Abstract
Background
In Myanmar, civil unrest and the establishment of internally displaced person (IDP) settlements along the Myanmar-China border have impacted malaria transmission.
Methods
Microsatellite markers were used to examine source-sink dynamics for Plasmodium vivax between IDP settlements and surrounding villages in the border region. Genotypic structure and diversity were compared across the 3 years following the establishment of IDP settlements, to infer demographic history. We investigated whether human migration and landscape heterogeneity contributed to P. vivax transmission.
Results
P. vivax from IDP settlements and local communities consistently exhibited high genetic diversity within populations but low polyclonality within individuals. No apparent genetic structure was observed among populations and years. P. vivax genotypes in China were similar to those in Myanmar, and parasite introduction was unidirectional. Landscape factors, including distance, elevation, and land cover, do not appear to impede parasite gene flow.
Conclusions
The admixture of P. vivax genotypes suggested that parasite gene flow via human movement contributes to the spread of malaria both locally in Myanmar and across the international border. Our genetic findings highlight the presence of large P. vivax gene reservoirs that can sustain transmission. Thus, it is important to reinforce and improve existing control efforts along border areas.
Keywords: Plasmodium vivax, malaria transmission, cross-border migration, microsatellites, genetic diversity, landscape genetics
Plasmodium vivax infections along the Myanmar-China border exhibited high genetic diversity but low polyclonality. Frequent parasite gene flow via human movement contributes to local and cross-border spread of malaria. It is thus important to reinforce and improve existing control efforts along border areas.
Myanmar has been engaged in the world’s longest-running civil unrest. Commonly known as Burma, the country has succumbed to ethnic civil unrest and turmoil since its independence in 1948, and this conflict remains unresolved today [1]. Political instability and military conflicts have driven hundreds of thousands of citizens into relocation camps known as internally displacement person (IDP) settlements along the country’s borders, specifically the Myanmar-China and Myanmar-Thailand borders. Large-scale human movements in addition to poor health infrastructure have led to intensive transmission of malaria [2–3]. Myanmar has the highest malaria burden among Southeast Asian countries, with approximately 200000 cases per year [4]. The number of malaria cases and the malaria-induced mortality rate have been consistently high for the past 3 decades [5–6] and present a major challenge for malaria control and elimination, especially to neighboring countries such as China and Thailand, in the Greater Mekong Subregion.
Cross-border malaria transmission is a complex challenge to countries trying to eliminate the disease. For instance, along the Brazil–French Guiana border, approximately 5000 malaria cases are reported annually, of which 82% are due to Plasmodium vivax infections associated with frequent cross-border migration, large-scale public works, or economic shifts in the region [7]. In Thailand, >45% of malaria cases reported from 2008 to 2012 were P. vivax that occurred in high-risk areas along the Thailand-Cambodia border [8]. In China, a national malaria elimination program was launched in 2011, with a goal of elimination by 2020. Although malaria cases have declined dramatically through years of control efforts, provinces such as Yunnan, which borders Myanmar, currently has the highest malaria burden among other Southeast Asian countries. In 2014, 333871 confirmed malaria cases and 236 malaria-related deaths were reported [9]. Cases of imported malaria in China increased from 16.2% in 2004 to 97.9% in 2013 [10]. Because of the continuous conflict in Myanmar, the current magnitude of and genetic diversity within the populations undergoing movement are unprecedented. The regional seasonality of population movements, coupled with movement along country borders, can heavily impact malaria transmission. A better way of tracking infections and implementation of concerted control interventions along the border are thus needed. Population genetic studies help guide malaria control by tracing routes of transmission and sources of epidemics.
This study examined the sources and spreading patterns of P. vivax along the international border between eastern Myanmar and western China. Specifically, we tested the hypothesis of whether P. vivax from the IDP settlements showed reduced diversity and a genetic bottleneck over the years; this pattern was observed in Plasmodium falciparum and was potentially due to malaria control interventions [11]. Here, P. vivax genotypes were compared over a 3-year period between the IDP settlements, which were established in 2011, and the surrounding villages. Information on the parasite diversity and the extent of malaria spread are keys to targeting disease control efforts in high-risk areas and will be of particular relevance when most other parts of Southeast Asia enter the malaria elimination phase.
MATERIALS AND METHODS
Ethics Statement
Scientific and ethical clearance was given by the institutional scientific and ethical review boards of Kunming Medical University (China), the University of California–Irvine, Pennsylvania State University (University Park), and the Ministry of Health of Kachin State (Myanmar). Written informed consent/assent for study participation was obtained from all consenting heads of households or parents/guardians (for minors aged <18 years) and each individual who participated in this study.
Sample Collection
A total of 895 samples from individuals with a diagnosis of P. vivax infection were collected from clinics/hospitals located in 2 IDP settlements (Je Yang Hka [JYH] and Hpum Lum Yang [HLY]), a nearby military base (CMH), and 4 surrounding villages/towns (Ja Htu Kawng [JHK], Laiza [LZCH], Mai Sak Pa [MSP], and Mung Seng Yang [MSY]) in Myanmar and from 2 town hospitals (Tengchong [TC] and Yingjiang [YJ]) in Yunnan, China (Supplementary Table 1). Among the Myanmar sites, LZCH is a regional hospital that has a larger catchment area of nearly 25000 people, whereas CMH, JHK, MSP, and MSY have smaller catchment areas of approximately 3000 people each. The populations of the IDP settlements, HLY and JYH, are about 1600 and 8600, respectively. Because of uneven sample sizes among sites, genetic variation was compared among 3 localities: a regional hospital in Myanmar (LZCH), the military base and villages in Myanmar (CMH, JHK, MSP, and MSY), and the IDP settlements (JYH and HLY). Samples from most sites were collected during 3 consecutive years (2011, 2012, and 2013) for comparison. All studied individuals showed fever or malaria-related symptoms at the time of sampling and received a diagnosis of P. vivax malaria by microscopy and polymerase chain reaction (PCR) analysis. For each individual, 30–50 μL of blood was blotted on Whatman 3MM filter paper. Parasite DNA was extracted by the Saponin/Chelex method [12].
Microsatellite Genotyping
Eleven single-copy microsatellites (SSRs) with trinucleotide or tetranucleotide repeats were typed for P. vivax. Alleles were amplified with fluorescence-labeled primers in accordance with the published protocol [13]. After PCR, amplification products were pooled on the basis of size difference (Supplementary Table 2), and the pooled products were separated on an ABI 3730 sequencer. All allele sizes were determined and visualized in Peak Scanner.
Data Analyses
Linkage Disequilibrium and Genetic Diversity
To examine whether the SSR loci represent an independent set of markers in P. vivax, linkage disequilibrium was tested by the Fisher exact test for each pair of loci with GenePop v4.2, using the Markov chain with 100 batches and 10000 iterations per batch [14]. Significance values were adjusted by sequential Bonferroni correction for multiple comparisons.
Genotypic variation was calculated in GenoDive v2.0b27 [15]. We first calculated squared Euclidean distances, based on the number of times a certain allele was found in the 2 individuals. The minimal distance class was set as threshold to identify the number of multilocus genotypes, the Simpson diversity index, and genotype evenness. Additionally, the number of effective alleles and expected heterozygosity were estimated for each locality setting.
Population Structure and Isolation by Distance
A model-based Bayesian method implemented in STRUCTURE, version 2.3.4, was performed to examine partitioning of individuals to genetic clusters [16]. All samples from the different years were included in a single analysis. The number of clusters (K) was determined by simulating K values from 1 (no genetic differentiation among sites) to 22 (all sites of different years were genetically differentiated). The posterior probability values of each K estimated from a Markov Chain Monte Carlo (MCMC) method were used to detect the modal value of ΔK [17]. A burn-in period of 500000 iterations followed by 106 iterations of each chain was performed to ensure MCMC convergence. Each MCMC chain for each value of K was run 8 times with the independent allele frequency option.
An FST analysis was conducted using θ, an FST estimator, in SPAGeDi, version 1.2e [18]. FST values were tested for significance, using 10000 permutations. Genetic differentiation among sites was displayed by multidimensional scaling plot based on DS values (an analog of FST) in R, version 3.3.0. Furthermore, an analysis of molecular variance was used to determine the hierarchical distribution of genetic variance within and among populations of the same year and among years, using GENALEX [19]. The relationships between genetic distances (DS values) and Euclidean geographical distance (estimated from spatial coordinates, using R [20]) were examined by Mantel tests (10000 randomizations) and reduced major axis regression in Isolation by Distance, version 3.23 [21].
Bottlenecks and Migration Rates
Signature of genetic bottleneck was detected with BOTTLENECK, version 1.2.02 [22] on the IDP settlement and village samples. Two tests were performed using 3 different mutation models: the infinite alleles model (IAM), the stepwise mutation model (SMM), and a combination of the 2 models, the 2-phase model. First was the overall distribution of allele frequency classes. Second was the Wilcoxon signed rank test to compare the number of loci that present a heterozygosity excess to the number of such loci expected by chance only.
Gene flow among IDP settlement and village populations was estimated for each year by maximum likelihood analyses implemented in Migrate-N, version 2.4.4 [23]. Parameters, including Θ (calculated as 4Neμ, where Ne is the effective population size, and μ is the mutation rate per generation and site), M (calculated as m/μ, where m is the immigration rate size ), and the number of effective migrants per generation Nem (calculated as ΘM/2 for haploid individuals), were estimated. Four independent runs were conducted with the Brownian motion model by using 10 short chains with 5000 sampled genealogies and 3 long chains with 50000 sampled genealogies to obtain the mean and range of Θ, M, and Nem values. Additionally, we inferred migration rates by using a Bayesian approach implemented in BayesAss, version 3 [24], which uses a MCMC algorithm to estimate the posterior probability distribution of the proportion of migrants from one population to another without assuming genetic equilibrium. We used 9 × 106 iterations, with a burn-in of 106 iterations and a sampling frequency of 2000 to ensure that the parameters of the model were converged.
Landscape Genetics
To test for the effects of landscape factors on gene flow, we used ResistanceGA, a package in R, to optimize resistance surfaces to our genetic data (pair-wise FST values) [25]. A resistance surface is defined as a spatial layer in which each cell in a grid is assigned a value that represents the degree to which that cell constrains gene flow or movement [25]. These values were often based on numerous assumptions about relationships between a landscape or environmental feature and the ability of a given organism to move through that feature. In contrast, ResistanceGA uses a genetic algorithm to unbiasedly optimize the values of landscape features to best fit the genetic data. Here, resistance surface raster files were created in ArcGIS 10, using land cover data from MODIS [26–27], elevation data from the SRTM Digital Elevation Database, version 4.1 [28], and distance to roads data from the World Roads shapefile (available at: http://www.arcgis.com), the 3 landscape factors that are most heterogeneous in the study region. We used Circuitscape, version 4.05, to measure the pair-wise landscape resistance distance between populations [29]. Circuitscape uses electrical circuit theory to predict landscape connectivity. By using circuit theory, as opposed to the commonly used least-cost path method, the model incorporates all possible pathways between populations into the analysis. The Akaike information criterion (AIC) with a penalty for extra parameters (AICc) was used as the measure of model fitness to genetic data and means for model selection. AICc was calculated from the linear mixed effects models with maximum likelihood population effects in lme4 [30]. Optimizations were run twice for each resistance surface to verify consistency in output.
RESULTS
Genetic Diversity in IDP Settlements and Local Communities
No significant linkage disequilibrium was detected for all pair-wise combinations of the 11 microsatellite loci (Bonferroni corrected P > .05) among the P. vivax samples. Approximately 29% of samples (263 of 895) were associated with polyclonal infections. Of these, 102 were biclonal, with 2 equally dominant alleles detected in a single locus (Supplementary Figure 1). We separated the genotypes of the 2 strains and included them in the analyses. However, 161 samples showed >1 allele in ≥2 loci. Owing to the challenge in unambiguously differentiating the genotypes of the different strains, these samples were discarded in the analyses.
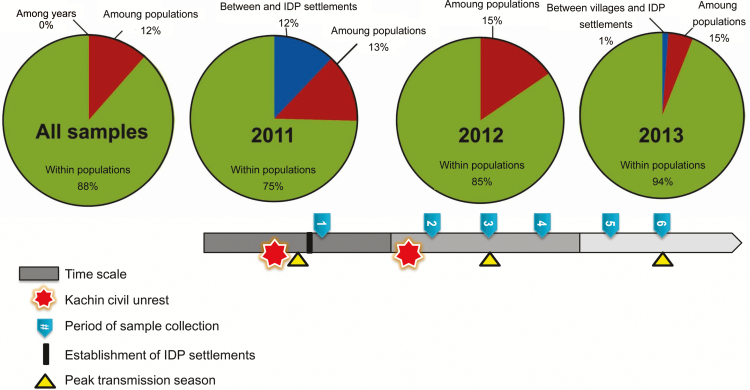
Genotypic diversity and expected heterozygosity were consistently high over the 3 years and showed no significant difference between samples from the local community (including regional hospital and village clinics) and those from IDP settlements (P > .05, by the 2-tailed t test; Table 1). Nevertheless, there was a significant decrease in genotypic evenness in the IDP samples in 2012 and 2013 (P < .05; Table 1), indicative of the preponderance of genotypes, whereas genotypic evenness remained similar among the community samples. Analysis of molecular variance indicated that most of the genetic variation (>75%) was within populations (Figure 1). No variation was detected among the 3 study years. The greatest partition of genetic variation between IDP settlements and local villages was in 2011 (12%), but this was drastically reduced in subsequent years (0% in 2012 and 1% in 2013).
Table 1.
Comparison of Plasmodium Vivax Genetic Diversity Measures Based on 14 Microsatellite Loci
| Locality, Year | Sample Size | Genotypic Diversitya | Gene Diversity, Mean ± SDb | |||
|---|---|---|---|---|---|---|
| G | D | E | Ne | He | ||
| Regional hospital | ||||||
| 2011 | 140 | 79.03 | 0.99 | 0.73 | 7.23 ± 1.34 | 0.82 ± 0.03 |
| 2012 | 57 | 43.32 | 0.98 | 0.85 | 5.77 ± 0.72 | 0.81 ± 0.03 |
| 2013 | 37 | 33.39 | 0.98 | 0.95 | 5.01 ± 0.78 | 0.74 ± 0.06 |
| Village | ||||||
| 2011 | 67 | 53.22 | 0.99 | 0.98 | 3.16 ± 0.25 | 0.80 ± 0.03 |
| 2012 | 58 | 44.55 | 0.99 | 0.98 | 2.51 ± 0.17 | 0.79 ± 0.03 |
| 2013 | 37 | 35.18 | 0.98 | 0.97 | 2.17 ± 0.17 | 0.81 ± 0.06 |
| IDP settlement | ||||||
| 2011 | 30 | 29 | 0.99 | 0.98 | 2.59 ± 0.34 | 0.75 ± 0.09 |
| 2012 | 132 | 44.59 | 0.95 | 0.47 | 4.18 ± 0.37 | 0.85 ± 0.03 |
| 2013 | 133 | 57.84 | 0.97 | 0.49 | 3.94 ± 0.36 | 0.78 ± 0.04 |
a“G” denotes the number of multilocus genotypes corrected for sample size. “D” denotes the Simpson diversity index, also known as the Nei genetic diversity, corrected for sample size and ranging from 0 (where 2 randomly chosen individuals in a population share a single genotype) to 1 (where individuals have different genotypes). “E” denotes the genotypic evenness, which ranges from 0 (where one or a few genotypes dominate in a population) to 1 (where all genotypes are of equal frequency in a population).
Figure 1.
Results of analyses of molecular variance of Plasmodium vivax samples obtained across years in different localities. The timing of major events and activities is indicated in the bar. The greatest partition of genetic variation between internally displaced person (IDP) settlements and local villages was in 2011 but decreased drastically in subsequent years, likely because of parasite gene flow.
Genetic Clustering of Samples
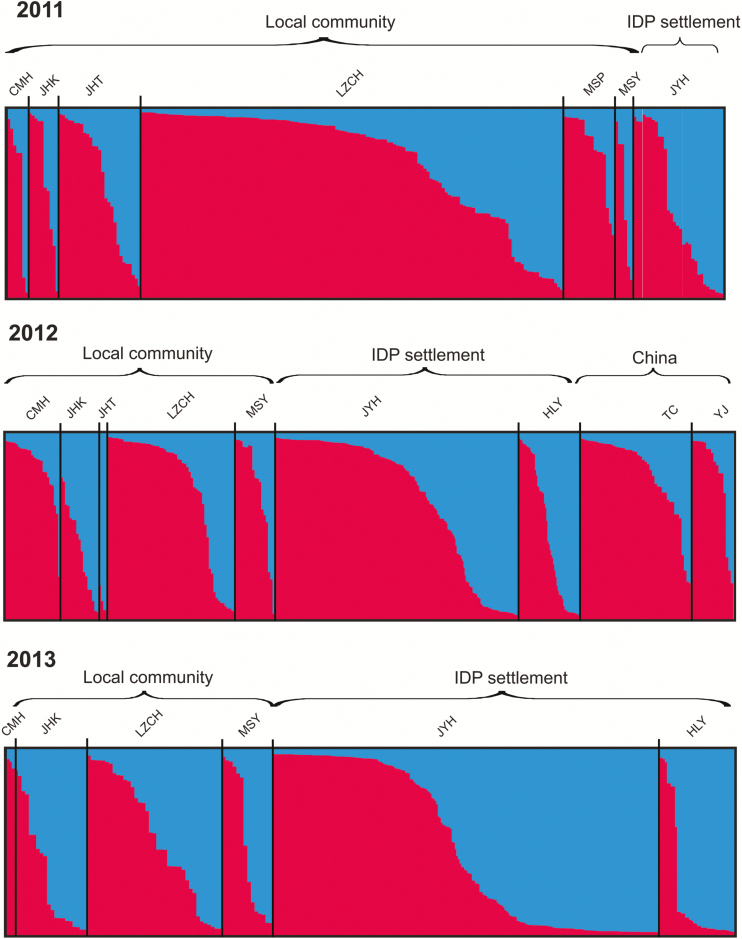
Two genetic clusters were most probable among all P. vivax samples, without clear population distinction (Figure 2 and Supplementary Table 3). The genetic composition between samples from local communities and those from IDP settlements was largely similar in 2011 and 2012. The red cluster was more predominant than the blue cluster among these samples. An apparent difference was seen in 2013, when both the community and IDP settlement samples constituted almost equal proportions of red and blue clusters (Figure 2). Genotypes varied slightly among seasons in the IDP samples (Supplementary Figure 2). A dominant red cluster was detected in samples before (January–April) and during (May–August) the peak transmission season. However, samples collected after the peak transmission season (September–December) constituted similar proportions of red and blue clusters. Most P. vivax samples from individuals in Chinese hospitals (TC and YJ) constituted the red cluster, similar to samples from individuals in Myanmar (Figure 2).
Figure 2.
Bayesian inferences of the K clusters estimated by STRUCTURE among Plasmodium vivax samples collected from clinics/hospitals located in 2 internally displaced person (IDP) settlements (Je Yang Hka [JYH] and Hpum Lum Yang [HLY]), a nearby military base (CMH), and 4 surrounding villages/towns (Ja Htu Kawng [JHK], Laiza [LZCH], Mai Sak Pa [MSP], and Mung Seng Yang [MSY]) in Myanmar and from 2 town hospitals (Tengchong [TC] and Yingjiang [YJ]) in Yunnan, China, during 2011–2013. The 2 most probable clusters are labeled red and blue, and individuals are represented as columns. For each column, the extent of the component colors indicates the magnitude of the membership coefficient (Q) corresponding to each cluster. Q values of respective clusters are presented in Supplementary Table 3.
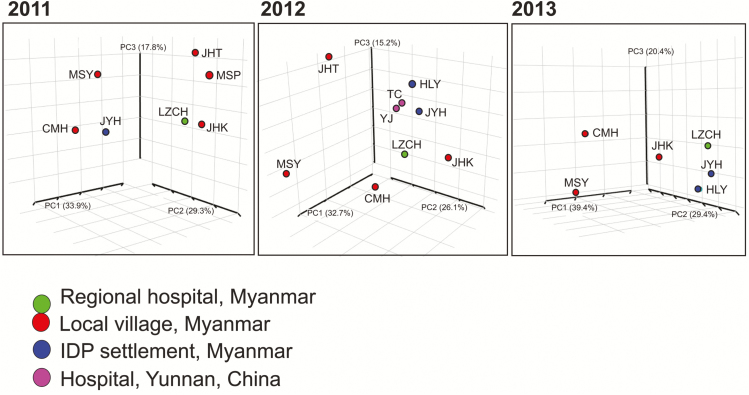
Mantel tests indicated no significant associations between geographical and genetic distances among sites with respect to different years (in 2011, R2 = 0.11 and P > .05; in 2012, R2 = 0.23 and P > .05; and in 2013, R2 = 0.14 and P > .05). No clear clustering was observed between samples from the IDP settlements and villages in Myanmar (Figure 3). Although samples from TC and YJ, in China, were geographically distant from those from the Myanmar sites, they were genetically close to samples from the IDP settlements (JYH and HLY) and regional hospital (LZCH) in Myanmar (Figure 3). These findings suggest that parasite gene flow via human movement was not limited by physical distance.
Figure 3.
Three-dimensional scatterplots of pair-wise DS values (an analog of FST), showing the genetic relatedness of Plasmodium vivax collected from clinics/hospitals located in 2 internally displaced person (IDP) settlements (Je Yang Hka [JYH] and Hpum Lum Yang [HLY]), a nearby military base (CMH), and 4 surrounding villages/towns (Ja Htu Kawng [JHK], Laiza [LZCH], Mai Sak Pa [MSP], and Mung Seng Yang [MSY]) in Myanmar and from 2 town hospitals (Tengchong [TC] and Yingjiang [YJ]) in Yunnan, China, during 2011–2013. The first 3 axes that contain >90% of the total variation are shown. Locations of the studied sites are presented in Supplementary Table 1.
Impact of Landscape Factors
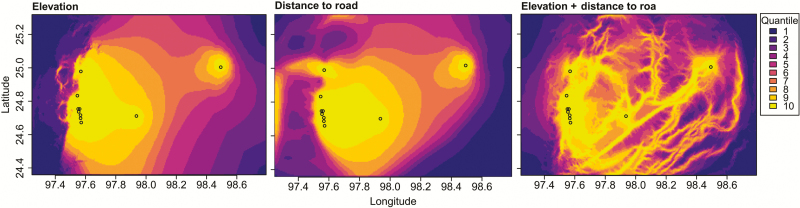
Distance to roads, elevation, and land cover resistance surfaces were not significantly associated with pair-wise FST values, although elevation and distance to roads surfaces were the best fitting models in 2012, as measured by AICC (Supplementary Table 4). The elevation and distance to roads surface model fitted best to pair-wise FST values after an inverse monomolecular transformation (Figure 4). However, neither optimized surface model was statistically significant (P = .08 for both, by a maximum likelihood population effects MODEL). In 2011 and 2013, no surface model performed better than the null model (Supplementary Table 4). The land cover surface model performed worst among all tested models in all years.
Figure 4.
Potential Plasmodium vivax gene flow across the Myanmar-China border area as constructed in Circuitscape, based on the highest-performing resistance surface models in 2012 from ResistanceGA analysis. Akaike information criterion values of the tested models to pair-wise FST values are presented in Supplementary Table 4. A higher quartile number indicates higher current flow. Circles indicate study sites.
Demographic Change and Migrations
Both the village and IDP samples showed a normal L-shape distribution in allele frequency (Table 2), suggesting that these P. vivax populations did not experience a recent severe bottleneck. The significant excess of heterozygosity observed in the IDP settlements over the 3 years under IAM and SMM suggested that these samples deviated from mutation-drift equilibrium, possibly because of small founder populations.
Table 2.
Parameters and Results of BOTTLENECK Analyses
| Locality, Year, Mutation Model | Heterozygote Excess, P |
|---|---|
| Regional hospital | |
| 2011 (n = 140) | |
| IAM | .002 |
| SMM | .004 |
| TPM | .36 |
| 2012 (n = 57) | |
| IAM | .002 |
| SMM | .006 |
| TPM | .73 |
| 2013 (n = 37) | |
| IAM | .02 |
| SMM | .82 |
| TPM | .13 |
| Village | |
| 2011 (n = 44) | |
| IAM | .006 |
| SMM | .03 |
| TPM | .82 |
| 2012 (n = 46) | |
| IAM | .12 |
| SMM | .20 |
| TPM | .49 |
| 2013 (n = 23) | |
| IAM | .15 |
| SMM | .43 |
| TPM | .43 |
| IDP settlement | |
| 2011 (n = 30) | |
| IAM | .04 |
| SMM | .03 |
| TPM | .64 |
| 2012 (n = 132) | |
| IAM | .002 |
| SMM | .004 |
| TPM | .49 |
| 2013 (n = 133) | |
| IAM | .03 |
| SMM | .001 |
| TPM | .03 |
The mode shift was normal, L-shaped for all analyses.
Abbreviations: IAM, infinite alleles model; SMM, stepwise mutation model; TPM, 2-phase model (a combination of the IAM and SMM).
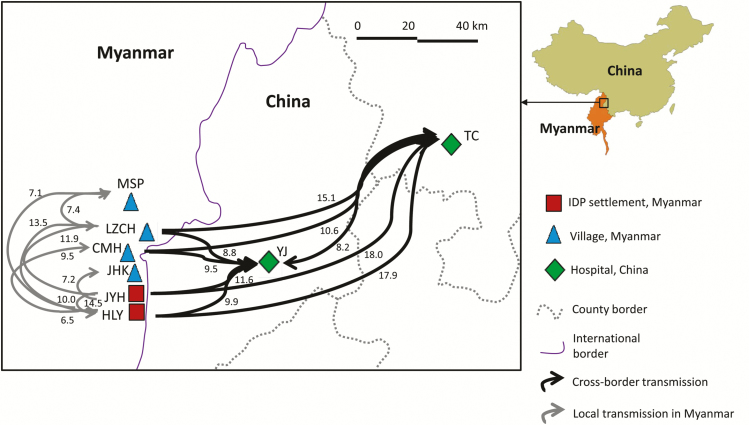
Maximum likelihood analyses revealed that all populations had relatively small effective population sizes (Θ), suggesting that the effect of drift was unequivocally as significant as migration (Figure 5 and Supplementary Table 5). Given that most values of M (calculated as m/μ) were >1, the effect of migration (m) was larger than the effect of mutation (μ). For the 2011 samples, the number of effective migrants per generation (calculated as Nem) ranged from 0.04 to 3.10. The greatest migration was observed among villages in Myanmar (from LZCH to JHK, Nem = 3.10). Migrations between the IDP settlement JYH and local villages were mostly symmetrical, based on their similar migration rates. For the 2012 samples, the greatest migration was observed from Myanmar to China (Figure 5; from JYH to TC, Nem = 6.02; from LZCH to TC, Nem = 5.84; from LZCH to YJ, Nem = 4.31; and from JYH to YJ, Nem = 3.88). By contrast, migration from China to Myanmar was relatively small (Nem = 0.07–1. 33; Supplementary Table 5). A greater migration rate from Myanmar to China than vice versa was also shown by Bayesian analyses (Supplementary Table 6). Migrations between the IDP settlements and local villages in Myanmar were not significantly different in 2012 (Nem = 0.04–4.01) and 2013 (Nem = 0.07–3.71).
Figure 5.
Migratory pathways (arrows) and rates (number above arrows, M) among samples collected from clinics/hospitals located in 2 internally displaced person (IDP) settlements (Je Yang Hka [JYH] and Hpum Lum Yang [HLY]), a nearby military base (CMH), and 4 surrounding villages/towns (Ja Htu Kawng [JHK], Laiza [LZCH], Mai Sak Pa [MSP], and Mung Seng Yang [MSY]) in Myanmar and from 2 town hospitals (Tengchong [TC] and Yingjiang [YJ]) in Yunnan, China, during 2012. Values of Θ (defined as 4Neμ, where Ne is the effective population size and μ is the mutation rate per generation and site), M (in m/μ, where m is the immigration rate scaled by the mutation rate), and the number of effective migrants per generation Nem (ΘM/2 for haploid individuals) are presented in Supplementary Table 5. Only pathways with an M value of >5 are shown.
DISCUSSION
Internal conflicts in Myanmar have led to tens of thousands of residents fleeing their home villages and displaced in resettlements located along international borders. The displacement of large numbers of people and their circulation favor malaria transmission and dissemination of the disease to neighboring countries [33]. This study showed that parasite gene flow via human migration between IDP settlements and local villages was bidirectional and frequent, contributing to vast genetic diversity within parasite populations but with little differentiation among populations. The mean He in our study populations (0.82) was comparable to that for other Southeast Asian areas and countries, such as central Vietnam (He = 0.88 [34]) and Cambodia (He = 0.84 [35]), with similar transmission intensities. While genetic diversity tends to be directly proportional to malaria transmission levels, other factors, such as geographical isolation, also affect genetic diversity. For instance, in the South Pacific, P. vivax diversity was intermediate despite high levels of transmission. This was mainly due to little human migration between island populations and thus limited parasite introduction [36]. Nevertheless, in some areas of low endemicity, such as Central Asia, P. vivax retains much of the preexisting diversity despite recent decrease in effective population sizes or reduced transmission levels [37]. In Cambodia, the eastern and western P. vivax populations lack clear differentiation [38]. The overall high genetic diversity illustrates the impact of human movements on mixing the parasite gene pools. Frequent inbreeding and recombination between parasite genotypes also play a role in contributing to high genetic diversity within populations [39].
Compared with other malaria-hypoendemic areas, such as Vietnam (71.4%; [40]), Sri Lanka (68%; [37]), Colombia (60–80%; [41]), and the Amazon Basin in Brazil (50%; [42]), the proportion of polyclonal infections among our samples was relatively low (29%). Our study sites represent a modest transmission area at the Myanmar-China border region. Malarial parasite infection rate ranges from <1% in local villages to 2% in the IDP settlements. Anopheles minimus sensu lato is the major vector in our study area, where nearly 20% of the mosquitoes harbored P. vivax sporozoites but only <0.5% harbored P. falciparum sporozoites (unpublished data). The low polyclonality of P. vivax may have resulted from inoculation of individuals with clonal infections by highly infectious mosquitoes. Moreover, our earlier study indicated that >95% of P. vivax infections showed clearance of parasitemia 2–3 days after chloroquine-primaquine treatment, which is currently the frontier therapy for radical cure of vivax malaria in Myanmar [43]. Rapid clearance of P. vivax from the time of diagnosis may imply a high efficacy of primaquine in treating polyclonal infections due to relapse and may contribute in part to the low polyclonality of the samples. Nevertheless, it is noteworthy that different molecular markers (single-nucleotide polymorphisms and microsatellites) and the number of gene loci can vary the proportion of polyclonal infections [40–41]. A standardized method would allow fair comparisons among studies. Also, the relapse rate of P. vivax malaria in the study area is not well known. The impact of relapse or recurrent infection on the extent of polyclonality within hosts remains to be investigated.
While P. falciparum revealed a lower genetic diversity in the IDP settlement samples than the community samples [11], P. vivax populations were more genetically diverse and less structured in Myanmar. Such a contrasting pattern was also reported in Papua, Indonesia, where P. falciparum and P. vivax occur in sympatry [44]. Myanmar has a considerably higher ratio of P. vivax to P. falciparum infections (9.91) than other neighboring countries, such as China (4.35) and Thailand (1.79) [45]. The higher ratio of P. vivax to P. falciparum infections implies a larger P. vivax reservoir that can sustain intense, stable transmission [46]. This could limit the impact of current control and elimination efforts. In addition, the observation of reduced genotypic evenness in 2012 and 2013 within the IDP settlements may suggest possible selection of P. vivax genotypes. In western Thailand and Papua, Indonesia, strong signals of recent selection have been seen in several regions of the P. vivax genomes that relate to drug resistance [47]. It remains unclear whether our parasite populations were subjected to strong selection, although chloroquine still appears to be effective in clearing P. vivax [43].
P. vivax infections detected in China were introduced from Myanmar, and the migration was asymmetrical. The migration analyses based on the coalescent method in Migrate-N were dependent on the assumptions of neutral evolution, steady population size, and mutation-drift equilibrium [23], Our P. vivax samples did not experience a recent severe bottleneck, and population size was likely similar. While low linkage disequilibrium may suggest high recombination or mutation rates of the microsatellite loci and violate some of model assumptions, the migration results were supported by the traveling record of the studied subjects indicating their recent visits to Myanmar before the diagnosis and confirmation of malaria [6]. Also, this finding was consistent with the pattern observed in sympatric populations of P. falciparum [11]. Frequent malaria introductions by migratory human populations are difficult to monitor [48]. Such migration was alarming because malaria spread not only at the local scale but also stretched over 100 km away from Myanmar, to China, with minimal hindrance by physical distance and landscape barriers. The movement of infected people from malaria-endemic areas to areas where the disease had been or nearly eliminated can heighten epidemic risk because people of all ages usually lack immunity and are susceptible to the parasites. In areas where malaria transmission is unstable, the risk of severe disease in children and adults is often greater [49]. Hence, it is imperative to identify sources of parasite infection in areas at risk of epidemics and to prevent reintroduction by tightening existing border surveillance through coordinated international efforts. Intensive control, including indoor/outdoor insecticide spraying and regular supply of effective antimalarial drugs, should be reinforced and maintained within the IDP settlements and local villages in Myanmar to combat vivax malaria.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Acknowledgments. We thank the field team, for their technical assistance; the communities and hospitals, for their support and willingness to participate in this research; and Drs Deirdre Joy, Daibin Zhong, and Xiaoguang Chen, for inspirational discussions of the data.
Disclaimer. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the National Institutes of Health.
Financial support. This work was supported by the National Institutes of Health (grant U19 AI089672).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Socheat D, Denis MB, Fandeur T, et al. Mekong malaria. II. Update of malaria, multi-drug resistance and economic development in the Mekong region of Southeast Asia. Southeast Asian J Trop Med Public Health 2003; 34:1–102. [PubMed] [Google Scholar]
- 2. Delacollette C, D’Souza C, Christophel E, et al. Malaria trends and challenges in the Greater Mekong Subregion. Southeast Asian J Trop Med Public Health 2009; 40:674–91. [PubMed] [Google Scholar]
- 3. Archavanitkul K, Vajanasara K. Health of migration workers from Myanmar, Cambodia and Laos. In: Kanchanachitra C, Podhisita C, Archavanikul Ket al. eds. Thai health: capitalism in crisis, opportunity for society? Bangkok: Amarin Pringting, 2010; 31–2. [Google Scholar]
- 4. World Health Organization South-East Asia Region. Malaria in the Greater Mekong Subregion: regional and country profiles. New Delhi: WHO press, 2010. [Google Scholar]
- 5. Cui L, Yan G, Sattabongkot J, et al. Malaria in the Greater Mekong Subregion: heterogeneity and complexity. Acta Trop 2012; 121:227–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhou G, Sun L, Xia R, et al. Clinical malaria along the China–Myanmar border, Yunnan Province, China, January 2011–August 2012. Emerg Infect Dis 2014; 20:675–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gomes Mdo S, Vieira JL, Machado RL, et al. Efficacy in the treatment of malaria by Plasmodium vivax in Oiapoque, Brazil, on the border with French Guiana: the importance of control over external factors. Malar J 2015; 14:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kaewpitoon N, Loyd RA, Kaewpitoon SJ, Rujirakul R. Malaria risk areas in Thailand border. J Med Assoc Thai 2015; 98:S17–21. [PubMed] [Google Scholar]
- 9. World Health Organization. World malaria report 2014. Geneva: WHO Press, 2014. [Google Scholar]
- 10. Feng J, Xiao H, Xia Z, et al. Analysis of malaria epidemiological characteristics in the People’s Republic of China, 2004–2013. Am J Trop Med Hyg 2015; 93:293–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lo E, Zhou G, Oo W, et al. Molecular inference of sources and spreading patterns of Plasmodium falciparum malaria parasites in internally displaced persons settlements in Myanmar-China border area. Infect Genet Evol 2015; 33:189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bereczky S, Mårtensson A, Gil JP, Färnert A. Short report: rapid DNA extraction from archive blood spots on filter paper for genotyping of Plasmodium falciparum. Am J Trop Med Hyg 2005; 72:249–51. [PubMed] [Google Scholar]
- 13. Karunaweera N, Ferreira M, Hartl D, et al. Fourteen polymorphic microsatellite DNA markers for the human malaria parasite Plasmodium vivax. Mol Ecol Notes 2007; 7:172–175. [Google Scholar]
- 14. Raymond M, Rousset F. GENEPOP: a population genetic software for exact test and ecumenicism. J Hered 1995; 86:248–249. [Google Scholar]
- 15. Meirmans PG, Van Tienderen PH. GenoType and GenoDive: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Note 2004; 4:792–4. [Google Scholar]
- 16. Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics 2000; 155:945–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 2005; 14:2611–20. [DOI] [PubMed] [Google Scholar]
- 18. Hardy OJ, Vekemans X. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Note 2002; 2:618–20. [Google Scholar]
- 19. Peakall R, Smouse PE. Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Note 2006; 6:288–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Casgrain P, Legendre P. The R package for multivariate and spatial analysis, v4.0d10. Montréal, Canada: Université de Montréal, 2004. http://www.bio.umontreal.ca/casgrain/en/labo/R/v4/. [Google Scholar]
- 21. Bohonak AJ. IBD (Isolation by Distance): a program for analyses of isolation by distance. J Hered 2002; 93:153–4. [DOI] [PubMed] [Google Scholar]
- 22. Piry S, Luikart G, Cornuet JM. BOTTLENECK: a computer program for detecting recent reductions in effective size using allele frequency data. J Hered 1999; 90:502–3. [Google Scholar]
- 23. Beerli P. Migrate: Documentation and program, part of LAMARC version 2.4.4 2004. http://evolution.gs.washington.edu/lamarc.html.
- 24. Wilson GA, Rannala B. Bayesian inference of recent migration rates using multilocus genotypes. Genetics 2003; 163:1177–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Spear SF, Balkenhol N, Fortin MJ, McRae BH, Scribner K. Use of resistance surfaces for landscape genetic studies: considerations for parameterization and analysis. Mol Ecol 2010; 19:3576–91. [DOI] [PubMed] [Google Scholar]
- 26. Friedl MA, Sulla-Menashe D, Tan B, et al. MODIS Collection 5 global land cover: Algorithm refinements and characterization of new datasets. Remote Sens Environ 2010; 114:168–182. [Google Scholar]
- 27. Channan S, Collins K, Emanuel WR. Global mosaics of the standard MODIS land cover type data. College Park, MD: University of Maryland and the Pacific Northwest National Laboratory, 2014. [Google Scholar]
- 28. Jarvis A, Reuter HI, Nelson A, et al. Hole-filled SRTM for the globe. Version 4.1 Washington DC: The CGIAR-CSI SRTM 90m Database, 2008. http://srtm.csi.cgiar.org
- 29. MaRae BH. Isolation by resistance. Evolution 2006; 60:1551–1561. [PubMed] [Google Scholar]
- 30. Bates D, Maechler M, Bolker B, et al. Fitting linear mixed-effects models using lme4. J Stat Softw 2015; 67:1–48. [Google Scholar]
- 31. Nielsen R, Tarpy DR, Kern-Reeve H. Estimating effective paternity number in social insects and the effective number of alleles in a population Mol Ecol 2003; 12:3157–3164. [DOI] [PubMed] [Google Scholar]
- 32. Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978; 89:53–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Martens P, Hall L. Malaria on the move: human population movement and malaria transmission. Emerg Infect Dis 2000; 6:103–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Van den Eede P, Erhart A, Van der Auwera G, et al. High complexity of Plasmodium vivax infections in symptomatic patients from a rural community in central Vietnam detected by microsatellite genotyping. Am J Trop Med Hyg 2010; 82:223–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Orjuela-Sánchez P, Sá JM, Brandi MC, et al. Higher microsatellite diversity in Plasmodium vivax than in sympatric Plasmodium falciparum populations in Pursat, Western Cambodia. Exp Parasitol 2013; 134:318–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Koepfli C, Rodrigues PT, Antao T, et al. Plasmodium vivax diversity and population structure across four continents. PLoS Negl Trop Dis 2015; 9:e0003872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gunawardena S, Ferreira MU, Kapilananda GM, Wirth DF, Karunaweera ND. The Sri Lankan paradox: high genetic diversity in Plasmodium vivax populations despite decreasing levels of malaria transmission. Parasitology 2014; 141:880–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Friedrich LR, Popovici J, Kim S, et al. Complexity of infection and genetic diversity in Cambodian Plasmodium vivax. PLoS Negl Trop Dis 2016; 10:e0004526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Koepfli C, Ross A, Kiniboro B, et al. Multiplicity and diversity of Plasmodium vivax infections in a highly endemic region in Papua New Guinea. PLoS Negl Trop Dis 2011; 5:e1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hong NV, Delgado-Ratto C, Thanh PV, et al. Population genetics of Plasmodium vivax in four rural communities in central Vietnam. PLoS Negl Trop Dis 2016; 10:e0004434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Imwong M, Snounou G, Pukrittayakamee S, et al. Relapses of Plasmodium vivax infection usually result from activation of heterologous hypnozoites. J Infect Dis 2007; 195:927–33. [DOI] [PubMed] [Google Scholar]
- 42. Batista CL, Barbosa S, Da Silva Bastos M, Viana SA, Ferreira MU. Genetic diversity of Plasmodium vivax over time and space: a community-based study in rural Amazonia. Parasitology 2015; 142:374–84. [DOI] [PubMed] [Google Scholar]
- 43. Lo E, Nguyen J, Oo W, et al. Examining Plasmodium falciparum and P. vivax clearance subsequent to antimalarial drug treatment in the Myanmar-China border area based on quantitative real-time polymerase chain reaction. BMC Infect Dis 2016; 16:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jennison C, Arnott A, Tessier N, et al. Plasmodium vivax populations are more genetically diverse and less structured than sympatric Plasmodium falciparum populations. PLoS Negl Trop Dis 2015; 9:e0003634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhou G, Lo E, Zhong D, et al. Impact of interventions on malaria in internally displaced persons along the China-Myanmar border: 2011–2014. Malar J 2016; 15:471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Koepfli C, Timinao L, Antao T, et al. A large Plasmodium vivax reservoir and little population structure in the South Pacific. PLoS One 2013; 8:e66041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pearson RD, Amato R, Auburn S, et al. Genomic analysis of local variation and recent evolution in Plasmodium vivax. Nat Genet 2016; 48:959–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jitthal N. Migration and malaria. Southeast Asian J Trop Med Public Health 2013; 44: 166–200. [PubMed] [Google Scholar]
- 49. Kiszewski AE, Teklehaimanot A. A review of the clinical and epidemiological burdens of epidemic malaria. Am J Trop Med Hyg 2004; 71(2 Suppl):128–35. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.