This study showed that simple tests based on measuring 3 host mRNAs or single host proteins can serve as pan-viral tests to identify patients with respiratory virus infection using a nasopharyngeal swab.
Keywords: CXCL11, host response, biomarker, respiratory virus, pan-viral test
Abstract
Background
Despite the high burden of respiratory infection and the importance of early and accurate diagnosis, there is no simple diagnostic test to rule in viral infection as a cause of respiratory symptoms.
Methods
We performed RNA sequencing on human nasal epithelial cells following stimulation of the intracellular viral recognition receptor RIG-I. Next, we evaluated whether measuring identified host mRNAs and proteins from patient nasopharyngeal swabs could predict the presence of a respiratory virus in the sample.
Results
Our first study showed that a signature of 3 mRNAs, CXCL10, IFIT2, and OASL, predicted respiratory virus detection with an accuracy of 97% (95% confidence interval [CI], 0.9–1.0), and identified proteins correlating with virus detection. In a second study, elevated CXCL11 or CXCL10 protein levels identified samples containing respiratory viruses, including viruses not on the initial test panel. Overall area under the curve (AUC) values were: CXCL11 AUC = 0.901 (95% CI, 0.86–0.94); CXCL10 AUC = 0.85 (95% CI, 0.80–0.91).
Conclusions
Host antiviral mRNAs and single host proteins detectable using nasopharyngeal swabs accurately predict the presence of viral infection. This approach holds promise for developing rapid, cost-effective tests to improve management of patients with respiratory illnesses.
Acute respiratory illnesses are extremely common, accounting for more than 500 million outpatient illnesses and 3.6 million hospitalizations per year in the USA alone [1, 2]. Viral infection is a common cause of these illnesses but it is usually a diagnosis of exclusion, because current tests to rule in viral infection are often prohibitive in cost and time. A simple, pan-viral test to rule in a viral cause for respiratory symptoms could have a tremendous positive impact by facilitating rapid diagnosis, improving patient care, and enabling more efficient use of medical resources for the millions of patients with respiratory illness [3, 4].
Current diagnostic strategies to rule in viral infection require testing for a number of distinct viruses that cause similar symptoms, because tests identify features specific to each virus. Common tests use polymerase chain reaction (PCR)-based identification of viral genomes or viral antigen detection [5]. Testing for a panel of suspected viruses can be time consuming and/or expensive, and falsely negative if the patient is infected with a virus that is not in the panel. Identifying which one of many clinically similar viruses is causing a respiratory illness usually does not impact treatment, because virus-specific therapies are only available for influenza. One promising approach to developing a pan-viral test is to focus on biomarkers indicating that the body is responding to a viral infection.
Several recent studies have demonstrated that gene expression patterns in blood cells or plasma can indicate the presence of viral infection [6–12], and a recent brief report showed that levels of certain host mRNAs detected on respiratory swabs correlated with symptomatic viral infection [13]. These findings demonstrate the promise of using the host response to develop a pan-viral diagnostic test. Therefore, we performed 2 studies to evaluate whether biomarkers of the antiviral response could identify virus-positive nasopharyngeal swabs, using swabs sent to our health care system for respiratory virus testing. We sought to identify host proteins as well as mRNAs that could indicate viral infection in this sample type, because immunoassay-based tests are in common use in laboratory and point-of-care testing.
Guided by in vitro RNA sequencing (RNAseq) experiments on nasal epithelial cells, in our first study we prospectively examined the performance of a signature of 3 host mRNAs for predicting viral infection, and we also used these samples to retrospectively identify promising potential protein biomarkers detectable in the swab-associated viral transport medium. In our second study, we prospectively evaluated CXCL10 and CXCL11 proteins and found a high correlation between levels of each of these proteins and the presence of viral infection. Here we report our findings, which indicate great potential for developing simple, pan-viral diagnostic tests to identify patients with respiratory virus infection.
METHODS
Nasal Epithelial Cell Culture and Stimulation With Viral-Mimetic Ligand SLR14
Primary human nasal epithelial cells were obtained commercially (Promocell) and grown in BEGM medium (Lonza). Hydrocortisone and epinephrine supplements were removed prior to stimulation. Cells were transfected with the RIG-I ligand SLR14 (also known as 14-hp), a generous gift from the Pyle lab, using Lipofectamine 2000 (Invitrogen) then incubated for 7 hours at 37°C [14].
RNAseq
The raw reads of RNAseq experiments were trimmed off sequencing adaptors and low-quality regions by btrim [15]. The trimmed reads were mapped to human genome (GRCh37) by tophat2 [16]. The counts of reads for each gene were based on Ensembl annotation (release 70) and differential expression analysis was done by DEseq2 [17], which calculated the adjusted P values. RNASeq data have been deposited in NCBI’s Gene Expression Omnibus (GEO), accession number GSE 107898.
Study Design
Sample Selection
Both studies used samples sent to the Yale-New Haven Hospital (YNHH) diagnostic virology laboratory by patients’ health care providers. Sample collection windows were selected during high test volume winter months, when multiple viruses were circulating, based on availability of personnel to process samples for research. For both studies, samples were included if: (1) direct fluorescent antigen (DFA) testing was ordered; (2) the 9-virus respiratory PCR panel was ordered (for Study 2, this was only required for DFA-negative samples); and (3) the samples were of adequate quality to perform DFA testing as determined by microscopy.
Sample Processing
Study 1 included 68 nasopharyngeal (NP) swabs collected in viral transport medium during 14 days between 12/2015 and 2/2016. Samples were stored for a maximum of 8 hours at 4°C prior to centrifugation and separate storage of cell pellets in lysis buffer and supernatants at −80°C. Study 2 included viral transport medium from 151 NP swabs sent to the YNHH lab during 1 week in December 2016 and stored at −80°C.
Human Subjects Oversight
All samples were de-identified and coded by the clinical laboratory. The protocol was approved by the Yale Human Investigations Committee.
Assessing Performance of Biomarker Tests
The investigator responsible for reverse-transcription quantitative PCR (RT-qPCR) testing and sample scoring (Study 1) or immunoassay testing (Study 2) (E. F. F.) was blinded to virology testing results until after the biomarkers were measured and samples were scored.
Clinical Virology Testing
Samples were collected by patients’ health care provider with flocked swabs placed into 3-mL universal viral transport medium (Becton Dickinson). Nucleic acids were isolated from 0.2 mL of transport medium using the Nuclisens (Boom method) on the Easy Mag instrument (bioMerieux). DFA tests were performed using commercial reagents (Light Diagnostics SimulFluor Respiratory Screen Reagent, Millipore Corporation). PCR and direct fluorescent antigen testing for a panel of 9 respiratory viruses (Table 1) were performed as described previously [18–24].
Table 1.
Respiratory Virus Tests in the Yale-New Haven Hospital Panel
| Adenovirus (Adeno)a,b |
| Human metapneumovirus (hMPV)a,b |
| Influenza A and B (Flu A, B)a,b |
| Parainfluenza 1, 2, and 3 (PIV 1–3)a,b |
| Respiratory syncytial virus A and B (RSV)a,b |
| Rhinovirus (RV)a |
aSemiquantitative qPCR
bDirect fluorescent antigen testing
Coronavirus and PIV4 Testing
Coronavirus multiplex PCR assay was adopted from Sultani et al, 2015 [25], for CoV-NL63, CoV-229E, CoV-OC43, and CoV-SARS, and confirmation of positive samples was performed using singleplex PCR. PIV4 testing was performed using a previously reported assay [26]. Sources of RNA were: Study 1, RNA from cell pellets; Study 2, nucleic acids for clinical virology testing, followed by genomic DNA digestion and RT (iScript gDNA clear, Bio-Rad Laboratories).
Quantitative RT-PCR for Host mRNAs, Study 1
RNA was isolated from cell pellets using the RNeasy kit (Qiagen) and reverse-transcribed RNA using iSCRIPT cDNA synthesis kit (Bio-Rad Laboratories), qPCR was performed using iTAQ Universal SYBR Green (Bio-Rad Laboratories). Primers were:
βACTIN (F:CCTGGCACCCAGCACAAT; R: GCCGAT CCACACGGAGTACT)
OASL (F: AAGGTAGTCAAGGTGGGCTC; R: CTCCTG GAAGCTGTGGAAAC)
IFIT2 (F: CCTCAAAGGGCAAAACGAGG; R: CTGATT TCTGCCTGGTCAGC)
CXCL10 (F: CCTGCAAGCCAATTTTGTCC R: ATGGC CTTCGATTCTGGATTC)
Immunoassays for Chemokines in Viral Transport Medium
Frozen viral transport medium was thawed on ice and centrifuged to remove cell debris. Chemokine levels were measured using the Bio-Plex or Luminex instrument with Milliplex MAP human cytokine panel III (HCYP3MAG-63K) or Bio-Plex Pro Human Chemokine panel following manufacturers’ instructions (Bio-Rad Laboratories, Millipore Corporation). CXCL10 and CXCL11 were measured in undiluted and 1:5 dilutions of all samples.
Data Analysis and Statistics
Data analysis employed SAS/STAT statistical analysis software and IBM SPSS statistical analysis software.
RESULTS
Identification of Potential Nasopharyngeal Pan-viral Biomarkers Using RIG-I Stimulation of Human Nasal Epithelial Cells In Vitro
Respiratory epithelial cells can mount vigorous antiviral defense responses upon viral recognition by the cell-intrinsic innate immune system, through rapid induction of antiviral genes [27]. To identify robust biomarkers of the antiviral response in human nasal epithelial cells (HNEC), we stimulated HNEC in vitro with a small molecule ligand of RIG-I, a cytoplasmic receptor for viral RNA. Although this treatment stimulated RIG-I mimicking RNA virus infection, analogous sensors for DNA viruses trigger many of the same downstream signals [28].
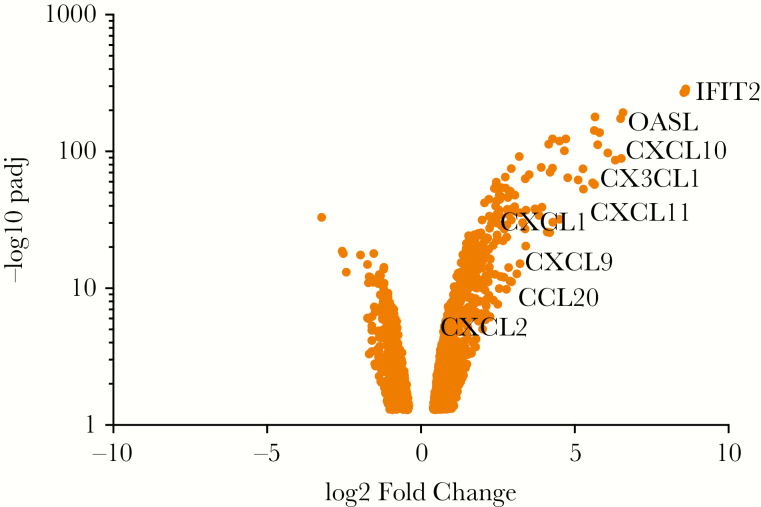
Consistent with previous studies of airway epithelial cells responding to viral infection, transcriptional changes were dominated by interferon-stimulated genes (ISGs; Figure 1 and Supplementary Table S1). These include mRNAs associated with respiratory virus infection in other sample types and/or patient populations, including IFIT genes, OAS genes, and RSAD2/viperin [6, 8, 13, 29]. Figure 1 highlights highly induced transcripts evaluated as potential biomarkers in this study. These include multiple chemokines, small secreted proteins that function as chemoattractants to recruit cells of the immune system to infected tissues. Transcripts that encode secreted proteins were of particular interest because they could potentially serve as biomarkers of viral infection at both the RNA and protein levels and therefore could be useful for developing both PCR-based and immunoassay-based diagnostic tests.
Figure 1.
Transcripts enriched by stimulation of primary human nasal epithelial cells with RIG-I ligand SLR14 to mimic viral infection. Primary human nasal epithelial cells (HNEC) were transfected with SLR14 for 1 hour, then incubated for 7 additional hours at 37°C, followed by RNA isolation and transcript analysis by RNAseq. Dots represent transcripts significantly different in control versus SLR14 stimulated HNEC (adjusted P-value of <0.05, log2 FoldChange >1 or <−1). Labels highlight transcripts examined in this study as potential nasopharyngeal biomarkers of viral respiratory infection. A more extensive list of differentially expressed transcripts can be found in Supplementary Table S1.
Transcriptional Signature Based on 3 mRNAs Predicts Respiratory Virus Infection from Nasopharyngeal Swabs
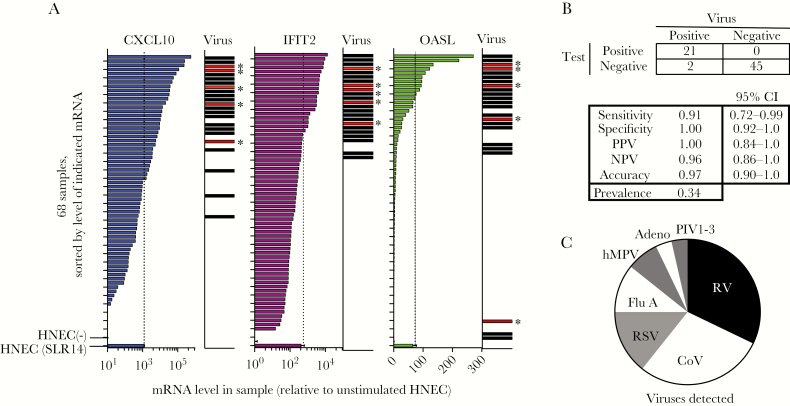
Next, we investigated whether a transcriptional signature based on 3 of the identified mRNAs could predict viral infection in nasopharyngeal swabs. We performed RT-qPCR for biomarkers on RNA isolated from 68 NP swabs sent to the YNHH lab for respiratory virus testing. Chart review revealed that most patients were older adults with multiple comorbidities, presenting with respiratory symptoms and/or fever, and/or altered mental status. Top comorbidities were cardiovascular disease (25/68), malignancy (24/68), and asthma/COPD (16/68) (see Supplementary Figure S1).
We measured levels of 3 mRNAs identified by the RNAseq experiment: 2 encoding intracellular proteins (OASL, IFIT2) and 1 encoding the chemokine CXCL10. Levels of each biomarker correlated highly with virus detection (Figure 2A and Supplementary Figure S2). For the mRNA signature test, we scored each mRNA level as above or below a cutoff determined by the mRNA level detected in SLR14 stimulated HNEC. If at least 2 mRNA were above the cutoff for a given sample, the sample was scored as positive for viral infection.
Figure 2.
Reverse-transcription quantitative polymerase chain reaction (RT-qPCR) test for 3 host mRNAs and correlation with respiratory virus detection in 68 nasopharyngeal swabs (Study 1). A, Patient cells were pelleted from the transport medium containing patient nasopharyngeal swabs, RNA was isolated, and RT-qPCR was performed for β-actin and 3 mRNAs associated with the antiviral response as identified in Figure 1 (OASL, IFIT2, CXCL10). Plots show mRNA levels for 68 samples relative to the mRNA level in resting human nasal epithelial cells (HNEC (−)), sorted by mRNA level for each transcript. Biomarker mRNA was normalized to β-actin mRNA in the same sample. Each mRNA biomarker was scored positive if level was above the level observed in SLR14-stimulated HNEC, indicated by the dotted line and lowest bar on each graph (HNEC bars show mean and SD of 3 replicates). mRNA levels were normalized to the level of β-actin mRNA in each sample. Bars in virus column indicate samples that tested positive for respiratory virus by qPCR. Bars marked with asterisk indicate the 5 samples that were negative on the initial virology test panel but positive upon subsequent testing for CoV. B, Test performance of mRNA biomarker signature. Samples were scored as biomarker test positive if 2/3 mRNA biomarkers were above the cutoff (dotted lines). C, Pie chart shows relative abundance of the viruses detected in this sample set, representing 28 detections (23 virus positive samples with 5 codetections). Abbreviations: virus names are listed in Table 1; CoV, coronavirus OC43 (only CoV detected); NPV, negative predictive value; PPV, positive predictive value.
All but 5 samples scoring positive on the host response test also tested positive for known respiratory viruses on the 9-virus YNHH panel (Table 1; Figure 2A). The YNHH virus panel does not include coronaviruses (CoV). Therefore, we tested RNA from all patient samples using a previously described multiplex assay for 4 CoV genotypes [25]; 8/68 specimens were positive for coronavirus OC43. Strikingly, all 5 of the biomarker-positive, virus panel-negative samples were positive for CoV-OC43 (indicated by asterisks next to bars in virus panel, Figure 2A), and the other CoV detections were codetections in samples positive for other viruses.
As shown in Figure 2B, the overall diagnostic accuracy of the mRNA biomarker test was 97% (95% confidence interval [CI], 0.9–1.0) and the positive predictive value was 100% (95% CI, 0.84–1.0). Two of 68 samples represented false negatives (virus detected in the absence of ISG signature), including 1 rhinovirus (RV)+ and 1 human metapneumovirus (hMPV)+ sample. Of the 16 samples from patients with asthma or COPD, 5 were virus positive; 4 were biomarker test positive and 1 was biomarker test negative (hMPV+ sample). All 11 virus-negative samples from asthma or COPD patients were biomarker test negative. Seven distinct viruses were detected in the samples by both conventional and the mRNA signature testing (Figure 2C), indicating the great potential for a single host-based test to capture infection by diverse viruses.
Based on chart review, 8 patients had findings indicative of nonviral infection (see Supplementary Table S2). Seven were virus negative. These 7 patients were also negative for the host mRNA signature described in Figure 2. In addition, 1 patient had radiological evidence of bacterial infection and sputum culture was positive for Pseudomonas. This patient also tested positive for coronavirus OC43 and for the host signature for viral infection described in Figure 2. Although this study was not designed to assess nonviral infection, these data suggest that the host response signature in Figure 2 is specific for viral infection and is not triggered by or inhibited by other infection types.
CXCL10 and CXCL11 Protein Levels Correlate With the Presence of Respiratory Virus
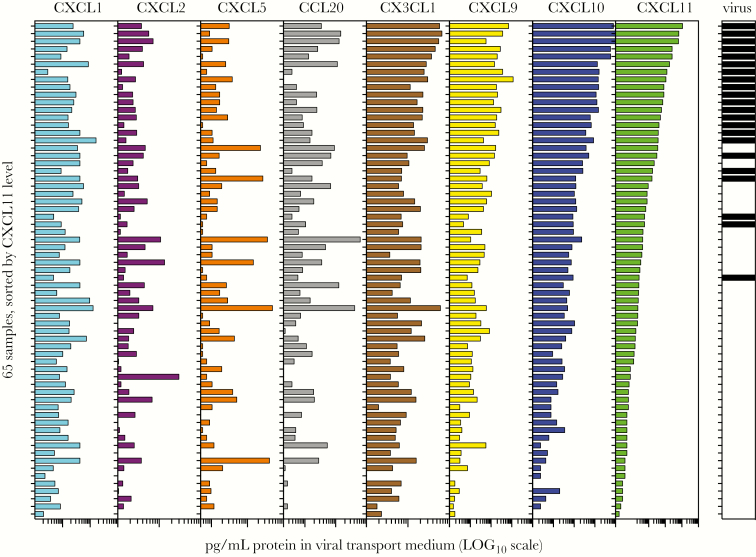
As indicated in Figure 1 and Supplementary Table S1, some of the most highly induced mRNAs triggered by RIG-I ligand in nasal cells in vitro encode chemokines. To identify which of these chemokines might serve as robust biomarkers of viral infection at the protein level, we measured levels of 8 chemokines identified in the RNAseq experiment, using immunoassay of the viral transport medium. Of the 8 chemokines tested, only CXCL9, CXCL10, and CXCL11 levels correlated with the presence of virus, with CXCL10 and CXCL11 showing the most robust correlation (Figure 3).
Figure 3.
Protein levels of 8 chemokines in the nasopharyngeal (NP) swab associated viral transport medium and relationship to virus detection (Study 1). Levels of 8 chemokines were measured in the viral transport medium associated with 65 NP swabs from the study described in Figure 2, using magnetic bead immunoassays. For graph, all samples (1–65) are sorted based on CXCL11 level. Black bars (right panel) indicate which samples also tested positive for a respiratory virus. Protein concentrations are plotted on a log scale across the following ranges: CXCL1(102–105), CXCL2 (10–105), CXCL5 (102–104), CCL20 (1–103), CX3CL1 (10–103), CXCL9 (10–105), CXCL10 (10–105), CXCL11 (1–105).
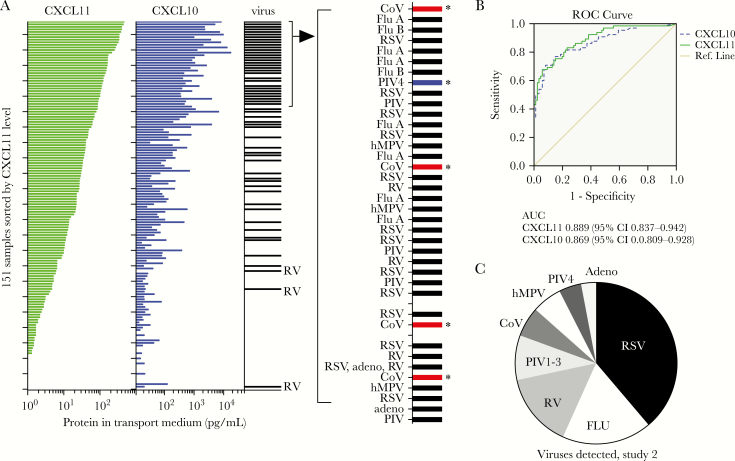
CXCL10 and CXCL11 Proteins Each Predicted Presence of Respiratory Virus in the Second Set of Nasopharyngeal Swabs
To further evaluate the usefulness of CXCL10 and/or CXCL11 proteins in predicting respiratory virus infection, we measured levels of both proteins in viral transport medium in a second set of stored samples from December 2016. Patients were primarily older adults (72% of patients over the age of 50) and also included young children (15% of patients vs 6% in Study 1; Supplementary Figures S1 and S3). CXCL10 and CXCL11 concentrations correlated highly with viral infection (Figure 4A). Samples were initially tested for the 9-virus panel and CoVs as in Study 1, then results were compared to biomarker levels. Because several virus-negative samples had very high levels of CXCL10 or CXCL11, we further expanded the test panel by testing for parainfluenza virus type 4 (PIV4). Three samples tested positive for PIV4, including 2 samples with high levels of CXCL10 and CXCL11 in which no other virus was detected. Figure 4A shows an expanded view of the 40 samples with the highest levels of CXCL11. Detected viruses that were not in the original test panel (Table 1) are indicated with an asterisk. We also observed CXCL11-low samples with viruses detected (Figure 4A). Rhinovirus was detected in the 3 virus-positive samples with the lowest concentrations of CXCL11. Further analysis of RV-positive samples in Study 2 revealed a correlation between [CXCL11] and [CXCL10] and the level of RV RNA detected by RT-qPCR, with the chemokine-low samples also having lower levels of RV (Supplementary Figure S4).
Figure 4.
Predictive value of CXCL10 or CXCL11 protein level for detection of respiratory virus in 151 patient nasopharyngeal swabs (Study 2). A, Correlation between NP swab CXCL10 or CXCL11 protein level and detection of respiratory virus in 151 nasopharyngeal swabs tested for respiratory viruses at Yale-New Haven Hospital in December 2016. CXCL10 and CXCL11 levels were measured using magnetic bead immunoassays. Nucleic acids isolated from the viral transport medium were used to test for viruses not on the original test panel, including 4 CoVs and PIV4. Plots show CXCL11 and CXCL10 levels for 151 samples, sorted by CXCL11 level. Horizontal bar in column labeled virus indicates detection of respiratory virus in the sample; asterisks indicate samples that tested positive for viruses not in the original test panel (Table 1). B, Receiver operating characteristic (ROC) curves for CXCL10 (dashed line) and CXCL11 (solid line) concentrations as predictors of virus detection in this sample set, calculated using SPSS software. C, Viruses detected in the 65/151 virus positive samples in Study 2. Ten distinct viruses were detected, including both influenza A and B and 2 CoV (OC43 and 229E).
Overall, 65/151 of samples (43%) were positive for 10 distinct viruses, including influenza A and B and CoV-OC43 and CoV-229E, and either CXCL11 or CXCL10 level was an excellent predictor of virus-positive and virus-negative status (CXCL11; area under the curve [AUC] = 0.889, 95% CI, 0.837–0.942; CXCL10, AUC 0.869 95% CI, 0.809–0.928; Figures 4B and C). Prevalence of individual viruses differed significantly from Study 1, with respiratory syncytial virus most prevalent in Study 2 and RV most prevalent in Study 1 (Figures 2C and 4C). These findings reveal that CXCL10 or CXCL11 concentration can predict infection with diverse viruses.
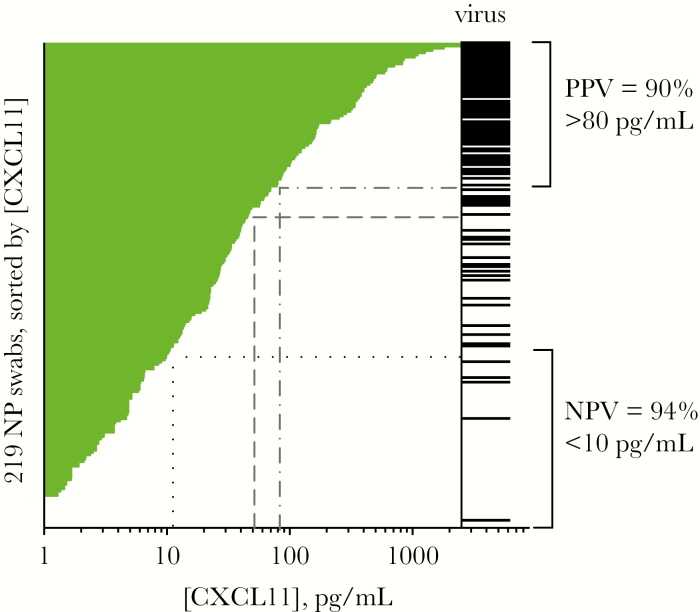
Figure 5 shows the CXCL11 protein concentration in 219 samples (Study 1 plus Study 2), sorted from highest to lowest concentration, with bars in the virus column indicating virus detections. As shown, high CXCL11 levels correlated strongly with virus detection and low levels correlated with absence of virus, with intermediate levels being indeterminate. Based on this pattern, it is possible to envision developing a rule-in/rule-out test using a high cutoff above which samples are predicted to be virus positive, and below which samples are predicted to be virus negative. Dashed grey lines in Figure 5 represent cutoffs of >80 pg/mL for rule-in, which in this sample set would have a positive predictive value (PPV) of 90% or a cutoff of >50 pg/mL, with a PPV of 86%; dotted black lines indicate a low cutoff of 10 pg/mL, below which this test has a negative predictive value of 94% for absence of virus infection. Using cutoffs of >80 pg/mL and <10 pg/mL, about 1/3 of the samples (35%) fall into the indeterminate zone. Consistent with the observations from each study (Figures 3 and 4A), we observed a high degree of correlation between CXCL10 and CXCL11 protein concentrations (Supplementary Figure S5; for 219 samples, R2 = 0.80) and a high correlation of each biomarker with virus detection (in set of 219 samples, CXCL11 AUC = 0.901; 95% CI, 0.86–0.94; CXCL10 AUC = 0.83; 95% CI, 0.80–0.91; Supplementary Figure S6). Although only CXCL11 is shown in Figure 5, either CXCL11 or CXCL10 measurements could be used to rule in/rule out virus infection. In sum, these results demonstrate how an immunoassay-based test measuring a single host protein could have high diagnostic utility for managing patients with suspected respiratory infection.
Figure 5.
Possible rule in/rule out test for viral respiratory infection based on CXCL11 protein level, using data from 219 nasopharyngeal (NP) swabs (Study 1 plus Study 2). Demonstration of how CXCL11 level could be used to create a rule-in/ rule-out test for respiratory virus infection. Plot shows CXCL11 concentrations measured in 219 samples (Study 1 plus Study 2), sorted by CXCL11 level. Black bars represent presence of virus. Brackets show how cutoffs could be used to rule in or rule out viral infection at the upper and lower ends of [CXCL11], with an intermediate indeterminate zone. Grey dashed lines show cutoff of >80 pg/mL to rule in viral infection with positive predictive value (PPV) of 90%, or >50 pg/mL to rule in viral infection with PPV of >86%. Brackets demonstrate that for approximately 2/3 of the samples (65%), virus infection can be ruled in with a PPV of 90% and ruled out with a negative predictive value (NPV) of 94% (dotted lines); for 1/3 of samples, test is indeterminate.
DISCUSSION
In this study, we tested whether mRNAs and proteins associated with NP swabs could be used to predict the detection of a respiratory virus in samples sent to our hospital laboratory for conventional virology testing. We chose possible biomarkers based on transcripts strongly induced by a virus-like stimulus in primary nasal epithelial cells in vitro. The 3 mRNAs we selected all showed predictive value for viral infection in NP swabs. Surprisingly, we were also able to identify high-performing biomarkers detectable at the protein level by testing chemokines indicated by the in vitro experiment.
The mRNA and protein biomarkers we identified are all molecules known to be associated with the antiviral interferon response, a key host defense response to both RNA and DNA viruses in which viral recognition leads to interferon secretion and induction of approximately 300 different antiviral effectors [30]. While this result is not unexpected, interferon-stimulated genes have diverse regulatory mechanisms, are differentially expressed in different host tissues, and are differentially antagonized by different viruses. Therefore, identifying which one(s) performed best as pan-viral infection biomarkers in the upper respiratory tract required empirical testing.
Interestingly, although many different chemokines were induced by RIG-I stimulation in vitro, only 1 family of chemokines correlated highly with viral infection in NP swab samples: the CXCR3 ligands CXCL9, CXCL10, and CXCL11. These ligands mediate chemotaxis of T cells to sites of viral infection [31]. The consistent correlation of these chemokines with presence of diverse respiratory viruses in NP swabs suggests a particularly robust and conserved role for these chemokines in local antiviral defense of the upper respiratory tract.
We were able to identify accurate mRNA and protein-based tests. We defined an mRNA signature with very high diagnostic accuracy by combining the information from 3 biomarkers (Figure 2; accuracy of 97% (95% CI, 90–100%); sensitivity 91%; specificity 100%). This test compares favorably with a recently reported mRNA-based index correlating Viperin mRNA levels to viral infection [13]. Technologies to support practical tests based on measuring host transcripts are under active development [32]. Importantly, our findings showed that NP swab-associated CXCL10 or CXCL11 protein concentrations can also serve as biomarkers of viral infection. Immunoassays are already widely used in diagnostic testing, and these findings open up new possibilities for developing practical tests for diverse patient care settings, including point-of-care.
Notably, we did observe some discordant samples. Biomarker-negative, virus-positive samples could represent infections that are not currently part of an active disease process. Recent studies show that while host antiviral responses can be detected in the respiratory tract of asymptomatic virus-infected subjects, the magnitude of transcriptional response (and possibly the presence of a response at the protein level) is greater in symptomatic than asymptomatic subjects [13, 33–35]. Consistently, analysis of rhinovirus-positive samples in Study 2 showed a clear correlation between low [CXCL11] and low viral load (Supplementary Figure S4). Biomarker-positive, virus-negative samples could represent infections with untested viruses. Applying technologies such as deep sequencing for viral identification could provide greater estimates of biomarker accuracy in future studies [36]. It will also be important to rule out nonviral biological processes leading to biomarker elevation. For example, elevated CXCR3 chemokines have been reported in chronic respiratory diseases [37]. Encouragingly, our initial results indicate specificity of the biomarker test for virus infection in this population. In Study 1, 16/68 (23.5%) of patients had COPD or asthma and 11 of these patients were virus negative; these patients also tested virus negative by the host mRNA biomarker signature (Figure 2).
In future studies, it will be important to explore the performance of these biomarkers in different patient populations and clinical settings. This study focused on the patients undergoing respiratory virus testing in our health care system, largely older adults and some young children. In this patient population, biomarker-based testing could provide a simple, cost-effective method to rule in/rule out of viral infection as the cause of symptoms and/or determine which patients require additional virology testing. If these biomarkers perform well in outpatients with acute upper respiratory infection, it is possible to imagine transformative new tests to aid in rapid diagnosis of these common illnesses and promote antimicrobial stewardship. For example, a pan-viral biomarker could potentially be paired with an analogous biomarker of bacterial respiratory infection to distinguish among viral infection, bacterial infection, and viral/bacterial coinfection. In Study 1, we observed that nonviral infections did not trigger the virus host response signature (Supplementary Table S2) indicating the potential feasibility of this approach.
In addition, biomarker-based tests offer other advantages over traditional virology panels. This includes identifying patients infected with unexpected viruses. For example, in this study, biomarkers identified samples positive for viruses that were not in the original 9-virus test panel (see Figures 2 and 4; bars indicated by asterisks in virus column). This illustrates how biomarker tests could be employed for surveillance for unexpected viruses, including emerging respiratory viruses. Also, biomarker-based tests offer promise for distinguishing incidental virus detections from active viral infection, an issue coming into focus as incidental respiratory virus detections are increasingly recognized to be highly prevalent in asymptomatic subjects [13, 33, 35, 38, 39].
In sum, our results show that biomarkers of the antiviral response are robustly detected using nasopharyngeal swabs, including protein biomarkers detected with immunoassay, and that even single biomarkers detected using this minimally invasive sample type offer high diagnostic accuracy. These results compel further study of using nasopharyngeal biomarkers for improving our understanding of host/virus interactions, and for improving the diagnosis and management of patients with respiratory illness.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
The authors would like to thank Valia Mihaylova for dedicated technical assistance with immunoassays, Shannon Smith, Maureen Owen, and Sandy Cohen for assistance with sample collection and testing, Veronika Shabanova for assistance with statistical analyses, Akiko Iwasaki and Yong Kong for support with RNAseq data analysis, Peter Tattersall and the Tattersall lab for helpful assistance, and Anna Pyle for the SLR14 RIG-I ligand.
Notes
Author contributions. Conceptualization, E. F. F. and M. L. L.; Methodology, E. F. F.; Investigation, E. F. F. and M. L. L.; Resources, M. L. L.; Formal Analysis, E. F. F.; Writing—Original draft, E. F. F.; Writing—Reviewing and Editing, E. F. F. and M. L. L., Funding Acquisition, E. F. F.
Financial support. This work was supported by the National Institutes of Health (NIH) and Biomedical Advanced Research and Development Authority (BARDA) (prize to E. F. F.) and by the Yale Department of Laboratory Medicine.
Potential conflicts of interest.E. F. F. reports other support from NIH/BARDA, during the conduct of the study; In addition, E. F. F. and M. L. L. have a patent US2017/056076 pending. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: 52nd Annual Meeting of the Academy of Clinical Laboratory Physicians and Scientists, New Haven, CT, June 15–17, 2017.
References
- 1. Fendrick AM, Monto AS, Nightengale B, Sarnes M. The economic burden of non-influenza-related viral respiratory tract infection in the United States. Arch Intern Med 2003; 163:487–94. [DOI] [PubMed] [Google Scholar]
- 2. Pfunter A, Wier LM, Stocks C.. Most frequent conditions in U.S. Hospitals 2010. Rockville, M.D: Agency for Healthcare Research and Quality, 2013. [PubMed] [Google Scholar]
- 3. Tsalik EL, McClain M, Zaas AK. Moving toward prime time: host signatures for diagnosis of respiratory infections. J Infect Dis 2015; 212:173–5. [DOI] [PubMed] [Google Scholar]
- 4. The White House. National Action Plan for combating antibiotic resistant bacteria. https://obamawhitehouse.archives.gov/sites/default/files/docs/national_action_plan_for_combating_antibotic-resistant_bacteria.pdf. Accessed 21 December 2017.
- 5. Ginocchio CC, McAdam AJ. Current best practices for respiratory virus testing. J Clin Microbiol 2011Suppl.:S44–8. [Google Scholar]
- 6. Andres-Terre M, McGuire HM, Pouliot Y et al. Integrated, multi-cohort analysis identifies conserved transcriptional signatures across multiple respiratory viruses. Immunity 2015; 43:1199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ioannidis I, McNally B, Willette M et al. Plasticity and virus specificity of the airway epithelial cell immune response during respiratory virus infection. J Virol 2012; 86:5422–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Suarez NM, Bunsow E, Falsey AR, Walsh EE, Mejias A, Ramilo O. Superiority of transcriptional profiling over procalcitonin for distinguishing bacterial from viral lower respiratory tract infections in hospitalized adults. J Infect Dis 2015; 212:213–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Woods CW, McClain MT, Chen M et al. A host transcriptional signature for presymptomatic detection of infection in humans exposed to influenza H1N1 or H3N2. PLoS One 2013; 8:e52198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zaas AK, Burke T, Chen M et al. A host-based RT-PCR gene expression signature to identify acute respiratory viral infection. Sci Transl Med 2013; 5:203ra126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zaas AK, Chen M, Varkey J et al. Gene expression signatures diagnose influenza and other symptomatic respiratory viral infections in humans. Cell Host Microbe 2009; 6:207–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sweeney TE, Wong HR, Khatri P. Robust classification of bacterial and viral infections via integrated host gene expression diagnostics. Sci Transl Med 2016; 8:346ra91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yahya M, Rulli M, Toivonen L, Waris M, Peltola V. Detection of host response to viral respiratory infection by measurement of messenger RNA for MxA, TRIM21, and viperin in nasal swabs. J Infect Dis 2017; 216:1099–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rawling DC, Fitzgerald ME, Pyle AM. Establishing the role of ATP for the function of the RIG-I innate immune sensor. Elife 2015; 4: e09391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kong Y. Btrim: a fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics 2011; 98:152–3. [DOI] [PubMed] [Google Scholar]
- 16. Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013; 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Heim A, Ebnet C, Harste G, Pring-Akerblom P. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J Med Virol 2003; 70:228–39. [DOI] [PubMed] [Google Scholar]
- 19. Landry ML, Ferguson D. SimulFluor respiratory screen for rapid detection of multiple respiratory viruses in clinical specimens by immunofluorescence staining. J Clin Microbiol 2000; 38:708–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van Elden LJ, van Loon AM, van der Beek A et al. Applicability of a real-time quantitative PCR assay for diagnosis of respiratory syncytial virus infection in immunocompromised adults. J Clin Microbiol 2003; 41:4378–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li H, McCormac MA, Estes RW et al. Simultaneous detection and high-throughput identification of a panel of RNA viruses causing respiratory tract infections. J Clin Microbiol 2007; 45:2105–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lu X, Holloway B, Dare RK et al. Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses. J Clin Microbiol 2008; 46:533–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Landry ML, Ferguson D. Cytospin-enhanced immunofluorescence and impact of sample quality on detection of novel swine origin (H1N1) influenza virus. J Clin Microbiol 2010; 48:957–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shu B, Wu KH, Emery S et al. Design and performance of the CDC real-time reverse transcriptase PCR swine flu panel for detection of 2009 A (H1N1) pandemic influenza virus. J Clin Microbiol 2011; 49:2614–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sultani M, Mokhtari Azad T, Eshragian M et al. Multiplex SYBR green real-time PCR assay for detection of respiratory viruses. Jundishapur J Microbiol 2015; 8:e19041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Arvia R, Corcioli F, Ciccone N, Della Malva N, Azzi A. Detection of 12 respiratory viruses by duplex real time PCR assays in respiratory samples. Mol Cell Probes 2015; 29:408–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Whitsett JA, Alenghat T. Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat Immunol 2015; 16:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol 2016; 17:1142–9. [DOI] [PubMed] [Google Scholar]
- 29. Zhai Y, Franco LM, Atmar RL et al. Host transcriptional response to influenza and other acute respiratory viral infections--a prospective cohort study. PLoS Pathog 2015; 11:e1004869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 2014; 32:513–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Groom JR, Luster AD. CXCR3 in T cell function. Exp Cell Res 2011; 317:620–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. St John A, Price CP. Existing and emerging technologies for point-of-care testing. Clin Biochem Rev 2014; 35:155–67. [PMC free article] [PubMed] [Google Scholar]
- 33. Wolsk HM, Følsgaard NV, Birch S et al. Picornavirus-induced airway mucosa immune profile in asymptomatic neonates. J Infect Dis 2016; 213:1262–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wesolowska-Andersen A, Everman JL, Davidson R et al. Dual RNA-seq reveals viral infections in asthmatic children without respiratory illness which are associated with changes in the airway transcriptome. Genome Biol 2017; 18:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Heinonen S, Jartti T, Garcia C et al. Rhinovirus detection in symptomatic and asymptomatic children: value of host transcriptome analysis. Am J Respir Crit Care Med 2016; 193:772–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Graf EH, Simmon KE, Tardif KD et al. Unbiased detection of respiratory viruses by use of RNA sequencing-based metagenomics: a systematic comparison to a commercial PCR panel. J Clin Microbiol 2016; 54:1000–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Saetta M, Mariani M, Panina-Bordignon P et al. Increased expression of the chemokine receptor CXCR3 and its ligand CXCL10 in peripheral airways of smokers with chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2002; 165:1404–9. [DOI] [PubMed] [Google Scholar]
- 38. Byington CL, Ampofo K, Stockmann C et al. Community surveillance of respiratory viruses among families in the Utah Better Identification of Germs-Longitudinal Viral Epidemiology (BIG-LoVE) study. Clin Infect Dis 2015; 61:1217–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jartti T, Jartti L, Peltola V, Waris M, Ruuskanen O. Identification of respiratory viruses in asymptomatic subjects: asymptomatic respiratory viral infections. Pediatr Infect Dis J 2008; 27:1103–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.