Capsid sequence changes in norovirus GII.17 strains result in loss of blockade antibody binding, indicating that viral evolution, specifically at residues 393–396, may have contributed to the emergence of cluster IIIb strains and the persistence of GII.17 in human populations.
Keywords: Norovirus, blockade antibody, antibody neutralization, viral evolution, GII.17
Abstract
Background
Human norovirus is a significant public health burden, with >30 genotypes causing endemic levels of disease and strains from the GII.4 genotype causing serial pandemics as the virus evolves new ligand binding and antigenicity features. During 2014–2015, genotype GII.17 cluster IIIb strains emerged as the leading cause of norovirus infection in select global locations. Comparison of capsid sequences indicates that GII.17 is evolving at previously defined GII.4 antibody epitopes.
Methods
Antigenicity of virus-like particles (VLPs) representative of clusters I, II, and IIIb GII.17 strains were compared by a surrogate neutralization assay based on antibody blockade of ligand binding.
Results
Sera from mice immunized with a single GII.17 VLP identified antigenic shifts between each cluster of GII.17 strains. Ligand binding of GII.17 cluster IIIb VLP was blocked only by antisera from mice immunized with cluster IIIb VLPs. Exchange of residues 393–396 from GII.17.2015 into GII.17.1978 ablated ligand binding and altered antigenicity, defining an important varying epitope in GII.17.
Conclusions
The capsid sequence changes in GII.17 strains result in loss of blockade antibody binding, indicating that viral evolution, specifically at residues 393–396, may have contributed to the emergence of cluster IIIb strains and the persistence of GII.17 in human populations.
Human noroviruses are the leading cause of acute viral gastroenteritis [1–3], justifying substantial investment in understanding the mechanisms of molecular evolution, determining the epidemiology and persistence of these viruses in populations, and vaccine development [1–5]. Extensive genetic diversity (there are >30 known genotypes) and antigenic drift within the predominant genotype (GII.4) make determining which strains to target in a multivalent virus-like particle (VLP) vaccine platform a primary challenge. For the past 2 decades, GII.4 strains have accounted for 70%–80% of norovirus outbreaks, providing a key target for the vaccine [6]. Recently, in parts of Asia, GII.17 strains have replaced GII.4.2012 strains as the predominant cause of norovirus outbreaks [7]. Phylogenetic analyses of these emergent strains suggest that they are evolving via an epochal pattern of evolution, perhaps in response to herd immunity, similarly to the GII.4 strains [7–9].
Norovirus Hu/GII.17/C142/1978/GUF (GII.17.1978) is the first GII.17 strain reported in GenBank and defines cluster I of the genotype. Strains remained genetically static until 2005, when the cluster II strains emerged [10]. Despite significant capsid sequence changes between clusters I and II, GII.17 strains were rarely detected in outbreak investigations [7]. The cluster III strains subsequently diverged into subclusters IIIa and IIIb, based on the sequence of the major capsid protein [11, 12]. In the winter season of 2014–2015, cluster IIIb strains became the predominant cause of norovirus outbreaks in China [8]. The IIIb strains spread, causing epidemic levels of disease in parts of Southeast Asia but only sporadic infections in Europe, Australia, and North America [7].
Compared with cluster I and II strains, cluster III strains have a unique RNA-dependent RNA polymerase (RDRP; GII.P17) and both insertions and deletions in the capsid gene, particularly in the P2 subdomain, that could affect both ligand binding and antigenicity profiles [7, 12, 13]. P particles of a reported cluster III strain bind to the saliva of blood-type O, A, and B individuals, and secretor-positive persons of all blood types experienced symptomatic infection during an outbreak, indicating broad histo-blood group antigen binding by this strain. Antigenicity differences between the clusters, determined using GII.17-derived antisera, have only been evaluated by a single convalescent serum sample, which preferentially blocked binding of the homotypic cluster III strain, compared with heterologous strain P particles [14].
Here, we empirically evaluated whether genetic differences in the major capsid gene between GII.17 clusters translated into antigenic changes by comparing the blockade antibody (Ab) response to a panel of VLPs representing cluster I, II, and IIIb strains, using polyclonal sera from adult humans and immunized mice. While blockade Ab titers in human sera were similar between the different GII.17 clusters, sera from mice immunized with a single GII.17 strain from each cluster demonstrated clear antigenic differences between the GII.17 strains, particularly the emergent cluster IIIb strain, GII.17.2015. These data confirm that the capsid sequence of the GII.17 genocluster is evolving at Ab epitopes, potentially allowing for escape from herd immunity and widespread disease. These findings demonstrate that norovirus GII genotypes other than GII.4 are capable of epochal evolution and antigenic drift, enhancing their pandemic potential. Our data support the hypothesis that enhanced strain-specific surveillance and vaccine efficacy trials, similar to those in place for GII.4 norovirus strains, are warranted against these newly defined, rapidly evolving GII.17 strains.
MATERIALS AND METHODS
Ethics Statement
Healthy human adult volunteers provided samples in June 2016, according to established institutional review board guidelines, after providing informed consent. Peripheral blood specimens were collected by venipuncture without anticoagulant and allowed to clot. Serum was removed, aliquoted, and stored at −80°C. Before use, sera underwent heat inactivation for 30 minutes at 56°C. Experimentation guidelines of the US Department of Health and Human Services were followed.
Ethics Statement on Mouse Studies
Mouse studies were executed in accordance with the recommendations for the care and use of animals by the Office of Laboratory Animal Welfare at the National Institutes of Health. The Institutional Animal Care and Use Committee at the University of North Carolina–Chapel Hill approved the animal studies performed here (protocol 14–090). Eight-week-old Balb/c mice (Charles River Laboratories) were immunized intramuscularly with 0.2 µg of VLPs adjuvanted with 5 µg of monophosphoryl lipid A and 50 µg of alhydrogel, mimicking the human norovirus vaccine [15]. Booster vaccinations were performed on days 28 and 62. Mice were euthanized with isoflurane on day 69, and terminal blood specimens were collected.
Modeling of GII.17 Amino Acid Changes Over Time
For this study, 4 GII.17 P2 subdomain capsid sequences were compared and analyzed, including GII.17.1978 (KC597139, cluster I), GII.17.2002 (KJ196286, cluster I), GII.17.2005 (DQ438972, cluster II), and GII.17.2015 (KP698930, cluster IIIb). For visual purposes, residue changes between the strains were highlighted onto the homology model of GII.17.2002 in Figure 1 [13]. The individual P2 subdomain monomer models were converted into dimers in Pymol, and the structural models were analyzed and colored to highlight epitopes and amino acid changes that occurred over time. For Figure 2, homology models of these 4 sequences were generated using the SWISS-MODEL server (available at: https://swissmodel.expasy.org/interactive) with default settings. Briefly, the entire capsid sequence was uploaded into the server in Fasta format, and the “Select Templates” option was selected. For each of the 4 sequences, the template with the highest identity was chosen. For GII.17.1987, GII.17.2002, and GII.17.2005, the template identified was Protein Data Bank accession number 5F4J [13], which is the crystal structure for the GII.17.2002 virus capsid. For GII.17.2015, the template identified was Protein Data Bank accession number 5LKC [16], which is the crystal structure for the GII.17.2015 capsid. Homology models were built using these templates and exported in pdb format for rendering in MacPymol, version 1.8.0.4 (available at: https://www.pymol.org/). Electrostatic potential was predicted using the vacuum electrostatics function in MacPymol, which warned that the results were only qualitatively useful. The amino acid sequences were aligned and compared using Clustal X, version 2.1 [17].
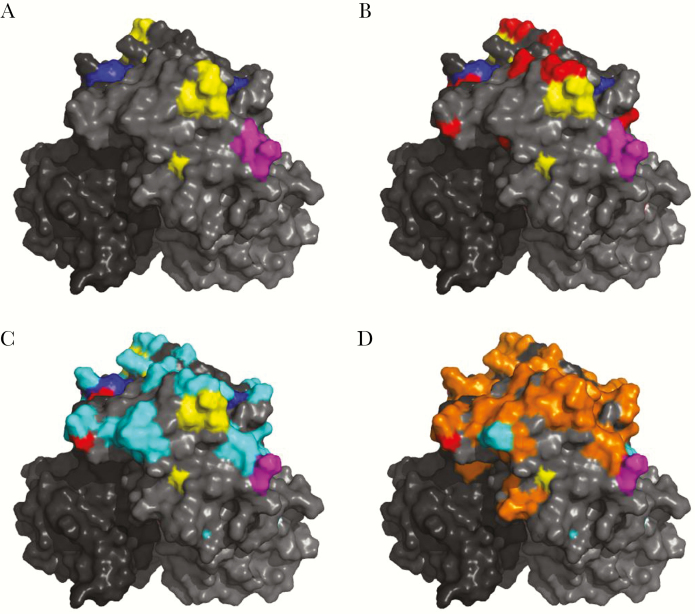
Figure 1.
GII.17 capsid P2 domain residue changes over time. A, Structural model of GII.17.2002 P domain dimer (shades of gray) with GII.4 surface-exposed blockade epitope A (yellow), D (blue), and E (magenta) highlighted. B–D, Residue changes between 1978 and 2002 (red; B), between 2002 (cluster I) and 2005 (cluster II; cyan; C), and between 2005 (cluster II) and 2015 (cluster IIIb; orange; D).
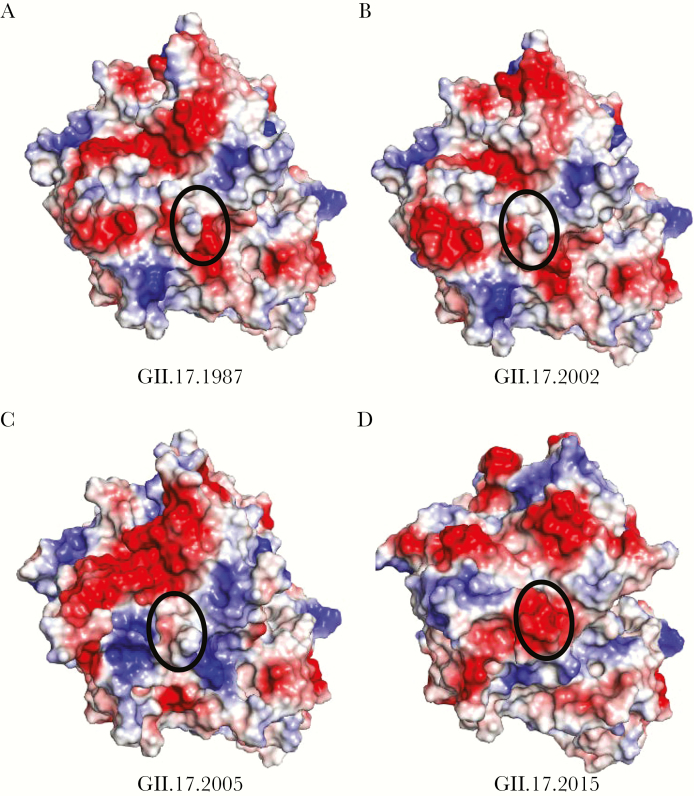
Figure 2.
Surface-exposed charge changes over time. The electrostatic potential (blue, positive; red, negative) of the top surface of the P2 subdomain is shown for GII.17.1978 (A), GII.17.2002 (B), GII.17.2005 (C), and GII.17.2015 (D). Surface-exposed charge changes in 2015 likely drove antigenic changes, particularly a 4 aspartic acid motif at positions 393–396 (circled).
GII.17 VLPs
Capsid sequences were synthesized by Bio–Basic (Amherst, NY), and VLPs (Table 1) were expressed in baby hamster kidney cells from Venezuelan equine encephalitis virus replicons expressing GII.17 open reading frame 2 protein, as described previously [18]. Particle integrity was confirmed by visualization of approximately 40-nm particles, using electron microscopy. VLP binding to pig gastric mucin (PGM) type III (Sigma Aldrich, St. Louis, MO) and human type B saliva was detected by polyclonal antiserum to GII.17 strains (Cocalico Biologicals, Stevens, PA), as described elsewhere [19, 20]. Half-maximum binding was defined in comparison to a VLP concentration of 4 µg/mL.
Table 1.
Characteristics of GII.17 Strain Virus-Like Particles (VLPs) Used in This Study
| VLP | GenBank Accession No. | ORF2 Classificationa | EC50 PGM Binding, µg/mL, Mean (95% CI) |
|---|---|---|---|
| GII.17.1978 | KC597139 | Cluster I | 1.23 (1.15–1.34)b |
| GII.17.2002 | KJ196286 | Cluster I | 1.43 (1.29–1.58)b |
| GII.17.2005 | DQ438972 | Cluster II | 0.58 (0.51–0.66) |
| GII.17.2015 | KP698930 | Cluster IIIb | 0.60 (0.55–0.61) |
| GII.17.1978.393- 396 chimeric | … | … | >4b |
Abbreviations: CI, confidence interval; EC50, half-maximum effective concentration; ORF2, open reading frame 2; PGM, pig gastric mucin.
aBased on work by Dang Thanh et al [33].
bSignificantly different from GII.17.2015, by analysis of variance.
IgG Enzyme Immunoassay (EIA) and Blockade Ab
EIA and blockade Ab assays were performed at 37°C. VLPs were used at concentrations of 0.25 µg/mL for EIA and 0.5 µg/mL for blockade assays [19, 21]. Mean half-maximum effective concentrations (EC50 values) and 95% confidence intervals (CIs) were determined from dose-response sigmoidal curve fits, using GraphPad 6.02 [22, 23]. To be positive by EIA, a sample had to have a minimum OD450 of >3 times the background level after background subtraction. Samples determined to be below this limit by EIA (half-maximum binding was defined in comparison to a VLP concentration of 2 µg/mL) or that did not block at least 50% of VLP binding to PGM at the lowest serum dilution tested were assigned a titer equal to 0.5 times the limit of detection, for statistical analysis.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism 6.02 [22, 24]. EC50 values were log transformed and compared by the Dunn multiple comparisons test (performed after analysis of variance) [24]. A difference was considered significant if it had a P value of <.05.
RESULTS
Representative strains of GII.17 clusters I (1978, 2002), II (2005), and IIIb (2015) were selected for analysis of antigenic properties (Table 1). Amino acid sequence alignment of the strains suggested that each cluster likely has unique antigenic properties based on residue changes in known GII.4 blockade Ab epitopes (Figure 1). However, these residues have not been established to comprise Ab epitopes in any strains other than GII.4. In addition, vacuum electrostatics was used to predict the surface-exposed charge of each of the 3 clusters over time (Figure 2). Charge rearrangements occurred between each of the 3 clusters. Cluster IIIb exhibited a major charge change with extensive negative potential in an area that was not strongly positive or negative in previous clusters, indicating a potential antigenic change. This single motif, spanning positions 393–396 and comprising 4 aspartic acids, is unique to this cluster. Notably, this region corresponds to a known carbohydrate-binding site in GII.4 noroviruses [24, 25].
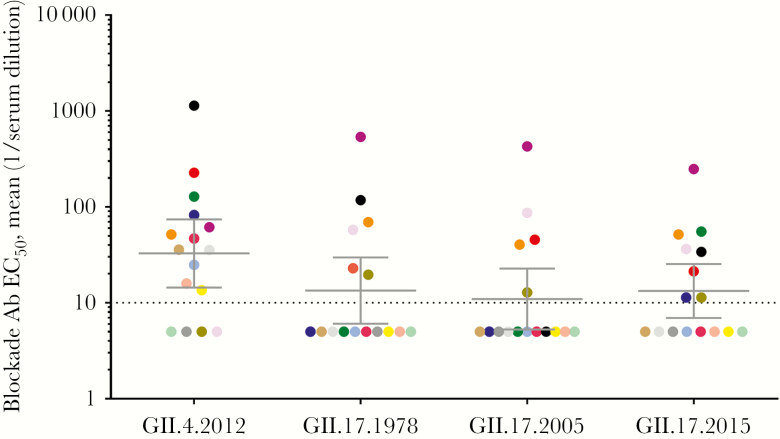
To test whether these charge changes correlate with antigenic changes, an Ab blockade of VLP binding to ligand assay was developed for the GII.17 VLPs. Each GII.17 VLP bound to PGM, although cluster I VLPs (1978 and 2002) required more VLPs to reach half-maximum binding than cluster II and IIIb VLPs (Table 1). First, human donor sera were measured for potential neutralization activity against each GII.17 cluster VLP and an additional VLP with wide population exposure, GII.4.2012 (Figure 3). Of 16 sera, 12 (75%) blocked GII.4.2012 (geometric mean titer [GMT], 32.8; 95% confidence interval [CI], 14.5–74.2), 6 (38%) blocked GII.17.1978 (GMT, 13.4; 95% CI, 6.1–29.7), 5 (31%) blocked GII.17.2005 (GMT, 11.0; 95% CI, 5.3–22.8), and 8 (50%) blocked GII.17.2015 (GMT, 13.3; 95% CI, 6.9–25.4). GMTs were similar between GII.17 strains and GII.4.2012, indicating preexposure history in this group of adults. GMTs were also similar between the GII.17 strains, indicating that GII.17 strains share common blockade Ab epitopes either with each other or with other prevalent GII strains or that the population has been exposed to the panel of strains over time.
Figure 3.
Adults have similar blockade antibody (Ab) titers to a time-ordered panel of GII.17 strains. Serum collected for donation from 16 adults (colored markers) living in the United States was evaluated for blockade Ab titer to GII.4.2012 and time-ordered GII.17 virus-like particles from clusters I (1978), II (2005), and IIIb (2015). Bars denote geometric mean titers, and whiskers denote 95% confidence intervals. EC50, half-maximum effective concentration. Dashed line equals the limit of detection.
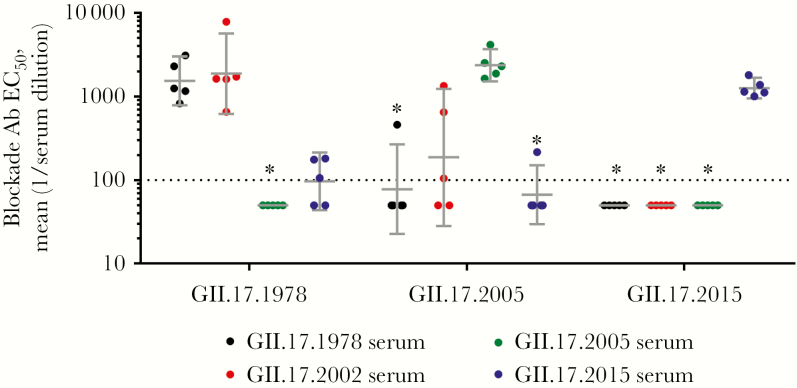
To determine whether the GII.17 strains share blockade epitopes, mice were immunized with either GII.17.1978, 2002, 2005, or 2015 VLPs, and serum reactivity was compared across the VLP panel by IgG EIA and blockade Ab. In support of human sera blockade Ab cross-reactivity findings, we observed that sera from mice immunized with a single GII.17 strain VLP reacted similarly across the GII.17 clusters, as revealed by IgG EIA (data not shown), indicating a degree of conserved intercluster Ab epitopes. To measure the potential for cross-strain protection as the virus evolves, we analyzed each serum sample for blockade activity across the 3 GII.17 clusters (Figure 4). GII.17.1978 VLP was blocked similarly by sera from 1978 (GMT, 1544; 95% CI, 788.9–3021), 2002 (GMT, 1886; 95% CI, 622.2–5718), and 2015 (GMT, 96.78; 95% CI, 43.73–214.2) and less well by GII.17.2005 sera (GMT, below the limit of detection). Although titer differences were not significant, 2015 sera GMTs were 16-fold less than 1978 GMTs. GII.17.2005 VLP was blocked similarly by homotypic (GMT, 2378; 95% CI, 1527–3701) and GII.17.2002 (GMT, 187.3; 95% CI, 28.33–1239) sera and less well by GII.17.1978 (GMT, 77.97; 95% CI, 22.71–267.7) and 2015 (GMT, 67.02; 95% CI, 29.71–151.1) sera, supporting the trend of reduced cross-cluster serum reactivity [26]. Most notably, only sera from mice immunized with GII.17.2015 (GMT, 1265; 95% CI, 951.1–1684) were able to block GII.17.2015 VLP binding to PGM (Figure 4). None of the serum samples from mice immunized with cluster I or II VLPs blocked the cluster IIIb VLP binding to ligand. These data demonstrate progressive antigenic change culminating in the cluster IIIb GII.17 strains, which are antigenically distinct from clusters I and II strains that circulated prior to cluster IIIb emergence.
Figure 4.
GII.17.2015 is antigenically distinct from earlier GII.17 strains at blockade antibody (Ab) epitopes. Balb/c mice were immunized intramuscularly with 0.2 µg of a single GII.17 virus-like particle (VLP) adjuvanted with monophosphoryl lipid A and alhydrogel and sera were evaluated for blockade Ab titer to VLPs from clusters I (1978), II (2005), and IIIb (2015). Bars denote geometric mean titers, and whiskers denote 95% confidence intervals. Dashed line equals the limit of detection. *Significantly different from homotypic serum (by analysis of variance). EC50, half-maximum inhibitory concentration.
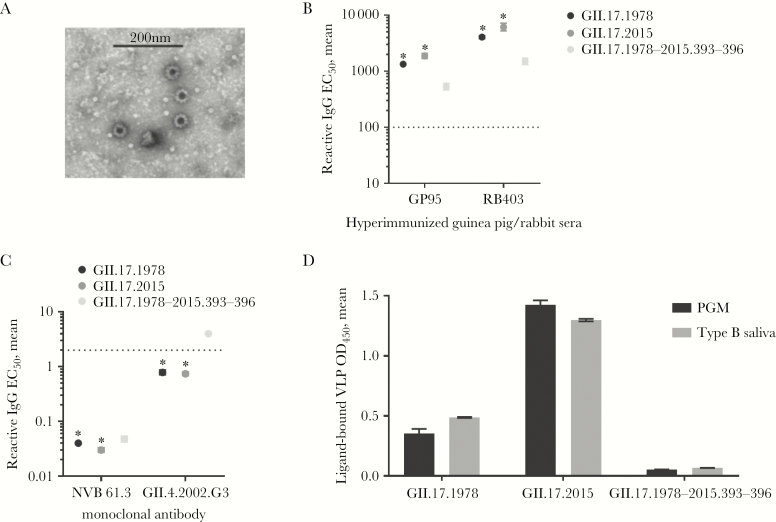
Cross-reactivity of blockade Ab responses between clusters agreed with electrostatic potential comparisons, indicating more antigenic similarity between clusters I and II and less similarity with cluster IIIb. Next, we tested whether residues 393–396 of GII.17.2015 (Figure 2) may be contributing to the antigenic divergence of GII.17.2015, by developing a chimeric VLP composed of the GII.17.1978 sequence with the corresponding residues replaced with the 4 aspartic acid residues found at positions 393–396 of GII.17.2015. The chimeric VLP, GII.17.1978–2015.393-396, formed approximately 40-nm particles, according to negative-stain electron microscopy (Figure 5A). Polyclonal sera from an immunized rabbit and guinea pig had strong but reduced reactivity to the chimeric particles (Figure 5B). GII.17-type–specific monoclonal antibodies are necessary to distinguish the exact epitope variation encoded within the time-ordered GII.17 strain VLP panel. As these antibodies are not available, we compared reactivity patterns between GII.17.1978, 2015, and the chimeric VLP with monoclonal antibodies known to recognize conserved genogroup II epitopes. Human monoclonal Ab NVB 61.3 recognizes a conformation dependent epitope [24] and reacted strongly with each GII.17 VLP tested but had reduced reactivity to the GII.17.1978–2015.393-396 chimeric VLP. Mouse monoclonal Ab GII.4.2002.G3 [27] reacted less potently with GII.17.1978 and 2015 and lost reactivity with GII.17.1978–2015.393-396 (Figure 5C). Both polyclonal and monoclonal antibodies differentiated the chimeric VLP from the parental VLPs. Exchange of 393–396 had a robust effect on VLP binding to ligand. The chimeric VLP had reduced binding to both PGM and human type B saliva as compared to the parental VLPs. At the titer used for Ab blockade assays, GII.17.1978–2015.393-396 chimeric VLP binding was not detected above background binding for PGM or type B saliva (Figure 5D). These data suggest that the exchange of the 4 aspartic acid residues of GII.17.2015 with the corresponding residues in GII.17.1978 either disrupted residues that affect both antigenicity and ligand binding of GII.17 strains or compromised particle integrity. Visual confirmation of particle size by microscopy and reactivity to conformation-dependent human monoclonal Ab NVB 61.3 indicate that the chimeric VLP was intact and that residues 393–396 regulated blockade Ab and ligand binding properties.
Figure 5.
Exchange of residues 393–396 from GII.17.2015 to GII.17.1978 decreases antibody reactivity and virus-like particle (VLP) binding to ligand while maintaining particle structure. A–C, Chimeric VLPs maintain particle integrity, as visualized by electron microscopy (A) but reduced reactivity to polyclonal sera from hyperimmunized guinea pig or rabbit (B) and monoclonal antibodies to conserved genogroup II epitopes (C). D, Ligand binding to both pig gastric mucin (PGM) and human type B saliva also was reduced as compared to parental VLPs. Dashed line equals the limit of detection. *Significantly different from the chimeric VLP (by analysis of variance). EC50, half-maximum effective concentration; IgG, immunoglobulin G.
DISCUSSION
Genome sequencing and analyses are powerful tools for global surveillance of RNA viruses, including influenza virus and human norovirus, which evolve into new serotypes and effectively abrogate protective herd immunity. Although predictive tools continue to improve, the translation of genome sequences into antigenicity characteristics will continue to be largely an empirical exercise as features of Ab and T-cell epitopes and the role these epitopes play in protection from infection are further developed. The observed pattern of epochal evolution in the capsid gene of the GII.17 strains has led to speculation that the genotype has the same potential for pandemic spread, as has been documented for the GII.4 norovirus strains [14, 25]. However, none of the identified changing residues have been established to be Ab epitopes in GII.17 or any other norovirus strain except GII.4 [24, 28]; thus, immunological assays are required for determining the effects of sequence change on antigenicity. This study is the first to empirically test the antigenic relationship between GII.17 strain VLPs and provide direct evidence that epochal evolution of the major capsid gene, likely driven by herd immunity, results in antigenic drift and novel emergent GII.17 strains.
To do this, we developed a panel of VLPs presenting GII.17 clusters I, II, and IIIb to evaluate whether the genetic clusters translate into antigenic clusters. Human sera blockade Ab responses were similar across the panel of GII.17 VLPs, indicating that either the VLPs share blockade epitopes or strain-specific Ab responses have formed after repeat exposures to genogroup II strains. Alternatively, donors may have experienced a recent GII.17 infection resulting in short-term cross-blockade Ab responses, as has been characterized following norovirus vaccination [22]. Responses in single-strain–immunized mice demonstrated that limited cross-cluster blockade Ab is generated between clusters after a single GII.17 exposure, indicating that the cross-cluster blockade Ab response in human sera is more likely the result of multiple exposures, not conserved blockade Ab epitopes. However, these studies cannot differentiate whether participants have been exposed to GII.17 strains from all 3 clusters or whether exposure to multiple GII.17 strains or other GII strains results in blockade Ab to epitopes that are conserved between strains, as has been demonstrated following GII.4 norovirus and influenza vaccination [22, 29]. Development of either of these mechanisms would be unlikely in young children, leaving them susceptible to infection with an emergent GII.17 strain.
Ligand binding and polymerase activity differences may have contributed to the epidemiologic characteristics of GII.17.2015 [7, 12–14]. At this time, it is not possible to determine the effect of polymerase changes on viral fitness or antigenicity, but preliminary neutralization assays using a novel virus cultivation system support a correlation between blockade Ab titer and virus neutralization titer. Serum that blocked VLP-ligand binding also neutralized virus [30]. The observation that GII.17.2015 VLP binding to PGM was uniquely resistant to serum Ab blockade from mice immunized with VLP from other GII.17 clusters indicates that antigenic divergence at key residues likely played a role in cluster IIIb strain emergence. Although blockade Ab levels were reduced, cluster I and II strains remained susceptible to cross-cluster Ab blockade, agreeing with modeling data indicating that the GII.17 strains remained relatively static over time until the cluster III strains emerged [10]. Similar to GII.4 norovirus VLPs, cross-reactive blockade Ab is more potent for previously circulating strains than for future strains, compared with the immunizing VLP [26]. Bioinformatics analyses identified the most prominent changes on the surface of the cluster IIIb P2 subdomain to be a 4–aspartic acid motif that added a negative patch on the surface at positions 393–396. Exchange of these residues with corresponding residues in GII.17.1978 reduced Ab reactivity and PGM binding, suggesting that the observed charge changes disrupted functional domains. The primary limitation of this study is the lack of GII.17-specific monoclonal antibodies that could definitively identify epitopes that are strain specific, as has been demonstrated for GII.4 norovirus [24]. The identified epitope likely involves additional residues outside of 393–396.
Changes in residues 393–396 also negatively affected ligand binding, supporting a role for the ligand affinity and antigenicity of these amino acids, as has been described for residues 393–395 (epitope D) of GII.4 strains [24, 25]. Epitope D is both a blockade Ab epitope and a regulator of ligand binding. Although not part of the carbohydrate-binding pocket, epitope D residues modulate affinity for different histo-blood group antigens [25, 31, 32]. Bioinformatics studies have suggested that the carbohydrate binding patterns may have changed between the early GII.17 strains and the cluster IIIb strains [13, 14]. This study did not characterize the carbohydrate binding profiles of the different cluster VLPs by specific carbohydrates, but each cluster VLP retained binding to PGM, our carbohydrate ligand source for comparing antigenicity. Cluster I strains had less avidity for PGM than cluster II and IIIb VLPs, supporting previous observations with cluster I P particles [14]. It is unknown whether this difference could impact infection rates between GII.17 strains.
The emergence of GII.17 strains emphasizes the need for enhanced surveillance systems that include, at a minimum, complete P2 sequences and, ideally, the entire capsid and polymerase regions for virus typing. Only continued surveillance will answer the most important questions concerning GII.17 strains: will GII.17 replace GII.4 as the leading cause of acute viral gastroenteritis? Will GII.17 cocirculate with GII.4, increasing the global burden of norovirus disease or will GII.17 strains return to endemic levels of infection? Will the GII.4 continue to evolve by epochal evolution in human populations? From 3 decades of GII.4 pandemics, we have learned that viruses with broad ligand binding patterns that undergo epochal evolution in response to herd immunity are a potential public health threat. As such, GII.17 should be considered as a component of a norovirus multivalent VLP vaccine or as a target of a cross-reactive norovirus vaccine.
Notes
Acknowledgments. We thank Victoria Madden of Microscopy Services Laboratory, Department of Pathology and Laboratory Medicine, University of North Carolina–Chapel Hill, for expert technical support.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (grants R56 A15-0756 and U19 AI109761 CETR); and the NIH (grant P30 AI50410 to the University of North Carolina–Chapel Hill Center for AIDS Research) and the Wellcome trust (A17-0915-001).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: 6th International Calicivirus Meeting, 10–13 October 2016, Savannah, Georgia.
References
- 1. Riddle MS, Walker RI. Status of vaccine research and development for norovirus. Vaccine 2016; 34:2895–9. [DOI] [PubMed] [Google Scholar]
- 2. Pringle K, Lopman B, Vega E, Vinje J, Parashar UD, Hall AJ. Noroviruses: epidemiology, immunity and prospects for prevention. Future Microbiol 2015; 10:53–67. [DOI] [PubMed] [Google Scholar]
- 3. Hall AJ, Glass RI, Parashar UD. New insights into the global burden of noroviruses and opportunities for prevention. Expert Rev Vaccines 2016; 15:949–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bartsch SM, Lopman BA, Hall AJ, Parashar UD, Lee BY. The potential economic value of a human norovirus vaccine for the United States. Vaccine 2012; 30:7097–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bartsch SM, Lopman BA, Ozawa S, Hall AJ, Lee BY. Global economic burden of norovirus gastroenteritis. PLoS One 2016; 11:e0151219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. CDC. Updated norovirus outbreak management and disease prevention guidelines. MMWR Recomm Rep 2011; 60:1–18. [PubMed] [Google Scholar]
- 7. de Graaf M, van Beek J, Vennema H et al. Emergence of a novel GII.17 norovirus—end of the GII.4 era? Euro Surveill 2015; 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lu J, Fang L, Zheng H et al. The evolution and transmission of epidemic GII.17 noroviruses. J Infect Dis 2016; 214:556–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang XF, Huang Q, Long Y et al. An outbreak caused by GII.17 norovirus with a wide spectrum of HBGA-associated susceptibility. Sci Rep 2015; 5:17687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Parra GI, Squires RB, Karangwa CK et al. Static and evolving norovirus genotypes: implications for epidemiology and immunity. PLoS Pathog 2017; 13:e1006136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang HB, Wang Q, Zhao JH et al. Complete nucleotide sequence analysis of the norovirus GII.17: A newly emerging and dominant variant in China, 2015. Infect Genet Evol 2016; 38:47–53. [DOI] [PubMed] [Google Scholar]
- 12. Chen H, Qian F, Xu J et al. A novel norovirus GII.17 lineage contributed to adult gastroenteritis in Shanghai, China, during the winter of 2014–2015. Emerg Microbes Infect 2015; 4:e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Singh BK, Koromyslova A, Hefele L, Gürth C, Hansman GS. Structural evolution of the emerging 2014-2015 GII.17 noroviruses. J Virol 2015; 90:2710–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jin M, Zhou YK, Xie HP et al. Characterization of the new GII.17 norovirus variant that emerged recently as the predominant strain in China. J Gen Virol 2016; 97:2620–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Treanor JJ, Atmar RL, Frey SE et al. A novel intramuscular bivalent norovirus virus-like particle vaccine candidate–reactogenicity, safety, and immunogenicity in a phase 1 trial in healthy adults. J Infect Dis 2014; 210:1763–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Koromyslova A, Tripathi S, Morozov V, Schroten H, Hansman GS. Human norovirus inhibition by a human milk oligosaccharide. Virology 2017; 508:81–9. [DOI] [PubMed] [Google Scholar]
- 17. Larkin MA, Blackshields G, Brown NP et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007; 23:2947–8. [DOI] [PubMed] [Google Scholar]
- 18. Debbink K, Costantini V, Swanstrom J et al. Human norovirus detection and production, quantification, and storage of virus-like particles. Curr Prot in Micro 2013; 31:15K 1 1–K 1 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lindesmith LC, Donaldson EF, Beltramello M et al. Particle conformation regulates antibody access to a conserved GII.4 norovirus blockade epitope. J Virol 2014; 88:8826–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lindesmith L, Moe C, Marionneau S et al. Human susceptibility and resistance to Norwalk virus infection. Nat Med 2003; 9:548–53. [DOI] [PubMed] [Google Scholar]
- 21. Lindesmith LC, Beltramello M, Swanstrom J et al. Serum immunoglobulin a cross-strain blockade of human noroviruses. Open Forum Infect Dis 2015; 2:ofv084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lindesmith LC, Ferris MT, Mullan CW et al. Broad blockade antibody responses in human volunteers after immunization with a multivalent norovirus VLP candidate vaccine: immunological analyses from a phase I clinical trial. PLoS Med 2015; 12:e1001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lindesmith LC, Mallory ML, Jones TA et al. Impact of pre-exposure history and host genetics on antibody avidity following norovirus vaccination. J Infect Dis 2017; 215:984–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lindesmith LC, Beltramello M, Donaldson EF et al. Immunogenetic mechanisms driving norovirus GII.4 antigenic variation. PLoS Pathog 2012; 8:e1002705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lindesmith LC, Donaldson EF, Lobue AD et al. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med 2008; 5:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lindesmith LC, Donaldson EF, Baric RS. Norovirus GII.4 strain antigenic variation. J Virol 2011; 85:231–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lindesmith LC, Debbink K, Swanstrom J et al. Monoclonal antibody-based antigenic mapping of norovirus GII.4-2002. J Virol 2012; 86:873–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Debbink K, Donaldson EF, Lindesmith LC, Baric RS. Genetic mapping of a highly variable norovirus GII.4 blockade epitope: potential role in escape from human herd immunity. J Virol 2012; 86:1214–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li GM, Chiu C, Wrammert J et al. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc Natl Acad Sci U S A 2012; 109:9047–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ettayebi K, Crawford SE, Murakami K et al. Replication of human noroviruses in stem cell-derived human enteroids. Science 2016; 353:1387–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. de Rougemont A, Ruvoen-Clouet N, Simon B et al. Qualitative and quantitative analysis of the binding of GII.4 norovirus variants onto human blood group antigens. J Virol 2011; 85:4057–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shanker S, Choi JM, Sankaran B, Atmar RL, Estes MK, Prasad BV. Structural analysis of histo-blood group antigen binding specificity in a norovirus GII.4 epidemic variant: implications for epochal evolution. J Virol 2011; 85:8635–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dang Thanh H, Than VT, Nguyen TH, Lim I, Kim W. Emergence of norovirus GII.17 variants among children with acute gastroenteritis in South Korea. PLoS One 2016; 11:e0154284. [DOI] [PMC free article] [PubMed] [Google Scholar]