Systems analysis of freeze-quenched mesophyll and bundle sheath tissues enriched under liquid nitrogen provides a reliable alternative to previously used separation techniques that showed bias in RNA quality.
Keywords: C4, cell separation, maize, meta-analysis, transcriptomics
Abstract
The high efficiency of C4 photosynthesis relies on spatial division of labor, classically with initial carbon fixation in the mesophyll and carbon reduction in the bundle sheath. By employing grinding and serial filtration over liquid nitrogen, we enriched C4 tissues along a developing leaf gradient. This method treats both C4 tissues in an integrity-preserving and consistent manner, while allowing complementary measurements of metabolite abundance and enzyme activity, thus providing a comprehensive data set. Meta-analysis of this and the previous studies highlights the strengths and weaknesses of different C4 tissue separation techniques. While the method reported here achieves the least enrichment, it is the only one that shows neither strong 3′ (degradation) bias, nor different severity of 3′ bias between samples. The meta-analysis highlighted previously unappreciated observations, such as an accumulation of evidence that aspartate aminotransferase is more mesophyll specific than expected from the current NADP-ME C4 cycle model, and a shift in enrichment of protein synthesis genes from bundle sheath to mesophyll during development. The full comparative dataset is available for download, and a web visualization tool (available at http://www.plant-biochemistry.hhu.de/resources.html) facilitates comparison of the the Z. mays bundle sheath and mesophyll studies, their consistencies and their conflicts.
Introduction
Specialization and coordination between two cell types improves photosynthetic efficiency in most C4 photosynthetic plants. Specifically, most C4 plants shuttle carbon from a surrounding mesophyll (M) tissue into a surrounded bundle sheath (BS) tissue (Hatch, 1987). The shuttling concentrates CO2 around the carbon fixing enzyme, Rubisco, thereby suppressing photorespiration and increasing photosynthetic efficiency. This lends selective advantage to C4 plants in photorespiration-inducing (e.g. hot and arid) environments (Schulze et al., 1996). The high photosynthetic efficiency and stress tolerance of C4 species has led to interest in engineering the trait. However, the complexity of the trait—with many changes to anatomy and metabolism beyond the core biochemical pump—makes this an ambitious goal, which will require a full systems-level understanding of both the mature C4 trait and its development to be achieved (Sage and Zhu, 2011).
BS and M cells show extensive specialization in metabolism and anatomy in C4 plants. In the classic C4 arrangement—Kranz anatomy—enlarged BS cells form a ring around the vascular bundle and are in turn surrounded by M cells (Hatch, 1987). Narrow vein spacing means each M cell borders a BS cell, allowing direct transfer of metabolites between them. Compared with a C3 leaf, there is a massive increase in the relative amount of BS tissue, allowing for a division of labor between cell types that includes both photosynthesis and major facets of other metabolism (Majeran et al., 2010; Friso et al., 2010). Following Rubisco, most enzymes in the Calvin–Benson–Bassham Cycle (CBBC) and the linked photorespiratory cycle are restricted to the BS (Broglie et al., 1984; Rawsthorne et al., 1988; Döring et al., 2016). In Z. mays, distribution of photosystem II and therefore linear electron transport and reducing equivalent regeneration are restricted to the M, while the BS relies on ATP from cyclic electron transport around photosystem I and biochemical shuttles that transfer reducing equivalents to the BS for energy (Romanowska et al., 2008; Wang et al., 2014; Bellasio and Griffiths, 2014). Subsets of metabolism are divided up between the two cell types with, for instance, amino acid, nucleotide, and isoprenoid synthesis in the M, and sulfur metabolism and starch synthesis in the BS (Majeran et al., 2005; Friso et al., 2010).
Information on anatomical and metabolic changes has been gained through comparative proteomic and transcriptomic studies both between C3 and C4 species (e.g. Bräutigam et al., 2011, 2014; Gowik et al., 2011; Wang et al., 2014; Covshoff et al., 2016), and between isolated tissue types (Majeran et al., 2005; Friso et al., 2010; Li et al., 2010; Chang et al., 2012; Tausta et al., 2014; John et al., 2014; Aubry et al., 2014). Many of the differences between cell types are set up early in development, and tissue maturation studies have obtained mechanistic insights. For instance, comparison of C4 and C3 Cleomaceae species linked delayed photosynthetic differentiation to extended vein proliferation and ultimately closer vein spacing in the C4 species (Külahoglu et al., 2014). In Z. mays carefully comparing the primordia of Kranz leaf tissue with non-Kranz husk tissue implicated the recruitment of the ScareCrow regulatory module from the root epidermis to BS cells (Wang et al., 2013). Potentially due to the difficulties of isolating cell types, to date there has only been one transcriptomics (Tausta et al., 2014) and one proteomics (Majeran et al., 2010) study that have looked at immature M and BS tissue. These studies have shown the early establishment of tissue specificity of major C4 enzymes and the roles of M and BS cells in sink vs source tissue to logically reflect the broader changes between source and sink tissue. As neither of the above studies could look at metabolites, and interstudy comparisons have produced distinct results on cell specificity—particularly of transcription factors (Tausta et al., 2014)—we judged further analysis to be warranted.
Here we successfully perform an ‘omics’-scale analysis on developmental tissue separated by a method developed by Stitt and Heldt (1985), and thus simultaneously capture changes in the transcriptome, enzymatic activities, and the metabolome. A subsequent meta-analysis of this and other BS and M separation studies highlights the strengths and weaknesses of each of the various separation methods, and the advantages of using complementary techniques. The comparative dataset has been made available for visual exploration or download, and can assist both in experimental design both for BS/M related studies and for studies in the broader category of tissue separation.
Materials and methods
Plant genome data
Genome and gene-model data was downloaded for Setaria viridis (v1.1/v311; Bennetzen et al., 2012) and Panicum virgatum (v1.1/v273; DOE-JGI, 2016) from Phytozome 11.0 (Goodstein et al., 2012). The AGPv3.22 release of the Zea mays genome with the 5b+ filtered gene set was obtained from ensemble plants (Kersey et al., 2016) and Gramene (Tello-Ruiz et al., 2016), respectively. Orthologs were identified by best BLAST (Altschul et al., 1997) hit from Z. mays to S. viridis or P. virgatum.
External RNAseq data
Complementary RNAseq data were downloaded from the sequence read archives (Kodama et al., 2012) and European nucleotide archives (Leinonen et al., 2010). We included two additional Z. mays BS and M separation studies (Chang et al., 2012: SRP009063; Tausta et al., 2014: SRP035577); corresponding whole developmental leaf sections (Li et al., 2010; SRP002265); Z. mays tissue atlas (Sekhon et al., 2013; SRP010680); and primordial leaf and husk tissue (Wang et al., 2013; SRP028231). The non-Z. mays studies were separation of BS and M cells in S. viridis (John et al., 2014; ERA275647) and P. virgatum (Rao et al., 2016; SRP062667).
Note that as the original authors included the same precise set of sequences for BS and M tissues in section 14 (Li et al., 2010; Tausta et al., 2014), and reported the same plant growth conditions, we’ve considered these studies broadly comparable. However, to avoid redundancy, the BS and M samples for section 14 are only included with Tausta et al. (2014).
Plant growth conditions and harvest
Z. mays B73 was grown in the summer of 2012 under conditions previously described (Pick et al., 2011). The third leaf was harvested when it measured 18 cm from the second ligule to the leaf tip. Two different harvesting methods were performed. In the first, a leaf gradient with five sequential developmental slices (4 cm each) was harvested with the ‘leaf guillotine’ (see Fig. S1A available at Dryad Digital Repository http://dx.doi.org/10.5061/dryad.tf6q6;Pick et al., 2011). This method required 10 s to extract the third leaf and properly align it, which does not allow for reliable estimates of the high-turnover photosynthetic metabolite distributions. Therefore, a second harvesting method was performed, in which the plants were positioned above two liquid nitrogen containers and two 8 cm slices were cut with connected scissors (see Fig. S1A, B at Dryad) achieving a delay of less than 1 s between slicing and flash-freezing. Metabolite abundance and enzyme activity were measured from both harvest sets; the full five-slice gradient was used for RNAseq.
Tissue enrichment
M and BS tissues were enriched using a method modified from Stitt and Heldt (1985). Ground material was filtered through 250, 80, and 41 μm meshes on liquid nitrogen. Three fractions were selected for further analysis. The ‘BS-e’ fraction showed the most enrichment of BS tissue (it did not pass through 80 μm mesh); the ‘M-e’ fraction showed most enrichment in M tissue (it passed through 41 μm mesh); and the ‘I-e’ fraction showed intermediate, but consistent, proportions of tissues (it did not pass through 41 μm mesh).
Extraction and abundance measurements metabolites and enzymes
Enzymes were extracted and desalted as described in Bräutigam et al. (2014), and the enzyme activity was measured through colorimetric assays as described in Hatch and Mau (1977) and Walker et al. (1995). Metabolites were extracted and quantified via gas chromatography–electron-impact time-of-flight mass spectrometry as described in Rudolf et al. (2013). Both low-signal metabolites and individual replicates with a percentage abundance in BS more than 3 standard deviations from the mean were excluded. The integrated peaks were divided by the area of the ribitol (internal standard) peak and the fresh weight, and to further reduce noise and compensate for FW/DW differences between the cell types by the mean abundance for the replicate. Therefore, normalized differences between metabolites represent not absolute distribution, but distribution relative to the other metabolites, particularly sucrose and the other highly abundant metabolites.
Sequencing and estimating transcriptional abundances
RNA was extracted with QIAGEN RNeasy Plant kits, according to the manufacturer’s instructions except for an extra wash step in 80% ethanol after the standard wash steps. Libraries were prepped from RNA with an RNA integrity number >8 and sequenced with the Illumina HiSeq 2000 platform. The quality was checked with FastQC (Andrews, 2010). Quality and adapter trimming was performed with Trimmomatic (Bolger et al., 2014). Trimmed reads were mapped to their respective genomes with Tophat2 (Kim et al., 2013) and the unique counts per locus were quantified with HTSeq (Anders et al., 2015); transcripts per million (TPM) was calculated from the unique counts and gene length. Coverage metrics including 3′ bias were calculated with PicardTools 2.4.1: CollectRnaSeqMetrics (Wysoker et al., 2012). Non-default parameters used for bioinformatics programs are provided (see Table S1 at Dryad). The same pipeline was used for all studies except as necessitated by experimental differences (e.g. paired vs single end reads), or otherwise noted.
Differential expression and tissue specificity normalization
Differential expression P-values and log2 fold changes were calculated with EdgeR (Robinson et al., 2009). Where no replicates were available (Chang et al., 2012), the mean common dispersion from the remaining studies was used. Additionally, due to the low level of enrichment achieved in this study, ContamDE (Shen et al., 2016), a cross-contamination tolerant package for RNAseq statistics, was employed for the data generated here. As necessary for interstudy comparisons in Z. mays, log2 fold changes from edgeR (Chang et al., 2012; Tausta et al., 2014) and ContamDE (this study) were quantile normalized, and the fully normalized TPM back calculated from the quantile normalized log2 fold change and mean TPM.
Estimation of initial tissue specificity by ‘deconvolution’
The distribution of metabolites and enzyme activities was compared with the distribution of markers to estimate the original tissue specificity in a method modified from Stitt and Heldt (1985). First, all data were converted into fraction of total by developmental slice. Second, marker enzyme activities were used as proxies for the amount of M (phosphoenolpyruvate carboxylase (PEPC) activity) and BS (NADP-malic enzyme (ME) activity) tissue in each enrichment fraction. The slope of a regression line between the ln(target/M) against ln(BS/M) estimated the fraction of the target found in pure BS (see Fig. S1C at Dryad). P-values were calculated with a null hypothesis of slope=0.5 (50% M, 50% BS). This was automated with a linear regression in R and calculated for every metabolite and non-marker enzyme. To estimate the ‘pure’ abundance values, the estimated fraction in BS and M (1–fraction BS) were multiplied by 2× the average abundance value for the developmental slice.
Functional category enrichment testing
Functional categories were assigned with Mercator (Lohse et al., 2014). Enrichment was tested with Fisher’s exact test, and the false discovery rate calculated according to (Benjamini and Yekutieli, 2001).
Statistics
Unless otherwise noted, all statistical analysis was performed in the R statistical environment (R Development Core Team, 2011) and whenever a test was performed more than 20 times, the false discovery rate (Benjamini and Hochberg, 1995) was calculated from the resulting P-values.
Accession numbers
The reads related to this article have been deposited in the Sequence Read Archives under the accession number SRP052802.
Results
Validation of separation method
Here, we enriched BS and M cells along a developing Z. mays leaf by grinding and serial filtration (Stitt and Heldt, 1985). Two harvesting methods were used, the first using a ‘guillotine’ (Pick et al., 2011) to sample five contiguous 4 cm slices from tissue just emerging from the ligule (slice 5) to the leaf tip (slice 1). In the second, targeted at capturing unadulterated metabolite levels, two 8 cm slices were harvested in full illumination and quenched in liquid nitrogen within a second of cutting. M and BS tissues were enriched using a method modified from Stitt and Heldt (1985) that capitalizes on the distinct physical properties of M and BS cells to enrich them in different separation fractions as ground tissue is filtered through serially smaller meshes over liquid nitrogen. The activity of C4 enzymes and the metabolite levels were measured from both harvests, and RNAseq was performed on material from the five-slice gradient.
The distribution of tissue specific markers indicated BS and M tissue were successfully enriched (see Fig. S1D and Dataset S1 at Dryad). The classic BS marker is NADP-ME, the enzyme responsible for releasing the carbon from C4 acids in the BS. NADP-ME activity and transcripts were both higher in the coarsest (from here on, BS-e for bundle sheath enriched) separation fraction; in between in the middle (from here on, I-e for intermediate enrichment) separation fraction; and lowest in the finest (from here on, M-e, for mesophyll enriched) fraction (see ‘Materials and methods’ for details). The classic M marker, PEPC, the C4 fixing enzyme, showed the opposite pattern, with highest activity and transcript abundance in the fine, M-e fraction. While the enrichment was strongest in mature tissue, it was also apparent in the youngest tissue (slice 5). For non-marker enzymes and metabolites, the original distribution was estimated based on the marker enzymes (see ‘Materials and methods’; Fig S1C at Dryad).
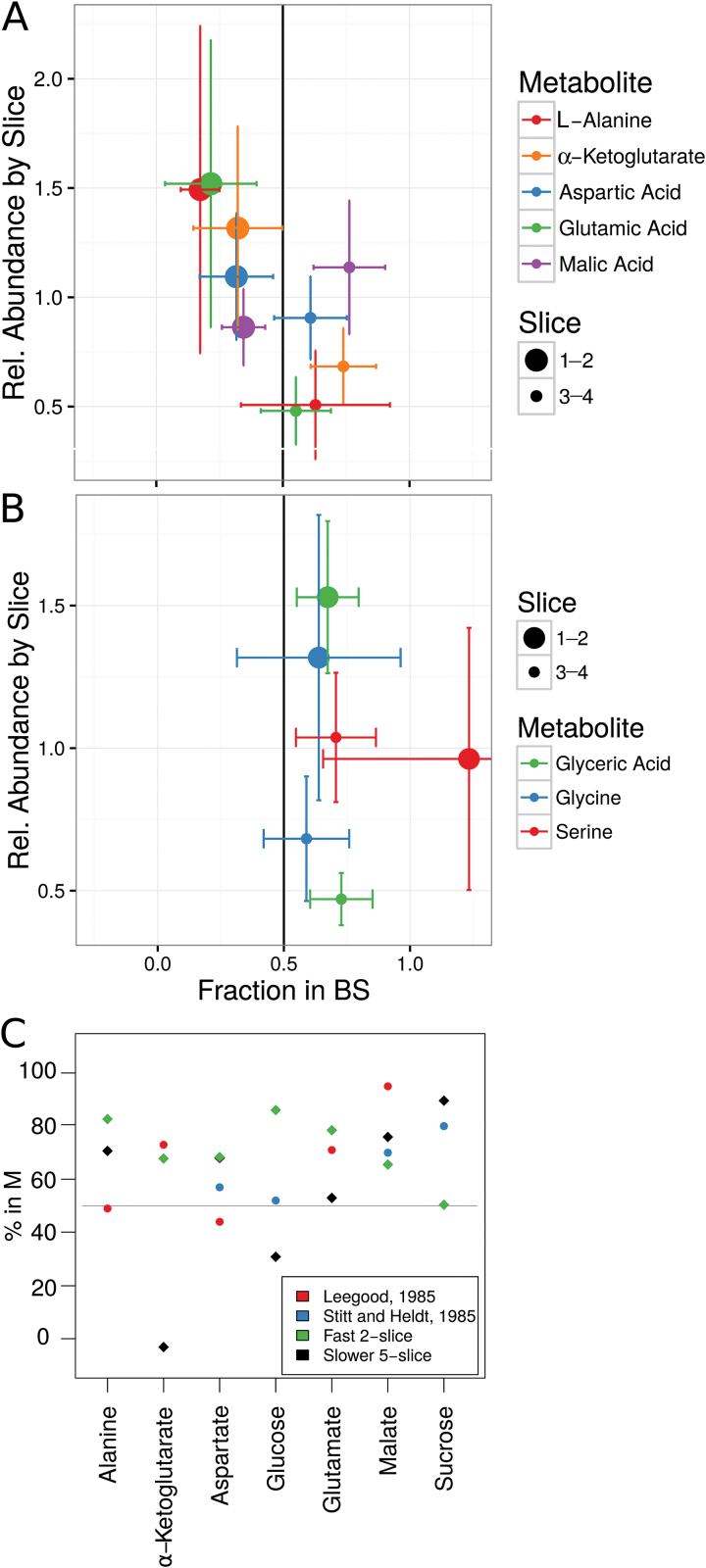
This enrichment method was chosen over other separation methods both for sample integrity and to obtain data on metabolite abundance, enzyme activity, and transcript abundance from the same material. However, in the rapid harvest (with less than 1 s between cutting and quenching in liquid nitrogen), very few significant differences were found between metabolite levels in M and BS ((iso)-citric acid and malonic acid were both enriched in BS slice 3–4; FDR<0.05; Fig. S2C at Dryad). In contrast, many metabolites showed significant differences based on leaf age (10 metabolites with FDR<0.05 between slice 3–4 and slice 1–2 in the sub-1 s harvest, and 20 with FDR<0.05 between at least one of the neighboring slices in the 10 s harvest; Fig. S2E and Dataset S1 at Dryad). The observed developmental changes were very similar between the sub-1 s and 10 s harvest; however, there were a few exceptions. One example is phenylalanine, which increased in abundance with leaf age in the fast harvest, but decreased in the slow harvest (Fig. S2C, D at Dryad). Although not statistically significant, the BS vs M trend of several metabolites corresponded with expectations. Notably, serine and the other photorespiratory metabolites were higher in the BS, where they are expected to be produced, both in the faster (Fig. 1B) and, to a lesser extent, also in the slower (Fig.S2B at Dryad) harvest. Malate, which presumably moves from M to BS entirely based on a diffusion gradient, tended towards enrichment in the mature M (slices 1–2, 1–3; Fig. 1A and Fig. S2A at Dryad). Further, there is a modest consistency between previous studies measuring distribution of metabolites and that measured here (Fig. 1C). All measured core C4 metabolites shift from putative BS towards putative M enrichment between slice 3–4 and 1–2 (Fig. 1A). Such synchronized changes could relate to increasing flux (or changing rate-limiting steps) in the C4 cycle. The differences between harvest speeds highlights how labile these metabolites can be, and discrepancies between studies or low enrichment values may simply reflect response to conditions and the readiness with which they pass the plasmodesmata, respectively. Higher confidence in metabolite distribution will require more replicates, and, potentially, more defined conditions.
Fig. 1.
Metabolites. (A, B) The estimated tissue enrichment and abundance of measurable metabolites associated with the photorespiratory cycle (A) and the C4 cycle (B). Error bars indicate standard error. (C) Comparison of metabolite tissue enrichment measured by Leegood (1985) and Stitt and Heldt (1985) with the average of slice 1 and 2 in the slower five-slice harvest and slice 1–2 in the faster two-slice harvest.
Comparison with other separated transcriptomes
Quantitative study comparison
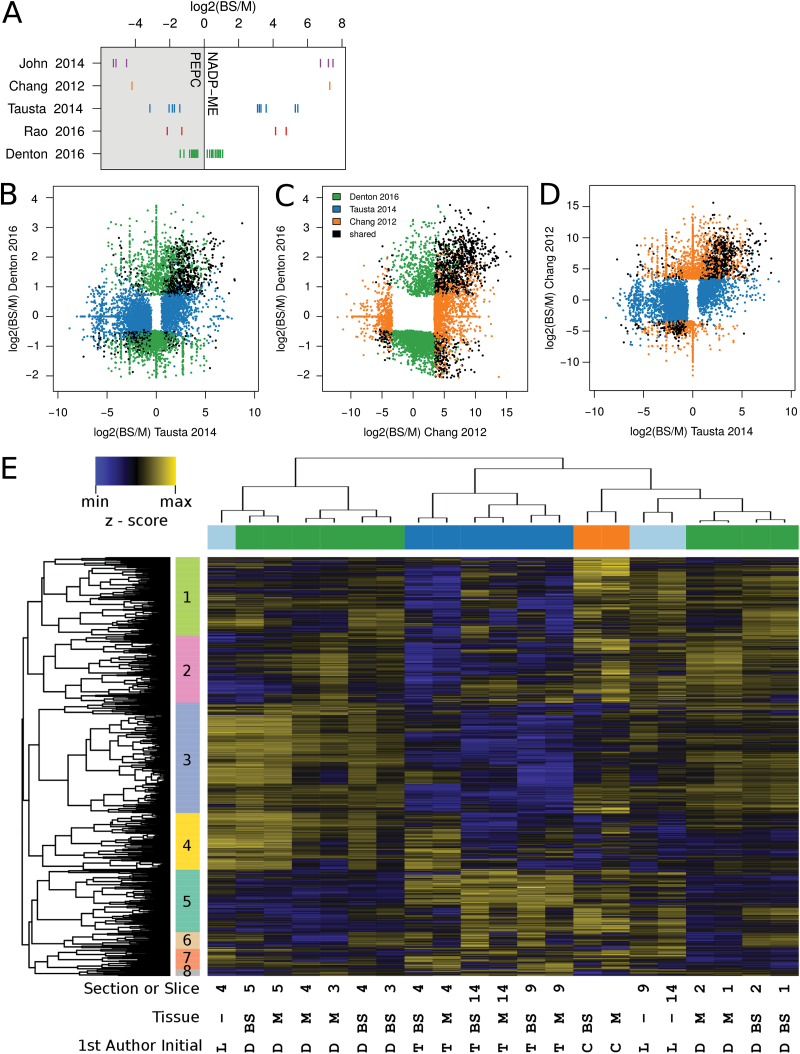
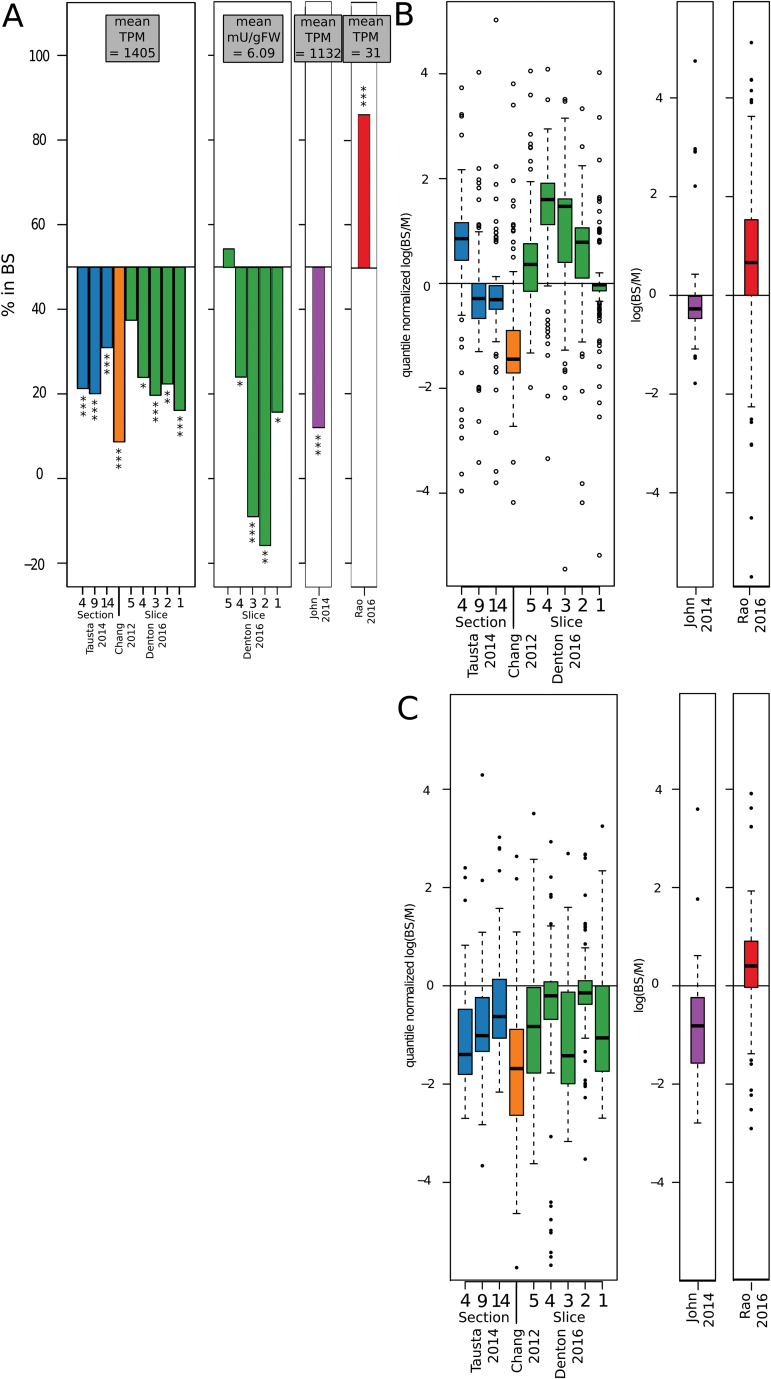
While this separation method provides high integrity and allowed us to simultaneously measure transcripts, metabolites, and enzyme activities, it comes with its own caveats due to the limited enrichment. As separation studies will likely continue, either in new species or with variations such as separating the husk (Huang and Brutnell, 2016), we evaluated the advantages and disadvantages of different separation methods and their effect on biological results. We compiled a comparative dataset from all existing M/BS specific full RNAseq experiments in monocots. These covered mechanical and enzymatic separation in Z. mays (Chang et al., 2012); mechanical separation in S. viridis (John et al., 2014); laser micro-dissection in Z. mays (Li et al., 2010; Tausta et al., 2014); mechanical micro-dissection in P. virgatum (Rao et al., 2016); and the serial filtration performed here (referred to as ‘Denton 2016’ in figures). While the data encompass three origins and two subtypes of C4 photosynthesis, and BS and M cell specificity is not expected to match perfectly, previous studies have found substantial conservation even between monocots and dicots (Aubry et al., 2014). Overall, the combination of mechanical BS preparation and enzymatic (Chang et al., 2012) or leaf rolling (John et al., 2014) M separation achieved the highest marker enrichment, followed by the micro-dissection studies (Li et al., 2010; Tausta et al., 2014; Rao et al., 2016), while the method used here, as anticipated from the original report (Stitt and Heldt, 1985), showed the least enrichment (Fig. 2A). Consistent with the lower enrichment, this study showed the lowest statistical power of the various methods with an average of 2100 discoveries (FDR<0.05) per slice, compared with 4030–12 777 discoveries for the other (biological-replicate-including) studies when computed with edgeR. Therefore, a cross contamination aware R-package, contamDE, which includes a factor for the relative tissue enrichment of each replicate, was employed. With contamDE an average of 4479 discoveries (BS-e vs M-e FDR<0.05) were made per slice, and this was used for further analysis (see Table S2 at Dryad).
Fig. 2.
Interstudy comparison. (A) Enrichment of the classic BS (NADP-ME) and M (PEPC) marker genes in each study. (B–D) Log2 fold change of genes that were significantly enriched in BS or M in at least one of the paired Z. mays studies. (E) Hierarchical clustering of fully normalized log2 (TPM) for Z. mays samples, with Pearson and Spearman correlation-based distance for genes and samples, respectively. Genes filtered to those with TPM min>0, max>50. Side colors included to help delineate studies on the x-axis and major clusters on the y-axis. C, Chang et al. (2012); D, this study; L, Li et al. (2010); T, Tausta et al. (2014).
Tissues were matched to achieve a more in-depth comparison between the Z. mays studies. For mature tissues, the sample from Chang et al. (2012) was most similar to section 14 from Tausta et al. (2014) and to slice 2, here, while the youngest section in Tausta et al. (2014) was most similar to slice 4, here (Spearman correlation, Fig. S3A at Dryad). The Tausta et al. (2014) study was able to detect genes with a lower log fold change (relative to the total log fold change distribution) than either Chang et al. (2012), with just one replicate, or this study, with low enrichment. However, examining log fold change indicated the differences between studies ran deeper than statistical power, with many genes significant in one study not enriched or even significantly enriched in the opposite direction in another study (Fig 2B–D).
Qualitative study comparison
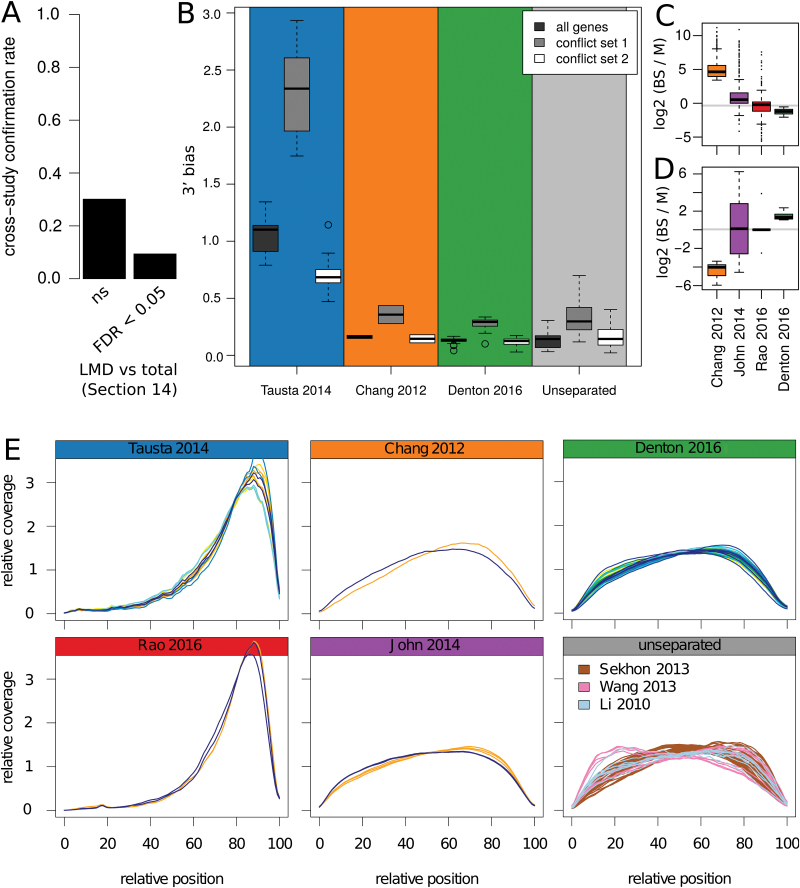
For a more qualitative look at the differences between studies we performed a hierarchical clustering of samples from this study, those from Chang et al. (2012) and Tausta et al. (2014), and the unseparated sections from Li et al. (2010) that corresponded to Tausta et al. (2014). The samples clustered primarily by study, followed by leaf age and then M and BS, with some mixing (Fig. 2E). Between-study differences could in theory come from growth conditions and plant age, from differences in separation method or from a combination thereof, and all studies but Li et al. (2010) and Tausta et al. (2014) used distinct growth and harvest conditions (see Table S3 at Dryad). Notably, the unseparated sections from Li et al. (2010), which were grown comparably to those from Tausta et al. (2014) clustered not with the associated leaf sections of Tausta et al. (2014), but with the respective older or younger serial filtration data here, indicating a substantial role of separation method in clustering. Indeed, one of the gene clusters (3) was primarily expressed at a lower level across the laser micro-dissection (Tausta et al., 2014) samples compared with all the other samples (including Li et al., 2010). RNA is known for its degradability under procedures like laser micro-dissection, and Li et al. (2010) clearly reported the 3′ bias in the laser micro-dissection section 14, but did not at that time have the comparative studies to evaluate how this would globally affect the results. A list of genes most dramatically affected by laser micro-dissection was obtained by looking for genes with significantly different abundance between unseparated (Li et al., 2010) and the laser micro-dissection separated section 14 (Li et al., 2010; Tausta et al., 2014). The majority (3298 of 3362) of the differentially regulated genes were downregulated in the laser micro-dissection samples. These laser micro-dissection ‘downregulated’ genes were depleted in BS vs M, differences shared with this study (Fig. 3A; Fisher’s exact test, P<0.001). Further, these genes showed several functional enrichments (MapMan categories), including major categories such as transport and signaling; and minor categories such as minor CHO metabolism.callose, GARP G2-like transcription factor family and Class XI Myosin (Dataset S2 at Dryad). Finally, the strong 3′ bias resulted in a low diversity library compared with the other studies (see Fig. S3B, C at Dryad).
Fig. 3.
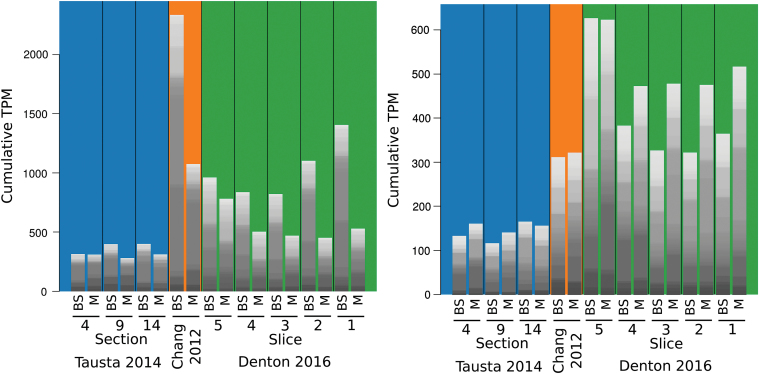
Technical bias. (A) The fraction of significant differences discovered here (slice 2) that were shared with the Tausta et al. (2014; section 14) study broken up based on whether these genes were of significantly lower abundance in the laser micro-dissected section 14 compared with whole section 14 (Li et al., 2010; Tausta et al., 2014). (B) The 3′ bias observed in the coverage for the genomic background and the two conflict sets in each Z. mays separation study, and all the unseparated samples of Li et al. (2010), Wang et al. (2013), and Sekhon et al. (2013). (C, D) The tissue enrichment of the Z. mays genes and the S. viridis (John et al., 2014) and P. virgatum (Rao et al., 2016) orthologs where the Z. mays gene was significantly more abundant in the BS in Chang et al. (2012), and the M in slice 2 (this study) (C), or vice versa (D). (E) Transcript coverage by study. For all BS and M separation studies, blue represents BS, yellow represents M, and tissue maturity increases from light to dark. Green represents I-e in this study (Denton 2016).
Considering the effect degraded RNA can have, we evaluated the 3′ bias across studies to see how the other separation methods compared. The three prime bias was highest in the laser (Tausta et al., 2014) and mechanical (Rao et al., 2016) micro-dissection studies; however, it was present to various degrees in at least some samples of the other separation studies and in multiple other Z. mays studies without separation (Li et al., 2010; Sekhon et al., 2013; Wang et al., 2013; Fig. 3C). Notably, both studies that used distinct methods for isolation of BS strands and M cells (John et al., 2014; Chang et al., 2012) showed minor 3′ bias, but each M sample showed more than its corresponding BS sample (Fig. 3C). The 3′ bias was not spread evenly across all genes, but was higher in the 199 genes where Chang et al. (2012) and slice 2 (this study) were significantly, but oppositely, enriched in the BS and M, respectively (see Fig. S4 at Dryad). Overall mild increases in 3′ bias between samples are prominent in these 199 genes and their orthologs, notably including the M samples in Chang et al. (2012) and Tausta et al. (2014) and one BS replicate from this study. The orthologs of these 199 genes, measured by John et al. (2014) mostly (138 of 184; 75%) were enriched in the same direction as Chang et al. (2012), while those measured by Rao et al. (2016) mostly (101 of 152; 66%) agreed with this study (Fig. 3C). In contrast, neither cross-species comparison showed a notable BS or M bias in orthologs of the opposite gene set—the 14 genes where Chang et al. (2012) and slice 2 (this study) were significantly enriched in the M and BS, respectively (Fig. 3D). In summary, despite evolutionary distance between Z. mays and S. viridis, the studies with higher, degradation-marking 3′ bias in the M than BS (Chang et al., 2012; John et al., 2014) share a set of ‘BS enriched’ genes that conflict with the M enrichment seen in Z. mays (this study) and P. virgatum (Rao et al., 2016).
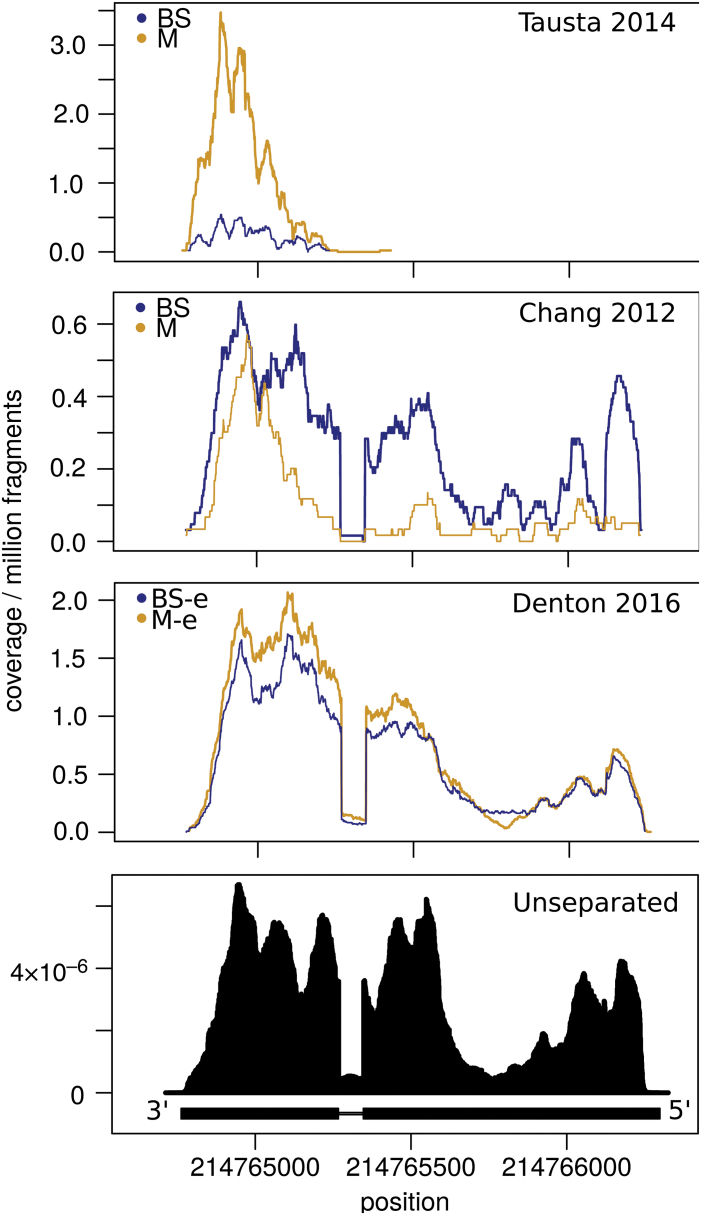
To determine if different RNA quality and 3′ bias relate to some of the discrepancies between the Z. mays studies, we quantified the level of 3′ bias on genes in two different conflict sets—conflict set 1: BS specific in Chang et al. (2012) or Tausta et al. (2014) and M specific in the comparable tissue here, or BS (Chang et al., 2012) and M (Tausta et al., 2014, section 14); conflict set 2: as conflict set 1 but with BS and M switched). This showed that conflict set 1 had the most 3′ bias across all studies while conflict set 2 had the same or even less bias than the whole gene set (Fig. 3B). One of the genes in ‘conflict set 1’ is related to the C4 cycle, namely phosphoenolpyruvate carboxylase kinase (PPCK; GRMZM2G178074), which regulates PEPC in the M (Vidal and Chollet, 1997). The coverage across the PPCK locus shows a mild 3′ bias in unseparated studies and in both BS and M samples here, with higher coverage in the M (Fig. 4). In the laser micro-dissection study, there is a strong 3′ bias in both samples, with more remaining coverage in the M sample, while in the Chang et al. (2012) sample, there is a mild 3′ bias in the BS sample, but a strong 3′ bias in the M sample, causing PPCK to appear higher in the BS based on total read count. While not all genes in ‘conflict set 1’ looked like this (e.g. many had a very strong 3′ bias across every sample and study; not shown), other similar examples were not hard to find (see Fig. S5 at Dryad). Further, components of ‘conflict set 1’ were enriched in several MapMan categories. These included three transcription factor sub-categories (PHD finger, pseudo ARR, and putative), and minor CHO metabolism.callose in genes BS specific in Chang et al. (2012), and M specific in slice 2 of this study (Dataset S2 at Dryad).
Fig. 4.
Coverage of example gene PPCK. Read depth across genomic region of PPCK (GRMZM2G178074; which is in conflict set 1) in the various Z. mays separation studies, and in the unseparated samples of Li et al. (2010), Wang et al. (2013), and Sekhon et al. (2013).
Another likely artifact of the separation method is the residual contamination with non-M and non-BS tissue types. The mechanical separation methods are expected to co-purify the vascular bundle with the BS cells, while the serial grinding and filtration used here presumably includes all cell types in at least one of the enrichment fractions. To confirm and quantify these expectations would require unambiguous markers that were known to, for instance, be highly specific to the vascular tissues and absent from M or BS. In the absence of fully characterized markers in Z. mays, we tested a variety of candidates, largely known from other species.
Putative vascular markers were initially selected from the literature based on functions expected to be highly vascular specific. Enzymes associated with lignification of proto-xylem elements (LAC17) were more abundant in the BS base sample (Fig. S6A at Dryad; FDR<0.05 for three of the four expressed). Similarly, homologs to Arabidopsis XYLEM CYSTEIN PROTEASE (XCP) 1 (GRMZM2G066326) and 2 (GRMZM2G367701), involved in programmed cell death in the xylem, were higher in the BS base sample (Fig. S6B at Dryad; FDR<0.001 for all three). These markers, however, were not expressed in older tissue and thus could not be used for interstudy comparisons. SUCROSE TRANSPORTER 2 (SUT2), frequently used as a companion cell marker in Arabidopsis (AT2G02860; Meyer et al., 2000), has five homologs in Z. mays, for which the cumulative expression was enriched in the BS across all Z. mays studies (see Fig. S6C at Dryad). A study on phloem transported RNAs in Arabidopsis (Deeken et al., 2008) provided a larger list of potential vascular markers; however, the cumulative expression was again higher in the BS across studies (Fig. S6D at Dryad). We further examined sets of genes that included ‘phloem’ (Fig. 5A), ‘xylem’ (Fig. S6E at Dryad), or ‘vascular’ (Fig. S6F at Dryad) in their descriptions. Cumulative expression of these keyword gene sets was largely higher in the BS across studies; however, for ‘phloem’ and ‘vascular’ genes, BS enrichment in the laser micro-dissected samples was less than BS enrichment of these genes in the mechanical separation studies.
Fig. 5.
Co-purification of additional tissues. Fully normalized abundance of genes that included the word ‘phloem’ (A) or ‘epidermal’ (B) in their MapMan description.
We further evaluated the distribution of putative epidermal markers. A previous study using laser micro-dissection to separate epidermal and M tissues identified two epidermal specific genes in Z. mays (Javelle et al., 2010). The more highly expressed of these, GRMZM2G345700, was consistently higher in the M samples (Fig. S6G at Dryad; FDR<0.05 in six of nine comparisons), while the less highly expressed GRMZM2G387360 was not significantly enriched. A broader look at all genes including the words ‘epidermal’ in their descriptions (Fig. 5B) showed higher cumulative expression in the M in most comparisons, while the most substantial M enrichment appeared to be in this study (Fig. 5B). It is hard to draw a firm conclusion in the absence of unambiguous markers, as expression patterns in epidermal cells may be more similar to M than BS, and vice versa for vascular expression. However, both expectation and a view on the broader patterns support co-purification of vascular tissues with the mechanical BS purification methods, co-purification of epidermal tissue with the M in the serial filtration method used here, and generally less co-purification using laser micro-dissection.
The strengths of interstudy comparison
Multiple study comparisons allow for confidence in results that would seem dubious alone. In this study, aspartate aminotransferase stood out as having transcript enrichment in M cells (Fig. 6A) that was contrary to the expected even distribution between cell types in the current Z. mays C4 model (Furbank, 2011; Pick et al., 2011). Comparison with the other datasets confirmed the same pattern in all NADP-ME studies (Z. mays and S. viridis). Previous studies (Chang et al., 2012; Tausta et al., 2014) have mentioned a low-expression BS specific AspAT paralog, or the detection of AspAT in both BS and M proteomic studies as balancing explanations. However, in both transcriptomics and proteomics (Friso et al., 2010; Majeran et al., 2010) the total abundance is much higher in the M. This is further supported by the high M specificity of the AspAT enzyme activity (Fig. 6A). A similar but less pronounced pattern in transcripts could be found for alanine aminotransferase (see Fig. S7A at Dryad).
Fig. 6.
Biological insights drawn from interstudy comparison. (A) Tissue enrichment of AspAT of transcripts in maize (left), enzyme activity in Z. mays (mid-left), transcripts in S. viridis (mid-right) and transcripts in P. virgatum (right). (B, C) Tissue enrichment of transcripts in the MapMan functional category for 60S ribosomal protein (B) and photosystem II (C) in Z. mays (left), S. italica (middle), and P. virgatum (right). In (A) asterisks denote significance of FDR for transcripts and P-values for enzyme activity (*P<0.05, **P<0.01, ***P<0.001).
Consistent BS or M enrichment as the leaf develops helps increase confidence, both as a repeat observation and as a simple explanation consistent with the gradual nature of changes in transcript abundance during leaf development (Pick et al., 2011). On the flip side, however, it seems less likely that a gene changed from BS specific to M specific or vice versa during development. We used the interstudy comparison to evaluate the reliability of observed switches in enrichment across leaf development. As expected, genes that were significantly enriched in M-then-BS or BS-then-M in sections 4 and 14 of Tausta et al. (2014) were much less likely to find cross-study support (same enrichment direction in slice 4 and 2 of this study) than their M-then-M or BS-then-BS enriched counterparts (19% vs 78%, Fisher’s exact test P<0.001). However, the 48 genes that were significantly enriched in the BS in section 4 (Tausta et al., 2014) and in the M in section 14 (Tausta et al., 2014) with support from this study showed enrichment in the functional category ‘protein.synthesis.ribosomal protein.eukaryotic.60S subunit’ and all parental categories there of (Dataset S2 at Dryad). Further investigation showed that both the 60S and 40S ribosomal subunits have a clear pattern with strong BS enrichment in young but entirely unsheathed tissue (section 4, Tausta et al., 2014; slice 4, here). As the leaf develops the strong BS enrichment fades, and even switches to a mild M enrichment (Fig. 6B and Fig. S7B at Dryad). To determine if the mature M enrichment could be related to supporting the high turnover of photosystem II components, we included the data for S. viridis (John et al., 2014) and P. virgatum (Rao et al., 2016) in the analysis. Notably the 60S and 40S ribosomal subunits showed M enrichment in S. viridis (Fig. 6B and Fig. S7B at Dryad), in which photosystem II, like in Z. mays, is primarily localized to the M (Fig. 6C). In contrast, these subunits showed BS enrichment in P. virgatum (Fig. 6B and Fig. S7B at Dryad), in which photosystem II is not primarily localized to the M (Fig. 6C).
Data accessibility and visualization
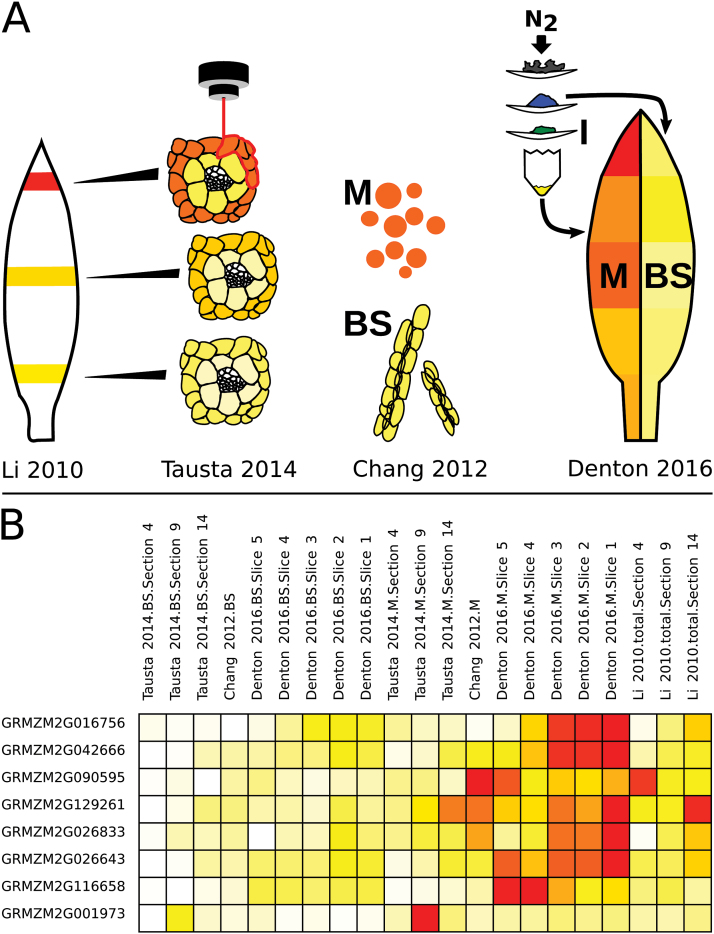
To facilitate public comparison of these transcriptomes, we are providing (i) a Z. mays gene browser with gene-specific or gene-group visualization of the data from BS/M separation studies in Z. mays; and (ii) all the data analysed in this study (including non-Z. mays BS vs M comparisons, and unseparated Z. mays studies) in tabular format (Dataset S3 and S4 at Dryad). The Z. mays gene browser aims to facilitate comparison and critical evaluation of the similarities and differences between these studies. To this end, the graphics include the separation method in the display and necessary contextual data (e.g. unseparated samples from Li et al. (2010) corresponding to the laser micro-dissection samples from Tausta et al. (2014), and 3′ bias (Fig. 7). Further, the browser includes several pre-loaded gene sets to help users compare studies (Fig. 7B). These sets include, for example ‘conflict set 1’ described above. Further gene sets include three gradations of highly supported M or BS specific genes across studies (735, 365, and 126 significant differences; shared between 7+, 8+, or all 9 of the comparisons, respectively), and highly supported M or BS specific transcription factors (52 significant differences shared between 7+ comparisons), and transcription factors of special interest in immature tissue (36 significant differences in two of the three youngest comparisons (Tausta et al., 2014 section 4, and slice 4 and 5, here) and higher in foliar than husk primordia in Wang et al. (2013). Full lists and descriptions are provided in Dataset S5 at Dryad and with the visualization tool at http://www.plant-biochemistry.hhu.de/resources.html.
Fig. 7.
Web visualization resource. (A) Comparative BS and M separation targeted graphical heatmap view of example gene (GRMZM2G129261). (B) Example gene set visualization of highest confidence M transcription factors.
Discussion
Despite the variety of BS and M separation methods used and increasing number of studies, no method presents itself as a clear best option. Rather, the various methods come with advantages and disadvantages, which should be considered both when planning the experiment and evaluating the data.
The only fast methods for metabolite extraction are leaf rolling (Leegood, 1985) for M compared with whole tissue, and grinding and serial filtration on liquid nitrogen as performed here (Stitt and Heldt, 1985). Only a handful of metabolites measured in these studies overlap, and the correlation between studies is modest. Elements of this study might help clarify why and plan the next experiment. First, there were very substantial differences between the <1 s harvest and the 10 s harvest and between the different leaf slices. This highlights that the dynamics of abundance of metabolites make them extremely sensitive to both conditions and harvest methods. Considering the dominance of age, conditions and method over BS vs M differences in the clustering of RNAseq data, it is perhaps unsurprising that the even more labile metabolites continue to pose challenges. Similarly, the low absolute enrichment of this method and the Leegood (1985) method decreases the signal to noise ratio, particularly making identification of low log fold changes between cell types difficult (as seen in the RNAseq). This is likely exacerbated by the division of some metabolites, such as aspartate and malate, into active and inactive pools. These inactive pools can be substantial, accounting for about 60% and 80% of the total aspartate and malate, respectively, in the grass Chloris gayana (Hatch, 1979). In contrast, the high density of plasmodesmata between M and BS cells in C4 plants supports diffusion of C4-cycle metabolites at the rate of carbon fixation (Laisk and Edwards, 2000); it is thus implausible that any cytoplasmic metabolite could build up enrichment levels comparable to transcripts and enzymes. Therefore a study prioritizing understanding metabolic differences between BS and M cells should err on the side of a few more replicates than the five that is the ‘industry standard’ for metabolic studies (Sumner et al., 2007). Similarly, sequencing a few more than the typical two to three replicates for RNAseq may help compensate for the lower sensitivity of this method.
For any study not targeting metabolites, the higher purity achieved by any of the other methods over the method here has an obvious allure; however, the biases associated with lower quality RNA must be accounted for. As shown here and reported previously (Romero et al., 2014) RNA does not degrade at consistent rates, but rather some RNA molecules, often including transcription factors (Yang et al., 2003), are much more sensitive to degradation. These degradation-sensitive genes are numerous (12.5% of detectable genes showed significantly lower abundance after laser micro-dissection; Li et al., 2010; Tausta et al., 2014). Further, shared genes with bias in Chang et al. (2012) and John et al. (2014) indicate degradation sensitivity is conserved across species and can masquerade as conserved tissue specificity. For the above reasons, care must be taken not to intermingle any biological signal sensitive to degradation and the biological signal between samples. For instance, the two callose synthases that Chang et al. (2012) discussed as being BS specific (GRMZM2G553532 and GRMZM2G004087) appear to be very sensitive to degradation as they are both among the genes significantly less abundant after laser micro-dissection, and one, GRMZM2G553532, is in the conflict set 1 list with strong 3′ bias. This raises the worrisome question of whether this is a case of differential expression, or differential degradation. Future studies may be able to circumvent such problems by including a third and unseparated sample that can be used to detect genes particularly affected by degradation—much as we’ve been using the unseparated section 14 from Li et al. (2010) as the context for the Tausta et al. (2014) separated section. This method has been employed by a recent study using SuperSage on mechanically separated BS and M protoplast in Sorghum (Döring et al., 2016).
A more ideal solution is of course to avoid mixing biological and technical signals by handling RNA in a fashion that preserves RNA quality or at least results in the same amount of degradation in the M and BS samples. Quality control must be performed carefully as a study using the same separation technique as John et al. (2014) for qPCR in sorghum achieved very comparable bioanalyser traces for their mechanical BS purified and their leaf-rolled M samples (Covshoff et al., 2013). Indeed, this method was specifically employed for its speed and lack of stress response compared with M protoplast isolation, but still showed distinctly higher 3′ bias in M than BS (John et al., 2014). Thus if a distinct method is to be used for M and BS purification, equivalent RNA needs to be confirmed for the particular species and particular researcher, and not simply assumed based on literature.
While the micro-dissection studies had the strongest overall 3′ bias, there was equivalent bias in the M and BS samples. This resulted in false negatives and lower library complexity, but had no clear link to false positives. In the microdissection studies, an alternative explanation for the 3′ bias is the synthesis of the first strand cDNA using an Arcturus Ribo Amp HS kit, which has been shown to induce a strong 3′ bias in housekeeping genes (Clément-Ziza et al., 2009). This does not, however, nullify the substantial differences and loss of transcript detection seen between the laser micro-dissected (Li et al., 2010; Tausta et al., 2014) and the unseparated samples (Li et al., 2010). There is ongoing research in improving laser-micro-dissection techniques in plants (Ludwig and Hochholdinger, 2014). We recommend that while techniques remain uncertain, researchers invest the necessary time and money in quality control steps and unseparated controls to assure that the bias that is there is traceable.
Use of a different bioinformatics workflow may make a small difference in the measured abundance of genes with a strong 3′ bias, but a perfect solution is not yet available, particularly as tools are not optimized for this. Small additions to a typical workflow, such as flagging discrepancy in 3′ bias between groups (e.g. Chang et al., (2012)’s samples in Fig. 4), could help avoid erroneous conclusions.
Where one study has weaknesses, interstudy comparison can provide a helpful additional opinion. The completion of a third Z. mays M and BS separation RNAseq study with a complementary technique here continued to yield new biological results. Particularly in areas where results may seem dubious, consensus between several studies (with different techniques or information gathered) is required to gain confidence. An example of this is AspAT’s consistent M localization, which while previously noted (Chang et al., 2012; Tausta et al., 2014), was not taken seriously without the supporting enzyme activity data. It may have a simple explanation such as a higher substrate to product ratio in the BS requiring less enzyme, or a more complex one such as an aspartate pool in the M simply adding stability to CO2 fixation should diffusion or decarboxylation of malate become temporarily limiting. Either way, this warrants further investigation. Similarly, the switch from BS to M specificity of ribosomal proteins is much easier to trust when identified in two independent studies. Differentiation of veins and the associated BS cells precedes that of the M, and signals from the BS are necessary for M differentiation in Arapidopsis (Kinsman and Pyke, 1998; Lundquist et al., 2014), and the C4 dicot Gynandropsis gynandra shows the same developmental trajectory (Külahoglu et al., 2014). Therefore, we hypothesize the initial BS enrichment in protein synthesis may reflect faster differentiation and photosynthetic ramp-up in the BS cells. As the photosynthetic rate increases along the developing leaf (Pick et al., 2011), the shifting of the protein synthesis towards the M likely supports the high turnover of photosystem II subunits (Rokka et al., 2005). Considering that photosynthesis-related proteins make up over half of mature leaf protein (Friso et al., 2010; Majeran et al., 2010), the distribution of protein synthesis in mature leaf may reflect the balance between the demand from synthesizing photosystem II (when in the M) and synthesizing Rubisco and the other BS-specific CBBC enzymes.
Altogether, the separation technique of choice depends upon the research question. In many cases the weaknesses of one study are compensated for by the strengths of another, particularly when biases are characterized and taken into consideration. This work provides a visual access tool summarizing this study and Li et al. (2010), Chang et al. (2012) and Tausta et al. (2014), tables of all data looked at here (above and Wang et al., 2013; Sekhon et al., 2013; John et al., 2014; Rao et al., 2016), and highlights biological observations drawn from the sum of many studies.
Data deposition
The following data are available at Dryad Digital Repository http://dx.doi.org/10.5061/dryad.tf6q6.
Datasets S1. Enzyme activity and metabolite abundance.
Datasets S2. Functional enrichments.
Datasets S3. Compiled RNAseq data.
Datasets S4. By gene 3′ bias.
Datasets S5. Gene sets of interest.
Fig. S1. Setup and confirmation of separation method.
Fig. S2. Metabolite enrichment.
Fig. S3. Contextual data for interstudy comparison.
Fig. S4. Coverage of BS (Chang et al., 2012) vs M (this study) conflict genes.
Fig. S5. Example read coverage.
Fig. S6. Co-purification of additional tissues.
Fig. S7. AlaAT and 40S ribosome distributions.
Table S1. Bioinformatics parameters.
Table S2. Counting significant differences.
Table S3. Harvest and growth conditions.
Competing interests
The authors declare that they have no competing financial interests.
Author contributions
AKD, AB and APMW designed the study. AKD performed wetlab measurements, computational, statistical and general data analysis and wrote the manuscript. JM developed the visualization tool, performed general data analysis, and assisted in wetlab measurements and writing the manuscript. CK, MJL, AB, and APMW contributed to data analysis, interpretation of the results, and writing the manuscript.
Acknowledgements
We acknowledge Katrin Weber for GC/MS assistance; Simon Schliesky for data management support; Björn Usadel and Shin-Han Shiu for helpful discussion; and the Deutsche Forschungsgemeinschaft (IRTG 1525 supporting JM, CK and AKD; EXC 1028 to MJL and APMW; and CRC 680 to MJL) as well as the European Union 7th Framework Program (EU 3to4 to APMW) for financial support.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC: A quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (last accessed 29 November 2016).
- Aubry S, Kelly S, Kümpers BM, Smith-Unna RD, Hibberd JM. 2014. Deep evolutionary comparison of gene expression identifies parallel recruitment of trans-factors in two independent origins of C4 photosynthesis. PLoS Genetics 10, e1004365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellasio C, Griffiths H. 2014. The operation of two decarboxylases, transamination, and partitioning of C4 metabolic processes between mesophyll and bundle sheath cells allows light capture to be balanced for the maize C4 pathway. Plant Physiology 164, 466–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B 57, 289–300. [Google Scholar]
- Benjamini Y, Yekutieli D. 2001. The control of the false discovery rate in multiple testing under dependency. Annals of Statistics 29, 1165–1188. [Google Scholar]
- Bennetzen JL, Schmutz J, Wang H, et al. 2012. Reference genome sequence of the model plant Setaria. Nature Biotechnology 30, 555–561. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A, Kajala K, Wullenweber J, et al. 2011. An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiology 155, 142–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A, Schliesky S, Külahoglu C, Osborne CP, Weber AP. 2014. Towards an integrative model of C4 photosynthetic subtypes: insights from comparative transcriptome analysis of NAD-ME, NADP-ME, and PEP-CK C4 species. Journal of Experimental Botany 65, 3579–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broglie R, Coruzzi G, Keith B, Chua NH. 1984. Molecular biology of C4 photosynthesis in Zea mays: differential localization of proteins and mRNAs in the two leaf cell types. Plant Molecular Biology 3, 431–444. [DOI] [PubMed] [Google Scholar]
- Chang Y-M, Liu W-Y, Shih AC-C, et al. 2012. Characterizing regulatory and functional differentiation between maize mesophyll and bundle sheath cells by transcriptomic analysis. Plant Physiology 160, 165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clément-Ziza M, Gentien D, Lyonnet S, Thiery JP, Besmond C, Decraene C. 2009. Evaluation of methods for amplification of picogram amounts of total RNA for whole genome expression profiling. BMC Genomics 10, 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covshoff S, Furbank RT, Leegood RC, Hibberd JM. 2013. Leaf rolling allows quantification of mRNA abundance in mesophyll cells of sorghum. Journal of Experimental Botany 64, 807–813. [DOI] [PubMed] [Google Scholar]
- Covshoff S, Szecowka M, Hughes TE, et al. 2016. C4 photosynthesis in the rice paddy: insights from the noxious weed Echinochloa glabrescens. Plant Physiology 170, 57–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeken R, Ache P, Kajahn I, Klinkenberg J, Bringmann G, Hedrich R. 2008. Identification of Arabidopsis thaliana phloem RNAs provides a search criterion for phloem-based transcripts hidden in complex datasets of microarray experiments. The Plant Journal 55, 746–759. [DOI] [PubMed] [Google Scholar]
- Denton AK, Maß J, Külahoglu C, Lercher MJ, Bräutigam A, Weber APM. 2016. Data from: Freeze-quenched maize mesophyll and bundle sheath separation uncovers bias in previous tissue-specific RNA-Seq data. Dryad Digital Repository http://dx.doi.org/10.5061/dryad.tf6q6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOE-JGI 2016. Panicum virgatum v1.1. https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Pvirgatum (last accessed 29 November 2016). [Google Scholar]
- Döring F, Streubel M, Bräutigam A, Gowik U. 2016. Most photorespiratory genes are preferentially expressed in the bundle sheath cells of the C4 grass Sorghum bicolor. Journal of Experimental Botany 67, 3053–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso G, Majeran W, Huang M, Sun Q, van Wijk KJ. 2010. Reconstruction of metabolic pathways, protein expression, and homeostasis machineries across maize bundle sheath and mesophyll chloroplasts: large-scale quantitative proteomics using the first maize genome assembly. Plant Physiology 152, 1219–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furbank RT. 2011. Evolution of the C4 photosynthetic mechanism: are there really three C4 acid decarboxylation types?Journal of Experimental Botany 62, 3103–3108. [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, et al. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Research 40, D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowik U, Bräutigam A, Weber KL, Weber AP, Westhoff P. 2011. Evolution of C4 photosynthesis in the genus Flaveria: how many and which genes does it take to make C4?The Plant Cell 23, 2087–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch MD. 1979. Mechanism of C4 photosynthesis in Chloris gayana: pool sizes and kinetics of 14CO2 incorporation into 4-carbon and 3-carbon intermediates. Archives of Biochemistry and Biophysics 194, 117–127. [DOI] [PubMed] [Google Scholar]
- Hatch MD. 1987. C4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Biochimica et Biophysica Acta 895, 81–106. [Google Scholar]
- Hatch MD, Mau S. 1977. Properties of phosphoenolpyruvate carboxykinase operative in C4 pathway photosynthesis. Functional Plant Biology 4, 207–216. [Google Scholar]
- Huang P, Brutnell TP. 2016. A synthesis of transcriptomic surveys to dissect the genetic basis of C4 photosynthesis. Current Opinion in Plant Biology 31, 91–99. [DOI] [PubMed] [Google Scholar]
- Javelle M, Vernoud V, Depège-Fargeix N, Arnould C, Oursel D, Domergue F, Sarda X, Rogowsky PM. 2010. Overexpression of the epidermis-specific homeodomain-leucine zipper IV transcription factor Outer Cell Layer1 in maize identifies target genes involved in lipid metabolism and cuticle biosynthesis. Plant Physiology 154, 273–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John CR, Smith-Unna RD, Woodfield H, Covshoff S, Hibberd JM. 2014. Evolutionary convergence of cell-specific gene expression in independent lineages of C4 grasses. Plant Physiology 165, 62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey PJ, Allen JE, Armean I, et al. 2016. Ensembl Genomes 2016: more genomes, more complexity. Nucleic Acids Research 44, D574–D580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsman EA, Pyke KA. 1998. Bundle sheath cells and cell-specific plastid development in Arabidopsis leaves. Development 125, 1815–1822. [DOI] [PubMed] [Google Scholar]
- Kodama Y, Shumway M, Leinonen R. 2012. The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Research 40, D54–D56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Külahoglu C, Denton AK, Sommer M, et al. 2014. Comparative transcriptome atlases reveal altered gene expression modules between two Cleomaceae C3 and C4 plant species. The Plant Cell 26, 3243–3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laisk A, Edwards GE. 2000. A mathematical model of C4 photosynthesis: The mechanism of concentrating CO2 in NADP-malic enzyme type species. Photosynthesis Research 66, 199–224. [DOI] [PubMed] [Google Scholar]
- Leegood RC. 1985. The intercellular compartmentation of metabolites in leaves of Zea mays L. Planta 164, 163–171. [DOI] [PubMed] [Google Scholar]
- Leinonen R, Akhtar R, Birney E, et al. 2010. The European nucleotide archive. Nucleic Acids Research 39, D28–D31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Ponnala L, Gandotra N, et al. 2010. The developmental dynamics of the maize leaf transcriptome. Nature Genetics 42, 1060–1067. [DOI] [PubMed] [Google Scholar]
- Lohse M, Nagel A, Herter T, et al. 2014. Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data. Plant, Cell & Environment 37, 1250–1258. [DOI] [PubMed] [Google Scholar]
- Ludwig Y, Hochholdinger F. 2014. Laser microdissection of plant cells. Methods in Molecular Biology 1080, 249–258. [DOI] [PubMed] [Google Scholar]
- Lundquist PK, Rosar C, Bräutigam A, Weber AP. 2014. Plastid signals and the bundle sheath: mesophyll development in reticulate mutants. Molecular Plant 7, 14–29. [DOI] [PubMed] [Google Scholar]
- Majeran W, Cai Y, Sun Q, van Wijk KJ. 2005. Functional differentiation of bundle sheath and mesophyll maize chloroplasts determined by comparative proteomics. The Plant Cell 17, 3111–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W, Friso G, Ponnala L, et al. 2010. Structural and metabolic transitions of C4 leaf development and differentiation defined by microscopy and quantitative proteomics in maize. The Plant Cell 22, 3509–3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Melzer M, Truernit E, HuÈmmer C, Besenbeck R, Stadler R, Sauer N. 2000. AtSUC3, a gene encoding a new Arabidopsis sucrose transporter, is expressed in cells adjacent to the vascular tissue and in a carpel cell layer. The Plant Journal 24, 869–882. [DOI] [PubMed] [Google Scholar]
- Pick TR, Bräutigam A, Schlüter U, et al. 2011. Systems analysis of a maize leaf developmental gradient redefines the current C4 model and provides candidates for regulation. The Plant cell 23, 4208–4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team 2011. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Rao X, Lu N, Li G, Nakashima J, Tang Y, Dixon RA. 2016. Comparative cell-specific transcriptomics reveals differentiation of C4 photosynthesis pathways in switchgrass and other C4 lineages. Journal of Experimental Botany 67, 1649–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawsthorne S, Hylton CM, Smith AM, Woolhouse HW. 1988. Photorespiratory metabolism and immunogold localization of photorespiratory enzymes in leaves of C3 and C3-C4 intermediate species of Moricandia. Planta 173, 298–308. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2009. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokka A, Suorsa M, Saleem A, Battchikova N, Aro EM. 2005. Synthesis and assembly of thylakoid protein complexes: multiple assembly steps of photosystem II. The Biochemical Journal 388, 159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowska E, Kargul J, Powikrowska M, Finazzi G, Nield J, Drozak A, Pokorska B. 2008. Structural organization of photosynthetic apparatus in agranal chloroplasts of maize. The Journal of Biological Chemistry 283, 26037–26046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero IG, Pai AA, Tung J, Gilad Y. 2014. RNA-seq: impact of RNA degradation on transcript quantification. BMC Biology 12, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf M, Machettira AB, Groß LE, et al. 2013. In vivo function of Tic22, a protein import component of the intermembrane space of chloroplasts. Molecular Plant 6, 817–829. [DOI] [PubMed] [Google Scholar]
- Sage RF, Zhu XG. 2011. Exploiting the engine of C4 photosynthesis. Journal of Experimental Botany 62, 2989–3000. [DOI] [PubMed] [Google Scholar]
- Schulze E-D, Ellis R, Schulze W, Trimborn P, Ziegler H. 1996. Diversity, metabolic types and δ13C carbon isotope ratios in the grass flora of Namibia in relation to growth form, precipitation and habitat conditions. Oecologia 106, 352–369. [DOI] [PubMed] [Google Scholar]
- Sekhon RS, Briskine R, Hirsch CN, Myers CL, Springer NM, Buell CR, de Leon N, Kaeppler SM. 2013. Maize gene atlas developed by RNA sequencing and comparative evaluation of transcriptomes based on RNA sequencing and microarrays. PloS One 8, e61005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Hu J, Jiang N, Hu X, Luo Z, Zhang H. 2016. contamDE: differential expression analysis of RNA-seq data for contaminated tumor samples. Bioinformatics 32, 705–712. [DOI] [PubMed] [Google Scholar]
- Stitt M, Heldt HW. 1985. Control of photosynthetic sucrose synthesis by fructose 2,6-bisphosphate: VI. Regulation of the cytosolic fructose 1,6-bisphosphatase in spinach leaves by an interaction between metabolic intermediates and fructose 2,6-bisphosphate. Plant Physiology 79, 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner LW, Amberg A, Barrett D, et al. 2007. Proposed minimum reporting standards for chemical analysis. Metabolomics 3, 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tausta SL, Li P, Si Y, Gandotra N, Liu P, Sun Q, Brutnell TP, Nelson T. 2014. Developmental dynamics of Kranz cell transcriptional specificity in maize leaf reveals early onset of C4-related processes. Journal of Experimental Botany 65, 3543–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tello-Ruiz MK, Stein J, Wei S, et al. 2016. Gramene 2016: comparative plant genomics and pathway resources. Nucleic Acids Research 44, D1133–D1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal J, Chollet R. 1997. Regulatory phosphorylation of C4 PEP carboxylase. Trends in Plant Science 2, 230–237. [Google Scholar]
- Walker RP, Trevanion SJ, Leegood RC. 1995. Phosphoenolpyruvate carboxykinase from higher plants: purification from cucumber and evidence of rapid proteolytic cleavage in extracts from a range of plant tissues. Planta 196, 58–63. [Google Scholar]
- Wang Y, Bräutigam A, Weber AP, Zhu XG. 2014. Three distinct biochemical subtypes of C4 photosynthesis? A modelling analysis. Journal of Experimental Botany 65, 3567–3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Kelly S, Fouracre JP, Langdale JA. 2013. Genome-wide transcript analysis of early maize leaf development reveals gene cohorts associated with the differentiation of C4 Kranz anatomy. The Plant Journal 75, 656–670. [DOI] [PubMed] [Google Scholar]
- Wysoker A, Tibbetts K, Fennell T. 2012. Picard. http://broadinstitute.github.io/picard/ (last accessed 29 November 2016).
- Yang E, van Nimwegen E, Zavolan M, Rajewsky N, Schroeder M, Magnasco M, Darnell JE., Jr 2003. Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Research 13, 1863–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
To facilitate public comparison of these transcriptomes, we are providing (i) a Z. mays gene browser with gene-specific or gene-group visualization of the data from BS/M separation studies in Z. mays; and (ii) all the data analysed in this study (including non-Z. mays BS vs M comparisons, and unseparated Z. mays studies) in tabular format (Dataset S3 and S4 at Dryad). The Z. mays gene browser aims to facilitate comparison and critical evaluation of the similarities and differences between these studies. To this end, the graphics include the separation method in the display and necessary contextual data (e.g. unseparated samples from Li et al. (2010) corresponding to the laser micro-dissection samples from Tausta et al. (2014), and 3′ bias (Fig. 7). Further, the browser includes several pre-loaded gene sets to help users compare studies (Fig. 7B). These sets include, for example ‘conflict set 1’ described above. Further gene sets include three gradations of highly supported M or BS specific genes across studies (735, 365, and 126 significant differences; shared between 7+, 8+, or all 9 of the comparisons, respectively), and highly supported M or BS specific transcription factors (52 significant differences shared between 7+ comparisons), and transcription factors of special interest in immature tissue (36 significant differences in two of the three youngest comparisons (Tausta et al., 2014 section 4, and slice 4 and 5, here) and higher in foliar than husk primordia in Wang et al. (2013). Full lists and descriptions are provided in Dataset S5 at Dryad and with the visualization tool at http://www.plant-biochemistry.hhu.de/resources.html.
Fig. 7.
Web visualization resource. (A) Comparative BS and M separation targeted graphical heatmap view of example gene (GRMZM2G129261). (B) Example gene set visualization of highest confidence M transcription factors.