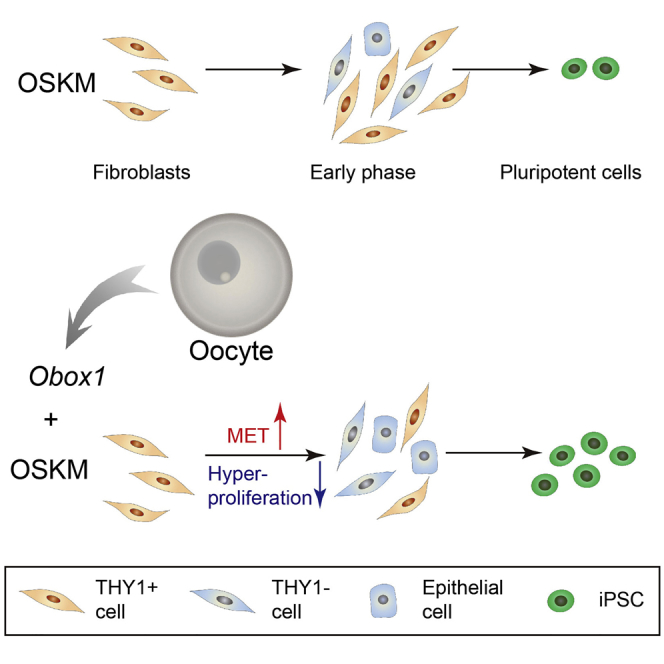
Summary
Mammalian oocytes possess fascinating unknown factors, which can reprogram terminally differentiated germ cells or somatic cells into totipotent embryos. Here, we demonstrate that oocyte-specific homeobox 1 (Obox1), an oocyte-specific factor, can markedly enhance the generation of induced pluripotent stem cells (iPSCs) from mouse fibroblasts in a proliferation-independent manner and can replace Sox2 to achieve pluripotency. Overexpression of Obox1 can greatly promote mesenchymal-to-epithelial transition (MET) at early stage of OSKM-induced reprogramming, and meanwhile, the hyperproliferation of THY1-positive cells can be significantly mitigated. Subsequently, the proportion of THY1-negative cells and Oct4-GFP-positive cells increased dramatically. Further analysis of gene expression and targets of Obox1 during reprogramming indicates that the expression of Obox1 can promote epithelial gene expression and modulate cell-cycle-related gene expression. Taken together, we conclude that the oocyte-specific factor Obox1 serves as a strong activator for somatic cell reprogramming through promoting the MET and mitigating cell hyperproliferation.
Keywords: oocyte factor, reprogramming, mesenchymal-to-epithelial transition, hyperproliferation, iPSCs, cell cycle, M phase, Obox1
Graphical Abstract
Highlights
-
•
Oocyte factor Obox1 enhances the efficiency of somatic cell reprogramming
-
•
Obox1 can replace Sox2 to complete somatic cell reprogramming
-
•
Obox1 promotes MET in the early stage of reprogramming
-
•
Obox1 mitigates cell hyper-proliferation during somatic cell reprogramming
Mammalian oocytes possess unknown factors, which can reprogram differentiated cells into totipotent embryos and thereafter give rise to a new organism. In this article, Gao and colleagues found that Obox1, an oocyte factor, can enhance somatic cell reprogramming to iPSCs by promoting mesenchymal-to-epithelial transition and mitigating cell hyperproliferation.
Introduction
Terminally differentiated somatic cells can be reprogrammed to become pluripotent either by somatic cell nuclear transfer (SCNT) (Gurdon et al., 1958, Wilmut et al., 1997) or by the forced expression of reprogramming factors, Oct4 (O), Sox2 (S), Klf4 (K), and c-Myc (M) (Takahashi et al., 2007, Takahashi and Yamanaka, 2006) to generate induced pluripotent stem cells (iPSCs). Benefits by technical simplification and free of ethical concerns, iPSCs make a significant step forward for patient-specific stem cells and individualized treatment. At the same time, the iPSC generation process is more likely a stochastic event, resulting in very low efficiency (<1%) while being time-consuming (2–3 weeks) and highly dependent on cell proliferation (Kawamura et al., 2009, Li et al., 2009, Ruiz et al., 2011, Utikal et al., 2009). On the other hand SCNT, whereby a somatic nucleus is reprogrammed by oocyte cytosolic factors in a deterministic manner, is rapid, relatively efficient, and cell division independent (Jullien et al., 2011, Jullien et al., 2014). The different efficiency between SCNT and iPSC technology (Le et al., 2014) implies that some magical factors present in the oocyte might be able to promote iPSC induction. In fact, growing evidence suggests that some oocyte-specific factors can enhance the efficiency and quality of iPSC reprogramming (Gaspar-Maia et al., 2013, Huynh et al., 2016, Jiang et al., 2013, Khaw et al., 2015, Kunitomi et al., 2016, Maekawa et al., 2011, Shinagawa et al., 2014, Singhal et al., 2010). However, although many transcription factors have been shown to enhance the generation of iPSCs, the majority of oocyte factors remain poorly investigated.
To investigate the role of oocyte factors in cellular reprogramming, we selected several highly expressed factors in oocytes based on our previously reported mass spectrometry-identified oocyte protein composition pool (Wang et al., 2010) and RNA sequencing (RNA-seq) data (Liu et al., 2016). In the present study, we focused on the maternal factor Obox1 because it is an extremely poorly studied oocyte-specific factor in development and somatic cell reprogramming. There are eight members in the Obox family, six of which were reported to express in germ cells specifically (Rajkovic et al., 2002). Obox1 was found exclusively expressed in mouse oocytes as early as one-layer follicles and throughout folliculogenesis (Rajkovic et al., 2002). In mouse stem cells, Obox genes were negatively regulated by Lin28 (Park et al., 2012). CPEB, a sequence-specific RNA binding protein, binds to Obox1 mRNA and may regulate its polyadenylation-induced translation (Racki and Richter, 2006). Recently, it was reported that Setd1b can promote the expression of the major oocyte transcription factors including Obox1, 2, 5, and 7 (Brici et al., 2017). However, the function of Obox1 remains unknown, especially in embryo development and somatic cell reprogramming. Here, we show that the overexpression of Obox1 can significantly promote the generation of iPSCs together with OSKM and can even replace Sox2 to achieve pluripotency. Further molecular analysis indicated that the overexpression of Obox1 can promote mesenchymal-to-epithelial transition (MET) and mitigate cell hyperproliferation, which can in turn selectively increase the proportion of THY1− cells dramatically in the early stage of somatic cell reprogramming.
Results
Obox1 Can Facilitate iPSC Induction
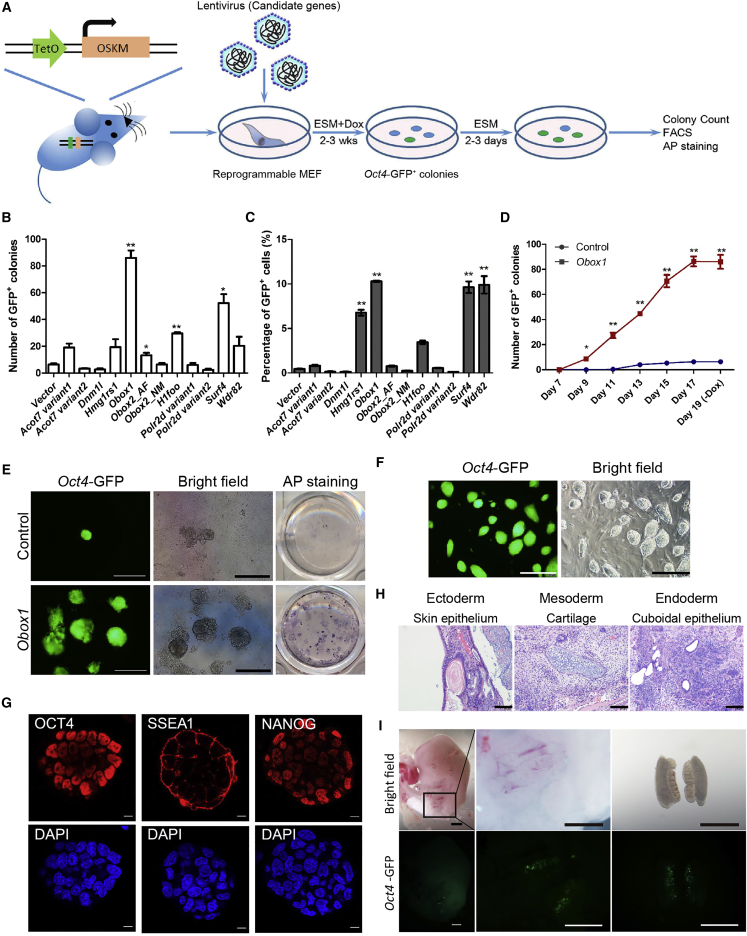
During the induction of iPSCs from somatic cells using transcription factors, only a very small proportion of cells can be reprogrammed successfully. In contrast, oocyte-based reprogramming is considered more efficient and synchronous. Recently, it has been shown that some oocyte-derived factors can indeed enhance the efficiency and quality of iPSC induction (Gonzalez-Munoz et al., 2014, Jiang et al., 2013, Khaw et al., 2015, Kunitomi et al., 2016, Maekawa et al., 2011, Shinagawa et al., 2014). We also found several highly expressed factors in oocytes in our previous study (Wang et al., 2010), after which we aimed to illustrate their roles in somatic reprogramming. To this end, we utilized reprogrammable mouse embryonic fibroblasts (MEFs) derived from the transgenic mice carrying the tetO-OSKM transgene and Oct4-GFP/Rosa26-M2rtTA (Carey et al., 2010). The induced expression of O, S, K, and M under the addition of doxycycline (Dox) was able to reprogram the MEFs into Oct4-GFP+ iPSCs (Figure 1A). We found that Obox1, Surf4, H1foo, Wdr82, and Hmg1rs1 can facilitate somatic cell reprogramming to various extent, as judged by Oct4-GFP+ colony numbers and the percentage of Oct4-GFP+ cells (Figures 1B and 1C). Among these factors, Obox1 exhibited the most dramatic positive effect on iPSC generation. Obox1 was exclusively expressed in oocytes and early embryos before the 2-cell stage (Figure S1A). Overexpression of Obox1 accelerated the formation of Oct4-GFP+ colonies and resulted in a 13-fold increase of Oct4-GFP+ colony numbers (Figures 1B and 1D). Notably, the percentage of Oct4-GFP+ cells also increased up to 10% at day 17 by exogenous Obox1 along with OSKM (Figure 1C). The alkaline phosphatase-positive (AP+) colonies were also multiplied (Figure 1E, right panel). The OSKM + Obox1-iPSCs exhibited typical embryonic stem cell (ESC) morphology (Figures 1E [left and middle panel], 1F, and S1B) with a compact appearance and a well-defined border and normal karyotype (Figure S1C). Quantitative real-time PCR (qRT-PCR) and immunofluorescence staining indicated that OSKM + Obox1-iPSCs exhibited expression of pluripotent genes at the mRNA and protein levels comparable with that of ESCs (Figures S1D and 1G). We then conducted in vitro and in vivo differentiation assays to examine the differentiation potential of OSKM + Obox1-iPSCs (Figures S1E, 1H, and 1I). Through embryoid body (EB)-mediated in vitro differentiation, the differentiated cells showed an upregulation of markers of three germ layers (Figure S1E). Teratomas also formed after subcutaneous injection of OSKM + Obox1-iPSCs into nude mice, with tissues of three germ layers, including skin epithelium (ectoderm), cartilage (mesoderm), and cuboidal epithelium (endoderm) (Figure 1H). Furthermore, the chimera formation assay was performed, whereby the OSKM + Obox1-iPS cell lines could integrate into the gonads of the chimeric mice (Figure 1I). Obox1 is specifically expressed in rodents, and we further investigated whether mouse Obox1 can promote human iPSC induction, and no positive effects could be observed (data not shown).
Figure 1.
Exogenous Expression of Obox1 Promotes iPSC Generation
(A) Strategy for functional studies of candidate genes in reprogramming. Parallel experiments were performed using individual candidate genes and the empty vector as a control.
(B) The number of Oct4-GFP+ colonies was counted at day 19 after induction.
(C) The percentage of Oct4-GFP+ cells was analyzed by FACS at day 19 after induction.
(D) Kinetics of the Oct4-GFP+ colonies formation are facilitated by Obox1.
(E) Morphology of Oct4-GFP+ primary colonies (left and middle panels). Representative AP-stained plates are shown 19 days after induction (right panel). Scale bars, 400 μm.
(F) Morphology of OSKM + Obox1-iPSC lines. Scale bars, 400 μm.
(G) Immunostaining of pluripotent marker genes OCT4 (red), SSEA1 (red), and NANOG (red) in OSKM + Obox1-iPSC lines. Nuclear staining by DAPI (blue). Scale bars, 50 μm.
(H) H&E staining of teratoma generated from OSKM + Obox1-iPSCs showing representative ectodermal (epidermis), mesodermal (cartilage), and endodermal (cuboidal epithelium) tissues. Scale bars, 100 μm.
(I) Representative photos of the contribution and spatial distribution of Oct4-GFP+ cells in the gonads from E12.5 OSKM + Obox1-iPSC-derived chimera embryo. Scale bars, 1 mm.
Data are presented as the mean ± SEM (n = 3); ∗p < 0.05, ∗∗p < 0.01 by Student's t test for comparison and empty vector as control. See also Figure S1 and Table S1.
Obox1 Can Replace Sox2 to Accomplish Successful Somatic Cell Reprogramming
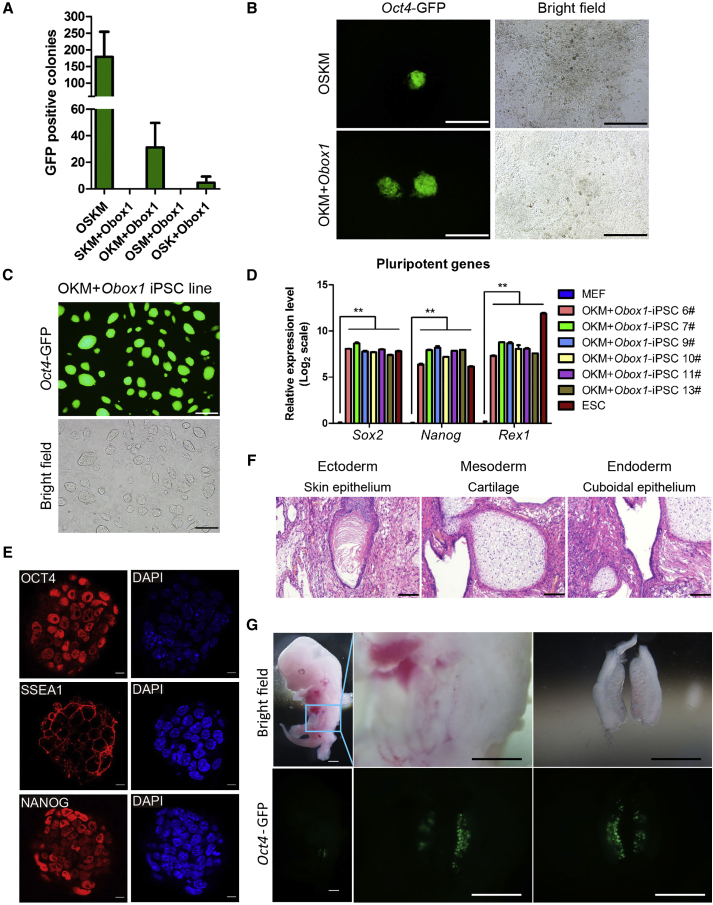
The drastic enhancement of somatic cell reprogramming by Obox1 promoted us to further explore whether Obox1 can replace Yamanaka factors. We then substituted individual factors with Obox1 and evaluated reprogramming efficiency using the MEFs derived from transgenic mice carrying Oct4-GFP/Rosa26-M2rtTA (OG2-MEF). Finally, we found that Obox1 can replace Sox2 to yield Oct4-GFP+ colonies with typical ESC colony morphology (Figures 2A and 2B). The OKM + Obox1-iPSCs exhibited typical ESC morphology (Figure 2C) and positive AP activity (Figure S2A). Besides, they expressed pluripotent markers and cell-surface markers of mouse ESCs (Figures 2D and 2E). Karyotype analysis showed that OKM + Obox1-iPSCs maintained the normal 40 chromosomes (Figure S2B). Moreover, OKM + Obox1-iPSCs also exhibited differentiation ability both in vitro and in vivo. EBs were formed using OKM + Obox1-iPSCs, and marker genes of the three germ layers were detected in the plated EBs (Figure S2C). Teratomas with three germ layers could be generated by OKM + Obox1-iPSCs (Figure 2F). Additionally, chimeric mice could be generated by germline transmission (Figure 2G). To understand the mechanism underlying the substitution, we collected the samples on day 3 of the reprogramming process and performed RNA-seq. Compared with OKM + empty vector, the OSKM and OKM + Obox1 possessed differential expression genes (DEGs) (fold change >2) sharing similar pathways by gene ontology (GO) analysis (Figures S2D and S2E). Thus, oocyte factor Obox1 not only facilitated iPSC induction, but was also able to replace the Yamanaka factor Sox2 to accomplish the reprogramming process.
Figure 2.
Obox1 Can Replace Sox2 during Somatic Cell Reprogramming
(A) The number of Oct4-GFP+ colonies was counted at the end of induction.
(B) Morphology of primary colonies. Scale bars, 400 μm.
(C) Morphology of OKM + Obox1-iPSC lines. Scale bars, 400 μm.
(D) qRT-PCR analysis shows pluripotency gene expression in OKM + Obox1-iPSCs. Relative mRNA expression was normalized to hypoxanthine-guanine phosphoribosyltransferase (Hprt) mRNA and represented relative to expression in MEFs.
(E) Immunostaining of pluripotent markers OCT4 (red), SSEA1 (red), and NANOG (red) in OKM + Obox1-iPSC lines. Nuclear staining by DAPI (blue). Scale bars, 50 μm.
(F) H&E staining of teratoma generated from OKM + Obox1-iPSCs showing representative ectodermal (epidermis), mesodermal (cartilage), and endodermal (cuboidal epithelium) tissues. Scale bars, 100 μm.
(G) Representative photos of the contribution and spatial distribution of Oct4-GFP+ cells in the gonads from E12.5 OKM + Obox1-iPSC-derived chimera embryo. Scale bars, 1 mm.
Data are presented as the mean ± SEM (n = 3); ∗∗p < 0.01 by Student's t test. See also Figure S2.
Obox1 Mitigates Cell Hyper-proliferation and Functions at the Early Stage of Reprogramming
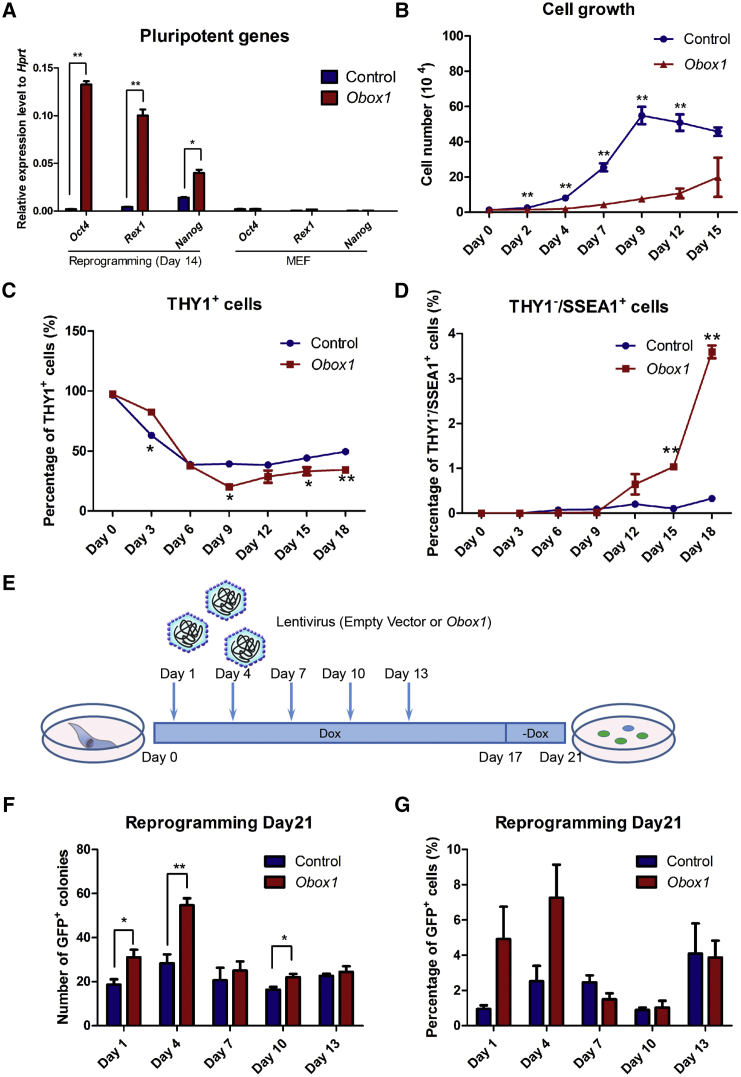
The positive role of Obox1 on reprogramming and its ability to replace Sox2 prompted us to investigate the influence of Obox1 overexpression on the activation of reprogramming factors and pluripotent genes. Although Oct4, Rex1, and Nanog were expressed remarkably higher under the ectopic expression of Obox1 in reprogramming compared with the control group (Figure 3A), overexpression of Obox1 alone could hardly activate the expression of these genes in MEFs (Figure 3A). Instead, we noticed that the cell proliferation under Obox1 overexpression was dramatically different from that of the control. The growth curve during reprogramming showed that the overexpression of Obox1 resulted in significant reduction of cell proliferation rate as early as the first 2 days (Figure 3B). Then we analyzed the intermediate population progression at successive time points during reprogramming. Flow cytometry showed that addition of Obox1 promoted the transition from the THY1+ cell population to the THY1− cell population (Figure 3C), which is accepted as one of the prerequisite characteristics of successful reprogramming in the early stage. Furthermore, SSEA1+ and subsequent Oct4-GFP+ cell numbers were significantly increased by forced expression of Obox1 (Figures 3D and 1D). We then investigated the time window during which Obox1 can enhance somatic cell reprogramming. We introduced Obox1 at different time points during the somatic cell reprogramming process and examined the number of Oct4-GFP+ colonies and the percentage of Oct4-GFP+ cells at the end of reprogramming (Figures 3E–3G). Early introduction of exogenous Obox1 before day 4 significantly increased the reprogramming efficiency while Obox1 overexpression after day 7 showed no obvious effect. These results suggested that Obox1 plays an important role in promoting somatic cell reprogramming at the initiation phase of reprogramming.
Figure 3.
Obox1 Mitigates Cell Hyper-proliferation and Functions at the Early Stage of Reprogramming
(A) qRT-PCR analysis shows pluripotent gene expression in reprogrammable cells on day 14 post induction with or without Obox1 or Obox1-infected MEFs on day 4, and the empty vector-infected cells as control for each group.
(B) Proliferation curves of reprogrammable cells transduced with or without Obox1.
(C) Kinetic changes of percentage of THY1+ population at indicated time points during reprogramming by FACS analysis.
(D) Percentage of kinetic changes of percentage of THY1−/SSEA1+ population during reprogramming by FACS analysis.
(E) Strategy of time-course study of Obox1 during reprogramming.
(F) Reprogrammable cells were infected with Obox1 at indicated time points and the number of Oct4-GFP+ colonies were counted at 21 days post induction.
(G) Reprogrammable cells were infected with Obox1 at indicated time points and Oct4-GFP+ cells were analyzed by FACS at 21 days post induction.
Data are presented as the mean ± SEM (n = 3); ∗p < 0.05, ∗∗p < 0.01 by Student's t test for comparison and empty vector at indicated time points as control.
Genome-wide Analysis of the Effects of Obox1 Overexpression on Somatic Reprogramming
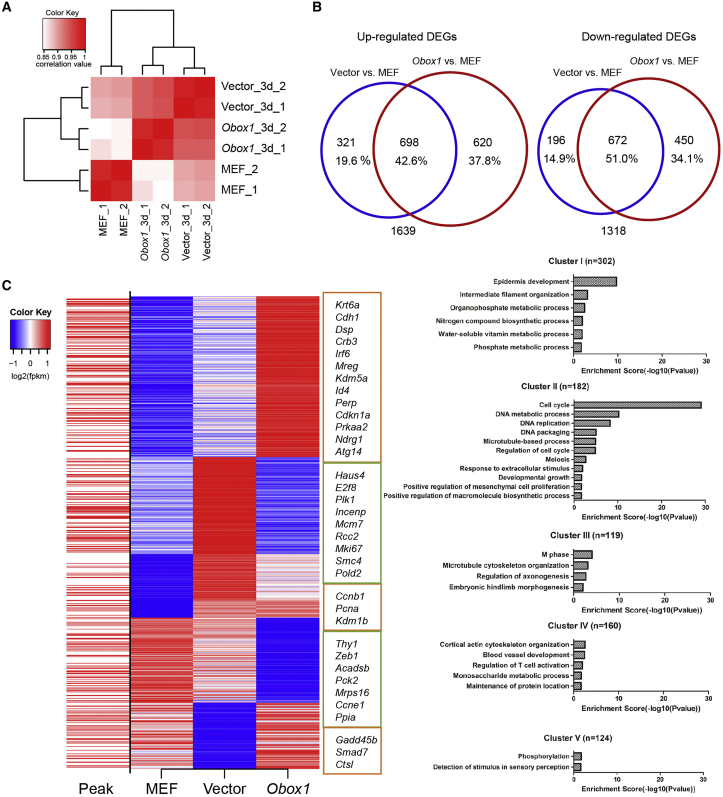
To understand how Obox1 promotes the reprogramming process, we employed the reprogrammable system with or without Obox1 overexpression, and performed RNA-seq and chromatin immunoprecipitation sequencing (ChIP-seq) on day 3. Unsupervised hierarchical clustering analysis (Figure 4A) and principal component analysis (Figure S3A) based on the RNA-seq data showed high similarity among replicates of the same treatment, while the MEF sample was highly distinguishable from other samples, regardless of whether the exogenous Obox1 was introduced or not (Figures 4A and S3A). Under the overexpression of Obox1, the number of DEGs (fold change ≥1.5 and false discovery rate ≤0.05) compared with the MEF was much more than that in the empty vector (control) (Figure 4B). ChIP-seq exhibited a significantly enriched binding of Obox1 at promoters (Figure S3B). Moreover, 36.7% of the DEGs had Obox1 binding within 2 kb of their transcription start sites (Figure 4C, left column). Most Obox1 targeted genes were also the targets of Sox2 by comparing our Obox1 ChIP-seq dataset and the previously reported Sox2 ChIP-seq data (Figures S3C and S3D) (Chronis et al., 2017), which provides another explanation for the ability of Obox1 to replace Sox2 in reprogramming.
Figure 4.
Genome-wide Analysis of the Effects of Obox1 Overexpression on Somatic Reprogramming
(A) Heatmap shows the Pearson correlation of gene transcription profiles of MEFs, reprogrammable cells transduced with OSKM or OSKM plus Obox1 on day 3 post induction.
(B) Venn diagram shows the overlap of different expression genes between samples (Vector and Obox1) on reprogramming day 3 and MEFs. Different expression genes were calculated by EdgeR (see Experimental Procedures).
(C) Different expression genes between samples (Vector and Obox1) on reprogramming day 3 were grouped into five clusters by K-means clustering based on RNA-seq data. Heatmap shows different expression genes (red, upregulated genes; blue, downregulated genes), which are targeted by Obox1 through ChIP-seq (left column). Gene ontology analysis of each cluster is shown in the right column.
According to the differential expression patterns in the three samples, we obtained several major gene clusters (Figures 4C and S3E; Table S2). A large number of genes increased in the early stage of reprogramming were further upregulated by Obox1 (cluster I; ∼300 genes), and GO analysis showed that they were mainly involved in epithelial cell differentiation and intermediate filament organization. Among these genes, 119 of them possess Obox1 binding sites. Another impressive cluster (cluster II) was composed of the genes that were significantly increased in the control group compared with MEF, but strikingly decreased under Obox1 overexpression; around half of them have Obox1 binding on their promoters, and the pathways associated with the cell cycle and DNA replication were highly enriched in this cluster. There was a similar case in cluster III, where genes had greatly increased control than in the Obox1 overexpression group, and genes were found to be enriched during M phase and microtubule cytoskeleton organization. Such a retraction in cell division-associated pathways was consistent with the slowdown in cell proliferation rate by Obox1 overexpression in reprogramming. Those genes (cluster IV, Figure 4C) that were downregulated early in reprogramming were dramatically decreased under Obox1 overexpression, including Thy1 and Zeb1, among others. Target analysis by integration of transcriptome and ChIP-seq data with BETA showed that Obox1 more likely acts as a repressor for these genes (Figure S3F).
Obox1 Promotes MET in the Initiation Stage of Reprogramming
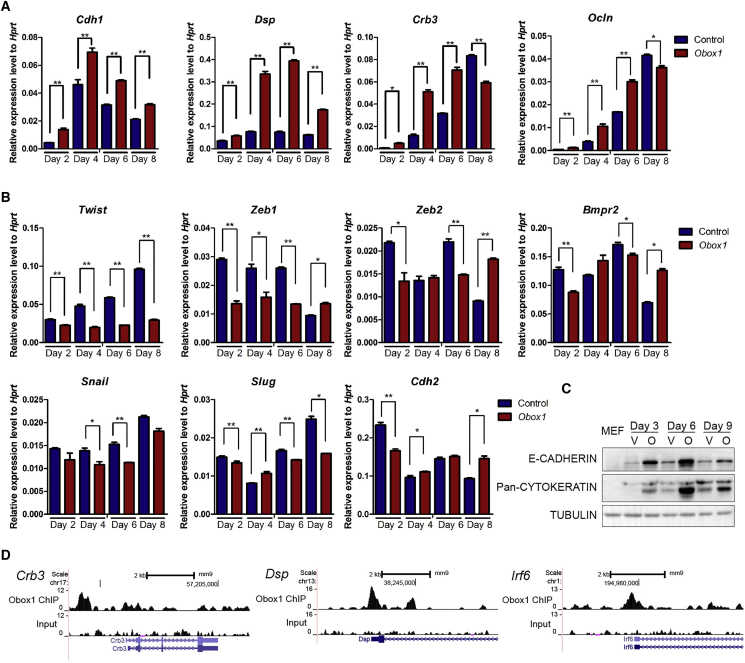
The MET was identified as a hallmark at the initiation phase of MEF reprogramming (Li et al., 2010, Samavarchi-Tehrani et al., 2010). To further explore the role of Obox1 on MET, we checked the levels of the key epithelial and mesenchymal regulators and fibroblast markers in our RNA-seq dataset. The low expression level of the epithelial genes was found in MEFs and in the empty vector control groups, while Obox1 overexpression lead to significant upregulation of these genes, including Cdh1, Ocln, Epcam, and Crb3 (Figure S4 and Table S3). Consistent with the activation of epithelial-like markers, the expression level of mesenchymal regulators, such as Snail, Slug, Zeb1, and Zeb2, were markedly downregulated in the Obox1 group, and the fibroblast markers Cdh2 and Thy1 were also significantly downregulated (Figure S4). These findings suggest that Obox1 can efficiently stimulate MET at the initiation stage of MEF reprogramming. To confirm this, we examined the RNA expression level of epithelial- and mesenchymal-associated genes during reprogramming by qRT-PCR. As shown in Figure 5A, concomitant with the Dox induction, the upregulation of epithelial regulators such as E-cadherin, Dsp, Crb3, and Ocln can be further elevated by Obox1 from day 2 to day 6, compared with the control at indicated time points, while the expression level of mesenchymal-associated genes, such as Twist, Zeb1, and Zeb2, were significantly downregulated (Figure 5B). Western blot analysis further validated the increased expression of E-CADHERIN and CYTOKERATINS (Figure 5C). Prominent Obox1 peaks in the promoter regions of epithelial-associated regulators Dsp, Crb3, and Irf6 suggested direct binding and regulation (Figure 5D). Therefore, Obox1 significantly promotes MET, which may in turn contribute to its role in enhancing somatic cell reprogramming.
Figure 5.
Obox1 Promotes MET at the Initiation Stage of Reprogramming
(A) qRT-PCR analysis of epithelial genes expression in Obox1-infected reprogrammable cells at indicated time points. The expression levels were normalized to Hprt.
(B) qRT-PCR analysis of mesenchymal genes expression in Obox1-infected reprogrammable cells at indicated time points. The expression levels were normalized to Hprt.
(C) Western blot analysis the protein levels of E-CADHERIN and Pan-CYTOKERATIN during reprogramming; α-TUBULIN was used as a loading control (V, vector; O, Obox1).
(D) ChIP density profiles of Obox1 at epithelial-related gene promoters.
Data are presented as the mean ± SEM (n = 3); ∗p < 0.05, ∗∗p < 0.01 by Student's t test for comparison and empty vector at indicated time points as control. See also Figure S4 and Table S3.
Obox1 Mitigates Cell Hyper-proliferation by Modulating Cell-Cycle-Related Gene Expression
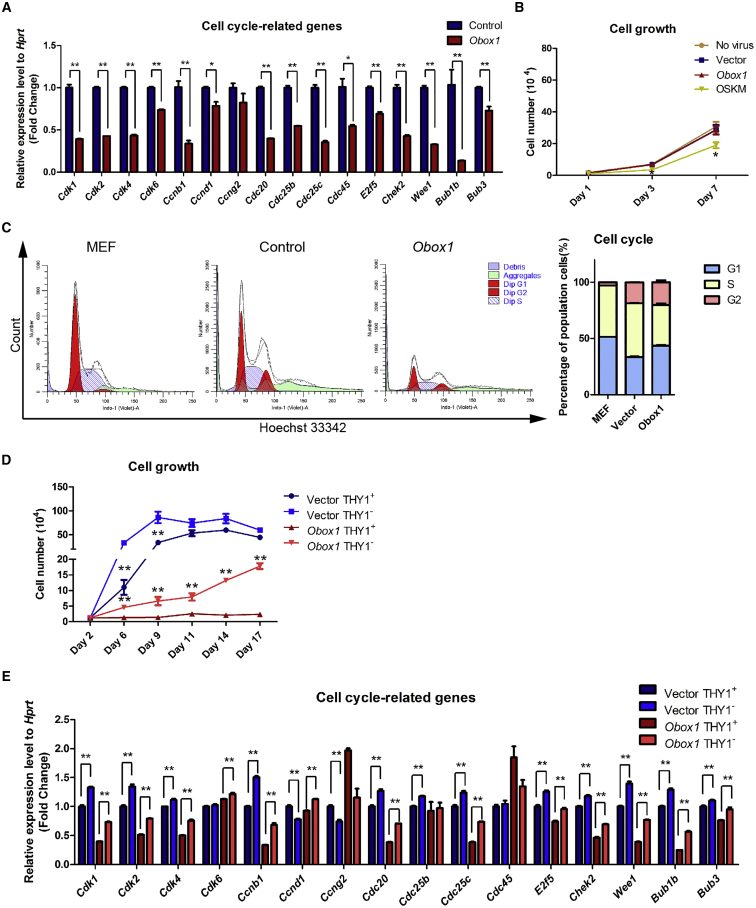
As mentioned above, OSKM reprogramming cells displayed a much higher proliferation rate compared with that of OSKM + Obox1 reprogramming cells (Figure 3B). Transcriptome profile showed that the genes associated with cell-cycle regulation remained unchanged or downregulated in the Obox1 group while these genes were markedly upregulated in the control group (Figure 4C). As shown in Figure S5A, the cyclin-dependent kinases (CDKs), cell division cycle proteins, and other cell-cycle-related genes were dramatically upregulated in OSKM reprogramming cells compared with those of OSKM + Obox1 reprogramming cells (Figure S5A and Table S4). Next, we performed qRT-PCR to validate expression levels of the genes associated with the cell cycle during reprogramming (Figure 6A). In addition, ChIP-seq analysis showed that Obox1 was enriched at the promoter regions of these genes, indicating direct regulation of the cell cycle by Obox1 (Figure S5B). More interestingly, overexpression of Obox1 alone does not affect cell number (Figure 6B), which suggests that the slowdown in cell proliferation rate by Obox1 might depend on OSKM expression. Furthermore, the impact of Obox1 on cell proliferation during reprogramming was also reflected in the cell-cycle properties. Distinct cell-cycle stage composition is reconstructed at the initial phase in OSKM-mediated reprogramming. Hoechst 33342 DNA staining assay and bromodeoxyuridine (BrdU) incorporation assay showed a markedly shortened G1 phase in OSKM reprogramming cells while this shortened G1 was not obvious in Obox1 + OSKM reprogramming cells (Figures 6C and S5C).
Figure 6.
Obox1 Mitigates Cell Hyper-proliferation and Enlarges the Proportion of THY1− Cells during Reprogramming
(A) qRT-PCR analysis of cell-cycle-related gene expression in reprogrammable cells transduced with OSKM or OSKM plus Obox1 on day 2 post induction. The expression level was normalized to Hprt.
(B) Proliferation curves of MEFs transduced with or without Obox1 or OSKM, respectively (empty vector as control).
(C) Cell-cycle analysis by FACS in MEFs or the reprogramming cells on day 2 post infection and comparison of the indicated cell-cycle phase (empty vector versus MEF; Obox1 versus empty vector).
(D) Proliferation curve of THY1+ and THY1− cells sorted from reprogrammable cells on day 2 post induction. The cells were seeded in 24-well plates at a density of 1.2 × 104 cells per well and counted at indicated time points, and the THY1+ population and THY1− population compared in the same group.
(E) qRT-PCR analysis of cell-cycle-related gene expression in THY1+ and THY1− cell populations sorted from reprogrammable cells on day 2 post induction. The expression levels were normalized to Hprt. THY1+ in vector or Obox1 group as control.
Data are presented as the mean ± SEM (n = 3); ∗p < 0.05, ∗∗p < 0.01 by Student's t test for comparison. See also Figure S5 and Table S4.
Since Obox1 accelerated the reduction in the THY1+ cell population (Figure 3C), we next asked whether Obox1 has a different effect on THY1+ and THY1− cell growth. After Dox induction for 2 days, we sorted the THY1+ and THY1− cells and performed a cell proliferation assay. Consistent with previous studies, OSKM induced a strong increase of cell proliferation rate in both the THY1+ and THY1− cell populations during reprogramming (Figure 6D). As shown in Figure 6D, in the OSKM + Obox1 group THY1− cells displayed increased proliferation similarly to OSKM-induced reprogramming, whereas the growth of THY1+ cells was dramatically reduced and eventually ceased. This result was consistent with the expression features of cell-cycle-related genes. Most cell-cycle-related genes were downregulated by Obox1 in both THY1+ and THY1− populations, but seemed much more obvious in the THY1+ population (Figure 6E).
Taken together, our data suggested that Obox1 can modulate the expression of genes associated with the cell cycle and that a proportion of THY1− cells shows selective growth advantage over THY1+ cells.
Discussion
Our study aims to discover more native oocyte-derived factors that can facilitate somatic cell reprogramming (Gurdon and Melton, 2008). By performing small-scale screening, we demonstrated that the oocyte factor Obox1 can markedly facilitate iPSC induction together with OSKM and can even replace Sox2 to accomplish somatic cell reprogramming. Furthermore, we found that overexpression of Obox1 during reprogramming can promote MET and mitigate cell hyper-proliferation, which can in turn increase the proportion of THY1− cells dramatically, thus benefiting the reprogramming efficiency.
As reported previously, MET is one of the critical early events in reprogramming of MEFs into iPSCs (Li et al., 2010, Samavarchi-Tehrani et al., 2010). As our ChIP-seq and RNA-seq analyses of OSKM + Obox1 in the reprogramming showed, Obox1 upregulated the epithelial-associated genes such as Dsp and Crb3, as well as Smad7, which is a key repressor of transforming growth factor β (TGF-β) signaling. TGF-β is a strong signal for EMT and the inhibition of this pathway (by small-molecule compounds or Smad7 overexpression) have been proved able to enhance reprogramming and replace c-Myc and Sox2 (Ichida et al., 2009, Li et al., 2010, Maherali and Hochedlinger, 2009). Targeting Smad7 and the epithelial factors and subsequently promoting MET in early reprogramming by overexpression of Obox1 may be the explanation for its ability to replace the role of Sox2 to achieve reprogramming together with OKM.
Cell proliferation is another important characteristic in reprogramming. While some factors advanced the iPSC derivation by increased cell growth, others facilitated the reprogramming independent of proliferation (Hanna et al., 2009). Until now, the role of cell proliferation on reprogramming has remained controversial. It is reported that rapid proliferation facilitates reprogramming (Hong et al., 2009, Kawamura et al., 2009, Ruiz et al., 2011), but excessive proliferation also decreased the reprogramming efficiency of both mouse and human fibroblasts (Gupta et al., 2015, Xu et al., 2013), while inhibition of cell proliferation by chemotherapeutic drugs at an early reprogramming stage could help to improve efficiency (Xu et al., 2013). In our study an appropriate dose of ectopic Obox1 negatively regulated cell proliferation, including downregulating cyclins, CDKs, and the genes involved in DNA replication, chromatin condensation, and cytokinesis, and upregulating cyclin-dependent kinase inhibitors. Moreover, the effect of Obox1 was shown to be much greater on the hyper-proliferation of THY1+ cells, leading to an increased proportion of THY1− cells thus able to facilitate the reprogramming process.
It is predicted that Obox1 may function as a repressor rather than an activator (Figure S3F). This could be due to its homeodomain-related and helix-turn-helix motif, lambda-like repressor domains (Gallardo et al., 2007). Although Obox1 and Obox2 share 97% similarity (only six different amino acid residues are different), their effects on reprogramming efficiency were different (Figures 1B and 1C). The individual characteristics of the side chain lead to the different elaborate three-dimensional structure between Obox1 and Obox2, which endows them with unique functions. For instance, the amino acid Leu76 in Obox1, located near the first helix, may fold in a hydrophobic pocket together with other amino acids, while Pro76 in Obox2 may cause a turn and change the direction of the main chain of the protein (Harrison and Aggarwal, 1990, Luscombe et al., 2001, Schofield, 1987).
Obox1 is an oocyte-specific protein first induced in early follicle growth that maintains high expression in oocytes and early embryo until the 2-cell stage (Gallardo et al., 2007, Rajkovic et al., 2002), which is also supported by our study. Obox1 belongs to the Obox family, which possesses the conserved homeodomain (and hence potential transcription factors) preferentially expressed in germ cells (Rajkovic et al., 2002). Knockdown of Obox4 in oocytes resulted in abnormal metaphase I arrest via STAT3 and MPF/MAPK signaling pathways (Lee et al., 2010, Lee et al., 2016). Obox6 knockout mice undergo normal early embryonic development and are fertile (Cheng et al., 2007). The functional study of Obox1 is thus far blank. The reduction of cell proliferation rate by Obox1 in somatic cell reprogramming mimics the early embryo state to some extent. In fact, nearly half of the DEGs specifically upregulated by Obox1 (288/620) in our data are also highly expressed in zygote/2-cell embryo (Macfarlan et al., 2012). Taken together, this evidence suggests that Obox1 is an extremely important factor in embryo development and reprogramming, and the functional exploration of its role during the folliculogenesis, oogenesis, and early embryo development would be very helpful in unveiling the mysterious oocyte factors and reprogramming process.
Experimental Procedures
Candidate Genes’ Lentiviral Vector Construction and iPSC Derivation
Full-length mouse Acot7 (NM_001146057, NM_133348), Dnm1l (NM_152816.3), H1foo (NM_138311), Hmga1-rs1 (NM_001166476), Polr2d (NM_027002, NM_027101), Surf4 (NM_011512), Wdr82 (NM_029896), Obox1 or FLAG-tagged-Obox1 (NM_027802), and Obox2 (NM_145708, AF461107) were cloned and inserted into the FUW-TET-On vector. Inducible iPSCs were generated as previously described (Brambrink et al., 2008, Stadtfeld et al., 2008). ESC-like colonies appeared 2–3 weeks after induction. The colonies were picked and propagated after Dox withdrawal.
The analyses of iPSCs, such as qRT-PCR, AP staining, in vitro differentiation, teratoma formation, and chimera experiments, were performed as previously described (Kang et al., 2009), as detailed in the Supplemental Experimental Procedures. All primer sequences are available in Table S1.
RNA Sequencing
Total RNA from independent biological replicates of each uninduced MEF, induced MEFs with or without Obox1 at 72 hr for OSKM + Obox1 system, and of overexpressing OSKM, OKM + Obox1, or OKM + empty vector at 72 hr for replacement system, was isolated using the QIAGEN RNeasy Kit (Germantown, USA). The RNA samples were subject to mRNA fragmentation, cDNA synthesis, and library preparation using a KAPA Stranded RNA-Seq Kit Illumina platform (KK8440; Kapa, Wilmington, USA). All adapters were diluted from the adapters offered by TruSeq Library Prep Pooling kit (Illumina, USA). Single-end 50-bp sequencing was further performed on HiSeq 2500 (Illumina) at Berry Genomics.
ChIP Sequencing
ChIP experiments were performed using the MAGnify Chromatin Immunoprecipitation System (Invitrogen, Carlsbad, USA) according to the manufacturer's recommendations. Ten million cells on reprogramming day 3 were used for per immunoprecipitation reaction. In brief, cells were chemically crosslinked at room temperature by the addition of formaldehyde to 1% final concentration for 10 min and quenched with 0.125 M final concentration glycine. Crosslinked cells were resuspended in lysis buffer and chromatin was sonicated to 100–400 bp with a Covaris M220 system. The sonicated chromatin was then immunoprecipitated with 3 μg of FLAG antibody (Sigma F1804; St. Louis, USA) for each immunoprecipitation reaction. A fraction of “whole-cell extract” obtained without antibody was retained as an input control. DNA was eluted by elution buffer and purified through phenol-chloroform extraction and isopropanol precipitation. The sequence libraries were generated using a KAPA HyperPlus Library Preparation kit (KK8510; Kapa), following the manufacturer's instructions. Single-end 50-bp sequencing was further performed on a HiSeq 2500 (Illumina) at Berry Genomics.
Trimming and Alignment of Sequencing Reads
All of the RNA-seq reads were first mapped to hg19 reference genome using TopHat (v 2.1.1) with default parameters (Trapnell et al., 2009). Gene expression for each sample was quantified to FPKM (fragments per kilobase of transcript per million mapped reads) using Cufflinks (v 2.2.1) to eliminate the effects of sequencing depth and transcript length (Trapnell et al., 2010). All ChIP-seq samples were mapped to mm9 reference genome using the bowtie 2 (v 2.2.9) command with default parameters (Li and Durbin, 2009).
Cell Growth Curve, BrdU Assay, and Cell-Cycle Analysis
For growth curve analysis, the cells were plated onto 12-well plates at a density of 1.2 × 104 cells per well, and were harvested every 48 or 72 hr and counted in a cell-counting chamber. Each group contained two or three replicates. For BrdU incorporation assay, cells were treated with BrdU for 60 min. The cells were stained following the manufacturer's instructions (FITC BrdU Flow Kit, Becton Dickinson, San Jose, USA). For cell-cycle analysis, cells were stained with Hoechst 33342 (Sigma) and analyzed using a Beckmam Coulter CytoFLEX S flow cytometer or BD FACSAria II (for sorting).
Mice
All of our study procedures were consistent with those in the Tongji University guide for the care and use of laboratory animals.
Statistical Analysis
Results are presented as the mean ± SEM of independent experiments. Significance was determined by Student's t test.
Author Contributions
L.W. designed and performed the experiments, data analysis, discussion, and writing; Y.W. performed bioinformatics analysis; B.P., Z.H., Y.D., K.C., M.G., H.L., X.C., X.K., Y.Z., Y.B., Y.W., and H.W. contributed to experimental work and discussion; S.G., L.K., and R.L. supervised the study and contributed to writing.
Acknowledgment
We would like to thank Yang Shi from the Yong Zhang laboratory for help with the bioinformatics analysis and Professor Jiuhong Kang from Tongji University for providing the E-CADHERIN antibodies. We are also grateful to our laboratory colleagues, especially Dr. Jiayu Chen, for their assistance with experiments and advice. This work was supported by the National Natural Science Foundation of China (31430056, 31501183, 31371512, 81322029, and 31721003), the Ministry of Science and Technology of China (2016YFA0100400), the Shanghai Subject Chief Scientist Program (15XD1503500), and the Chenguang Program (15CG19) from the Shanghai Education Development Foundation and the Shanghai Municipal Education Commission.
Published: October 12, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, five figures, and four tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2017.09.012.
Contributor Information
Rongrong Le, Email: lerongrong@tongji.edu.cn.
Lan Kang, Email: lank0305@126.com.
Shaorong Gao, Email: gaoshaorong@tongji.edu.cn.
Accession Numbers
The sequencing datasets have been deposited in NCBI's Gene Expression Omnibus and are accessible through the accession number GEO: GSE97859.
Supplemental Information
Different expression genes between samples (Vector and Obox1) on reprogramming day 3 were grouped into five clusters by K-means clustering based on RNA-seq data. The potential targets of Obox1 based on the ChIP-seq data are presented as 1 in the binding column.
References
- Brambrink T., Foreman R., Welstead G.G., Lengner C.J., Wernig M., Suh H., Jaenisch R. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brici D., Zhang Q., Reinhardt S., Dahl A., Hartmann H., Schmidt K., Goveas N., Huang J., Gahurova L., Kelsey G. Setd1b, encoding a histone 3 lysine 4 methyltransferase, is a maternal effect gene required for the oogenic gene expression program. Development. 2017;144:2606–2617. doi: 10.1242/dev.143347. [DOI] [PubMed] [Google Scholar]
- Carey B.W., Markoulaki S., Beard C., Hanna J., Jaenisch R. Single-gene transgenic mouse strains for reprogramming adult somatic cells. Nat. Methods. 2010;7:56–59. doi: 10.1038/nmeth.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W.C., Hsieh-Li H.M., Yeh Y.J., Li H. Mice lacking the Obox6 homeobox gene undergo normal early embryonic development and are fertile. Dev. Dyn. 2007;236:2636–2642. doi: 10.1002/dvdy.21261. [DOI] [PubMed] [Google Scholar]
- Chronis C., Fiziev P., Papp B., Butz S., Bonora G., Sabri S., Ernst J., Plath K. Cooperative binding of transcription factors orchestrates reprogramming. Cell. 2017;168:442–459.e20. doi: 10.1016/j.cell.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo T.D., John G.B., Shirley L., Contreras C.M., Akbay E.A., Haynie J.M., Ward S.E., Shidler M.J., Castrillon D.H. Genomewide discovery and classification of candidate ovarian fertility genes in the mouse. Genetics. 2007;177:179–194. doi: 10.1534/genetics.107.074823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A., Qadeer Z.A., Hasson D., Ratnakumar K., Leu N.A., Leroy G., Liu S., Costanzi C., Valle-Garcia D., Schaniel C. MacroH2A histone variants act as a barrier upon reprogramming towards pluripotency. Nat. Commun. 2013;4:1565. doi: 10.1038/ncomms2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Munoz E., Arboleda-Estudillo Y., Otu H.H., Cibelli J.B. Cell reprogramming. Histone chaperone ASF1A is required for maintenance of pluripotency and cellular reprogramming. Science. 2014;345:822–825. doi: 10.1126/science.1254745. [DOI] [PubMed] [Google Scholar]
- Gupta M.K., Teo A.K.K., Rao T.N., Bhatt S. Excessive cellular proliferation negatively impacts reprogramming efficiency of human fibroblasts. Stem Cells Transl. Med. 2015;4:1101–1108. doi: 10.5966/sctm.2014-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon J.B., Elsdale T.R., Fischberg M. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature. 1958;182:64–65. doi: 10.1038/182064a0. [DOI] [PubMed] [Google Scholar]
- Gurdon J.B., Melton D.A. Nuclear reprogramming in cells.pdf. Science. 2008;322:5. doi: 10.1126/science.1160810. [DOI] [PubMed] [Google Scholar]
- Hanna J., Saha K., Pando B., van Zon J., Lengner C.J., Creyghton M.P., van Oudenaarden A., Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S.C., Aggarwal A.K. DNA recognition by proteins with the helix-turn-helix motif. Annu. Rev. Biochem. 1990;59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- Hong H., Takahashi K., Ichisaka T., Aoi T., Kanagawa O., Nakagawa M., Okita K., Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh L.M., Shinagawa T., Ishii S. Two histone variants TH2A and TH2B enhance human induced pluripotent stem cell generation. Stem Cells Dev. 2016;25:251–258. doi: 10.1089/scd.2015.0264. [DOI] [PubMed] [Google Scholar]
- Ichida J.K., Blanchard J., Lam K., Son E.Y., Chung J.E., Egli D., Loh K.M., Carter A.C., Di Giorgio F.P., Koszka K. A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell. 2009;5:491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Lv W., Ye X., Wang L., Zhang M., Yang H., Okuka M., Zhou C., Zhang X., Liu L. Zscan4 promotes genomic stability during reprogramming and dramatically improves the quality of iPS cells as demonstrated by tetraploid complementation. Cell Res. 2013;23:92–106. doi: 10.1038/cr.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien J., Pasque V., Halley-Stott R.P., Miyamoto K., Gurdon J.B. Mechanisms of nuclear reprogramming by eggs and oocytes: a deterministic process? Nat. Rev. Mol. Cell Biol. 2011;12:453–459. doi: 10.1038/nrm3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien J., Miyamoto K., Pasque V., Allen G.E., Bradshaw C.R., Garrett N.J., Halley-Stott R.P., Kimura H., Ohsumi K., Gurdon J.B. Hierarchical molecular events driven by oocyte-specific factors lead to rapid and extensive reprogramming. Mol. Cell. 2014;55:524–536. doi: 10.1016/j.molcel.2014.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang L., Wang J., Zhang Y., Kou Z., Gao S. iPS cells can support full-term development of tetraploid blastocyst-complemented embryos. Cell Stem Cell. 2009;5:135–138. doi: 10.1016/j.stem.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Kawamura T., Suzuki J., Wang Y.V., Menendez S., Morera L.B., Raya A., Wahl G.M., Izpisua Belmonte J.C. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaw S.L., Min-Wen C., Koh C.G., Lim B., Shyh-Chang N. Oocyte factors suppress mitochondrial polynucleotide phosphorylase to remodel the metabolome and enhance reprogramming. Cell Rep. 2015;12:1080–1088. doi: 10.1016/j.celrep.2015.07.032. [DOI] [PubMed] [Google Scholar]
- Kunitomi A., Yuasa S., Sugiyama F., Saito Y., Seki T., Kusumoto D., Kashimura S., Takei M., Tohyama S., Hashimoto H. H1foo has a pivotal role in qualifying induced pluripotent stem cells. Stem Cell Reports. 2016;6:825–833. doi: 10.1016/j.stemcr.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le R., Kou Z., Jiang Y., Li M., Huang B., Liu W., Li H., Kou X., He W., Rudolph K.L. Enhanced telomere rejuvenation in pluripotent cells reprogrammed via nuclear transfer relative to induced pluripotent stem cells. Cell Stem Cell. 2014;14:27–39. doi: 10.1016/j.stem.2013.11.005. [DOI] [PubMed] [Google Scholar]
- Lee H.S., Kim E.Y., Kim K.H., Moon J., Park K.S., Kim K.S., Lee K.A. Obox4 critically regulates cAMP-dependent meiotic arrest and MI-MII transition in oocytes. FASEB J. 2010;24:2314–2324. doi: 10.1096/fj.09-147314. [DOI] [PubMed] [Google Scholar]
- Lee H.S., Kim K.H., Kim E.Y., Lee S.Y., Ko J.J., Lee K.A. Obox4-silencing-activated STAT3 and MPF/MAPK signaling accelerate nuclear membrane breakdown in mouse oocytes. Reproduction. 2016;151:369–378. doi: 10.1530/REP-15-0020. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Collado M., Villasante A., Strati K., Ortega S., Canamero M., Blasco M.A., Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Liu X., Wang C., Liu W., Li J., Li C., Kou X., Chen J., Zhao Y., Gao H., Wang H. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature. 2016;537:558–562. doi: 10.1038/nature19362. [DOI] [PubMed] [Google Scholar]
- Luscombe N.M., Laskowski R.A., Thornton J.M. Amino acid–base interactions a three-dimensional analysis of protein–DNA interactions at an atomic level.pdf. Nucleic Acids Res. 2001;29:15. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan T.S., Gifford W.D., Driscoll S., Lettieri K., Rowe H.M., Bonanomi D., Firth A., Singer O., Trono D., Pfaff S.L. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa M., Yamaguchi K., Nakamura T., Shibukawa R., Kodanaka I., Ichisaka T., Kawamura Y., Mochizuki H., Goshima N., Yamanaka S. Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature. 2011;474:225–229. doi: 10.1038/nature10106. [DOI] [PubMed] [Google Scholar]
- Maherali N., Hochedlinger K. Tgfbeta signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr. Biol. 2009;19:1718–1723. doi: 10.1016/j.cub.2009.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G.T., Seo Y.M., Lee S.Y., Lee K.A. Lin28 regulates the expression of neuropeptide Y receptors and oocyte-specific homeobox genes in mouse embryonic stem cells. Clin. Exp. Reprod. Med. 2012;39:87–93. doi: 10.5653/cerm.2012.39.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racki W.J., Richter J.D. CPEB controls oocyte growth and follicle development in the mouse. Development. 2006;133:4527–4537. doi: 10.1242/dev.02651. [DOI] [PubMed] [Google Scholar]
- Rajkovic A., Yan C., Yan W., Klysik M., Matzuk M.M. Obox, a family of homeobox genes preferentially expressed in germ cells. Genomics. 2002;79:711–717. doi: 10.1006/geno.2002.6759. [DOI] [PubMed] [Google Scholar]
- Ruiz S., Panopoulos A.D., Herrerias A., Bissig K.D., Lutz M., Berggren W.T., Verma I.M., Izpisua Belmonte J.C. A high proliferation rate is required for cell reprogramming and maintenance of human embryonic stem cell identity. Curr. Biol. 2011;21:45–52. doi: 10.1016/j.cub.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P., Golipour A., David L., Sung H.K., Beyer T.A., Datti A., Woltjen K., Nagy A., Wrana J.L. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Schofield P.N. Patterns, puzzles and paradigms the riddle of the homeobox.pdf. Trends Neurosci. 1987;10:3. [Google Scholar]
- Shinagawa T., Takagi T., Tsukamoto D., Tomaru C., Huynh L.M., Sivaraman P., Kumarevel T., Inoue K., Nakato R., Katou Y. Histone variants enriched in oocytes enhance reprogramming to induced pluripotent stem cells. Cell Stem Cell. 2014;14:217–227. doi: 10.1016/j.stem.2013.12.015. [DOI] [PubMed] [Google Scholar]
- Singhal N., Graumann J., Wu G., Araúzo-Bravo M.J., Han D.W., Greber B., Gentile L., Mann M., Schöler H.R. Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell. 2010;141:943–955. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
- Stadtfeld M., Maherali N., Breault D.T., Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utikal J., Polo J.M., Stadtfeld M., Maherali N., Kulalert W., Walsh R.M., Khalil A., Rheinwald J.G., Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Kou Z., Jing Z., Zhang Y., Guo X., Dong M., Wilmut I., Gao S. Proteome of mouse oocytes at different developmental stages. Proc. Natl. Acad. Sci. USA. 2010;107:17639–17644. doi: 10.1073/pnas.1013185107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmut I., Schnieke A.E., McWhir J., Kind A.J., Campbell K.H. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- Xu Y., Wei X., Wang M., Zhang R., Fu Y., Xing M., Hua Q., Xie X. Proliferation rate of somatic cells affects reprogramming efficiency. J. Biol. Chem. 2013;288:9767–9778. doi: 10.1074/jbc.M112.403881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Different expression genes between samples (Vector and Obox1) on reprogramming day 3 were grouped into five clusters by K-means clustering based on RNA-seq data. The potential targets of Obox1 based on the ChIP-seq data are presented as 1 in the binding column.