Summary
Although diagnostic research assays for Mycoplasma genitalium exist, none are cleared by the Food and Drug Administration. Because of associated diseases, commercial assays to diagnose this organism and guide treatment choices should be developed and made available through regulatory approval.
Keywords: Mycoplasma genitalium, Diagnostic tests, macrolide resistance, azithromycin resistance
Abstract
Background
Mycoplasma genitalium is very difficult to grow in culture but has been more able to be studied for disease associations since the advent of research molecular amplification assays. Polymerase chain reaction (PCR) and other molecular assays have demonstrated an association with adverse disease outcomes, such as urethritis or nongonococcal urethritis in men and adverse reproductive sequelae in women—for example, cervicitis, endometritis, and pelvic inflammatory disease (PID), including an association with risk for human immunodeficiency virus. The lack of commercially available diagnostic assays has limited widespread routine testing. Increasing reports of high rates of resistance to azithromycin detected in research studies have heightened the need available commercial diagnostic assays as well as standardized methods for detecting resistance markers. This review covers available molecular methods for the diagnosis of M. genitalium and assays to predict the antibiotic susceptibility to azithromycin.
Methods
A PubMed (US National Library of Medicine and National Institutes of Health) search was conducted for literature published between 2000 and 2016, using the search terms Mycoplasma genitalium, M. genitalium, diagnosis, and detection.
Results
Early PCR diagnostic tests focused on the MPa adhesion gene and the 16S ribosomal RNA gene. Subsequently, a transcription-mediated amplification assay targeting ribosomes was developed and widely used to study the epidemiology of M. genitalium. Newer methods have proliferated and include quantitative PCR for organism load, AmpliSens PCR, PCR for the pdhD gene, a PCR-based microarray for multiple sexually transmitted infections, and multiplex PCRs. None yet are cleared by the Food and Drug Administration in the United States, although several assays are CE marked in Europe. As well, many research assays, including PCR, gene sequencing, and melt curve analysis, have been developed to detect the 23S ribosomal RNA gene mutations that confer resistance to azithromycin. One recently developed assay can test for both M. genitalium and azithromycin resistance mutations at the same time.
Conclusions
It is recommended that more commercial assays to both diagnose this organism and guide treatment choices should be developed and made available through regulatory approval. Research is needed to establish the cost-effectiveness of routine M. genitalium testing in symptomatic patients and screening in all individuals at high risk of acquiring and transmitting sexually transmitted infections.
The mollicute Mycoplasma genitalium is a fastidious obligate intracellular bacterium and is the smallest prokaryote capable of self-replication [1]. Its tiny genome is 580 kb long and contains 482 protein-coding genes [1, 2]. Although very difficult to grow, M. genitalium was first cultured by Tully et al [3] from urethral specimens obtained from men with urethritis (nongonococcal) in 1981. Tissue culture is used to enhance the recovery of the organism, but slow growth and cumbersome detection methods preclude the method’s routine use for diagnosis [4]. The bacterium has been able to be better studied for disease associations since the advent of research molecular amplification assays, such as polymerase chain reaction (PCR) and transcription-mediated amplification assays (TMAs) [5–10].
Since the availability of such molecular assays, the organism has been associated with many adverse disease outcomes, such as urethritis or nongonococcal urethritis in men and many adverse reproductive sequelae in women, including cervicitis, endometritis, and PID [11–16]. There is also a strong association with risk for human immunodeficiency virus [17]. The lack of a Food and Drug Administration (FDA)–cleared diagnostic assay, however, has severely limited the availability of testing in symptomatic and asymptomatic high-risk individuals. Increasing the concerns for treatment of the diseases caused by M. genitalium are reports of rising rates of resistance to azithromycin [18]. The emerging role for M. genitalium as a sexually transmitted infection (STI) pathogen has been comprehensively reviewed by Taylor-Robinson and Jensen [19] and Manhart [11]. This review focuses on new molecular methods for the diagnosis of M. genitalium and assays to predict antibiotic susceptibility to azithromycin.
METHODS
A PubMed (US National Library of Medicine and the National Institutes of Health) search was conducted for literature published between 2000 and 2016 using the following search terms: Mycoplasma genitalium, M. genitalium, diagnosis, and detection. A list of 1003 published articles was retrieved. Articles were reviewed and analyzed for diagnostic methods by amplification and molecular methods, as well as for diagnosis of resistance markers.
RESULTS AND DISCUSSION
Diagnosis of Syndromes Associated With M. genitalium
Urethritis in men was the first syndrome associated with Mycoplasma. The diagnosis and causes of urethritis are well established and has been recently reviewed by Bachmann et al [20]. That review covered M. genitalium as an emerging cause of urethritis. Although many studies have used the urethral Gram stain criterion of ≥5 polymorphonuclear leukocytes per high-power field to diagnose urethritis, many chlamydia and possibly M. genitalium infections may be missed by this criterion, and now the Centers for Disease Control and Prevention recommend using ≥2 polymorphonuclear leukocytes per high-power field as the diagnostic threshold [20]. Persistence of urethritis after therapy has been especially associated with M. genitalium and may be due to suspected antibiotic resistance [21–22]. However, there are other possible causes, including reinfection, noncompliance with therapy, drug resistance, or postinfectious immunologic reaction. Diagnosis of Mycoplasma infection and other STIs in women are less likely to be associated with specific symptomatic states and are often asymptomatic. Mycoplasma can be found in the vagina, cervix, and endometrium. Much new evidence has accumulated in the past few years implicating M. genitalium as an important cause of morbid effects in women [11, 12, 15]. (See article on the subject in this supplement.)
Diagnosis of M. genitalium by Molecular Methods
Although there are no FDA-cleared M. genitalium assays currently available in the United States, research assays based on nucleic acid amplification tests (NAATs) have been in use since the early 1990s and are highly sensitive and specific for detecting many infectious organisms, such as chlamydia, gonorrhea, and trichomonas, as well as M. genitalium [5–9]. The assay formulated by Jensen and colleagues [6–8] targeting a conserved region of the 16S ribosomal RNA (rRNA) gene have been widely used in research-based studies (Table 1).
Table 1.
Selected Types of Amplified Molecular Tests for Detection of Mycoplasma genitalium
| Type of Test | Name of Test | Gene Target | Author | CE Mark |
|---|---|---|---|---|
| PCR | NA | MgPA | Deguchi et al [5] | No |
| PCR | NA | MgPA; 16S rRNA |
Jensen et al [6–8] | No |
| PCR | Duplex PCR | MgPA; 16S rRNA |
Hardick et al [9] | No |
| TMA | Aptima MG | 23S rRNA | Hologic 23]/Getman et al [24] | Yes |
| Multiplex PCR | STD Finder | MLPA | Muvunyi et al [25] | No |
| Multiplex PCR | Bio-Rad Dx | NA | Le Roy et al [26] | No |
| qPCR | Real-time PCR | pdhD | Müller et al [27] | No |
| Multiplex qPCR | AmpliSense | gyrB | Rumyantseva et al [28] | Yes |
| DNA microarray | Multiplex PCR-based microarray | 16S rRNA | Cao et al [29] | No |
| qPCR | Diagenode S-DiaMGTV | Mg219 gene | van Alphen et al [30] | Yes |
| Real-time PCR comparison |
TIB MOLBIOL/ Roche and Diagenode M. genitalium |
Mg219 and gap gene fragments | Le Roy et al [31] | No |
| Mulitplex qPCR PlexZyme | SpeeDX | 23S rRNA; detects resistant mutations |
Tabrizi et al [44] | Yes |
Abbreviations: MLPA, multiplex ligation-dependent probe amplification; NA, not applicable; PCR, polymerase chain reaction; qPCR, quantitative PCR; rRNA, ribosomal RNA; TMA, transcription-mediated amplification.
The process for a laboratory to develop and use a research diagnostic test within a single laboratory requires extensive testing to demonstrate its accuracy and is known as a laboratory-developed test (LDT). LDTs are laboratory tests that hospitals, academic, and clinical laboratories develop as testing services according to their own procedures. They are often created in response to unmet clinical needs and may be used for precise diagnosis and for monitoring and guiding patient treatment. LDTs are also used to diagnose and assess diseases and disorders for which no FDA-authorized test kit currently exists, such as for rare diseases.
For laboratories in the United States to use research NAATs developed by a company, often termed “research-use only” (RUO) tests, for diagnosis and treatment of disease in patients seen in clinical settings, each laboratory must be internally validate the assay, similar to that procedure required for a LDT. This process involves each individual laboratory performing its own study to compare the RUO assay with another assay for the analyte or comparing the laboratory’s individual results with those obtained by another laboratory, much like what the FDA would require for an LDT. This is an expensive process and not easily accomplished.
The next step on the path for performing commercialization is for a company to submit its test reagents to the FDA to obtain an analyte-specific reagent (ASR) status. This means that the company can sell the reagents but not as a “kit” with controls or controls. A laboratory would still have to perform a validation study, similar to the process for developing a LDR, to use the results of this assay clinically, that is, for treating patients Very few laboratories have done this. Only 1 company has achieved the ASR status in the United States with a TMA assay originally designed and offered as an RUO assay by the GenProbe/Hologic Company [9, 10]. Hardick et al reported the overall sensitivity and specificity of the RUO product in 607 persons as 98.1% and 98.1%, respectively, compared with 2 research assays (100% and 97.9% in urine samples from male subjects, respectively and 98.4% in vaginal swab samples) [9]. This M. genitalium TMA ASR assay is also now CE marked in Europe [23, 24]. It has been widely used for much Mycoplasma research and to study the epidemiology of M. genitalium [10, 13, 14, 21, 22]. The next step would be the performance of a FDA clinical trial, which is a costly process.
The term CE originated as an abbreviation of conformité Européenne, meaning European conformity, but the term is not defined as such in the relevant legislation. The CE marking is a symbol of free marketability in the European Economic Area (internal market). The CE mark on an assay in Europe is a manufacturer’s declaration that the product complies with the essential requirements of the relevant European protection legislations. It signifies that products sold in the European Economic Area have been assessed to meet high safety, health, and environmental protection requirements. It ensures the free movement of the product within the European Union. No clinical validation against a reference standard is needed.
Fortunately, a large group of other diagnostic molecular methods, described below, have been developed both internationally and nationally, and some are beginning to receive the CE mark. Some of these hold great promise for diagnostic options for both research and clinical use. Several very new assays have also been developed (Table 1).
When a new multiplex PCR assay, the STDFinder assay, was studied, it was reported to be 100% specific and 100% sensitive in detecting Mycoplasma, but only 3 cases were detected [25]. The multiplex ligation-dependent probe amplification technology uses a single assay, which is able to amplify up to 45 different targets simultaneously using 1 universal primer set in the final amplification.
An early evaluation of the performance of the Bio-Rad Dx CT/NG/MG assay for the detection of Chlamydia trachomatis, M. genitalium, and Neisseria gonorrhoeae in urogenital samples indicated that, compared with an in-house TaqMan PCR assay for M. genitalium, the new assay had a specificity for detecting M. genitalium of 99.5% for swab samples and 100% for urine samples from female subjects, but very few cases of Mycoplasma were detected [26].
Müller et al developed and validated a real-time quantitative PCR (qPCR) assay to determine the M. genitalium bacterial load in endocervical swab samples, based on amplification of the pdhD gene that encodes dihydrolipoamide dehydrogenase, using the Rotor-Gene platform [27]. The assay demonstrated a detection limit of 300 genome copies/mL using the G37 M. genitalium genomic DNA (ASTCC 33530D). The assay amplified an 82–base pair fragment of the pdhD gene of M. genitalium (nucleotides corresponding to the sequence with GenBank No. L43967.2). The assay was specific for M. genitalium DNA and did not amplify the DNA from other Mycoplasma and Ureaplasma species, compared with a commercial assay (Sacace Biotechnologies). Only endocervical swab samples were tested. In 119 positive endocervical swab samples, from a total of 1600, the M. genitalium loads ranged between <300 and 3 240000 copies/mL, with a limit of detection of 10 copies per reaction (or 300 copies/mL of cervical swab sample suspension). This limit was similar to that found by others in urine samples from male subjects [8]. This qPCR assay demonstrated good detection, reproducibility, and specificity and is useful for qualitative and quantitative analyses of M. genitalium in endocervical and probably other genital specimens [27].
The new CE-marked multiplex real-time AmpliSens N. gonorrhoeae/C.trachomatis/M.genitalium/T.vaginalis-MULTIPRIME-FRT PCR assay was evaluated and compared with 4 Aptima tests: the COMBO 2 assay, the Trichomonas vaginalis assay (FDA approved), and 2 different APTIMA M. genitalium RUO assays [28], one of which was used for for discrepancy analysis. Overall, for M. genitalium, the multiplex AmpliSens PCR assay demonstrated 100% specificity, but suboptimal sensitivity (81.9%; 95% confidence interval, 70.7%–89.7%). These results were in concordance with characteristics of a singleplex AmpliSens M. genitalium PCR assay evaluated earlier. It is of interest that a lower sensitivity (76.5%) was obtained for vaginal samples, is in contrast to findings of many previous studies using other NAATs [28].
A 2015 study documented a PCR-based microarray including 31 specific probes to simultaneously detect a diverse panel of 17 STI-associated infections, including N. gonorrhoeae, C. trachomatis, M. genitalium, M. hominis, Ureaplasma, herpes simplex virus types 1 and 2, and 10 human papillomavirus types [29]. The microarray was specific, because commensal and other closely related urogenital microbes did not cross-react with the microarray probes. It was 10 times more sensitive than multiplex PCR. Of 158 suspected human papillomavirus specimens evaluated, Ureaplasma was detected in 21, M. hominis in 15, C. trachomatis in 4, and N. gonorrhoeae in 1. However, no M. genitalium seemed to be detected. This research may be one of the first reports of a high-throughput detection system that identifies common pathogens associated with STIs from clinical samples [29].
The fairly new Diagenode S-DiaMGTV qPCR commercial kit allows qualitative detection and differentiation of M. genitalium and T. vaginalis by qPCR in clinical specimens [30]. In a small study where the prevalence of Mycoplasma was 3.7%, it demonstrated almost perfect agreement with reference assays, and more studies will be welcome. One report compared the TIB MOLBIOL LightMix Kit and the Diagenode real-time PCR kit for M. genitalium with the in-house TaqMan RTPCR used routinely for the M. genitalium diagnostic [31]. Concordances of 96% and 93% were found for the TIB MOLBIOL and the Diagenode assays, respectively, compared with the results of the in-house assay.
Detection and Diagnosis of Antibiotic Resistance in M. genitalium
Mutations that confer antibiotic resistance to macrolides for M genitalium are concentrated in nucleotide positions 2058 and 2059 in region V of the 23S rRNA gene and are adenine to guanine, cytosine or thymidine transitions [18]. The locations of the 5 mutations that confer resistance are A2058G, A2058C, A2058T, A2059G, and A2059C. Jensen et al [18, 32] first described azithromycin failure in patients with M. genitalium associated with induced macrolide resistance and mutations in the region 23S rRNA gene. There have been several published research assays that can detect macrolide resistance in samples known to be positive for M. genitalium, using detection of the 23s rRNA gene mutations that are associated with resistance by PCR and melt curve analysis [32–36]. However, none of these assays are available commercially. All of these resistance studies confirm the high prevalence rate of mutations seen in other studies [24, 37].
Normally, macrolide antibiotics such as azithromycin inhibit bacterial multiplication by binding to the 50S subunit of the ribosome, which includes both the 5S and 23S subunits to prevent translation of messenger RNA and prevent protein synthesis. Resistance occurs by the mutational changes that prevent binding of the antibiotic, and thus protein synthesis is not prevented, as would happen when the organism is sensitive to the antibiotic.
Getman et al [24] reported on the use of the CE-marked TMA assay (GenProbe/Hologic) to determine prevalence of M. genitalium infections. and coinfections with other sexually transmitted organisms and the frequency of a macrolide antibiotic resistance phenotype were determined in urogenital specimens collected from 946 care-seeking female and male subjects in the United States. Neither the ASR nor the CE-marked TMA assay detects the resistance mutations, but sequencing was used to assess macrolide antibiotic resistance among M. genitalium–positive subjects in the study by Getman et al [24]. Prevalence rates for M. genitalium were 16.1% for female and 17.2% for male subjects. The macrolide-resistant phenotype was found in 50.8% of female and 42% of male subjects, confirming the high prevalence of resistance in other studies [32–37]
Need for Detection and Resistance Testing
With the rapid emergence of macrolide-resistant strains of M. genitalium [37–43], it is recommended that all positive M genitalium assays be reflexed to a test to determine mutations for macrolide resistance. This can be performed by Sanger sequencing [32], pyrosequencing [37], high-resolution melt analysis [32], or use of FRET probes [34, 35, 36],
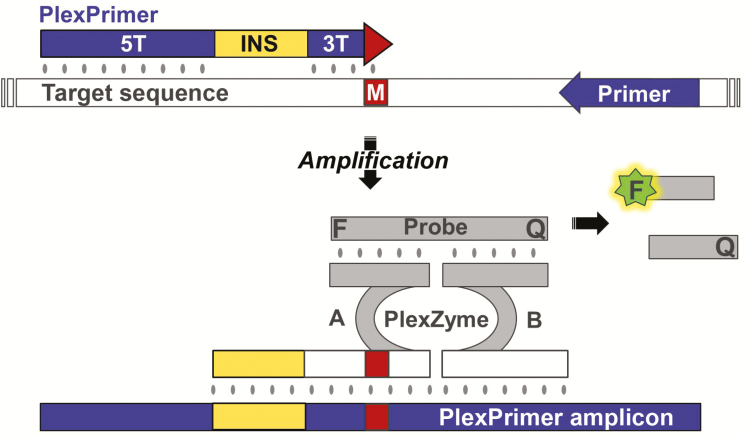
A combination assay that can rapidly detect the organism and at the same time determine the molecular resistance markers for azithromycin, a standard antibiotic used for therapy, is desperately needed. One such assay recently developed is the 23S assay, which uses novel PlexZyme and PlexPrime technology (SpeeDX) [44]. A multiplex qPCR assay for detection of M. genitalium and the 5 mutations associated with macrolide resistance uses novel PlexZyme (formerly known as MNAzyme) technology and PlexPrimers that selectively amplify target sequences, coupled to enable high-level multiplexing of mutant detection (Figure 1).
Figure 1.
PlexPrimers are combined with PlexZymes for mutation-specific amplification and detection with quantitative polymerase chain reaction (qPCR). The PlexPrimer contains 3 sections; a long 5’ target-recognition sequence (5T), a short 3’ target-specific region (3T) matched to the mutation sequence (M) and an intervening insert sequence (INS), which does not bind to the target rather increases the specificity of the 3T region for the mutant over the wild type sequence. During amplification, the INS is incorporated into the PlexPrimer amplicons, and these can be detected in real time using PlexZymes, nucleic acid enzymes that form, from their component partzymes (A and B), only when matched PlexPrimer amplicons are present. Each partzyme contains a probe-binding arm, a partial catalytic core, and a target-binding arm, oriented such that partzyme A binds to the amplicon in the region containing the INS, and partzyme B binds adjacently downstream. Catalytically active PlexZymes bind and cleave reporter probes separating a fluorophore (F) and quencher (Q), resulting in signal generation.
The assay was evaluated in 400 samples from 254 consecutively infected participants, undergoing tests of cure. Of the M. genitalium–positive samples in the test of cure, 56% showed a macrolide resistance mutation. The MG 23S assay was compared with a reference qPCR method for M. genitalium detection and high-resolution melt analysis and sequencing for detection of macrolide resistance mutations; its sensitivity and specificity were 99.1% and 98.5% for M. genitalium detection and 97.4% and 100% for macrolide resistance. Recent use of this assay in our United States research studies has demonstrated similar high prevalence of macrolide resistance patterns: 50% in a prospective study and 36% in a retrospective study in over a total of 400 urogenital samples from female subjects (Justin Hardick, personal communication). The ability to diagnose M. genitalium infections and genetic markers for macrolide resistance simultaneously is a clinically important advance in new diagnostic test technology.
Moxifloxin is often recommended for treatment for patients in whom azithromycin fails, but fluoroquinolone resistance has now been well documented [39, 43, 45–47]. Microbiological failure of moxifloxacin treatment in patients has been associated with the detection of fluoroquinolone resistance–associated mutations in their clinical specimens [27]. If the resistance to moxifloxacin increases to high levels, it may be necessary to further develop diagnostic tests for detecting these resistance markers as well.
CONCLUSIONS
Currently, treatment of most M. genitalium infections occurs mainly in the context of syndromic management for urethritis, cervicitis, and PID, owing to the lack of diagnostic test availability in the United States. Testing is readily available in Europe but it is not clear how commonly it is used. A substantial body of clinical research based on RUO diagnostic assays has revealed much about the epidemiology and disease associations of M. genitalium. Now that we have learned that this organism is significantly associated with urethritis in men and significant morbid effects in women, including PID and increased susceptibility to human immunodeficiency virus infection, and that treatment with recommended antibiotics for the common STI syndromes often fail, it is time for further development of commercially available assays for use in diagnosing M. genitalium infection and guiding treatment decisions [45]. Determining the cost-effectiveness of widespread diagnosis of M. genitalium in symptomatic and asymptomatic high-risk individuals is urgently needed to drive this agenda forward, as are more commercially available, FDA-cleared assays.
Notes
Acknowledgment. This work is an outcome of a Mycoplasma genitalium Experts Technical Consultation that was supported by the Division of Microbiology and Infectious Diseases of the National Institute of Allergy and Infectious Diseases, Contract HHSN272201300012I, with the University of Alabama at Birmingham Sexually Transmitted Infections Clinical Trials Group.
Financial support. This work was supported by the National Institute of Biomedical Imaging and Bioengineering (grant U54EB007958) and the National Institute of Allergy and Infectious Diseases (grant U-01068613), National Institutes of Health.
Supplement sponsorship. This work is part of a supplement sponsored by the University of Alabama at Birmingham Sexually Transmitted Infections Clinical Trials Unit and the National Institute of Allergy and Infectious Diseases.
Potential conflicts of interest. C. A. G. received M. genitalium test kits from GenProbe//Hologic and SpeeDX companies for research studies. The author has submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Fraser CM, Gocayne JD, White O et al. . The minimal gene complement of Mycoplasma genitalium. Science 1995; 270:397–403. [DOI] [PubMed] [Google Scholar]
- 2. Glass JI, Assad-Garcia N, Alperovich N et al. . Essential genes of a minimal bacterium. Proc Natl Acad Sci U S A 2006; 103:425–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tully JG, Taylor-Robinson D, Cole RM, Rose DL. A newly discovered Mycoplasma in the human urogenital tract. Lancet 1981; 1:1288–91. [DOI] [PubMed] [Google Scholar]
- 4. Hamasuna R, Osada Y, Jensen JS. Isolation of Mycoplasma genitalium from first-void urine specimens by coculture with Vero cells. J Clin Microbiol 2007; 45:847–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Deguchi T, Yoshida T, Yokoi S et al. . Longitudinal quantitative detection by real-time PCR of Mycoplasma genitalium in first-pass urine of men with recurrent nongonococcal urethritis. J Clin Microbiol 2002; 40:3854–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jensen JS, Uldum SA, Søndergård-Andersen J, Vuust J, Lind K. Polymerase chain reaction for detection of Mycoplasma genitalium in clinical samples. J Clin Microbiol 1991; 29:46–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jensen JS, Borre MB, Dohn B. Detection of Mycoplasma genitalium by PCR amplification of the 16S rRNA gene. J Clin Microbiol 2003; 41:261–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jensen JS, Björnelius E, Dohn B, Lidbrink P. Use of TaqMan 5’ nuclease real-time PCR for quantitative detection of Mycoplasma genitalium DNA in males with and without urethritis who were attendees at a sexually transmitted disease clinic. J Clin Microbiol 2004; 42:683–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hardick J, Giles J, Hardick A, Hsieh YH, Quinn T, Gaydos C. Performance of the gen-probe transcription-mediated [corrected] amplification research assay compared to that of a multitarget real-time PCR for Mycoplasma genitalium detection. J Clin Microbiol 2006; 44:1236–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Munson E, Bykowski H, Munson KL et al. . Clinical laboratory assessment of Mycoplasma genitalium transcription-mediated amplification using primary female urogenital specimens. J Clin Microbiol 2016; 54:432–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Manhart LE. Mycoplasma genitalium: an emergent sexually transmitted disease? Infect Dis Clin North Am 2013; 27:779–92. [DOI] [PubMed] [Google Scholar]
- 12. Manhart LE, Holmes KK, Hughes JP, Houston LS, Totten PA. Mycoplasma genitalium among young adults in the United States: an emerging sexually transmitted infection. Am J Public Health 2007; 97:1118–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gaydos C, Maldeis NE, Hardick A, Hardick J, Quinn TC. Mycoplasma genitalium compared to chlamydia, gonorrhoea and trichomonas as an aetiological agent of urethritis in men attending STD clinics. Sex Transm Infect 2009; 85:438–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gaydos C, Maldeis NE, Hardick A, Hardick J, Quinn TC. Mycoplasma genitalium as a contributor to the multiple etiologies of cervicitis in women attending sexually transmitted disease clinics. Sex Transm Dis 2009; 36:598–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lis R, Rowhani-Rahbar A, Manhart LE. Mycoplasma genitalium infection and female reproductive tract disease: a meta-analysis. Clin Infect Dis 2015; 61:418–26. [DOI] [PubMed] [Google Scholar]
- 16. McGowin CL, Anderson-Smits C. Mycoplasma genitalium: an emerging cause of sexually transmitted disease in women. PLoS Pathog 2011; 7, e1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Napierala Mavedzenge S, Weiss HA. Association of Mycoplasma genitalium and HIV infection: a systematic review and meta-analysis. AIDS 2009; 23:611–20. [DOI] [PubMed] [Google Scholar]
- 18. Jensen JS, Bradshaw CS, Tabrizi SN, Fairley CK, Hamasuna R. Azithromycin treatment failure in Mycoplasma genitalium-positive patients with nongonococcal urethritis is associated with induced macrolide resistance. Clin Infect Dis 2008; 47:1546–53. [DOI] [PubMed] [Google Scholar]
- 19. Taylor-Robinson D, Jensen JS. Mycoplasma genitalium: from chrysalis to multicolored butterfly. Clin Microbiol Rev 2011; 24:498–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bachmann LH, Manhart LE, Martin DH et al. . Advances in the understanding and treatment of male urethritis. Clin Infect Dis 2015; 61:S763–9. [DOI] [PubMed] [Google Scholar]
- 21. Schwebke JR, Rompalo A, Taylor S et al. . Re-evaluating the treatment of nongonococcal urethritis: emphasizing emerging pathogens—a randomized clinical trial. Clin Infect Dis 2011; 52:163–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Seña AC, Lensing S, Rompalo A et al. . Chlamydia trachomatis, Mycoplasma genitalium, and Trichomonas vaginalis infections in men with nongonococcal urethritis: predictors and persistence after therapy. J Infect Dis 2012; 206:357–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aptima Mycoplasma genitalium assay [package insert]. CE marked, 20 January 2015. Document AW-14170-001, Rev002. San Diego, CA: Hologic, 2015.
- 24. Getman D, Jiang A, O’Donnell M, Cohen S. Mycoplasma genitalium prevalence, coinfection, and macrolide antibiotic resistance frequency in a multicenter clinical study cohort in the United States. J Clin Microbiol 2016; 54:2278–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Muvunyi CM, Dhont N, Verhelst R et al. . Evaluation of a new multiplex polymerase chain reaction assay STDFinder for the simultaneous detection of 7 sexually transmitted disease pathogens. Diagn Microbiol Infect Dis 2011; 71:29–37. [DOI] [PubMed] [Google Scholar]
- 26. Le Roy C, Le Hen I, Clerc M et al. . The first performance report for the Bio-Rad Dx CT/NG/MG assay for simultaneous detection of Chlamydia trachomatis, Neisseria gonorrhoeae and Mycoplasma genitalium in urogenital samples. J Microbiol Methods 2012; 89:193–7. [DOI] [PubMed] [Google Scholar]
- 27. Müller EE, Venter JM, Magooa MP, Morrison C, Lewis DA, Mavedzenge SN. Development of a rotor-gene real-time PCR assay for the detection and quantification of Mycoplasma genitalium. J Microbiol Methods 2012; 88:311–5. [DOI] [PubMed] [Google Scholar]
- 28. Rumyantseva T, Golparian D, Nilsson CS et al. . Evaluation of the new AmpliSens multiplex real-time PCR assay for simultaneous detection of Neisseria gonorrhoeae, Chlamydia trachomatis, Mycoplasma genitalium, and Trichomonas vaginalis. APMIS 2015; 123:879–86. [DOI] [PubMed] [Google Scholar]
- 29. Cao B, Wang S, Tian Z et al. . DNA microarray characterization of pathogens associated with sexually transmitted diseases. PLoS One 2015; 10: e0133927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van Alphen PTW, van Herk CMC, Roymans RTJM, van de Bovenkamp JHB. Clinical validation of the Diagenode S-DiaMGTV qPCR kit for detection of Mycoplasma genitalium and Trichomonas vaginalis ECCMID poster; 2016. http://www.diagenodediagnostics.com/en/list-products-3.php. Accessed 23 January 2017. [Google Scholar]
- 31. Le Roy C, Pereyre S, Bébéar C. Evaluation of two commercial real-time PCR assays for detection of Mycoplasma genitalium in urogenital specimens. J Clin Microbiol 2014; 52:971–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jensen JS. Protocol for the detection of Mycoplasma genitalium by PCR from clinical specimens and subsequent detection of macrolide resistance- mediating mutations in region V of the 23S rRNA gene. Methods Mol Biol 2012; 903:129–39. [DOI] [PubMed] [Google Scholar]
- 33. Twin J, Jensen JS, Bradshaw CS et al. . Transmission and selection of macrolide resistant Mycoplasma genitalium infections detected by rapid high resolution melt analysis. PLoS One 2012; 7:e35593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Touati A, Peuchant O, Jensen JS, Bébéar C, Pereyre S. Direct detection of macrolide resistance in Mycoplasma genitalium isolates from clinical specimens from France by use of real-time PCR and melting curve analysis. J Clin Microbiol 2014; 52:1549–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wold C, Sorthe J, Hartgill U, Olsen AO, Moghaddam A, Reinton N. Identification of macrolide-resistant Mycoplasma genitalium using real-time PCR. J Eur Acad Dermatol Venereol 2015; 29:1616–20. [DOI] [PubMed] [Google Scholar]
- 36. Kristiansen GQ, Lisby JG, Schønning K. A 5’ nuclease genotyping assay for identification of macrolide-resistant Mycoplasma genitalium in clinical specimens. J Clin Microbiol 2016; 54:1593–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Salado-Rasmussen K, Jensen JS. Mycoplasma genitalium testing pattern and macrolide resistance: a Danish nationwide retrospective survey. Clin Infect Dis 2014; 59:24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Manhart LE. Diagnostic and resistance testing for Mycoplasma genitalium: what will it take? Clin Infect Dis 2014; 59:31–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tagg KA, Jeoffreys NJ, Couldwell DL, Donald JA, Gilbert GL. Fluoroquinolone and macrolide resistance-associated mutations in Mycoplasma genitalium. J Clin Microbiol 2013; 51:2245–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nijhuis RH, Severs TT, Van der Vegt DS, Van Zwet AA, Kusters JG. High levels of macrolide resistance-associated mutations in Mycoplasma genitalium warrant antibiotic susceptibility-guided treatment. J Antimicrob Chemother 2015; 70:2515–8. [DOI] [PubMed] [Google Scholar]
- 41. Lau A, Bradshaw CS, Lewis D et al. . The efficacy of azithromycin for the treatment of genital Mycoplasma genitalium: a systematic review and meta-analysis. Clin Infect Dis 2015; 61:1389–99. [DOI] [PubMed] [Google Scholar]
- 42. Walker J, Fairley CK, Bradshaw CS et al. . Mycoplasma genitalium incidence, organism load, and treatment failure in a cohort of young Australian women. Clin Infect Dis 2013; 56:1094–100. [DOI] [PubMed] [Google Scholar]
- 43. Deguchi T, Yasuda M, Horie K et al. . Drug resistance-associated mutations in Mycoplasma genitalium in female sex workers, Japan. Emerg Infect Dis 2015; 21:1062–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tabrizi SN, Tan LY, Walker S et al. . Multiplex assay for simultaneous detection of Mycoplasma genitalium and macrolide resistance using PlexZyme and PlexPrime technology. PLoS One 2016; 11: e0156740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pond MJ, Nori AV, Witney AA, Lopeman RC, Butcher PD, Sadiq ST. High prevalence of antibiotic-resistant Mycoplasma genitalium in nongonococcal urethritis: the need for routine testing and the inadequacy of current treatment options. Clin Infect Dis 2014; 58:631–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kikuchi M, Ito S, Yasuda M et al. . Remarkable increase in fluoroquinolone-resistant Mycoplasma genitalium in Japan. J Antimicrob Chemother 2014; 69:2376–82. [DOI] [PubMed] [Google Scholar]
- 47. Couldwell DL, Tagg KA, Jeoffreys NJ, Gilbert GL. Failure of moxifloxacin treatment in Mycoplasma genitalium infections due to macrolide and fluoroquinolone resistance. Int J STD AIDS 2013; 24:822–8. [DOI] [PubMed] [Google Scholar]