Summary
Intestinal helminth infection increases small-intestinal colonization by Salmonella, which occurs independently of the induction of T regulatory or Th2 cells following helminth infection. Helminth infection disrupts the intestinal metabolome, and the resulting shift in metabolites directly enhances Salmonella virulence.
Keywords: co-infection, immunomodulation, helminths, bacterial infection, intestinal metabolites.
Abstract
Intestinal helminth infections occur predominantly in regions where exposure to enteric bacterial pathogens is also common. Helminth infections inhibit host immunity against microbial pathogens, which has largely been attributed to the induction of regulatory or type 2 (Th2) immune responses. Here we demonstrate an additional 3-way interaction in which helminth infection alters the metabolic environment of the host intestine to enhance bacterial pathogenicity. We show that an ongoing helminth infection increased colonization by Salmonella independently of T regulatory or Th2 cells. Instead, helminth infection altered the metabolic profile of the intestine, which directly enhanced bacterial expression of Salmonella pathogenicity island 1 (SPI-1) genes and increased intracellular invasion. These data reveal a novel mechanism by which a helminth-modified metabolome promotes susceptibility to bacterial coinfection.
Chronic helminth infections occur predominantly in regions of poor sanitation, where the risk of coinfection with microbial pathogens is high [1]. Ongoing helminth infections have been associated with increased susceptibility to secondary microbial infections in mice and people. In mice, helminths impair host resistance to Citrobacter rodentium, Salmonella enterica serovar Typhimurium, Escherichia coli strain UTI89 (uropathogenic E. coli), norovirus, and γ-herpesvirus [2–6]. In humans, helminth infections correlate with increased severity of tuberculosis, higher Plasmodium burdens during malaria, impaired immunity to Vibrio cholerae, and greater incidence of human immunodeficiency virus (HIV) infection [7–12].
It is currently thought that helminth-mediated immunomodulation underpins increased susceptibility to secondary microbial infections. Helminth infections characteristically induce a robust T helper (Th) 2 immune response, marked by the cytokines interleukin 4 (IL-4), interleukin 5 (IL-5), and interleukin 13 (IL-13), and a strong regulatory T-cell (Treg) response [13]. Both Th2 and Treg responses have been proposed to impair the generation of protective Th1 or Th17 immunity against bacterial or viral pathogens [2–5, 14–16]. In this regard, widespread IL-4 signaling during helminth infection can impair the production of antimicrobial interferon-γ from CD4+ T cells and invariant natural killer (NK) T cells [2, 5, 17], and helminth-elicited IL-4 and IL-10 can block effector differentiation of CD8+ T cells after challenge with irradiated Toxoplasma gondii parasites [16]. Moreover, Th2 cytokines can directly promote viral replication: IL-4 switches on signal transducer and activator of transcription 6 (Stat6), which binds to and activates γ-herpesvirus viral promotors controlling latent-lytic switch genes [4]. Alongside Th2 cytokines, Tregs can impede antimicrobial immunity through the suppression of effector T-cell responses [14, 15]. In this manner, helminth-induced Tregs can also reduce inflammation during allergic airway inflammation and graft-vs-host disease [18–20]. Thus, direct modulation of host immunity is one pathway by which helminths facilitate secondary microbial infections.
Here, we use a mouse model of coinfection to identify an additional mechanism by which intestinal helminths alter host immunity to concurrent bacterial pathogens. We show that the greater susceptibility of helminth-infected mice to the bacterial pathogen S. Typhimurium occurs independently of the induction of Treg or Th2 cells following helminth infection. Instead, we reveal a previously unidentified pathway of interkingdom interaction between helminths and bacteria. We show that helminths disrupt the metabolic profile of the small intestine, and that the resulting metabolites directly affect the virulence of S. Typhimurium to enhance bacterial colonization.
METHODS
Mice
All mouse experiments were performed at the University of British Columbia (UBC) and approved by UBC’s Animal Care Committee and the Canadian Council on Animal Care. Wild-type C57BL/6 mice were purchased from the Jackson Laboratory, and wild-type 129S1/SvImJ, rag1-/-, C57BL/6 4get [21], C57BL/6 KN2/KN2 (il4-/-) [22], and C57BL/6 4get stat6-/- [23] mice were bred in-house. All mice were housed in individually ventilated cages in specific-pathogen-free conditions, with ad libitum access to food and water. Experimental mice were age- and sex-matched and used at 6–12 weeks old. Littermates were randomized between test groups prior to the start of each experiment.
Infections
Mice were left naive or infected by oral gavage with 200 Heligmosomoides polygyrus third-stage larvae. Where indicated, naive or day 14 H. polygyrus–infected mice were infected with streptomycin-resistant S. Typhimurium wild-type or aroA mutant strain SL1344. 129S1/SvImJ mice were orally gavaged with 3 × 106 colony-forming units (CFU) of stationary-phase S. Typhimurium in phosphate-buffered saline (PBS), from overnight cultures grown in Luria-Bertani (LB) broth. C57BL/6 mice were orally gavaged with 3 × 108 CFU of stationary-phase aroA mutant S. Typhimurium [24].
In Vivo Salmonella Burden Quantification
Serial dilutions of homogenized tissues were plated onto LB plates containing 100 μg/mL streptomycin (Sigma-Aldrich). The following day, S. Typhimurium colonies were counted and CFU per gram of tissue was calculated.
Metabolite Collection
Intestinal contents were collected from the proximal 6 cm of the small intestine of naive or day 14 H. polygyrus–infected mice, and weights were recorded. One hundred microliters of acetonitrile (VWR International) was added for each 10 mg of intestinal content, and samples were shaken at 4°C overnight. Samples were spun at 13000 rpm at 4°C for 15 minutes, and supernatants containing small molecules were collected. Supernatants were sterile-filtered and aliquoted into 250-μL (containing 25 mg intestinal contents) fractions, and acetonitrile was evaporated using a speed vacuum concentrator. Samples were stored at –80°C prior to the use of metabolites for compositional analysis or functional assays. Control tubes were generated where acetonitrile was used as described above to do mock extractions in an empty tube.
Metabolite Analysis
In brief, metabolites were extracted as above from naive or day 14 H. polygyrus–infected 129S1/SvImJ mice and sent to the University of Victoria–Genome BC Proteomics Centre for untargeted metabolomics by ultra-high-performance liquid chromatography–Fourier transform mass spectrometry (UPLC-FTMS) analysis. The positive-ion and negative-ion UPLC-FTMS datasets were processed individually, and the output of the data processing was the retention time, mass- to-charge ratio (m/z), and peak area of each detected metabolite or metabolite feature. Welch t test with unequal variances was applied for the statistical analysis. Multivariate and clustering analyses were carried out using Metaboanalyst version 3.0 software (http://www.metaboanalyst.ca/faces/home.xhtml) [25, 26], using an m/z tolerance of 0.0005 and a retention time tolerance of 30 seconds. Data were log-transformed and auto-scaled. Principal components analysis was performed and plots were generated showing separation of data based on the first 2 principal components. Heat maps were generated showing relative abundance of all small-intestinal metabolites. Maximum abundance was reported in red and minimum abundance in blue. Clustering of samples from different mice was shown using a Euclidean distance and Ward clustering algorithm. Where possible, putative identities of identified metabolite features were assigned using the Metlin database (https://metlin.scripps.edu/metabo_batch.php) based on the m/z value of each feature.
Statistical analyses were performed separately on datasets from positive and negative ion detection datasets. Full details are provided in the Supplementary Methods.
Incubation of Salmonella With Intestinal Metabolites
Mock-extracted metabolites (control), metabolites from small-intestinal contents of naive mice, or metabolites from small-intestinal contents of day 14 H. polygyrus–infected mice were resuspended in 1 mL of LB media and sterile-filtered using a 0.22-μΜ pore filter unit (Sigma-Aldrich). Thirty microliters of stationary-phase overnight cultures of S. Typhimurium or aroA mutant S. Typhimurium grown in LB was diluted into the 1 mL of LB-containing metabolites, and shaken at 37°C for 3 hours. Bacteria were then pelleted and resuspended in PBS twice, to wash cells.
Salmonella Typhimurium Gene Expression
After incubation with metabolites as described above, S. Typhimurium was resuspended in RNAprotect Bacteria Reagent (Qiagen), and RNA was extracted using an RNeasy Mini Kit (Qiagen). Genomic DNA was removed using a DNA-Free kit (Ambion), and cDNA was prepared using a QuantiTect Reverse Transcription Kit (Qiagen). Quantitative PCR (qPCR) was performed on a 7500 Fast Real-Time PCR System (Applied Biosystems). Cycling conditions used were as follows: 2 minutes at 50°C, 15 minutes at 95°C, followed by 40 cycles of 95°C for 15 seconds, 58°C for 30 seconds, and 72°C for 30 seconds. Primers used were (5′-3′) hilA: F-ACACCTGCAGGATAATCCAA, R-ATTTTCGTGCCAGTTCATGT; sipA: F-GTCATAATGCCAGGTATGCAGACCG, R-CCTTTAATTTCCCCTGACAGCGTCG; sprB: F-CATTAACTGCACTTTTGCATTCCCTATCCG, R-GCCACTACCAAAACTTTACGGTTCTGCA; and glyceraldehyde-3-phosphate dehydrogenase (gapA): F-GGCGCTAACTTTGACAAATACGAAGG, R-AGTCATCAGACCTTCGATGATGCCG. Normalized expression units of test genes were calculated using the ΔΔCt method relative to gapA. Expression levels were normalized to that of the control group (S. Typhimurium cultured with mock-extracted metabolites), which was set to an expression level of 1.
In Vitro S. Typhimurium Invasion Assay
HeLa cells were purchased from the American Type Culture Collection (ATCC) and cultured in Dulbecco’s modified Eagle’s medium (DMEM; HyClone) containing 10% heat-inactivated FBS (HyClone), 1% GlutaMAX (ThermoFisher), and 1% nonessential amino acids (Gibco) (complete DMEM), at 37°C with 5% CO2. Cultures of S. Typhimurium that had been incubated with intestinal metabolites (described above) were used to infect HeLa cells at a multiplicity of infection of 50. Bacterial inoculates were plated on LB plates containing 100 μg/mL streptomycin (Sigma-Aldrich) to confirm the CFU of S. Typhimurium present in inoculates. Twenty minutes postinfection, HeLa cells were washed with PBS, and complete DMEM containing 50 μg/mL gentamicin (Gold Biotechnology) was added. After a further 70-minute incubation, HeLa cells were lysed using PBS containing 1% Triton X-100 (Sigma-Aldrich) and 0.1% sodium dodecyl sulfate (Sigma-Aldrich). Serial dilutions of lysate were plated onto LB plates containing 100 μg/mL streptomycin (Sigma-Aldrich). The following day, S. Typhimurium colonies were counted and invasion level was calculated as the percentage of S. Typhimurium CFU present in inoculates that invaded HeLa cells. The control group (S. Typhimurium cultured with mock-extracted metabolites) invasion level was set to 1, and invasion levels of other groups shown relative to this value.
Statistical Analysis
Data were analysed for normality using a D’Agostino-Pearson omnibus normality test. For assessing differences between 2 groups, an unpaired t test was used for normally distributed data, and a Mann–Whitney test was used for data that were not normally distributed. When >2 test groups were being assessed, a 1-way analysis of variance followed by a Tukey multiple comparisons test was used for normally distributed data, and a Kruskal–Wallis test followed by a Dunn multiple comparisons test was used for data that were not normally distributed. A P value ≥.05 was considered statistically significant.
RESULTS
Helminth-Coinfected Mice Exhibit Elevated S. Typhimurium Burdens in the Small Intestine
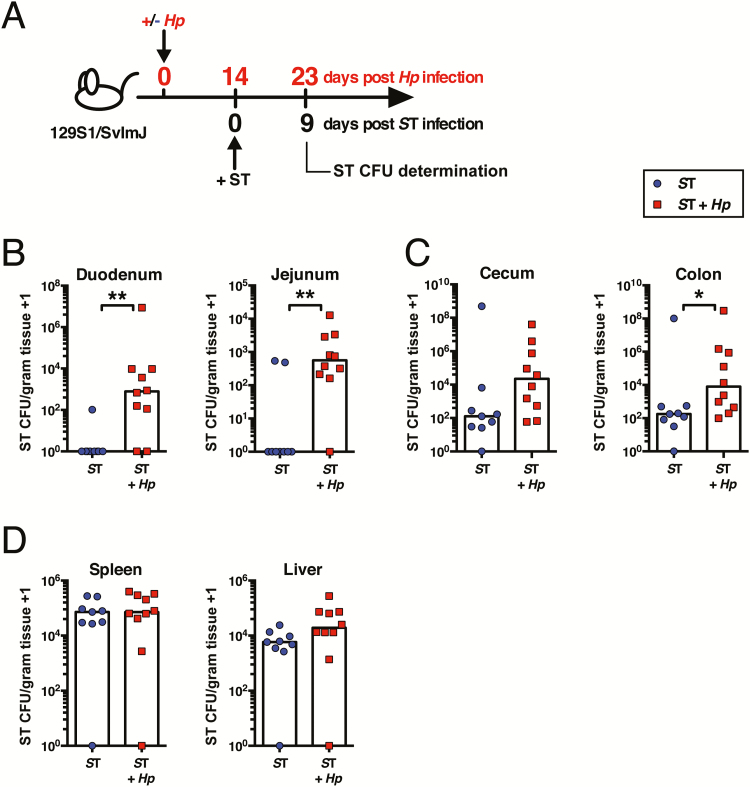
To test the effects of intestinal helminth infection on bacterial co-colonization, we developed a model of coinfection using the murine helminth H. polygyrus and the bacteria S. Typhimurium. 129S1/SvImJ mice were orally infected with H. polygyrus, a strictly enteric pathogen which establishes a chronic infection in the small intestine [27]. At day 14 following H. polygyrus infection, by which time adult worms are present in the lumen of the duodenum and jejunum, mice were orally challenged with S. Typhimurium, alongside mice infected with S. Typhimurium alone (Figure 1A). The majority of singly infected mice were able to clear S. Typhimurium from the small intestine within 9 days, yet H. polygyrus–coinfected mice maintained high bacterial burdens (Figure 1B). In the cecum and colon, sites distal to helminth infection, the effect of helminth coinfection on S. Typhimurium clearance was less marked, although helminth coinfection did result in significantly higher S. Typhimurium levels in the colon (Figure 1C). Levels of systemic S. Typhimurium were unaffected by helminth coinfection (Figure 1D). Together, these data suggest that H. polygyrus exerts a local effect to promote S. Typhimurium colonization.
Figure 1.
Helminth-coinfected 129S1/SvlmJ mice exhibit elevated Salmonella Typhimurium (ST) burdens in the small intestine. A, Experimental setup. 129S1/SvImJ mice were left naive or infected with Heligmosomoides polygyrus (Hp). Fourteen days later, all mice were orally infected with ST. Nine days later, ST colony-forming unit (CFU) counts were determined. ST CFU counts in the duodenum and jejunum (B), cecum and colon (C), and spleen and liver (D) are shown. Data shown are pooled from 2 independent experiments and are representative of the results from 3 independent experiments. *P ≤ .05; **P ≤ .01.
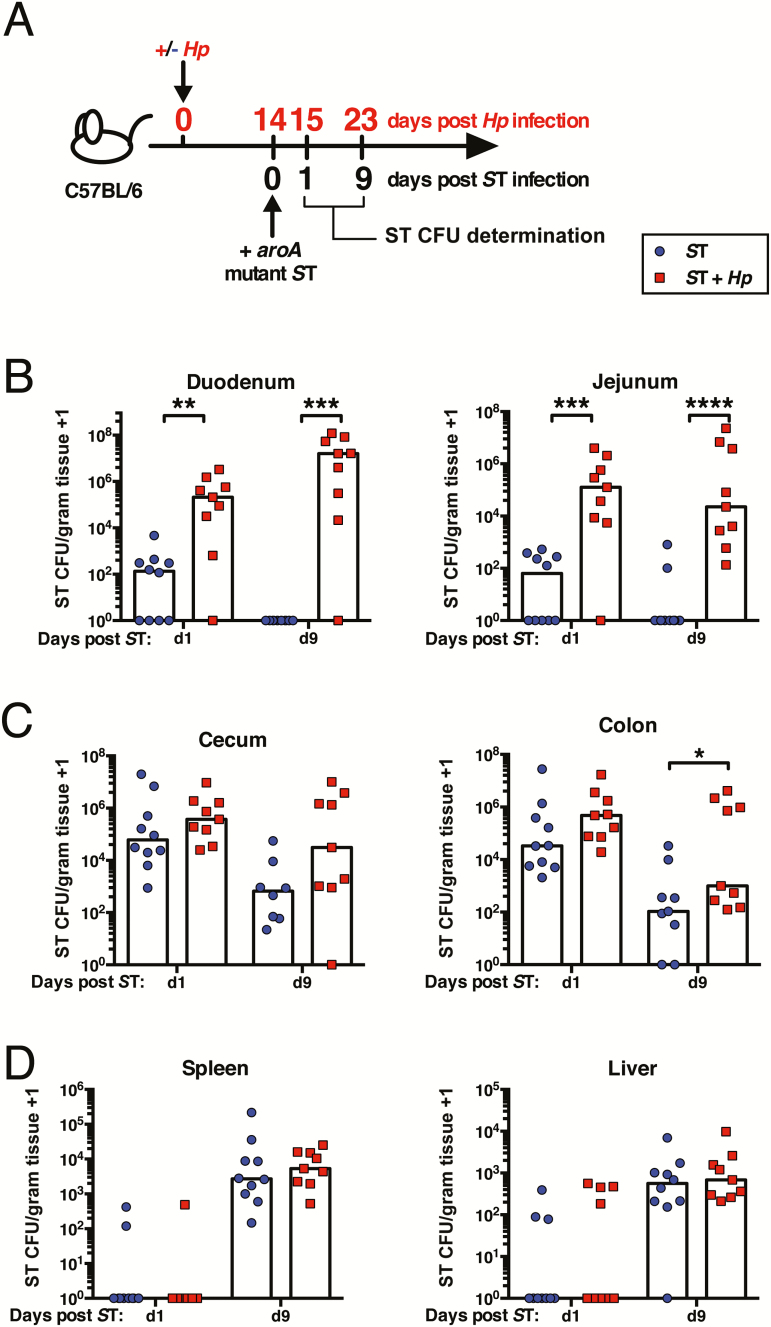
To test the effect of helminth infection on bacterial colonization was affected by genetic background, we also coinfected C57BL/6 mice (Figure 2A). Unlike 129S1/SvImJ mice, C57Bl/6 mice lack the natural resistance-associated macrophage protein 1 (Nramp1), and rapidly succumb to infection with doses of wild-type S. Typhimurium that 129S1/SvImJ mice survive [28]. For this reason, all experiments with C57BL/6 mice were conducted with an attenuated strain of S. Typhimurium (aroA mutant [24]). Similar to 129S1/SvImJ mice, C57BL/6 mice infected with S. Typhimurium alone were able to clear this pathogen from the small intestine within 9 days, whereas helminth-coinfected mice maintained high S. Typhimurium burdens in the small intestine (Figure 2B). The greatest impact of helminth coinfection was at sites proximal to H. polygyrus colonization (Figure 2C and D). Notably, small-intestinal bacterial burdens were significantly higher in helminth-coinfected mice as early as 24 hours following S. Typhimurium infection (Figure 2B). Therefore, in 2 different inbred strains of mice, the presence of an intestinal helminth enhances local bacterial colonization following challenge infection.
Figure 2.
Helminth-coinfected C57BL/6 mice exhibit elevated Salmonella Typhimurium (ST) burdens in the small intestine. A, Experimental setup. C57BL/6 mice were left naive or infected with Heligmosomoides polygyrus (Hp). Fourteen days later, all mice were orally infected with aroA mutant ST. One and 9 days later, ST colony-forming unit (CFU) counts were determined. ST CFU counts in the duodenum and jejunum (B), cecum and colon (C), and spleen and liver (D) are shown. Data shown are pooled from 2 independent experiments and are representative of the results from 4 independent experiments. *P ≤ .05; **P ≤ .01; ***P ≤ .001; ****P ≤ .0001.
Elevated S. Typhimurium Burdens in Helminth-Coinfected Mice Are Independent of Induction of Th2 or Treg Cells
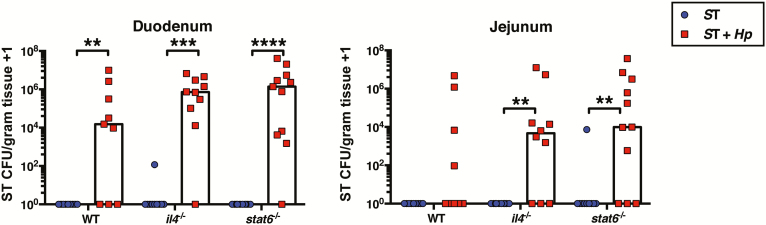
Th2 cells induced by helminths have been previously shown to impair immunity to microbial infections [2–5]. We hypothesized, therefore, that the potent Th2 response induced by H. polygyrus [27] may be inhibiting effective bacterial clearance, leading to S. Typhimurium persistence. To test this, we coinfected mice that are unable to mount a Th2 response. Surprisingly, similar to coinfected wild-type mice, both il4-deficient and stat6-deficient coinfected mice failed to clear S. Typhimurium from the small intestine by day 9 postinfection (Figure 3), suggesting that during coinfection increased susceptibility to secondary bacterial infection is independent of Th2 cells.
Figure 3.
Elevated Salmonella Typhimurium (ST) burdens in helminth-coinfected mice are independent of induction of Th2 cells. C57BL/6 (wild-type [WT]), il4-/-, and stat6-/- mice were left naive or infected with Heligmosomoides polygyrus (Hp). Fourteen days later, all mice were orally infected with aroA mutant ST. Nine days later, ST colony-forming unit (CFU) counts were determined in the duodenum and jejunum. Data shown are pooled from 2 independent experiments. **P ≤ .01; ***P ≤ .001; ****P ≤ .0001.
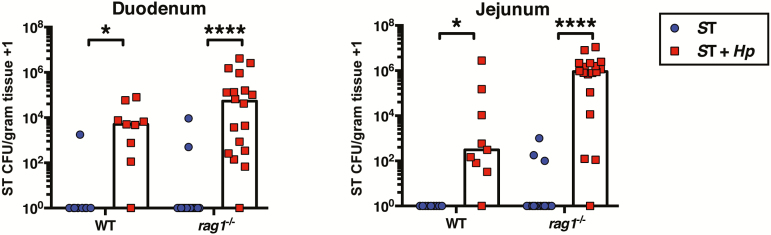
In addition to induction of Th2 cells, H. polygyrus also stimulates expansion and activation of Tregs [29]. Therefore, Treg-mediated immunosuppression of antibacterial responses could account for elevated bacteria burdens in coinfected mice. To test this, we coinfected rag1-deficient mice, which lack all mature T cells including Tregs, as well as all mature B cells [30]. We compared S. Typhimurium burdens between singly infected and helminth-coinfected rag1-deficient mice 1 day after S. Typhimurium infection, before differences in susceptibility to S. Typhimurium between wild-type and rag1-deficient mice emerge [31]. Similar to wild-type mice, at 1 day following S. Typhimurium infection, helminth-coinfected rag1-deficient mice had dramatically elevated S. Typhimurium burdens in the small intestine, compared to rag1-deficient mice infected with S. Typhimurium alone (Figure 4). This suggests that during coinfection suppression of immunity to S. Typhimurium is not mediated by helminth-induced Tregs. Together these data demonstrate that helminth infection can impair resistance to bacterial pathogens independently of Th2 or Treg conditioning of host immunity.
Figure 4.
Elevated Salmonella Typhimurium (ST) burdens in helminth-coinfected mice are independent of induction of regulatory T cells. C57BL/6 and rag1-/- mice were left naive or infected with Heligmosomoides polygyrus (Hp). Fourteen days later, all mice were orally infected with aroA mutant ST. One day later, ST colony-forming unit (CFU) counts were determined in the duodenum and jejunum. Data shown are pooled from 3 independent experiments. *P ≤ .05; ****P ≤ .0001.
Helminth Infection Alters the Metabolic Profile of the Small Intestine
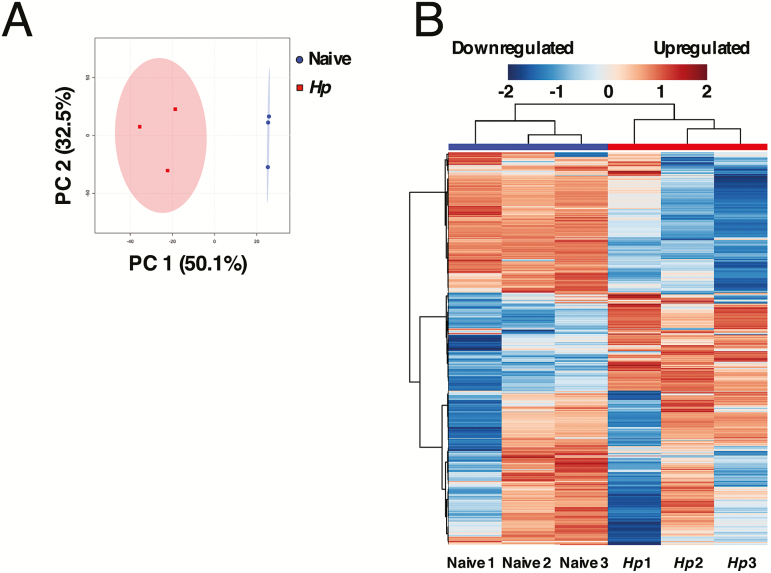
Colonization with helminth parasites has been associated with changes to the intestinal microbiota in both mice and humans [32]. Heligmosomoides polygyrus infection causes profound shifts in the composition of the small-intestinal microbiota [33–35]. Alterations to the microbiota imposed by H. polygyrus have recently been demonstrated to enhance production of microbiota-derived short-chain fatty acids, which can alleviate allergic airway inflammation [36]. Helminth infection has also been shown to inhibit the development of inflammatory bowel disease by reducing the prevalence of specific inflammatory species within the microbiota [37]. In both these cases, helminth-induced microbiota changes were seen to target host pathways. We hypothesized that there may also be a direct effect of intestinal metabolic changes on concurrent microbial pathogens. We identified metabolites present in the small intestine by UPLC-FTMS, and assessed the relative abundance of each metabolite between naive and H. polygyrus–infected mice. Helminth infection significantly altered the metabolic profile of the small intestine (Figure 5A and B; Supplementary Figure 1A and 2B; Supplementary Figure 2A and 2B). Of 4593 metabolite features detected, 362 were significantly altered in abundance during H. polygyrus infection (P ≤ .01, ≥2-fold difference), with 41 upregulated and 321 suppressed during H. polygyrus infection (m/z, retention time, and putative identities of metabolite features reported in Supplementary Figure 2A and 2B and Supplementary Tables 1–4).
Figure 5.
Helminth infection alters the metabolic profile of the small intestine. A, The differential abundance of small-intestinal metabolites from naive or day 14 Heligmosomoides polygyrus–infected (Hp) 129S1/SvImJ mice was determined by ultra-high-performance liquid chromatography–Fourier transform mass spectrometry. A principal components (PC) analysis plot was generated from metabolites detected in positive ion mode. B, Heatmap showing the relative abundance of all metabolites detected in naive and day 14 Hp-infected 129S1/SvImJ mice, detected in positive ion mode. Clustering of naive and Hp-infected mice is shown using a Euclidean distance and Ward clustering algorithm.
Helminth-Modulated Small-Intestinal Metabolites Promote S. Typhimurium Intracellular Invasion
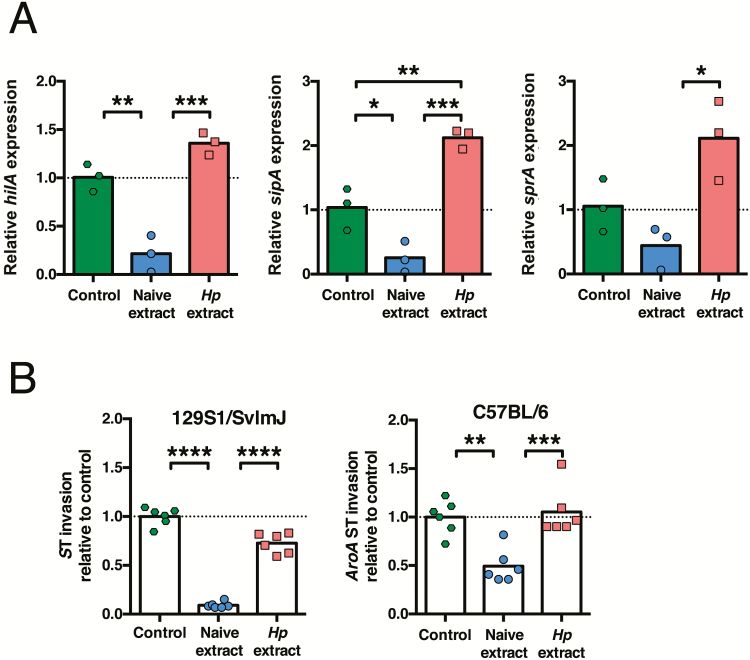
We next aimed to determine whether a helminth-altered metabolome had an impact on the growth or invasive capacity of S. Typhimurium. We first tested whether helminth- altered small-intestinal metabolites affected the growth rate of S. Typhimurium, and found no evidence that helminth- altered small-intestinal metabolites promoted the growth of S. Typhimurium in an in vitro growth assay (Supplementary Figure 3A and B). We next investigated whether exposure to helminth-modulated small-intestinal metabolites altered the expression levels of S. Typhimurium virulence genes. HilA is a bacterial transcription factor that plays a central role in regulating expression of S. Typhimurium genes controlling intracellular invasion, which are encoded within Salmonella pathogenicity island 1 (SPI-1) [38]. We found that hilA expression was inhibited in S. Typhimurium cultured with metabolites from naive mice but not by metabolites from H. polygyrus–infected mice (Figure 6A). Expression levels of sipA and sprB, which are also found within the SPI-1 locus, were likewise elevated after culture with small-intestinal metabolites from H. polygyrus–infected mice compared to after culture with small-intestinal metabolites from naive mice (Figure 6A). These data indicate that small-intestinal metabolites altered in abundance during helminth infection promote the expression of virulence genes in S. Typhimurium. To determine whether the altered expression of S. Typhimurium virulence genes after exposure to helminth-modulated metabolites corresponded with an altered invasive capacity of S. Typhimurium, we tested the effect of metabolites from naive or H. polygyrus–infected mice on S. Typhimurium intracellular invasion. Metabolites extracted from the small intestine of naive mice, either 129S1/SvImJ or C57BL/6, significantly suppressed the ability of S. Typhimurium to invade human epithelial (HeLa) cells (Figure 6B). In contrast, metabolites extracted from the small intestine of H. polygyrus–infected mice did not suppress the ability of S. Typhimurium to invade HeLa cells (Figure 6B). These data reveal for the first time that modulation of intestinal metabolites during helminth infection directly affects the invasive capacity of pathogenic S. Typhimurium bacteria. Together our data reveal a new interaction in helminth-bacterial coinfection, in which helminth infection disrupts the protective composition of the intestinal metabolome, allowing for increased intracellular invasion and colonization by pathogenic bacteria.
Figure 6.
Helminth-modified small-intestinal metabolites promote intracellular invasion by Salmonella Typhimurium (ST). A, aroA mutant ST bacteria were cultured without metabolites (control) or with metabolites extracted from the small intestine of naive or Heligmosomoides polygyrus (Hp)–infected C57BL/6 mice. Expression levels of ST hilA, sipA, and sprB were then determined. Each data point represents gene expression levels of 3 ST cultures that were split and cultured with metabolites from each group. Data shown are representative of results from 4 independent experiments that each used independent mice and ST cultures. B, Wild-type or aroA mutant ST bacteria were cultured without metabolites (control) or with metabolites extracted from the small intestine of naive or Hp–infected 129S1/SvImJ or C57BL/6 mice, prior to infection of HeLa cells. Each data point represents a technical replicate of HeLa cells infected with ST that had been cultured with metabolites pooled from 3–5 naive or 3 Hp-infected mice. Data are representative of results from 2 (129S1/SvImJ) or 3 (C57BL/6) independent experiments that each used independent mice and ST cultures. *P ≤ .05; **P ≤ .01; ***P ≤ .001; ****P ≤ .0001.
DISCUSSION
A number of studies have demonstrated that an ongoing helminth infection can result in heightened susceptibility to microbial pathogens. This was attributed to helminth-mediated immunomodulation that acted to compromise the development of antimicrobial immune responses [1–5, 13]. In this study, we describe a novel mechanism by which helminths increase susceptibility to a microbial pathogen, independently of immune conditioning toward Th2 or Treg responses. We provide evidence to show that the presence of helminths disrupts the metabolic composition of the intestine, and the resultant shift in metabolites directly alters the invasive capacity of the intracellular bacterial pathogen S. Typhimurium.
Salmonella invasion gene expression is strongly repressed by metabolites extracted from the feces of naive mice or humans [39]. These inhibitory metabolites are likely derived from both the microbiota as well as the mammalian host, as metabolites extracted from the feces of both germ-free and conventionally raised mice inhibited Salmonella invasion gene expression, although to a lesser extent by metabolites from germ-free mice [39]. Disruption of the intestinal metabolic environment by antibiotic treatment can promote S. Typhimurium expansion in mice [40]. Antibiotic treatment induces host expression of inducible nitric oxide synthase, which mediates elevated carbohydrate oxidation, releasing galactarate and glucarate that can promote the expansion of S. Typhimurium [40]. Heligmosomoides polygyrus infection could disrupt the intestinal metabolome by shifting the composition of the microbiota, thereby altering the abundance of microbiota-derived products [33–35], or by interfering with host metabolism. Additionally, it is possible that metabolites produced directly by helminths [36, 41] are responsible for promoting Salmonella virulence.
It is becoming increasingly clear that complex communication between kingdoms occurs in the mammalian intestine. The bacterial microbiota and intestinal helminths share a niche within the host, and can influence each other’s fitness and persistence [32]. For example, the presence of the microbiota is critical for the establishment of Trichuris muris, a murine whipworm whose eggs hatch in the ceca [42]. For hatching, the eggs require direct contact with structural components of microbes within the intestinal microbiota [42]. Specific species within the microbiota can also affect the chronicity of adult helminths. Lactobacillus species promote the persistence of adult H. polygyrus and T. muris worms, likely through the induction of Tregs and inhibition of Th2 responses directed against the parasites [33, 43]. A common pathway of interbacterial communication in the intestine is through the production of bacterial-derived metabolites, which can shape the population dynamics of the bacterial microbiota, as well as influencing the ability of pathogenic bacteria to colonize the intestine [44]. Our work provides the first example of how helminths can influence the virulence of pathogenic bacteria through an altered small-intestinal metabolome.
A helminth-altered cecal metabolome has been shown to mediate, at least in part, the suppression of airway inflammation during helminth infection in a murine model of allergic asthma [36]. It has been previously demonstrated that helminth infection results in elevated levels of short-chain fatty acids (SCFAs) in the ceca of mice, which enhanced the suppressive function of Tregs and protected against airway inflammation [36]. Helminths may have evolved the ability to shift the metabolite profile toward one which promotes their own chronicity, for example, through inducing SCFAs, which have been shown to induce and enhance the suppressive function of Tregs [45–47]. There is a current interest in the use of helminths, or helminth-derived products, for the therapeutic treatment of inflammatory diseases including allergy and inflammatory bowel disease [48]. As our and others’ data suggest that helminth infection can also increase susceptibility to microbial infections [2–6], it will be important to fully characterize the pathways by which helminths affect host physiology, such that susceptibility to pathogenic microbes during therapeutic administration of helminths can be predicted and controlled.
Helminths are potent immunomodulators, which can alter host immunity to infectious and immune-mediated diseases through multiple mechanisms, dependent on the disease context, host genetics, and the microbiota [13, 36, 37]. Both helminths and the microbiota have potent immunomodulatory effects during inflammatory and infectious diseases [13, 32, 49, 50]. Our data identify a novel mechanism by which a helminth-modified metabolic environment can promote the ability of a bacterial pathogen to colonize the intestine. Understanding the mechanisms by which helminths promote susceptibility to microbial coinfections will aid disease treatment and prevention strategies in the world regions where helminths are prevalent.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Acknowledgments. We thank Dr Kelly McNagny at UBC for provision of rag1-/- mice.
Financial support. Metabolomics analysis was performed at the UVic-Genome BC Proteomics Centre and was supported by funding to the Metabolomics Innovation Centre through the Genome Innovations Network from Genome Canada, Genome Alberta, and Genome British Columbia for operations (250MET and 7203) and technology development (215MET and MC3T). L. A. R. was supported by postdoctoral fellowship awards from the Canadian Institutes of Health Research (CIHR) and the Michael Smith Foundation for Health Research (MSFHR) in partnership with AllerGen. N. G. was supported by postdoctoral fellowship awards from CIHR and MSFHR. E. M. B. was supported by a CIHR doctoral research award and a 4-year doctoral fellowship (4YF) from UBC. N. C. M. was supported by a Vanier Canada Graduate Scholarship and a 4YF from UBC. Y. V. was supported by postdoctoral fellowship awards from CIHR and the MSFHR. C. H. B. was supported by the Leading Edge Endowment Fund; and the Segal McGill Family Chair in Molecular Oncology, the Warren Y. Soper Foundation, and the Alvin Segal Family Foundation, all at the Jewish General Hospital, Montreal, Quebec, Canada. Work in the laboratory of G. P.-W. was supported by CIHR (MOP-126061) and funds from UBC. G. P.-W. is a MSFHR Scholar and holds a CIHR New Investigator Award granted in partnership with the Crohn’s and Colitis Foundation Canada, and the Canadian Association of Gastroenterology. Work in the laboratory of B. B. F. was supported by operating grants from CIHR (MOP-133561 and MOP-299601), the Canadian Institute for Advanced Research, and Institut Mérieux. B. B. F. is a Peter Wall Distinguished Professor at UBC.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Salgame P, Yap GS, Gause WC. Effect of helminth-induced immunity on infections with microbial pathogens. Nat Immunol 2013; 14:1118–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen CC, Louie S, McCormick B, Walker WA, Shi HN. Concurrent infection with an intestinal helminth parasite impairs host resistance to enteric Citrobacter rodentium and enhances Citrobacter-induced colitis in mice. Infect Immun 2005; 73:5468–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Osborne LC, Monticelli LA, Nice TJ, et al. Coinfection. Virus-helminth coinfection reveals a microbiota-independent mechanism of immunomodulation. Science 2014; 345:578–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reese TA, Wakeman BS, Choi HS, et al. Helminth infection reactivates latent γ-herpesvirus via cytokine competition at a viral promoter. Science 2014; 345:573–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hsieh YJ, Fu CL, Hsieh MH. Helminth-induced interleukin-4 abrogates invariant natural killer T cell activation-associated clearance of bacterial infection. Infect Immun 2014; 82:2087–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Su L, Su CW, Qi Y, et al. Coinfection with an intestinal helminth impairs host innate immunity against Salmonella enterica serovar Typhimurium and exacerbates intestinal inflammation in mice. Infect Immun 2014; 82:3855–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Harris JB, Podolsky MJ, Bhuiyan TR, et al. Immunologic responses to Vibrio cholerae in patients co-infected with intestinal parasites in Bangladesh. PLoS Negl Trop Dis 2009; 3:e403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Elias D, Mengistu G, Akuffo H, Britton S. Are intestinal helminths risk factors for developing active tuberculosis? Trop Med Int Health 2006; 11:551–8. [DOI] [PubMed] [Google Scholar]
- 9. Resende Co T, Hirsch CS, Toossi Z, Dietze R, Ribeiro-Rodrigues R. Intestinal helminth co-infection has a negative impact on both anti-Mycobacterium tuberculosis immunity and clinical response to tuberculosis therapy. Clin Exp Immunol 2007; 147:45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Downs JA, Mguta C, Kaatano GM, et al. Urogenital schistosomiasis in women of reproductive age in Tanzania’s Lake Victoria region. Am J Trop Med Hyg 2011; 84:364–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Le Hesran JY, Akiana J, Ndiaye EHM, Dia M, Senghor P, Konate L. Severe malaria attack is associated with high prevalence of Ascaris lumbricoides infection among children in rural Senegal. Trans R Soc Trop Med Hyg 2004; 98:397–9. [DOI] [PubMed] [Google Scholar]
- 12. Degarege A, Legesse M, Medhin G, Animut A, Erko B. Malaria and related outcomes in patients with intestinal helminths: a cross-sectional study. BMC Infect Dis 2012; 12:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Maizels RM, Yazdanbakhsh M. Immune regulation by helminth parasites: cellular and molecular mechanisms. Nat Rev Immunol 2003; 3:733–44. [DOI] [PubMed] [Google Scholar]
- 14. Haeryfar SM, DiPaolo RJ, Tscharke DC, Bennink JR, Yewdell JW. Regulatory T cells suppress CD8+ T cell responses induced by direct priming and cross-priming and moderate immunodominance disparities. J Immunol 2005; 174:3344–51. [DOI] [PubMed] [Google Scholar]
- 15. Johanns TM, Ertelt JM, Rowe JH, Way SS. Regulatory T cell suppressive potency dictates the balance between bacterial proliferation and clearance during persistent Salmonella infection. PLoS Pathog 2010; 6:e1001043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kang PF, Wu WJ, Tang Y, et al. Activation of ALDH2 with low concentration of ethanol attenuates myocardial ischemia/reperfusion injury in diabetes rat model. Oxid Med Cell Longev 2016; 2016:6190504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Perona-Wright G, Mohrs K, Mohrs M. Sustained signaling by canonical helper T cell cytokines throughout the reactive lymph node. Nat Immunol 2010; 11:520–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Y, Chen HL, Bannick N, et al. Intestinal helminths regulate lethal acute graft-versus-host disease and preserve the graft-versus-tumor effect in mice. J Immunol 2015; 194:1011–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wilson MS, Taylor MD, Balic A, Finney CA, Lamb JR, Maizels RM. Suppression of allergic airway inflammation by helminth-induced regulatory T cells. J Exp Med 2005; 202:1199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kitagaki K, Businga TR, Racila D, Elliott DE, Weinstock JV, Kline JN. Intestinal helminths protect in a murine model of asthma. J Immunol 2006; 177:1628–35. [DOI] [PubMed] [Google Scholar]
- 21. Mohrs M, Shinkai K, Mohrs K, Locksley RM. Analysis of type 2 immunity in vivo with a bicistronic IL-4 reporter. Immunity 2001; 15:303–11. [DOI] [PubMed] [Google Scholar]
- 22. Mohrs K, Wakil AE, Killeen N, Locksley RM, Mohrs M. A two-step process for cytokine production revealed by IL-4 dual-reporter mice. Immunity 2005; 23:419–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity 1996; 4:313–9. [DOI] [PubMed] [Google Scholar]
- 24. Hoiseth SK, Stocker BA. Aromatic-dependent Salmonella Typhimurium are non-virulent and effective as live vaccines. Nature 1981; 291:238–9. [DOI] [PubMed] [Google Scholar]
- 25. Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0–making metabolomics more meaningful. Nucleic Acids Res 2015; 43:W251–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xia J, Psychogios N, Young N, Wishart DS. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res 2009; 37:W652–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reynolds LA, Filbey KJ, Maizels RM. Immunity to the model intestinal helminth parasite Heligmosomoides polygyrus. Semin Immunopathol 2012; 34:829–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Govoni G, Vidal S, Gauthier S, Skamene E, Malo D, Gros P. The Bcg/Ity/Lsh locus: genetic transfer of resistance to infections in C57BL/6J mice transgenic for the Nramp1 Gly169 allele. Infect Immun 1996; 64:2923–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grainger JR, Smith KA, Hewitson JP, et al. Helminth secretions induce de novo T cell Foxp3 expression and regulatory function through the TGF-β pathway. J Exp Med 2010; 207:2331–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 1992; 68:869–77. [DOI] [PubMed] [Google Scholar]
- 31. Kupz A, Bedoui S, Strugnell RA. Cellular requirements for systemic control of Salmonella enterica serovar Typhimurium infections in mice. Infect Immun 2014; 82:4997–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reynolds LA, Finlay BB, Maizels RM. Cohabitation in the intestine: interactions among helminth parasites, bacterial microbiota, and host immunity. J Immunol 2015; 195:4059–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reynolds LA, Smith KA, Filbey KJ, et al. Commensal-pathogen interactions in the intestinal tract: lactobacilli promote infection with, and are promoted by, helminth parasites. Gut Microbes 2014; 5:522–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Walk ST, Blum AM, Ewing SA, Weinstock JV, Young VB. Alteration of the murine gut microbiota during infection with the parasitic helminth Heligmosomoides polygyrus. Inflamm Bowel Dis 2010; 16:1841–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rausch S, Held J, Fischer A, et al. Small intestinal nematode infection of mice is associated with increased enterobacterial loads alongside the intestinal tract. PLoS One 2013; 8:e74026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zaiss MM, Rapin A, Lebon L, et al. The intestinal microbiota contributes to the ability of helminths to modulate allergic inflammation. Immunity 2015; 43:998–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ramanan D, Bowcutt R, Lee SC, et al. Helminth infection promotes colonization resistance via type 2 immunity. Science 2016; 352:608–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bajaj V, Lucas RL, Hwang C, Lee CA. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol 1996; 22:703–14. [DOI] [PubMed] [Google Scholar]
- 39. Antunes LCM, McDonald JAK, Schroeter K, et al. Antivirulence activity of the human gut metabolome. MBio 2014; 5:e01183–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Faber F, Tran L, Byndloss MX, et al. Host-mediated sugar oxidation promotes post-antibiotic pathogen expansion. Nature 2016; 534:697–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tielens AGM, van Grinsven KWA, Henze K, van Hellemond JJ, Martin W. Acetate formation in the energy metabolism of parasitic helminths and protists. Int J Parasitol 2010; 40:387–97. [DOI] [PubMed] [Google Scholar]
- 42. Hayes KS, Bancroft AJ, Goldrick M, Portsmouth C, Roberts IS, Grencis RK. Exploitation of the intestinal microflora by the parasitic nematode Trichuris muris. Science 2010; 328:1391–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dea-Ayuela MA, Rama-Iñiguez S, Bolás-Fernandez F. Enhanced susceptibility to Trichuris muris infection of B10Br mice treated with the probiotic Lactobacillus casei. Int Immunopharmacol 2008; 8:28–35. [DOI] [PubMed] [Google Scholar]
- 44. Vogt SL, Peña-Díaz J, Finlay BB. Chemical communication in the gut: effects of microbiota-generated metabolites on gastrointestinal bacterial pathogens. Anaerobe 2015; 34:106–15. [DOI] [PubMed] [Google Scholar]
- 45. Arpaia N, Campbell C, Fan X, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013; 504:451–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013; 341:569–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Furusawa Y, Obata Y, Fukuda S, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013; 504:446–50. [DOI] [PubMed] [Google Scholar]
- 48. Fleming JO, Weinstock JV. Clinical trials of helminth therapy in autoimmune diseases: rationale and findings. Parasite Immunol 2015; 37:277–92. [DOI] [PubMed] [Google Scholar]
- 49. McSorley HJ, Maizels RM. Helminth infections and host immune regulation. Clin Microbiol Rev 2012; 25:585–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell 2014; 157:121–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.