Plasma membrane-associated cation-binding protein 1-like protein of Nicotiana benthamiana plays a role in defense against Bamboo mosaic virus by binding the viral replicase and restricting viral intercellular movement.
Keywords: Bamboo mosaic virus, defense protein, NbPCaP1L, Nicotiana benthamiana, positive-sense RNA virus, replicase, viral RNA movement
Abstract
To establish a successful infection, a virus needs to replicate and move cell-to-cell efficiently. We investigated whether one of the genes upregulated in Nicotiana benthamiana after Bamboo mosaic virus (BaMV) inoculation was involved in regulating virus movement. We revealed the gene to be a plasma membrane-associated cation-binding protein 1-like protein, designated NbPCaP1L. The expression of NbPCaP1L in N. benthamiana was knocked down using Tobacco rattle virus-based gene silencing and consequently the accumulation of BaMV increased significantly to that of control plants. Further analysis indicated no significant difference in the accumulation of BaMV in NbPCaP1L knockdown and control protoplasts, suggesting NbPCaP1L may affect cell-to-cell movement of BaMV. Using a viral vector expressing green fluorescent protein in the knockdown plants, the mean area of viral focus, as determined by fluorescence, was found to be larger in NbPCaP1L knockdown plants. Orange fluorescence protein (OFP)-fused NbPCaP1L, NbPCaP1L-OFP, was expressed in N. benthamiana and reduced the accumulation of BaMV to 46%. To reveal the possible interaction of viral protein with NbPCaP1L, we performed yeast two-hybrid and co-immunoprecipitation experiments. The results indicated that NbPCaP1L interacted with BaMV replicase. The results also suggested that NbPCaP1L could trap the BaMV movement RNP complex via interaction with the viral replicase in the complex and so restricted viral cell-to-cell movement.
Introduction
To complete an infection cycle, a plant virus needs to not only replicate efficiently but also move cell-to-cell successfully in the form of a virion or viral ribonucleoprotein complex (RNP) via the plasmodesmata (PD) (Hofmann et al., 2007). The latter process requires both viral-encoded movement proteins (MPs) and host factors (Scholthof, 2005; Taliansky et al., 2008; Benitez-Alfonso et al., 2010; Niehl and Heinlein, 2011; Schoelz et al., 2011). In addition, the cytoskeleton system is involved in transporting the viral components to the PD (Prokhnevsky et al., 2005; Avisar et al., 2008). Although the microtubules do not play a major role in viral RNA/protein complex trafficking in tobacco mosaic virus (TMV), it is still a key player involved in delivering the complex to microfilaments for transportation (Li et al., 2011; Peña and Heinlein, 2012; Liu and Nelson, 2013). Potato mop-top virus (PMTV) possesses a triple gene block (TGB) that encodes three MPs for virus movement. Mutants with an N-terminal 84-amino acid deletion of TGB1 in PMTV, with lost activity in microtubule association, failed to show long distance movement (Agre et al., 1998). A host protein NbMPB2Cb from N. benthamiana, a homolog of TMV-MP30 binding protein 2C, MPB2C, which is involved in organizing the cortical microtubules (Chaumont et al., 2000), could regulate the movement of Potato virus X (PVX). The movement of PVX is restricted when NbMPB2Cb is overexpressed; by contrast the movement is enhanced when the expression of NbMPB2Cb is silenced (Hachez et al., 2013).
Bamboo mosaic virus (BaMV) is a member of the Potexvirus genus of α-Flexiviridae. The genome of BaMV has one single-stranded positive-sense RNA approximately 6.4 kb long with a 5′ m7GpppG structure and 3′ adenylates numbering ~250−300 ( Lin et al., 1992; Chen et al., 2005). The genomic RNA of BaMV contains five open reading frames, ORFs 1 to 5. ORF 1 encodes a 155 kDa polypeptide comprising three functional domains. The N-terminal capping enzyme domain possesses S-adenosylmethionine–dependent guanylytransferase activity (Li et al., 2001a; Huang et al., 2004; Huang et al., 2005); the middle part is a helicase-like domain with nucleoside triphosphatase (NTPase) and RNA 5′-triphosphatase activities (Li et al., 2001b) and the C-terminal RNA-dependent RNA polymerase (RdRp) domain (Li et al., 1998; Huang et al., 2001) has viral RNA replication activity. The overlapped ORFs 2 to 4, TGBp1 to 3, encode MPs for cell-to-cell movement (Lin et al., 2004; Lin et al., 2006). ORF 5 encodes a 25 kDa viral capsid protein (CP) required for virion assembly, symptom development, and movement in plant cells (Lan et al., 2010; Hung et al., 2014a; Hung et al., 2014b). Infected protoplasts and plants show two major subgenomic RNAs, 2 and 1 kb long (Tsai et al., 1999; Yeh et al., 1999).
Several host factors positively or negatively regulate the infection cycle of BaMV. The chloroplast phosphoglycerate kinase (Cheng et al., 2013a), heat-shock protein 90 (Huang et al., 2012), a glutathione transferase NbGSTU4 (Chen et al., 2013), an exoribonuclease NbXRN4 (Lee et al., 2015), and a Rab-GTPase NbRABG3f (Huang et al., 2016) are involved in assisting viral RNA replication. A Ser/Thr kinase-like protein NbSTKL (Cheng et al., 2013b) and a Rab-GTPase activation protein NbRabGAP1 (Huang et al., 2013) support viral movement. By contrast, a cytoplasmic form of glyceraldehyde 3-phosphate dehydrogenase (Prasanth et al., 2011) and a putative methyltransferase (Cheng et al., 2009) suppress BaMV RNA replication by interacting with the 3′ UTR and replicase, respectively.
A group of proteins on the plasma membrane containing six transmembrane domains termed plasma membrane intrinsic proteins (PIPs) play roles in regulating the diffusion of water and small uncharged solutes (Agre et al., 1998; Schäffner, 1998; Chaumont et al., 2000; Hachez et al., 2013). Some PIPs can even transport hydrogen peroxide in response to stresses (Jang et al., 2012). A new class of PIPs, without any cross-membrane domain and associated with the plasma membrane via myristoylation, bind Ca2+, Mg2+, and Cu2+ and are called plasma membrane-associated cation-binding proteins (PCaPs) (Ide et al., 2007; Nagasaki-Takeuchi et al., 2008). Arabidopsis PCaP1 incorporates [3H]myristic acid during in vitro transcription and translation and interacts with phosphatidylinositol phosphates (PtdInsPs). Furthermore, AtPCaP1 could bind calmodulin in a Ca2+-dependent manner (Nagasaki et al., 2008). Hence, AtPCaP1 could be involved in intracellular signaling via interaction with PtdInsPs and calmodulin (Kato et al., 2010).
The 25 kDa protein AtPCaP1 is also called microtubule-destabilizing protein 25 (MDP25) and was found to regulate the elongation of hypocotyl cells by destabilizing cortical microtubules dependent on calcium (Li et al., 2011). When cytoplasmic calcium increases, MDP25 binds directly to and destabilizes microtubules to enhance its depolymerization (Qin et al., 2012). In addition, AtPCaP1 can interact with the movement-required novel protein P3N-PIPO of Turnip mosaic virus (TuMV) and promote viral cell-to-cell movement (Vijayapalani et al., 2012). Disrupted expression of AtPCaP1 reduced the accumulation and cell-to-cell movement of TuMV, which led to enhanced plant resistance.
In this study, we investigated one gene with upregulated expression from a previous study of BaMV inoculation (Cheng et al., 2010) that may play a negative role in regulating the accumulation of BaMV. This gene, designated NbPCaP1L, is an ortholog of PCaP1 in N. benthamiana. BaMV accumulation was increased when NbCaP1L expression was knocked down but was decreased when NbCaP1L was transiently overexpressed. Further analysis indicated that NbCaP1L acts on the movement of BaMV. The mechanism of this regulation is discussed.
Materials and methods
Viruses and plants
BaMV, PVX, and Cucumber mosaic virus (CMV) were used for inoculation. N. benthamiana were grown at 28°C in a growth room with a 16 h light/8 h dark cycle.
Virus particle purification
The infected leaves were homogenized with extraction buffer of 0.5 M borate at pH 9.0, 1 mM EDTA, and 0.1% β-mercaptoethanol, in a 1 g/2 ml ratio, filtrated through a miracloth, and centrifuged at 12000×g at 4°C for 10 min. The supernatant was stirred at 4℃ for 10 min with the addition of 4M K2HPO4 and 2M CaCl2 to 1% and 2%, respectively, and was centrifuged at 12000×g at 4°C for 10 min. The supernatant was stirred at 4℃ for 30 min with the addition of triton X-100 and PEG 6000 to 2% and 6%, respectively, and was centrifuged at 12000×g at 4°C for 10 min. The pellet was resuspended in BE buffer of 50 mM borate at pH 8.0 and 1 mM EDTA, and centrifuged at 14300×g at 4°C for 1 h on a cushion of 20 % sucrose. The virus pellet was resuspended in 10 ml BE buffer and quantification carried out using a spectrophotometer.
Virus-induced gene silencing (VIGS) and virus challenge
Tobacco rattle virus (TRV)-based VIGS was used to knock down the expression of NbPCaP1L. The cDNA fragment ACGT11 derived from NbPCaP1L was obtained from cDNA-AFLP and cloned into pGEM-T Easy vector (Promega, Madison, WI, USA) previously (Cheng et al., 2010). The 118 bp cDNA was isolated by digestion with EcoRI and subcloned into the pTRV2 vector. The resulting plasmid was electroporated into the Agrobacterium tumefaciens C58C1 for knockdown experiments as described previously (Huang et al., 2013). Approximately 500 ng of viral particles was mechanically inoculated into the fourth leaf above the infiltrated leaves at 10 d post-infiltration (dpi). Total proteins were extracted from these virus-inoculated leaves at 1, 3, 5, and 7 dpi with BaMV inoculation and 5 dpi with CMV and PVX inoculation.
Cloning the NbPCaP1L gene
To clone the full-length NbPCaP1L gene, the 3′ primer (5′ CTCGAGTCAGTCG ACAGCTTTTGGTGGTTCCGGT 3′) and 5′ primer (5′ TGCTAGC GGATCCAT GATGGGTTATTGGCAAGC 3′) were used for PCR. The 3′ primer contains the XhoI site (underlined) and the 5′ primer contains the NheI and BamHI sites (underlined) for cloning. Full-length NbPCaP1L was cloned into the pGEM-T Easy vector (Promega). The sequence was verified by sequencing on an IR2 System (LI-COR Biosciences, Lincoln, NE, USA). Finally, the full-length NbPCaP1L gene was subcloned into the pBIN-OFP vector containing orange fluorescent protein (OFP) driven by the CMV 35S promoter (reconstructed from pmKO2-S1; MBL international, Woburn, MA, USA).
Protoplast isolation and inoculation
Protoplasts were isolated from the fourth leaf above N. benthamiana leaves infiltrated with TRV/Luc, containing part of the luciferase gene from nucleotide 110 to 508, and TRV/NbPCaP1L at 10 d post Agro-infiltration. The detailed protocol for protoplast isolation was previously described (Tsai et al., 1999). In brief, approximately 4 g of tissue was digested with 25 ml of enzyme solution at 25°C overnight. Intact mesophyll protoplasts were collected from the interphase of the 0.55 M mannitol-MES solution and the 0.55 M sucrose cushion. Finally, protoplasts were washed and re-suspended in mannitol-MES solution. Approximately 2.5 × 105 protoplasts were inoculated with 1 µg BaMV RNA with 20% polyethyleneglycol-6000. The inoculated protoplasts were incubated at 25°C under constant light. Total protein and RNA were extracted from protoplasts after 24 h of incubation.
Western blot assay
Total protein harvested from inoculated protoplasts or leaves was separated in 12% SDS-PAGE and transferred to nitrocellulose membranes (PROTRAN BA 85 Schleicher & Schnell), which were probed with the rabbit antibodies for BaMV, PVX, or CMV CP, then incubated with IRDye 800-conjugated affinity-purified anti-rabbit IgG antibody (Rockland Immunochemicals, Gilbertsville, PA, USA). The fluorescence density on membranes was determined using LI-COR Odyssey (LI-COR Biosciences).
RNA extraction and real-time quantitative RT-PCR
Inoculated leaves of NbPCaP1L knockdown N. benthamiana were collected and ground. Total RNA was extracted as described (Lin et al., 2007). First-strand cDNA was synthesized with 20 pmole 39d(T) oligo primer and reverse transcriptase (Promega) according to the manufacturer’s protocol. Real-time quantitative PCR was performed in a 20 μl-reaction containing a 1000X dilution of SYBR green I (Cambrex Bio Science Rockland, ME, USA) with primer GT11_5′ (5′- AAGGTTGTTCCAAAATTAAAGC-3′) and GT11_3′ (5′- TTCAATCCTGAAACCTTTGGTCCC-3′) in 0.2-ml PCR tubes. The conditions began with an initial hold at 95°C for 5 min, followed by 30 cycles of 94°C for 30 sec, 56°C for 30 sec and 72°C for 30 sec. The expression of β-actin was amplified with the primer pair actin_5′ (5′ GATGAAGATACTCACAGAAAGA 3′) and actin_3′ (5′ GTGGTTTCATGAATGCCAGCA 3′) as an internal control for normalization.
Detection of BaMV cell-to-cell movement
Approximately 10 d after VIGS infiltrated with TRV/luciferase or TRV/NbPCaP1L, the fourth and fifth leaves above infiltrated leaves of N. benthamiana plants were mechanically inoculated with 7.5 μg pCBG (Lin et al., 2006), the infectious cDNA clone carrying the green fluorescent protein (GFP) gene driven by the BaMV subgenomic promoter. The fluorescence derived from GFP accompanied by BaMV movement was captured by inverted fluorescent microscopy (Olympus IX71) with an excitation wavelength of 460 to 495 nm, an emission wavelength of 510 nm, and a dichromatic mirror at 505 nm at 6 dpi. The area of each lesion was measured using Image J (http://rsbweb.nih.gov/ij/).
Laser scanning confocal microscopy
Plasmids pBIN-OFP (vector only) and pBIN-NbPCaP1L-OFP were transformed into A. tumefaciens C58C1. A. tumefaciens containing pBIN-OFP, pBIN-NbPCaP1L-OFP or pBIN61-HcPro was cultured to OD600=1; and the cells were resuspended in 10 mM MgCl2 with 500 μM acetosyringone for induction after being spun down. The pBIN-OFP or pBIN-NbPCaP1L culture was mixed with that of pBIN61-HcPro in a 1:1 volume ratio and infiltrated into N. benthamiana leaves. The images were taken by laser scanning confocal microscopy (Olympus Fluoview FV1000) with 405 nm and 543 nm laser excitation for cyan fluorescent protein and OFP, respectively, 2 d after infiltration.
Yeast two-hybrid interaction
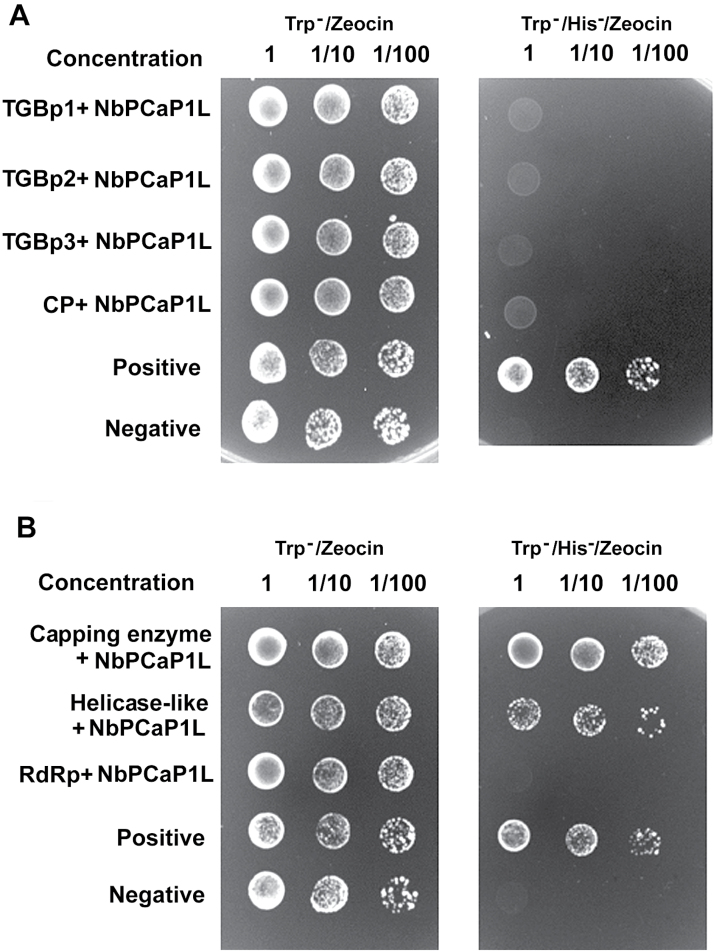
The gene fragments encoding the MP and CP of BaMV were constructed into the prey plasmid pYESTrp2 and designated pYES-TGBp1, -2, -3, and -CP. The replication-related DNA fragments were constructed into the bait plasmid pHybLex/Zeo and designated pLEX-Capping, -RdRp, and -Helicase (Cheng et al., 2009; Lee et al., 2011). The DNA fragment for cloning NbPCaP1L into yeast prey and bait plasmids was amplified with two sets of primers: for the prey plasmid, YES5′PCaP1HindIII (5′ GGGAAGCTTATGATGGGTTATTGGCAAGC 3′) and YES3′PCaP1XhoI (5′ GTCGACTCAGTCGACAGCTTTTG GTGGTTCCGGT 3′), and for the bait plasmid, LEX5′P CaP1EcoRI (5′ GAATTCATGATGGGTTATTGGCAAGC 3′) and LEX3′PCaP1PstI (5′ GCTGCAGTCAGTCG ACAGCTTTTGGTGGTTCCGGT 3′) (restriction enzyme site underlined). The amplified DNAs were cloned into pGEM-T Easy vector (Promega) and verified by sequencing. The full-length NbPCaP1L was then subcloned into the yeast prey plasmid with HindIII and XhoI sites and into the bait plasmid with EcoRI and PstI sites. Saccharomyces cerevisiae strain L40, harboring the bait plasmid (pLEX-Capping, -RdRp, or -Helicase) or prey plasmid (pYES-TGBp1, -2, -3, and -CP), was transformed with the prey plasmid pYES-NbPCaP1L or bait plasmid pLEX-NbPCaP1L, respectively. Positive colonies were grown on Trp-/His-/Zeocin selection agar plates.
Co-immunoprecipitation
N. benthamiana leaves were infiltrated with the Agrobacteria mixture containing the plasmids pKBRepHA21 plus pEpyon-OFP or pKBRepHA21 plus pBIN-NbPCaP1L-OFP. Total proteins were extracted using an extraction buffer of 20 mM Tris–HCl at pH 7.5, 2 mM MgCl2, 300 mM NaCl, 5 mM DTT, and 1% protease inhibitor cocktail (Roche), at 3 dpi. The supernatant was collected after centrifugation at 4000×g at 4°C for 10 min and underwent immunoprecipitation with anti-HA antibody (Sigma-Aldrich, H9658) at 4°C for 4 h. The interacting proteins were incubated with 20 μl protein A magnetic beads (GE Healthcare) at 4°C for 2 h. Finally, the samples were washed, eluted, and underwent western blot analysis with anti-HA (Sigma-Aldrich, H6908), anti-OFP, or anti-BaMV CP antibodies.
Results
Cloning the plasma membrane–associated cation-binding protein 1-like gene NbPCaP1L from N. benthamiana
A cDNA fragment ACGT11 118 bp in length was found upregulated after BaMV inoculation by cDNA-AFLP and was revealed to have an effect on BaMV accumulation in N. benthamiana (Cheng et al., 2010). This 118 bp cDNA fragment had 94% identity to that of PIP from N. tabacum (GenBank accession Y08609.1). This PIP was defined as a new family of PIP because it lacks a transmembrane domain, whereas conventional PIPs have six membrane-spanning domains with cytosolic N- and C-termini (Lin et al., 2011; Zhao et al., 2013). In a further analysis, PIP from N. tabacum showed 57% identity to the PCaP1 of Arabidopsis thaliana (GenBank accession NM_118145) (see Supplementary Fig. S1 at JXB online). The identity of this PIP from N. tabacum is more similar to PCaP1 than conventional PIP.
To clone the full-length ACGT11 from N. benthamiana, we designed primers according to the conserved region among the PCaP1 orthologs from different species (Supplementary Fig. S1) and the nucleotide sequence of PIP from N. tabacum. The coding sequence of the PCaP1-like gene from N. benthamiana, designated NbPCaP1L (GenBank accession MF346700), is 711 nucleotides long encoding a 236-amino acid polypeptide and has 89% and 88% nucleotide and amino acid identity to NtPIP (Supplementary Fig. S1).
Reducing the expression of NbPCaP1L increases BaMV accumulation in N. benthamiana plants but not protoplasts
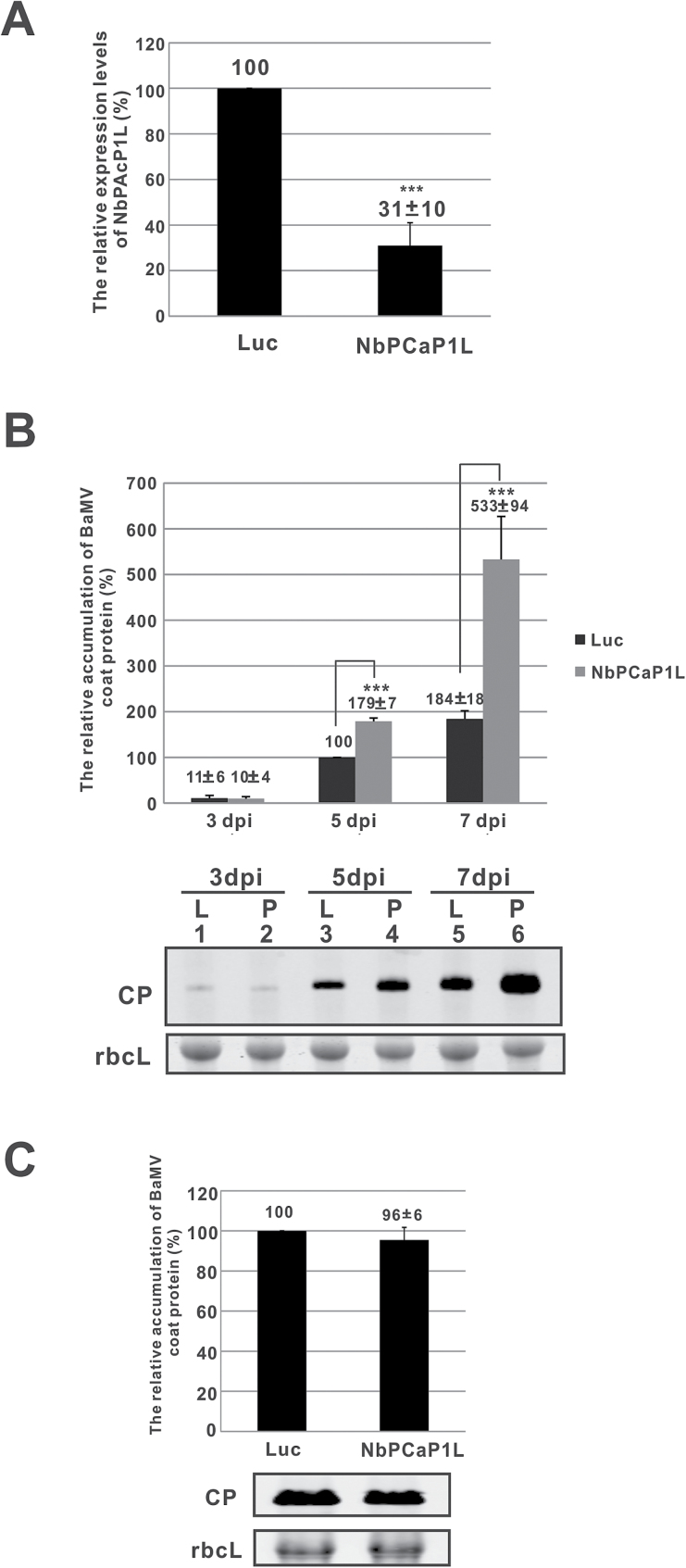
To characterize the role of NbPCaP1L in BaMV infection, we used TRV-based VIGS to knock down the expression of NbPCaP1L and inoculate BaMV. Knockdown of NbPCaP1L expression by 70% compared with control plants, Luc-knockdown plants, as determined by real-time qRT-PCR (Fig. 1A), conferred no morphologic alterations (Supplementary Fig. S2). After normalization with the large subunit of RuBisCO stained with Coomassie blue, western blot analysis revealed that BaMV CP levels increased in NbPCaP1L knockdown plants to 180% that of control plants at 5 dpi but not at earlier infection times (Fig. 1B). Furthermore, BaMV levels were strikingly increased up to 2.9-fold in NbPCaP1L knockdown plants at 7 dpi (Fig. 1B). Hence, NbPCaP1L may suppress the accumulation of BaMV CP in plants, especially during late infection.
Fig. 1.
The relative accumulation of BaMV coat protein in NbPCaP1L knockdown plants and protoplasts. (A) The expression of NbPCaP1L mRNA was determined by real-time quantitative RT-PCR in Luc and NbPCaP1L knockdown leaves. (B) Western blot analysis of coat protein (CP) accumulation on the inoculated leaves. The accumulation of control plants was set to 100%. Data are mean ± standard deviation relative levels of BaMV CP from at least three independent experiments with at least three plants in each experiment. Representative western blot results of CP levels with triplicate results and rbcL as a control. (C) Western blot analysis of CP accumulation. The accumulation in control protoplasts at 24 h post inoculation (hpi) was set to 100%. Representative results are shown. Data are mean ± standard deviation relative levels of BaMV CP from at least three independent experiments. Luc, luciferase knockdown control plants or protoplasts; NbPCaP1L, NbPCaP1L knockdown plants or protoplasts; CP, BaMV coat protein; rbcL, RuBisCO large subunit (the loading control for normalization). **P<0.01, ***P<0.001 by Student’s t-test.
To investigate whether increased BaMV CP accumulation in NbPCaP1L knockdown plants was caused by enhanced viral RNA replication or viral movement in plants, we prepared NbPCaP1L knockdown protoplasts to exclude the movement effect. NbPCaP1L knockdown and control protoplasts showed no difference in BaMV accumulation (Fig. 1C). Hence, reducing the expression of NbPCaP1L did not impede the replication process of BaMV RNA. NbPCaP1L could be involved in regulating BaMV cell-to-cell movement. To determine whether reduced NbPCaP1L expression could affect other viruses, we inoculated NbPCaP1L knockdown plants with PVX and CMV. NbPCaP1L knockdown and control plants did not differ in CP accumulation of these viruses at 5 dpi (Supplementary Fig. S3). The effect of NbPCaP1L on viral accumulation may be specific to BaMV but not PVX or CMV.
NbPCaP1L regulates BaMV cell-to-cell movement
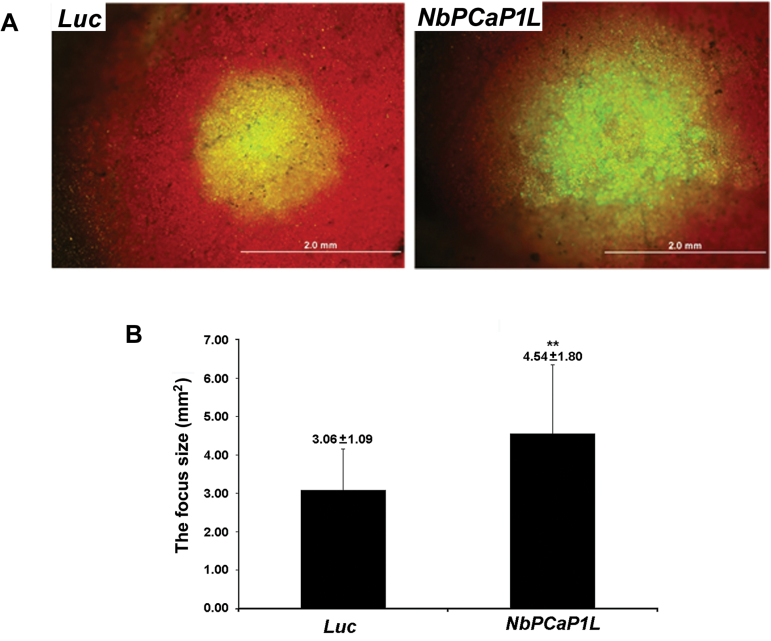
As the results of the NbPCaP1L knockdown experiments indicated that NbPCaP1L had no effect on BaMV accumulation in protoplasts but a marked effect in plants, NbPCaP1L may be involved in BaMV cell-to-cell movement. To test this, NbPCaP1L- and Luc-knockdown plants were inoculated with the full-length infectious BaMV cDNA plasmid pCBG expressing GFP with the BaMV subgenomic promoter (Lin et al., 2004), with measurements taken at 6 dpi (Fig. 2A). The mean GFP focus area in Luc and NbPCaP1L knockdown plants was 3.06 ± 1.09 and 4.54 ± 1.80 mm2, respectively (Fig. 2B). Reducing the expression of NbPCaP1L in N. benthamiana plants made them more accessible to BaMV infection, with more and larger foci than in control plants.
Fig. 2.
Cell-to-cell movement of BaMV in Luc and NbPCaP1L knockdown plants. (A) Fluorescent microscopy of the area of fluorescent foci in inoculated leaves of luciferase knockdown control (Luc) and NbPCaP1L knockdown (NbPCaP1L) plants after inoculation with the BaMV infectious plasmid carrying green fluorescent protein (GFP). Scale bar, 2 mm. (B) Statistical analysis. Data are mean ± standard deviation of 19 and 29 foci from Luc and NbPCaP1L knockdown plants, respectively. **P<0.01 by Student’s t-test.
BaMV accumulation is reduced in NbPCaP1L- expressing leaves
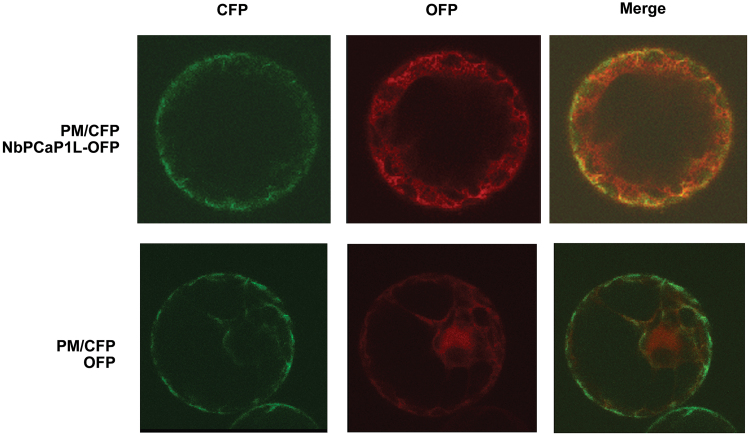
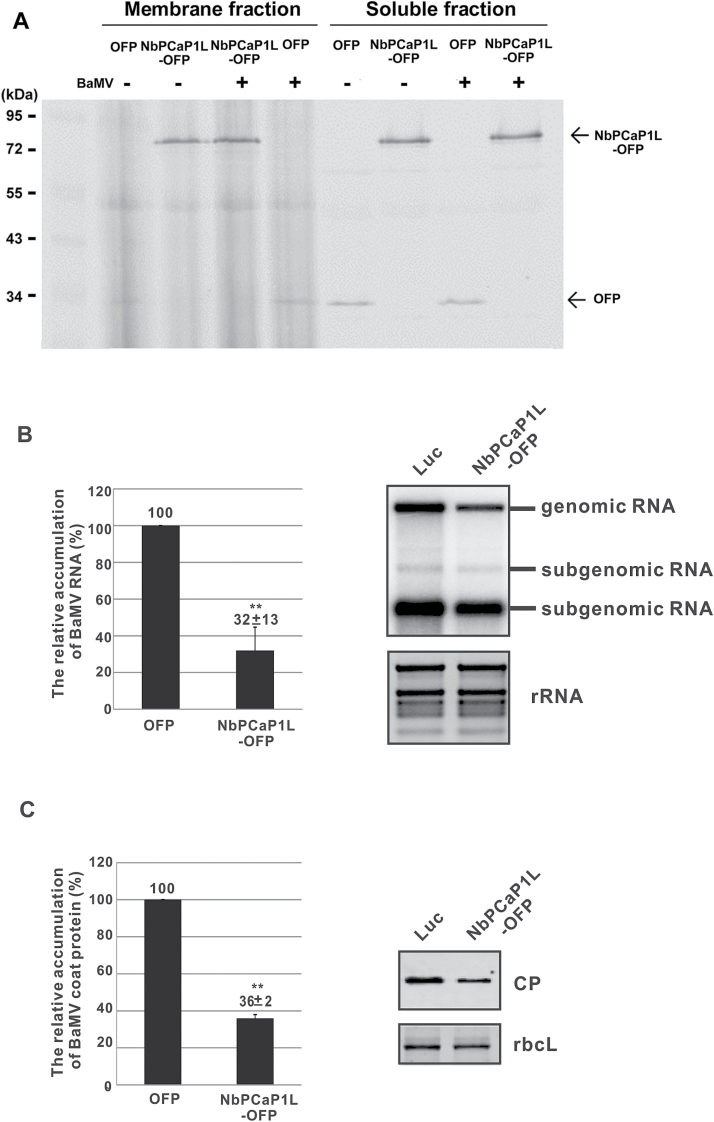
To visualize the subcellular localization of NbPCaP1L protein in cells, we fused OFP at the C-terminus of NbPCaP1L and transiently expressed it in N. benthamiana plants. NbPCaP1L-OFP was clearly present in both the plasma membrane, as it co-localized with a plasma membrane marker, and cytoplasm in protoplasts (Fig. 3). Further analysis of protein extracts after fractionation revealed that NbPCaP1L-OFP was equally distributed in the membrane and soluble fractions. This distribution profile of NbPCaP1L-OFP in N. benthamiana cells in the membrane and soluble fraction was not changed after BaMV infection (Fig. 4A). The subcellular localization of AtPCaP1 was reported to target the cytoskeleton, cell junction, plasmodesmata, and cell membrane via lipid-anchoring (Nühse et al., 2003; Marmagne et al., 2004; Ide et al., 2007; Nagasaki et al., 2008; Li et al., 2011; Vijayapalani et al., 2012). AtPCaP1 shuttled from the plasma membrane to cytoplasm with increased calcium levels (Kato et al., 2010; Li et al., 2011). The subcellular localization of NbPCaP1L-OFP in both the cytoplasm and plasma membrane was therefore similar to that of AtPCaP1.
Fig. 3.
The localization at the plasma membrane (PM) and cytoplasm of NbPCaP1L with OFP fused at the C-terminus and transiently expressed in N. benthamiana protoplasts. Cyan fluorescence protein (CFP) fused with a plasma membrane aquaporin AtPIP2A is transiently expressed in protoplasts and is used as the PM marker.
Fig. 4.
Effect of expression of NbPCaP1L on BaMV infection. (A) Western blot analysis of the fractionation of transiently expressed NbPCaP1L-OFP and OFP alone. Relative accumulation of BaMV CP (B) and RNAs (C) at 5 d post-inoculation (dpi) on inoculated leaves, with NbPCaP1L-OFP or OFP only (as a control) transiently expressed on the same leaves at 3 dpi. Data are mean ± standard error of at least three independent experiments. **P<0.01 by Student’s t-test.
To validate whether NbPCaP1L plays a negative role in BaMV infection, we inoculated NbPCaP1L-OFP transiently expressing leaves with BaMV. The BaMV RNA and CP levels were significantly reduced to 32% and 36% of that of control plants, respectively (Fig. 4B, C). Hence, NbPCaP1L negatively regulates BaMV movement.
NbPCaP1L interacts with BaMV replicase
To examine whether the host protein NbPCaP1L interacts with any viral-encoded proteins, we used the yeast two-hybrid system. The genes encoding the MPs and CP of BaMV were constructed in a yeast prey plasmid; full-length NbPCaP1L was cloned into the bait plasmid and transformed into yeast containing each of the viral-encoding genes, TGBp1, 2, 3, and CP. No yeast could grow on the selective medium (Trp-/His-/Zeocin) except the positive control yeast (Fig. 5A).
Fig. 5.
Interaction of NbPCaP1L with BaMV-encoded polypeptides in yeast cells. Yeast strain L40 co-transformed with the prep plasmid, pYES-TGBp1, -2, -3, or -CP, and bait plasmid pLEX-NbPCaP1L in (A) or prey plasmid pYES-NbPCaP1L and bait plasmid, pLEX-Capping, -RdRp, or -Helicase in (B). Positive control, yeast containing pLEX-Fos2 and pYES-Jun; negative control, yeast containing pHybLex/Zeo and pYESTrp2. Positive colonies were grown on Trp-/His-/Zeocin selection agar plates. The yeast concentrations with the dilution factor are indicated at the top of each panel.
Another set of constructs containing each domain of the BaMV-encoded replicase, namely the capping, helicase-like, and RdRp domains, was cloned into the bait plasmid of the yeast two-hybrid system (Cheng et al., 2009; Lee et al., 2011). NbPCaP1L was then constructed into the prey plasmid for testing of the interaction with BaMV replicase. The capping and helicase-like domains could interact with NbPCaP1L (Fig. 5B). Overall, these results suggest that the host protein NbPCaP1L could interact with the N-terminal portion of BaMV replicase, that is the capping and helicase-like domains, in the yeast cells.
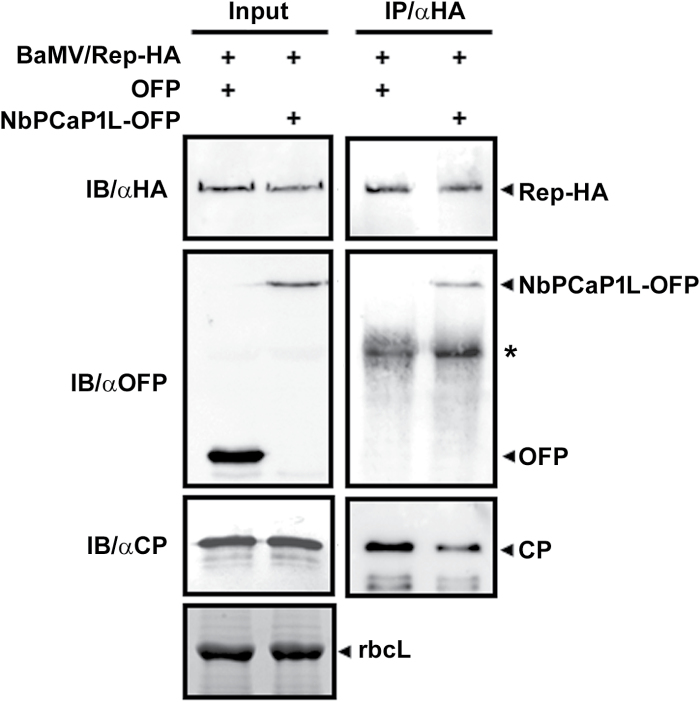
To validate the results of the yeast two-hybrid finding that NbPCaP1L interacts with BaMV replicase, we used co-immunoprecipitation. BaMV modified with a HA-tag at the C-terminus of the replicase, BaMV/Rep-HA, was agro-infiltrated onto transiently expressed OFP or NbPCaP1L-OFP leaves. Total proteins were immunoprecipitated with anti-HA antibody. NbPCaP1L-OFP could interact with replicase-HA but not OFP alone (Fig. 6).
Fig. 6.
Co-immunoprecipitation (IP) assay to verify the interaction between BaMV replication protein and NbPCaP1L. Total proteins (input) were extracted from N. benthamiana leaves co-infiltrated with Agrobacteria containing the infectious BaMV/Rep-HA and OFP vector or NbPCaP1L-OFP. Equal amounts of input proteins were used for immunoblotting (IB) with antibodies against HA, OFP, and BaMV CP as indicated. rbcL, RuBisCo large subunit is used as the loading control. After IP with anti-HA antibody, the co-IP of Rep-HA, NbPCaP1L-OFP, and CP was detected using corresponding antibodies for IB.
Discussion
The process of viral movement is critical for plant viruses to establish a successful infection, even with effective replication in cells. Here we investigated whether one host gene from N. benthamiana, a plasma membrane-associated cation-binding protein 1-like protein designated NbPCaP1L, which shows upregulated expression after BaMV inoculation, was involved in regulating virus movement.
Plants have evolved a few different strategies, such as post-transcriptional gene silencing (Hammond et al., 2001) and hypersensitive response (Baurès et al., 2008), to protect themselves against pathogen infection. However, pathogens can also develop a counter-strike mechanism to break down the host defense systems, such as virus-encoded silencing suppressors. The host could therefore have evolved some other unidentified defense mechanisms to resist the pathogen infection. These types of offense/defense strategies must be identified to gain knowledge into plant protection, especially for important crops.
These unidentified defense genes might express their genes differentially during pathogen infection. We previously used cDNA-AFLP to isolate the differentially expressed genes from N. benthamiana plants after BaMV infection (Cheng et al., 2010). NbPCaP1L was one of the potential upregulated defense genes based on our results in this study, which could act on viral movement.
The movement complex of potexviruses, including PVX and BaMV, was proposed to assemble on the perinuclear endoplasmic reticulum-derived membrane-bound body (Tian et al., 2004; Verchot-Lubicz et al., 2010). The complex, comprising the membrane-associated TGBp2 and TGBp3 translated at the endoplasmic reticulum (Verchot-Lubicz et al., 2010) and ribonucleoparticles containing viral RNA, CP, TGBp1, and replicase (Tian et al., 2004; Lee et al., 2011; Wu et al., 2011; Park et al., 2013), was transported to the endoplasmic reticulum network directly or to actin filaments associated with membrane vesicles toward the PD (Tian et al., 2004). The cellular vesicle formation factor Rab-GTPase activation protein, (NbRabGAP1, regulates BaMV cell-to-cell and systemic movement (Huang et al., 2013), so the membrane vesicle could be involved in trafficking the BaMV movement complex.
The association of viral replicase in the movement complex was first proposed by observing the differential time required for the spread of TMV from primary inoculated cells to secondary cells, at 18 to 20 h post inoculation, and secondary cells to tertiary cells, as 2 to 4 h post inoculation (Kawakami et al., 2004). Viral RNA in primary inoculated cells was suggested to need more time to set up the movement complex for trafficking to secondary cells, whereas the movement complex containing the replicase could initiate replication on entry to secondary cells and speed up the assembly of the movement complex to tertiary cells. The interaction between BaMV CP and replicase is needed for efficient viral movement. Mutations in CP that fail to interact with viral replicase diminish cell-to-cell movement in plants (Lee et al., 2011). The anti-TGBp3 immunopurified movement complex from BaMV-infected N. benthamiana harbors endogenous RdRp activity (Chou et al., 2013), so BaMV replicase may be included in the movement complex.
The host protein NbPCaP1L could interact with the replicase of BaMV (Figs 5 and 6), so the mechanism of NbPCaP1L retarding the movement of BaMV could involve competing for replicase availability with the competent viral movement complex. Although we did not observe the interaction between NbPCaP1L with viral membrane-associated MPs TGBp2 and TGBp3 in the yeast two-hybrid system (Fig. 5), which might not be a good way to detect this interaction, NbPCaP1L constraining viral movement via interaction with MPs cannot be ruled out. We did however demonstrate that the interaction of NbPCaP1L with BaMV replicase could be the major effector for restricting virus movement.
Arabidopsis PCaP1 could assist the viral movement of TuMV via interaction with the potyviral protein P3N-PIPO of the viral movement complex to target the plasma membrane and localize to the plasmadesmata (Vijayapalani et al., 2012). By contrast, the homolog protein NbPCaP1L in N. benthamiana for BaMV may play a different role in retarding viral movement by interacting with the replication protein in the movement complex. NbPCaP1L is a membrane-associated protein that harbors a novel activity by interacting with the viral RdRp and trapping the viral movement complex to restrict viral cell-to-cell movement.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Sequence alignment and the expression profile of NbPCaP1L after BaMV inoculation.
Fig. S2. The morphology of control and NbPCaP1L knockdown plants.
Fig. S3. The relative accumulation of viral coat protein (CP) in NbPCaP1L-knockdown and control plants.
Supplementary Material
Acknowledgements
We are grateful for help from the Bioimage core lab of the Graduate Institute of Biotechnology at National Chung Hsing University for providing confocal laser scanning microscopy. This research was supported by Ministry of Science and Technology of Taiwan (Grant 99-2313-B-005-019-MY3).
Glossary
Abbreviations:
- BaMV
bamboo mosaic virus
- CMV
cucumber mosaic virus
- CP
capsid protein
- dpi
days post infiltration
- GFP
green fluorescent protein
- MPs
movement proteins
- OFP
orange fluorescent protein
- PCaPs
plasma membrane-associated cation-binding proteins
- PD
plasmodesmata
- PIPs
plasma membrane intrinsic proteins
- PMTV
potato mop-top virus
- PVX
potato virus X
- RdRp
RNA-dependent RNA polymerase
- RNP
ribonucleoprotein complex
- TGB
triple gene block
- TMV
tobacco mosaic virus
- TRV
tobacco rattle virus
- TuMV
Turnip mosaic virus
- VIGS
virus-induced gene silencing.
References
- Agre P, Bonhivers M, Borgnia MJ. 1998. The aquaporins, blueprints for cellular plumbing systems. The Journal of Biological Chemistry 273, 14659–14662. [DOI] [PubMed] [Google Scholar]
- Avisar D, Prokhnevsky AI, Dolja VV. 2008. Class VIII myosins are required for plasmodesmatal localization of a closterovirus Hsp70 homolog. Journal of Virology 82, 2836–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baurès I, Candresse T, Leveau A, Bendahmane A, Sturbois B. 2008. The Rx gene confers resistance to a range of potexviruses in transgenic Nicotiana plants. Molecular Plant-Microbe Interactions 21, 1154–1164. [DOI] [PubMed] [Google Scholar]
- Benitez-Alfonso Y, Faulkner C, Ritzenthaler C, Maule AJ. 2010. Plasmodesmata: gateways to local and systemic virus infection. Molecular Plant-Microbe Interactions 23, 1403–1412. [DOI] [PubMed] [Google Scholar]
- Chaumont F, Barrieu F, Jung R, Chrispeels MJ. 2000. Plasma membrane intrinsic proteins from maize cluster in two sequence subgroups with differential aquaporin activity. Plant Physiology 122, 1025–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IH, Chiu MH, Cheng SF, Hsu YH, Tsai CH. 2013. The glutathione transferase of Nicotiana benthamiana NbGSTU4 plays a role in regulating the early replication of Bamboo mosaic virus. New Phytologist 199, 749–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IH, Chou WJ, Lee PY, Hsu YH, Tsai CH. 2005. The AAUAAA motif of Bamboo mosaic virus RNA is involved in minus-strand RNA synthesis and plus-strand RNA polyadenylation. Journal of Virology 79, 14555–14561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CW, Hsiao YY, Wu HC et al. . 2009. Suppression of bamboo mosaic virus accumulation by a putative methyltransferase in Nicotiana benthamiana. Journal of Virology 83, 5796–5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SF, Huang YP, Chen LH, Hsu YH, Tsai CH. 2013. Chloroplast phosphoglycerate kinase is involved in the targeting of Bamboo mosaic virus to chloroplasts in Nicotiana benthamiana plants. Plant Physiology 163, 1598–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SF, Huang YP, Wu ZR, Hu CC, Hsu YH, Tsai CH. 2010. Identification of differentially expressed genes induced by Bamboo mosaic virus infection in Nicotiana benthamiana by cDNA-amplified fragment length polymorphism. BMC Plant Biology 10, 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SF, Tsai MS, Huang CL, Huang YP, Chen IH, Lin NS, Hsu YH, Tsai CH, Cheng CP. 2013. Ser/Thr kinase-like protein of Nicotiana benthamiana is involved in the cell-to-cell movement of Bamboo mosaic virus. PLoS ONE 8, e62907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou YL, Hung YJ, Tseng YH et al. . 2013. The stable association of virion with the triple-gene-block protein 3-based complex of Bamboo mosaic virus. PLoS Pathogens 9, e1003405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachez C, Besserer A, Chevalier AS, Chaumont F. 2013. Insights into plant plasma membrane aquaporin trafficking. Trends in Plant Science 18, 344–352. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Caudy AA, Hannon GJ. 2001. Post-transcriptional gene silencing by double-stranded RNA. Nature Reviews. Genetics 2, 110–119. [DOI] [PubMed] [Google Scholar]
- Hofmann C, Sambade A, Heinlein M. 2007. Plasmodesmata and intercellular transport of viral RNA. Biochemical Society Transactions 35, 142–145. [DOI] [PubMed] [Google Scholar]
- Huang CY, Huang YL, Meng M, Hsu YH, Tsai CH. 2001. Sequences at the 3’ untranslated region of Bamboo mosaic potexvirus RNA interact with the viral RNA-dependent RNA polymerase. Journal of Virology 75, 2818–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YL, Han YT, Chang YT, Hsu YH, Meng M. 2004. Critical residues for GTP methylation and formation of the covalent m7GMP-enzyme intermediate in the capping enzyme domain of Bamboo mosaic virus. Journal of Virology 78, 1271–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YL, Hsu YH, Han YT, Meng M. 2005. mRNA guanylation catalyzed by the S-adenosylmethionine-dependent guanylyltransferase of Bamboo mosaic virus. The Journal of Biological Chemistry 280, 13153–13162. [DOI] [PubMed] [Google Scholar]
- Huang YP, Chen JS, Hsu YH, Tsai CH. 2013. A putative Rab-GTPase activation protein from Nicotiana benthamiana is important for Bamboo mosaic virus intercellular movement. Virology 447, 292–299. [DOI] [PubMed] [Google Scholar]
- Huang YP, Jhuo JH, Tsai MS, Tsai CH, Chen HC, Lin NS, Hsu YH, Cheng CP. 2016. NbRABG3f, a member of Rab GTPase, is involved in Bamboo mosaic virus infection in Nicotiana benthamiana. Molecular Plant Pathology 17, 714–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YW, Hu CC, Liou MR, Chang BY, Tsai CH, Meng M, Lin NS, Hsu YH. 2012. Hsp90 interacts specifically with viral RNA and differentially regulates replication initiation of Bamboo mosaic virus and associated satellite RNA. PLoS Pathogens 8, e1002726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CJ, Hu CC, Lin NS, Lee YC, Meng M, Tsai CH, Hsu YH. 2014. Two key arginine residues in the coat protein of Bamboo mosaic virus differentially affect the accumulation of viral genomic and subgenomic RNAs. Molecular Plant Pathology 15, 196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CJ, Huang YW, Liou MR, Lee YC, Lin NS, Meng M, Tsai CH, Hu CC, Hsu YH. 2014. Phosphorylation of coat protein by protein kinase CK2 regulates cell-to-cell movement of Bamboo mosaic virus through modulating RNA binding. Molecular Plant-Microbe Interactions 27, 1211–1225. [DOI] [PubMed] [Google Scholar]
- Ide Y, Nagasaki N, Tomioka R, Suito M, Kamiya T, Maeshima M. 2007. Molecular properties of a novel, hydrophilic cation-binding protein associated with the plasma membrane. Journal of Experimental Botany 58, 1173–1183. [DOI] [PubMed] [Google Scholar]
- Jang JY, Rhee JY, Chung GC, Kang H. 2012. Aquaporin as a membrane transporter of hydrogen peroxide in plant response to stresses. Plant Signaling & Behavior 7, 1180–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Nagasaki-Takeuchi N, Ide Y, Tomioka R, Maeshima M. 2010. PCaPs, possible regulators of PtdInsP signals on plasma membrane. Plant Signaling & Behavior 5, 848–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami S, Watanabe Y, Beachy RN. 2004. Tobacco mosaic virus infection spreads cell to cell as intact replication complexes. Proceedings of the National Academy of Sciences, USA 101, 6291–6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan P, Yeh WB, Tsai CW, Lin NS. 2010. A unique glycine-rich motif at the N-terminal region of Bamboo mosaic virus coat protein is required for symptom expression. Molecular Plant-Microbe Interactions 23, 903–914. [DOI] [PubMed] [Google Scholar]
- Lee CC, Ho YN, Hu RH, Yen YT, Wang ZC, Lee YC, Hsu YH, Meng M. 2011. The interaction between Bamboo mosaic virus replication protein and coat protein is critical for virus movement in plant hosts. Journal of Virology 85, 12022–12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CC, Lin TL, Lin JW, Han YT, Huang YT, Hsu YH, Meng M. 2015. Promotion of Bamboo mosaic virus accumulation in Nicotiana benthamiana by 5’→3’ exonuclease NbXRN4. Frontiers in Microbiology 6, 1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang X, Qin T et al. . 2011. MDP25, a novel calcium regulatory protein, mediates hypocotyl cell elongation by destabilizing cortical microtubules in Arabidopsis. The Plant Cell 23, 4411–4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YI, Chen YJ, Hsu YH, Meng M. 2001a. Characterization of the AdoMet-dependent guanylyltransferase activity that is associated with the N terminus of Bamboo mosaic virus replicase. Journal of Virology 75, 782–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YI, Cheng YM, Huang YL, Tsai CH, Hsu YH, Meng M. 1998. Identification and characterization of the Escherichia coli-expressed RNA-dependent RNA polymerase of Bamboo mosaic virus. Journal of Virology 72, 10093–10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YI, Shih TW, Hsu YH, Han YT, Huang YL, Meng M. 2001b. The helicase-like domain of plant potexvirus replicase participates in formation of RNA 5’ cap structure by exhibiting RNA 5’-triphosphatase activity. Journal of Virology 75, 12114–12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JW, Ding MP, Hsu YH, Tsai CH. 2007. Chloroplast phosphoglycerate kinase, a gluconeogenetic enzyme, is required for efficient accumulation of Bamboo mosaic virus. Nucleic Acids Research 35, 424–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Luo Z, Yan F, Lu Y, Zheng H, Chen J. 2011. Interaction between potyvirus P3 and ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) of host plants. Virus Genes 43, 90–92. [DOI] [PubMed] [Google Scholar]
- Lin MK, Chang BY, Liao JT, Lin NS, Hsu YH. 2004. Arg-16 and Arg-21 in the N-terminal region of the triple-gene-block protein 1 of Bamboo mosaic virus are essential for virus movement. The Journal of General Virology 85, 251–259. [DOI] [PubMed] [Google Scholar]
- Lin MK, Hu CC, Lin NS, Chang BY, Hsu YH. 2006. Movement of potexviruses requires species-specific interactions among the cognate triple gene block proteins, as revealed by a trans-complementation assay based on the Bamboo mosaic virus satellite RNA-mediated expression system. The Journal of General Virology 87, 1357–1367. [DOI] [PubMed] [Google Scholar]
- Lin NS, Lin FZ, Huang TY, Hsu YH. 1992. Genome properties of Bamboo mosaic virus. Phytopathology 82, 731–734. [Google Scholar]
- Liu C, Nelson RS. 2013. The cell biology of Tobacco mosaic virus replication and movement. Frontiers in Plant Science 4, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmagne A, Rouet MA, Ferro M, Rolland N, Alcon C, Joyard J, Garin J, Barbier-Brygoo H, Ephritikhine G. 2004. Identification of new intrinsic proteins in Arabidopsis plasma membrane proteome. Molecular & Cellular Proteomics 3, 675–691. [DOI] [PubMed] [Google Scholar]
- Nagasaki-Takeuchi N, Miyano M, Maeshima M. 2008. A plasma membrane-associated protein of Arabidopsis thaliana AtPCaP1 binds copper ions and changes its higher order structure. Journal of Biochemistry 144, 487–497. [DOI] [PubMed] [Google Scholar]
- Nagasaki N, Tomioka R, Maeshima M. 2008. A hydrophilic cation-binding protein of Arabidopsis thaliana, AtPCaP1, is localized to plasma membrane via N-myristoylation and interacts with calmodulin and the phosphatidylinositol phosphates PtdIns(3,4,5)P(3) and PtdIns(3,5)P(2). The FEBS Journal 275, 2267–2282. [DOI] [PubMed] [Google Scholar]
- Niehl A, Heinlein M. 2011. Cellular pathways for viral transport through plasmodesmata. Protoplasma 248, 75–99. [DOI] [PubMed] [Google Scholar]
- Nühse TS, Boller T, Peck SC. 2003. A plasma membrane syntaxin is phosphorylated in response to the bacterial elicitor flagellin. The Journal of Biological Chemistry 278, 45248–45254. [DOI] [PubMed] [Google Scholar]
- Park MR, Seo JK, Kim KH. 2013. Viral and nonviral elements in potexvirus replication and movement and in antiviral responses. Advances in Virus Research 87, 75–112. [DOI] [PubMed] [Google Scholar]
- Peña EJ, Heinlein M. 2012. RNA transport during TMV cell-to-cell movement. Frontiers in Plant Science 3, 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KR, Huang YW, Liou MR, Wang RY, Hu CC, Tsai CH, Meng M, Lin NS, Hsu YH. 2011. Glyceraldehyde 3-phosphate dehydrogenase negatively regulates the replication of Bamboo mosaic virus and its associated satellite RNA. Journal of Virology 85, 8829–8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhnevsky AI, Peremyslov VV, Dolja VV. 2005. Actin cytoskeleton is involved in targeting of a viral Hsp70 homolog to the cell periphery. Journal of Virology 79, 14421–14428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin T, Li J, Yuan M, Mao T. 2012. Characterization of the role of calcium in regulating the microtubule-destabilizing activity of MDP25. Plant Signaling & Behavior 7, 708–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäffner AR. 1998. Aquaporin function, structure, and expression: are there more surprises to surface in water relations?Planta 204, 131–139. [DOI] [PubMed] [Google Scholar]
- Schoelz JE, Harries PA, Nelson RS. 2011. Intracellular transport of plant viruses: finding the door out of the cell. Molecular Plant 4, 813–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholthof HB. 2005. Plant virus transport: motions of functional equivalence. Trends in Plant Science 10, 376–382. [DOI] [PubMed] [Google Scholar]
- Taliansky M, Torrance L, Kalinina NO. 2008. Role of plant virus movement proteins. Methods in Molecular Biology 451, 33–54. [DOI] [PubMed] [Google Scholar]
- Tian C, Wan P, Sun S, Li J, Chen M. 2004. Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Molecular Biology 54, 519–532. [DOI] [PubMed] [Google Scholar]
- Tsai CH, Cheng CP, Peng CW, Lin BY, Lin NS, Hsu YH. 1999. Sufficient length of a poly(A) tail for the formation of a potential pseudoknot is required for efficient replication of Bamboo mosaic potexvirus RNA. Journal of Virology 73, 2703–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot-Lubicz J, Torrance L, Solovyev AG, Morozov SY, Jackson AO, Gilmer D. 2010. Varied movement strategies employed by triple gene block-encoding viruses. Molecular Plant-Microbe Interactions 23, 1231–1247. [DOI] [PubMed] [Google Scholar]
- Vijayapalani P, Maeshima M, Nagasaki-Takekuchi N, Miller WA. 2012. Interaction of the trans-frame potyvirus protein P3N-PIPO with host protein PCaP1 facilitates potyvirus movement. PLoS Pathogens 8, e1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Lee SC, Wang CW. 2011. Viral protein targeting to the cortical endoplasmic reticulum is required for cell-cell spreading in plants. The Journal of Cell Biology 193, 521–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh TY, Lin BY, Chang YC, Hsu YH, Lin NS. 1999. A defective RNA associated with Bamboo mosaic virus and the possible common mechanisms for RNA recombination in potexviruses. Virus Genes 18, 121–128. [DOI] [PubMed] [Google Scholar]
- Zhao J, Liu Q, Zhang H, Jia Q, Hong Y, Liu Y. 2013. The rubisco small subunit is involved in tobamovirus movement and Tm-2²-mediated extreme resistance. Plant Physiology 161, 374–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.