Flaveria bidentis carbonic anhydrase 3 catalyses the first step in C4 photosynthesis, with its cognate gene containing an element that shares homology and function with the C4Flaveria MEM1 motif.
Keywords: C4 photosynthesis, carbonic anhydrase, evolution of C4 photosynthesis, Flaveria, gene expression, MEM1, mesophyll cell expression, translatome
Abstract
The first two reactions of C4 photosynthesis are catalysed by carbonic anhydrase (CA) and phosphoenolpyruvate carboxylase (PEPC) in the leaf mesophyll (M) cell cytosol. Translatome experiments using a tagged ribosomal protein expressed under the control of M and bundle-sheath (BS) cell-specific promoters showed transcripts encoding CA3 from the C4 species Flaveria bidentis were highly enriched in polysomes from M cells relative to those of the BS. Localisation experiments employing a CA3-green fluorescent protein fusion protein showed F. bidentis CA3 is a cytosolic enzyme. A motif showing high sequence homology to that of the Flaveria M expression module 1 (MEM1) element was identified approximately 2 kb upstream of the F. bidentis and F. trinervia ca3 translation start sites. MEM1 is located in the promoter of C4Flaveria ppcA genes, which encode the C4-associated PEPC, and is necessary for M-specific expression. No MEM1-like sequence was found in the 4 kb upstream of the C3 species F. pringlei ca3 translation start site. Promoter–reporter fusion experiments demonstrated the region containing the ca3 MEM1-like element also directs M-specific expression. These results support the idea that a common regulatory switch drives the expression of the C4Flaveria ca3 and ppcA1 genes specifically in M cells.
Introduction
The C4 photosynthetic pathway is an extraordinary example of convergent evolution with more than 65 independent origins among the angiosperms (Sage et al., 2012; Sage, 2016). The pathway functions as a CO2 concentrating mechanism (CCM) by increasing the levels of CO2 around Rubisco, thereby enhancing the likelihood of CO2, rather than O2, landing in the active site of the enzyme. This results in C4 plants demonstrating reduced photorespiration, increased photosynthetic rates, and greater photosynthetic water and nitrogen use efficiencies relative to C3 plants in hot, dry, high light environments (Ghannoum et al., 2011).
Two major groups of land plants using the C4 pathway have been described – one group employs both mesophyll (M) and bundle-sheath (BS) cells to fix atmospheric CO2, while species in the other group operate a C4 cycle in a single cell type (Edwards and Voznesenskaya, 2011). C4 species using M and BS cells exhibit Kranz anatomy (Haberlandt 1896), for which at least 25 different forms have been described (Edwards and Voznesenskaya, 2011), but which generally is recognised as vascular tissue surrounded by BS cells, which in turn are surrounded by M cells. Kranz C4 species have been categorised further as one of three subtypes (Gutierrez et al., 1974; Hatch et al., 1975; Hatch, 1987; Kanai and Edwards, 1999), based on the decarboxylase that shows the greatest activity in the BS: NADP-malic enzyme (NADP-ME), NAD-malic enzyme or phosphoenolpyruvate carboxykinase (PCK). In all these C4 subtypes, the M cells constitute the photosynthetic carbon acquisition tissue and contain all of the C4 form of phosphoenolpyruvate carboxylase (PEPC), the primary carboxylase in C4 plants. All the Rubisco in a C4 leaf is in the BS, and therefore, these cells compose the photosynthetic carbon reduction tissue.
While C4 plants may differ in their anatomy, primary decarboxylases, and the species of three- and four-carbon acids transferred between the M and BS, the first two reactions of the C4 pathway are invariant, and take place in the M cell cytosol. The reactions involve the conversion of atmospheric CO2 to bicarbonate (HCO3−) by the enzyme carbonic anhydrase (CA), and the subsequent utilisation of HCO3− by PEPC to form oxaloacetate through the carboxylation of phosphoenolpyruvate. Depending on the decarboxylase(s) present, the oxaloacetate is rapidly converted to malate and/or aspartate, which then diffuse into the BS where they are decarboxylated, and the released CO2 is re-fixed by Rubisco. The three-carbon organic acids resulting from the decarboxylation reaction diffuse back into the M where they can be used in another round of the C4 acid transfer cycle.
Flaveria is one of a small number of taxa containing species that are C3, others that are C4, and still others that are C3–C4 intermediates (Powell, 1978; Edwards and Ku, 1987). This dicotyledonous group has been at the forefront of research into the evolution of C4 photosynthesis, not only because it contains multiple C3 and C4 species, but also because of numerous C3–C4 intermediate species that essentially form a continuum, representing the stages along the path to the C4 syndrome from an ancestral C3 state (McKown et al., 2005; McKown and Dengler, 2007; Sage et al., 2012; Heckmann et al., 2013; Lyu et al., 2015). The PEPC and CA isoforms in the leaves of a number of Flaveria species are some of the best characterised C4-associated proteins with respect to the molecular mechanisms used during evolution that distinguish their cognate gene expression patterns, inter- and intracellular locations, and biochemistry from the ancestral C3 homologues (reviewed in Westhoff and Gowik, 2004; Ludwig, 2011).
In Flaveria, the gene family coding for PEPC consists of three classes, ppcA, ppcB and ppcC (Hermans and Westhoff, 1992; Ernst and Westhoff 1997), with the C4-associated PEPC encoded by the ppcA gene (Hermans and Westhoff, 1992; Westhoff and Gowik, 2004). The proteins encoded by orthologous ppcA genes from C3 and C4Flaveria congeners show different kinetic and regulatory properties (Svensson et al., 1997; Bläsing et al., 2000). The expression of the C4Flaveria ppcA gene in the M cytosol requires the M expression module 1 (MEM1), a 41 bp element located in the 2.2 kb region upstream of the ppcA translation start site (Gowik et al., 2004). The element is composed of A and B segments, with a guanine residue in the first position distinguishing C4 and C4-like ppcA MEM1 A segments from the orthologues of C3 and C3–C4 intermediate Flaveria species, which contain an adenine in the homologous position (Gowik et al., 2004; Akyildiz et al., 2007). Interestingly, a CACT tetranucleotide in the B segment is found in Flaveria C4, C4-like and C3–C4 intermediate ppcA MEM1 elements, but is absent in the upstream region of C3Flaveria ppcA genes (Gowik et al., 2004; Akyildiz et al., 2007). The MEM1 acts as an enhancer element, conferring M cell-specific reporter gene expression, and in combination with a proximal promoter region (PR) leads to high M expression (Gowik et al., 2004). It also represses gene activity, inhibiting ppcA expression in the BS cells and vascular bundles of the leaf (Akyildiz et al., 2007).
Three cDNAs encoding distinct CA isoforms, CA1, CA2, and CA3, have been isolated from the leaves of the C3F. pringlei and C4F. bidentis (Tetu et al., 2007; Tanz et al., 2009). F. bidentis plants genetically transformed with an antisense construct recognising CA3 mRNA showed reduced levels of total leaf CA activity (von Caemmerer et al., 2004), with transformants containing less than 10% of wild type activity exhibiting a compromised CCM, and a growth requirement for high CO2. Transcripts encoding CA3 in F. bidentis are at least 50 times more abundant than those coding for CA1 or CA2 (Tetu et al., 2007), and are an order of magnitude greater on a leaf total RNA basis than the transcripts coding for any of the CA isoforms in F. pringlei (Ludwig, 2011). Although these transgenic and quantitative analyses suggested CA3 is the C4-associated CA in Flaveria, and preferential expression is expected in the M to ensure high concentrations of HCO3− for PEPC function (Gutierrez et al., 1974; Ku and Edwards, 1975; Burnell and Hatch, 1988), the studies did not resolve whether the high level of ca3 expression in F. bidentis was in fact in a specific leaf cell type. At the protein level, studies using radiolabelled CA precursor proteins indicated that while F. pringlei CA3 was imported into isolated pea chloroplasts (Tanz et al., 2009), where CA is required for lipid biosynthesis and stress responses (DiMario et al., 2016), CA3 from F. bidentis was not, and was presumed to be a cytosolic form of the enzyme (Tetu et al., 2007), again a result in keeping with the earlier work indicating a cytosolic location of C4-associated CA isoforms (Gutierrez et al., 1974; Ku and Edwards, 1975; Burnell and Hatch, 1988). Support for a cytosolic location of CA3 also came from sequence analyses that showed relative to the N-terminus of CA3 from F. pringlei, the F. bidentis isoform lacks 72 amino acids, which have characteristics of a chloroplast targeting sequence (Tetu et al., 2007; Tanz et al., 2009). Immunocytochemistry using an anti-CA antiserum also supported a location in M cytosol in F. bidentis; however, the antiserum was not specific to CA3 (Tetu et al., 2007). CA2, which shows similar transcript abundance in leaves, roots and flowers, and is therefore unlikely to be associated with C4 photosynthesis, also localises to the cytosol in F. bidentis (Tetu et al., 2007) and may have been immunolabelled. Nevertheless, all these results led to the working hypothesis that the C4-associated CA in Flaveria evolved via the loss of the sequence coding for the chloroplast transit peptide from the C3 CA3 orthologue (Tanz et al., 2009).
Here we present unequivocal evidence that the ca3 gene of C4F. bidentis encodes the C4-associated CA isoform. We show the ca3 gene is preferentially expressed in M cells, and the encoded protein localises to the cytosol of M cells. Moreover, our initial experiments on the identification of regulatory sequences controlling ca3 gene expression show the 2.1 kb region upstream of the translation start of the genes encoding CA3 in C4Flaveria spp. contains a sequence similar to the MEM1 motif found in the promoter regions of C4Flaveria ppcA genes. The ca3 MEM1-like motif directs M cell-specific expression of the β-glucuronidase (GUS) reporter gene and, in combination with other elements in the upstream region, confers relatively high levels of reporter gene expression.
Materials and methods
Transformation of Flaveria bidentis
Flaveria bidentis was transformed as described by Chitty et al. (1994) using Agrobacterium tumefaciens strain AGL1 (Lazo et al., 1991). Integration of the chimerical genes into the F. bidentis genome was examined by PCR.
Mesophyll and bundle-sheath translatomes
F. bidentis plants were transformed with constructs that contained either the M-specific ppcA promoter of Flaveria trinervia (Stockhaus et al., 1997) or the BS-specific promoter of the gene encoding the glycine decarboxylase P subunit (GLDPA) from F. trinervia (Engelmann et al., 2008) fused to a His(6)-FLAG-tag and the coding sequence of one of the two ribosomal protein RPL18 genes of F. bidentis in the binary vector pBI121 (Jefferson et al., 1987).
The ppcA-L-Ft and GLDPA-Ft constructs described previously by Stockhaus et al. (1994) and Engelmann et al. (2008), respectively, were used as the starting points for the generation of the translatome constructs. Both were digested with XmaI and SacI to remove the uidA gene from the vector backbone. The His(6)-FLAG tagged FbRPL18 sequence was generated via PCR and the primers FbRPL18_fw and FbRPL18_rv (Supplementary Table S1 at JXB online). His(6)-FLAG-tag and restriction sites were added using PCR and overlapping extended primers (FbRPL18_rv_SacI, Tag1_FbRPL18_fw and Tag2_FbRPL18_fw_XmaI; Supplementary Table S1). The final PCR fragment was inserted into pJet1.2/blunt with the CloneJET PCR Cloning Kit (Clontech), and its sequence confirmed. The plasmids were then digested with XmaI and SacI, and the inserts introduced into pBI121 containing either the ppcA or the GLDPA promoter.
Purification of cell-specific polysomes and RNA isolation from mature leaves harvested before the onset of flowering were performed as described previously (Zanetti et al., 2005; Mustroph et al., 2009; Reynoso et al., 2015). The polysome extraction buffer, bead wash buffer, wash buffer and elution buffer were prepared as described by Reynoso et al. (2015). RNA isolation was performed by adding 2 volumes of 8 M guanidine-HCl and 3 volumes of 100% ethanol to the eluate, followed by an overnight incubation at −20 °C and 45 min of centrifugation at 15 000 g at 4 °C. After washing with 70% ethanol and resuspension in 100 µl H2O, a subsequent purification of the RNA with the RNeasy Plant Mini Kit (Qiagen) was performed as described by Mustroph et al. (2009).
RNA concentrations were measured with the NanoDrop ND-1000 (NanoDrop Technologies), and 20 ng was reverse transcribed with the QuantiTect Reverse Transcription Kit (Qiagen), following the manufacturer’s protocol. Reverse transcription quantitative PCR (RT-qPCR) was performed with a 7500 Fast Real Time machine (Applied Biosystems), and the KAPA SYBR® FAST qPCR Kit (KAPA Biosystems) using a 100-fold dilution of the cDNA and gene specific primers for CA (CAS_fw and CAS_rv; Supplementary Table S1), PPDK (PPDK_fw and PPDK_rv; Supplementary Table S1), and GLDPA (GLDPA_fw and GLDPA_rv; Supplementary Table S1). The denaturation step was for 3 min at 95 °C, followed by 40 cycles with a two-step setting of 95 °C for 3 s and 60 °C for 30 s. The delta-delta-Ct method (Livak and Schmittgen, 2001) was used to analyse the relative amount of cDNAs in M-enriched, BS-enriched RNA, and total leaf RNA (from the same isolation as the cell-type-enriched RNAs). The F. bidentis actin gene was used as an internal reference gene (Actin_fw and Actin_rv; Supplementary Table S1). Reactions were done in triplicate.
Flaveria bidentis CA3 subcellular localisation
The sequence encoding the ORF (stop codon removed) of F. bidentis CA3 was amplified from a pBluescript-CA3 template (Tanz et al., 2009) using the primers MS33-XbaI-F and MS34-AscI-R (Supplementary Table S1). The product was digested with XbaI and AscI and subcloned into the corresponding sites of the binary vector pMDC83 (Curtis and Grossniklaus, 2003) to produce the plasmid pMDC83-CA3Fbid:GFP.
Transformation and growth of Agrobacterium tumefaciens GV3101(pMP90) (Koncz and Schell, 1986) cells, as well as the growth, Agrobacterium-infiltration of Nicotiana benthamiana, subsequent protoplast preparation and confocal microscopy were carried out as described by Rolland et al. (2016). Green fluorescent protein (GFP) and chlorophyll were excited at 488 nm and emission was recorded at 499–535 and 630–735 nm, respectively.
Flaveria spp. genome walking
Genomic DNA was isolated from F. bidentis and Flaveria pringlei following the method of Marshall et al. (1996), and that from F. trinervia was isolated according to Gowik et al. (2004). Genome walking libraries for F. bidentis and F. pringlei (Universal Genomewalker), and F. trinervia (Universal GenomeWalker 2.0) were constructed according to the manufacturer’s instructions (Clontech).
To obtain the upstream regions of the F. bidentis and F. pringlei ca3 genes, adaptor primers (Clontech), and the F. bidentis and F. pringlei gene specific primers SAN15 and SAN14 (Supplementary Table S1), respectively, were used in the initial genome walking PCRs according to the manufacturer’s instructions (Clontech). Both primers hybridised to the coding regions of the respective ca3 genes, between 60 and 85 bp downstream of the translation start sites. Subsequent genome walking assays were done using the Clontech adaptor primers and gene specific primers designed from the 5′-sequences of fragments obtained in previous walking steps. Fragments of 4333 and 2256 bp upstream of the ca3 translation start codons were isolated for F. pringlei and F. bidentis, respectively.
An initial 900 bp fragment of the F. trinervia ca3 gene upstream region was isolated using adaptor primers (Clontech) and the primer EN3-R (Supplementary Table S1), which hybridised in the coding region of the F. trinervia ca3 gene. A forward primer was then designed, based on the sequence 5′ to the F. bidentis MEM1-like element (MS106-F; Supplementary Table S1), and used in combination with EN6-R, which hybridised at the 5′-end of the product of the first walk. This resulted in the amplification of a 1540 bp fragment, which included the sequence encoding the F. trinervia ca3 MEM1-like motif. To confirm the isolated fragments were contiguous, a PCR using MS112-R and MS113-F primers (Supplementary Table S1) resulted in a 1320 bp fragment that shared a 420 bp overlap with the fragment amplified with MS106 and EN6-R, and extended to 10 bp upstream of the F. trinervia ca3 translation start.
Cloning of promoter–reporter gene constructs
A 2114 bp fragment upstream of the translation start site of the F. bidentis ca3 gene was amplified with primers CA3-1 and CA3-2 (Supplementary Table S1). The primers contained the restriction sites SmaI (CA3-1) and HindIII (CA3-2) that were used to fuse the promoter to the gene encoding GUS in the plant transformation vector pBI121 (construct ca3Fb). For the construct ca3Fb-1.8, which did not contain the MEM1-like motif, a 1872 bp fragment of the F. bidentis ca3 upstream region was amplified with primers CA3_1 and CA3_3 (Supplementary Table S1), and inserted into pBI121. To fuse the MEM1-like motif to the PR of the F. trinervia ppcA promoter (construct ca3Fb-ppcAFtPR), a 74 bp fragment of the ca3 upstream region containing the MEM1-like motif, from −1943 to −1869, with respect to the ca3 AUG, was amplified with primers CA3_4 and CA3_5. The primers contained the restriction sites XbaI (CA3_4) and HindIII (CA3_5) that were used to insert the fragment adjacent to the F. trinervia ppcA PR in the construct ppcA-S-Ft (in pBI121) described in Stockhaus et al. (1994).
In situ detection of β-glucuronidase and fluorimetric activity measurements
Fluorimetric measurements of GUS activity were performed according to Jefferson et al. (1987) and Kosugi et al. (1990). The statistical significance of the difference between two data sets was analysed using the Mann–Whitney U test (Mann and Whitney, 1947). Before the onset of flowering, the fifth leaf of 40- to 50-cm tall T0 F. bidentis plants was harvested for the analyses. Histochemical GUS staining and light microscopy were performed as described by Engelmann et al. (2008).
Accession numbers
Sequence information reported in this manuscript can be found in GenBank at the National Center for Biotechnology Information under accession numbers KY239618, KY239617, and KY239619 for the upstream regions of F. pringlei, F. bidentis, and F. trinervia, respectively.
Results
Flaveria bidentis carbonic anhydrase 3 is expressed in the cytosol of mesophyll cells
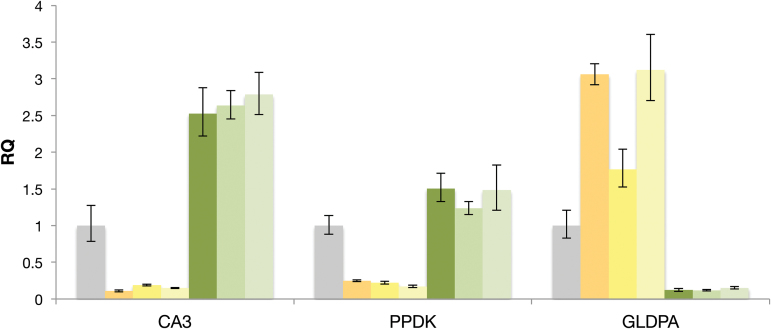
Previous work showed ca3 transcripts are the most abundant CA mRNAs in F. bidentis leaves (Tetu et al., 2007); however, the cell type in which the transcripts accumulated was not resolved. F. bidentis plants transformed with a construct encoding an epitope tagged ribosomal protein combined with affinity chromatography showed that the mRNA coding for CA3 is highly enriched in polysome complexes from leaf M cells (Fig. 1). The relative enrichment in the three individual plants examined was at least 45% greater than that of transcripts encoding PPDK, which were used as the control for M cell translation complexes. In contrast, the ca3 transcripts captured in association with leaf BS polysomes from three individual F. bidentis plants are depleted to 10% or less, whereas transcripts encoding GLDPA, show up to a three-fold enrichment in epitope-tagged polysomes isolated from BS cells (Fig. 1).
Fig. 1.
Relative quantification of Flaveria bidentis carbonic anhydrase 3 transcripts in leaf cell types. Relative quantification (RQ) of F. bidentis transcripts encoding carbonic anhydrase 3 (CA3), pyruvate orthophosphate dikinase (PPDK) and glycine decarboxylase P protein (GLDPA) associated with polysomes from the bundle-sheath cells of three individuals (yellow columns) and mesophyll cells of three individuals (green columns). Transcripts of the reference sample, i.e. polysome-associated RNA from whole leaves, were set to 1 (grey columns). Error bars represent three technical replicates.
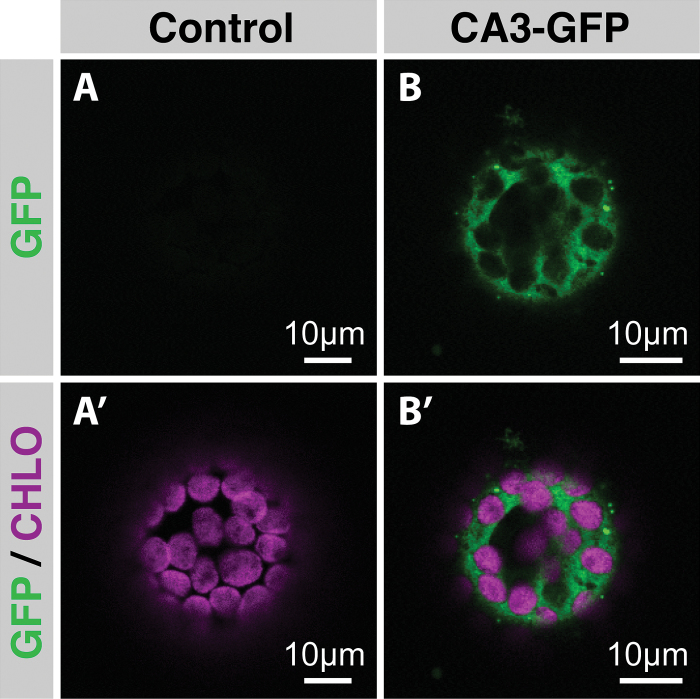
Earlier studies using radiolabelled CA3 proteins from F. bidentis and its congener the C3F. pringlei, and isolated pea chloroplasts demonstrated that unlike F. pringlei CA3, the isoform from F. bidentis was not recovered in the chloroplast fraction after the import period (Tetu et al., 2007; Tanz, et al., 2009). It was concluded that F. bidentis CA3 is a cytosolic M protein; however, it could not be ruled out that it localised to another organelle or a membrane system in M cells. To definitively show its subcellular location, N. benthamiana leaves were transformed via infiltration with Agrobacterium containing constructs encoding the ORF of F. bidentis CA3 fused to that of GFP. Protoplasts isolated 2 days post-infiltration from untransformed N. benthamiana leaves showed only chlorophyll autofluorescence (Fig. 2A, A′). In contrast, protoplasts expressing the CA3–GFP fusion protein showed a GFP signal that did not co-localise with the chlorophyll autofluorescence of the chloroplasts, but instead clearly surrounded each of the chloroplasts, indicating a cytosolic location (Fig. 2B, B′).
Fig. 2.
Subcellular localisation of Flaveria bidentis carbonic anhydrase 3. (A, A′) Single plane through a protoplast isolated from an N. benthamiana leaf that does not express GFP (A), but shows chlorophyll autofluorescence from each chloroplast (A′). (B, B′) Single plane through a protoplast from an N. benthamiana leaf expressing CA3–GFP, demonstrating that the GFP signal (B) is cytosolic and does not overlap the chlorophyll autofluorescence emitted from the chloroplasts (B′).
The upstream regions of ca3 from C4 Flaveria species contain a MEM1-like element
Genome walking was used to isolate the 5′-region of the ca3 genes from F. bidentis and F. pringlei, with the aim of identifying cis-acting motifs responsible for the differences in expression levels and patterns of the C3 and C4 orthologues. Sequence determination of the ~2.1 kb region upstream of the translation start site of the F. bidentis ca3 gene revealed a 41 bp fragment with segments showing high sequence identity to the MEM1 element responsible for M-specific expression of the ppcA gene, which codes for the C4-associated PEPC (Fig. 3; Gowik et al., 2004). The F. bidentis ca3 MEM1-like sequence consists of A and B segments homologous to those of the ppcA MEM1; however, the sequence of the ca3 A segment is inverted relative to that of the ppcA element (Fig. 3). The F. bidentis ca3 MEM1-like B segment shows little sequence identity to the ppcA Mem1 B segment, except for the CACT tetranucleotide (Fig. 3; Gowik et al., 2004). A MEM1-like element is also found in the comparable upstream region of the ca3 gene from another C4Flaveria species, F. trineriva; however, while the sequences of the two ca3 MEM1-like A segments are identical, the tetranucleotide in the B segment in F. trinervia is CATT (Fig. 3).
Fig. 3.
Structures and sequences of mesophyll expression module 1 (MEM1) and MEM1-like elements. (A) The mesophyll expression module 1 (MEM1) and MEM1-like elements of C4Flaveria ppcA and ca3 genes, respectively, are located within the first 2 kb upstream of the translation start sites of the proteins. The 41 bp elements consist of A and B segments, with the sequence of the ca3 A segment inverted relative to that of ppcA. (B) The B segments of the F. bidentis ca3 MEM1-like element, like the C4Flaveria ppcA MEM1, encodes a CACT tetranucleotide; however, the corresponding region of the MEM1-like B segment from F. trinervia ca3, is a CATT tetranucleotide. Little sequence homology is seen in the comparable upstream region of the ca3 gene from the C3 species F. pringlei. (This figure is available in colour at JXB online.)
In contrast to the two C4Flaveria species, the 2 kb upstream of the translation start site of ca3 from the C3 species, F. pringlei, does not contain a sequence with homology to either the MEM1 A or B segments (Fig. 3 and Supplementary Fig. S1). As a consequence, the sequence of the F. pringlei ca3 upstream region was extended a further 2 kb upstream; however, still no homology was found with C4 MEM1 or MEM1-like elements (data not shown). In fact, the F. pringlei ca3 upstream region shares only limited blocks of sequence homology with the corresponding regions of the two C4 species (Supplementary Fig. S1).
The MEM1-like element of the Flaveria bidentis carbonic anhydrase 3 gene directs expression in mesophyll cells
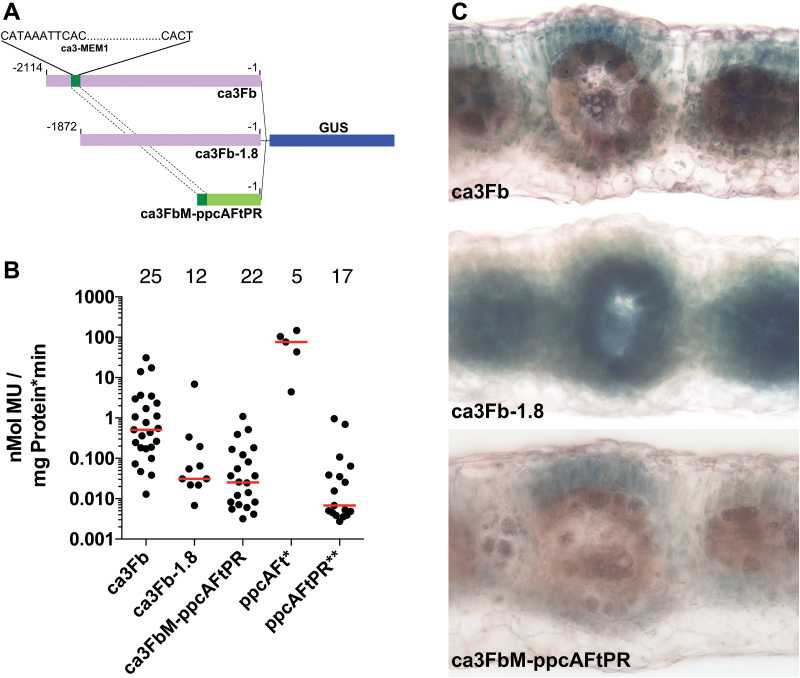
To test whether the MEM1-like element of the F. bidentis ca3 upstream region, like the ppcA MEM1, is capable of conferring M cell-specific expression, F. bidentis wild type plants were transformed with constructs containing parts of the ~2.1 kb upstream region from the F. bidentis ca3 gene fused with the GUS reporter gene (Fig. 4A). When the entire ~2.1 kb fragment, which contained the MEM1-like element (ca3Fb), was used in the reporter construct, GUS activity in the leaves of transformants was approximately 16 times greater than when the upstream fragment without the MEM-1-like sequence (ca3Fb-1.8) was fused to GUS (Fig. 4B). This difference is significant as judged by the Mann–Whitney U test (P = 0.0007). By comparison, GUS activity in leaves of F. bidentis plants that were transformed with the F. trinervia ppcA promoter containing the MEM1 sequence (ppcAFt; Stockhaus et al., 1997) was more than two orders of magnitude and significantly (P < 0.0001) greater than the activity found with ca3Fb (Fig. 4B). Approximately 3.5 times more GUS activity was found in the leaves of transformants when the F. bidentis ca3 MEM1-like sequence was fused to the PR of the ppcA gene (ca3FbM-ppcAFtPR; Fig. 4B) relative to the PR alone (ppcAFtPR; Fig. 4B), although this difference was not significant (P = 0.2289). However, the level of activity was of the same magnitude as the relatively low activity found for the ca3Fb-1.8 construct (Fig. 4B).
Fig. 4.
Analysis of Flaveria bidentis promoter–reporter gene constructs. (A) Structures of the ca3–β-glucuronidase (GUS) chimerical genes: ca3Fb represents the 2114 bp region upstream of the ca3 translation start site, and includes the MEM1-like element; ca3Fb1.8 denotes the 1872 bp upstream of the ca3 translation start site; ca3FbM-ppcAFtPR designates the F. bidentis ca3 MEM1-like element fused to the proximal promoter region (PR) of the F. trinervia ppcA gene. (B) GUS activities in leaves of transgenic F. bidentis plants. Data for constructs ppcAFt (*) and ppcAFtPR (**) were taken from Gowik et al. (2004). Median values are indicated as red bars. Numbers above the values represent the number of individual plants assayed. Mu, 4-methylumbelliferone. (C) Histochemical localisation of GUS activity in leaf sections of transgenic F. bidentis plants transformed with the ca3Fb, ca3-1.8 and ca3Fb-ppcAFtPR constructs. Incubation times for GUS staining were 8 h (ca3Fb), 24 h (ca3Fb-1.8.) and 26 h (ca3FbM-ppcAFtPR).
Histochemical staining of GUS activity in the leaves of ca3Fb transformants was detected only in M cells (Fig. 4C). In contrast, no cell-specific GUS staining was seen in the leaves of plants transformed with ca3Fb-1.8; instead staining was detected in M and BS cells, as well as in the vascular tissue (Fig. 4C). Although the fluorometric assays indicated relatively low GUS activity in the leaves of plants transformed with the ca3 MEM1-like sequence fused to the PR of the ppcA gene (ca3FbM-ppcAFtPR), histochemical localisation of GUS activity in the leaves was detected only in the M (Fig. 4C). In contrast, transformants containing constructs with only the ppcA PR also demonstrated GUS activity in the BS and vasculature (Akyildiz et al., 2007).
We conclude from these initial promoter analyses that the MEM1-like motif of the F. bidentis ca3 gene acts as an enhancer of gene expression preferentially in the M cells of F. bidentis as an increase in GUS activity was observed when the motif was present in the transformation construct relative to its absence (Fig. 4B). However, the element also appears to inhibit GUS activity in the BS and vascular tissues when it is present (Fig. 4C). While the levels of GUS activity in plants transformed with ca3FbM-ppcAFtPR were similar to those of the ca3 upstream region without the MEM1-like element, histochemical staining showed the presence of the element conferred M-specific GUS expression (Fig. 4B, C). Taken together, these results indicate that the MEM1-like element of the F. bidentis ca3 gene is a cis-acting element that directs M cell-specific expression.
Discussion
The C4 photosynthetic pathway has evolved independently from C3 ancestors in at least 65 different angiosperm lineages (Sage et al., 2012; Sage, 2016). This suggests, in terms of molecular genetics, that it is a relatively easy conversion (Gowik et al., 2004). In a current model of C4 evolution, the steps that include the strict compartmentation of enzymes between M and BS and the optimisation of the pathway, with the accompanying evolution of the regulatory elements controlling these processes, are considered to occur during the later stages of the transition (Sage et al., 2012). Increasing evidence indicates that distinct mechanisms control the expression patterns and levels of genes encoding C4 isoforms. Modifications to sequences in ancestral C3 promoter and untranslated regions (UTRs), as well as introns, control levels of gene expression, while different motifs in promoters, exons and UTRs direct cell-specific patterns of expression (Ludwig, 2013; Heimann et al., 2013; Williams et al., 2016).
Flaveria bidentis ca3 encodes the carbonic anhydrase associated with the C4 pathway
In this study, we have focused on the absolute identification of the gene encoding the CA isoform that catalyses the first step in the C4 pathway in Flaveria and the elements controlling its expression. Previous work on CA in the C4 species F. bidentis strongly supported a C4-associated role for the CA3 isoform (von Caemmerer et al., 2004; Tetu et al., 2007; Tanz et al., 2009). In the present study, we have extended these results and have shown unequivocally that the F. bidentis ca3 gene is expressed preferentially in leaf M cells and it encodes a cytosolic form of CA.
Previous results of RT-qPCR assays indicated that mRNAs encoding CA3 are at least 50 times more abundant than those coding for CA1 or CA2 in F. bidentis leaves (Tetu et al., 2007), and more than 10 times greater than any of the CA transcripts from the C3F. pringlei on a leaf total RNA basis (Ludwig, 2011). While these high expression levels argued that the ca3 gene most likely encoded the CA isoform associated with the C4 pathway in Flaveria, they did not show whether the accumulation of ca3 transcripts was specifically in the M cells, as anticipated for a C4 species (Gutierrez et al., 1974; Ku and Edwards, 1975; Burnell and Hatch 1988). Results of translatome experiments in the present study conclusively demonstrated that ca3 transcripts are highly enriched in the polysome fraction of F. bidentis M cells, being about 15 times greater than in the translation complexes of BS cells, and about twice the abundance of the mRNA encoding the C4-associated PPDK that functions in M cells (Fig. 1).
Having established the F. bidentis ca3 gene is preferentially expressed in M cells, we then set out to definitively show that its cognate protein has a cytosolic location, which is essential for the provision of HCO3− to PEPC and C4 pathway function. Tetu et al. (2007) showed, with import studies using isolated pea chloroplasts, that the F. bidentis CA3 was not imported into the isolated organelles, unlike the CA3 homologue from the C3 species F. pringlei (Tanz et al., 2009). It was concluded that F. bidentis CA3 is a cytosolic protein; however, as cytosolic fractions could not be isolated in these import studies, there was no direct evidence for this conclusion. Here we demonstrated that when the coding region of F. bidentis CA3 is fused to that of GFP and used to transform N. benthamiana leaves, GFP fluorescence is unequivocally cytosolic in protoplasts from these leaves (Fig. 2). GFP signal clearly surrounds the chlorophyll autofluorescence emitted from the chloroplasts, with no overlap in these fluorescence signals.
The translatome and the localisation results of the present study substantiate the proposal that the loss of the sequence encoding the chloroplast transit peptide of the ancestral C3Flaveria CA3 protein enabled the evolution of the C4 form of the enzyme, by trapping it in the M cytosol (Tanz et al., 2009). In addition, they corroborate the finding that reduction of CA3 in F. bidentis through antisense technology leads to a significant impairment of the CCM in this C4 species (von Caemmerer et al., 2004).
The MEM1-like element of the F. bidentis ca3 gene, like the C4Flaveria ppcA MEM1, directs mesophyll cell-specific expression
Like the MEM1 element of the F. trinervia and F. bidentis ppcA genes (Gowik et al., 2004; Akyildiz et al., 2007), the MEM1-like motif of C4Flaveria ca3 genes is located about 2 kb upstream of the translation start site. In contrast, while homologous sequences can be identified 2–2.5 kb upstream of the translation start sites of C3 and C4Flaveria ppcA orthologues (Gowik et al., 2004; Akyildiz et al., 2007), the ca3 upstream region from the C3 species F. pringlei shows no sequence similarity to the C4F. bidentis and F. trinervia 5′-regions in the vicinity of the MEM1-like motif (Supplementary Fig. S1). As the upstream regions of the ppcA genes from Flaveria congeners show insertions and deletions relative to one another, we determined the sequence of a further 2 kb upstream of the F. pringlei ca3 gene; however, we found no evidence of a MEM1-like sequence in this part of the genome.
The structures of the ppcA MEM1 and ca3 MEM1-like motifs are highly similar, with recognisable A and B segments in the MEM1-like elements (Fig. 3); however, the sequence of the MEM1-like A segments is the reverse complement of the sequence encoding the A segments of the C4ppcA MEM1. The MEM1 B segment of C4, C4-like and C3–C4Flaveria ppcA genes has an invariant CACT tetranucleotide, which is not seen in the orthologues of C3 congeners (Gowik et al., 2004). A CACT sequence is found 23 bp downstream of the MEM1-like A segment in the F. bidentis ca3 upstream region while a CATT tetranucleotide is found in the corresponding position of the C4F. trinervia MEM1-like motif. The distal promoter region encoding the ppcA MEM1, in combination with the PR of the ppcA promoter, was found to direct M-specific GUS expression in both sequence orientations, characteristic of a transcriptional enhancer (Gowik et al., 2004). This activity supports the evidence presented here that shows the MEM1-like motif also acts as an enhancer, conferring a higher level of GUS expression when present with the ca3Fb-1.8 region or when fused to the F. trinervia ppcA PR (Fig. 4).
Histochemical localisation of GUS activity showed preferential staining of the M cells in leaves of transformed F. bidentis plants when the ~2.1 kb upstream region of the F. bidentis ca3 gene was included in the transformation construct (ca3Fb; Fig. 4C). This M-specific staining pattern was also found when only the MEM1-like region was used in combination with the PR of the F. trinervia ppcA gene (ca3FbM-ppcAFtPR; Fig. 4C). In contrast, no cell-specificity in GUS staining was seen when the F. bidentis ca3 upstream region without the MEM1-like motif (ca3Fb-1.8; Fig. 4C), or just the PR of the F. trinervia ppcA gene (Akyildiz et al., 2007) was used to transform F. bidentis plants.
From these experiments we can conclude that the ca3 MEM1-like motif resembles the C4Flaveria ppcA MEM1 element not only in its structure but also in its function as it preferentially directs M expression of the GUS reporter gene and acts as an enhancer of expression in the M. Moreover, the MEM1-like motif also functions to repress transcriptional activity in the BS cells, as well as in other leaf cell types.
The GUS activity levels in the leaves of ca3Fb transformants are at least two orders of magnitude less than those of F. bidentis transformants containing the promoter region of the F. trinervia ppcA gene (ppcAFt; Fig. 4B). This difference may be attributed to additional promoter elements not in the ca3 ~2.1 kb 5′-region. Alternatively it may reflect a true difference in the strengths of the two promoters that could imply additional post-transcriptional regulation of transcript levels. An antisense construct targeted against F. bidentis ca3 transcripts showed that although CA activity in wild type F. bidentis plants does not limit photosynthesis, relatively high activity levels are required for the CCM to function properly in this C4 dicot (von Caemmerer et al., 2004). Although care needs to be taken in extending transcriptional activity with either protein abundance or activity (Vélez-Bermúdez and Schmidt, 2014), it is likely that the MEM1-like element and its associated transcription factors are not the only mechanism ensuring sufficient CA activity is present to support the provision of HCO3− for PEPC. As our current understanding of C4 gene expression expands, we need to consider control at the transcriptional level involving epigenetic marks, and post-transcriptional mechanisms at the level of both the transcript and the protein.
Evolution of C4 related cis-regulatory elements and gene regulation
It is well known that the expression patterns of most of the genes encoding proteins involved in C4 photosynthesis changed during C4 evolution as overall expression was enhanced and many of these genes acquired either M- or BS-specific expression. However, the modifications in gene structure responsible for these changes in expression have been identified at the molecular level for only a few of these genes (Rosche et al., 1998; Nomura et al., 2000; Gowik et al., 2004; Brown et al., 2011; Heimann et al., 2013; Williams et al., 2016).
Interestingly, recent studies have shown that in different C4 lineages, several genes encoding C4-associated proteins appear to be controlled, at least partially, by common mechanisms and cis-regulatory motifs. Common histone modifications that control the expression of genes encoding multiple C4-associated proteins have been identified in different grass C4 lineages. In maize, Sorghum bicolor (sorghum) and Setaria italica, light-regulated acetylation of histone H3 at K9 was found to be a shared histone mark in the promoter regions of genes encoding the C4-associated PEPC and NADP-ME, and in maize this modification was also observed in the promoter regions of genes encoding the C4 forms of CA, PCK and PPDK (Heimann et al., 2013). Cell-specific regulation of trimethylation of K4 on histone H3 was a common modification in these lineages for a number of genes encoding C4-associated enzymes, including maize CA (Heimann et al., 2013).
In the coding regions of NAD-malic enzyme and NADP-ME subunit genes from different C4 lineages, homologous sequences have been isolated that confer BS-specific reporter gene expression (Brown et al., 2011). More recently, Williams et al. (2016) described a nine-nucleotide motif that is found in the 3′- and 5′-UTRs of GgCA4, the C4-associated CA of Gynandropsis gynandra. This sequence, designated MEM2, in combination with an element in the G. gynandra PR is sufficient to direct high levels of the GUS reporter gene preferentially in M cells. MEM2 motifs are also found in the 3′- and 5′-UTRs of the gene encoding the C4-associated PPDK in G. gynandra, as well as in the 3′-end of the gene coding for GgCA2 (Williams et al., 2016).
Here we demonstrated that the MEM1-like element of the F. bidentis ca3 gene shares the regulatory function of directing M cell-specific expression with the C4Flaveria ppcA gene MEM1 motif. This implies that these motifs were already established within the promoter sequences when these genes were recruited to the C4 pathway, bringing both genes under the control of a common trans-regulatory network that might have also existed in the last non-C4Flaveria ancestors.
In case of the Flaveria ppcA promoter it appears that MEM1 evolved step by step from an ancestral C3 motif via point mutations as well as insertions and deletions of short DNA stretches (Gowik et al., 2004; Akyildiz et al., 2007). Sequences very similar to that of MEM1 and the regions surrounding it, but not functional in M-specific gene expression, are found in the ppcA promoters of C3Flaveria species (Gowik et al., 2004; Akyildiz et al., 2007), indicating that the C4 MEM1 evolved from a C3 predecessor (Gowik et al., 2004). The ppcA genes of Flaveria are thought to have originated from the duplication of an ancestral ppcB-like gene long before the emergence of C4 photosynthesis in this genus (Svensson et al., 2003). Importantly, sequences with obvious similarity to MEM1 have been identified in the promoter regions of ppcB genes from C3 and C4Flaveria species (Akyildiz et al., 2007). This implies that a MEM1-related sequence in Flaveria ppc promoters was an ancestral motif that was recruited for function in C4 photosynthesis after some modification and optimisation.
The situation is quite different for the Flaveria ca3 genes. The sequences surrounding the MEM1-like motifs are highly conserved in the two C4 species, but cannot be identified in the 4 kb ca3 upstream region from the C3 species F. pringlei. This implies that the MEM1-like motif was not part of the ancestral Flaveria ca genes, but instead was acquired before or during C4 evolution in the genus by recombination. Alternatively, the motif may have been lost from the predecessor of C3ca3 genes after the relatively recent divergence of C3 and C4Flaveria species (Lyu et al., 2015).
Ascertaining the scenario by which the MEM1-like element was acquired for C4Flaveria ca3 gene expression will be possible once genome sequences of C3 and C4Flaveria congeners are available. The distribution of MEM1-like motifs in the genomes could be examined, and putative recombination events could be reconstructed. Importantly, the possible spreading of cis-regulatory element precursors within the genome with subsequent modifications and recruitment to C4-related gene regulation could be investigated. These types of comparative studies will provide insights and a potential mechanism into how similar changes in the expression patterns of several genes during C4 evolution has been realised in multiple C4 lineages.
Conclusion
In the cytosol of C4 M cells, the enzymes CA and PEPC catalyse the first two reactions of the C4 photosynthetic pathway, regardless of C4 subtype (Hatch 1987; Hatch and Burnell, 1990), or whether a plant uses Kranz anatomy or a single-celled C4 system (Offermann et al., 2011). As the activity of PEPC is dependent on HCO3−, the product of CA catalysis, it is conceivable that during the evolution of C4 photosynthesis in 65 (or more) angiosperm lineages, a similar regulatory mechanism was adopted to ensure the coordinated expression of the cognate genes. As shown here, this appears to be the case in Flaveria.
The present study has built on previous work (von Caemmerer et al., 2004; Tetu et al., 2007, Tanz et al., 2009) to conclusively show the ca3 gene from F. bidentis encodes the CA associated with the C4 pathway. Our results indicate that the ca3 MEM1-like element, like the ppcA MEM1, is sufficient and required for M-specific promoter activity. They also suggest that distinct mechanisms control this cell-type expression pattern and the activity of the ca3 gene promoter. In all likelihood additional transcriptional as well as post-transcriptional control mechanisms are required to provide sufficient CA activity to support the F. bidentis C4 CCM.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Multiple sequence alignment of C3 and C4Flaveria carbonic anhydrase 3 upstream regions.
Table S1. Primers used in this study.
Supplementary Material
Acknowledgements
Funding from the Australian Research Council to ML (award number DP150101037) and the Deutsche Forschungsgemeinschaft through the Excellence Cluster EXC 1028 (From Complex Traits Towards Synthetic Modules) to PW is gratefully acknowledged. We also thank the Australian Research Council Centre of Excellence for Translational Photosynthesis, and Oliver Berkowitz for supplying the pMDC83 vector.
Glossary
Abbreviations:
- BS
bundle-sheath
- CA
carbonic anhydrase
- CCM
CO2 concentrating mechanism
- GFP
green fluorescent protein
- GLDPA
glycine decarboxylase P protein
- GUS
β-glucuronidase
- M
mesophyll
- MEM
mesophyll expression module
- NADP-ME
NADP-malic enzyme
- PCK
phosphoenolpyruvate carboxykinase
- PEPC
phosphoenolpyruvate carboxylase
- PPDK
pyruvate orthophosphate dikinase
- PR
proximal promoter region.
References
- Akyildiz M, Gowik U, Engelmann S, Koczor M, Streubel M, Westhoff P. 2007. Evolution and function of a cis-regulatory module for mesophyll-specific gene expression in the C4 dicot Flaveria trinervia. The Plant Cell 19, 3391–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bläsing OE, Westhoff P, Svensson P. 2000. Evolution of C4 phosphoenolpyruvate carboxylase in Flaveria, a conserved serine residue in the carboxyl-terminal part of the enzyme is a major determinant for C4-specific characteristics. The Journal of Biological Chemistry 275, 27917–27923. [DOI] [PubMed] [Google Scholar]
- Brown NJ, Newell CA, Stanley S, Chen JE, Perrin AJ, Kajala K, Hibberd JM. 2011. Independent and parallel recruitment of preexisting mechanisms underlying C4 photosynthesis. Science 331, 1436–1439. [DOI] [PubMed] [Google Scholar]
- Burnell JN, Hatch MD. 1988. Low bundle sheath carbonic anhydrase is apparently essential for effective C4 pathway operation. Plant Physiology 86, 1252–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitty JA, Furbank RT, Marshall JS, Chen Z, Taylor WC. 1994. Genetic transformation of the C4 plant, Flaveria bidentis. The Plant Journal 6, 949–956. [Google Scholar]
- Curtis MD, Grossniklaus U. 2003. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology 133, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMario RJ, Clayton H, Mukherjee A, Ludwig M, Moroney JV. 2016. Plant carbonic anhydrases: Structures, locations, evolution and physiological roles. Molecular Plant, doi: 10.1016/j.molp.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards GE, Ku MSB. 1987. Biochemistry of C3-C4 intermediates. In: Hatch MD, Boardman NK, eds. The biochemistry of plants, Vol. 10 New York: Academic Press, 275–325. [Google Scholar]
- Edwards GE, Voznesenskaya EV. 2011. C4 photosynthesis: Kranz forms and single-cell C4 in terrestrial plants. In Raghavendra AS, Sage RF, eds. C4 photosynthesis and related CO2 concentrating mechanisms. Dordrecht: Springer, 29–60. [Google Scholar]
- Engelmann S, Wiludda C, Burscheidt J, Gowik U, Schlue U, Koczor M, Streubel M, Cossu R, Bauwe H, Westhoff P. 2008. The gene for the P-subunit of glycine decarboxylase from the C4 species Flaveria trinervia: analysis of transcriptional control in transgenic Flaveria bidentis (C4) and Arabidopsis (C3). Plant Physiology 146, 1773–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst K, Westhoff P. 1997. The phosphoenolpyruvate carboxylase (ppc) gene family of Flaveria trinervia (C4) and F. pringlei (C3): molecular characterization and expression analysis of the ppcB and ppcC genes. Plant Molecular Biology 34, 427–443. [DOI] [PubMed] [Google Scholar]
- Ghannoum O, Evans JR, von Caemmerer S. 2011. Nitrogen and water use efficiency of C4 plants. In Raghavendra AS, Sage RF, eds. C4 Photosynthesis and related CO2 concentrating mechanisms. Dordrecht: Springer, 129–146. [Google Scholar]
- Gowik U, Burscheidt J, Akyildiz M, Schlue U, Koczor M, Streubel M, Westhoff P. 2004. cis-Regulatory elements for mesophyll-specific gene expression in the C4 plant Flaveria trinervia, the promoter of the C4 phosphoenolpyruvate carboxylase gene. The Plant Cell 16, 1077–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez M, Gracen VE, Edwards GE. 1974. Biochemical and cytological relationships in C4 plants. Planta 119, 279–300. [DOI] [PubMed] [Google Scholar]
- Haberlandt G. 1896. Physiologische Pflanzenanatomie, 2nd edn Leipzig: Wilhelm Engelman. [Google Scholar]
- Hatch MD. 1987. C4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Biochimica et Biophysica Acta 895, 81–106. [Google Scholar]
- Hatch MD, Burnell JN. 1990. Carbonic anhydrase activity in leaves and its role in the first step of C4 photosynthesis. Plant Physiology 93, 825–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch MD, Kagawa T, Craig S. 1975. Subdivision of C4-pathway species based on differing C4 decarboxylating systems and ultrastructural features. Australian Journal of Plant Physiology 2, 111–128. [Google Scholar]
- Heckmann D, Schulze S, Denton A, Gowik U, Westhoff P, Weber AP, Lercher MJ. 2013. Predicting C4 photosynthesis evolution: modular, individually adaptive steps on a Mount Fuji fitness landscape. Cell 153, 1579–1588. [DOI] [PubMed] [Google Scholar]
- Heimann L, Horst I, Perduns R, Dreesen B, Offermann S, Peterhansel C. 2013. A Common histone modification code on C4 genes in maize and its conservation in Sorghum and Setaria italica. Plant Physiology 162, 456–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans J, Westhoff P. 1992. Homologous genes for the C4 isoform of phosphoenolpyruvate carboxylase in a C3 and a C4Flaveria species. Molecular & General Genetics: MGG 234, 275–284. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai R, Edwards GE. 1999. The biochemistry of C4 photosynthesis. In: Sage RF, Monson RK, eds. C4 plant biology. London: Academic Press, 49–87. [Google Scholar]
- Koncz C, Schell J. 1986. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Molecular & General Genetics: MGG 204, 383–396. [Google Scholar]
- Kosugi S, Ohashi Y, Nakajima K, Arai Y. 1990. An improved assay for β-glucuronidase in transformed cells: Methanol almost completely suppresses a putative endogenous β-glucuronidase activity. Plant Science 70, 133–140. [Google Scholar]
- Ku SB, Edwards GE. 1975. Photosynthesis in mesophyll protoplasts and bundle sheath cells of various types of C4 plants. Zeitschift für Pflanzenphysiologie 77, 16–32. [Google Scholar]
- Lazo GR, Stein PA, Ludwig RA. 1991. A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology 9, 963–967. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Ludwig M. 2011. The molecular evolution of β-carbonic anhydrase in Flaveria. Journal of Experimental Botany 62, 3071–3081. [DOI] [PubMed] [Google Scholar]
- Ludwig M. 2013. Evolution of the C4 photosynthetic pathway: events at the cellular and molecular levels. Photosynthesis Research 117, 147–161. [DOI] [PubMed] [Google Scholar]
- Lyu MJ, Gowik U, Kelly S, et al. 2015. RNA-Seq based phylogeny recapitulates previous phylogeny of the genus Flaveria (Asteraceae) with some modifications. BMC Evolutionary Biology 15, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKown AD, Dengler NG. 2007. Key innovations in the evolution of Kranz anatomy and C4 vein pattern in Flaveria (Asteraceae). American Journal of Botany 94, 382–399. [DOI] [PubMed] [Google Scholar]
- McKown AD, Moncalvo JM, Dengler NG. 2005. Phylogeny of Flaveria (Asteraceae) and inference of C4 photosynthesis evolution. American Journal of Botany 92, 1911–1928. [DOI] [PubMed] [Google Scholar]
- Mann HB, Whitney DR. 1947. On a test of whether one of two random variables is stochastically larger than the other. Annals of Mathematical Statistics 18, 50–60. [Google Scholar]
- Marshall JS, Stubbs JD, Taylor WC. 1996. Two genes encode highly similar chloroplastic NADP-malic enzymes in Flaveria. Implications for the evolution of C4 photosynthesis. Plant Physiology 111, 1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJ, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J. 2009. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proceedings of the National Academy of Sciences, USA 106, 18843–18848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M, Sentoku N, Nishimura A, et al. 2000. The evolution of C4 plants: acquisition of cis-regulatory sequences in the promoter of C4-type pyruvate, orthophosphate dikinase gene. The Plant Journal 22, 211–221. [DOI] [PubMed] [Google Scholar]
- Offermann S, Okita TW, Edwards GE. 2011. Resolving the compartmentation and function of C4 photosynthesis in the single-cell C4 species Bienertia sinuspersici. Plant Physiology 155, 1612–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell AM. 1978. Systematics of Flaveria (Flaveriinae-Asteraceae). Annals of the Missouri Botanical Garden 65, 590–636. [Google Scholar]
- Reynoso MA, Juntawong P, Lancia M, Blanco FA, Bailey-Serres J, Zanetti ME. 2015. Translating ribosome affinity purification (TRAP) followed by RNA sequencing technology (TRAP-SEQ) for quantitative assessment of plant translatomes. Methods in Molecular Biology 1284, 185–207. [DOI] [PubMed] [Google Scholar]
- Rolland V, Badger MR, Price GD. 2016. Redirecting the cyanobacterial bicarbonate transporters BicA and SbtA to the chloroplast envelope: soluble and membrane cargos need different chloroplast targeting signals in plants. Frontiers in Plant Science 7, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosche E, Chitty J, Westhoff P, Taylor WC. 1998. Analysis of promoter activity for the gene encoding pyruvate orthophosphate dikinase in stably transformed C4 flaveria species. Plant Physiology 117, 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage RF. 2016. A portrait of the C4 photosynthetic family on the 50th anniversary of its discovery: species number, evolutionary lineages, and Hall of Fame. Journal of Experimental Botany 67, 4039–4056. [DOI] [PubMed] [Google Scholar]
- Sage RF, Sage TL, Kocacinar F. 2012. Photorespiration and the evolution of C4 photosynthesis. Annual Review of Plant Biology 63, 19–47. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: A laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Stockhaus J, Poetsch W, Steinmüller K, Westhoff P. 1994. Evolution of the C4 phosphoenolpyruvate carboxylase promoter of the C4 dicot Flaveria trinervia: an expression analysis in the C3 plant tobacco. Molecular & General Genetics: MGG 245, 286–293. [DOI] [PubMed] [Google Scholar]
- Stockhaus J, Schlue U, Koczor M, Chitty JA, Taylor WC, Westhoff P. 1997. The promoter of the gene encoding the C4 form of phosphoenolpyruvate carboxylase directs mesophyll-specific expression in transgenic C4Flaveria spp. The Plant Cell 9, 479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson P, Bläsing OE, Westhoff P. 1997. Evolution of the enzymatic characteristics of C4 phosphoenolpyruvate carboxylase: A comparison of the orthologous PPCA phosphoenolpyruvate carboxylases of Flaveria trinervia (C4) and Flaveria pringlei (C3). European Journal of Biochemistry 246, 452–460. [DOI] [PubMed] [Google Scholar]
- Svensson P, Bläsing OE, Westhoff P. 2003. Evolution of C4 phosphoenolpyruvate carboxylase. Archives of Biochemistry and Biophysics 414, 180–188. [DOI] [PubMed] [Google Scholar]
- Tanz SK, Tetu SG, Vella NG, Ludwig M. 2009. Loss of the transit peptide and an increase in gene expression of an ancestral chloroplastic carbonic anhydrase were instrumental in the evolution of the cytosolic C4 carbonic anhydrase in Flaveria. Plant Physiology 150, 1515–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetu SG, Tanz SK, Vella N, Burnell JN, Ludwig M. 2007. The Flaveria bidentis beta-carbonic anhydrase gene family encodes cytosolic and chloroplastic isoforms demonstrating distinct organ-specific expression patterns. Plant Physiology 144, 1316–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vélez-Bermúdez IC, Schmidt W. 2014. The conundrum of discordant protein and mRNA expression. Are plants special?Frontiers in Plant Science 5, 619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Caemmerer S, Quinn V, Hancock NC, Price GD, Furbank RT, Ludwig M. 2004. Carbonic anhydrase and C4 photosynthesis: a transgenic analysis. Plant, Cell and Environment 27, 697–703. [Google Scholar]
- Westhoff P, Gowik U. 2004. Evolution of C4 phosphoenolpyruvate carboxylase. Genes and proteins: a case study with the genus Flaveria. Annals of Botany 93, 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BP, Burgess SJ, Reyna-Llorens I, Knerova J, Aubry S, Stanley S, Hibberd JM. 2016. An untranslated cis-element regulates the accumulation of multiple C4 enzymes in Gynandropsis gynandra mesophyll cells. The Plant Cell 28, 454–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti ME, Chang IF, Gong F, Galbraith DW, Bailey-Serres J. 2005. Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant Physiology 138, 624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.