CIA8 is involved in bicarbonate uptake in the carbon dioxide-concentrating mechanism (CCM) of Chlamydomonas reinhardtii.
Keywords: Bile acid transporter, CCM, Chlamydomonas reinhardtii, CO2 assimilation, inorganic carbon uptake
Abstract
The supply of inorganic carbon (Ci) at the site of fixation by Rubisco is a key parameter for efficient CO2 fixation in aquatic organisms including the green alga, Chlamydomonas reinhardtii. Chlamydomonas reinhardtii cells, when grown on limiting CO2, have a CO2-concentrating mechanism (CCM) that functions to concentrate CO2 at the site of Rubisco. Proteins thought to be involved in inorganic carbon uptake have been identified and localized to the plasma membrane or chloroplast envelope. However, current CCM models suggest that additional molecular components are involved in Ci uptake. In this study, the gene Cia8 was identified in an insertional mutagenesis screen and characterized. The protein encoded by Cia8 belongs to the sodium bile acid symporter subfamily. Transcript levels for this gene were significantly up-regulated when the cells were grown on low CO2. The cia8 mutant exhibited reduced growth and reduced affinity for Ci when grown in limiting CO2 conditions. Prediction programs localize this protein to the chloroplast. Ci uptake and the photosynthetic rate, particularly at high external pH, were reduced in the mutant. The results are consistent with the model that CIA8 is involved in Ci uptake in C. reinhardtii.
Introduction
Green algae and other photosynthetic aquatic organisms are often exposed to low and fluctuating CO2 conditions in the natural environment. CO2 availability for these organisms can be restricted by, among other factors: slow diffusion of gases in water, slow interconversion of the two inorganic carbon (Ci) forms (CO2 and HCO3–), and pH changes. Consequently, almost all unicellular aquatic photosynthetic organisms have evolved a CO2-concentrating mechanism (CCM) inducible under limiting CO2 conditions, to concentrate Ci effectively for fixation by Rubisco (Giordano et al., 2005). In a current CCM model for the green alga Chlamydomonas reinhardtii (Jungnick et al., 2014; Wang and Spalding, 2014) it is thought that bicarbonate transporters on the plasma membrane and chloroplast envelope are key components of the CCM allowing the movement of Ci, particularly HCO3–, through the membranes. Other CCM components include carbonic anhydrase enzymes that interconvert CO2 and HCO3– (Mitra et al., 2005; Moroney et al., 2011), and a prominent compartment called the pyrenoid which is a dense protein complex in the chloroplast. The pyrenoid is where Rubisco is sequestered under limiting CO2 conditions (Kuchitsu et al., 1988; Rawat et al., 1996; Borkhsenious et al., 1998; Mackinder et al., 2016). An extensive network of thylakoid tubules and mini-tubules is associated with the pyrenoid (Engel et al., 2015; Mackinder et al., 2016) presumably to provide a pathway for HCO3– to enter into the pyrenoid. The carbonic anhydrase, CAH3, is found in these tubules and it is hypothesized that CAH3 converts the HCO3– within the lumen to CO2 for fixation (Karlsson et al., 1998; Moroney and Ynalvez, 2007; Blanco-Rivero et al., 2012).
Chlamydomonas reinhardtii cells grown under high CO2 conditions (5% v/v in air) exhibit a low affinity for Ci. When high-CO2-acclimated cells are exposed to low CO2 conditions (0.04% v/v), induction of high affinity transporters has been reported. While CO2 will readily diffuse across membranes in the cell (Gutknecht et al., 1977), numerous studies have since established the need for an active transport system to facilitate the movement of Ci (particularly HCO3–) to the point of fixation by Rubisco in low CO2 cells (Moroney et al., 1987; Sültemeyer et al., 1988; Badger et al., 1994; Ohnishi et al., 2010). In addition, molecular and physiological studies have also confirmed the occurrence of multiple forms of Ci transporters on the plasma membrane and chloroplast envelope of microalgal cells (Amoroso et al., 1998; Duanmu et al., 2009; Atkinson et al., 2015; Gao et al., 2015; Yamano et al., 2015). In marine cyanobacteria, HCO3– transport at the plasma membrane is often coupled to the external Na+ ion concentration (Price et al., 2008). In freshwater environments where C. reinhardtii is found, transport is thought to be H+ coupled since Na+ is relatively low (Morth et al., 2011; Taylor et al., 2012). Consequently, genomic studies with C. reinhardtii and Volvox carteri have revealed the presence of both H+- and Na+-coupled transporters at least for sulphate and phosphate (Pootakham et al., 2010). It is not yet clear whether these ions are also needed for bicarbonate uptake in C. reinhardtii.
To date, one low and two high affinity bicarbonate transport proteins in C. reinhardtii are characterized and known to be functional under low CO2 conditions. The first, a high-light-activated protein (HLA3) is an ATP-binding cassette (ABC)-type transporter of the Multi-Drug Resistance protein family localized to the plasma membrane (Im and Grossman, 2002). The Hla3 transcript is induced by both high light and low CO2 conditions and is controlled by the CCM1 ‘master regulator’ encoded by the Cia5 gene. Duanmu et al. (2009) showed in HLA3 RNAi knockdown mutants, a significant reduction in Ci affinity and Ci uptake, supporting the role of the protein in HCO3– transport. The second, the novel transporter LCI1, is a relatively small protein with little homology to other transmembrane proteins in the databases. LCI1 is strongly up-regulated under low CO2 conditions and has been localized to the plasma membrane (Ohnishi et al., 2010). In this study with LCI1, the authors also confirmed increased Ci uptake by overexpressing LCI1 protein in the Lcr1 (C. reinhardtii strain lacking a MYB transcription factor) background (Ohnishi et al., 2010). Thus, HLA3 and LCI1 are thought to be Ci transporters located on the plasma membrane.
The third transporter, NAR1.2 (also known as LCIA), is a chloroplast envelope protein of the Formate/Nitrite transporter family. Although the NAR1.2 protein has lower affinity for bicarbonate as revealed by the K(0.5) value which falls in the low millimolar range, a modest increase in HCO3– uptake is observed in Xenopus laevis oocytes when NAR1.2 is expressed in those cells (Mariscal et al., 2006; Atkinson et al., 2015). NAR1.2 has so far been attributed to Ci uptake on the chloroplast envelope even though there has been no direct evidence to support this. While the NAR1.2 protein plays an important role in Ci uptake, it may also have a regulatory function. This follows from a recent study in which Hla3 transcript did not accumulate in the absence of NAR1.2 protein; hence, the authors suggest that these proteins co-operate in bicarbonate accumulation (Yamano et al., 2015). In another proposed bicarbonate route into the chloroplast, NAR1.2 seems also to associate with a soluble protein LCIB (Wang and Spalding, 2014).
Two soluble proteins (LCIB/LCIC) form a complex, and are thought to be involved in recapture of CO2 leaking from the pyrenoid after observations that they closely associate with the pyrenoid when cells are acclimated to low CO2 (Yamano et al., 2010; Wang et al., 2011). Other putative transporters CCP1 and CCP2, now confirmed to be mitochondrial (Atkinson et al., 2015), are yet to be resolved. In light of all this, it is evident that the Ci transport system in C. reinhardtii remains to be further clarified in order to have a better understanding of the CCM.
In this study, we report for the first time the identification and characterization of a gene, which we designate Cia8 (Phytozome ID: Cre09.g395700), encoding a solute carrier protein, identified in a mutagenesis screen. The Cia8 gene encodes a transmembrane protein that belongs to the Na+/bile acid symporter family (SBF/BASS) also referred to as SLC10, a group of conserved transmembrane proteins (Pushkin and Kurtz, 2006). Our results show that the Cia8 gene product is needed for optimal growth on low CO2 and appears to be needed for bicarbonate uptake in C. reinhardtii.
Materials and methods
Cell culture and growth
Chlamydomonas reinhardtii culture conditions were the same as described in Ma et al. (2011). The D66 strain (nit2–, cw15, mt+) was obtained from Dr Rogene Schnell (University of Arkansas, Little Rock) and the cc124 strain (nit1–, nit2–, mt–) was obtained from the Chlamydomonas Genetics Center at Duke University (http://www.chlamy.org/). Tris-acetate-phosphate pH 7.3 (TAP) and minimal pH 6.8 (MIN) media (without acetate) were prepared according to Sueoka (1960). Both TAP and MIN plates for the growth medium were prepared by adding 1.2% (w/v) agar. Cell cultures were initiated by inoculating colonies from TAP plates into 100 ml of TAP liquid medium in Erlenmeyer flasks. Cultures were grown to early log phase with continuous illumination (100 µmol m–2 s–1) and shaking for 48 h. The TAP-grown cultures were harvested and washed with MIN medium, and re-suspended in MIN medium, connected to high CO2 (5% v/v CO2 in air) bubbling for 48 h to reach a cell density OD730 of between 0.2 and 0.3 (~2–3 × 106 cells ml–1). For CCM induction, the cells were transferred to low CO2 (0.01% v/v CO2 in air) bubbling for 4 h.
Mutagenesis, isolation of growth impaired CO2 mutants, and phenotypic screen
Mutants were generated in a mutagenesis screen according to Jungnick et al. (2014). A linear plasmid (pSL18) bearing the AphVIII gene conferring paromomycin resistance (paraR) was transformed into the D66 strain of C. reinhardtii by electroporation (Shimogawara et al., 1998). Transformed cells were selected on TAP plates containing the antibiotic paromomycin (7.5 µg ml–1; Invitrogen). Antibiotic-resistant strains were screened for growth in a high CO2 chamber (5% v/v CO2 in air) and a low CO2 chamber (0.01% v/v CO2 in air) with the same light conditions as mentioned above. Cells were streaked on MIN plates and placed in growth chambers. Spot tests were done by suspending actively growing cells in liquid MIN medium to the same cell densities (OD730 =0.15, 0.07, and 0.03) and spotting 15 µl of each suspension onto MIN plates. These were placed in high, ambient, and low CO2 chambers for 7 d. CO2 concentration in the growth chambers was measured using an Environmental Gas Monitor (EGM-4, PP systems, Amesbury, MA, USA).
Identification of the flanking region
An adaptor-mediated PCR method was used to identify the DNA region flanking the AphVIII insertion (Pollock et al., 2017). Homology searches were done using the Joint Genome Institute C. reinhardtii in the Phytozome database version 10.3: https://phytozome.jgi.doe.gov/ (Merchant et al., 2007; Blaby et al., 2014).
Linkage analysis
Genetic crosses and tetrad analysis were done as described previously (Moroney et al., 1986; Harris, 2009). Briefly, cia8 (mt+) and cc124 (mt–) cell cultures in log phase were transferred to nitrogen-deficient TAP medium in the light overnight to induce gametogenesis. The next morning, 3 ml of each culture were mixed to allow mating for 3 h in the light. Aliquots (0.5 ml) were plated on TAP minus-nitrogen medium containing 4% agar. The plates were stored in the dark for 2 weeks to allow zygote maturation. After 14 d, zygotes were transferred to 1.2% agar TAP medium plates for meiotic germination. Tetrad dissections were conducted and linkage determined by association of the paromomycin resistance gene with progeny that did not grow well on low CO2.
Photosynthetic assays
Chlamydomonas reinhardtii cultures were started heterotrophically in 100 ml of TAP medium for 48 h to reach log phase. The cells were centrifuged and transferred to 250 ml Erlenmeyer flasks and re-suspended in MIN medium, bubbled with 5% CO2 until they reached a cell density OD730=0.2–0.3 (3 × 106 cells ml–1). The cultures were then transferred to low CO2 (0.01%) for 4 h to allow CCM induction. The affinity for external Ci (K0.5[Ci]) was estimated according to Ma et al. (2011). In the method, cells with an equivalent of 100 µg of chlorophyll were suspended in 25 mM HEPES-KOH buffer (pH 7.3) or 25 mM CHES-KOH buffer (pH 9.0) bubbled with inert nitrogen gas (i.e CO2 free). The cells were transferred to an O2 electrode chamber (Rank Brothers, Cambridge, UK) illuminated at 300 µmol m–2 s–1, and left to deplete any remaining Ci in the buffer and intracellular spaces. Upon depletion of endogenous CO2, no net O2 evolution is observed. Known concentrations of NaHCO3 were injected into the chamber and the rate of O2 evolution was measured. The K0.5[Ci] was calculated as the Ci concentration required for half-maximal rates of oxygen evolution (Badger, 1985). Chlorophyll content was measured by combining Chl a and b. Chlorophyll was extracted in 100% methanol and measured using a spectrophotometer. The K0.5 (CO2) is taken as the CO2 concentration needed to reach half Vmax O2 evolution.
Intracellular localization
The whole Cre09.395700 gene (4846 bp) was amplified and introduced into a modified pSL18CrGFP vector using EcoRI and NdeI restriction sites. The Cre09.395700 gene was inserted such that it was in-frame with the Chlamydomonas codon-optimized CrGFP (C. reinhardtii green fluorescent protein) gene (Fuhrmann et al., 1999) already in the vector. The vector was linearized with KpnI digestion. Transformation of the wild-type strain D66 with the Cia8 gene fragment was achieved by electroporation with the linearized vector, and paromomycin-resistant colonies were analysed using PCR. For imaging CrGFP-tagged proteins, 5 μl of cells were mounted onto a slide with 1.5% low melting point agarose. Cells were imaged using a Leica Sp2 confocal microscope. The Kr/Ar laser was set at a wavelength of 488 nm to excite both CrGFP and chlorophyll, with the photomultiplier tube set to 500–520 nm to detect GFP fluorescence and 660–700 nm to detect chlorophyll autofluorescence. A ×20 lens was used to image the cells.
Gene expression analysis
RNA extraction was done using Triazol reagent following the guidelines provided (Invitrogen, Carlsbad, CA, USA). A 1 µg aliquot of total RNA was used as template for synthesis of cDNA. The Superscript First Strand Synthesis System for transcripts with high GC content (Invitrogen) was used to synthesize cDNA according to the manufacturer’s instructions. An aliquot of 100 ng of cDNA was used as the template with SYBR Select (Applied Biosystems, Foster City, CA, USA) for quantitative PCR in an ABI Prism 7000 sequence detection system following the manufacturer’s instructions (Applied Biosystems). Normalized primers were specific for the glyceraldehyde phosphate dehydrogenase (GAPDH) gene for quantitative real-time PCR (qPCR). For qPCR, the Cblp gene was used as a control (Schloss, 1990). The primers used for the respective cDNAs are listed in Supplementary Table S1 at JXB online.
Complementation of Cia8
Complementation of the cia8 mutant was achieved by transformation of cia8 cells with a construct containing a wild-type copy of the Cia8 gene. The construct consisted of the Cia8 coding sequence (CDS) fragment including the 5'- and 3'-untranslated regions (UTRs; 2949 bp) and a 1080 bp fragment of the promoter region. These fragments were ligated using the NotI restriction site, which was then ligated into pGEM-T. The sequenced fragment was cloned into the shuttle vector pSP124s using BamHI and SacI restriction sites. Sequences were obtained from Phytozome Version 10.2 (http://www.phytozome.net/). Primers used to amplify the Cia8 gene fragments are shown in Supplementary Table S1. The electroporation method was used for cell transformation, and strains were selected for bleomycin resistance (Shimogawara et al., 1998). The presence of complemented DNA in selected Chlamydomonas strains was confirmed by PCR.
Ci uptake assay
Active species uptake of H14CO3– was carried out at pH 9.0 (25 °C) with 15, 60, and 240 s uptake periods, terminated by silicone oil centrifugation–filtration principally following the protocol of Badger et al. (1980). Cells acclimated to high and low CO2 conditions were harvested and resuspended at 16 µg of chlorophyll ml–1 in 50 mM HEPES-NaOH, pH 7.8, and illuminated (300 μmol m–2 s–1 white light) to deplete endogenous Ci. Cells were then harvested and resuspended in the same volume of 50 mM HEPES-NaOH, pH 9, and 200 µl aliquots of cell suspension were layered on top of the silicon oil in 400 µl microfuge tubes in the light. A radioactive 25 mM bicarbonate solution (pH ~9.5) was made by mixing radioactive NaH14CO3 and NaHCO3. Uptake measurements were initiated by adding radioactive bicarbonate to a final concentration of 500 μM to the cells in the light. After centrifugation, each cell pellet was resuspended in 200 µl of 2 N NaOH and divided into two 100 µl aliquots. One aliquot was used to determine total NaHCO3 taken up, whereas the other was acidified with HCl to determine the proportion of photosynthetically fixed CO2. Intracellular bicarbonate pool sizes were estimated by subtraction of fixed bicarbonate from total bicarbonate in the samples. Amounts of bicarbonate were calculated from counts per million (cpm) and the specific activity of the radioactive bicarbonate solution determined with a liquid scintillation analyser (Perkin-Elmer TriCarb 2810 TR).
Results
The cia8 mutant requires high CO2 conditions to grow well
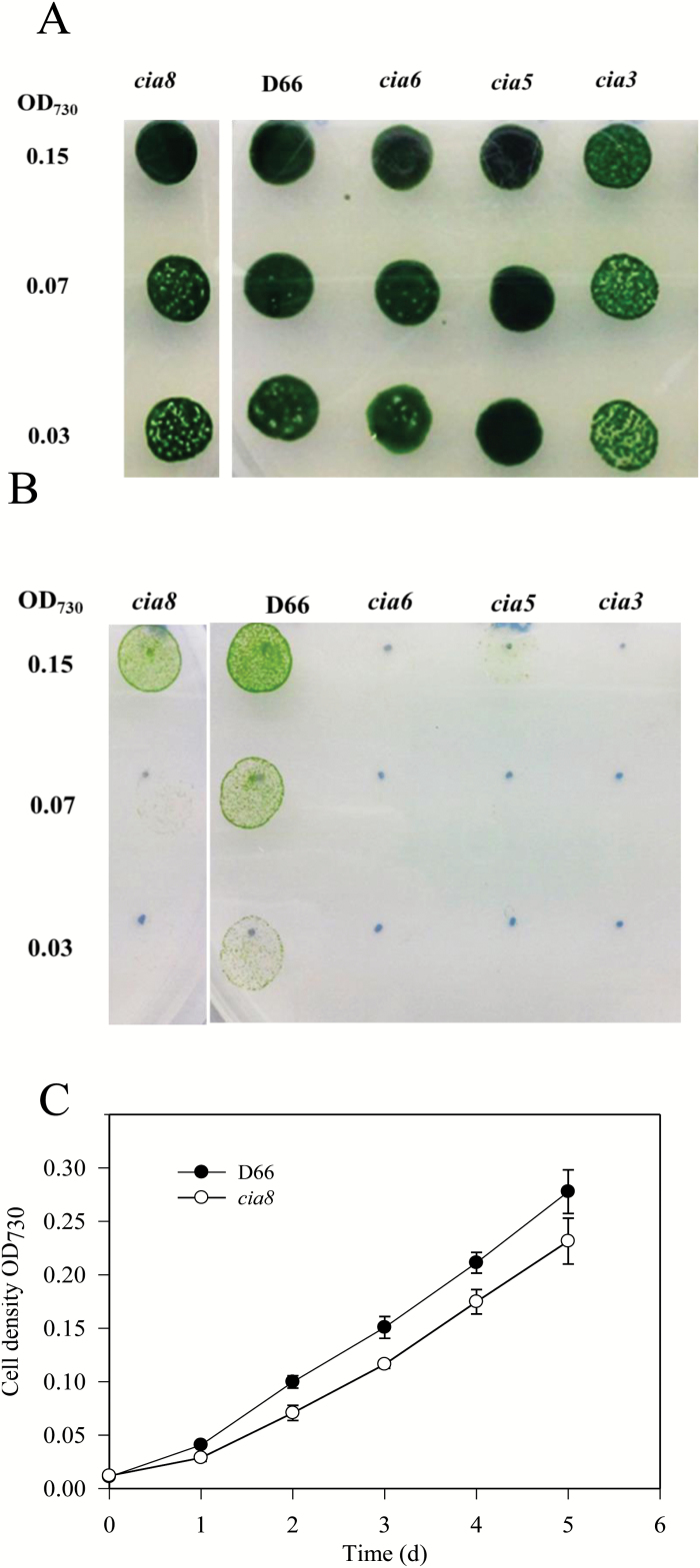
The cia8 mutant was identified following an insertional mutagenesis screen using a 1.8-kb AphVIII paraR cassette containing the aminoglycoside 3'-phosphotransferase type VIII-encoding gene (AphVIII) from Streptomyces rimosus which confers paromomycin resistance (Sizova et al., 2001). Mutants were screened by growing them on high CO2 (5%) and low CO2 (0.01%) on MIN plates in which potential CCM mutants would show slow growth when grown on low CO2 (Jungnick et al., 2014). While the cia8 mutant grew well in high CO2 conditions (Fig. 1A), it showed impaired growth in low CO2 (0.01–0.025% v/v CO2 in air) (Fig. 1B) and was thus taken for further investigation. The slow growing phenotype was also apparent when cells were grown in liquid (MIN) culture at 0.01% CO2, where the doubling time for the cia8 mutant was 30 h compared with 20 h for D66 (Fig. 1C).
Fig. 1.
Spot test for the growth of C. reinhardtii strains in (A) high CO2 (5% CO2 in air) and (B) low CO2 (0.01% CO2 in air) pH 7.3. The strains include wild-type D66, the cia8 mutant, and three known CCM mutants cia6, cia5, and cia3 used as controls. The numbers to the left represent the initial OD730 of 0.15 (~1.5 × 106 cells) and two serial dilutions. (C) Growth of wild-type strain D66 and the cia8 mutant in liquid culture (MIN). Cultures were grown in Erlenmeyer flasks blowing in low CO2 (0.01–0.025%). Values are expressed as the mean ± SE (n=4). (This figure is available in colour at JXB online.)
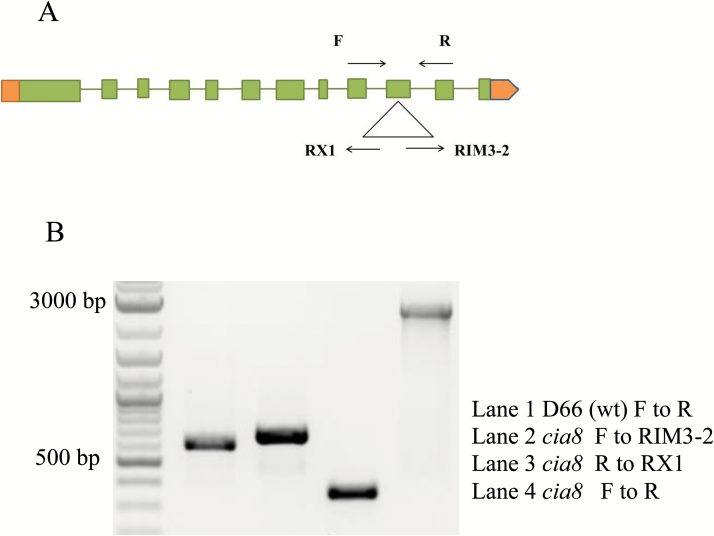
Identification of the Cia8 gene and confirmation of insertional inactivation
The location of the AphVIII insert in the Cia8 gene was determined using the adaptor PCR described by Pollock et al. (2017). The insertion is located in the 10th exon of the Cia8 gene as shown in the gene model in Fig. 2A. PCR amplification was done to confirm the presence of the insertion using two primer pairs that amplify the 5' and 3' ends of the insertion (Fig. 2B). Supplementary Table S1 has the list of primers used in the study. The genomic DNA flanking the insertion site in this mutant was isolated and sequenced. Analysis of the sequenced genomic DNA revealed a deletion of 16 bases of the gene sequence at the 5' end of the insertion, and a new region of 19 bases was inserted. However, the 3' end of the insertion was intact with no deletion or insertion. The analysis confirmed that the target gene sequence had been disrupted in the 10th exon and that no other large DNA deletions or insertions had occurred.
Fig. 2.
(A) Genomic structure of the Cia8 gene locus (ID: Cre09.g395700) showing the position of the AphVIII insertion (triangle). Note that the cassette is in reverse orientation in relation to the direction of the gene. Green bars, spaces, and orange bars represent exons, introns, and untranslated regions, respectively. The arrows represent the direction of the primers. (B) Confirmation of AphVIII insertion in exon 10 of the Cia8 gene. Lane 1: D66 is the control genomic DNA without the cassette and the other three are Cia8 genomic DNA. Lane 2 represents the 3' end of insertion and lane 3 represents the 5' end of insertion. Lane 4 spans the whole insertion region including the genomic region flanking the cassette using a primer pair in the gene. (This figure is available in colour at JXB online.)
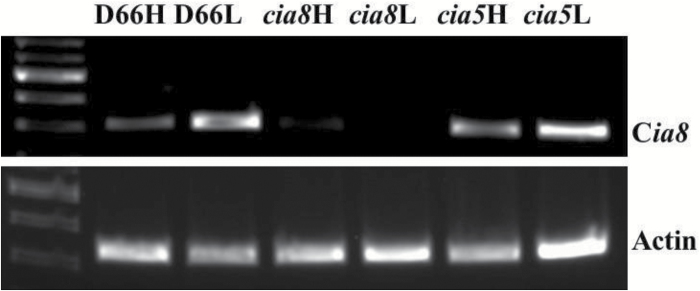
Disruption of transcription of the gene was also confirmed by reverse transcription–PCR (RT–PCR) using a gene-specific primer pair spanning the insertion site. RT–PCR results show that there is no detectable expression of the intact Cia8 transcript in the cia8 mutant (Fig. 3), confirming that the transcript has been disrupted and that the gene product in the mutant is below detectable levels. As expected, mRNA for Cia8 was detected in wild-type high CO2 cells. There was, however, a marked up-regulation of transcript in low CO2 cells (Fig. 3), suggesting that the Cia8 gene is inducible under low CO2 conditions. The result from this RT–PCR also shows that the gene encoding CIA8 does not appear to be regulated by the Cia5 gene, the master regulator of many CCM genes, as the transcript was expressed in the cia5 mutant equally to that in the wild type.
Fig. 3.
RT–PCR analysis of D66 and cia8 mutant strains using poly(A) RNA from high and low CO2-grown cells as templates for RT–PCR. For the cia8 mutant, the primers shown in Supplementary Table S1 amplify a 1500 bp product from cDNA. GAPDH was used as a loading control and a 1000 bp product was amplified.
Genetic crosses of the cia8 mutant with the wild-type strain demonstrated a 1:1 segregation ratio of paromomycin-resistant to paromomycin-sensitive progeny, illustrating that the mutant carries a single insertion (data not shown). Further investigation by PCR confirmed that the paromomycin-resistant strains carried the cassette (data not shown). Phenotypic analysis (data not shown) showed that the tetrads with resistance to paromomycin consistently demonstrated impaired growth in low CO2 conditions compared with the wild type. Taking the results together, we concluded that the Cia8 gene locus is segregating with, and therefore responsible for, the observed slow growth phenotype under low CO2 conditions.
Prediction of structure and transmembrane domains of the Cia8 gene
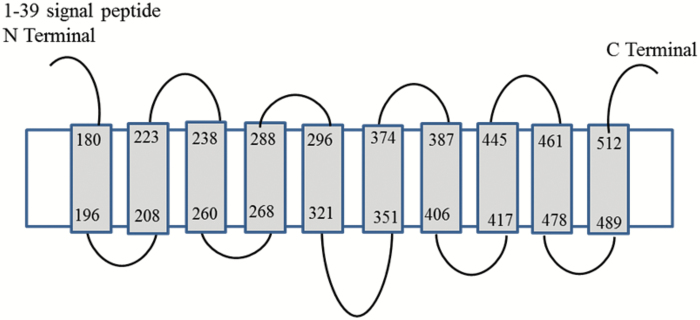
A comparison of cDNA and genomic sequences shows that Cia8, located on chromosome 9 in the genome, has 12 exons. On the basis of nucleotide sequence, Cia8 is predicted to encode a polypeptide of 529 amino acids and aligns best with the sodium/bile acid (SBF-like) solute carrier protein group. We used the PHYRE2 software (Kelley et al., 2015) to thread CIA8 to known protein structures. Using the core transmembrane part of the protein containing 370 amino acids (amino acids 160–529) gave the highest homology scores, and was predicted to have 10 putative transmembrane domains of 15–24 amino acids and relatively small extracellular loops between the transmembrane helices of 8–30 amino acids, as shown in Fig. 4. The protein threaded best to the crystal structure for a bacterial (Yersinia frederikseni) Na+/metabolite symporter (82%), but it also threaded well to HapA proteins (Na+/H+ antiporters) from several bacteria (56–98%).
Fig. 4.
Topology model predictions for CIA8 based on the alignment with Yersinia frederiksenii. The grey boxes are putative membrane-spanning helices connected by loops of variable lengths; 62% of the residues were modelled at a >90% confidence level. Predictions were generated by PHYRE2 (Kelley et al., 2015).
Similarity of CIA8 to other anion transporters
Searches in the C. reinhardtii protein database revealed eight other predicted proteins similar to CIA8: Cre02.g095085; Cre02.g095086; Cre02.g147450; Cre06.g250450; Cre09.g393250; Cre10.g448350; Cre12.g521950; and Cre12.g532500, all annotated as Na+/bile acid transporters. Sequence identity of these proteins to CIA8 varies between 19% and 31%. The number of Na+/bile acid transporter genes in C. reinhardtii is quite comparable with other species, with six having been reported in Arabidopsis (Sawada et al., 2009), seven in humans (He et al., 2009), and five in the marine diatom Phaeodactylum tricornutum (Ashworth et al., 2016). The numbers are not surprising considering the variety of ions that are transported by these types of transporter proteins. The best alignment using the core (amino acids 160–529) were to chloroplast BASS4 proteins (35–40% identity with >90% coverage). The CIA8 protein also shows considerable sequence similarity to several SBF proteins from higher plant species, many of which have not yet been characterized (Supplementary Fig. S1). The closest algal homologue was found in Volvox carteri with 76% identity. Our analysis suggests that the Cia8 gene product belongs to a family of conserved membrane transport proteins characterized by conserved domains.
CIA8 has a reduced affinity for Ci
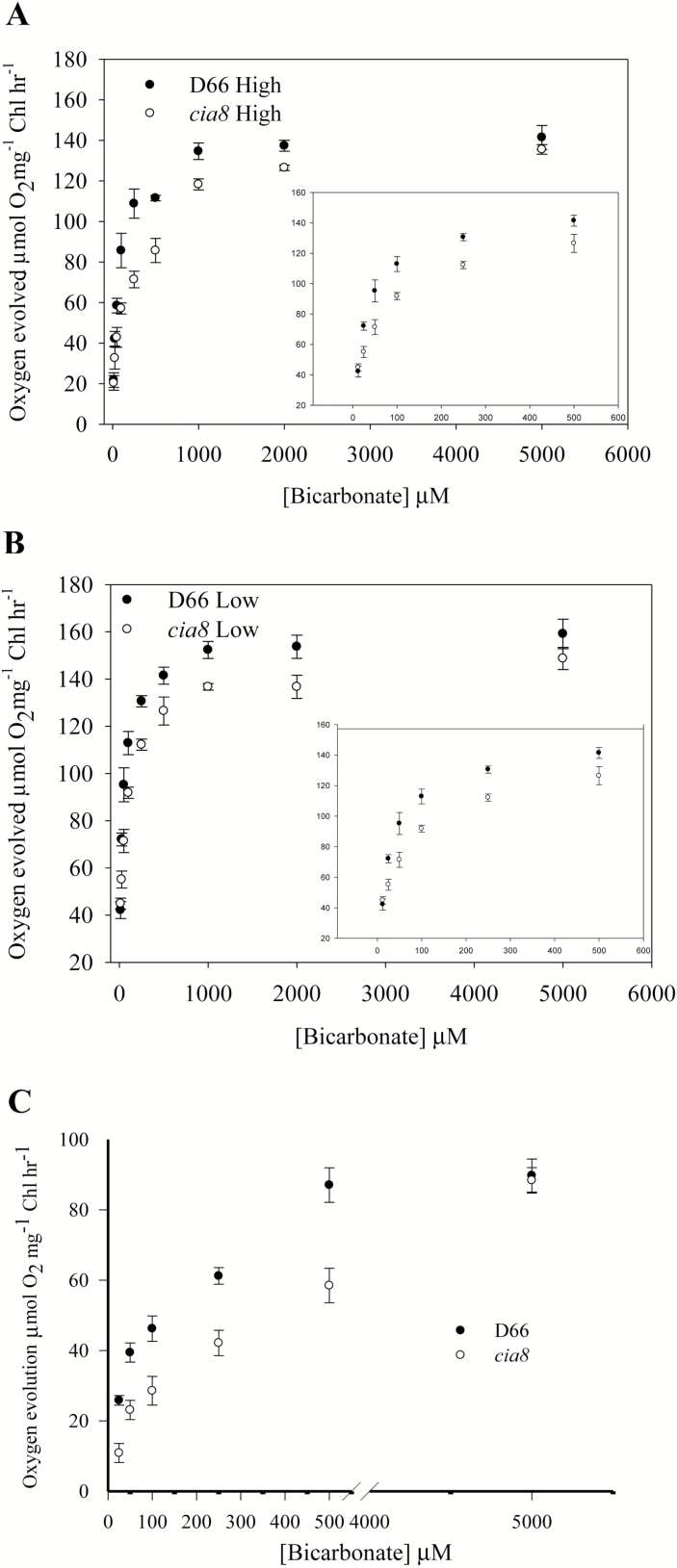
Since the expression of CIA8 is strongly responsive to CO2 limitation, we evaluated its possible function in the CCM using physiological assays. The strain missing CIA8 exhibited a higher K(0.5) (Ci), indicating that they had a lower affinity for inorganic carbon than wild-type cells (Fig. 5A, B). At pH 7.3, the K(0.5) (Ci) for wild-type cells was 25 μM. In contrast, cia8 mutant cells had a significantly higher K(0.5) (Ci) of 90 μM (Table 1), suggesting a reduced affinity for Ci in the mutant. Oxygen evolution was also measured at pH 9.0 where the predominant Ci species in the medium would be bicarbonate (87% HCO3–, 13% CO32–, <0.15% CO2), hence, oxygen evolution would be highly dependent on active bicarbonate uptake (Duanmu et al., 2009). The shift in the K(0.5) was also significant at higher pH, with cia8 mutant cells exhibiting a greater K(0.5) (Ci) of 350 μM as compared with 100 μM in the wild-type cells (Fig. 5C; Table 1). Thus, the reduced affinity for Ci is observed at both pH values. This reduced affinity for Ci in the mutant is consistent with the hypothesis that the Cia8 gene product is important in Ci uptake.
Fig. 5.
Ci-dependent photosynthetic oxygen evolution in the C. reinhardtii wild type and the cia8 mutant. (A) High CO2-grown cells and (B) low CO2-grown cells assayed at pH 7.3; (C) low CO2-grown cells assayed at pH 9.0. Cells were grown at high CO2 and transferred to low CO2 for 4 h. The insert in (A) and (B) represents lower concentrations >500 μM used for determination of K(0.5). Each point represents the mean and SE of three separate experiments.
Table 1.
Maximal oxygen evolution activity (V max) and Ci affinity, K (0.5) values for wild-type D66 and cia8 mutant cells
Cells were grown on high CO2 for 48 h and acclimated to low CO2 for 4 h.
| V max (μmol O 2 mg Chl –1 h –1 ) | K (0.5) Ci (μM) | |||
|---|---|---|---|---|
| D66 | Cia8 | D66 | Cia8 | |
| pH 7.3 high CO2 cells | 166 (±3.3) | 155 (±4.5) | 95(±2.8) | 158 (±2.9) |
| pH 7.3 low CO2 cells | 160 (±7.8) | 150 (±5.3) | 20 (±2.7) | 60 (±4.7) |
| pH 9.0 low CO2 cells | 92.7 (±1.5) | 90 (±5.2) | 110 (±5.0) | 320 (±10.5) |
For Vmax, all means are not significantly different P>0.1 (α=0.05). For K(0.5) Ci, the means are significantly different from each other P<0.001 (α=0.05). In parenthesis, ±SEM; n=3.
Localization of CIA8 to the chloroplast in C. reinhardtii
Analysis using YLoc (https://omictools.com/yloc-tool) (Briesemeister et al., 2010) suggested that the CIA8 peptide is a chloroplast membrane protein (score 81%). Another alignment of the N-terminal sequence of this protein was conducted using SCLpred - Bologna Biocomputing Group (schloro.biocomp.unibo.it). The software predicts that CIA8 has a chloroplast transit peptide (cTP; score 0.79) and thylakoid (0.81). It also predicts a location as thylakoid membrane (0.6). As a control, we used Kea3.3, an Arabidopsis thylakoid antiporter, and the software predicts a cTP (score 0.77) and a location as thylakoid membrane (0.51). These predictions give a strong indication that CIA8 is a chloroplast protein. To confirm this subcellular localization, a C. reinhardtii line expressing the Cia8 coding sequence fused to GFP (Cia8:CrGFP) under the control of the PsaD promoter was generated. The PsaD promoter is a high expression promoter which drives a nuclear gene encoding an abundant chloroplast protein located on the stromal side of PSI in C. reinhardtii (Fischer and Rochaix, 2001). RT–PCR analysis revealed that GFP transcript was detectable in this strain (Supplementary Fig. S2A). Although green fluorescence of the fused protein CIA8:CrGFP was detected in chloroplasts by live imaging on the confocal microscope (Supplementary Fig. S2B), the signal was weak and not sufficient to conclude chloroplast localization. The weak signal however, did not seem to be confined to the chloroplast envelope, but diffused throughout the organelle, suggesting a thylakoid localization.
CIA8 transport is not dependent on Na+
Several solute transporters are known to be Na+ dependent, including BicA, a low affinity bicarbonate transporter in cyanobacteria (Price et al., 2004; Price and Howitt, 2011). To determine whether Cia8 gene function could be dependent on Na+ ions, wild-type and mutant cells were grown on different concentrations of Na+ (<10 µM to 200 mM). Both cultures grew well on low Na+ concentrations (<10 µM) up to 100 mM on pH 6.8 plates (Supplementary Fig. S3). Growth of cells was, however, inhibited at 200 mM Na+ (data not shown), probably due to osmotic stress. The observation that wild-type and cia8 cells grew well at low Na+ concentrations (<10 µM) may imply that the Na+ ions are not required for the acquisition of HCO3– in C. reinhardtii. This was not an unexpected result since the external Na+ concentration in fresh water is generally low (<1 mM) (Pootakham et al., 2010). However, since some of the transporters, including CIA8, are internal, it is not clear that changing the external Na+ concentration would alter the internal concentration. Therefore, further analysis may be required to elucidate the role. if any, of Na+ in Ci uptake by C. reinhardtii cells.
Expression of Cia8 is up-regulated on low CO2 conditions
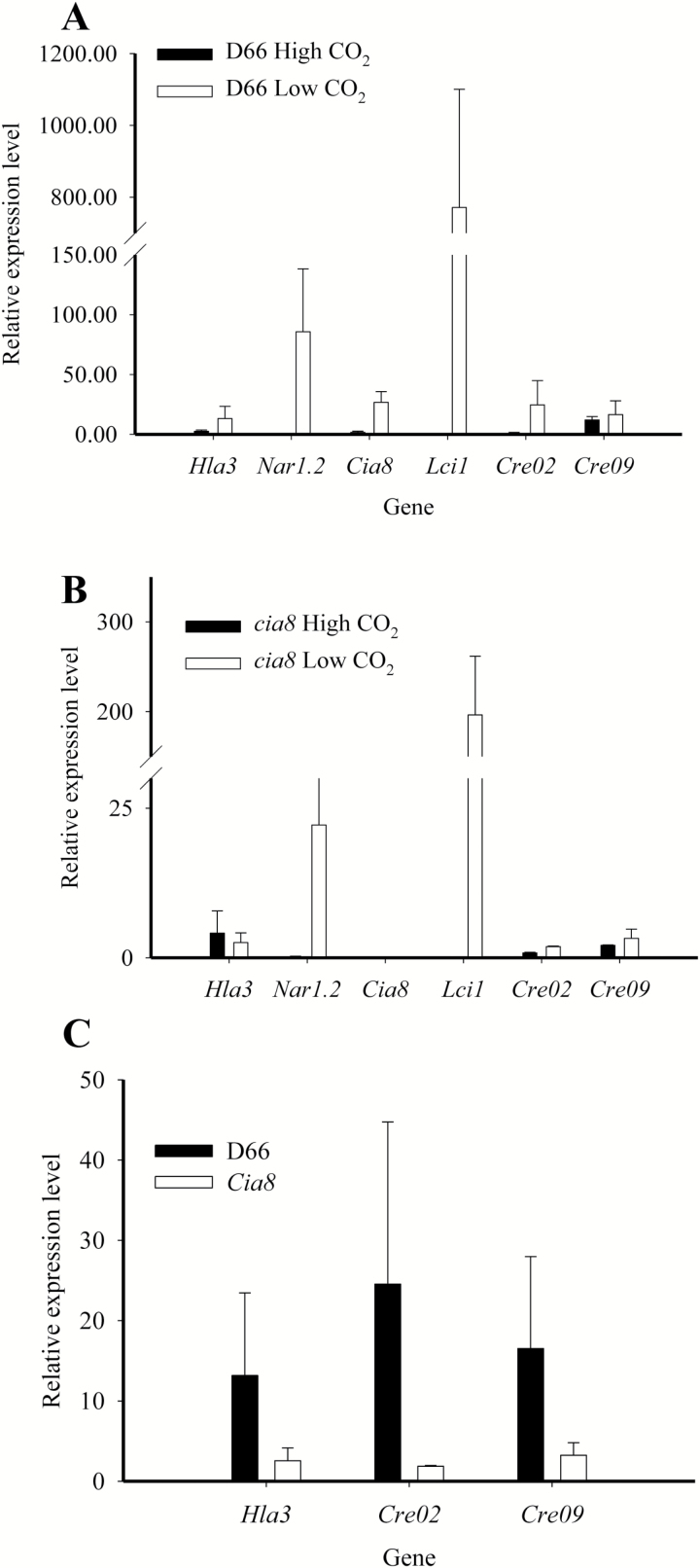
To quantify and compare the expression of the other known transporters in Chlamydomonas in the cia8 mutant and in the wild type, we investigated the expression of Hla3, Nar1.2, Lci1, and Cia8 as well as two other SBF genes (Gene ID: Cre02.g147450 and Cre09.g393250) by qPCR analysis. RNA samples were obtained from high CO2- and low CO2-acclimated cultures. The results show the level of expression of these transporter genes in the wild type (Fig. 6A) and in the cia8 mutant (Fig. 6B) relative to the Cplb gene which encodes a G-protein beta subunit-like polypeptide and also known to exhibit constitutive expression of transcript (Schloss, 1990). The results confirm up-regulation of the Cia8 gene and the other transporters upon acclimation to low CO2 conditions, except for Hla3 which usually increases in expression at times >4 h (Duanmu et al., 2009). In the wild type, there was a 4-fold up-regulation of the Cia8 gene on acclimation to low CO2 conditions. There was also a notable up-regulation of the two SBF genes in the wild-type cells. The relative expression levels of these two SBF genes were also somewhat lower in the cia8 mutant cells as compared with the wild type (Fig. 6C). While the data confirm that the Cia8 gene product is involved in the CCM, they also suggest that loss of CIA8 may affect the transcriptional control of other genes normally up-regulated under low CO2 conditions.
Fig. 6.
Quantitative real-time PCR results for transporter genes under high and low CO2 conditions in the wild-type (A) and cia8 mutant (B) strains. Wild-type D66 and mutant cells were grown on high CO2 for 48 h then subjected to low CO2 acclimation for 4 h. Lack of the Cia8 gene causes down-regulation of other CCM transporters and two other genes in the SBF family (C). The Cblp gene was used as the internal control. Relative expression level is expressed as 1000/2∆Ct where ∆Ct=Ctgene–Ctcblp.
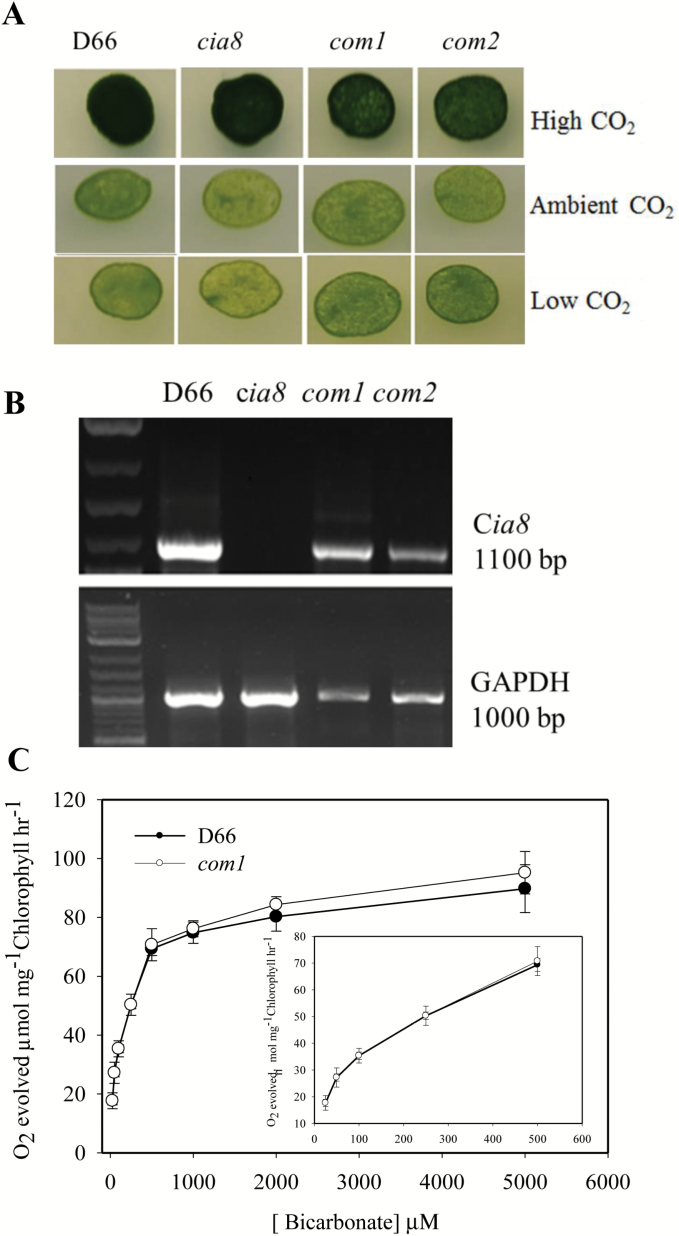
The Cia8 gene can be complemented
Complementation of CIA8 was achieved by expressing Cia8 cDNA (1869 bp) including the entire 5' UTR and 3' UTR in the pSP124 vector under the control of its own promoter and terminator in the cia8 mutant. Transformed cells were selected on bleomycin. Ten transformants were selected and subjected to further growth tests and photosynthesis assays. Results of growth experiments show that the wild-type phenotype in low CO2 conditions was restored in complemented lines (referred to as com1 and com2) (Fig. 7A). RNA was extracted from these two complemented strains and cDNA synthesized. The resulting RT–PCR analysis showed that Cia8 transcript had been restored, although to levels slightly lower than the wild type (Fig. 7B). In addition, the Ci-dependent oxygen evolution assay of com1 at pH 9.0 showed that the K(0.5) (Ci) in the complemented strain was the same as that of the wild type. Also, the maximal oxygen evolved (Vmax) as well as CO2 fixed were not significantly different from the wild type (Figs. 7C and 8, respectively). These results collectively confirm that this Cre09.g395700 coding sequence complemented growth of the cia8 mutant, restoring the expression of transcript and functionality in Ci uptake.
Fig. 7.
(A) Growth phenotype of complemented cia8 strains, com1 and com2. These strains had the wild-type Cia8 gene reintroduced into the cia8 mutant as described in the Materials and methods. (B) RT–PCR showing restoration of the Cia8 transcript in the complemented strains. GAPDH was used as the loading control. (C) Oxygen evolution for the wild type, D66, and one complemented line, com1, at pH 9.0. Each point represents the mean and SE of three separate experiments. (This figure is available in colour at JXB online.)
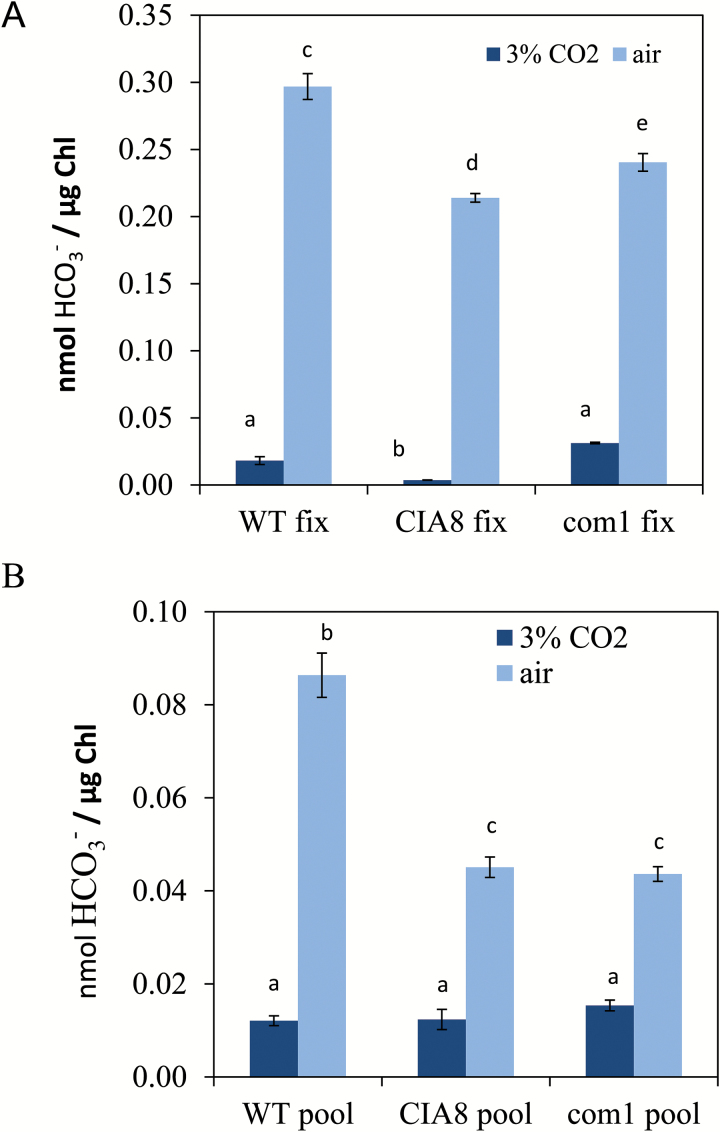
Fig. 8.
Accumulation of 14C in wild-type (WT) D66, in the cia8 mutant, and in the complemented cell line com1. Cells were grown on elevated CO2 (5% v/v CO2 in air) and acclimated to low CO2. Analysis was done at pH 9.0. (A) CO2 fixed and (B) CO2 remaining in the pool. Statistical analysis was done by Tukey’s HSD test. Different letters indicate that means are significantly different at P=0.01. (This figure is available in colour at JXB online.)
Ci uptake assays
To evaluate the contribution of CIA8 to Ci uptake activity, the silicone oil layer centrifugation method was used to measure accumulation of 14C in D66, in CIA8, and in the complemented cell line com1. Analysis was done at pH 9.0 to ensure the predominant Ci species was bicarbonate, using cells grown in elevated CO2 (3% v/v CO2 in air) or in air (low CO2) cells. In all cases, the cells acclimated to low CO2 had greater amounts of accumulated Ci in the cells than cells grown on elevated CO2 (Fig. 8). When Ci uptake and bicarbonate pools were compared, wild-type cells had greater Ci accumulation and greater Ci fixation than cia8 cells (Fig. 8). In addition, photosynthesis was nearly restored to wild-type levels in the com1 cells line, although Ci accumulation was not fully restored. These data support the hypothesis that Ci uptake, most probably in the form of bicarbonate, is reduced in cia8 cells.
Discussion
The need for the uptake and accumulation of bicarbonate is a key feature of the C. reinhardtii CCM. The putative bicarbonate transporters in C. reinhardtii are generally induced by limiting CO2 conditions. It also appears that the transporters have overlapping partially redundant functions. If the expression of only one transporter is reduced, there usually is not a strong growth phenotype, and a small effect on Ci uptake is observed. However, if two or more transporters are reduced, Ci uptake is affected and growth on low CO2 is reduced (Duanmu et al., 2009; Yamano et al., 2015). In this work, we generated new mutants by insertional mutagenesis and identified a mutant with an insertion in the gene (Phytozome ID: Cre09.g395700) which we have named Cia8 (for Ci accumulation). The main objective of this work was to determine whether the CIA8 protein participated in the CCM of C. reinhardtii. We characterized and demonstrated that the insertion in this gene caused poorer growth on low CO2 and resulted in a significant decrease in Ci affinity when cells were grown under limiting CO2 conditions. In complementation experiments, the expression of the Cia8 coding region from the wild-type gene was able to restore the normal growth phenotype and function in photosynthetic assays (Fig. 7).
The CIA8 protein belongs to the SBF/BASS subfamily which includes membrane-bound transporters designated as bile acid transporters. These proteins transport a diverse number of substrates. In Arabidopsis, two of these genes have been characterized, and BASS2 is involved in pyruvate transport across the chloroplast envelope in the methylerythritol 4-phosphate (MEP) pathway, whereas BASS4 transports 2-keto acids during glucosinolate metabolism (Gigolashvili et al., 2009; Furumoto et al., 2011). Bioinformatic searches in the PFAM database reveal that some SBF and SBF-like anion transporters may be involved in bicarbonate uptake. Despite being important transporters in other eukaryotic species, this subfamily of SBF/BASS gene products remains of unknown function in C. reinhardtii.
Physiological characterization of CIA8 in this study supports the idea that it is involved in CCM function and Ci uptake under limiting CO2 conditions. The CIA8 transporter seems to make a significant contribution to CCM function because it is the only transporter so far that shows a distinct phenotype in single knockout mutants (Fig. 1). The other known transporters HLA3 and NAR1.2 did not show a definitive growth phenotype as single knockout mutants (Yamano et al., 2015), and no growth differences were reported when LCI1 was expressed in the Lcr1 background (Ohnishi et al., 2010). In this study, the apparent affinity of mutant and wild-type cells for Ci at both pH 7.3 and 9.0 was tested. At pH 9.0, most of the Ci is in the form of bicarbonate. Under these conditions, a significant fraction of the Ci is thought to be transported as HCO3– at both the plasma membrane and the chloroplast envelope (Wang et al., 2014). The loss of CIA8 clearly reduces the apparent affinity of the cells for Ci at both pH values. The decreased rate of Ci-dependent photosynthesis and reduced affinity for Ci in the cia8 mutant at pH 7.3 and 9.0 (Fig. 5) provides evidence that CIA8 might be involved directly or indirectly in Ci uptake.
While SBF transporters are often associated with co-transport of Na+ ions as revealed in some animal proteins (Romero et al., 2013), the results in this study suggest that Na+ ions may not necessarily be essential for the functionality of the CIA8 protein (Supplementary Fig. S3). Low Na+ ion concentrations (i.e. <10 µM to 100 mM) did not affect the growth of cells in the mutant and wild type. This was not a surprising result since TAP and MIN media in which C. reinhardtii cultures are grown contain no added sodium. Thus, C. reinhardtii cells grow well with very low concentrations of sodium. While there is ample support for Na+-coupled transport in marine algae and freshwater cyanobacteria (Shibata et al., 2002; Price et al., 2004; Nakajima et al., 2013), H+-coupled transport has always been thought to play a role in freshwater algae. However, other ions such as Na+ or K+ might be coupled to bicarbonate movement. Another possibility could involve an exchange (antiport) of bicarbonate for an anion such as chloride.
Our analysis of the gene transcripts showed that the cia8 mutant is a knockout. In the wild type, the Cia8 transcript was strongly up-regulated (4-fold) when high-CO2 cells were acclimated to low CO2, and this induction is consistent with the other Ci transporters (Figs 3, 6) although the level of induction is not as high as some of the CCM proteins. Regulation of transporters by transcript abundance in C. reinhardtii is well documented (Ohnishi et al., 2010; Gao et al., 2015; Yamano et al., 2015). This type of regulation arguably prevents expression of HCO3– transporters under high CO2 conditions where they would not be required. It is interesting that the Cia8 gene is expressed, albeit at a low level, in high CO2 conditions, which may suggest a housekeeping function for the protein. Additionally, the relative expression of the other transporters is down-regulated in the cia8 mutant as compared with the wild type (Fig. 6). This could be explained by a negative feedback mechanism whereby the loss of CIA8, a putative chloroplast envelope protein, leads to accumulation of HCO3– in the cytosol, which negatively impacts the two plasma membrane transporters (HLA3 and LCI1), and later NAR1.2. Alternatively, it may be that Cia8 gene may have some form of regulatory control over these other genes, just like the Nar1.2 gene transcript which has recently been shown to regulate the expression of Hla3 (Yamano et al., 2015). However, the transcriptional changes we observed were relatively small and could simply reflect the fact that the cia8 cells are growing more slowly at the lower CO2 concentration. These results raise the possibility that the contribution of CIA8 to CCM function may thus be more than simply Ci uptake, an interesting aspect that requires further investigation.
Several studies in the past have hypothesized the occurrence of a putative HCO3– channel or transporter on the thylakoid membrane of the algal CCM (Raven, 1997; Wang et al., 2011; Jungnick et al., 2014). In this study, the signal of the GFP fused to CIA8 was too weak to provide a conclusive result, but seemed to be evenly diffused throughout the chloroplast (Supplementary Fig. S3). We speculate that the weak CrGFP signal may be attributed to a general low expression, poor folding, or poor targeting of this protein. It is also possible that the CIA8:CrGFP chimeric protein is not highly stable and is degraded. Nevertheless, the prediction programs did provide strong evidence for this localization to the organelle. Localization throughout the organelle, however, would suggest that the protein may be on the thylakoid membrane, a strategic location to pump HCO3– directly into the thylakoid lumen. At this point, it is not clear whether CIA8 is a symporter or antiporter. However, if it is on the thylakoid membrane it might transport an ion using the transmembrane H+ gradient set up in the light. If CIA8 is on the thylakoid membrane, the NAR1.2 protein localized on the chloroplast envelope might be unable to make up for the loss of CIA8. We expect that future analyses of Ci uptake in a double knockout Cia8/Nar1.2 mutant will be very informative. Also with this potential thylakoid localization, the Cia8 gene product could be making a significant contribution to CCM function, and perhaps to Ci uptake, possibly in conjunction with another protein with a function similar to CIA8.
In conclusion, the CIA8 protein is a new putative transporter that plays a role in the C. reinhardtii CCM. When the gene is knocked out, the mutant shows a compromised Ci uptake and growth inhibition when grown in limiting CO2 conditions. We propose that the Cia8 gene encodes a putative bicarbonate transporter that is involved in uptake of HCO3– in the chloroplast for CO2 fixation. The predicted CIA8 protein is hydrophobic and, according to prediction programs, has a topology comparable with other known transporters in the SBF/BASS subfamily. Further studies need to be carried out to quantify the contribution of the protein to Ci uptake in C. reinhardtii cells. The function of the CIA8 homologues in C. reinhardtii also remains to be determined. To our knowledge, this is the first report of an SBF/BASS gene in C. reinhardtii being involved in CCM function, and possibly in bicarbonate uptake.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Sequences of primers used in this study.
Fig. S1. Clustal Omega alignment of the CIA8 primary protein sequence with other SBF transporters from higher plants and other chlorophytes.
Fig. S2. (A) RT–PCR analysis of the cia8 mutant strain transformed with the fused CIA8:CrGFP protein. (B) Live imaging of the CIA8:CrGFP transformant cells showing the localization of the chimeric protein
Fig. S3. Growth of wild-type D66 and cia8 mutant cells of C. reinhardtii on MIN plates (pH 6.8) with <10 µM up to 100 mM NaCl.
Supplementary Material
References
- Amoroso G, Sültemeyer D, Thyssen C, Fock HP. 1998. Uptake of HCO3− and CO2 in cells and chloroplasts from the microalgae Chlamydomonas reinhardtii and Dunaliella tertiolecta. Plant Physiology 116, 193–201. [Google Scholar]
- Ashworth J, Turkarslan S, Harris M, Orellana MV, Baliga NS. 2016. Pan-transcriptomic analysis identifies coordinated and orthologous functional modules in the diatoms Thalassiosira pseudonana and Phaeodactylum tricornutum. Marine Genomics 26, 21–28. [DOI] [PubMed] [Google Scholar]
- Atkinson N, Feike D, Mackinder LC, Meyer MT, Griffiths H, Jonikas MC, Smith AM, McCormick AJ. 2015. Introducing an algal carbon-concentrating mechanism into higher plants: location and incorporation of key components. Plant Biotechnology Journal 14, 1302–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger MR. 1985. Photosynthetic oxygen exchange. Annual Review of Plant Physiology 36, 27–53. [Google Scholar]
- Badger MR, Kaplan A, Berry JA. 1980. Internal inorganic carbon pool of Chlamydomonas reinhardtii: evidence for a carbon dioxide-concentrating mechanism. Plant Physiology 66, 407–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger MR, Palmqvist K, Yu J-W. 1994. Measurement of CO2 and HCO3− fluxes in cyanobacteria and microalgae during steady-state photosynthesis. Physiologia Plantarum 90, 529–536. [Google Scholar]
- Blaby IK, Blaby-Haas CE, Tourasse N et al. 2014. The Chlamydomonas genome project: a decade on. Trends in Plant Science 19, 672–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Rivero A, Shutova T, Román MJ, Villarejo A, Martinez F. 2012. Phosphorylation controls the localization and activation of the lumenal carbonic anhydrase in Chlamydomonas reinhardtii. PLoS One 7, e49063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkhsenious ON, Mason CB, Moroney JV. 1998. The intracellular localization of ribulose-1,5-bisphosphate carboxylase/oxygenase in Chlamydomonas reinhardtii. Plant Physiology 116, 1585–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briesemeister S, Rahnenführer J, Kohlbacher O. 2010. YLoc—an interpretable web server for predicting subcellular localization. Nucleic Acids Research 38, W497–W502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duanmu D, Miller AR, Horken KM, Weeks DP, Spalding MH. 2009. Knockdown of limiting-CO2-induced gene HLA3 decreases HCO3– transport and photosynthetic Ci affinity in Chlamydomonas reinhardtii. Proceedings of the National Academy of Sciences, USA 106, 5990–5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel BD, Schaffer M, Kuhn Cuellar L, Villa E, Plitzko JM, Baumeister W. 2015. Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo-electron tomography. Elife 13, 04889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N, Rochaix JD. 2001. The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii. Molecular Genetics and Genomics 265, 888–894. [DOI] [PubMed] [Google Scholar]
- Fuhrmann M, Oertel W, Hegemann P. 1999. A synthetic gene coding for the green fluorescent protein (GFP) is a versatile reporter in Chlamydomonas reinhardtii. The Plant Journal 19, 353–361. [DOI] [PubMed] [Google Scholar]
- Furumoto T, Yamaguchi T, Ohshima-Ichie Y et al. 2011. A plastidial sodium-dependent pyruvate transporter. Nature 476, 472–475. [DOI] [PubMed] [Google Scholar]
- Gao H, Wang Y, Fei X, Wright DA, Spalding MH. 2015. Expression activation and functional analysis of HLA3, a putative inorganic carbon transporter in Chlamydomonas reinhardtii. The Plant Journal 82, 1–11. [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, Yatusevich R, Rollwitz I, Humphry M, Gershenzon J, Flügge UI. 2009. The plastidic bile acid transporter 5 is required for the biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana. The Plant Cell 21, 1813–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano M, Beardall J, Raven JA. 2005. CO2 concentrating mechanisms in algae: mechanisms, environmental modulation, and evolution. Annual Review of Plant Biology 56, 99–131. [DOI] [PubMed] [Google Scholar]
- Gutknecht J, Bisson MA, Tosteson FC. 1977. Diffusion of carbon dioxide through lipid bilayer membranes: effects of carbonic anhydrase, bicarbonate, and unstirred layers. Journal of General Physiology 69, 779–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EHS, Stern D, Witman G. eds 2009. The Chlamydomonas sourcebook. Amsterdam: Academic Press. [Google Scholar]
- He L, Vasiliou K, Nebert DW. 2009. Analysis and update of the human solute carrier (SLC) gene superfamily. Human Genomics 3, 195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im CS, Grossman AR. 2002. Identification and regulation of high light-induced genes in Chlamydomonas reinhardtii. The Plant Journal 30, 301–313. [DOI] [PubMed] [Google Scholar]
- Jungnick N, Ma Y, Mukherjee B, Cronan JC, Speed DJ, Laborde SM, Longstreth DJ, Moroney JV. 2014. The carbon concentrating mechanism in Chlamydomonas reinhardtii: finding the missing pieces. Photosynthesis Research 121, 159–173. [DOI] [PubMed] [Google Scholar]
- Karlsson J, Clarke AK, Chen ZY, Hugghins SY, Park YI, Husic HD, Moroney JV, Samuelsson G. 1998. A novel α-type carbonic anhydrase associated with the thylakoid membrane in Chlamydomonas reinhardtii is required for growth at ambient CO2. EMBO Journal 17, 1208–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols 10, 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchitsu K, Tsuzuki M, Miyachi S. 1988. Changes of starch localization within the chloroplast induced by changes in CO2 concentration during growth of Chlamydomonas reinhardtii: independent regulation of pyrenoid starch and stroma starch. Plant and Cell Physiology 29, 1269–1278. [Google Scholar]
- Ma Y, Pollock SV, Xiao Y, Cunnusamy K, Moroney JV. 2011. Identification of a novel gene, CIA6, required for normal pyrenoid formation in Chlamydomonas reinhardtii. Plant Physiology 156, 884–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinder LCM, Meyer MT, Mettler-Altmann T et al. 2016. A repeat protein links Rubisco to form the eukaryotic carbon-concentrating organelle. Proceedings of the National Academy of Sciences, USA 113, 5958–5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariscal V, Moulin P, Orsel M, Miller AJ, Fernández E, Galván A. 2006. Differential regulation of the Chlamydomonas Nar1 gene family by carbon and nitrogen. Protist 157, 421–433. [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O et al. 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra M, Mason CB, Xiao Y, Ynalvez RA, Lato SM, Moroney JV. 2005. The carbonic anhydrase gene families of Chlamydomonas reinhardtii. Canadian Journal of Botany 83, 780–795. [Google Scholar]
- Moroney JV, Kitayama M, Togasaki RK, Tolbert NE. 1987. Evidence for inorganic carbon transport by intact chloroplasts of Chlamydomonas reinhardtii. Plant Physiology 83, 460–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroney JV, Ma Y, Frey WD, Fusilier KA, Pham TT, Simms TA, DiMario RJ, Yang J, Mukherjee B. 2011. The carbonic anhydrase isoforms of Chlamydomonas reinhardtii: intracellular location, expression, and physiological roles. Photosynthesis Research 109, 133–149. [DOI] [PubMed] [Google Scholar]
- Moroney J, Tolbert NE, Sears B. 1986. Complementation analysis of the inorganic carbon concentrating mechanism of Chlamydomonas reinhardtii. Molecular and General Genetics 204, 199–203. [Google Scholar]
- Moroney JV, Ynalvez RA. 2007. Proposed carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii. Eukaryotic Cell 6, 1251–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morth JP, Pedersen BP, Buch-Pedersen MJ, Andersen JP, Vilsen B, Palmgren MG, Nissen P. 2011. A structural overview of the plasma membrane Na+,K+-ATPase and H+-ATPase ion pumps. Nature Reviews. Molecular Cell Biology 12, 60–70. [DOI] [PubMed] [Google Scholar]
- Nakajima K, Tanaka A, Matsuda Y. 2013. SLC4 family transporters in a marine diatom directly pump bicarbonate from seawater. Proceedings of the National Academy of Sciences, USA 110, 1767–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi N, Mukherjee B, Tsujikawa T, Yanase M, Nakano H, Moroney JV, Fukuzawa H. 2010. Expression of a low CO2-inducible protein, LCI1, increases inorganic carbon uptake in the green alga Chlamydomonas reinhardtii. The Plant Cell 22, 3105–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock SV, Mukherjee B, Bajsa-Hirschel J, Machingura MC, Mukherjee A, Grossman AR, Moroney JV. 2017. A robust protocol for efficient generation, and genomic characterization of insertional mutants of Chlamydomonas reinhardtii. Plant Methods 13, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pootakham W, Gonzalez-Ballester D, Grossman AR. 2010. Identification and regulation of plasma membrane sulfate transporters in Chlamydomonas. Plant Physiology 153, 1653–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price GD, Badger MR, Woodger FJ, Long BM. 2008. Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation, and prospects for engineering into plants. Journal of Experimental Botany 859, 1441–1461. [DOI] [PubMed] [Google Scholar]
- Price GD, Howitt SM. 2011. The cyanobacterial bicarbonate transporter BicA: its physiological role and the implications of structural similarities with human SLC26 transporters. Biochemistry and Cell Biology 89, 178–188. [DOI] [PubMed] [Google Scholar]
- Price GD, Woodger FJ, Badger MR, Howitt SM, Tucker L. 2004. Identification of a SulP-type bicarbonate transporter in marine cyanobacteria. Proceedings of the National Academy of Sciences, USA 101, 18228–18233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushkin A, Kurtz I. 2006. SLC4 base (HCO3–, CO32–) transporters: classification, function, structure, genetic diseases, and knockout models. American Journal of Physiology. Renal Physiology 290, F580–F599. [DOI] [PubMed] [Google Scholar]
- Raven J. 1997. Putting the C in phycology. European Journal of Phycology 32, 319–333. [Google Scholar]
- Rawat M, Henk MC, Lavigne LL, Moroney JV. 1996. Chlamydomonas reinhardtii mutants without ribulose-1,5-bisphosphate carboxylase-oxygenase lack a detectable pyrenoid. Planta 198, 263–270. [Google Scholar]
- Romero MF, Chen AP, Parker MD, Boron WF. 2013. The SLC4 family of bicarbonate (HCO3–) transporters. Molecular Aspects of Medicine 34, 159–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada Y, Toyooka K, Kuwahara A, Sakata A, Nagano M, Saito K, Hirai MY. 2009. Arabidopsis bile acid:sodium symporter family protein 5 is involved in methionine-derived glucosinolate biosynthesis. Plant and Cell Physiology 50, 1579–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss JA. 1990. A Chlamydomonas gene encodes a G protein beta subunit-like polypeptide. Molecular and General Genetics 221, 443–452. [DOI] [PubMed] [Google Scholar]
- Shibata M, Katoh H, Sonoda M, Ohkawa H, Shimoyama M, Fukuzawa H, Kaplan A, Ogawa T. 2002. Genes essential to sodium-dependent bicarbonate transport in cyanobacteria: function and phylogenetic analysis. Journal of Biological Chemistry 277, 18658–18664. [DOI] [PubMed] [Google Scholar]
- Shimogawara K, Fujiwara S, Grossman A, Usuda H. 1998. High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 148, 1821–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova I, Fuhrmann M, Hegemann P. 2001. A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene 277, 221–229. [DOI] [PubMed] [Google Scholar]
- Sueoka N. 1960. Mitotic replication of de-oxyribonucleic acid in Chlamydomonas reinhardi. Proceedings of the National Academy of Sciences, USA 46, 83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sültemeyer DF, Klöck G, Kreuzberg K, Fock HP. 1988. Photosynthesis and apparent affinity for dissolved inorganic carbon by cells and chloroplasts of Chlamydomonas reinhardtii grown at high and low CO2 concentrations. Planta 176, 256–260. [DOI] [PubMed] [Google Scholar]
- Taylor AR, Brownlee C, Wheeler GL. 2012. Proton channels in algae: reasons to be excited. Trends in Plant Science 17, 675–684. [DOI] [PubMed] [Google Scholar]
- Wang Y, Duanmu D, Spalding MH. 2011. Carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii: inorganic carbon transport and CO2 recapture. Photosynthesis Research 109, 115–122. [DOI] [PubMed] [Google Scholar]
- Wang Y, Spalding MH. 2014. Acclimation to very low CO2: contribution of limiting CO2 inducible proteins, LCIB and LCIA, to inorganic carbon uptake in Chlamydomonas reinhardtii. Plant Physiology 166, 2040–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano T, Sato E, Iguchi H, Fukuda Y, Fukuzawa H. 2015. Characterization of cooperative bicarbonate uptake into chloroplast stroma in the green alga Chlamydomonas reinhardtii. Proceedings of the National Academy of Sciences, USA 112, 7315–7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano T, Tsujikawa T, Hatano K, Ozawa S, Takahashi Y, Fukuzawa H. 2010. Light and low-CO2-dependent LCIB–LCIC complex localization in the chloroplast supports the carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant and Cell Physiology 51, 1453–1468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.