Implementing new methodology for sampling rhizosecreted glucosinolates from Arabidopsis, we discovered that import from apoplast is a prerequisite for the translocation of stele-synthesized phytochemicals across the endodermis barrier and into the rhizosphere.
Keywords: Arabidopsis, biosynthesis, exudate, glucosinolates (GLS), glucosinolates catabolites, GTR, import, rhizosphere, rhizosecretion, stele-synthesized phytochemicals, trans-cellular transport
Abstract
Casparian strip-generated apoplastic barriers not only control the radial flow of both water and ions but may also constitute a hindrance for the rhizosecretion of stele-synthesized phytochemicals. Here, we establish root-synthesized glucosinolates (GLS) are in Arabidopsis as a model to study the transport routes of plant-derived metabolites from the site of synthesis to the rhizosphere. Analysing the expression of GLS synthetic genes in the root indicate that the stele is the major site for the synthesis of aliphatic GLS, whereas indole GLS can be synthesized in both the stele and the cortex. Sampling root exudates from the wild type and the double mutant of the GLS importers GTR1 and GTR2 show that GTR-mediated retention of stele-synthesized GLS is a prerequisite for the exudation of both intact GLS and their catabolites into the rhizosphere. The expression of the GTRs inside the stele, combined with the previous observation that GLS are exported from biosynthetic cells, suggest three possible routes of stele-synthesized aliphatic GLS after their synthesis: (i) GTR-dependent import to cells symplastically connected to the cortical cells and the rhizosphere; (ii) GTR-independent transport via the xylem to the shoot; and (iii) GTR-dependent import to GLS-degrading myrosin cells at the cortex. The study suggests a previously undiscovered role of the import process in the rhizosecretion of root-synthesized phytochemicals.
Introduction
The rhizosphere, the zone closely surrounding the roots where the exchange of micronutrients and organic matter occurs, is pivotal for both symbiotic and pathogenic interactions between the plant and soil organisms. There is an increased awareness of the importance of root exudation for the establishment of the microbial community in the rhizosphere (Bais et al., 2006; Badri et al., 2009b; Baetz and Martinoia, 2014; Carvalhais et al., 2015) and in the response to environmental signals (Hoehenwarter et al., 2016; Ziegler et al., 2016). Depending on the species, the rhizosphere is apoplastically isolated from the rest of the root by the endodermis and/or exodermis with their Casparian strip-impregnated cells. Compounds synthesized outside or within the endodermis and/or exodermis can be released to the rhizosphere by passive diffusion or through specific exporters (Badri et al., 2009a; Weston et al., 2012). Compounds that are synthesized and released into the apoplast behind the apoplastic barriers (endodermis or exodermis) can follow one of two transport routes from the inside to the outside of the endodermal/exodermal cells. The first route is mediated by plasmodesmata (PD) connecting biosynthetic cells symplastically to the endodermis from which they are released by exporters to the rhizosphere. The second route is trans-cellular and involves the repeated efflux and influx through consecutively arranged and polarized carrier proteins (Andersen et al., 2015). Even though several transporters have been identified for releasing compounds from root cells, these are all involved in the efflux of metabolites from cells outside the Casparian strip (Sugiyama et al., 2007; Badri et al., 2009a; Fourcroy et al., 2014; Sasse et al., 2015). By contrast, little is known about the transport route to the rhizosphere of specialized metabolites synthesized within the stele.
Arabidopsis thaliana (hereafter Arabidopsis) with glucosinolates (GLS) as major defence compounds has become an ideal model system to study the mechanism of transport of specialized metabolites because of its powerful genetic toolbox (Jørgensen et al., 2015). Arabidopsis produces tryptophan-derived indole GLS and methionine-derived aliphatic GLS which are further divided into short-chained (with three to five methylene groups, C3–C5) and long-chained (with six to eight methylene groups, C6–C8) aliphatic GLS (Grubb and Abel, 2006), of which the latter are primarily synthesized and stored in the roots (Andersen et al., 2013). Previous studies showed that root cells synthesizing long-chained aliphatic GLS by default export these GLS to the apoplast from where the two plasma-membrane localized glucosinolate importers, GTR1 and GTR2 (Nour-Eldin et al., 2012), are suggested to import them into local storage cells (Andersen et al., 2013). Indeed, gtr1 gtr2 roots contain drastically reduced levels of long-chained, aliphatic GLS whereas rosettes over-accumulate them. This over-accumulation is caused by translocation of the exported GLS via the xylem (Andersen et al., 2013; Madsen et al., 2014). However, the total levels of GLS in gtr1 gtr2 roots and rosettes are higher compared with wild-type levels. This shows that, in the absence of GTR1 and GTR2 which are both expressed in the root, higher amounts of GLS remain within the plant. It is conceivable that these higher amounts of GLS accumulating in the gtr1 gtr2 mutant were destined for rhizosecretion. This hypothesis proposes that GTR-mediated GLS import plays an important role in the exudation pathway of stele-synthesized GLS. Noticeably, the tissue-specific localization of GLS biosynthesis has not previously been determined in Arabidopsis roots using non-invasive approaches. In addition, although exudation of the GSLs and/or their catabolites has been observed (Schreiner et al., 2011) and suggested in Brassica crops (Choesin and Boerner, 1991; McCully et al., 2008), intact GLS have not previously been identified in the root exudates of the model plant Arabidopsis, whereas biosynthetic intermediates and catabolites of GLS have been detected in different metabolomics studies (Badri et al., 2013; Strehmel et al., 2014, 2016; Monchgesang et al., 2016; Ziegler et al., 2016).
It is shown here that long-chained, aliphatic GLS are synthesized in the stele of Arabidopsis roots and exuded into the rhizosphere. A new sampling method has enabled the detection of both intact GLS and their catabolites in Arabidopsis root exudates. Therefore, we propose that elucidating the transport pathway of long-chained, aliphatic GLS provides a model system for understanding the root exudation of stele-synthesized compounds in general. As a first step towards investigating the transport route of GLS from the site of synthesis to the rhizosphere, we compared root exudates of gtr1 gtr2 with the wild type and determined the localization of GTR at the cellular level. Our data indicate that GTR1 and GTR2 import into the symplasm is a prerequisite for the translocation of stele-synthesized GLS across the apoplastic barrier of the endodermis into the rhizosphere.
Materials and methods
Plant growth and sampling root exudates
A. thaliana Col-0 and mutants with a T-DNA insertion in both GTR1 (At3g47960) and GTR2 (At5g62680; gtr1 gtr2 dKO) were grown on a non-sterile standardized plant growth substrate (Fruhstorfer Erde type P, Germany) with a pH value of 6.0 in a climate chamber under short-day conditions (8/16h light/ dark, 22 °C, 40–60% humidity). After 2 weeks, single plants were transferred to sand-filled pots and watered with nutrient solution as described by Gibeaut et al. (1997). After an additional 4 weeks, the sand was carefully removed from the roots and the plants were transferred to tubes filled with distilled water. After 1h, plants were transferred to fresh tubes (two plants per tube) filled with bi-distilled water and exudates were collected for 4h. In order to determine whether enzymatic hydrolysis of exuded GLS takes place, 100 µl of 0.1mM 2-propenyl GLS (Carl Roth GmbH, Germany) was added to each tube. During the whole procedure, plants were kept under the cultivation conditions. Exudates were filtered through a mixed cellulose ester membrane filter of pore size 0.22 µm (Carl Roth GmbH, Germany) to remove cellular debris and external microorganisms. Exudates from approximately 20 plants were pooled to give one sample and three samples were collected for each harvest. After collection of exudates, the roots were harvested and the fresh weight was determined to relate the amount of exudates to root biomass.
Construct cloning
Derivatives of the USER™ compatible pCambia1300U and pCambia2300U plant expression vectors (Nour-Eldin et al., 2006) were used in this work. The PCR product was then introduced into the PacI and Nt.BbvCI digested pCambia2300U vector by USER™ cloning. The mVenus variant of the yellow fluorescent protein was amplified with the same primer pair and introduced into the opened pCambia1300U vector.
The promoter sequences including upstream genomic sequences and the 5′ UTR of the CYP83A1, CYP83B1 genes (1033bp and 1001bp, upstream of the start codon, respectively) were amplified from genomic DNA of Arabidopsis thaliana ecotype Columbia-0. The corresponding coding sequences were amplified from Arabidopsis thaliana ecotype Columbia-0 cDNA. The promoter and coding sequences were seamlessly fused by USER™ fusion (Geu-Flores et al., 2007) and, at the same time, inserted into the USER™ cassette of the opened pCambia1300-mVenus (CYP83A1, CYP83B1) vectors. The proper fusion and insertion of sequences was confirmed by DNA sequencing.
Cellular localization
Localization experiments were done on 12–14-d-old plants using a Leica SP5-X confocal microscope. Fluorophores were excited at the respective absorption maxima (514nm for YFP and 546nm for mOrange). Emission was collected at 525–540nm (YFP), and 560–580nm (mOrange).
GUS assay and histology
Detection of GUS expression was performed with 5-bromo-4-chloro-3-indolyl-β-d-glucuronidase (X-Gluc). Tissues were vacuumed in 50mM sodium phosphate buffer, pH 7.0 (1% Triton X-100/1% DMSO/10mM EDTA/0.5mM potassium ferrocyanide/0.5mM potassium ferricyanide/1mM X-Gluc) and then incubated at 37 °C. The stained samples were washed twice with 70% ethanol, fixed in 4% formaldehyde, and cleared with 70% ethanol. Tissues were mounted in 50% glycerol, and microscopy was conducted. Histological analysis followed by sections was performed as described with some modifications. Briefly, tissues were gently fixed in 90% acetone, washed with water, incubated in 50mM sodium phosphate buffer, pH 7.0 (0.2% Triton X-100Z/10mM EDTA/10mM potassium ferrocyanide/10mM potassium ferricyanide/1mM X-Gluc), at 37 °C. The stained tissues were fixed in 4% paraformaldehyde, dehydrated with acetone then embedded in Spurr Resin. Resin sections (5 μm) were microtomed (Leica) and were observed after staining.
GLS analysis
Desulfo-GLS profiles and concentrations of lyophilized plant tissue were determined as described before (by Witzel et al. (2015) with slight modifications: For the determination of GLS in exudates, the total exudate volume (500ml) was first subjected to catabolites analysis (see below). Then the exudate was spiked with 20 nmol of the internal standard (IST) 4-hydroxybenzyl GLS (Carl Roth GmbH, Karlsruhe, Germany) and loaded onto a 1ml DEAE-Sephadex A-25 ion-exchanger column (Sigma-Aldrich Chemie GmbH, Steinheim, Germany), further treated and analysed with the UHPLC-system as reported previously (Witzel et al., 2015).
Analysis of GLS breakdown products
For the determination of enzymatically formed breakdown products of the GLS in fresh plant tissues, benzonitrile (≥99.9%, Sigma-Aldrich Chemie GmbH) was added as an IST and catabolites were extracted with dichloromethane (Carl Roth GmbH; GC Ultra Grade) as described previously (Witzel et al., 2015). For the analysis of glucosinolate breakdown products in exudates, the total volume of each exudate (500ml) was extracted with 50ml of dichloromethane in a separating funnel. The separated dichloromethane layer was dried using anhydrous NaSO4 (VWR International GmbH, Darmstadt, Germany; ≥99%) and the aqueous phase was re-extracted twice using 25ml of dichloromethane. The combined dichloromethane extract was dried under nitrogen to 300 µl and subjected to GC-MS analysis as described above.
Statistical data analysis
To represent the changes in GLS levels, two-tailed Student’s t tests and one-way ANOVA followed by Tukey HSD Calculator multiple comparison post-hoc analysis (P <0.05) were performed using Origin Pro 2015.
In silico analysis
Cell-type-specific expression was derived from microarray studies of RNA bound to ribosomes which were immunoprecipitated by use of epitope-tagged ribosomal protein from seedlings (Mustroph et al., 2009; http://efp.ucr.edu/).
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or EMBL/GenBank data libraries under accession numbers: At3g47960 (GTR1); At5g62680 (GTR2); At AT4G13770 (CYP83A1); AT4G31500 (CYP83B1).
Results
Sampling of Arabidopsis root exudate
Towards our goal of understanding the root exudation process using Arabidopsis and GLS as a model system, we developed a method to measure GLS and their catabolites in Arabidopsis root exudates under physiological conditions (see Supplementary Fig. S1A–C at JXB online). Based on previous reports, we optimized several steps in the procedure. First, we optimized root growth conditions by growing plants in sand instead of using a hydroponics system (Badri et al., 2013; Strehmel et al., 2014). Secondly, the exudation was carried out in de-ionized water instead of growth media. Thirdly, dichloromethane (CH2Cl2) was used to denature the myrosinase (Sah, 1999) while partitioning intact GLS to the aqueous phase and catabolites to the organic phase. Finally, we applied DEAE-Sephadex anion exchange chromatography to enrich GLS, followed by targeted HPLC and LC-MS detection methods to measure intact GLS (Brown et al., 2003; Mikkelsen et al., 2003).
We first tested the recovery efficiency of GLS in exudates by adding exogenous 2-propenyl GLS standard (a GLS not found in Arabidopsis Col-0) to the medium prior to sampling. We were able to recover 37±8% of the added 2-propenyl GLS while another 8±3% was recovered as the corresponding catabolite product allyl-ITC (AITC) (Supplementary Fig. S1D). As 2-propenyl GLS can be taken up and translocated from the root to the shoot (Francisco et al., 2016), 2-propenyl GLS was analysed in the whole plant. No 2-propenyl GLS or AITC in either shoot or root tissue was detected (Supplementary Tables S1, S2). We concluded that, in the current sampling method, approximately 50% GLS was recovered from the exudate, of which approximately 75% GLS remained intact and approximately 25% was degraded during sample preparation.
GLS and their catabolites in root exudates of the wild type
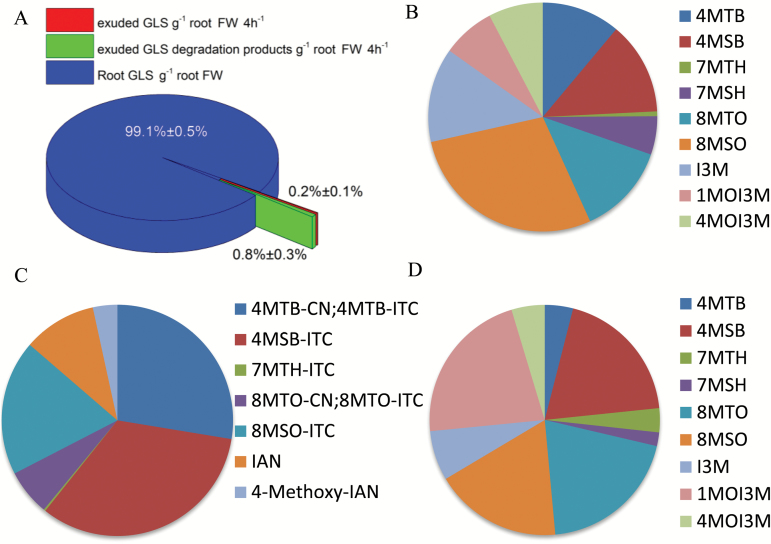
Implementing our method, we set out to isolate both intact GLS and their catabolites in root exudates from Arabidopsis Col-0. After 4h, the total amount of collected GLS and their catabolites per gram of root tissue represented approximately 1% of GLS per gram of root tissue (Fig. 1A). The ratio of GLS to their catabolites was 1:4. In root exudates from the wild type, all three classes of GLS, i.e. short-chained and long-chained aliphatic GLS as well as indole GLS, were present (Fig. 1B; Table 1). The profile of aliphatic GLS in the root exudate was similar to the endogenous root profile, whereas the profile of indole GLS differed between the root and the exudate (Fig. 1B, D; Table 1). For several of the GLS catabolites, the profile in root exudates differed from the profile of the GLS in root exudates (Fig. 1B, C; Table 1) and from the catabolite profile of homogenized root tissue (Supplementary Table S2). The difference in profile of GLS and their catabolites between root and exudate (Fig. 1; Table 1) suggests that hydrolysis does not occur during sampling and extraction. Consistent with this observation, in the root exudate the ratio of catabolites to intact GLS is 4:1 while the ratio of catabolites to intact GLS for the exogenous 2-propenyl GLS is 1:4.
Fig. 1.
Profiling of GLS and their catabolites in Arabidopsis Col-0 root exudates. (A) Abundance of exuded GLS and catabolites per gram of root tissue collected in 4h. (B–D) Comparison of the profiles of GLS and their catabolites in different samples. The average percentage of each compound of at least two independent experiments are shown; for the details see Table 1. (B) GLS profile in the root exudate. (C) Profile of GLS catabolites in the root exudate. (D) GLS profile in the root.
Table 1.
Comparison of the profiles of GLS and their catabolites in Arabidopsis Col-0 roots and root exudates.
Quantities are derived from the mean of at least two biological repeats each with three batches of plants (each n=20). GLS profiles of roots and root exudates, as well as the catabolite profile in root exudates were determined. To enable comparison between the amount of GLS and catabolites, the catabolites were converted into their corresponding GLS precursors. The values were determined by one-way ANOVA followed by Tukey HSD Calculator multiple comparison post-hoc analysis (P <0.05). The significance of differences between the profiles of roots and exudates in the respective comparisons are indicated by letters. Errors denote standard deviation. 4MTB, 4-(methylthio)butyl; 7MTH, 7-(methylthio)heptyl; 8MTO, 8-(methylthio)octyl; 3MSP, 3-(methylsulfinyl)propyl; 4MSB, 4-(methylsulfinyl)butyl; 7MSH, 7-(methylsulfinyl)heptyl; 8MSO, 8-(methylsulfinyl)octyl; I3M, 3-indolylmethyl; 1MOI3M, 1-methoxy-3-indolylmethyl; 4MOI3M, 4-methoxy-3-indolylmethyl glucosinolate. n.d. Not detected.
| Root | Exudate | Exudate | |
|---|---|---|---|
| GLS(%)a | GLS(%)a | GLS precursors of catabolites (%)b | |
| Short-chained aliphatic GLS | |||
| 4MTB | 3.9±1.5 a | 11.1±8.8 a | 27.6±9.2 b |
| 4MSB | 19.4±4.6 a | 13.1±16.3 a | 33.1±17.1 a |
| Long-chained aliphatic GLS | |||
| 7MTH | 3.4±1.7 a | 0.7±0.7 a | 0.3±0.4 a |
| 7MSH | 1.9±1.6 a | 5.4±2.9 b | n.d. |
| 8MTO | 19.9±8.9 a | 12.9±11.5 a | 6.4±8.4 b |
| 8MSO | 17.9±5.0 a | 28.3±17.2 a | 18.9±30.6 a |
| Indole GLS | |||
| I3M | 7.0±0.9 a | 13.3±5.4 a | 10.2±9.6 a |
| 1MOI3M | 21.9±7.7 a | 7.5±6.3 b | n.d. |
| 4MOI3M | 4.7±2.2 a | 7.7±5.9 a | 3.4±5.9 a |
a Percentage of total GLS.
b Percentage of total GLS catabolites.
Cellular localization of the GLS synthesis enzymes CYP83A1 and CYP83B1 in the root
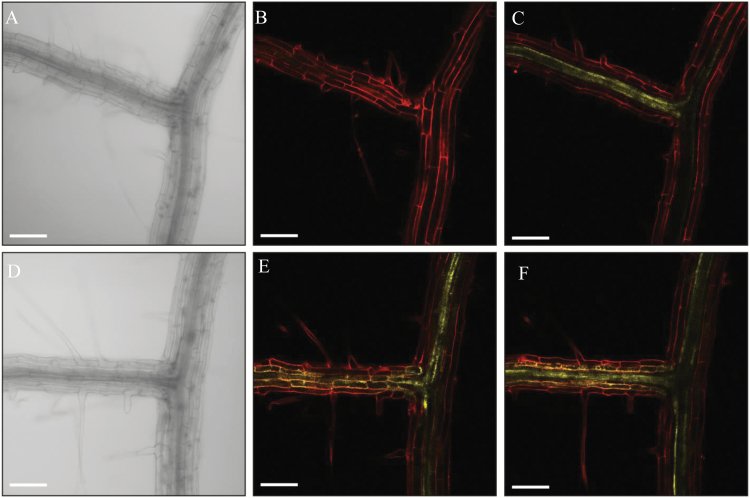
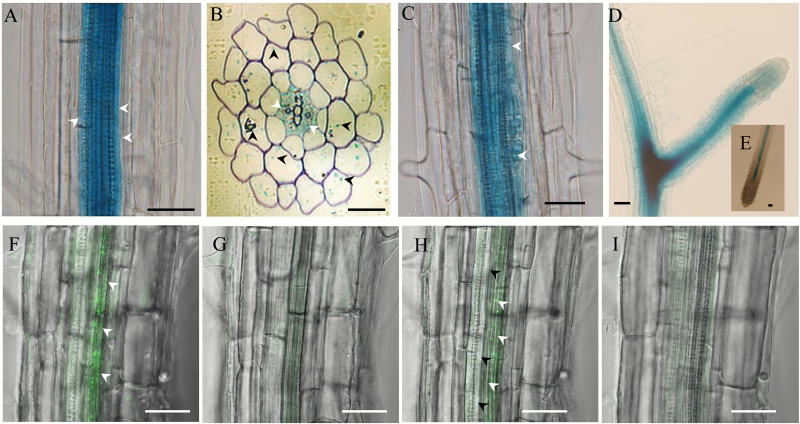
In order to define the rhizosecretion route of GLS from the site of synthesis to the rhizosphere, we first analysed the root cell type-specific localization of the fluorophore-tagged biosynthetic enzymes CYP83A1 and CYP83B1 representative of aliphatic and indolic GLS biosynthesis, respectively (Naur et al., 2003). We monitored fluorescence throughout a z-stack panel from the outer towards the inner layers of the root tissue. The CYP83A1-YFP signal was detected exclusively in the stele along the main root with relatively higher expression in the vasculature at the lateral root branch site (Fig. 2A–C). By contrast, the CYP83B1 signal was detected in both the outer and inner layers of root cells at both the main root and the lateral root branch sites (Fig. 2D–F). The stele-specific localization of aliphatic biosynthetic enzymes, combined with the presence of GLS in the rhizosphere, indicate that the transport route of aliphatic GLS from the site-of-synthesis tothe rhizosphere requires a coupled trans-cellular transport pathway and/or symplastic pathway to pass the Casparian strip. By contrast, a trans-cellular independent, but symplastic transport-dependent pathway may exist for the rhizosecretion of indole GLS.
Fig. 2.
Propidium iodide-stained roots of 12-d-old Arabidopsis seedlings expressing ProCYP83A1:CYP83A1-mVenus (A–C) or ProCYP83B1:CYP83B1-mVenus (D–F). (A, D) Bright field channel. (B, E) Focused on the cortex layer of a lateral root branch point. (C, F) Focused on the stele tissue of a lateral root branch point. Scale bar: 100 μm.
Role of GTR importers in the rhizosecretion of GLS
To explore the root exudation process of GLS, we investigated whether GTR1 and GTR2 play a role in this process. Previously, GTR1 and GTR2 have been shown to play a key role in the loading of GLS into seeds. In that study, there was no significant effect on GLS in seeds of the gtr1 single mutants and only about a 50% reduction in the gtr2 single mutants, whereas the gtr1 gtr2 double mutants had zero GLS left in seeds (Nour-Eldin et al., 2012). Due to redundancy in GTR1 and GTR2 function we have, in the present study, chosen to investigate gtr1 gtr2 double mutants to maximize any observed effect on root exudation. We first analysed the GLS distribution pattern in the rosette and roots of 6-week-old wild type and gtr1 gtr2 mutants. In gtr1 gtr2, we observed a dramatic reduction of aliphatic GLS in roots and an over-accumulation in the rosette (Supplementary Fig. S2; Table S1), in agreement with previous observations for 3-week-old hydroponically grown plants (Andersen et al., 2013). This over-accumulation of root-synthesized GLS in the rosette suggested that, in the wild type, root storage cells and/or the rhizosphere (and not the rosette) were the major sinks for those GLS.
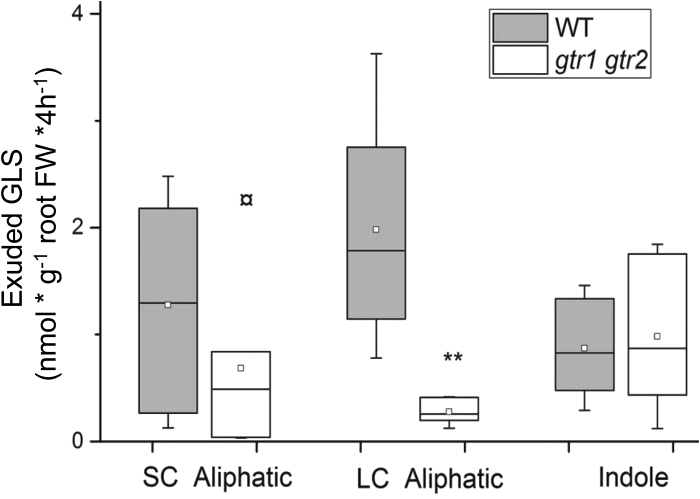
When we measured the exudates from 6-week-old wild type and gtr1 gtr2, we found that exudates of gtr1gtr2 root contained strongly reduced levels of particularly long-chained aliphatic GLS (Fig. 3; Table 2). The levels of 4MSB—the major short-chained aliphatic GLS—varied substantially between the different biological repeats, but was not significantly changed in gtr1 gtr2 compared with the wild type, as was also observed for indole GLS (Table 2). This showed that exudation of stele-synthesized, long-chained aliphatic GLS requires a GTR-mediated import process of GLS from the sites of synthesis to a symplastic domain for exudation.
Fig. 3.
Exudation of GLS in the wild type (Col-0) and gtr1 gtr2. GLS exudates were collected after 4h in water with 6-week-old, sand-grown Arabidopsis wild-type (Col-0) and gtr1gtr2 mutant plants. GLS are grouped into short-chained (SC) (C3–C5) and long-chained (LC) (C6–C8) aliphatic and indole GLS (Indole). For individual GLS data see Supplementary Table S1 and Table 1. The box is determined by the 25th and 75th percentiles. The whiskers are determined by the 5th and 95th percentiles. Median and mean are shown as a line and a square. ** Statistically significant differences of gtr1 gtr2 compared with equivalent Col-0 parts (two-tailed Students t test, P <0.05).
Table 2.
GLS present in the exudates of Arabidopsis Col-0 and the gtr1 gtr2 mutantThe quantities shown in nmol g-1 root fresh weight are derived from the mean of three biological experiments with three batches of plants (each n=20). Errors denote standard deviation. 4MTB, 4-(methylthio)butyl; 7MTH, 7-(methylthio)heptyl; 8MTO, 8-(methylthio)octyl; 3MSP, 3-(methylsulfinyl)propyl; 4MSB, 4-(methylsulfinyl)butyl; 7MSH, 7-(methylsulfinyl)heptyl; 8MSO, 8-(methylsulfinyl)octyl; I3M, 3-indolylmethyl; 1MOI3M, 1-methoxy-3-indolylmethyl; 4MOI3M, 4-methoxy-3-indolylmethyl glucosinolate. n.d. Not detected. Significant differences between Col-0 and gtr1 gtr2 in the respective tissue comparisons are indicated by asterisks (* P <0.05).
| Col-0 | gtr1 gtr2 | |
|---|---|---|
| Methylthioalkyl GLS | ||
| 4MTB | 0.49±0.39 | 0.02±0.02* |
| 7MTH | 0.03±0.03 | 0.01±0.01 |
| 8MTO | 0.57±0.51 | 0.07±0.10* |
| Methylsulfinylalkyl GLS | ||
| 4MSB | 0.58±0.72 | 0.69±1.05 |
| 7MSH | 0.24±0.13 | 0.01±0.02* |
| 8MSO | 1.25±0.76 | 0.23±0.15* |
| Indole GLS | ||
| I3M | 0.59±0.24 | 1.39±1.21 |
| 1MOI3M | 0.33±0.28 | 0.66±0.55 |
| 4MOI3M | 0.34±0.26 | 0.56±0.44 |
Role of GTR importers in rhizosecretion of GLS catabolites
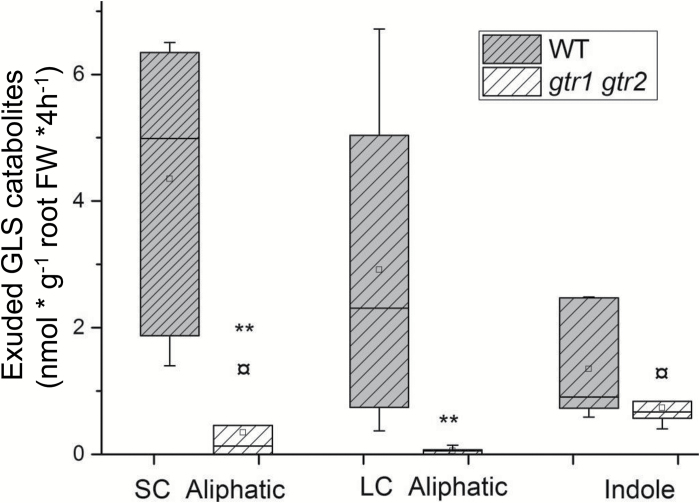
Catabolites of both short-chained and long-chained aliphatic GLS in root exudates in the gtr1 gtr2 mutants constituted 10% of the concentration in wild-type exudates (Fig. 4; Table 3). Although GLS levels in the roots of gtr1 gtr2 themselves were also decreased, the decrease was more pronounced for the catabolites than for intact GLS which suggests that GTR-mediated import into, for example, myrosin cells is a critical step prior to hydrolysis and exudation. Puzzlingly, a major part of total aliphatic GLS catabolites in root exudates of gtr1 gtr2 mutants was from short-chained aliphatic GLS (Table 3) that was not detectable in the roots of gtr1 gtr2 (Supplementary Table S1) proposing a complete hydrolysis of short-chained aliphatic GLS in the mutant. Catabolites of indole GLS were not significantly reduced in root exudates of gtr1 gtr2 mutants compared with the wild type (Fig. 4; Table 2).
Fig. 4.
Exudation of GLS catabolites in wild-type (Col-0) and gtr1 gtr2 plants. Exudate of GLS catabolites was collected after 4h in water with 6-week-old, sand-grown Arabidopsis wild-type (Col-0) and gtr1 gtr2 plants. Catabolites of GLS are grouped according to their precursors of short-chained (SC) (C3–C5) and long-chained (LC) aliphatic (C6–C8) and indole (Indole) GLS. For individual GLS catabolite data see Table 2. The whiskers are determined by the 5th and 95th percentiles. Median and mean are shown as a line and a square. ** Statistically significant differences of gtr1 gtr2 compared with equivalent Col-0 parts (two-tailed Students t test, P <0.05).
Table 3.
GLS catabolites present in the exudates of Arabidopsis Col-0 and the gtr1 gtr2 mutant
Quantities shown in nmol g-1 root fresh weight are derived from the mean of three biological experiments with three batches of plants (each n=20). Errors denote standard deviation. 4MTB-CN, 5-(methylthio)pentylnitrile; 4MTB-ITC, 4-(methylthio)butyl ITC; 7MTH-ITC, 7-(methylthio)heptyl ITC; 8MTO-CN, 9-(methylthio)nonylnitrile; 8MTO-ITC, 8-(methylthio)octyl ITC; 4MSB-ITC, 4-(methylsulfinyl)butyl ITC; 8MSO-ITC, 8-(methylsulfinyl)octyl ITC; IAN, indole-3-acetonitrile; 4-Methoxy-IAN, 4-methoxyindole-3-acetonitrile; ITC, isothiocyanate. n.d. Not detected. Significant differences between Col-0 and the gtr1 gtr2 mutant in the respective tissue comparisons are indicated by asterisks (* P <0.05).
| Col-0 | gtr1 gtr2 | |
|---|---|---|
| Methylthioalkyl catabolites | ||
| 4MTB-CN | 0.91±0.22 | 0.003±0.005* |
| 4MTB-ITC | 1.19±0.48 | n.d. |
| 7MTH-ITC | 0.02±0.03 | n.d. |
| 8MTO-CN | 0.22±0.38 | n.d. |
| 8MTO-ITC | 0.27±0.26 | 0.02±0.04 |
| Methylsulfinylalkyl catabolites | ||
| 4MSB-ITC | 2.52±1.30 | 1.53±2.54 |
| 8MSO-ITC | 1.44±2.33 | n.d. |
| Indole catabolites | ||
| IAN | 0.78±0.73 | 0.31±0.27 |
| 4-Methoxy-IAN | 0.26±0.45 | n.d. |
Cellular expression of GTR2 in the root
In order to investigate the localization of GTR-mediated import for stele-synthesized aliphatic GLS, we analysed the cell type-specific expression of GTR2. In agreement with microarray and translatome databases (Mustroph et al., 2009), a previous study indicated that GTR1 and GTR2 have similar tissue expression patterns in the root (Andersen et al., 2013). Thus, we used GTR2 reporter lines for further analysis. We first analysed the cellular expression patterns by promoter GUS fusion in Arabidopsis root (Nour-Eldin et al., 2012). Promoter activity was detected at the pericycle and inner cell layers in the stele of the elongation and maturation regions (Fig. 5A, B, arrows). Cross-sections of the root within the same region confirmed the stele accumulation and also indicated weak GTR2 expression in the cortical and epidermal cells (Fig. 5B, arrow heads). In the lateral root initiation region, GTR2 was expressed in lateral root founder cells (Fig. 5C, arrow heads), which are differentiated from specific pericycle cells from the xylem pole. At a later developmental stage of the lateral roots, the signal spread to the whole root primordia (stage III–V) and accumulated at later root branch sites (Fig. 5D). In the mature lateral root it was restricted to the vasculature and no signal was detected in the endodermal cells and root tip of the mature roots (Fig. 5A–E). Next, we analysed the cellular distribution of GTR2 protein using fluorescent protein fusion (Andersen et al., 2013). In agreement with the GUS signal, in the main root of Arabidopsis seedlings GTR2-mOrange signals were detected in the stele (Fig. 5F, G–I), of which cells next to pro-xylem cells (Fig. 5F, H) showed the strongest expression. The expression of the GTRs inside the stele (including the pericycle cells), and sporadically in the cortex, shows where the import of GLS occurs prior to exudation.
Fig. 5.
Cellular localization of GTR2 in the root. Transgenic Arabidopsis plants expressing a GTR2-mOrange fusion and GFP-GUS fusion under the control of a 2-kb promoter of GTR2 were analysed by confocal microscopy or GUS staining. (A) GUS signal shown in pericycle cells (white arrowheads) and the inner cell layers. (B) Cross-section of a root region similar to the one shown in (A). GUS signals in the stele (white arrowheads) and sporadically distributed in the cortex and epidermal cells (black arrowheads). (C) GUS signals in a lateral root initiation site (arrowheads showed lateral root founder cells). (D) GUS signals in a lateral root mature stage. (E) GUS signal in the main root tip. (f–I) mOrange signal from longitudinal optical sections along the x-y axis of intact roots of GTR2-mOrange. The mOrange signals are in green. (F) Z-stack of optical sections of intact roots of GTR2-mOrange. (G–I) Serial optical sections along the z-axis from the top to the bottom of the same region of root as in (F). The GTR2-mOrange was predominantly expressed in cells next to proxylem cells (F, H, white arrowheads) but not in proxylem cells (I, black arrowheads). Bars in (A, C-E and F–I)=25 μm. Bars in (B)=100 μm.
Discussion
Rhizosecretion of GLS and their catabolites in Arabidopsis
One prerequisite for using GLS in Arabidopsis as a model system for studying root exudation is that GLS are exuded and detectable. Accordingly, we developed a method that enables the detection of intact GLS and their catabolites simultaneously in root exudates from Arabidopsis. Previous studies indicated that other plants within the Brassicaceae family can exude both intact GLS and their catabolites (Schreiner et al., 2011; Ngala et al., 2015). Hitherto, in Arabidopsis, however, only GLS catabolites and GLS biosynthetic intermediates were identified in root exudates (Badri et al., 2013; Strehmel et al., 2014, 2016; Monchgesang et al., 2016). In our method, we collected root exudates from sand-grown plants in de-ionized water instead of nutrient medium which was previously used for hydroponically cultured plants (Badri et al., 2013; Strehmel et al., 2014). By replacing the artificial hydroponic growth conditions with sand, we reduced the hypoxia stress often seen in insufficiently aerated hydroponics. Furthermore, we collected the exudate from plants in de-ionized water instead of nutrient medium, thereby reducing the amount of salt in the samples. The previously-reported desalination step may have caused the loss of GLS and be the reason why intact GLS are rarely (if at all) detected in root exudate analyses (Strehmel et al., 2014). In addition, the previous inability to detect intact GLS in exudates may be caused by myrosinase-mediated hydrolysis during the isolation process. The use of dichloromethane (CH2Cl2) instead of AgNO3 (Schreiner et al., 2011)—to inactivate myrosinase and simultaneously partition intact GLS to the aqueous phase and catabolites to the organic phase—appears to represent an improvement. Based on the proportion of recovered intact exogenous 2-propenyl GLS, we estimate that approximately 20% of the total exuded GLS is degraded during the 4h of sampling.
It is noticeable that the profiles of GLS and GLS catabolites in root exudates differed significantly (Fig.1; Table 1), in spite of the large variance of individual GLS between biological repeats (Table 1). Assuming that GLS are hydrolysed at the same rate, our data suggest that hydrolysis does not occur during sampling and extraction, as this would have given the same profile for intact and hydrolysed GLS in the root exudate. All three classes of GLS and their catabolites, i.e. short-chained and long-chained aliphatic GLS as well as indole GLS, were present (Table 1) which, to the best of our knowledge, is the first time this has been reported.
The total amount of GLS and catabolites collected in wild-type root exudate after 4h represented approximately 1% of GLS per gram of root tissue (Fig. 1A). Hence, over a longer time, Arabidopsis roots may exudate a considerable amount of GLS and/or their catabolites into the rhizosphere where a vast diversity of plant-associated microbiota live. PEN2-dependent indole glucosinolate metabolism is required for the plant growth promotion effect by the root endophyte Colletotrichum tofieldiae in phosphate-limited conditions (Hiruma et al., 2016). Despite substantial knowledge about the defensive roles of GLS, the physiological significance of their role in the rhizosphere is largely unknown. The measurement of GLS in root exudates of the model plant Arabidopsis is a tool for dissecting the role of GLS in plant–rhizosphere interactions.
Import-dependent rhizosecretion of stele-synthesized metabolites
A key finding in this study is that GTR1- and GTR2-mediated import is critical for the rhizosecretion of stele-synthesized GLS. Intuitively, one would have anticipated that the rhizosecreted phytochemicals were produced in the outer cell layers of the root rather than in the central stele. The formation of GLS in the stele might benefit from the carbon supply from leaf-active primary metabolism via the phloem. Concurrently, the vascular-associated synthesis followed by export to the apoplast enables the efficient mobilization of defence compounds via the ascending xylem, in addition to loading on to the endodermis symplastically for exudation. This suggests that the GLS importers GTR1 and GTR2 are essential in orchestrating the flow of stele-synthesized GLS to balance above- and below-ground defence in response to environmental challenge. Many other defence compounds like nicotine (Morita et al., 2009), tropane alkaloids (Ziegler and Facchini, 2008), and pyrrolizidine alkaloids (Hartmann and Ober, 2000) are synthesized in roots and stored in leaves. Many of these compounds are also involved in rhizosphere communication (Zhuang et al., 2013; Tsednee et al., 2014). This suggests that modulating specific transport activities (in our case, plasma-membrane importers) are essential for shaping the distribution pattern of the defence compounds at the organismal level, for example, between shoot, root, and rhizosphere.
In contrast to the stele-specific synthesis of long-chained aliphatic GLS, indole GLS are synthesized in both the stele and cortex and their rhizosecretion occurs in a GTR1- and GTR2-independent manner (Figs 2, 3). As GTR1 and GTR2 show broad substrate specificity towards GLS (Nour-Eldin et al., 2012), the presence of indole GLS at similar levels in root exudates of gtr1 gtr2 and the wild type suggests that an additional and indole-specific GLS importer(s) exist which functions redundantly with GTR1 and GTR2.
The similar level of exuded 4MSB from gtr1 gtr2 mutant and wild-type roots (Fig. 4; Table 2) suggests that exudation of this glucosinolate is not dependent on GTR transport activity. Interestingly, 4MSB was detected inside cortical cells in a cell type-specific metabolite profiling of root cells (Moussaieff et al., 2013) and expression of 4MSB-specific biosynthesis genes, for example, CYP79F2 and FMOGS-OX1, includes cortical cells (Supplementary Fig. S3B) (Chen et al., 2003; Hruz et al., 2008; Li et al., 2011). Therefore, we suggest that root-synthesized 4MSB is continuously exported to the rhizosphere from cells outside the Casparian strip which agreed with the GLS distribution in canola roots (Brassica napus) by cell-specific sulphur analyses (McCully et al., 2008).
Exudation of catabolites of aliphatic GLS relies on GTR1 and GTR2 activity. A possible scenario is that the GTRs import aliphatic GLS to myrosin cells for hydrolysis prior to exudation. The expression pattern of the major root myrosinases TGG4 and TGG5 (Supplementary Fig. S3C, D) overlaps with that of GTR1 and GTR2 in the cortex (data not shown and Fig. 5B). This supports the loading of GLS into myrosin cells by GTRs.
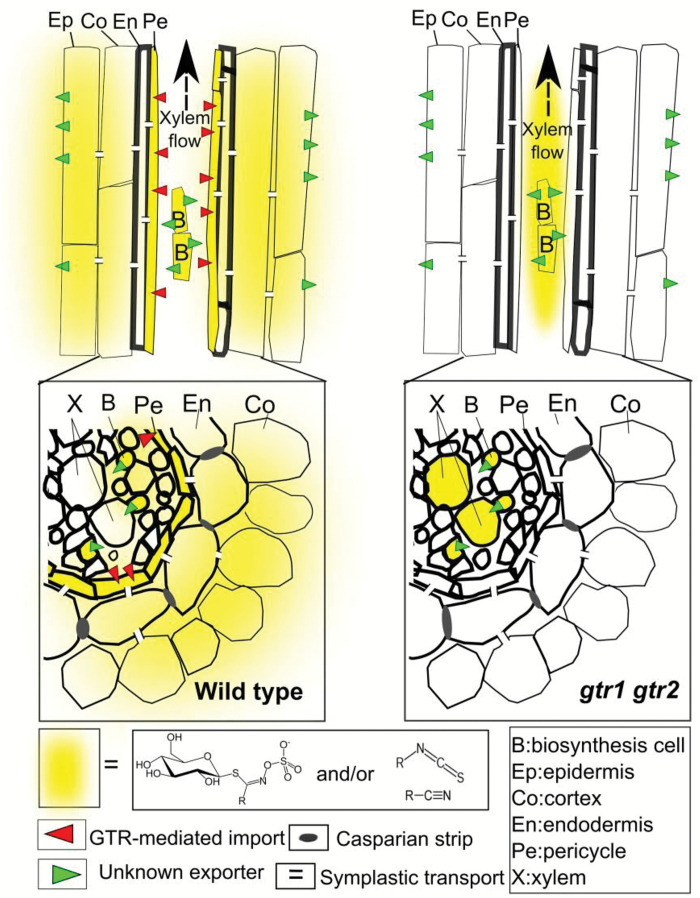
In summary, our data indicate that two import processes are critical for rhizosecretion: (i) the import of stele-synthesized long-chained aliphatic GLS into an endodermis-connected symplastic domain, and (ii) the import of short-chained aliphatic GLS from the apoplast in the cortex into myrosin cells (Fig. 6). This is an important first step in understanding the root exudation process. Future studies will reveal how intact GLS and their catabolites are exported from root cells.
Fig. 6.
Model for the transport route of stele-synthesized GLS and their catabolites to the rhizosphere in Arabidopsis. Upon synthesis in the cells adjacent to the root vasculature, aliphatic GLS are exported out of the biosynthetic cells by unknown exporter(s). A GTR1- and GTR2-dependent uptake of aliphatic GLS from the apoplast is likely to occur in cells adjacent to the xylem, For example, in the pericycle cells which are symplastically connected with the endodermal cells for exudation, as well as into phloem companion cells for long-distance transport. From endodermal cells, GLS are either exported by an unknown transporter to the cortex and rhizosphere or enter a symplastic route toward the rhizosphere via plasmodesmata. GTRs are essential for GLS entering myrosin cells which produce GLS catabolites for secretion by unknown mechanism. The GLS that are not taken up by GTRs are translocated to the shoot through the xylem. In gtr1 gtr2 mutants, the reduced levels of GLS in root exudates are due to a defect in the loading of GLS into the symplastic domain for exudation in the mutant. A consequence of the lack of this import is the translocation of the apoplastic localized, stele-synthesized GLS into the shoot through ascending xylem sap and thus a reduction of root GLS and an over accumulation of shoot GLS in the mutant.
Supplementary data
Supplementary data can be found at JXB online.
Figure S1. Sampling and profiling of GLS and their catabolites in Arabidopsis root exudates.
Figure S2. GLS concentration of rosettes and roots of 6-week-old sand-grown Arabidopsis wild type (Col-0) and gtr1gtr2.
Figure S3. Cell type-specific expression of MAM3, FMOGS-OX1, TGG4, and TGG5 in Arabidopsis roots.
Table S1. GLS in the leaf and root of Arabidopsis Col-0 and the gtr1 gtr2 mutant.
Table S2. GLS catabolites in the leaf and root of Arabidopsis.
Supplementary Material
Acknowledgements
We are grateful to Andrea Jankowsky, Rebecca Klopsch, Andrea Maikath, and Annett Platalla from the Leibniz Institute of Vegetable and Ornamental Crops for their excellent technical assistance. We thank the members of the DynaMo Center for fruitful discussions. The work was supported by the Danish National Research Foundation (grant no. DNRF99).
References
- Andersen TG, Barberon M, Geldner N. 2015. Suberization–the second life of an endodermal cell. Current Opinion in Plant Biology 28, 9–15. [DOI] [PubMed] [Google Scholar]
- Andersen TG, Nour-Eldin HH, Fuller VL, Olsen CE, Burow M, Halkier BA. 2013. Integration of biosynthesis and long-distance transport establish organ-specific glucosinolate profiles in vegetative Arabidopsis. The Plant Cell 25, 3133–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badri DV, Chaparro JM, Zhang R, Shen Q, Vivanco JM. 2013. Application of natural blends of phytochemicals derived from the root exudates of Arabidopsis to the soil reveal that phenolic-related compounds predominantly modulate the soil microbiome. The Journal of Biological Chemistry 288, 4502–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badri DV, Quintana N, El Kassis EG, Kim HK, Choi YH, Sugiyama A, Verpoorte R, Martinoia E, Manter DK, Vivanco JM. 2009a An ABC transporter mutation alters root exudation of phytochemicals that provoke an overhaul of natural soil microbiota. Plant Physiology 151, 2006–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badri DV, Weir TL, van der Lelie D, Vivanco JM. 2009b Rhizosphere chemical dialogues: plant-microbe interactions. Current Opinion in Biotechnology 20, 642–650. [DOI] [PubMed] [Google Scholar]
- Baetz U, Martinoia E. 2014. Root exudates: the hidden part of plant defense. Trends in Plant Science 19, 90–98. [DOI] [PubMed] [Google Scholar]
- Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. 2006. The role of root exudates in rhizosphere interations with plants and other organisms. Annual Review of Plant Biology 57, 233–266. [DOI] [PubMed] [Google Scholar]
- Brown PD, Tokuhisa JG, Reichelt M, Gershenzon J. 2003. Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 62, 471–481. [DOI] [PubMed] [Google Scholar]
- Carvalhais LC, Dennis PG, Badri DV, Kidd BN, Vivanco JM, Schenk PM. 2015. Linking jasmonic acid signaling, root exudates, and rhizosphere microbiomes. Molecular Plant–Microbe Interactions 28, 1049–1058. [DOI] [PubMed] [Google Scholar]
- Chen SX, Glawischnig E, Jorgensen K, Naur P, Jorgensen B, Olsen CE, Hansen CH, Rasmussen H, Pickett JA, Halkier BA. 2003. CYP79F1 and CYP79F2 have distinct functions in the biosynthesis of aliphatic glucosinolates in Arabidopsis. The Plant Journal 33, 923–937. [DOI] [PubMed] [Google Scholar]
- Choesin DN, Boerner REJ. 1991. Allyl isothiocyanate release and the allelopathic potential of Brassica napus (Brassicaceae). American Journal of Botany 78, 1083–1090. [Google Scholar]
- Fourcroy P, Siso-Terraza P, Sudre D, Saviron M, Reyt G, Gaymard F, Abadia A, Abadia J, Alvarez-Fernandez A, Briat JF. 2014. Involvement of the ABCG37 transporter in secretion of scopoletin and derivatives by Arabidopsis roots in response to iron deficiency. New Phytologist 201, 155–167. [DOI] [PubMed] [Google Scholar]
- Francisco M, Joseph B, Caligagan H, Li B, Corwin JA, Lin C, Kerwin R, Burow M, Kliebenstein DJ. 2016. The defense metabolite, allyl glucosinolate, modulates Arabidopsis thaliana biomass dependent upon the endogenous glucosinolate pathway. Frontiers in Plant Science doi: 10.3389/fpls.2016.00774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geu-Flores F, Nour-Eldin HH, Nielsen MT, Halkier BA. 2007. USER fusion: a rapid and efficient method for simultaneous fusion and cloning of multiple PCR products. Nucleic Acids Research doi: 10.1093/nar/gkm106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibeaut DM, Hulett J, Cramer GR, Seemann JR. 1997. Maximal biomass of Arabidopsis thaliana using a simple, low-maintenance hydroponic method and favorable environmental conditions. Plant Physiology 115, 317–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb CD, Abel S. 2006. Glucosinolate metabolism and its control. Trends in Plant Science 11, 89–100. [DOI] [PubMed] [Google Scholar]
- Hartmann T, Ober D. 2000. Biosynthesis and metabolism of pyrrolizidine alkaloids in plants and specialized insect herbivores. Biosynthesis: Aromatic Polyketides, Isoprenoids, Alkaloids 209, 207–243. [Google Scholar]
- Hiruma K, Gerlach N, Sacristan S, et al. 2016. Root endophyte Colletotrichum tofieldiae confers plant fitness benefits that are phosphate status dependent. Cell 165, 464–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehenwarter W, Monchgesang S, Neumann S, Majovsky P, Abel S, Muller J. 2016. Comparative expression profiling reveals a role of the root apoplast in local phosphate response. BMC Plant Biology doi: 10.1186/s12870-016-0790-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. 2008. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Advances in Bioinformatics doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen ME, Nour-Eldin HH, Halkier BA. 2015. Transport of defense compounds from source to sink: lessons learned from glucosinolates. Trends in Plant Science 20, 508–514. [DOI] [PubMed] [Google Scholar]
- Li J, Kristiansen KA, Hansen BG, Halkier BA. 2011. Cellular and subcellular localization of flavin-monooxygenases involved in glucosinolate biosynthesis. Journal of Experimental Botany 62, 1337–1346. [DOI] [PubMed] [Google Scholar]
- Madsen SR, Olsen CE, Nour-Eldin HH, Halkier BA. 2014. Elucidating the role of transport processes in leaf glucosinolate distribution. Plant Physiology 166, 1450–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCully ME, Miller C, Sprague SJ, Huang CX, Kirkegaard JA. 2008. Distribution of glucosinolates and sulphur-rich cells in roots of field-grown canola (Brassica napus). New Phytologist 180, 193–205. [DOI] [PubMed] [Google Scholar]
- Mikkelsen MD, Petersen BL, Glawischnig E, Jensen AB, Andreasson E, Halkier BA. 2003. Modulation of CYP79 genes and glucosinolate profiles in Arabidopsis by defense signaling pathways. Plant Physiology 131, 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monchgesang S, Strehmel N, Schmidt S, Westphal L, Taruttis F, Muller E, Herklotz S, Neumann S, Scheel D. 2016. Natural variation of root exudates in Arabidopsis thaliana–linking metabolomic and genomic data. Scientific Reports doi:10.1038/srep29033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Shitan N, Sawada K, Van Montagu MCE, Inzé D, Rischer H, Goossens A, Oksman-Caldentey KM, Moriyama Y, Yazaki K. 2009. Vacuolar transport of nicotine is mediated by a multidrug and toxic compound extrusion (MATE) transporter in Nicotiana tabacum. Proceedings of the National Academy of Sciences, USA 106, 2447–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussaieff A, Rogachev I, Brodsky L, Malitsky S, Toal TW, Belcher H, Yativ M, Brady SM, Benfey PN, Aharoni A. 2013. High-resolution metabolic mapping of cell types in plant roots. Proceedings of the National Academy of Sciences, USA 110, E1232–E1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJH, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J. 2009. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proceedings of the National Academy of Sciences, USA 106, 18843–18848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naur P, Petersen BL, Mikkelsen MD, Bak S, Rasmussen H, Olsen CE, Halkier BA. 2003. CYP83A1 and CYP83B1, two nonredundant cytochrome P450 enzymes metabolizing oximes in the biosynthesis of glucosinolates in Arabidopsis. Plant Physiology 133, 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngala BM, Haydock PPJ, Woods S, Back MA. 2015. Biofumigation with Brassica juncea, Raphanus sativus and Eruca sativa for the management of field populations of the potato cyst nematode Globodera pallida. Pest Management Science 71, 759–769. [DOI] [PubMed] [Google Scholar]
- Nour-Eldin HH, Andersen TG, Burow M, Madsen SR, Jorgensen ME, Olsen CE, Dreyer I, Hedrich R, Geiger D, Halkier BA. 2012. NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature 488, 531–534. [DOI] [PubMed] [Google Scholar]
- Nour-Eldin HH, Hansen BG, Norholm MHH, Jensen JK, Halkier BA. 2006. Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Research 34, e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah H. 1999. Stabilization of proteins against methylene chloride/water interface-induced denaturation and aggregation. Journal of Controlled Release 58, 143–151. [DOI] [PubMed] [Google Scholar]
- Sasse J, Simon S, Gubeli C, Liu GW, Cheng X, Friml J, Bouwmeester H, Martinoia E, Borghi L. 2015. Asymmetric localizations of the ABC transporter PaPDR1 trace paths of directional strigolactone transport. Current Biology 25, 647–655. [DOI] [PubMed] [Google Scholar]
- Schreiner M, Krumbein A, Knorr D, Smetanska I. 2011. Enhanced glucosinolates in root exudates of Brassica rapa ssp rapa mediated by salicylic acid and methyl jasmonate. Journal of Agricultural and Food Chemistry 59, 1400–1405. [DOI] [PubMed] [Google Scholar]
- Strehmel N, Böttcher C, Schmidt S, Scheel D. 2014. Profiling of secondary metabolites in root exudates of Arabidopsis thaliana. Phytochemistry 108, 35–46. [DOI] [PubMed] [Google Scholar]
- Strehmel N, Monchgesang S, Herklotz S, Kruger S, Ziegler J, Scheel D. 2016. Piriformospora indica stimulates root metabolism of Arabidopsis thaliana. International Journal of Molecular Sciences 17, 1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama A, Shitan N, Yazaki K. 2007. Involvement of a soybean ATP-binding cassettetype transporter in the secretion of genistein, a signal flavonoid in legume–Rhizobium symbiosis. Plant Physiology 144, 2000–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsednee M, Yang SC, Lee DC, Yeh KC. 2014. Root-secreted nicotianamine from Arabidopsis halleri facilitates zinc hypertolerance by regulating zinc bioavailability. Plant Physiology 166, 839–U553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston LA, Ryan PR, Watt M. 2012. Mechanisms for cellular transport and release of allelochemicals from plant roots into the rhizosphere. Journal of Experimental Botany 63, 3445–3454. [DOI] [PubMed] [Google Scholar]
- Witzel K, Hanschen FS, Klopsch R, Ruppel S, Schreiner M, Grosch R. 2015. Verticillium longisporum infection induces organ-specific glucosinolate degradation in Arabidopsis thaliana. Frontiers in Plant Science doi: 10.3389/fpls.2015.00508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang XL, Gao J, Ma AZ, Fu SL, Zhuang GQ. 2013. Bioactive molecules in soil ecosystems: masters of the underground. International Journal of Molecular Sciences 14, 8841–8868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler J, Facchini PJ. 2008. Alkaloid biosynthesis: metabolism and trafficking. Annual Review of Plant Biology 59, 735–769. [DOI] [PubMed] [Google Scholar]
- Ziegler J, Schmidt S, Chutia R, Muller J, Bottcher C, Strehmel N, Scheel D, Abel S. 2016. Non-targeted profiling of semi-polar metabolites in Arabidopsis root exudates uncovers a role for coumarin secretion and lignification during the local response to phosphate limitation. Journal of Experimental Botany 67, 1421–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.