Summary
This study describes Zika virus seroprevalence of 6.2% in Senegal and Nigeria over a 20-year period, demonstrating previously unrecognized persistence in human populations and concurrent infection with HIV/malaria.
Keywords: Zika virus, Seroprevalence, HIV, Malaria, Senegal, Nigeria, Africa.
Abstract
First identified in 1947 in Uganda, Zika virus (ZIKV) has remained largely unstudied until the recent outbreak in Latin America. This study aimed to measure the prevalence of ZIKV in febrile patients in Senegal and Nigeria in samples collected from 1992 to 2016. The seroprevalence of ZIKV was 6.2% based on ZIKV immunoglobulin M and negative for dengue reactivity. ZIKV envelope was amplified from 4 samples. Phylogenetic analysis showed that the ZIKVs belonged to the African lineage, grouping with either the Nigerian or MR766 sublineages. This study provides evidence that ZIKV has been silently circulating in West Africa for 2 decades.
The recent unprecedented outbreak of Zika virus (ZIKV) in the Americas has established it as a globally relevant human pathogen. A flavivirus closely related to dengue virus (DENV), ZIKV is transmitted by mosquitoes of the genus Aedes, and infection in humans can lead to a wide range of clinical manifestations, from mild fever and rash to severe neurological disorders including transmitted microcephaly and Guillain-Barré syndrome in adults [1, 2]. Moreover, ZIKV has recently been reported to be transmitted via maternofetal and sexual routes. No effective antiviral agent or vaccine yet exists to treat or control ZIKV infection and, therefore, treatment remains supportive.
Despite considerable global efforts to control or eliminate the mosquito vector in response to other viral and parasitic pathogens, ZIKV has expanded in its geographical distribution over the past 70 years, with limited published research until recently. ZIKV was first isolated in 1947 from a sentinel rhesus monkey stationed in the Zika forest in Uganda [2]. In 1948, the virus was isolated from Aedes africanus mosquitoes, and in 1954 human disease related to ZIKV was reported in Nigeria [3]. ZIKV remained obscure with only 14 documented human cases confined to Africa and Asia, until a 2007 ZIKV fever epidemic in Yap island in Micronesia [4]. Larger epidemics followed in Oceania, including the 2013–2014 French Polynesian outbreak, and subsequently in Latin America and the Cape Verde archipelago in Africa [2, 5]. In May 2015, ZIKV was first reported in Brazil, where nearly 1.3 million people were subsequently infected [2]. By June 2016, autochthonous transmission of ZIKV was confirmed in 40 countries and territories throughout Latin America [6]. Simultaneously, >7500 suspected cases were reported in Cape Verde between October 2015 and May 2016 [5].
Although ZIKV was responsible for significant neuropathology throughout French Polynesia, Latin America, and Cape Verde, human infections prior to 2015 were not associated with neurological disease. Recent hypotheses suggest that the genetic diversification of ZIKV may be responsible for its emergence, neurotropism, and expansion. Phylogenetic analyses using partial and complete genome regions of ZIKV revealed the evolution of the virus into 2 distinct groups, designated African and Asian lineages, with the recent outbreak in Latin America traced to the Asian lineage [4, 7]. Additionally, disease severity in humans may be correlated with enhanced ZIKV infection caused by preexisting DENV antibodies, a phenomenon known as antibody-dependent enhancement [8].
The explosive transcontinental spread of ZIKV highlights the need to quantify the prevalence of ZIKV in regions endemic for the insect vector. Understanding the geographical distribution and burden of disease attributable to ZIKV, in the context of other flaviviruses, across endemic and high-risk regions is also critical. Minimal data exist on the prevalence of ZIKV in humans in West Africa, a region with approximately 450 million people inhabiting areas environmentally suitable for the transmission of multiple mosquito-borne pathogens [9]. In this study, we evaluated previously collected samples from febrile patients from 3 different study cohorts in Senegal and Nigeria for potential ZIKV infection.
METHODS
Study Population and Study Sites
This retrospective study used samples and data from 3 cohorts: (1) a Senegalese human immunodeficiency virus (HIV) female sex worker (FSW) cohort; (2) a Senegalese malaria cohort; and (3) a Nigerian HIV treatment cohort.
The Senegalese FSW cohort, described previously [10], included 119 samples prospectively collected between 1992 and 2004 from self-identified FSW regularly visiting a health care clinic in Dakar. Participants underwent annual blood tests for various sexually transmitted infections, including HIV-1 and HIV-2. This HIV-1/2 natural history study helped define some of the in vivo characteristics of HIV-2 infection in West Africa. The Senegalese malaria cohort, also described previously [11, 12], included 80 samples originally collected under passive surveillance from febrile patients who had positive malaria smear/rapid diagnostic test results during various malaria transmission seasons (August to December). Twenty-seven samples were collected from a health facility in Velingara (2005), a highly malaria-endemic rural area in southern Senegal, and 53 samples from a facility in Thiès (2013), a hypoendemic urban area 70 km from Dakar. The Nigerian HIV cohort included 188 samples collected between 2004 and 2016 from adult patients (≥15 years of age) receiving HIV care and treatment at Jos University Teaching Hospital, Jos, Nigeria, in the Harvard T.H. Chan School of Public Health/AIDS Prevention Initiative in Nigeria (APIN) HIV treatment program, as previously described [13]. Clinical visit records from all 3 studies were queried for fever ≥37.5°C, and of these, excess stored blood specimens that were collected within 7 days of the fever visit were included in the study.
Ethical Considerations
All patients from each of the 3 cohorts provided informed consent for the original collection of samples. The primary studies under which the samples and data were collected received ethical clearance from the Harvard Institutional Review Board (IRB) and the local research ethics committees at Cheikh Anta Diop University, Dakar, Senegal, or Jos University Teaching Hospital, Jos, Nigeria. All excess samples and corresponding data were banked and de-identified prior to the analyses. This study received an exemption determination from the Harvard IRB and from the Senegal Ministry of Health IRB committee.
Serologic Testing
Serum or plasma were screened for the presence of immunoglobulin M (IgM) antibodies by ZV-IgM enzyme-linked immunosorbent assay (ELISA; MyBioSource, San Diego, California), according to the manufacturer’s instructions. Samples that tested ZIKV IgM positive were subsequently screened by DENV Detect IgM Capture ELISA (InBios, Seattle, Washington), according to the manufacturer’s instructions. Assay performances were monitored by using internal controls, and cutoffs were determined as specified by the manufacturer for individual kits.
Nucleic Acid Testing and Sequencing
RNA was extracted from all ZIKV IgM–positive samples using the QIAamp RNA Viral Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. RNA was reverse transcribed using ZIKV-specific primers for a 364-nucleotide region of the ZIKV envelope (E) gene as previously described [4]. Reverse-transcription polymerase chain reaction (RT-PCR) products were purified using the Wizard SV Gel and PCR Clean-Up System (Promega, Madison, Wisconsin). PCR products of expected size were purified from agarose gels and the complete nucleotide sequence of ZIKV E was determined by Sanger sequencing (Genewiz, Cambridge, Massachusetts).
Phylogenetic Analysis
Sequences were aligned with full-length or partial reference ZIKV E genes from GenBank using MEGA6 [14]. Phylogenetic trees were constructed using the maximum-likelihood method logarithm based on the Tamura-Nei model with 1000 bootstrap resampling.
RESULTS
A total of 387 samples collected over a 24-year period were included in the study (Table 1). The age of study participants ranged from 1 to 70 years with an overall median of 35 years (interquartile range, 27–41 years), and 70% were female. The average fever temperature was highest in the Senegalese malaria cohort (39.1°C), followed by the Nigerian HIV cohort (38.3°C) and the Senegalese FSW cohort (37.8°C).
Table 1.
Cohort Characteristics and Zika Virus (ZIKV)/Dengue Virus Serology and ZIKV Reverse-Transcription Polymerase Chain Reaction
| Characteristic | Senegal FSW Cohort (n = 119) |
Senegal Malaria Cohort (n = 80) | Nigeria HIV Cohort (n = 188) |
Total (N = 387) |
|---|---|---|---|---|
| Demographics | ||||
| Sample year range | 1992–2004 | 2005 and 2013 | 2004–2016 | … |
| Age, y, median (range) | 37 (21–58) | 15 (1–50) | 37 (20–70) | 35 (1–70) |
| % Female | 100 | 33 | 66 | 70 |
| Temperature, average, °C | 37.8 | 39.1 | 38.3 | … |
| Malaria status, No. (%) | ||||
| Malaria positive | NT | 80 (100) | NT | 80 (20.7) |
| Malaria negative | NT | … | NT | … |
| HIV status, No. (%) | ||||
| HIV negative | 80 (67.2) | NT | … | 80 (20.7) |
| HIV-1 | 23 (19.3) | NT | 188 (100) | 211 (54.5) |
| HIV-2 | 12 (10.1) | NT | … | 12 (3.1) |
| HIV-dual | 4 (3.4) | NT | … | 4 (1.0) |
| Serology and ZIKV RT-PCR, No. (%) | ||||
| ZIKV IgM | 6 (5.0) | 6 (7.5) | 12 (6.4) | 24 (6.2) |
| DENV IgMa | 0 | 0 | 0 | 0 |
| ZIKV RT-PCRa | 1 | 1 | 2 | 4 |
Abbreviations: DENV, dengue virus; FSW, female sex worker; HIV, human immunodeficiency virus; IgM, immunoglobulin M; NT, not tested; RT-PCR, reverse-transcription polymerase chain reaction; ZIKV, Zika virus.
aDENV IgM and ZIKV RT-PCR were performed on 24 ZIKV IgM–positive samples.
Of the total number of samples tested for ZIKV IgM antibodies, 24 tested positive, resulting in an overall ZIKV seroprevalence of 6.2%. The highest ZIKV seroprevalence was observed in the Senegalese malaria cohort (6/80 [7.5%]), followed by the Nigerian HIV cohort (12/188 [6.4%]) and the Senegalese FSW cohort (6/119 [5%]). All 24 ZIKV IgM–positive samples tested negative for DENV IgM.
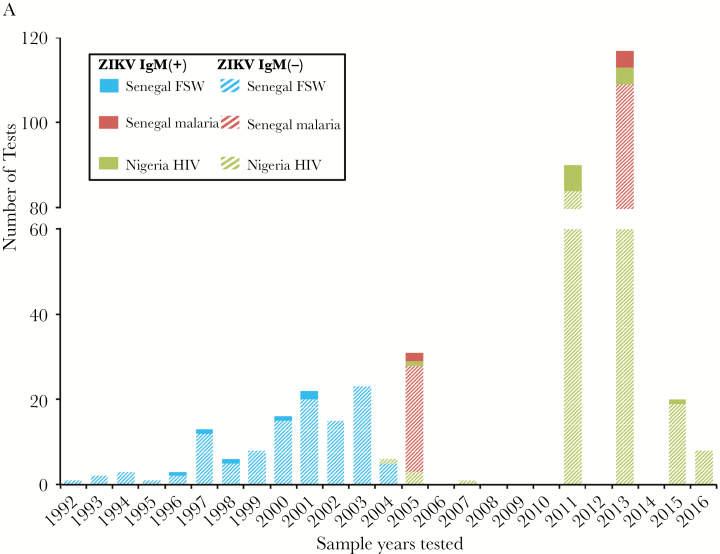
ZIKV IgM–positive samples were found throughout a 2-decade span between 1992 and 2016 (Figure 1A). While we tested samples from 1992 to 2004 in the Senegalese FSW cohort, ZIKV was detected in samples collected between 1996 and 2001. Positive samples were detected in both the 2005 and 2013 groups in the Senegalese malaria cohort. While we tested samples from 7 years between 2004 and 2016 in the Nigerian HIV cohort, positive samples were detected in 2005, 2011, 2013, and 2015. A majority of ZIKV IgM–positive patients did not report signs or symptoms corresponding to ZIKV infection. Additionally, logistic regression using decade (1992–1999, 2000–2009, 2010–2016), rainy season (Nigeria: April–October; Senegal: June–October), age, sex, and temperature as predictors, clustering by cohort, found a lower odds of ZIKV IgM positivity after 2009 compared with before 2000 (odds ratio, 0.80; P = .003; data not shown).
Figure 1.
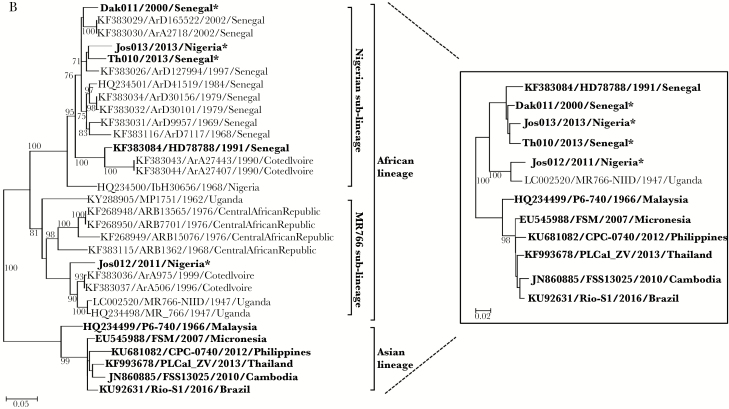
A, Frequency of Zika virus (ZIKV) results between 1992 and 2016. Blue, Senegalese female sex worker (FSW) cohort; red, Senegalese malaria cohort; green, Nigerian human immunodeficiency virus (HIV) cohort. The solid bars represent the number of immunoglobulin M (IgM)–positive samples and the hashed bars represent the number of negative samples. B, Phylogenetic analysis of ZIKV E genes of 4 ZIKVs from Senegal and Nigeria using reference ZIKV sequences isolated from humans, monkeys, and mosquitoes. The evolutionary history was inferred by using the maximum-likelihood method based on the Tamura-Nei model with 1000 boot resampling. Spondweni virus was used to root the tree. The boxed maximum-likelihood tree was produced as above using ZIKVs isolated from only humans and the MR766 monkey ZIKV. The 4 ZIKVs identified in this study are indicated with an asterisk (*).
Amplicons corresponding to a specific 364-nucleotide region of the ZIKV E gene were amplified by RT-PCR in 4 of 24 ZIKV IgM–positive samples (Table 1). Of the RT-PCR–positive samples, 1 was from the Senegalese FSW cohort (Dak011) collected in 2000, 1 from the Senegalese malaria cohort (Th010) collected in 2013, and 2 from the Nigerian HIV cohort (Jos012, Jos013), collected in 2011 and 2013, respectively. ZIKV RT-PCR–positive samples from the Senegalese FSW and Nigerian HIV cohorts were HIV-1 positive, while the positive sample from the Senegalese malaria cohort screened positive for malaria.
Full-length ZIKV E was sequenced for the 4 ZIKV RT-PCR–positive samples. Using reference E genes from human, monkey, and mosquito ZIKVs, nearly identical phylogenetic trees were generated by several methods (maximum likelihood, neighbor-joining, minimum evolution). A final maximum-likelihood tree was generated and analyzed with 1000 replicates for bootstrap testing (Figure 1B). Dak011, Jos013, and Th010 clustered with the Nigerian sublineage, whereas Jos012 clustered with the MR766 sublineage. Within the MR766 sublineage, Jos012 was most closely related by nucleotide sequencing to reference mosquito ZIKVs from Cote d’Ivoire.
DISCUSSION
Although these 3 study cohorts were distinct, we achieved a relatively homogenous high-risk study population by focusing on febrile patients with temperature ≥37.5°C at the time of sample collection. This study revealed a previously unmeasured ZIKV IgM seroprevalence among febrile patients in Senegal and Nigeria of 6.2%, demonstrating the continued transmission of ZIKV in these countries over 2 decades [15]. It is noteworthy that while our study utilized the ZV-IgM ELISA (MyBioSource), a recent US Food and Drug Administration (FDA) alert suggested higher false positives than expected with the FDA Emergency Use Authorization–approved ZIKV Detect IgM Capture ELISA (InBios). Additionally, uncertainty revolves around the persistence of ZIKV IgM after infection and whether ZIKV IgM seropositivity indicates recent infection. For other flaviviruses such as DENV and West Nile virus, IgM may persist up to 6 or 7 months, respectively.
DENV is thought to circulate throughout West Africa, and due to extensive cross-reactivity between the flaviviruses, we tested the ZIKV IgM–positive samples for DENV IgM, a Centers for Disease Control and Prevention–recommended protocol. All of the ZIKV IgM–positive samples tested negative for DENV IgM, indicating that DENV cross-reactivity is unlikely; however, these results do not exclude cross-reactivity between other flaviviruses that may circulate in Senegal and Nigeria.
Of the ZIKV IgM–positive samples, 70% were from females. Because our study population was comprised of 70% females, this is proportionate to our study population. Our logistic regression analysis revealed a 20% lower odds of a positive ZIKV IgM result during 2010–2016 compared with 1992–1999; however, this correlation could be an artifact of the small number of ZIKV IgM–positive samples (n = 24) or the small number of samples tested between 1992 and 1999 (n = 38).
Prior to this study, there were published ZIKV sequence data from only 1 human infection in Africa, isolated in 1991 in Senegal (KF383084/HD78788) [4]. Our study identified 4 additional human African ZIKVs from both Senegal and Nigeria. These newly characterized ZIKVs grouped with the African lineage, as expected. Interestingly, Jos012 clustered with the MR766 sublineage, comprised of mostly East African ZIKVs, and was most closely related to mosquito ZIKVs from Cote d’Ivoire, whereas the other 3 ZIKVs clustered within the Nigerian sublineage, comprised of mostly West African mosquito ZIKVs. Similar results were observed when a maximum-likelihood tree was generated using E genes of human ZIKVs and the 1947 MR766 monkey ZIKV. These results corroborate the presence of 2 African ZIKV sublineages circulating throughout West Africa [4].
Although ZIKV appears to be endemic in Senegal and Nigeria, data regarding the prevalence of the virus in humans in West Africa have been lacking. Though not population based, our study found a 6.2% prevalence of ZIKV in febrile patients presenting to clinics, providing evidence that ZIKV, though largely unreported, has been circulating in West Africa for the past 2 decades. Furthermore, we have shown that concurrent infection of HIV/malaria and ZIKV does occur. Our study emphasizes the need for improved detection methods for ZIKV, to distinguish among fever-causing pathogens. Further studies will help elucidate the impact of ZIKV on patient outcomes, leading to improved understanding of the pathogenesis of ZIKV disease in Africa.
Notes
Acknowledgments. We thank Professor Dyann Wirth, Courtney Edison, Amanda Lukens, and Karell Pelle (Harvard T.H. Chan School of Public Health) for assistance with the malaria sample selection; the Jos University Teaching Hospital laboratory staff (Yetunde Isa, Titus Obadiah, Chindak Lekuk, and Mangai Yakubu) for their assistance with the project; Professor Wei-Kung Wang (University of Hawaii at Manoa) for scientific advice and invaluable discussion; and Seema Meloni for critical reading of the manuscript.
Financial support. The Senegalese FSW study was supported in part by the US Department of Defense (grant number DAMD-17-87-7072) and the National Institutes of Health (NIH; grant numbers AI301795 and CA39805). The Senegalese malaria sample collection in Thiès was supported by NIH grants R01GM061351 and D43TW001503, the Malaria Diversity grant from the Bill & Melinda Gates Foundation, and the Ellison Medical Foundation. The Senegalese sample collection in Velingara was supported in part by grant 5D43TW001503 from the Fogarty International Center, grant U19AI089696 from the International Centers of Excellence for Malaria Research, and NIH grants K23AI072033 and AI034969. The Nigerian HIV cohort was supported in part by the US Department of Health and Human Services, Health Resources and Services Administration (U51HA02522) and the Centers for Disease Control and Prevention through a cooperative agreement with APIN (PS001058).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Simpson DI. Zika virus infection in man. Trans R Soc Trop Med Hyg 1964; 58:335–8. [PubMed] [Google Scholar]
- 2. Weaver SC, Costa F, Garcia-Blanco MA, et al. Zika virus: history, emergence, biology, and prospects for control. Antiviral Res 2016; 130:69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Macnamara FN. Zika virus: a report on three cases of human infection during an epidemic of jaundice in Nigeria. Trans R Soc Trop Med Hyg 1954; 48:139–45. [DOI] [PubMed] [Google Scholar]
- 4. Faye O, Freire CC, Iamarino A, et al. Molecular evolution of Zika virus during its emergence in the 20(th) century. PLoS Negl Trop Dis 2014; 8:e2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. World Health Organization. The history of Zika virus. Available at: http://www.who.int/emergencies/zika-virus/history/en/ Accessed 2 February 2017. [Google Scholar]
- 6. Armstrong P, Hennessey M, Adams M, et al. ; Zika Virus Response Epidemiology and Laboratory Team Travel-associated Zika virus disease cases among U.S. residents—United States, January 2015–February 2016. MMWR Morb Mortal Wkly Rep 2016; 65:286–9. [DOI] [PubMed] [Google Scholar]
- 7. Lanciotti RS, Lambert AJ, Holodniy M, Saavedra S, Signor Ldel C. Phylogeny of Zika virus in Western Hemisphere, 2015. Emerg Infect Dis 2016; 22:933–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dejnirattisai W, Supasa P, Wongwiwat W, et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with Zika virus. Nat Immunol 2016; 17:1102–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Messina JP, Kraemer MU, Brady OJ, et al. Mapping global environmental suitability for Zika virus. Elife 2016; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kanki PJ, Travers KU, MBoup S, et al. Slower heterosexual spread of HIV-2 than HIV-1. Lancet 1994; 343:943–6. [DOI] [PubMed] [Google Scholar]
- 11. Daily JP, Scanfeld D, Pochet N, et al. Distinct physiological states of Plasmodium falciparum in malaria-infected patients. Nature 2007; 450:1091–5. [DOI] [PubMed] [Google Scholar]
- 12. Daniels RF, Schaffner SF, Wenger EA, et al. Modeling malaria genomics reveals transmission decline and rebound in Senegal. Proc Natl Acad Sci U S A 2015; 112:7067–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Meloni ST, Chang CA, Eisen G, et al. Long-term outcomes on antiretroviral therapy in a large scale-up program in Nigeria. PLoS One 2016; 11:e0164030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28:2731–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sow A, Loucoubar C, Diallo D, et al. Concurrent malaria and arbovirus infections in Kedougou, southeastern Senegal. Malar J 2016; 15:47. [DOI] [PMC free article] [PubMed] [Google Scholar]