The kinases responsible for phosphorylation of phytochrome-interacting factors (PIFs) in plants undergoing photomorphogenesis are largely unknown. We show that the MKK10-MPK6 cascade regulates red light-induced photomorphogenesis through phosphorylation of PIF3.
Keywords: Arabidopsis thaliana, cotyledon opening, MKK10-MPK6, phytochrome-interacting factor, phosphorylation, photomorphogenesis
Abstract
Photomorphogenesis is an important process in which seedlings emerge from soil and begin autotrophic growth. Mechanisms of photomorphogenesis include light signal perception, signal transduction, and the modulation of expression of light-responsive genes, ultimately leading to cellular and developmental changes. Phytochrome-interacting factors (PIFs) play negative regulatory roles in photomorphogenesis. Light-induced activation of phytochromes triggers rapid phosphorylation and degradation of PIFs, but the kinases responsible for the phosphorylation of PIFs are largely unknown. Here, we show that Arabidopsis MPK6 is a kinase involved in phosphorylating PIF3 and regulating red light-induced cotyledon opening, a crucial process during seedling photomorphogenesis. MPK6 was activated by red light, and the cotyledon opening angle in red light was reduced in mpk6 seedlings. MKK10, a MAPKK whose function is currently unclear, appears to act as a kinase upstream of MPK6 in regulating cotyledon opening. Activation of MPK6 by MKK10 led to the phosphorylation of PIF3 and accelerated its turnover in transgenic seedlings. Accordingly, the overexpression of PIF3 suppressed MKK10-induced cotyledon opening. MKK10 and MPK6 function downstream of phyB in regulating seedling cotyledon opening in red light. Therefore, the MKK10-MPK6 cascade appears to mediate the regulation of red-light-controlled seedling photomorphogenesis via a mechanism that might involve the phosphorylation of PIF3.
Introduction
Light is the primary energy source and an important signal for plant growth, development, and stress responses. The wavelength, intensity, duration, and direction of light are sensed by plants through a series of photoreceptors, including phytochromes (Li et al., 2011), cryptochromes (Chaves et al., 2011), phototropins (Christie, 2007), LOV/F-Box/Kelch repeat proteins (Ito et al., 2012), and UV-B receptors (Jenkins, 2014). The signals from these photoreceptors are then transduced through distinct signaling pathways and influence many developmental and physiological processes throughout the plant’s life cycle, such as seed germination, seedling photomorphogenesis, shade avoidance, circadian rhythms, and flowering time.
Phytochromes are red (R) and far-red (FR) light photoreceptors in plants (Franklin and Quail, 2010; Li et al., 2011) that are present in the inactive red-light-absorbing form (Pr) in darkness and convert to the active far-red-absorbing form (Pfr) upon exposure to R light (Fankhauser, 2001; Quail, 1997). The translocation of active phytochromes from the cytosol to the nucleus is a key event in the phytochrome signaling pathway (Kevei et al., 2007; Nagatani, 2004). The Arabidopsis thaliana genome encodes five phytochromes, namely, phyA to phyE, which play overlapping but distinct roles in various responses to light (Bae and Choi, 2008). After germination, seedlings undergo skotomorphogenesis (etiolation) in darkness and photomorphogenesis (de-etiolation) in light. Skotomorphogenic phenotypes include elongated hypocotyls, exaggerated apical hooks, unopened cotyledons, and the development of the proplastids into etioplasts, whereas photomorphogenic phenotypes include inhibited hypocotyl elongation, opened and expanded cotyledons, and the development of chloroplasts (Arsovski et al., 2012; Li et al., 2011; Seluzicki et al., 2017). FR-light-promoted seedling photomorphogenesis is primarily under the control of phyA, whereas R-light-induced photomorphogenesis is predominantly controlled by phyB (McCormac et al., 1993; Parks and Quail, 1993; Reed et al., 1994).
Phytochromes regulate light responses through downstream intermediates (Chen and Chory, 2011; Li et al., 2011; Quail, 2010; Xu et al., 2015). Phytochrome-interacting factors (PIFs), a subfamily of basic helix-loop-helix (bHLH) transcription factors, are central players in phytochrome-mediated light signaling networks that regulate the expression of over 1000 light-responsive genes (Castillon et al., 2007; Leivar and Quail, 2011). In darkness, the inactivation of phytochromes allows PIFs to accumulate and promote skotomorphogenesis, whereas upon exposure to light, the activation of phytochromes induces phosphorylation and degradation of PIFs to initiate photomorphogenesis (Leivar and Monte, 2014; Leivar and Quail, 2011). In Arabidopsis, PIF1 is phosphorylated by Casein Kinase 2 (CK2), whereas PIF4 and PIF3 are phosphorylated by Brassinosteroid Insensitive 2 (BIN2) (Bernardo-García et al., 2014; Bu et al., 2011; Ling et al., 2017); however, mutated PIF1 and PIF4 proteins in which the known phosphorylation sites were substituted by alanine residues continued to display robust light-induced phosphorylation. The kinase AsphyA phosphorylates PIF1, PIF3, and PIF4 in oat (Avena sativa); however, transgenic plants expressing mutant AsphyA protein with significantly lower kinase activity continued to show light-induced phosphorylation of PIF3 (Shin et al., 2016). Photoregulatory protein kinases (PPKs), a small family of CK1-like proteins, were recently shown to phosphorylate PIF3; however, knockout of multiple PPK genes reduced but did not abolish light-induced PIF3 phosphorylation and degradation (Ni et al., 2017). Over 20 phosphorylation sites have been identified in Arabidopsis PIF3 (Ni et al., 2013), suggesting that other kinase(s) might also mediate the phosphorylation of PIFs and PIF-regulated light responses (Xu et al., 2015).
Mitogen-activated protein kinase (MAPK) cascades are highly conserved signaling modules in eukaryotic cells composed of three types of kinases that are sequentially activated by phosphorylation: MAPKK kinases (MAPKKKs), MAPK kinases (MAPKKs), and MAPKs (MAPK Group, 2002). MAPK cascades are activated by upstream kinase(s) or receptor(s). Active MAPK phosphorylates a variety of substrate proteins, thereby altering their activity, localization, stability, and transcriptional level (Xu and Zhang, 2015). MAPK cascades in plants play important roles in regulating growth, development, and stress responses. Although the role of MAPK cascades in regulating plant light responses is far from clear, several lines of evidence suggest that they are involved in this process: two MAPKs in cucumber (Cucumis sativus) cotyledons are activated during FR- and R-light-induced seedling de-etiolation (Alvarez-Flórez et al., 2013); the transcription of several MAPK cascade genes in Arabidopsis is regulated by light, and among these, MKKK14 is up-regulated by R light and its null mutant shows a short-hypocotyl phenotype in FR- but not R-light-induced seedling de-etiolation (Khanna et al., 2006; López-Juez et al., 2008; Tepperman et al., 2006); and the Arabidopsis MKK3-MPK6 cascade is activated by blue (B) light and regulates hypocotyl growth through phosphorylation of MYC2 (Sethi et al., 2014). Arabidopsis contains 60 MAPKKKs, 10 MAPKKs, and 20 MAPKs (nominated as MKKKs, MKKs, and MPKs, respectively) (MAPK Group, 2002). MKKs function in the following processes: MKK1 in defense, abscisic acid, and reactive oxygen species responses (Pitzschke et al., 2009; Qiu et al., 2008; Xing et al., 2008); MKK2 in cold and salt tolerance (Teige et al., 2004), MKK3 in hypocotyl growth and jasmonic acid responses (Sethi et al., 2014; Takahashi et al., 2007); MKK4/MKK5 in H2O2 (Ren et al., 2002) and NO production (Wang et al., 2010a), stomatal and ovule development (Wang et al., 2008; Wang et al., 2007), and defense responses (Asai et al., 2002; Wang et al., 2010b); MKK6 in cytokinesis (Soyano et al., 2003); MKK7 in polar auxin transport (Dai et al., 2006; Jia et al., 2016); and MKK4/MKK5/MKK9 in ethylene and camalexin biosynthesis (Ren et al., 2008; Xu et al., 2008). However, the activities and biological functions of MKK8 and MKK10 remain obscure.
In this study, we show that MPK6 is activated by R light. MPK6 loss-of-function mutant seedlings displayed less pronounced cotyledon opening in R light than did wild-type seedlings. MKK10 functions upstream of MPK6 in regulating cotyledon opening. After MKK10-mediated activation of MPK6, this latter kinase phosphorylated PIF3 in vitro and in vivo. Phosphorylation of PIF3 by MKK10-MPK6 accelerated the turnover of PIF3 in transgenic seedlings in darkness. Overexpression of PIF3 suppressed MKK10-induced cotyledon opening in dark-grown seedlings. Genetic analysis demonstrated that the MKK10-MPK6 cascade functions downstream of phyB in regulating cotyledon opening in seedlings exposed to R light. Our findings suggest that MKK10-MPK6-PIF3 is a module that regulates phyB-mediated seedling photomorphogenesis in R light.
Materials and methods
Plant materials
Seeds of Arabidopsis thaliana Col-0 wild type, mutants, and transgenic lines were surface sterilized, incubated in the dark for 4 days at 4 °C, and sown on 0.8% agar plates containing 0.5×Murashige and Skoog (MS) medium, pH 5.7, and 1% sucrose. Seven-day-old seedlings were transferred from the plates to soil and grown at 22 °C in a growth room under a 16 h light/8 h dark photoperiod at a photon flux density of 100 μmol m –2 s–1. To analyze seedling phenotypes, seeds were treated at 4 °C and sown on 0.8% agar plates containing 1×MS, pH 5.7. After exposure to 100 μmol m–2 s–1 white light for 3 h, plates were transferred to darkness, R (1, 8, or 30 μmol m–2 s–1), FR (30 μmol m–2 s–1), or B (8 μmol m–2 s–1) light conditions and grown for 4 days. To induce expression of the transgenes 0.02 or 0.05 μmol dexamethasone (DEX) was added to the medium.
Vector construction
Total RNA was isolated from seedlings using Trizol reagent (Invitrogen). First-strand cDNA was synthesized with M-MLV virus reverse transcriptase (Promega) using dT(16) as the primer and RNA as the template.
The coding regions of MKKs and MPKs with an NdeI site added before the first ATG codon were obtained by PCR and cloned into a pBluescript vector. Point mutations were introduced into the MKKs with a Quick-Change Site-directed Mutagenesis Kit (Stratagene). The NdeI/XhoI fragments of MKKs were cloned into a modified pBluescript vector with an Ω sequence and Flag-epitope tag coding sequence at the 5ʹ end (Xu et al., 2008). SpeI/XhoI fragments of MKKs were ligated into a pTA7002 vector (Aoyama and Chua, 1997). A 2.5 kb fragment before the first ATG codon of MKK10 was amplified by PCR, using genomic DNA as the template, and used as the MKK10 native promoter. The promoter fragment was inserted into a pCAMBIA 1300-221 vector (Invitrogen) to substitute the CaMV35S promoter sequence. The PIF3 coding region with the stop codon removed was added before the Green Fluorescent Protein (GFP) sequence in the modified pCAMBIA 1300 vector; the MPK6-mCherry coding sequence was inserted into the modified pCAMBIA 1300 vector to substitute for the GFP coding sequence (Cao et al., 2010). The resulting constructs were transformed into Agrobacterium tumefaciens strain GV3101.
The coding regions of PIF3 with an NdeI site added before the first ATG codon and an XhoI site added to its 3ʹ end were obtained by PCR and cloned into a pBluescript vector. The NdeI/XhoI fragment of PIF3 and the NdeI/SalI fragment of MKK10 mutants were cloned into a pGEX4T-2 vector. The NdeI/SalI fragments of MPK3 and MPK6, and BamHI/SalI fragments of MPK12 and MPK10 were cloned into a pET28a(+) vector. The resulting constructs were transformed into Escherichia coli strain BL21.
Primers used are listed in Table S1 available at the Dryad Digital Repository, http://dx.doi.org/10.5061/dryad.hq7b8.
Agrobacterium-mediated transformation
Transgenic Arabidopsis plants were generated using the floral dip method (Clough and Bent, 1998). MKKs and MKK10 promoter-GUS transgenic plants were screened with 15 mg/l hygromycin, and expression of the transgene was detected using an immunoblot analysis and β-glucuronidase (GUS) staining.
Transient transformation of tobacco (Nicotiana benthamiana) leaves was performed as described previously (Xu et al., 2008). PIF3-GFP and MPK6-mCherry fluorescence signals were viewed under a Zeiss LSM510 Meta Confocal Laser-Scanning System.
Mutant generation and genetic crosses
To generate the mkk10 mutant, an MKK10-specific target for Cas9 was selected to mutate MKK10 using CRISPR-PLANT (http://www.genome.arizona.edu/crispr/CRISPRsearch.html). The target sequence was cloned into a CRISPR/Cas9 vector, pHEE 401, as described previously (Wang et al., 2015). The resulting construct was transformed into A. tumefaciens strain GV3101. Arabidopsis transformation was performed using the floral dip method (Clough and Bent, 1998). Transgenic seedlings were screened using hygromycin selection, genomic PCR, and sequencing as described previously (Wang et al., 2015). The mkk10 mutant obtained had two insertions, an A after base 136 and a T after base 386 of the start codon in the MKK10 gene. An MluI site in the MKK10 genomic sequence was mutated in mkk10 due to the insertion of a T base, and was used for mkk10 mutant genotyping.
T-DNA insertional mutants, including pif3 (Salk_030753), mpk3 (SALK_100651), mpk6-3 (SALK_127507), mpk6-4 (SALK_062471), and mkk9 (SAIL_060_H06), were obtained from the Arabidopsis Biological Resource Center. The homozygous mutants were screened using genomic PCR and reverse transcription (RT)-PCR. phyA (phyA-211) and phyB (phyB-9) were the point mutation mutants. cry1 (cry1-304) was a fast-neutron mutation mutant. PIF3 OE was a gift from Dr Giltsu Choi (Park et al., 2004). The homozygous mutants were identified as described previously (Mockler et al., 1999; Reed et al., 1993; Reed et al., 1994).
Genetic crossing was performed to generate MKK10D/PIF3 OE, MKK10D/mpk6, MKK10D/phyA, MKK10D/phyB, MKK10D/cry1, mkk9/mkk10, and mpk6/pif3 mutants. The mpk6-4 mutant was used for the kinase activity assay and for genetic crossing.
Preparation of recombinant proteins
Escherichia coli cells transformed with pGEX4T-2 or pET28a(+) constructs were cultured, and isopropyl-D-thiogalactopyranoside was added to induce the recombinant protein expression. His-MPK proteins were purified using a Ni2+-Cheating Sepharose Fast Flow column (GE Healthcare), and GST-PIF3 or GST-MKK10 mutant proteins were purified using a Glutathione Sepharose 4B column (GE Healthcare).
Protein extraction and immunoblot analysis
Proteins were extracted from seedlings using kinase buffer as previously described (Xu et al., 2008) and used for in-gel kinase assays and immunoblot analysis of the MPKs, MKKs (except for MKK7), and tubulin. For the MKK7 expression assay, proteins were extracted using 1×SDS sample buffer (diluted with kinase pre-mix buffer: 100 mM HEPES, pH 7.5, 5 mM EDTA, 5 mM EGTA, 10 mM Na3VO4, 10 mM NaF, 50 mM β-glycerophosphate). For the MYC-PIF3 expression assay, proteins were extracted using kinase buffer containing 40 μM MG132 for co-immunoprecipitation (Co-IP) and boiling extraction buffer for Phos-tag mobility shift assays (Al Sady et al., 2006). Protein separation and immunoblotting were performed as described previously (Xu et al., 2008). The primary antibodies included anti-Flag, anti-MYC, anti-MPK3, anti-MPK6, and anti-β-tubulin antibodies. Secondary antibodies included horseradish peroxidase-conjugated goat anti-mouse or anti-rabbit antibodies.
Co-immunoprecipitation assay
For the MYC-PIF3 and MPK6 Co-IP assay, protein extracts were incubated with anti-MYC beads for 3.5 h at 4 °C in the dark on a rotating mixer. The beads were collected and washed four times with kinase pre-mix buffer containing 0.02% Triton X-100. Bound proteins were eluted in 1 M NH3OH, pH 11.5, with 5% SDS, and separated by 10% SDS-PAGE. Immunoblot assays were performed using anti-MPK6 and anti-MYC antibodies as the primary antibodies.
Pull-down assay
GST or GST-PIF3 proteins were incubated with Glutathione Sepharose 4B beads pre-balanced with extraction buffer (0.1 M Tris-HCl, pH 7.5, 150 mM NaCl) at 4 °C for 1 h. The beads were collected, washed three times with washing buffer (0.1 M Tris-HCl, pH 7.5, 150 mM NaCl, 0.02% Triton X-100), and incubated with His-MPK6 protein in binding buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.02% Triton X-100) at 4 °C for 2 h. After centrifugation, the beads were washed five times with washing buffer. The beads were boiled in 1×SDS buffer, and the proteins were separated by 10% SDS-PAGE. Immunoblot assays were performed using anti-GST and anti-His antibodies as the primary antibodies.
Kinase assays
GST-MKK10 mutant proteins were incubated with His-MPKs in phosphorylation buffer (20 mM Tris-HCl, pH 7.5, 10 mM MgCl2, and 50 µM ATP) at 30 °C for 30 min. A 0.5 µg sample of His-MPK was taken from each reaction and added into a new tube with 5 µg myelin basic protein (MBP) and 1 μCi [γ-32P]-ATP in phosphorylation buffer. After incubation at 30 °C for 30 min, the reactions were stopped by adding 4×SDS buffer. The reaction mixture was heated, and the proteins were separated by 10% SDS-PAGE. The gels were dried, and phosphorylation of MBP was detected by autoradiography.
An in-gel kinase assay was performed as described previously (Ren et al., 2002).
Cotyledon opening assay in seedlings
Cotyledon opening angles were measured and analyzed using ImageJ software (http://rsb.info.nih.gov/ij/). Cotyledon angle represents the angle between the lines drawn through two cotyledons. An angle of 0° indicates fully folded cotyledons.
Real-time quantitative RT-PCR
Real-time quantitative RT-PCR (Q-PCR) was performed using a SYBR Premix EX TaqTM Kit (Takara) in the presence of SYBR Green I. Amplification was monitored in real time with a 7500 real-time PCR system (Applied Biosystems). The expression levels of selected genes were normalized to the UBQ10 control. Primers used for Q-PCR are listed in Table S1 (available at the Dryad Digital Repository, http://dx.doi.org/10.5061/dryad.hq7b8.
Accession number
The accession numbers of genes used in this study are listed in Table S2 at Dryad.
Results
MPK6 is activated by red light and mediates red-light-regulated cotyledon opening
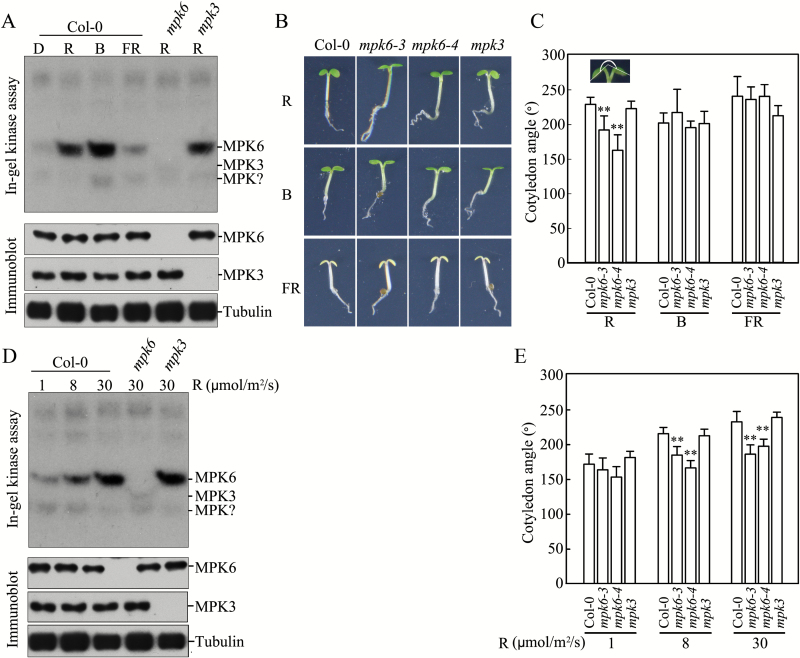
MPK6 is activated in seedlings transferred from darkness to B light, and mpk6 mutant seedlings have shorter hypocotyls than wild-type seedlings in white, R, FR, and B light (Sethi et al., 2014). Therefore, the activation of a MAPK cascade might mediate the signaling pathway underlying inhibited hypocotyl elongation, an important aspect of seedling photomorphogenesis. To explore whether a MAPK cascade (and, if so, which one) is involved in regulating the cotyledon opening process, another crucial aspect of seedling photomorphogenesis, we analyzed MAPK activity and cotyledon opening in seedlings grown in R, FR, and B light. MAPK activity in total proteins extracted from seedlings grown in darkness, 8 μmol m–2 s–1 of R light, 30 μmol m–2 s–1 of FR light, or 8 μmol m–2 s–1 of B light for 4 days was detected using in-gel kinase activity assays, with MBP as an artificial MAPK substrate. The extract from wild-type (Col-0) seedlings grown in the dark contained three kinases with lower basal-level activity (with molecular weights of ~ 60, 46, and 40 kD) (Fig. 1A). However, the activity of the 46 kD kinase increased strongly in seedlings grown in R and B light, and slightly in seedlings grown in FR light, compared with dark-grown seedlings. The activity of the 40 kD kinase was slightly decreased in seedlings grown in R light and increased in seedlings grown in B and FR light compared with seedlings grown in the dark. Finally, the activity of the 60 kD kinase did not significantly differ among seedlings grown in the dark or in R, B, or FR light. We also analyzed kinase activity using extracts from seedlings of MPK3 and MPK6 T-DNA insertional null mutants. The lack of MPK3 protein in mpk3 seedlings and MPK6 protein in mpk6 seedlings was confirmed by immunoblot analysis with MPK3- or MPK6-specific antibodies. The 46 kD kinase was not activated in mpk6 seedlings grown in R light, indicating that this kinase was MPK6 (Fig. 1A).
Fig. 1.
MPK6 is activated by different light conditions and loss of MPK6 activity inhibits cotyledon opening in seedlings in R light. (A) Kinase activity in Col-0, mpk3, and mpk6 seedlings detected in an in-gel kinase assay. MPK3, MPK6, and tubulin were detected by immunoblot analysis. (B) Photographs of Col-0, mpk3, and mpk6 seedlings showing the cotyledon phenotypes. White bar=2 mm. (C) Cotyledon opening angles in Col-0, mpk3, and mpk6 seedlings in R, B, and FR light. (D) Kinase activity in Col-0 seedlings grown in different R light conditions detected in an in-gel kinase assay. (E) Cotyledon opening angles of Col-0, mpk3, and mpk6 seedlings in different R light conditions. Data are means±SD (n=30). Asterisks indicate significant differences between Col-0 and mpk6 seedlings (Student’s t-test, **P<0.01). D, dark; R, red light; FR, far-red light; B, blue light.
Since R, FR, and B light activated MPK6 in seedlings, we compared the hypocotyl lengths and cotyledon opening angles of Col-0, mpk6, and mpk3 seedlings grown under these different light conditions. We used two alleles of mpk6 (mpk6-3 and mpk6-4) to exclude any possible effects of an additional mutation in the mpk6 mutants. As shown in Fig. 1B and C, the cotyledon opening angles of mpk6-3 and mpk6-4 seedlings grown in R light were 16% and 28% lower, respectively, than those of Col-0 seedlings, whereas no significant differences were observed in FR or B light. Under all light conditions, the cotyledon opening angles of mpk3 seedlings did not significantly differ from those of Col-0 seedlings. In addition, the hypocotyl lengths of Col-0, mpk6, and mpk3 seedlings did not differ significantly under darkness or under the different light conditions (Fig. S1 at Dryad). These results suggest that the activation of MPK6 by R light might play a role in R-light-induced cotyledon opening.
To confirm the R-light-dependent activation of MPK6 and to explore the role of MPK6 in regulating cotyledon opening, we analyzed MAPK activity and cotyledon-opening angles in seedlings grown under different fluence rates of R light (1, 8, and 30 μmol m–2 s–1). In wild-type seedlings, MPK6 activity increased in R light in an intensity-dependent manner (Fig. 1D). The cotyledon opening angles of mpk6-3 and mpk6-4 seedlings were significantly reduced when grown in 8 and 30 μmol m–2 s–1 R light compared with Col-0 seedlings, but these angles were not significantly different in 1 μmol m–2 s–1 R light. The cotyledon opening angles of mpk6-3 and mpk6-4 seedlings were 14% and 23% lower in 8 μmol m–2 s–1 R light and 20% and 15% lower in 30 μmol m–2 s–1 R light, respectively, than those of Col-0 seedlings (Fig. 1E). The cotyledon opening angles of Col-0 and mpk3 seedlings grown under the same fluence rate of R light did not differ significantly. These results suggest that a MAPK cascade involving MPK6 mediates the R-light-regulated seedling photomorphogenesis signaling pathway.
MKK10 is the predominant Arabidopsis MAPKK that regulates cotyledons opening
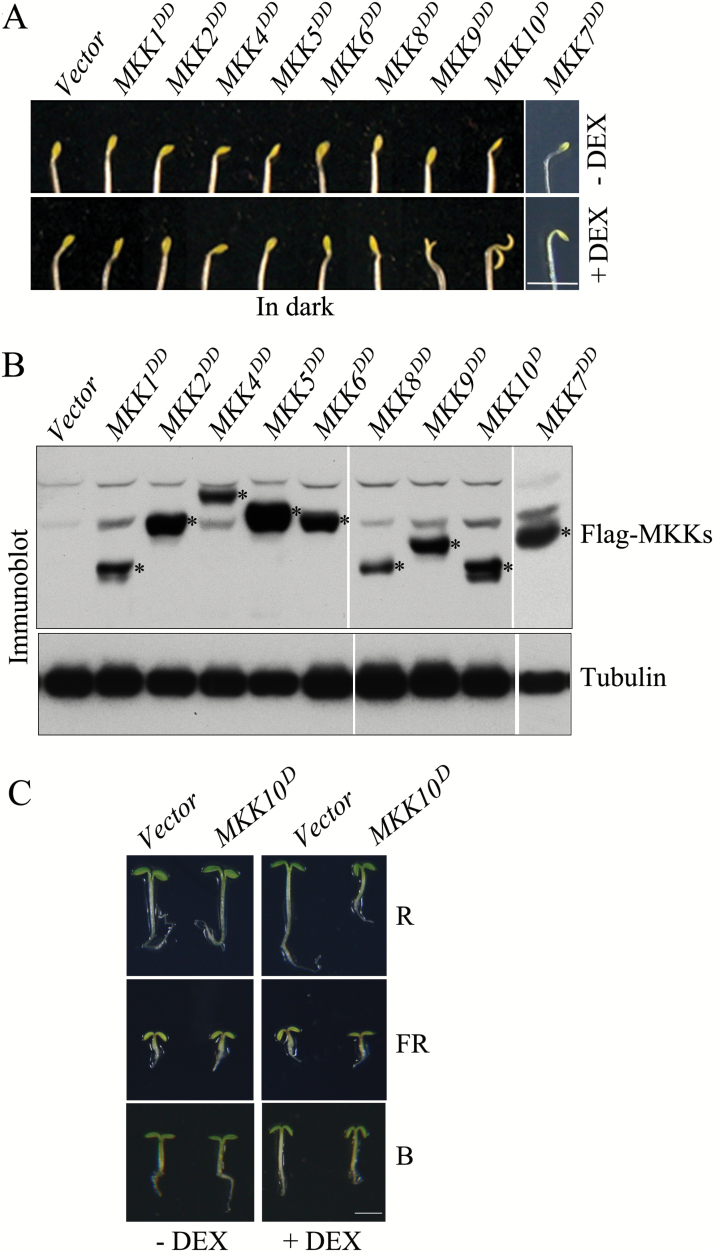
Since the activation of MPK6 is involved in regulating cotyledon opening in seedlings, we speculated that the activation of an MKK upstream of MPK6 might cause the same phenotype. To support this hypothesis, we generated transgenic kinase-active Arabidopsis lines for various MKKs and used them to identify MAPKK(s) that function upstream of MPK6 in regulating seedling cotyledon opening. Based on information from previous reports (Jia et al., 2016; Matsuoka et al., 2002; Ren et al., 2002; Takahashi et al., 2007; Teige et al., 2004; Xu et al., 2008) and sequence analyses, we generated kinase-active lines for all 10 MKKs in Arabidopsis. For MKK1 to MKK9, the Thr (T) and Ser (S) residues in the T/SxxxxxS/T motif in the activation loops were substituted with Asp (D) (designated as MKK1DD to MKK9DD). Owing to the absence of the typical T/SxxxxxS/T motif in the MKK10 activation loop, and since no MKK10-active mutant has been reported previously, we substituted Ser197 in its putative activation domain with Asp (D) to generate MKK10D, a constitutive mutant for MKK10 (Fig. S2A at Dryad). The mutated MKK1 to MKK10 genes were placed under the control of the DEX-inducible promoter in the pTA7002 vector (Aoyama and Chua, 1997) and transformed into Arabidopsis. For MKK3DD, over 20 transgenic lines showing hygromycin resistance were obtained; unfortunately, MKK3DD protein expression was not detected in any of these lines in an immunoblot analysis. In contrast to MKK3DD, we obtained transgenic plants with normal induction of MKK10D protein for MKK10D and with normal induction of MKKxDD proteins for the eight MKKxDD genes.
Homozygous MKKxDD and MKK10D transgenic seeds were germinated and grown on plates with or without DEX in dark. Uninduced (–DEX) MKKs transgenic seedlings did not exhibit cotyledon opening in dark, and nor did induced (+DEX) MKK1DD to MKK8DD seedlings. However, MKK10D seedlings had large cotyledon opening angles, and MKK9DD seedlings had slightly open cotyledons after MKK10D or MKK9DD protein induction (+DEX) in the dark. When germinated and grown under light conditions, the induction of MKK10D led to seedlings with large cotyledon opening angles in both R and B light, whereas FR light did not affect cotyledon opening angles compared with the Vector control. All of the MKKs transgenic seedlings expressed the transgenes effectively, as shown by immunoblot analysis (Fig. 2). These results suggest that MKK10 is the MKK that plays a major role in regulating cotyledon opening and that MKK9 may function redundantly with MKK10 in this process.
Fig. 2.
Analysis of cotyledon opening in MKKs active mutant transgenic seedlings. (A) Photographs showing cotyledon opening phenotypes in MKKs transgenic seedlings with (+DEX) or without (–DEX) induction of transgenes. Scale bar=2 mm. (B) Protein levels of MKKs in transgenic seedlings detected by immunoblot analysis with anti-Flag antibody. Asterisks indicate Flag-MKK proteins. (C) Cotyledon opening phenotypes in MKK10D and Vector transgenic seedlings in R, FR, and B light with (+DEX) or without (–DEX) induction of transgenes.
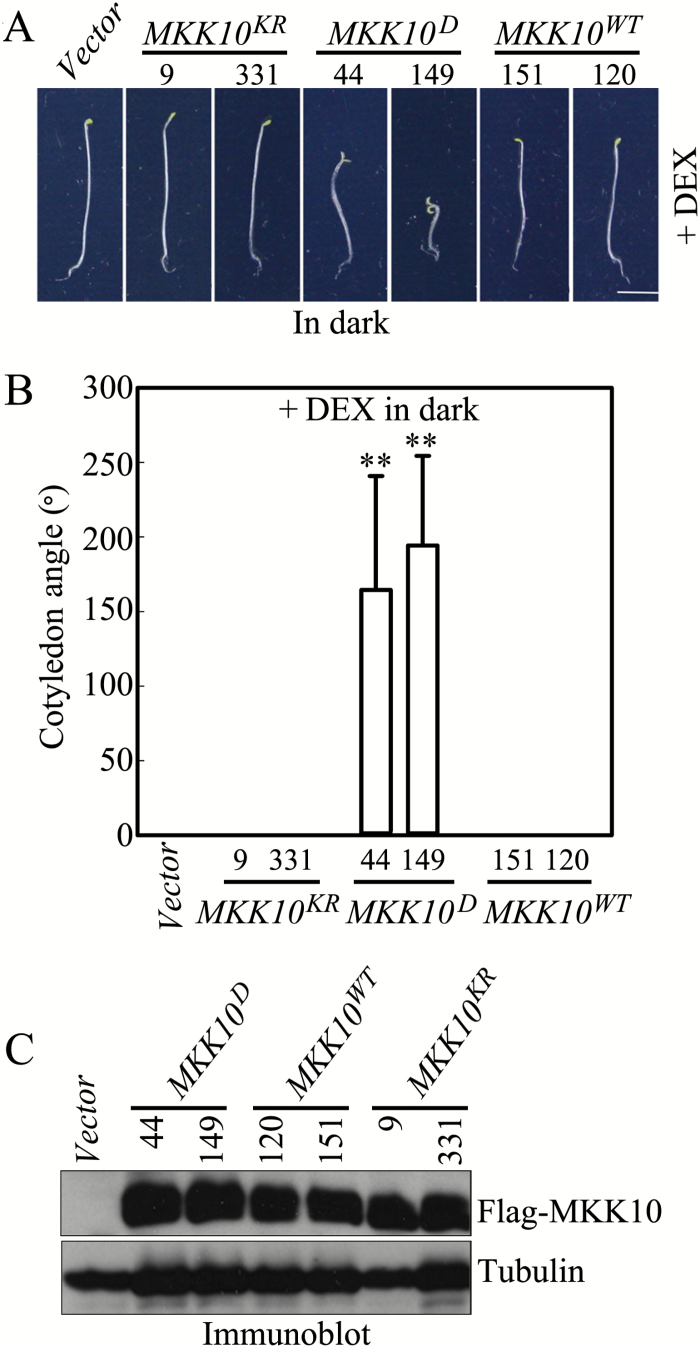
To confirm the importance of MKK10 activity in inducing cotyledon opening, we generated MKK10KR, a kinase-inactive form of MKK10 with the Lys77 (K) in its ATP-binding site substituted with Arg (R) (Fig. S2B at Dryad). We generated MKK10KR and MKK10WT transgenic plants and analyzed independent transgenic lines for MKK10D, MKK10KR, and MKK10WT to exclude any possible effects of the transgene insertion on phenotype. Consistent with the results shown in Fig. 2A, both MKK10D lines displayed markedly increased cotyledon opening after transgene induction in the dark, but the Vector, MKK10KR, and MKK10WT seedlings did not (Fig. 3A). Statistical analyses revealed that cotyledon opening angles in both MKK10D lines were significantly larger than those in the Vector, MKK10KR, and MKK10WT lines (Fig. 3B). Immunoblot analysis showed that all MKK10 variant transgenic lines expressed comparable levels of Flag-MKK10 protein (Fig. 3C). These results suggest that only the expression of the MKK10 active form caused cotyledons to open in darkness. The MKK10D lines also had shortened hypocotyls, and in both lines, the hypocotyl length was negatively correlated with MPK6 activity (Figs 2C, 3A, and S3 at Dryad). To explore where MKK10 normally functions, we expressed GUS under the control of the native MKK10 promoter in seedlings of both independent transgenic lines to examine the expression pattern of MKK10. GUS activity staining was detected only in the cotyledons of seedlings and not in hypocotyls (Fig. S4 at Dryad). This result implies that MKK10 normally regulates cotyledon opening but not hypocotyl length at the seedling stage. Perhaps the inhibited hypocotyl elongation in MKK10D seedlings is due to the strong expression MKK10D in overexpression lines.
Fig. 3.
Expression of active mutant MKK10 induces cotyledon opening in seedlings. (A) Photographs showing cotyledon opening phenotypes in MKK10WT, MKK10D, and MKK10KR seedlings. Scale bar=2 mm. (B) Cotyledon opening angles in transgenic seedlings. Data are means±SD (n=30). Asterisks indicate significant differences between MKK10D and Vector transgenic seedlings (Student’s t-test, **P<0.01). (C) Protein levels of MKK10WT, MKK10D, and MKK10KR in transgenic seedlings detected by immunoblot analysis with anti-Flag antibody.
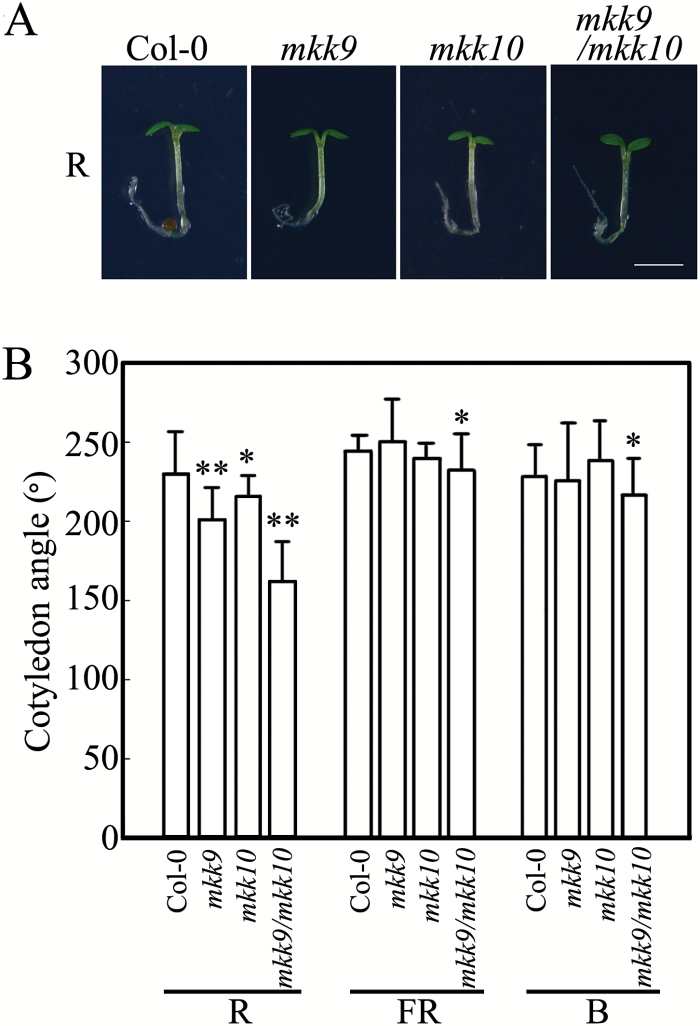
We examined the cotyledon opening phenotype in the mkk10 and mkk9 mutants. mkk9 is a T-DNA insertional null mutant, and mkk10 is a mutant that we generated in this study using the CRISPR/Cas9 method (Fig. S5 at Dryad). Sequence analysis revealed two single nucleotide insertions in the coding region of MKK10 in mkk10, resulting in an early stop codon in MKK10 mRNA. As shown in Fig. 4A and B, the cotyledon opening angles of mkk9 and mkk10 seedlings were 12% and 6% lower than those of Col-0 wild-type seedlings, respectively, when grown in R light. However, the cotyledon opening angles of mkk9/mkk10 double mutant seedlings were strongly reduced, to 30% lower than those of Col-0 seedlings. When grown in FR and B light, differences among the cotyledon opening angles of Col-0, mkk9, and mkk10 seedlings were negligible, and the cotyledon opening angles of mkk9/mkk10 seedlings were reduced by only 4% compared with Col-0 seedlings. These results suggest that MKK10 and MKK9 function redundantly in R-light-regulated cotyledon opening, which is consistent with the results of overexpression analysis.
Fig. 4.
Loss of function of MKK9 and MKK10 reduces R-light-induced cotyledon opening angles in seedlings. (A) Photographs of Col-0, mkk9, mkk10, and mkk9/mkk10 seedlings showing cotyledon phenotypes. Scale bar=2 mm. (B) Cotyledon opening angles in Col-0, mkk9, mkk10, and mkk9/mkk10 seedlings in R, FR, and B light. Data are means±SD (n=30). Asterisks indicate significant differences between Col-0 and mutant seedlings (Student’s t-test, *P<0.05, **P<0.01).
MKK10 functions upstream of MPK6 in regulating cotyledon opening
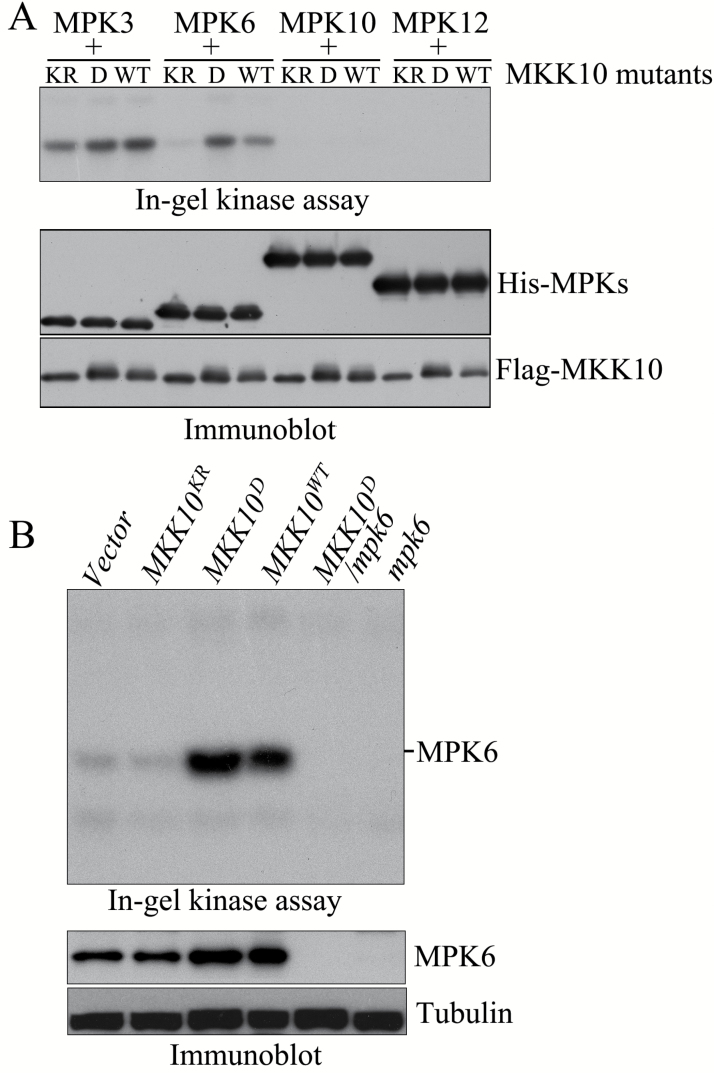
We next performed a series of experiments to determine whether MKK10 is the MKK that functions upstream of MPK6. First, the recombinant MKK10 variant proteins were used to phosphorylate MPK6, and then MPK6 kinase activity was examined using MBP as substrate. MPK3, MPK10, and MPK12 were used as controls. As shown in Fig. 5A, MKK10D strongly activated MPK6, while MKK10WT moderately activated MPK6 and MKK10KR did not activate this kinase. MPK10 and MPK12 were not activated by MKK10, but MPK3 was weakly activated by MKK10. We then generated transgenic Arabidopsis plants harboring MKK10WT, MKK10D, and MKK10KR and subjected them to an in-gel kinase assay. As shown in Fig. 5B, after transgene induction, MKK10D seedlings showed strong activation of a 46 kD kinase, MKK10WT seedlings showed moderate activation of this kinase, and MKK10KR and Vector seedlings showed only lower basal levels of 46 kD kinase activity. Therefore, following previously developed rules for the functions of MKKs (Jia et al., 2016; Matsuoka et al., 2002; Ren et al., 2002; Takahashi et al., 2007; Teige et al., 2004; Xu et al., 2008), MKK10D is considered to be the active MKK10 and MKK10KR the inactive MKK10. Third, to confirm that the 46 kD kinase activated by MKK10 in transgenic plants is MPK6, we generated a MKK10D/mpk6 mutant by genetic crossing. An in-gel kinase assay showed that the activation of the 46 kD kinase by MKK10D induction was abolished in MKK10D/mpk6 seedlings, demonstrating that the 46 kD kinase is indeed MPK6 (Fig. 5B). These results suggest that MKK10 is an MKK that functions upstream of MPK6.
Fig. 5.
MKK10 activates MPK6 in vitro and in vivo. (A) Activation of His-MPKs by Flag-MKK10 mutant proteins in vitro. His-MPK activity was detected using an in-gel kinase assay (upper panel). Flag-MKK10 mutant proteins and His-MPK proteins in the reaction were detected by immunoblot analysis with anti-Flag and anti-His antibodies (lower panel). (B) MPK6 activity in MKK10 mutant transgenic lines and in lines crossed with these plants detected by in-gel kinase assays (upper panel). MPK6 protein were detected by immunoblot analysis with anti-MPK6 antibody(lower panel).
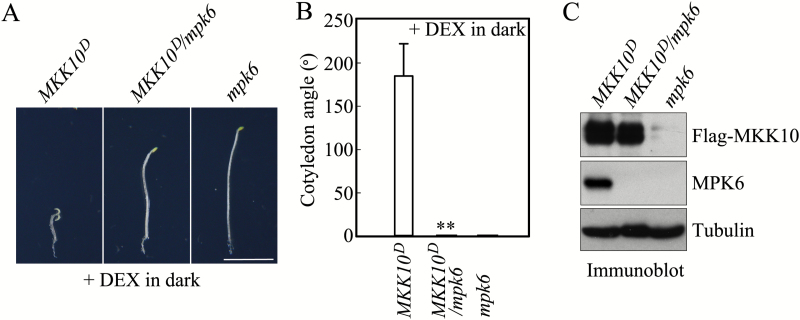
We analyzed cotyledon opening in MKK10D and MKK10D/mpk6 seedlings. As shown in Fig. 6A, after MKK10D induction in the dark (+DEX in dark), MKK10D seedlings displayed a strong cotyledon opening phenotype, whereas MKK10D/mpk6 seedlings did not (Fig. 6B). Immunoblot analysis showed that comparable levels of MKK10D were present in MKK10D and MKK10D/mpk6 seedlings (Fig. 6C). These results suggest that activation of MPK6 by MKK10 causes the cotyledon opening phenotype of MKK10D seedlings.
Fig. 6.
Activation of MPK6 by MKK10 is required for MKK10-regulated cotyledon opening in seedlings. (A) Photographs showing the cotyledon opening phenotypes in MKK10D, MKK10D/mpk6, and mpk6 seedlings. Scale bar=2 mm. (B) Cotyledon opening angles in MKK10D, MKK10D/mpk6, and mpk6 seedlings. Data are mean ±SD (n=30). Asterisks indicate significant differences between MKK10D and MKK10D/mpk6 seedlings (Student’s t-test, **P<0.01). (C) Protein levels of MKK10D and MPK6 detected by immunoblot with anti-Flag and anti-MPK6 antibodies.
MKK10-MPK6 functions downstream of phyB in regulating cotyledon opening
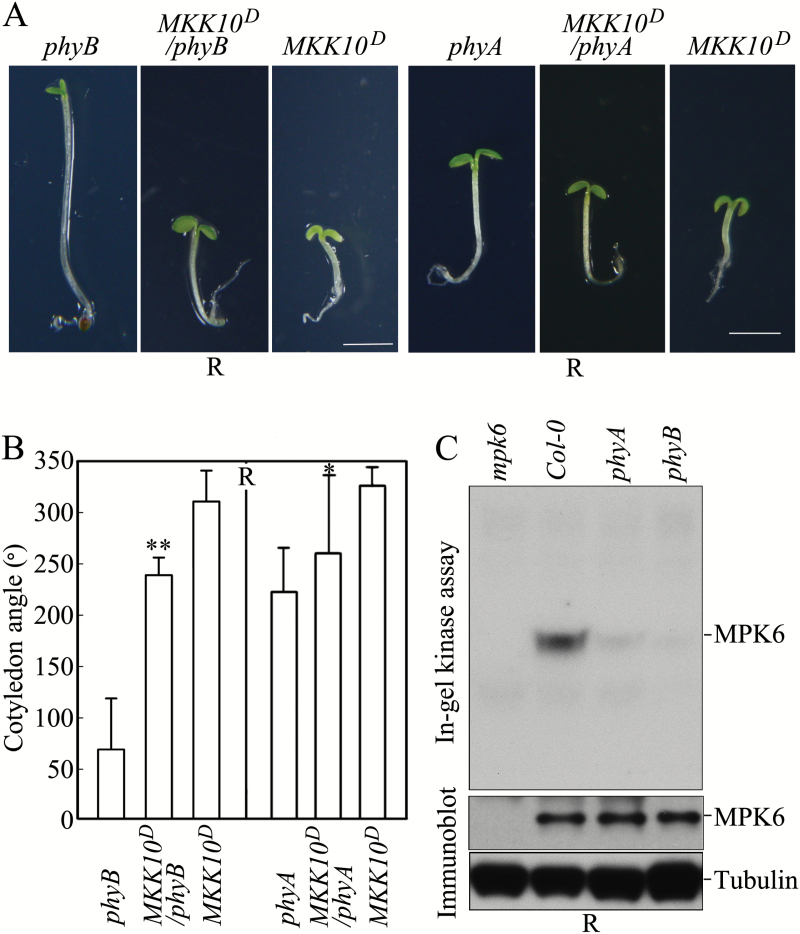
The phytochrome phyB is the predominant phytochrome that regulates seedling photomorphogenesis in R light, and phyA has partially overlapping functions with phyB (Li et al., 2011; Reed et al., 1994). We therefore analyzed MPK6 activity in phy mutant seedlings grown in R light. As shown in Fig. 7, MPK6 was strongly activated in Col-0 seedlings grown in R light; however, R light only weakly activated MPK6 in the phyA mutant and did not activate MPK6 in the phyB mutant. We crossed phyA and phyB into the MKK10D background. The reduced cotyledon opening angle in phyB mutant seedlings grown in R light was rescued (increased by 253%) in MKK10D/phyB after MKK10D induction. By contrast, the cotyledon opening angle was only slightly (~17%) higher in MKK10D/phyA seedlings in R light compared with phyA mutant seedlings (Fig. 7A and B). The induction of MKK10D in MKK10D/cry1 seedlings in B light also led to a strong increase (~290%) in cotyledon opening angle compared with cry1 mutant seedlings (Fig. S7 at Dryad). These results suggest that MKK10-MPK6 acts downstream of phyB in regulating cotyledon opening and that phyA or CRY1 share partially overlapping functions with phyB (Casal and Boccalandro, 1995; Casal and Mazzella, 1998).
Fig. 7.
phyB functions upstream of MKK10-MPK6 in regulating cotyledon opening in seedlings in R light. (A) Photographs showing the cotyledon opening phenotype in MKK10D, MKK10D/phyB, and MKK10D/phyA seedlings in R light. Scale bar=2 mm. (B) Cotyledon opening angles in MKK10D, MKK10D/phyB, and MKK10D/phyA seedlings in R light. Data are means±SD (n=30). Asterisks indicate significant differences between phyB and MKK10D/phyB or between phyA and MKK10D/phyA (Student’s t-test, *P<0.05, **P<0.01). (C) Kinase activity in Col-0, mpk6, phyA, and phyB seedlings in R light detected using an in-gel kinase assay (upper pane;). MPK6 protein was detected by immunoblot analysis with anti-MPK6 antibody (lower panel).
PIF3 regulates MKK10-MPK6-mediated cotyledon opening
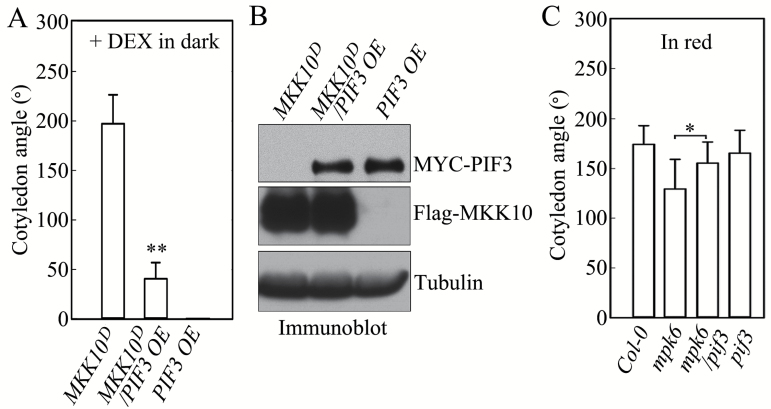
PIF3, a repressor of photomorphogenesis, plays a pivotal role in phyB-mediated R light responses (Leivar and Monte, 2014; Leivar and Quail, 2011). We investigated whether PIF3 is involved in MKK10-MPK6-regulated cotyledon opening by generating and analyzing MKK10D/PIF3 OE and mpk6/pif3 lines (Fig. S6 at Dryad). As shown in Fig. 8A, the MKK10D-induced cotyledon opening phenotype in the dark was repressed when MYC-PIF3 was overexpressed in MKK10D/PIF3 OE seedlings. The increased cotyledon opening angle caused by the induction of MKK10D was 79% smaller in MKK10D/PIF3 OE seedlings in the dark than in MKK10D seedlings. Immunoblot analysis revealed comparable levels of MKK10D in MKK10D and MKK10D/PIF3 OE seedlings, and MYC-PIF3 in MKK10D/PIF3 OE and PIF3 OE seedlings (Fig. 8B). The reduced cotyledon opening angle caused by the loss of function of MPK6 was partially rescued in mpk6/pif3 seedlings in R light (Fig. 8C). These results suggest that PIF3 mediates the cotyledon opening process regulated by the MKK10-MPK6 cascade. However, overexpression of PIF3 did not completely abolish the MKK10D-induced cotyledon opening phenotype, implying that additional factors other than PIF3 (e.g. other PIFs) are also involved in this process.
Fig. 8.
PIF3 functions downstream of MKK10-MPK6 in regulating cotyledon opening in seedlings in R light. (A) Cotyledon opening angles of MKK10D, MKK10D/PIF3 OE, and PIF3 OE seedlings in the dark. Data are means±SD (n=30). Asterisks indicate significant differences between MKK10D and MKK10D/PIF3 OE. (B) Protein levels of MKK10D and MYC-PIF3 detected by immunoblot analysis with anti-Flag and anti-MYC antibodies. (C) Cotyledon opening angles of Col-0, mpk6, pif3, and mpk6/pif3 seedlings in R light. Data are means±SD (n=30). Asterisks indicate significant differences between mpk6 and mpk6/pif3 seedlings. (Student’s t-test, *P<0.05, **P<0.01).
MKK10-MPK6 induces PIF3 phosphorylation and accelerates PIF3 degradation
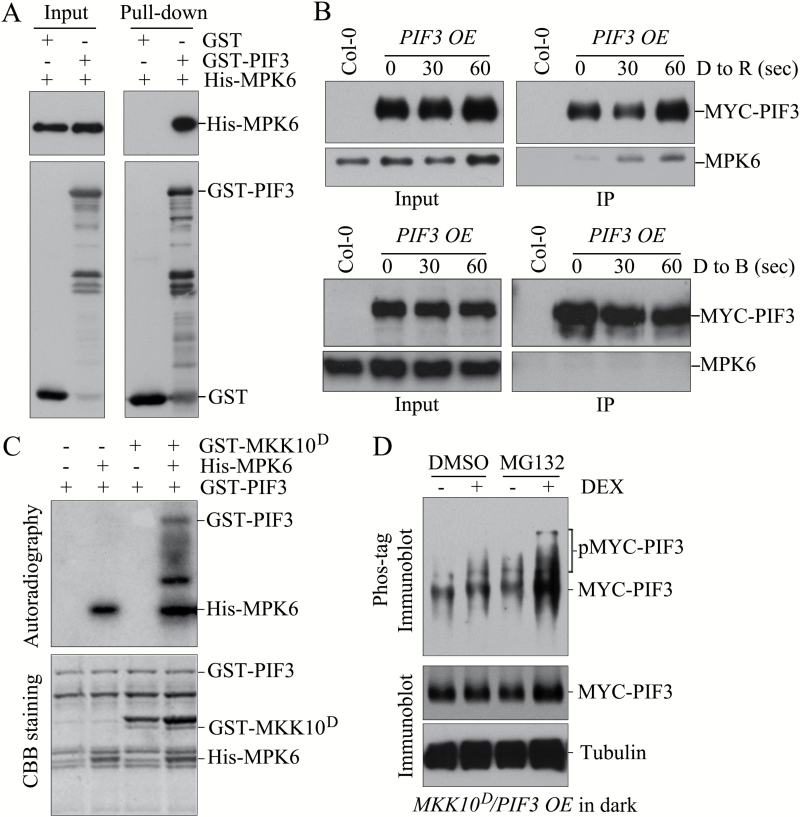
The phosphorylation and subsequent degradation of PIF3 are required for the promotion of photomorphogenesis when seedlings are exposed to light (Leivar and Monte, 2014; Leivar and Quail, 2011); however, little is known about the kinase(s) that phosphorylate PIF3. Since PIF3 is involved in MKK10-MPK6-regulated cotyledon opening, we investigated whether MKK10-MPK6 phosphorylates PIF3. The interaction between MPK6 and PIF3 and the phosphorylation of PIF3 by MKK10-MPK6 were examined.
We performed pull-down experiments using His-MPK6 with GST-PIF3 or GST as input proteins to detect the interaction between MPK6 and PIF3 in vitro. As shown in Fig. 9A, anti-GST antibody detected both GST and GST-PIF3, and anti-His antibody detected His-MPK6 in both input samples, indicating the successful loading of GST, GST-PIF3, and His-MPK6. After incubation and pull-down with GST-affinity beads, we found that anti-His antibody detected His-MPK6 in the GST-PIF3 pull-down sample but not in the GST pull-down sample. The His-MPK6 was pulled down by GST-PIF3, suggesting that MPK6 interacted with PIF3 in vitro. We performed Co-IP experiments with MPK6 and PIF3 using PIF3 OE seedlings that were grown in the dark and transferred to the R light condition for 0, 30, and 60 s to test the interaction of PIF3 with MPK6 in vivo and the regulation of this interaction by R light. As shown in Fig. 9B, a lower level of MPK6 co-immunoprecipitated with MYC-PIF3 in the samples from seedlings before transfer to R light (0 s); however, after the seedlings were exposed to R light for 30 or 60 s, the levels of MPK6 that co-immunoprecipitated with MYC-PIF3 increased significantly. Co-IP of PIF3 with MPK6 suggested that MPK6 interacted with PIF3 in vivo. Exposure of seedlings to B light did not enhance the interaction of PIF3 with MPK6. To confirm the interaction of these proteins, we investigated the transient co-expression of MPK6-mCherry and PIF3-GFP in tobacco leaves. MPK6 and PIF3 co-localized in nuclear bodies (Fig. S8 at Dryad). Our findings that R light treatment increased the interaction of MPK6 with PIF3 and that MPK6 was activated by R light suggest that activation of MPK6 strengthens its interaction with PIF3.
Fig. 9.
MPK6 phosphorylates PIF3 and accelerates its degradation. (A) GST pull-down assay. GST or GST-PIF3 was bound to Glutathione Sepharose 4B beads and then incubated with His-MPK6. Proteins bound to beads were analyzed by immunoblot analysis with anti-His antibody (upper panel) and anti-GST antibody (lower panel). (B) Co-immunoprecipitation of MPK6 and MYC-PIF3. Dark-grown PIF3 OE seedlings were exposed to 30 μmol m–2 s–1 R light or 8 μmol m–2 s–1 B light for 30 and 60 s. Proteins were extracted from the seedlings, and an immunoprecipitation (IP) assay was performed using anti-MYC beads. Proteins bound to beads were analyzed by immunoblot analysis with anti-MYC antibody (upper panels) and anti-MPK6 antibody (lower panels). (C) Phosphorylation of PIF3 by MKK10-MPK6 in vitro. His-MPK6 was activated by GST-MKK10D and used to phosphorylate GST-PIF3. After the reaction, the proteins were separated by SDS-PAGE and visualized by Coomassie Brilliant Blue (CBB) staining (lower panel). The phosphorylated proteins were detected by autoradiography (upper panel). (D). Phosphorylation of PIF3 by MKK10-MPK6 in vivo. MKK10D/PIF3 OE seedlings were grown on medium with or without DEX in the dark and then treated with MG132 (50 μM final concentration). Protein extracts from seedlings were separated on an SDS-PAGE gel with Phos-tag reagent and transferred to a nitrocellulose membrane. MYC-PIF3 was detected by immunoblot analysis with anti-MYC antibody (upper panel). A standard immunoblot assay was performed to detect total MYC-PIF3 protein in the extracts.
We performed phosphorylation experiments via in vitro kinase activity assays using GST-MKK10D, His-MPK6, and GST-PIF3. As shown in Fig. 9C, GST-PIF3 was phosphorylated in the presence of both GST-MKK10D and His-MPK6 in the reaction, whereas GST-PIF3 was not phosphorylated in the presence of GST-MKK10D or His-MPK6 alone. These results suggested that activation of MPK6 by MKK10 phosphorylated PIF3 in vitro. We also examined the phosphorylation of MYC-PIF3 in MKK10D/PIF3 OE seedlings grown in the dark before or after MKK10D induction using Phos-tag mobility shift assays. Because the phosphorylation of PIFs is degraded through the 26S proteasome-mediated pathway (Al-Sady et al., 2006; Ni et al., 2013; Park et al., 2004; Shen et al., 2005; Shen et al., 2007), we also used MG132, a 26S proteasome inhibitor, in this experiment. As shown in Fig. 9D, in the absence of MG132, the strengths and patterns of the MYC-PIF3 bands in seedlings before and after MKK10D induction (DMSO with or without DEX) did not significantly differ. However, when seedlings were pretreated with MG132, multiple newly emerged bands were observed and the strength of the formerly existed bands of MYC-PIF3 in the sample of seedlings after MKK10D induction (MG132 with DEX) increased. These results demonstrate that MYC-PIF3 is phosphorylated and that the phosphorylation of PIF3 by MKK10-MPK6 accelerates its degradation in vivo.
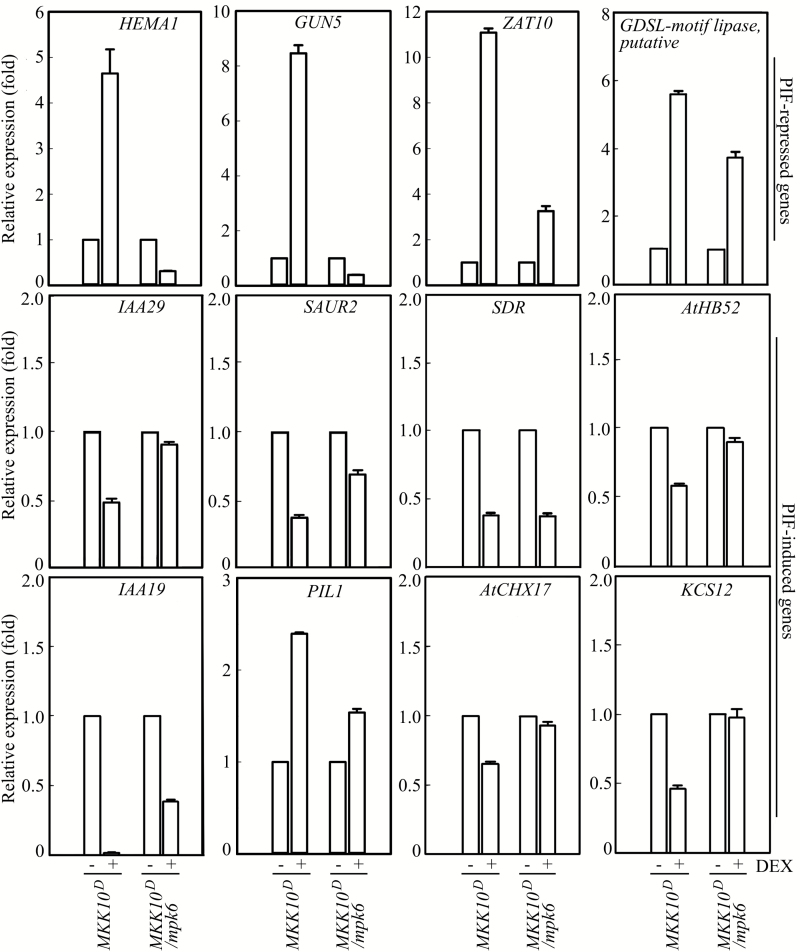
Activation of MKK10-MPK6 regulates transcription of light-responsive genes
The transcription of many genes is regulated in seedlings during photomorphogenesis (Jiao et al., 2007; Quail, 2007). PIFs such as PIF1, PIF3, PIF4, and PIF5 play a central role in regulating the transcription of these genes (Leivar et al., 2009; Zhang et al., 2013). Because MKK10-MPK6 phosphorylated PIF3 and accelerated its degradation (Fig. 9), we speculated that the transcription of some PIF-regulated genes would be altered after MKK10-MPK6 activation. We performed Q-PCR to measure the transcript levels of several genes that are induced or repressed by PIFs, as shown previously, in MKK10D and MKK10D/mpk6 seedlings with or without MKK10D induction. As shown in Fig. 10, the transcription of some PIF-repressed genes, e.g. HEMA1, GUN5, ZAT10, and a gene encoding a protein with a putative GDSL motif (GDSL-motif lipase putative), was strongly induced in MKK10D seedlings after MKK10D induction, whereas the induction levels of ZAT10 and GDSL-motif lipase putative were partially compromised and HEMA1 and GUN5 transcript levels were reduced in MKK10D/mpk6 seedlings. The transcription of PIF-induced genes, including IAA29, SAUR2, SDR, AtHB52, IAA19, AtCHX17, and KCS12, was reduced in MKK10D seedlings after MKK10D induction, whereas the reduced transcription of these genes (except for SDR) was partially or fully reversed in MKK10D/mpk6 seedlings. However, the transcription of some genes, e.g. PIL1, which is induced by PIFs (Luo et al., 2014; Zhang et al., 2013), was induced in MKK10D seedlings and reduced in MKK10D/mpk6 seedlings. These results suggest that the activation of MKK10-MPK6 regulates the transcription of some PIF-regulated genes.
Fig. 10.
Q-PCR detection of PIF-induced or PIF-repressed gene transcription in MKK10D and MKK10D/mpk6 seedlings. Seedlings were grown on medium with or without DEX in the dark for 4 days. The transcription levels of the indicated genes were monitored by Q-PCR. Data are means±SD of three biological replicates.
Discussion
Genetic screening and molecular studies have revealed that many genes are involved in the regulation of seedling photomorphogenesis. The mutation of photoreceptor genes suppresses seedling photomorphogenesis under specific spectral compositions of light, leading to the following phenotypes: phyA seedlings show elongated hypocotyls in FR and B light and exaggerated cotyledons in FR light; phyB seedlings display elongated hypocotyls and reduced cotyledon opening in R and white light; cry1 seedlings show elongated hypocotyls and reduced cotyledon opening in B and white light; phyA/phyB double mutant seedlings exhibit increased hypocotyl elongation in FR, R, B, and white light, exaggerated cotyledons in FR and R light, and reduced cotyledon opening in white light; phyA/phyB/cry1 triple mutant seedlings show severe cotyledon-opening and hypocotyl-elongation phenotypes (Arsovski et al., 2012; Li et al., 2011; Mockler et al., 1999; Neff and Chory, 1998; Reed et al., 1994; Yu et al., 2010). Mutation analysis showed that some transcription factors act downstream of photoreceptors as either positive or negative regulators of seedling photomorphogenesis. For example, HY5 (a bZIP transcription factor) and HFR1 (an atypical bHLH transcription factor) are positive regulators of this process. Whereas hy5 mutant seedlings show elongated hypocotyls in the light, seedlings overexpressing truncated HY5 (COP1-interactive domain deleted) exhibit shortened hypocotyls (Ang et al., 1998; Ang and Deng, 1994; Arsovski et al., 2012; Lee et al., 2007; Li et al., 2011; Oyama et al., 1997; Ulm et al., 2004). Moreover, hfr1 mutant seedlings have elongated hypocotyls and reduced cotyledon opening in FR and B light (Duek and Fankhauser, 2003; Fairchild et al., 2000; Fankhauser and Chory, 2000; Soh et al., 2000), whereas seedlings overexpressing truncated HFR1 (N-terminal 105 amino acids deleted) have shortened hypocotyls in the dark and FR light and open cotyledons in the dark (Yang et al., 2003). A small subset of PIFs, including PIF1, PIF3, PIF4 and PIF5, are key negative regulators of photomorphogenesis (Leivar and Monte, 2014; Leivar and Quail, 2011; Monte et al., 2007; Seluzicki et al., 2017; Zhang et al., 2013). In the dark, quadruple mutant (pifq) seedlings with these four PIF genes mutated exhibit a partially constitutive photomorphogenic phenotype, that is, shortened hypocotyls and open cotyledons (Leivar et al., 2008). However, the signaling modules that link photoreceptors and these transcription factors are far from clear.
MAPK cascades are well-known signaling modules that perceive signals from receptors/sensors and transduce these signals to elicit cellular responses (Andreasson and Ellis, 2010; Colcombet and Hirt, 2008; Meng and Zhang, 2013). In this study, we explored whether MAPK cascades are involved in regulating the responses of Arabidopsis seedlings to R light and, if so, which MAPK cascades are involved, and how. We used cotyledon opening angle in seedlings as the primary parameter for phenotypic observations. The strong activation of MPK6 by R light and the reduced cotyledon opening in mpk6 mutant seedlings compared with Col-0 seedlings under R light suggested that MPK6 activity is involved in R-light-regulated cotyledon opening in seedlings. To demonstrate the sufficiency of MPK6 activity for the regulation of cotyledon opening, seedlings with long-lasting MPK6 activation must be examined in the dark. The active forms of MKK1 to MKK5, MKK7, and MKK9 have previously been reported to activate MPK6, with the activation of MPK6 by different MKKs in plants affecting distinct biological functions (Jia et al., 2016; Ren et al., 2002; Sethi et al., 2014; Takahashi et al., 2007; Teige et al., 2004; Xing et al., 2008; Xu et al., 2008); however, none of these MKKs has been implicated in the activation of MPK6 to regulate cotyledon opening. We therefore generated transgenic plants overexpressing active MKKs (MKK1DD to MKK9DD and MKK10D) and observed cotyledon opening in seedlings in the dark. The overexpression of MKK1DD to MKK8DD did not induce cotyledon opening, and the overexpression of MKK9DD only weakly induced cotyledon opening in the dark; however, surprisingly, the overexpression of MKK10D strongly induced cotyledon opening in the dark. Because the biological function of MKK10 has not previously been reported, our results provide the first evidence that MKK10 is the primary MKK that regulates cotyledon opening in seedlings.
GUS driven by the MKK10 promoter was specifically expressed in cotyledon, further supporting the role of MKK10 in cotyledon opening (Fig. S3 at Dryad). The activation of MPK6 by MKK10 in vitro and in vivo, and the abolishment of the MKK10D-induced cotyledon opening phenotype in MKK10D/mpk6 seedlings, demonstrated that MKK10 indeed functions upstream of MPK6 in R-light-regulated cotyledon opening in seedlings. Considering the function of MKK10-MPK6 in R-light-regulated cotyledon opening in seedlings and the predominant role of phyB in R light perception, we reasoned that the MKK10-MPK6 cascade might function downstream of phyB. This speculation was further supported by the findings that R-light-induced MPK6 activation was significantly reduced in phyB mutant seedlings and that overexpression of MKK10D rescued the suppressed cotyledon opening in the phyB mutant background. Overexpression of MKK10D increased the cotyledon opening angle in the cry1 or phyA mutant background, whereas the mpk6 and mkk10 mutants did not show reduced cotyledon opening angles in FR or B light, suggesting that phyA and CRY1 play a minor role in the MKK10-MPK6-mediated cotyledon opening signaling pathway. The conditional synergistic interaction between phyB and CRY1 or phyA in have been reported previously (Casal and Boccalandro, 1995; Casal and Mazzella, 1998).
Phosphorylation of target proteins is a primary mechanism by which MAPK cascades transduce signals. The phosphorylation and subsequent degradation of PIFs are required for seedling photomorphogenesis (Bu et al., 2011; Lorrain et al., 2008; Ni et al., 2013; Shen et al., 2008; Shen et al., 2007). Multiple phosphorylation residues have been identified in PIF proteins; for example, over 20 phosphorylation sites are present in PIF3, a founding member of the PIF subset (Bu et al., 2011; Ni et al., 2013). Multiple light-regulated kinases are likely responsible for the phosphorylation of PIF3 (Xu et al., 2015); however, to date, the only kinases that have been shown to phosphorylate PIF3 are Avena sativa AsphyA and Arabidopsis BIN2 and PPKs (Ling et al., 2017; Shin et al., 2016). Our results indicate that MPK6 is another kinase responsible for the phosphorylation and functional regulation of PIF3: MPK6 was activated by R light; MPK6 interacted with PIF3 in vitro and in vivo, and the interaction in seedlings was enhanced by R light; the activation of MPK6 by MKK10 resulted in PIF3 phosphorylation and accelerated its degradation; MKK10-induced cotyledon opening in MKK10D seedlings was greatly suppressed by the overexpression of PIF3; and the MKK10-MPK6 cascade regulates the transcription of several PIF3-regulated genes. These findings suggest that MKK10-MPK6-PIF3 is a module that functions downstream of phyB to regulate cotyledon opening in R light. PIF3 has been linked to ethylene signaling (Zhong et al., 2014). However, ethylene signaling was not conclusively shown to be involved in MKK10-MPK6-PIF3-regulated cotyledon opening in this study, as the MKK10 variant with the cotyledon opening phenotype (MKK10D) only showed lower levels of induction of ERFs (less than 2-fold for all four ERFs) (Fig. S9 at Dryad).
Inhibited hypocotyl elongation was also observed in seedlings overexpressing MKK10D and in lines crossed with these plants. However, we believe that this phenotype does not reflect a true biological function of MKK10 for two reasons: first, MKK10 was not expressed in hypocotyls, as revealed by examining the expression of GUS driven by the native promoter of MKK10; second, no obvious hypocotyl phenotype was detected in mpk6 or mkk10 seedlings.
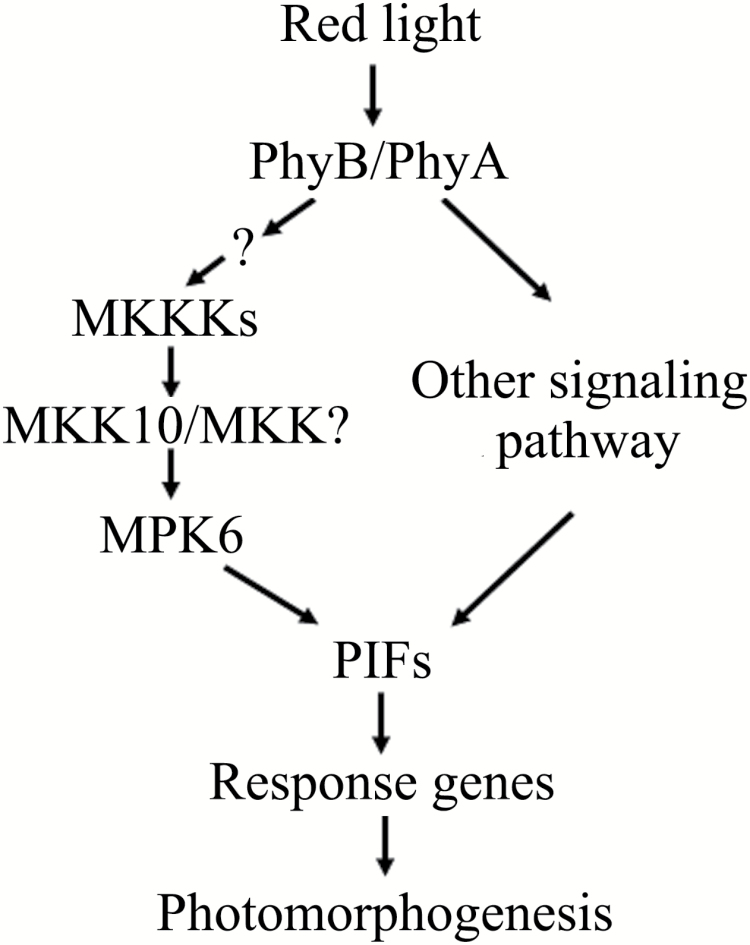
Based on the current and previous results, we propose a working model for the role of a MAPK cascade in R-light-regulated cotyledon opening (Fig. 11). When seedlings are exposed to R light, phyB perceives the light signal and is activated; active phyB in turn activates a currently unidentified MAPKKK (labeled ‘MKKKs’ in Fig. 11) directly or through an additional mediator, and the unidentified MKKK(s)-MKK10-MPK6 cascade is sequentially phosphorylated and activated; the active MPK6 phosphorylates PIF3 and accelerates its degradation; and PIF3-repressed or -induced genes are inversely regulated to promote cotyledon opening in seedlings. Perhaps the phosphorylation and degradation of other PIFs or transcription factors that are functionally redundant with PIF3 are also involved in this process. Identifying the unknown MKKK(s) and exploring the MPK6 phosphorylation sites in PIF3 and the effect of MPK6 phosphorylation on PIF3 function in future studies will help uncover the mechanisms by which the MKK10-MPK6 cascade regulates cotyledon opening in R light.
Fig. 11.
Proposed working model for MKK10-MPK6 cascade-mediated regulation of cotyledon opening in seedlings.
Data deposition
The following tables and figures are available at Dryad Data Repository: http://dx.doi.org/10.5061/dryad.hq7b8.
Table S1. Oligonucleotides used in this study.
Table S2. Accession numbers of genes used in this study.
Fig. S1. Hypocotyl length of Col-0, mpk3, and mpk6 seedlings under different light conditions.
Fig. S2. Sequence alignment of Arabidopsis MKKs.
Fig. S3. Kinase activity assay of MKK10D seedlings.
Fig. S4. MKK10 gene expression pattern in seedlings.
Fig. S5. The CRISPR construct used to create the mkk10 mutant.
Fig. S6. pif3 mutant identification.
Fig. S7. Analysis of cotyledon opening in MKK10D/cry1 mutant seedling.
Fig. S8. Co-localization analysis of MPK6 and PIF3.
Fig. S9. Q-PCR detection of ERF gene transcription in transgenic seedlings harboring MKK10 variants.
Acknowledgements
We thank Dr Giltsu Choi in the Department of Biological Sciences, KAIST, Korea, for providing us with the PIF3 OE seeds. This work was supported by grants from the National Natural Science Foundation of China (grant no. 31470351 and no. 31125006) and the State Basic Research Program (grant no. 2014CB138200) to DR.
References
- Al-Sady B, Ni W, Kircher S, Schäfer E, Quail PH. 2006. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Molecular Cell 23, 439–446. [DOI] [PubMed] [Google Scholar]
- Alvarez-Flórez F, Vidal D, Simón E. 2013. MAP-kinase activity in etiolated Cucumis sativus cotyledons: the effect of red and far-red light irradiation. Plant Physiology and Biochemistry 63, 1–7. [DOI] [PubMed] [Google Scholar]
- Andreasson E, Ellis B. 2010. Convergence and specificity in the Arabidopsis MAPK nexus. Trends in Plant Science 15, 106–113. [DOI] [PubMed] [Google Scholar]
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW. 1998. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Molecular Cell 1, 213–222. [DOI] [PubMed] [Google Scholar]
- Ang LH, Deng XW. 1994. Regulatory hierarchy of photomorphogenic loci: allele-specific and light-dependent interaction between the HY5 and COP1 loci. The Plant Cell 6, 613–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T, Chua NH. 1997. A glucocorticoid-mediated transcriptional induction system in transgenic plants. The Plant Journal 11, 605–612. [DOI] [PubMed] [Google Scholar]
- Arsovski AA, Galstyan A, Guseman JM, Nemhauser JL. 2012. Photomorphogenesis. The Arabidopsis Book 10, e0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. 2002. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983. [DOI] [PubMed] [Google Scholar]
- Bae G, Choi G. 2008. Decoding of light signals by plant phytochromes and their interacting proteins. Annual Review of Plant Biology 59, 281–311. [DOI] [PubMed] [Google Scholar]
- Bernardo-García S, de Lucas M, Martínez C, Espinosa-Ruiz A, Davière JM, Prat S. 2014. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes & Development 28, 1681–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q, Zhu L, Dennis MD, Yu L, Lu SX, Person MD, Tobin EM, Browning KS, Huq E. 2011. Phosphorylation by CK2 enhances the rapid light-induced degradation of phytochrome interacting factor 1 in Arabidopsis. The Journal of Biological Chemistry 286, 12066–12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Yang KZ, Xia C, Zhang XQ, Chen LQ, Ye D. 2010. Characterization of DUF724 gene family in Arabidopsis thaliana. Plant Molecular Biology 72, 61–73. [DOI] [PubMed] [Google Scholar]
- Casal JJ, Boccalandro H. 1995. Co-action between phytochrome B and HY4 in Arabidopsis thaliana. Planta 197, 213–218. [DOI] [PubMed] [Google Scholar]
- Casal JJ, Mazzella MA. 1998. Conditional synergism between cryptochrome 1 and phytochrome B is shown by the analysis of phyA, phyB, and hy4 simple, double, and triple mutants in Arabidopsis. Plant Physiology 118, 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon A, Shen H, Huq E. 2007. Phytochrome Interacting Factors: central players in phytochrome-mediated light signaling networks. Trends in Plant Science 12, 514–521. [DOI] [PubMed] [Google Scholar]
- Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen LO, van der Horst GT, Batschauer A, Ahmad M. 2011. The cryptochromes: blue light photoreceptors in plants and animals. Annual Review of Plant Biology 62, 335–364. [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J. 2011. Phytochrome signaling mechanisms and the control of plant development. Trends in Cell Biology 21, 664–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM. 2007. Phototropin blue-light receptors. Annual Review of Plant Biology 58, 21–45. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Colcombet J, Hirt H. 2008. Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. The Biochemical Journal 413, 217–226. [DOI] [PubMed] [Google Scholar]
- Dai Y, Wang H, Li B, Huang J, Liu X, Zhou Y, Mou Z, Li J. 2006. Increased expression of MAP KINASE KINASE7 causes deficiency in polar auxin transport and leads to plant architectural abnormality in Arabidopsis. The Plant Cell 18, 308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek PD, Fankhauser C. 2003. HFR1, a putative bHLH transcription factor, mediates both phytochrome A and cryptochrome signalling. The Plant Journal 34, 827–836. [DOI] [PubMed] [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH. 2000. HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes & Development 14, 2377–2391. [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C. 2001. The phytochromes, a family of red/far-red absorbing photoreceptors. The Journal of Biological Chemistry 276, 11453–11456. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Chory J. 2000. RSF1, an Arabidopsis locus implicated in phytochrome A signaling. Plant Physiology 124, 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Quail PH. 2010. Phytochrome functions in Arabidopsis development. Journal of Experimental Botany 61, 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Song YH, Imaizumi T. 2012. LOV domain-containing F-box proteins: light-dependent protein degradation modules in Arabidopsis. Molecular Plant 5, 573–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GI. 2014. The UV-B photoreceptor UVR8: from structure to physiology. The Plant Cell 26, 21–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W, Li B, Li S, et al. 2016. Mitogen-activated protein kinase cascade MKK7-MPK6 plays important roles in plant development and regulates shoot branching by phosphorylating PIN1 in Arabidopsis. PLoS Biology 14, e1002550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW. 2007. Light-regulated transcriptional networks in higher plants. Nature Reviews Genetics 8, 217–230. [DOI] [PubMed] [Google Scholar]
- Kevei E, Schafer E, Nagy F. 2007. Light-regulated nucleo-cytoplasmic partitioning of phytochromes. Journal of Experimental Botany 58, 3113–3124. [DOI] [PubMed] [Google Scholar]
- Khanna R, Shen Y, Toledo-Ortiz G, Kikis EA, Johannesson H, Hwang YS, Quail PH. 2006. Functional profiling reveals that only a small number of phytochrome-regulated early-response genes in Arabidopsis are necessary for optimal deetiolation. The Plant Cell 18, 2157–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Juez E, Dillon E, Magyar Z, Khan S, Hazeldine S, de Jager SM, Murray JA, Beemster GT, Bögre L, Shanahan H. 2008. Distinct light-initiated gene expression and cell cycle programs in the shoot apex and cotyledons of Arabidopsis. The Plant Cell 20, 947–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW. 2007. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. The Plant Cell 19, 731–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E. 2014. PIFs: systems integrators in plant development. The Plant Cell 26, 56–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH. 2008. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Current Biology 18, 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH. 2011. PIFs: pivotal components in a cellular signaling hub. Trends in Plant Science 16, 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Monte E, Calderon RH, Liu TL, Quail PH. 2009. Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. The Plant Cell 21, 3535–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li G, Wang H, Deng XW. 2011. Phytochrome signaling mechanisms. The Arabidopsis Book 9, e0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J-J, Li J, Zhu D, Deng XW. 2017. Noncanonical role of Arabidopsis COP1/SPA complex in repressing BIN2-mediated PIF3 phosphorylation and degradation in darkness. Proceedings of the National Academy of Sciences, USA 114, 3539–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. 2008. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. The Plant Journal 53, 312–323. [DOI] [PubMed] [Google Scholar]
- Luo Q, Lian HL, He SB, Li L, Jia KP, Yang HQ. 2014. COP1 and phyB physically interact with PIL1 to regulate its stability and photomorphogenic development in Arabidopsis. The Plant Cell 26, 2441–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAPK Group 2002. Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends in Plant Science 7, 301–308. [DOI] [PubMed] [Google Scholar]
- Matsuoka D, Nanmori T, Sato K, Fukami Y, Kikkawa U, Yasuda T. 2002. Activation of AtMEK1, an Arabidopsis mitogen-activated protein kinase, in vitro and in vivo: analysis of active mutants expressed in E. coli and generation of the active form in stress response in seedlings. The Plant Journal 29, 637–647. [DOI] [PubMed] [Google Scholar]
- McCormac AC, Wagner D, Boylan MT, Quail PH, Smith H, Whitelam GC. 1993. Photoresponses of transgenic Arabidopsis seedlings expressing introduced phytochrome B-encoding cDNAs: evidence that phytochrome A and phytochrome B have distinct photoregulatory functions. The Plant Journal 4, 19–27. [Google Scholar]
- Meng X, Zhang S. 2013. MAPK cascades in plant disease resistance signaling. Annual Review of Phytopathology 51, 245–266. [DOI] [PubMed] [Google Scholar]
- Mockler TC, Guo H, Yang H, Duong H, Lin C. 1999. Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126, 2073–2082. [DOI] [PubMed] [Google Scholar]
- Monte E, Al-Sady B, Leivar P, Quail PH. 2007. Out of the dark: how the PIFs are unmasking a dual temporal mechanism of phytochrome signalling. Journal of Experimental Botany 58, 3125–3133. [DOI] [PubMed] [Google Scholar]
- Nagatani A. 2004. Light-regulated nuclear localization of phytochromes. Current Opinion in Plant Biology 7, 708–711. [DOI] [PubMed] [Google Scholar]
- Neff MM, Chory J. 1998. Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiology 118, 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W, Xu SL, Chalkley RJ, Pham TN, Guan S, Maltby DA, Burlingame AL, Wang ZY, Quail PH. 2013. Multisite light-induced phosphorylation of the transcription factor PIF3 is necessary for both its rapid degradation and concomitant negative feedback modulation of photoreceptor phyB levels in Arabidopsis. The Plant Cell 25, 2679–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W, Xu SL, González-Grandío E, Chalkley RJ, Huhmer AFR, Burlingame AL, Wang ZY, Quail PH. 2017. PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nature Communications 8, 15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K. 1997. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes & Development 11, 2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Kim J, Lee Y, Shin J, Oh E, Chung WI, Liu JR, Choi G. 2004. Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant & Cell Physiology 45, 968–975. [DOI] [PubMed] [Google Scholar]
- Parks BM, Quail PH. 1993. hy8, a new class of Arabidopsis long hypocotyl mutants deficient in functional phytochrome A. The Plant Cell 5, 39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke A, Djamei A, Bitton F, Hirt H. 2009. A major role of the MEKK1-MKK1/2-MPK4 pathway in ROS signalling. Molecular Plant 2, 120–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JL, Zhou L, Yun BW, Nielsen HB, Fiil BK, Petersen K, Mackinlay J, Loake GJ, Mundy J, Morris PC. 2008. Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiology 148, 212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH. 1997. The phytochromes: a biochemical mechanism of signaling in sight?BioEssays 19, 571–579. [DOI] [PubMed] [Google Scholar]
- Quail PH. 2007. Phytochrome-regulated gene expression. Journal of Integrative Plant Biology 49, 11–20. [Google Scholar]
- Quail PH. 2010. Phytochromes. Current Biology 20, R504–R507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagatani A, Elich TD, Fagan M, Chory J. 1994. Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiology 104, 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. 1993. Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. The Plant Cell 5, 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Liu Y, Yang K-Y, Han L, Mao G, Glazebrook J, Zhang S. 2008. A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proceedings of the National Academy of Sciences, USA 105, 5638–5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Yang H, Zhang S. 2002. Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. The Journal of Biological Chemistry 277, 559–565. [DOI] [PubMed] [Google Scholar]
- Seluzicki A, Burko Y, Chory J. 2017. Dancing in the dark: darkness as a signal in plants. Plant, Cell & Environment 40, 2487–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi V, Raghuram B, Sinha AK, Chattopadhyay S. 2014. A mitogen-activated protein kinase cascade module, MKK3-MPK6 and MYC2, is involved in blue light-mediated seedling development in Arabidopsis. The Plant Cell 26, 3343–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Moon J, Huq E. 2005. PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. The Plant Journal 44, 1023–1035. [DOI] [PubMed] [Google Scholar]
- Shen H, Zhu L, Castillon A, Majee M, Downie B, Huq E. 2008. Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. The Plant Cell 20, 1586–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Khanna R, Carle CM, Quail PH. 2007. Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiology 145, 1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin AY, Han YJ, Baek A, et al. 2016. Evidence that phytochrome functions as a protein kinase in plant light signalling. Nature Communications 7, 11545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh MS, Kim YM, Han SJ, Song PS. 2000. REP1, a basic helix-loop-helix protein, is required for a branch pathway of phytochrome A signaling in Arabidopsis. The Plant Cell 12, 2061–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T, Nishihama R, Morikiyo K, Ishikawa M, Machida Y. 2003. NQK1/NtMEK1 is a MAPKK that acts in the NPK1 MAPKKK-mediated MAPK cascade and is required for plant cytokinesis. Genes & Development 17, 1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi F, Yoshida R, Ichimura K, Mizoguchi T, Seo S, Yonezawa M, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K. 2007. The mitogen-activated protein kinase cascade MKK3-MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. The Plant Cell 19, 805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teige M, Scheikl E, Eulgem T, Dóczi R, Ichimura K, Shinozaki K, Dangl JL, Hirt H. 2004. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Molecular Cell 15, 141–152. [DOI] [PubMed] [Google Scholar]
- Tepperman JM, Hwang YS, Quail PH. 2006. phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of Arabidopsis seedling de-etiolation. The Plant Journal 48, 728–742. [DOI] [PubMed] [Google Scholar]
- Ulm R, Baumann A, Oravecz A, Mate Z, Adam E, Oakeley EJ, Schafer E, Nagy F. 2004. Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proceedings of the National Academy of Sciences, USA 101, 1397–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Liu Y, Bruffett K, Lee J, Hause G, Walker JC, Zhang S. 2008. Haplo-insufficiency of MPK3 in MPK6 mutant background uncovers a novel function of these two MAPKs in Arabidopsis ovule development. The Plant Cell 20, 602–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S. 2007. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. The Plant Cell 19, 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Du Y, Li Y, Ren D, Song CP. 2010a. Hydrogen peroxide-mediated activation of MAP kinase 6 modulates nitric oxide biosynthesis and signal transduction in Arabidopsis. The Plant Cell 22, 2981–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li J, Hou S, Wang X, Li Y, Ren D, Chen S, Tang X, Zhou JM. 2010b. A Pseudomonas syringae ADP-ribosyltransferase inhibits Arabidopsis mitogen-activated protein kinase kinases. The Plant Cell 22, 2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZP, Xing HL, Dong L, Zhang HY, Han CY, Wang XC, Chen QJ. 2015. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biology 16, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Jia W, Zhang J. 2008. AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. The Plant Journal 54, 440–451. [DOI] [PubMed] [Google Scholar]
- Xin X, Chen W, Wang B, Zhu F, Li Y, Yanga H, Li J, Ren D. 2017. Data from: Arabidopsis MKK10-MPK6 mediates red light-regulated seedling cotyledon opening through phosphorylating PIF3. Dryad Digital Repository htpp://dx.doi.org/10.5061/dryad.hq7b8.
- Xu J, Li Y, Wang Y, Liu H, Lei L, Yang H, Liu G, Ren D. 2008. Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. The Journal of Biological Chemistry 283, 26996–27006. [DOI] [PubMed] [Google Scholar]
- Xu J, Zhang S. 2015. Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends in Plant Science 20, 56–64. [DOI] [PubMed] [Google Scholar]
- Xu X, Paik I, Zhu L, Huq E. 2015. Illuminating progress in phytochrome-mediated light signaling pathways. Trends in Plant Science 20, 641–650. [DOI] [PubMed] [Google Scholar]
- Yang KY, Kim YM, Lee S, Song PS, Soh MS. 2003. Overexpression of a mutant basic helix-loop-helix protein HFR1, HFR1-deltaN105, activates a branch pathway of light signaling in Arabidopsis. Plant Physiology 133, 1630–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Liu H, Klejnot J, Lin C. 2010. The cryptochrome blue light receptors. The Arabidopsis Book 8, e0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Mayba O, Pfeiffer A, Shi H, Tepperman JM, Speed TP, Quail PH. 2013. A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genetics 9, e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Shi H, Xue C, Wei N, Guo H, Deng XW. 2014. Ethylene-orchestrated circuitry coordinates a seedling’s response to soil cover and etiolated growth. Proceedings of the National Academy of Sciences, USA 111, 3913–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.