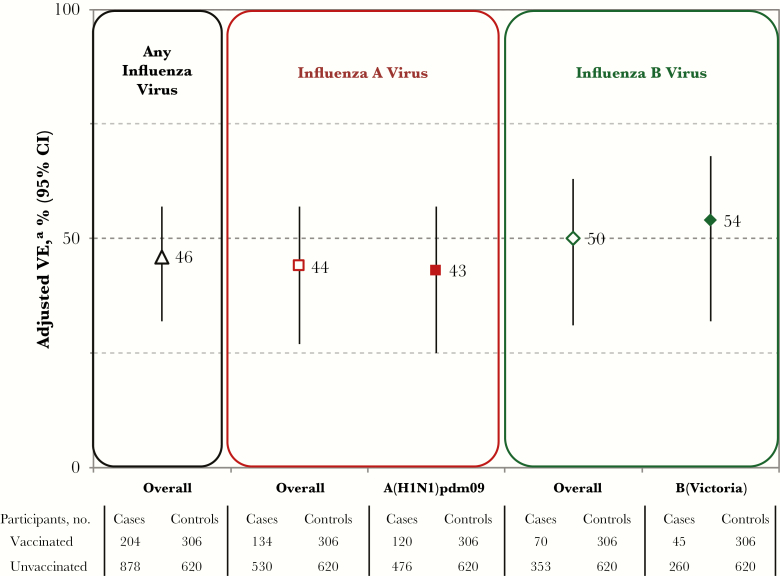
Adjusted VE estimates are shown in Figure 2, with model and participant details in Supplement 11. The VE was 43% (95% confidence interval [CI], 25%–57%) against A(H1N1)pdm09 and 54% (95% CI, 32%–68%) against B(Victoria). Sample size did not support adjustment of the VE by vaccine type in children; unadjusted estimates for live attenuated influenza vaccine (LAIV) were lower against A(H1N1)pdm09 but higher against influenza B as compared to inactivated influenza vaccine, with widely overlapping 95% CIs (Supplement 12).
Figure 2.
Adjusted vaccine effectiveness (VE) estimates for 2015–2016 seasonal influenza vaccine, by influenza type and subtype or lineage (primary analysis), Canadian Sentinel Practitioner Surveillance Network. Abbreviation: CI, confidence interval. aAdjusted for age group, sex, comorbidity, province, collection interval, and calendar time (week of specimen collection as modeled using cubic B spline functions with 3 equally spaced knots). Details are specified in Supplement 3 and Supplement 11.
Repeat Vaccination Effects
A(H1N1)pdm09
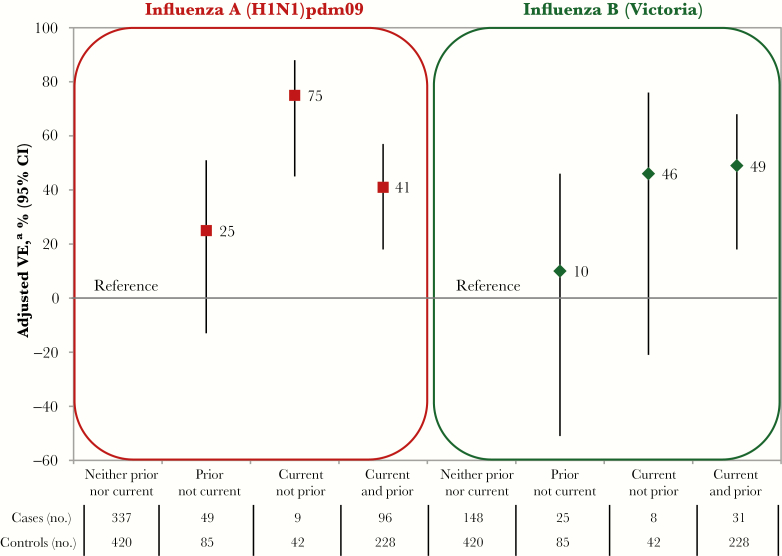
With homologous vaccines (v1≡v2) and viruses considered antigenically similar (v1≈e) but genetically variant, statistically significant negative interference by v1 on v2 was observed for A(H1N1)pdm09 (P = .01 for the interaction term). Adjusted VE was lower for v2+v1 than for v2 alone (41% [95% CI, 18%–57%] vs 75% [95% CI, 45%–88%]), with a similar pattern observed after incorporating participants who received vaccine during an additional prior season (Figure 3 and Supplement 13 and Supplement 14). Repeat vaccine recipients over two seasons had a significantly, 2-fold higher risk of A(H1N1)pdm09 illness than participants vaccinated during the current season only (adjusted OR, 2.33; 95% CI, 1.04–5.21; Supplement 13). There was no residual protection from 2009 monovalent A(H1N1)pdm09 vaccine alone (−4%; 95% CI, −58%–31%; data not shown).
Figure 3.
Adjusted vaccine effectiveness (VE) for 2015–2016 (current) and/or 2014–2015 (prior) seasonal influenza vaccines against 2009 pandemic influenza A(H1N1) virus (A[H1N1]pdm09) and influenza B(Victoria), during the 2015–2016 season, Canadian Sentinel Practitioner Surveillance Network. The exclusion criteria were the same as in the primary analysis but limited to participants ≥9 years old and with complete information for 2014–2015 and 2015–2016 vaccine receipt. CI, confidence interval. aAdjusted for age group, sex, comorbidity, province, collection interval, and calendar time (week of specimen collection was modeled using cubic B spline functions with 3 equally spaced knots). Details are specified in Supplement 3 and Supplement 13.
B(Victoria)
With B(Yamagata) v1 and v2 strains that were antigenically related (v1≈v2) but phylogenetically distinct (ie, clade 2 vs clade 3, respectively; Supplement 3) and lineage-level mismatched to the dominant B(Victoria) epidemic strain (ie, v1≠e and v2≠e), there was no significant interaction between v1 and v2 (P = .90 for the interaction term). Adjusted VE against B(Victoria) was comparable for recipients of v2+v1 and v2 alone (49% [95% CI, 18%–68%]) vs 46% [95% CI, −21%–76%]), with a similar pattern observed after incorporating individuals who received vaccine during an additional prior season (Figure 3 and Supplement 13 and Supplement 14).
Analysis Stratified by Age
VE against A(H1N1)pdm09 was higher for children 1–19 years old (67%; 95% CI, 31%–84%) as compared to nonelderly adults 20–64 years old (35%; 95% CI, 10%–52%) or elderly adults (43%; 95% CI, −26%–74%). Such a difference between children and nonelderly adults was not evident for B(Victoria); there were too few elderly B(Victoria) cases for reliable interpretation (Supplement 11).
A(H1N1)pdm09: Exploratory Cohort Analyses
For adults 32–76 years old (birth cohorts 1940–1984) and 37–51 years old (birth cohorts 1965–1979), for whom Linderman et al [18] predicted pronounced K163-priming specificity, VE was 41% (95% CI, 19%–57%) and 43% (95% CI, 4%–66%), respectively (Supplement 15).
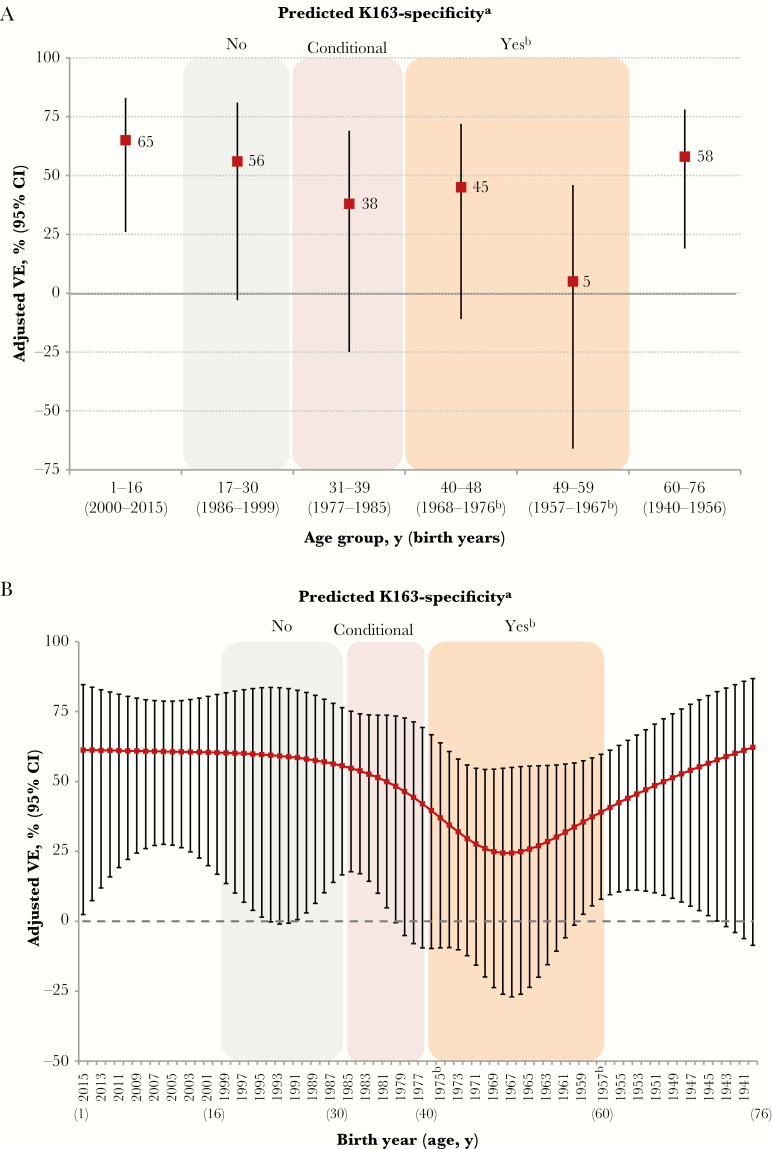
As shown in Figure 4A (analysis details are in Supplement 1 and Supplement 15), VE estimates with age regrouping were generally consistent with potential K163-priming effects. However, VE estimates were notably lower (25%; 95% CI, −16%–51%) among participants 40–59 years old (birth cohorts 1957–1976), a cohort of individuals for whom heterosubtypic A(H2N2) and/or A(H3N2) priming would have likely preceded A(H1N1) exposure (Supplement 1 and Supplement 15). The VE was lowest (5%; 95% CI, −66%–46%) for participants 49–59 years old (birth cohorts 1957–1967), among whom heterosubtypic priming to A(H2N2) in particular (or both A[H2N2] and A[H3N2]) was likely before reemergence of A(H1N1) viruses (K163 bearing) in 1977. Similar age-related patterns were evident in stratified analyses based on subsets of the data (Supplement 15) and in an analysis where age in years was smoothed as a restricted cubic spline with 5 knots based on percentiles (Figure 4B). The 5-knot model had slightly better fit (based on Akaike Information Criterion [AIC] values) than models with 3 or 7 knots; however, in the 7-knot model, VE remained lowest among those who were likely heterosubtypically primed with A(H2N2), while additional age-related variability consistent with the report by Linderman et al [18] was also suggested (eg, improved VE with a birth year after 1985; Supplement 16).
Figure 4.
Adjusted vaccine effectiveness (VE) for 2015–2016 seasonal influenza vaccine against 2009 pandemic influenza A(H1N1) virus (A[H1N1]pdm09), stratified by age group (A) and birth year (B), among Canadian Sentinel Practitioner Surveillance Network participants 1–76 years old. A, Adjusted VE stratified by age groups adapted to reflect potential variation in K163-priming specificity as per Supplement 1, with stratification by age group based on an interaction model as detailed in Supplement 3 and Supplement 15. B, Adjusted VE stratified by single year of age (or derived year of birth), with age smoothed as a restricted cubic-spline function with 5 knots based on percentiles as detailed in Supplement 3 and Supplement 16. Color shading was performed as per Supplement 1 and corresponds to birth year, as follows: orange, pronounced K163 specificity predicted; pink, K163 specificity anticipated but conditional upon age of first A(H1N1) exposure; and gray, K163 specificity not predicted. Birth years without color shading are for children or older adults for whom priming in relation to position 163 varies for additional reasons specified in Supplement 1. CI, confidence interval. aAdapted from the work by Linderman et al [18]. bPeriod of no A(H1N1) circulation (ie, 1957–1976) for which heterosubtypic priming with A(H2N2) (ie, 1957–1967) and/or A(H3N2) (ie, 1968–1976) is likely before reemergence of A(H1N1) viruses (K163 bearing) in 1977.
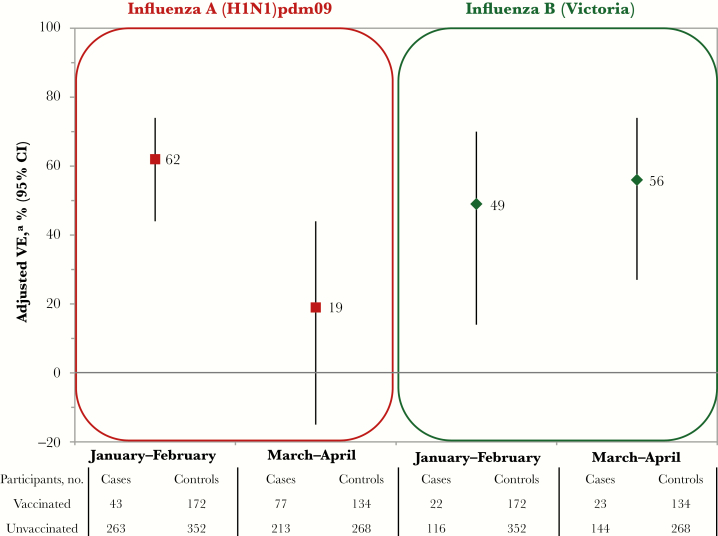
Analysis by Epidemic Period
There was significant decline in the adjusted VE against A(H1N1)pdm09 between January–February (62%; 95% CI, 44%–74%) and March–April (19%; 95% CI, −15%–44%; P < .01 for the interaction term; Figure 5 and Supplement 11). Adults 49–59 years old contributed slightly more A(H1N1)pdm09 cases in March–April (21%), compared with January–February (17%; Supplement 10), but the interaction between age group and epidemic period (P = .88) or month (P = .09) was not significant in VE models.
Figure 5.
Adjusted vaccine effectiveness (VE) for 2015–2016 seasonal influenza vaccine against 2009 pandemic influenza A(H1N1) virus (A[H1N1]pdm09) and B(Victoria) virus, by epidemic period, Canadian Sentinel Practitioner Surveillance Network. Stratified estimates were derived using an interaction model based on the entire analytic sample for A(H1N1)pdm09 and B(Victoria); sample sizes for epidemic subgroups are displayed for interest. Abbreviation: CI, confidence interval. aAdjusted for age group, sex, comorbidity, province, collection interval, epidemic period (based on month of specimen collection), and vaccination status*epidemic period interaction. Details are provided in Supplement 11.
There was no decline in VE between January–February and March–April for influenza B(Victoria) (49% [95% CI, 14%–70%] vs 56% [95% CI, 27%–74%]; Figure 5 and Supplement 11).
DISCUSSION
In this analysis for the delayed 2015–2016 A(H1N1)pdm09 and concurrent influenza B(Victoria) epidemics, we identified a VE of <50% against A(H1N1)pdm09, despite antigenic match, and a VE of >50% for B(Victoria), despite lineage-level mismatch, to the trivalent vaccine. To examine these discordant findings, we used the integrated virological and epidemiological databases of the Canadian SPSN, incorporating other agent-host considerations to understand vaccine performance, such as viral genomic variation, birth (or immunological) cohort effects, prior vaccination history and potential within-season waning of vaccine protection.
The VE of 43% (95% CI, 25%–57%) we report is the lowest measured by the Canadian SPSN against A(H1N1)pdm09 since its emergence in 2009 and was substantially lower than the VE of 71% (95% CI, 58%–80%) measured during the last A(H1N1)pdm09 epidemic, during 2013–2014 (Supplement 17) [4], and lower also than the average seasonal A(H1N1)pdm09 VE reported in recent meta-analysis (61%; 95% CI, 57%–65%) [6]. Decline in A(H1N1)pdm09 VE from 2013–2014 to 2015–2016 was also identified by networks in the United States (54% [95% CI, 46%–61%] vs 45% [95% CI, 34%–53%]) and Europe (48% [95% CI, 16%–67%] vs 33% [95% CI, 4%–52%]) but was less substantial as compared to Canada (with absolute decreases of 9% in the United States and 15% in Europe, compared with 28% in Canada) [24, 30–32]. In Canadian children 2–17 years old, the odds of vaccine failure against A(H1N1)pdm09 in 2015–2016 was greater (nonsignificantly) for LAIV recipients than for inactivated influenza vaccine recipients (unadjusted OR, 3.75; 95% CI, .65–21.74; Supplement 12), as also reported (significantly) by the United States (adjusted OR, 3.67; 95% CI, 1.86–7.31) [33]. The 2015–2016 unadjusted estimate of LAIV effectiveness as compared to no vaccine receipt (51%; 95% CI, −37%–83%; Supplement 12) in Canada was not as low as the adjusted estimate reported by the US FluVE network (−19%; 95% CI, −113%–33%) [30] but was more similar to adjusted estimates reported by the manufacturer (50%; 95% CI, −2%–75%) [34] and from the United Kingdom (42%; 95% CI, −9%–69%) [35]. Overall, however, children who received LAIV were not the main drivers of low VE against A(H1N1)pdm09 in Canada, the United States [30], or Europe [31].
During A(H1N1)pdm09 epidemics in both 2013–2014 and 2015–2016, circulating viruses were considered antigenically matched to the unchanged vaccine despite mutations in the pivotal antigenic site Sa distinguishing clade 6B viruses in 2013–2014 (bearing K163Q) and clade 6B.1 viruses in 2015–2016 (bearing K163Q and S162N) [4, 13]. The new S162N substitution in clade 6B.1 viruses in 2015–2016 may have conferred a gain of glycosylation that shielded not only antigenic site Sa of the same monomer, including Q163, but also part of antigenic sites Sb and Ca2 on the adjacent monomer of the HA trimer. Such mutations and associated glycosylation changes are anticipated to affect antigenicity and immunogenicity, including antibody binding and VE [36].
In 2013–2014, when A(H1N1)pdm09 had an earlier January peak, VE estimates in Canada were comparable between mid- and full-season analyses (74% [95% CI, 58%–83%] and 71% [95% CI, 58%–80%], respectively), which was expected because 70% of the A(H1N1)pdm09 cases had already accrued by mid-season analysis [4, 12]. Conversely, in 2015–2016, the published interim VE included specimens collected during December–February but missed more than half the season’s A(H1N1)pdm09 cases that accrued thereafter, notably during the delayed peak in March 2016 [13]. The VE we report here for specimens collected during January–February 2016 (62%; 95% CI, 44%–74%) is similar to interim published data from Canada (64%; 95% CI, 44%–77%) [13], whereas the VE for March–April 2016 dropped significantly (19%; 95% CI, −15%–44%). A similar but less pronounced decrease between mid-season (44%; 95% CI, −3%–70%) [37] and full-season (33%; 95% CI, 4%–52%) [31] VE was also reported from Europe in 2015–2016, revealing a consistent pattern of intra-season waning of protection that, absent notable differences in virologic or participant characteristics across that period, warrants further investigation.
With identical prior and current season’s vaccines and antigenically similar epidemic virus (v1≡v2≈e), a pattern of positive interference (ie, improved VE) was suggested for v2+v1 versus v2 alone against A(H1N1)pdm09 in the initial postpandemic seasons in Canada, although with overlapping 95% CIs [9, 10, 15]. This pattern was also noted in Spain, where, similar to Canada, the majority of monovalent A(H1N1)pdm09 vaccine distributed in 2009 was adjuvanted [38], the significance of which in interpreting these findings is unclear. By 2013–2014, residual protection from the 2009 AS03-adjuvanted vaccine was no longer evident in Canada [4]. Also beginning in 2013–2014, a marginally reduced VE for recipients of v2+v1 versus v2 alone was observed (72% [95% CI, 56%–82%] vs 80% [95% CI, 45%–92%])—a pattern that became more pronounced in 2015–2016 (41% [95% CI, 18%–57%] vs 75% [95% CI, 45%–88%]), with significant interaction between v1 and v2 (Supplement 18). Residual protection from the prior season’s v1 alone was also lower in 2015–2016 as compared to 2013–2014 (25% [95% CI, −13%–51%] vs 49% [95% CI, 15%–70%]; Supplement 18). Although VE against A(H1N1)pdm09 was also marginally reduced for v2+v1 versus v2 recipients in the United States in 2013–2014 (51% [95% CI, 37%–62%] vs 61% [95% CI, 40%–74%]) [24], that pattern was not observed in 2015–2016 (40% [95% CI, 24%–52%] vs 38% [95% CI, 18%–54%]) [30].
Since 2009, the same conditions of v1≡v2≈e have been maintained for A(H1N1)pdm09, based on antigenic characterization by the HI assay. However, gene sequencing analysis identified evolution away from vaccine viruses, including key epitope differences and glycosylation changes, that are also potentially relevant in interpreting repeat vaccination effects. Antigenic distance metrics conventionally based on fold reductions in HI titers, determined using antisera from influenza-naive ferrets, may otherwise lack the necessary resolution to reflect vaccine virus relatedness or reliably inform the impact of repeat vaccination. Findings for A(H1N1)pdm09 in 2015–2016 reinforce the need for updated models that more directly link genomic, immunological, and epidemiological information. Ideally, these should also take into account preexisting immunological landscapes established over an extended period and upon which single versus serial vaccination effects are then superimposed [17].
Linderman et al [18] have proposed that distant childhood priming with A(H1N1) viruses bearing the exposed K163 epitope may have induced preferential K163-specific memory responses in middle-aged adults. They further proposed that annual administration of K163-bearing A/California/07/2009 (H1N1)pdm09 vaccine, used consistently since 2009, may have preferentially reinforced K163 specificity, potentially limiting protection against contemporary Q163 clade 6B variants (Supplement 1). Our age-based explorations for 2015–2016 generally align with predicted patterns extrapolated from Linderman et al. However, our age-stratified VE findings for A(H1N1)pdm09 are also consistent with other more prominent epochal distinctions, notably heterosubtypic priming with ancestral A(H2N2) and/or A(H3N2) subtype viruses. The VE against A(H1N1)pdm09 was lowest and negligible in 2015–2016 (5%; 95% CI, −66%–46%) among participants 49–59 years old (birth cohorts 1957–1967), for whom heterosubtypic priming to A(H2N2) (group 1 HA) in particular and/or subsequently to A(H3N2) (group 2 HA) are likely to have preceded A(H1N1) (group 1 HA) exposure before its reemergence in 1977 [39, 40]. Much remains unknown about heterosubtypic interactions, but their influence on age-related risk has recently been highlighted in other contexts for influenza A virus [39, 40]. The potential benefits of early influenza virus infection on protection later in life are also important to acknowledge [19, 41], as demonstrated in the higher cross-reactive antibody levels and lower infection risk among very old individuals during the A(H1N1)pdm09 pandemic [42]. Protection of very old individuals against A(H1N1) strains related to those to which they were exposed during childhood may have a negative ecologic corollary in their greater vulnerability to A(H3N2) [43]. The opposite pattern of A(H1N1)pdm09 risk in cohorts originally primed with A(H2N2) and/or A(H3N2) also remains speculative: we highlight that pronounced variation in age-adjusted VE was not statistically significant and was not as evident in our data for 2013–2014 (Supplement 19). Other, more recent shifts in age-related serosusceptibility have also been proposed to explain the 2013–2014 epidemic resurgence [44].
The substantial cross-lineage VE (54%; 95% CI, 32%–68%) we report for predominantly B(Yamagata) vaccine (trivalent) against predominantly B(Victoria) epidemic viruses is similar to the United States VE estimate for 2015–2016 (49%; 95% CI, 30%–64%) and also suggests immunological interactions across antigenically distinct viruses—a previously recognized but poorly understood phenomenon [10, 45–47]. The lower median age for B(Victoria) versus B(Yamagata) infections in 2015–2016 (19 vs 39 years; Supplement 10) or for infections during the predominantly B(Victoria) epidemic in 2015–2016 as compared to those during the predominantly B(Yamagata) epidemic in 2013–2014 (23 vs 43.5 years) is another previously recognized but poorly understood pattern that likely also reflects complex immuno-epidemiological considerations [48]. It is unlikely that the VE we report for B(Victoria) in 2015–2016 is attributable to the quadrivalent B(Victoria) vaccine component, given its limited use (<15% of total doses distributed) in Canada in 2015–2016 (Supplement 3). Antigenic relatedness of circulating B(Victoria) viruses to the quadrivalent vaccine component is furthermore uncertain, given the loss of N-linked glycosylation with egg adaptation of the vaccine strain, a recognized issue for interpreting HI characterization [49]. Unlike A(H1N1)pdm09, we did not observe further variation in VE against influenza B(Victoria) based on repeat vaccination, age, or epidemic period. However, in the United States, where there has been greater quadrivalent vaccine use, negative interference was suggested in 2015–2016 with repeat vaccination against B(Victoria) [30].
There are limitations to these analyses, several of which are exploratory and lack statistical power for conclusive interpretation, notably with subgroup stratification. Ultimately, given the observational design, we cannot rule out random variation or residual confounding. Additionally, as elaborated in Supplement 1, discrete cohort effects are challenging to ascribe using age as a surrogate for original influenza virus (or epitope-specific) priming, especially since an individual’s first influenza virus exposure may lag several years around a particular birth annum, obscuring epochal distinctions. Most of the population is likely to have been immunologically primed before 6 years old [29], with variability at the individual level. We interpreted findings by using a maximum 9-year lag from birth to influenza virus priming, as used by expert committees in their 1- versus 2-dose vaccine recommendations for children [27, 28]. This outer range accounted for an extended period of uncertainty in delineating immunological cohorts and excluded individuals with K163 specificity from those without such priming after 1985 (Supplement 1). In sensitivity analyses, VE estimates were not identical, but overall age-related patterns were robust. Vaccination history was based on a combination of self-report and sentinel practitioner documentation that may be subject to information bias; however, vaccine status was recorded before the influenza virus test result is known, minimizing differential recall, and vaccine coverage among our test-negative controls is generally consistent with that in other Canadian surveys [50]. Most vaccinated participants are habitual recipients; findings for smaller subsets of current or prior vaccination only may be associated with greater statistical and epidemiological uncertainty.
In conclusion, complex agent-host and immuno-epidemiological interactions beyond antigenic match are relevant to understanding vaccine performance. This includes variation in the viral genome and shifting patterns of glycosylation; potential cohort effects induced by childhood priming and subsequent cross-reactive boosting exposures (including possible heterosubtypic or interlineage); preexisting immunological landscapes and superimposed refocusing effects of repeat vaccination; as well as within-season waning of immunity across early versus late epidemic periods. Such influences on VE estimates may be daunting to untangle but warrant further in-depth investigation to identify improved vaccine and program options.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Acknowledgments. We thank the sentinel sites, whose regular submission of specimens and data provide the basis of our analyses, for their contribution; the epidemiological and laboratory staff in all participating provinces, for coordination and technical support; Lisan Kwindt (British Columbia Centre for Disease Control), Elaine Douglas, Kinza Rizvi, and Virginia Goetz (University of Calgary), Romy Olsha (Public Health Ontario), and Sophie Auger and Isabelle Petillot (Institut national de santé publique du Québec), for network coordination and data entry activities in each province; those who provided laboratory support in the British Columbia Centre for Disease Control Public Health Laboratory, the Alberta Provincial Laboratory for Public Health (ProvLab), the Public Health Ontario Laboratory, and the Laboratoire de santé publique du Québec; Aimin Li, Janet Obando, and Narisha Shakuralli, for virus sequencing support; Alireza Eshaghi (Public Health Ontario Laboratory), for supportive genomic input; Dr Robert Balshaw, for statistical input and consultation; and the authors, originating, and submitting laboratories of the reference virus sequences from Global Initiative on Sharing All Influenza Data’s EpiFlu Database (available at: http://www.gisaid.org) that informed genetic comparisons.
Financial support. This work was supported by the British Columbia Centre for Disease Control, Alberta Health and Wellness, Public Health Ontario, Ministère de la santé et des services sociaux du Québec, l’Institut national de santé publique du Québec, and the Public Health Agency of Canada.
Potential conflicts of interest. G. D. S. has received grants from GSK and Pfizer for activities unrelated to influenza; has received travel reimbursement to attend an ad hoc advisory board meeting of GSK unrelated to influenza; and has provided paid expert testimony in a grievance against a vaccinate-or-mask healthcare worker influenza vaccination policy for the Ontario Nurse Association. J. G. has received a research grant from Pfizer to conduct microbiological surveillance of Streptococcus pneumoniae. M. K. has received research grants from Roche, Merck, Siemens, Hologic, and Boerhinger Ingelheim for unrelated studies. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: Canadian Immunization Conference 2016, Ottawa, Canada 6–8 December 2016.
References
- 1. Public Health Agency of Canada. FluWatch report: August 14 to August 27, 2016 (weeks 33–34) http://healthycanadians.gc.ca/publications/diseases-conditions-maladies-affections/fluwatch-2015-2016-33-34-surveillance-influenza/index-eng.php. Accessed 14 August 2017.
- 2. United States Centers for Disease Control and Prevention. Fluview 2015–2016 influenza season week 39 ending October 1, 2016 https://www.cdc.gov/flu/weekly/weeklyarchives2015-2016/Week39.htm. Accessed 14 August 2017.
- 3. Public Health Agency of Canada. FluWatch report: 27 July to 9 August, 2014 (weeks 31 & 32) http://publications.gc.ca/collections/collection-2014/aspc-phac/HP58-1-2014-32-eng.pdf. Accessed 14 August 2017.
- 4. Skowronski DM, Chambers C, Sabaiduc S et al. Integrated sentinel surveillance linking genetic, antigenic, and epidemiologic monitoring of influenza vaccine-virus relatedness and effectiveness during the 2013-2014 influenza season. J Infect Dis 2015; 212:726–39. [DOI] [PubMed] [Google Scholar]
- 5. Epperson S, Blanton L, Kniss K et al. Influenza activity—United States, 2013–14 season and composition of the 2014–15 influenza vaccines. MMWR Morb Mortal Wkly Rep 2014; 63:483–90. https://www.cdc.gov/mmwr/pdf/wk/mm6322.pdf. Accessed 14 August 2017. [PMC free article] [PubMed] [Google Scholar]
- 6. Belongia EA, Simpson MD, King JP et al. Variable influenza vaccine effectiveness by subtype: a systematic review and meta-analysis of test-negative design studies. Lancet Infect Dis 2016; 16:942–51. [DOI] [PubMed] [Google Scholar]
- 7. British Columbia Centre for Disease Control. Canadian Sentinel Practitioner Surveillance Network (SPSN) vaccine effectiveness (VE) estimates (95%CI), 2004–05 to 2015–16 seasons http://www.bccdc.ca/resource-gallery/Documents/Statistics%20and%20Research/Publications/Epid/Influenza%20and%20Respiratory/SPSN_VE_By_Year_Table.pdf. Accessed 14 August 2017.
- 8. Skowronski DM, Janjua NZ, De Serres G et al. Effectiveness of AS03 adjuvanted pandemic H1N1 vaccine: case-control evaluation based on sentinel surveillance system in Canada, autumn 2009. BMJ 2011; 342:c7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Skowronski DM, Janjua NZ, De Serres G et al. A sentinel platform to evaluate influenza vaccine effectiveness and new variant circulation, Canada 2010-2011 season. Clin Infect Dis 2012; 55:332–42. [DOI] [PubMed] [Google Scholar]
- 10. Skowronski DM, Janjua NZ, Sabaiduc S et al. Influenza A/subtype and B/lineage effectiveness estimates for the 2011-2012 trivalent vaccine: cross-season and cross-lineage protection with unchanged vaccine. J Infect Dis 2014; 210:126–37. [DOI] [PubMed] [Google Scholar]
- 11. Skowronski DM, Janjua NZ, De Serres G et al. Low 2012-13 influenza vaccine effectiveness associated with mutation in the egg-adapted H3N2 vaccine strain not antigenic drift in circulating viruses. PLoS One 2014; 9:e92153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Skowronski DM, Chambers C, Sabaiduc S et al. Interim estimates of 2013/14 vaccine effectiveness against influenza A(H1N1)pdm09 from Canada’s sentinel surveillance network, January 2014. Euro Surveill 2014; 19:pii=20690 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20690. Accessed 14 August 2017. [DOI] [PubMed] [Google Scholar]
- 13. Chambers C, Skowronski D, Sabaiduc S et al. Interim estimates of 2015/16 vaccine effectiveness against influenza A(H1N1)pdm09, Canada, February 2016. Euro Surveill 2016; 21:pii=30168 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=21415. Accessed 14 August 2017. [DOI] [PubMed] [Google Scholar]
- 14. Smith DJ, Forrest S, Ackley DH, Perelson AS. Variable efficacy of repeated annual influenza vaccination. Proc Natl Acad Sci U S A 1999; 96:14001–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Belongia EA, Skowronski DM, McLean HQ, Chambers C, Sundaram ME, De Serres G. Repeated annual influenza vaccination and vaccine effectiveness: review of evidence. Expert Rev Vaccines 2017; 16:1–14. [DOI] [PubMed] [Google Scholar]
- 16. Katz JM, Hancock K, Xu X. Serologic assays for influenza surveillance, diagnosis and vaccine evaluation. Expert Rev Anti Infect Ther 2011; 9:669–83. [DOI] [PubMed] [Google Scholar]
- 17. Fonville JM, Wilks SH, James SL et al. Antibody landscapes after influenza virus infection or vaccination. Science 2014; 346:996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Linderman SL, Chambers BS, Zost SJ et al. Potential antigenic explanation for atypical H1N1 infections among middle-aged adults during the 2013-2014 influenza season. Proc Natl Acad Sci U S A 2014; 111:15798–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cobey S, Hensley SE. Immune history and influenza virus susceptibility. Curr Opin Virol 2017; 22:105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hensley SE. Challenges of selecting seasonal influenza vaccine strains for humans with diverse pre-exposure histories. Curr Opin Virol 2014; 8:85–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang KY, Rijal P, Schimanski L et al. Focused antibody response to influenza linked to antigenic drift. J Clin Invest 2015; 125:2631–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Petrie JG, Parkhouse K, Ohmit SE, Malosh RE, Monto AS, Hensley SE. Antibodies against the current influenza A(H1N1) vaccine strain do not protect some individuals from infection with contemporary circulating influenza A(H1N1) virus strains. J Infect Dis 2016; 214:1947–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Morens DM, Burke DS, Halstead SB. The wages of original antigenic sin. Emerg Infect Dis 2010; 16:1023–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gaglani M, Pruszynski J, Murthy K et al. Influenza vaccine effectiveness against 2009 pandemic Influenza A(H1N1) virus differed by vaccine type during 2013-2014 in the United States. J Infect Dis 2016; 213:1546–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ohmit SE, Petrie JG, Malosh RE et al. Substantial influenza vaccine effectiveness in households with children during the 2013-2014 influenza season, when 2009 pandemic influenza A(H1N1) virus predominated. J Infect Dis 2016; 213:1229–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McLean HQ, Thompson MG, Sundaram ME et al. Influenza vaccine effectiveness in the United States during 2012-2013: variable protection by age and virus type. J Infect Dis 2015; 211:1529–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. National Advisory Committee on Immunization (NACI). Canadian Immunization Guide Chapter on Influenza and Statement on Seasonal Influenza Vaccine for 2015–2016 http://www.phac-aspc.gc.ca/naci-ccni/assets/pdf/flu-2015-grippe-eng.pdf. Accessed 14 August 2017.
- 28. Grohskopf LA, Sokolow LZ, Olsen SJ, Bresee JS, Broder KR, Karron RA. Prevention and control of influenza with vaccines: recommendations of the Advisory Committee on Immunization Practices, United States, 2015–16 influenza season. MMWR Morb Mortal Wkly Rep 2015; 64:818–25. https://www.cdc.gov/mmwr/preview/mmwrhtml/mm6430a3.htm?s-c. Accessed 14 August 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bodewes R, de Mutsert G, van der Klis FR et al. Prevalence of antibodies against seasonal influenza A and B viruses in children in Netherlands. Clin Vaccine Immunol 2011; 18:469–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jackson ML, Chung JR, Jackson LA et al. Influenza vaccine effectiveness in the United States during the 2015-2016 season. N Engl J Med 2017; 377:534–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kissling E, Valenciano M, Reuss A et al. Influenza vaccine effectiveness (VE) estimates from the I-MOVE multicentre case control study in Europe, 2015–16: Feasibility of representative clade-specific VE ESCAIDE conference 29th of November 2016; Stockholm, Sweden page 101 http://www.escaide.eu/sites/escaide/files/documents/ESCAIDE%20Abstract%20Book%202016.pdf. Accessed 14 August 2017. [Google Scholar]
- 32. Valenciano M, Kissling E, Reuss A et al. The European I-MOVE Multicentre 2013–2014 case-control study. Homogeneous moderate influenza vaccine effectiveness against A(H1N1)pdm09 and heterogenous results by country against A(H3N2). Vaccine 2015; 2813–22. [DOI] [PubMed] [Google Scholar]
- 33. Flannery B, Chung J. Influenza vaccine effectiveness, including LAIV vs. IIV in children and adolescents, US Flu VE Network, 2015–16 https://www.cdc.gov/vaccines/acip/meetings/meetings-info.html. Accessed 4 June 2017.
- 34. Ambrose C. 2015–2016 US influenza vaccine effectiveness – Influenza Clinical Investigation for Children (ICICLE) Study https://www.cdc.gov/vaccines/acip/meetings/meetings-info.html. Accessed 4 June 2017.
- 35. Pebody R, Warburton F, Ellis J et al. Effectiveness of seasonal influenza vaccine for adults and children in preventing laboratory-confirmed influenza in primary care in the United Kingdom: 2015/16 end-of-season results. Euro Surveill 2016; 21:pii=30348 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=22592. Accessed 14 August 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tate MD, Job ER, Deng YM, Gunalan V, Maurer-Stroh S, Reading PC. Playing hide and seek: how glycosylation of the influenza virus hemagglutinin can modulate the immune response to infection. Viruses 2014; 6:1294–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kissling E, Valenciano M. Early influenza vaccine effectiveness results 2015–16: I-MOVE multicentre case-control study. Euro Surveill 2016; 21:pii=30134 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=21378. Accessed 14 August 2017. [DOI] [PubMed] [Google Scholar]
- 38. Jiménez-Jorge S, Savulescu C, Pozo F et al. ; cycEVA Study Team; Spanish Influenza Sentinel Surveillance System Effectiveness of the 2010-11 seasonal trivalent influenza vaccine in Spain: cycEVA study. Vaccine 2012; 30:3595–602. [DOI] [PubMed] [Google Scholar]
- 39. Gostic KM, Ambrose M, Worobey M, Lloyd-Smith JO. Potent protection against H5N1 and H7N9 influenza via childhood hemagglutinin imprinting. Science 2016; 354:722–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Te Beest DE, de Bruin E, Imholz S, Koopmans M, van Boven M. Heterosubtypic cross-reactivity of HA1 antibodies to influenza A, with emphasis on nonhuman subtypes (H5N1, H7N7, H9N2). PLoS One 2017; 12:e0181093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Monto AS, Malosh RE, Petrie JG, Martin ET. The doctrine of original antigenic sin: separating good from evil. J Infect Dis 2017; 215:1782–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Skowronski DM, Hottes TS, McElhaney JE et al. Immuno-epidemiologic correlates of pandemic H1N1 surveillance observations: higher antibody and lower cell-mediated immune responses with advanced age. J Infect Dis 2011; 203:158–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Centers for Disease Control and Prevention. Estimates of deaths associated with seasonal influenza—United States, 1976–2007. MMWR Morb Mortal Wkly Rep 2010; 59:1057–62. https://www.cdc.gov/mmwr/preview/mmwrhtml/mm5933a1.htm. Accessed 14 August 2017. [PubMed] [Google Scholar]
- 44. Skowronski DM, Chambers C, Sabaiduc S et al. Pre- and postpandemic estimates of 2009 pandemic influenza A(H1N1) seroprotection to inform surveillance-based incidence, by age, during the 2013-2014 epidemic in Canada. J Infect Dis 2015; 211:109–14. [DOI] [PubMed] [Google Scholar]
- 45. Skowronski DM, Hamelin ME, Janjua NZ et al. Cross-lineage influenza B and heterologous influenza A antibody responses in vaccinated mice: immunologic interactions and B/Yamagata dominance. PLoS One 2012; 7:e38929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Skowronski DM, Hottes TS, De Serres G et al. Influenza Β/Victoria antigen induces strong recall of Β/Yamagata but lower Β/Victoria response in children primed with two doses of Β/Yamagata. Pediatr Infect Dis J 2011; 30:833–9. [DOI] [PubMed] [Google Scholar]
- 47. Tricco AC, Chit A, Soobiah C et al. Comparing influenza vaccine efficacy against mismatched and matched strains: a systematic review and meta-analysis. BMC Med 2013; 11:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Skowronski DM, Chambers C, De Serres G et al. Age-related differences in influenza B infection by lineage in a community-based sentinel system, 2010–11 to 2015–16, Canada. J Infect Dis 2017; 15:697–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Robertson JS, Naeve CW, Webster RG, Bootman JS, Newman R, Schild GC. Alterations in the hemagglutinin associated with adaptation of influenza B virus to growth in eggs. Virology 1985; 143:166–74. [DOI] [PubMed] [Google Scholar]
- 50. Public Health Agency of Canada. Influenza vaccine uptake: Results from the 2015/16 national influenza immunization coverage survey in Canada Government of Canada: Ottawa, 2017. https://www.canada.ca/en/public-health/services/publications/healthy-living/vaccine-uptake-results-2015-16-national-influenza-immunization-coverage-survey.html. Accessed 14 August 2017. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.