Three species of Entamoeba were common in South Africa: E. dispar, E. histolytica, and the recently described E. bangladeshi. In E. histolytica–positive samples, changes in both parasite and P. copri levels were associated with alterations in gastrointestinal status.
Keywords: Entamoeba bangladeshi, Entamoeba histolytica, Protozoa, microbiome, Prevotella copri
Abstract
Background
Diarrhea is frequent in communities without clean water, which include low-income South African populations in Giyani and Pretoria. In these populations, the amount of diarrhea caused by Entamoeba histolytica, inclusive of all ages, sexes, and human immunodeficiency virus status, is uncertain. Infection with E. histolytica can modulate the host microbiota, and a key species indicative of this is the Prevotella copri pathobiont.
Methods
A cross-sectional study of patients attending gastroenterology clinics was conducted to determine the frequency and burden of 4 Entamoeba species and P. copri.
Results
Entamoeba species were present in 27% of patients (129/484), with E. histolytica detected in 8.5% (41), E. dispar in 8% (38), E. bangladeshi in 4.75% (23), and E. moshkovskii in 0%. This is the first description of E. bangladeshi outside Bangladesh. In E. histolytica–positive samples, the levels of both the parasite and P. copri were lower in nondiarrheal samples, validating the results of a study in Bangladesh (P = .0034). By contrast, in E. histolytica–negative samples positive for either of the nonpathogenic species E. dispar or E. bangladeshi, neither P. copri nor Entamoeba levels were linked to gastrointestinal status.
Conclusions
Nonmorphologic identification of this parasite is essential. In South Africa, 3 morphologically identical Entamoeba were common, but only E. histolytica was linked to both disease and changes in the microbiota.
Entamoeba species are a group of unicellular, anaerobic, parasitic organisms found in humans, nonhuman primates, and other vertebrate and invertebrate species [1]. Entamoeba infections in humans can result in asymptomatic carriage or a wide range of symptomatic diseases. Of the subset of individuals developing symptoms, diarrhea and dysentery are the most common manifestations. Extraintestinal complications occur less frequently but can be associated with high mortality [2].
Species of Entamoeba that can infect and be found in the intestinal lumen of humans include Entamoeba histolytica, Entamoeba dispar, Entamoeba moshkovskii, Entamoeba coli, Entamoeba polecki, Entamoeba hartmanni, and Entamoeba bangladeshi [3]. Of these, E. bangladeshi is the most recent to be described, with the species name reflecting the geographic origin of the first patient it was isolated from. E. bangladeshi is indistinguishable by microscopy from E. histolytica, the prototypical pathogenic Entamoeba species, but it can be differentiated from other known Entamoeba species by immunologic and molecular techniques.
A recent study involving stool samples collected from infants (age range, 0–24 months) residing in Vhembe District, Limpopo, South Africa, failed, however, to detect E. histolytica by polymerase chain reaction (PCR) analysis [4]. However E. histolytica was common (18%) in an earlier 2006 study involving participants of all ages (ie, from 0 to >60 years) [5]. The cause for this discrepancy is not fully understood. In preliminary work, samples collected from patients of all ages visiting a gastroenterology clinic between November 2013 and June 2015 in both rural Giyani (Mopani District, Limpopo) and urban Pretoria (Soshanguve District, Gauteng, South Africa) were evaluated by microscopy for the presence of ameboid organisms, and 50% of the samples were Entamoeba positive. These findings might be due to the presence of other morphologically identical non–E. histolytica species of Entamoeba, such as E. dispar and E. bangladeshi, to geographical heterogeneity in the frequency of E. histolytica in South African populations, or to a much lower frequency of E. histolytica in the community-based surveys of enteric disease than in patients requiring clinical care [3, 6, 7].
To test these hypotheses, DNA was extracted from the Giyani and Pretoria samples, and a multiplex quantitative PCR (qPCR) assay was used to detect both E. histolytica and the other morphologically identical Entamoeba species, such as E. dispar, E. moshkovskii, and E. bangladeshi. This assay was also modified to include a general Entamoeba probe to capture data on the presence of novel Entamoeba species genetically similar to the pathogenic species E. histolytica that may be present in the South African population.
The qPCR assay data captured quantitative information, and this permitted us to examine the correlation between the parasite burden in these samples and the outcome of infection. A link between parasite burden and symptomatic disease has been found in previous studies [4, 8, 9]. Recent studies have also highlighted the relationship between Entamoeba species and the bacterial communities of the gut. The presence of Entamoeba organisms was associated with a decrease in the abundance of Prevotella copri in farmers and fishermen from Southwest Cameroon [10], and the abundance of P. copri increased in diarrheal E. histolytica cases [9]. Hence, we also sought to quantify this bacterium in the microbiome of Entamoeba-positive samples in our study population.
METHODS
Ethics Statement
The research and ethics committee of the University of Venda granted institutional approval. The study received ethical clearance from the Department of Health and Welfare, Polokwane, Limpopo Province, South Africa. We also obtained permission from the ethics committee of participating hospitals and clinics to collect samples. The objectives and concepts of the study were clearly explained in the language understood by the potential participants (ie, English, Sepedi, Xitsonga, and Tshivenda). A written, informed consent form was signed prior to study enrollment. In cases where the participant was either a non-English speaker or illiterate, a witness also signed the consent form.
Study Area and Population
The tested stool samples were predominantly from urban and rural populations of moderate-to-low socioeconomic status [11, 12]. They were collected between November 2013 and June 2015 from diarrheal and nondiarrheal patients in the rural Nkomo clinic (Giyani) and the urban clinic within the Dr George Mukhari Hospital (Soshanguve District).
The catchment area for the Dr Georges Mukhari Hospital includes Soshanguve, Ga-Rankuwa, Mabopane, and parts of Madibeng District. The Nkomo clinic serves households within the Greater Giyani Local Municipality, Mopani District. Household water and sewage access is summarized in Table 1 [11, 12].
Table 1.
Water and Sewage Arrangements Among Households, by Gastrointestinal Clinic and Districts
| Clinic, District | Piped Water, % | Flush Toilet Connected to Sewage System, % |
|---|---|---|
| Dr Georges Mukhari Hospital | ||
| Soshanguve | 58.7 | 85.3 |
| Ga-Rankuwa | 73.6 | 90.1 |
| Mabopane | 62.5 | 85.4 |
| Madibenga | 22.2 | 27.2 |
| Nkomo clinic | ||
| Greater Giyani Municipalityb | 13.4 | 11.9 |
Data are from the South African Government STATS SA Community Survey of 2016 [12].
aOnly part of Madibeng District is considered to be in the Dr Georges Mukhari Hospital catchment area.
bGreater Giyani Municipality lies within Mopani District.
Both adults and children of all ages were eligible for participation. A questionnaire was used to collect sociodemographic information, such as the age, sex, and origin of the study participants.
Sample Collection
After the patients were given a clear explanation of the stool sample collection process, they received screw-cap bottles into which they placed their samples. Stool samples were classified as diarrheal or nondiarrheal on the basis of the physical presentation of the sample, as defined by the Bristol stool form scale (diarrheal specimens, types 6 and 7; nondiarrheal specimens, types 1–5) [13]. The bottles were labelled with unique participant identifiers and then placed in a cooler box and transported to the University of Venda microbiology laboratory for further processing. Upon arrival to the laboratory, samples were aliquoted in 2-mL tubes and stored frozen at −20oC. The aliquoted samples were shipped to the University of Virginia Infectious Diseases Research laboratory for analysis.
Genomic DNA Purification
Genomic DNA from each patient’s sample was extracted using a QIAamp DNA Stool Mini Kit (Qiagen) according to the manufacturer’s recommended procedures, using approximately 200 mg of stool samples with the modifications described by Liu et al [14]. One stool sample from a healthy US child whose stool had previously been tested and found to be negative for all Entamoeba species was included in each batch, to monitor for the occurrence of contamination during extraction. The DNA was eluted in 200 μL of elution buffer (Qiagen) and stored at −80°C until further analysis.
Multiplex qPCR Assay for Detection of Entamoeba Species
A multiplex qPCR assay was used for the amplification and detection of all Entamoeba species. Genus-specific primers were used in combination with a 42-nucleotide probe that should hybridize to E. bangladeshi, E. dispar, E. histolytica, and E. moshkovskii amplicons. Owing to the length required to generate this probe in these A/T-rich genomes, a double quencher was included in the design of the probe (Biosearch Technologies; Figure 1). This probe recognizes E. histolytica, E. moshkovskii, E. dispar, E. bangladeshi, and E. hartmanni amplicons but was not similar to the ribosomal RNA (rRNA) region in the nonpathogenic species E. coli, E. polecki, Endolimax nana, Iodamoeba bütschlii and Entamoeba gingivalis. The PCR was performed with 25-µL reaction mixture containing Bio-Rad iQ powermix, 0.4 µM of primers, and 0.2 µM for each probe. Probes, primers, and reaction conditions are shown in Table 2.
Figure 1.
Alignment of the 18S ribosomal DNA sequences of the Entamoeba species that are known to infect humans. A, Quantitative polymerase chain reaction assay designs for detection of Entamoeba histolytica (GenBank accession no. KX528461.1), Entamoeba dispar (KP722600.1), Entamoeba moshkovskii (AF149906.1), and Entamoeba bangladeshi (KR025412.1) Sequences were aligned using the Clustal Omega program [41]. The target of the Entamoeba genus probe is highlighted in gray, and the sequences of the species-specific probes and the genus-specific primers are underlined.
Table 2.
Entamoeba Species and Prevotella copri Probes, Primers, and Cycling Conditions
| Oligonucleotide (Dye) | Probe/Primer Sequencea | Cycling Conditions |
|---|---|---|
| E. histolytica probe (FAM) | TCATT+GAATGAATTGGCCATTTb | 95°C for 3 min, then 40 cycles at 95°C for 10 s and 61°C for 20 s |
| E. bangladeshi probe (Texas Red) | CCTTACAGAG+TATGGCCAATTTb | 95°C for 3 min, then 40 cycles at 95°C for 10 s and 61°C for 20 s |
| E. dispar probe (HEX) | ACTTA+CATAAATTGGCCAACTTTb | 95°C for 3 min, then 40 cycles at 95°C for 10 s and 61°C for 20 s |
| E. moshkovskii probe (Quasar 670) | CCGTGAAGAGAGTGGCCGAb | 95°C for 3 min, then 40 cycles at 95°C for 10 s and 61°C for 20 s |
| Entamoeba genus probe (Quasar 705) | GGATAGCTT[I-X]TGTGAATGATAAAGATAATACTT GAGACGATCCc | 95°C for 3 min, then 40 cycles at 95°C for 10 s and 61°C for 20 s |
| Ehd-88R | GCGGACGGCTCATTATAACAd | 95°C for 3 min, then 40 cycles at 95°C for 10 s and 61°C for 20 s |
| EM-RT-F2 | GTCCTCGATACTACCAACe | 95°C for 3 min, then 40 cycles at 95°C for 10 s and 61°C for 20 s |
| P. copri probe (FAM) | TGCCCACCACT+TGG | 95°C for 3 min, then 40 cycles at 95°C for 10 s and 60°C for 1 min |
| P. copri F | AAGCTTGCTTTTGATGGGCG | 95°C for 3 min, then 40 cycles at 95°C for 10 s and 60°C for 1 min |
| P. copri R | TGATCGT+CGCCTTGG | 95°C for 3 min, then 40 cycles at 95°C for 10 s and 60°C for 1 min |
aEach “+” indicates the location of a “locked” nucleotide [43], and “[I-X]” indicates the location of the internal quencher.
bProbe from Arju and Gilchrist (unpublished data).
cProbe specific to this study.
dPrimer Ehd-88R from Verweij et al [44].
ePrimer EM-RT-F2 from Lau et al [22].
Analysis of qPCR Entamoeba-Positive Samples Uncharacterized at the Species Level by the qPCR Assay
Any sample that gave a positive signal with the Entamoeba probe but was negative for all 4 Entamoeba species of interest (ie, E. histolytica, E. moshkovskii, E. dispar, and E. bangladeshi) was characterized further by amplifying additional 18S regions and determining their sequences [3, 15]. This DNA was amplified by primers Ehd-88R and EM-RT-F2 (Table 2), using the high-fidelity Phusion polymerase as described by Royer et al (98°C for 30 seconds; 40 cycles at 98°C for 20 seconds, 56°C for 30 seconds, and 72°C for 30 seconds; and a final extension at 72°C for 10 minutes) [3]. A 2% agarose gel stained with 3 µL of ethidium bromide was used to separate the amplified DNA. The PCR products were extracted from the agarose gel by using the Qiagen QIAquick Gel Extraction Kit, and the purified amplicons were sequenced using the Sanger method (Genewiz).
Sequence and Phylogenetic Analysis
After sequence results were obtained, the ABI files were downloaded and trimmed using Geneious (version 7.0.6). A phylogenetic tree was created to examine the phylogenetic relationship between novel Entamoeba species and the known Entamoeba parasites by use of the neighbor-joining algorithm included in Geneious (Biomatters) [16].
P. copri qPCR Assay
To improve the specificity of qPCR detection of P. copri in clinical fecal samples, a new TaqMan assay was designed, using the National Center for Biotechnology Information (NCBI) reference sequence NR_113411.1. Optimal qPCR assay conditions were initially determined using DNA amplified from cultured P. copri (CB7, DSMZ; a gift from D. Littman). Assay specificity was determined by purification and sequencing of the amplicons from selected fecal samples and by comparison with both the NCBI reference sequence NR_113411.1 and the sequence obtained from the cultured P. copri DNA. The PCR was performed with a 25-µL reaction mixture containing Bio-Rad iQ powermix, 0.4 µM of primers, and 0.2 µM for each probe. Probes, primers, and reaction conditions are shown in Table 2. The previously described Enterobacteriaceae assay was used to normalize both the P. copri levels and as a measure of extracted bacterial DNA quality [9]. Enterobacteriaceae- and P. copri–negative samples were omitted from the quantitative analysis of P. copri (Supplementary Table 1).
Statistical Analysis
The Fisher exact test was used to analyze contingency tables. The D’Agostino and Pearson omnibus normality test and the nonparametric Mann-Whitney comparisons test were used to analyze and compare qualitative data. Tests were performed using GraphPad Prism, version 6. The differences were considered significant if the P value was <.05.
RESULTS
Demographic and Clinical Features
A total of 484 participants were recruited in this study, of whom 227 (47%) were from Giyani (a rural setting) and 257 (53%) were from Pretoria (an urban setting). Table 3 summarizes the demographic data of the study population.
Table 3.
Demographic and Clinical Features of the Study Population
| Characteristic | Rural Setting, Giyani (n = 227) |
Urban Setting, Pretoria (n = 257) |
|---|---|---|
| Sex (n = 395) | ||
| Male | 83 (37) | 104 (40) |
| Female | 105 (46) | 103 (40) |
| Not recorded | 39 (17) | 50 (19) |
| Age, y (n = 346) | ||
| Range | 2–73 | 1–90 |
| Mean ± SD | 19.2 ± 17.71 | 41.7 ± 22.13 |
| <5 | 37 (16.3) | 19 (7.4) |
| 6–64 | 93 (41) | 154 (60) |
| >65 | 6 (3) | 37 (14.4) |
| Not recorded | 91 (40) | 47 (19) |
| Stool typea (n = 484) | ||
| Diarrheal (type 6 and 7) | 61 (27) | 125 (49) |
| Nondiarrheal (types 1–5) | 166 (73) | 132 (51) |
Data are no. (%) of participants, unless otherwise stated.
aBased on the Bristol stool scale.
Rural Setting
Participants with recorded ages ranged between 2 and 73 years old, with the majority (41%) aged 6–64 years. Thirty-seven percent (83) were male, and 46% (105) were female; for 17% (39), data on sex were not recorded. Of the 227 stool samples collected, 61 (27%) were diarrheal and 166 (73%) were nondiarrheal.
Urban Setting
Participants with recorded ages ranged between 1 and 90 years old, with the majority (60%) aged 6–64 years. Male sex was recorded for 40% (104), and 40% (103) were female; data on sex were not recorded for 19.5% (50). Of the 257 stool samples collected, 125 (49%) were diarrheal and 132 (51%) were nondiarrheal.
Prevalence and Distribution of Entamoeba Species, by qPCR Analysis
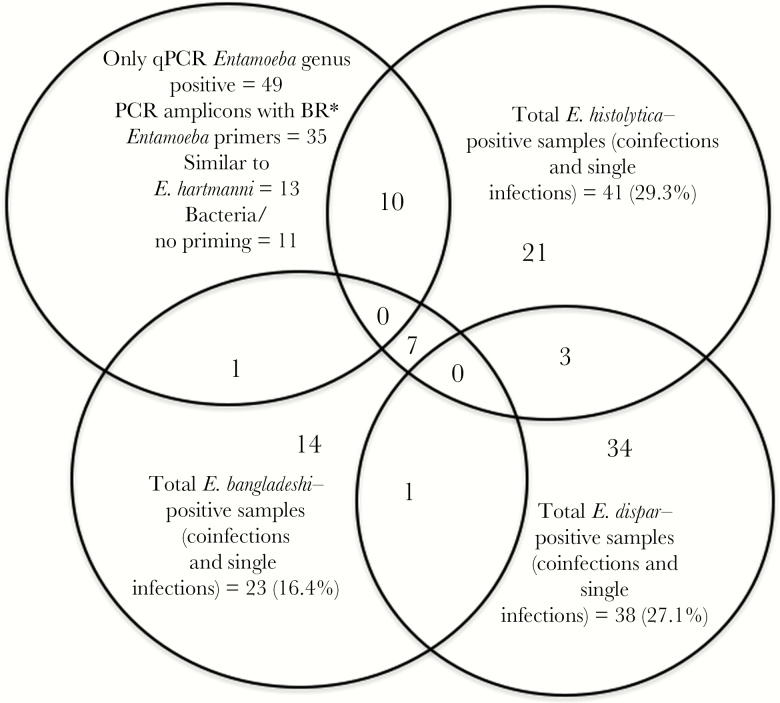
Of the 484 samples tested by qPCR analysis, 29% (140) were positive for Entamoeba species. The total frequency of E. dispar detected by qPCR in the study population was 8% (38), with E. histolytica detected in 6.4% (31), E. bangladeshi detected in 4.5% (22), and unknown Entamoeba species detected in 10% (49). In 11 samples, coinfections with different Entamoeba species were observed (7 were coinfected with E. histolytica and E. bangladeshi, 1 was coinfected with E. bangladeshi and E. dispar, and 3 were coinfected with E. histolytica and E. dispar). In line with previous results, E. moshkovskii was not identified in the South African populations studied (Figure 2).
Figure 2.
Entamoeba species found in South African populations. A total of 140 Entamoeba-positive samples were identified by quantitative polymerase chain reaction (qPCR) analysis. In 11 samples, the infecting species could not be identified. Thirty-eight samples were positive for Entamoeba dispar, 41 were positive for Entamoeba histolytica, 23 were positive for Entamoeba bangladeshi, and 0 were positive for Entamoeba moshkovskii. In 11 samples, coinfections with different Entamoeba species were observed (7 were coinfected with E. histolytica and E. bangladeshi, 1 was coinfected with E. bangladeshi and E. dispar, and 3 were coinfected with E. histolytica and E. dispar). E. hartmanni, Entamoeba hartmanni. *Broad range (BR).
Confirmatory Testing
To confirm the E. bangladeshi qPCR assay results, we sequenced the amplicon from 4 positive samples and compared the sequences to the E. bangladeshi sequence deposited in the NCBI GenBank (accession number KR025412.1). The South African sequences were identical to that of E. bangladeshi.
Characterization of Entamoeba Samples Not Identified by Species-Specific Probes
Entamoeba primers (Ehd-88R; EM-RT-F2) were used to amplify DNA fragments from the 49 samples that were qPCR positive for the broad range Entamoeba but negative for all the species-specific probes. The amplified DNA was separated by electrophoresis, and in the 35 cases where the bands of the size predicted for the Entamoeba species were identified, it was purified from the agarose by using the QIAquick Gel Extraction Kit (Qiagen). The SSU rRNA gene amplicon was detected in 35 samples. Sequencing of the purified amplicon identified 10 additional E. histolytica–positive samples (n = 41), with an adjusted frequency Entamoeba qPCR-positive samples of 29.3%, and 1 additional E. bangladeshi sample (n = 23), with an adjusted positivity frequency of 16.4% (Figure 2). This result suggested that in these samples the parasite level had simply fallen below the detection limit of the species-specific qPCR assay. These samples were not included in the later analysis. In 13 cases, the 18S rRNA amplicon sequences were similar to those of the nonpathogenic species E. hartmanni (all sequences were deposited in GenBank under accession numbers MF471201–MF471217), and in the remaining 11 cases either no useful sequence data were obtained or findings were similar to sequences from bacteria and had no significant similarity to any Entamoeba reference sequence in the NCBI database.
Parasite Burden in South African Samples
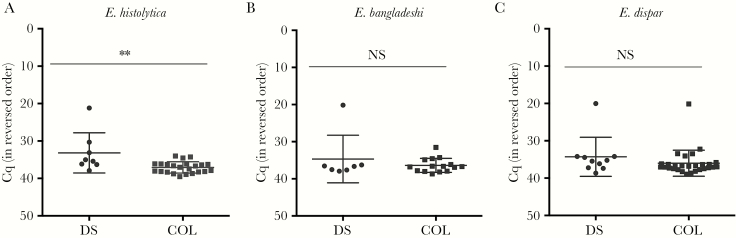
Other enteropathogens are common in this South African population, and in diarrheal samples coinfections can make it challenging to identify the causal organism [4]. Entamoeba were no more frequent in diarrheal samples than in controls (data not shown). No differences in Entamoeba frequency was observed between the rural and urban populations. The cycle value at which the (baseline-corrected) amplification curve exceeds the background fluorescence (Cq) is closely related to the amount of input DNA. The Cq data provided by the qPCR assay can therefore be used to determine whether the Entamoeba species burden was different in diarrheal and control fecal samples [4, 13, 17]. As the distribution of Cq values were non-Gaussian, significance was determined using the Mann-Whitney test. As expected, a significant difference in Entamoeba levels was observed in cases of E. histolytica–associated diarrhea (P = .0072; Figure 3A), but the level of the nonpathogenic species E. dispar was unchanged in control and diarrheal samples (Figure 3C). Interestingly, the level of E. bangladeshi was also unchanged (Figure 3B). Again, no significant differences were observed in the parasite burden in rural and urban samples.
Figure 3.
High parasite burden was associated with diarrhea due to Entamoeba histolytica (A) but not Entamoeba bangladeshi (B) and Entamoeba dispar (C). The y-axes indicate threshold values of quantitative polymerase chain reaction analyses positive for each parasite. Horizontal lines indicate mean values, and vertical lines indicate standard deviations. **P ≤ .0072, by the Mann-Whitney test. Cq, cycle value at which the (baseline-corrected) amplification curve exceeds the background fluorescence; NS, not significant.
Quantity of P. copri in Entamoeba-Positive Samples.
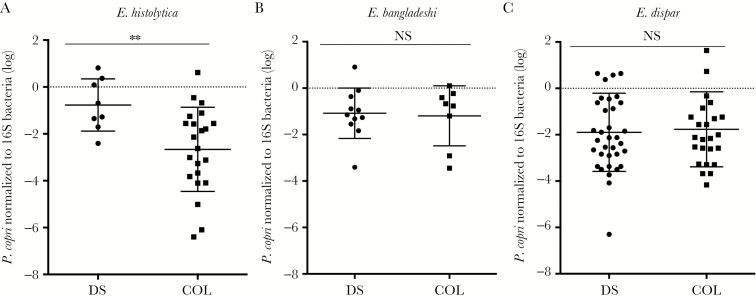
In a Bangladesh study, elevated levels of the pathobiont P. copri were associated with E. histolytica–associated diarrhea [9, 18]. The level of P. copri in Entamoeba-positive diarrheal and control samples was measured and, to control for variations in fecal bacterial numbers, was normalized using an Enterobacteriaceae bacterial reference [9, 19]. A fecal DNA standard was used to control for any differences in amplification efficiency in the P. copri and Enterobacteriaceae qPCR assays. Samples negative for either P. copri or Enterobacteriaceae were omitted from the quantitative analysis towing to concerns about sample quality (Supplementary Table 1). To convert the qPCR results to bacteria concentrations, DNA was extracted from a known amount of E. coli (ATCC 25922) and assayed. The relative level of P. copri was 1 log lower in E. histolytica–colonized samples as compared to the level in E. histolytica diarrheal samples (Figure 4A) but was unchanged in E. dispar or E. bangladeshi infections when diarrheal and nondiarrheal cases were compared (Figure 4B and 4C).
Figure 4.
Increased levels of Prevotella copri were associated with diarrhea due to Entamoeba histolytica (A) but not Entamoeba bangladeshi (B) and Entamoeba dispar (C). In the y-axes, the cycle values at which the (baseline-corrected) amplification curve exceed the background fluorescence were converted to bacteria numbers by use of a calibration curve and normalized to the Enterobacteriaceae levels. Horizontal lines indicate mean values and vertical lines indicate the standard deviations. **P = .0034, by the Mann-Whitney test. COL, asymptomatic colonizer samples; DS, diarrheal samples; NS, not significant.
DISCUSSION
The present study reports an overall frequency of Entamoeba species in our samples collected from gastrointestinal clinics as 27% (129/484), with E. histolytica being present in 6.4% of the cases (31/484). Differences in the assay used, as well as in age, geographic location, and the fact that these samples were collected from gastrointestinal clinics, make it difficult to compare these results to those obtained from previous population-based studies [4, 8]. A weakness in the current study was that information on human immunodeficiency virus status (expected to increase with age) was not available. In addition, the study was not adequately powered to analyze the susceptibility to Entamoeba among participants stratified by age [20, 21].
The Cq of the majority of our Entamoeba-positive asymptomatic samples was ≥35 and would have been missed by a less sensitive assay. Assay specificity at high Cq values was confirmed by amplicon sequencing of select samples (data not shown). In the work reported here, the assay included an Entamoeba general probe that acted as an independent control to identify any potential closely related novel South African Entamoeba species present in these samples (an in-depth surveillance of the Entamoeba species in the Mopani district of South Africa had not previously been done). All assay results were analyzed to be certain that the species-specific signal remained at a constant ratio to the result obtained from the broad range probe. The probe would have recognized any Entamoeba species similar to the pathogenic species E. histolytica, E. moshkovskii, and E. bangladeshi or the nonpathogenic species E. dispar (Figure 1A). The sequences of the closely related species also blocked nonspecific hybridization, allowing the higher assay Cq cutoff of ≤40 and increased assay sensitivity (Figure 1A) [22].
A higher E. histolytica parasite burden in samples increases the probability that the E. histolytica strain detected is responsible for diarrheal symptoms [9, 17]. In agreement with the previous studies, our results showed a statistically significant increase in the E. histolytica parasite load in South African diarrheal samples. The level of the nonpathogenic species E. dispar did not significantly change. This suggested that diarrhea coincident with E. dispar infections was due to other pathogens. The pathogenicity of the recently identified E. bangladeshi is still uncertain, but the level of E. bangladeshi was also the same in both diarrheal and nondiarrheal South African samples. Additional work is planned to identify whether other coinfecting enteric pathogens are present in these samples.
Novel Entamoeba species have been identified in different geographical contexts. Therefore, samples that were positive with the Entamoeba general probe but negative with the species-specific probes were characterized by amplicon sequencing (Figure 3) [1, 3, 23, 24]. While novel South African Entamoeba species were not identified, to our knowledge this study is the first to describe the presence of E. bangladeshi in samples collected outside Bangladesh. This species was first described in Bangladesh in 2011, but our results suggest that E. bangladeshi may actually have a broad geographical range and is prevalent in both Asian and African continents [3]. This finding also suggests that other members of the Entamoeba genus not identified in previous surveys may also be common in South Africa [5, 25–28]
In addition to the parasite burden, predisposition to diarrheal disease is thought to be influenced by the parasite environment [29–32]. Moreover, it has been suggested that specific components of the microbiota might be associated with symptomatic or asymptomatic E. histolytica colonization [33–35]. We examined the level of P. copri in the South African samples positive for Entamoeba species. Consistent with previous studies, the level of this bacterium was lower in asymptomatic E. histolytica–positive samples when compared to the level in E. histolytica–associated diarrheal samples [9]. Future work is needed to determine the significance of E. histolytica–associated changes in the microbiota. The gut Prevotella species are anaerobic bacilli predominant in the lumen of the colon [36]. Recent studies, however, suggest that disruption of the host mucosa can result in an increase in Prevotella species at mucosal sites and a subsequent increase in host inflammatory responses [18, 37]. Additional studies are needed to determine whether low P. copri levels could mitigate the host immune response occurring during amoebic colitis. It is possible that E. histolytica, unlike nonpathogenic Entamoeba species, disrupts the protective mucosal layer and exposes the host epithelium to the luminal microorganisms. This could expose the epithelium to high P. copri levels, as well as to E. histolytica, and result in an excessive inflammatory response with subsequent diarrhea [38–40]. In samples positive for the commensal E. dispar (which is not known to induce an inflammatory response or diarrhea), P. copri levels were not significantly different in either diarrheal or nondiarrheal samples [39].
In summary, an increase in the commensal bacterium P. copri was associated with diarrhea due to E. histolytica. The interplay between the pathogen, host, and host microbiota may be of importance in the development of symptomatic disease.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Acknowledgments. We thank the University of Venda, the Department of Health and Welfare, and participating hospitals and clinics, for giving us the permission to collect samples; and the patients, for their cooperation throughout the study.
Disclaimer. The funders had no role in study design, data collection and analysis, or decision to submit for publication.
Financial support. This work was supported by the University of Virginia (Global Infectious Disease Research Training Grant); the Fogarty Center, National Institutes of Health (NIH; award D43 TW006578); the NIH (grants R01 AI043596 [to W. P.] and R21 AI103536 [to C. G.]); and the National Research Foundation (award 95292 to R. N.).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: Annual Meeting of the American Society of Tropical Medicine and Hygiene, Atlanta, Georgia, November 2016; Global Heath Research in Africa Symposium, Thohoyandou, South Africa, March 2017.
References
- 1. Stensvold CR, Lebbad M, Victory EL et al. Increased sampling reveals novel lineages of Entamoeba: consequences of genetic diversity and host specificity for taxonomy and molecular detection. Protist 2011; 162:525–41. [DOI] [PubMed] [Google Scholar]
- 2. Shirley DA, Moonah S. Fulminant amebic colitis after corticosteroid therapy: a systematic review. PLoS Negl Trop Dis 2016; 10:e0004879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Royer TL, Gilchrist C, Kabir M et al. Entamoeba bangladeshi nov. sp., Bangladesh. Emerg Infect Dis 2012; 18:1543–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Platts-Mills JA, Babji S, Bodhidatta L et al. Pathogen-specific burdens of community diarrhoea in developing countries: a multisite birth cohort study (MAL-ED). Lancet Glob Health 2015; 3:e564–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Samie A, Obi LC, Bessong PO, Stroup S, Houpt E, Guerrant RL. Prevalence and species distribution of E. histolytica and E. dispar in the Venda region, Limpopo, South Africa. Am J Trop Med Hyg 2006; 75:565–71. [PubMed] [Google Scholar]
- 6. Kotloff KL, Nataro JP, Blackwelder WC et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 2013; 382:209–22. [DOI] [PubMed] [Google Scholar]
- 7. Faria CP, Zanini GM, Dias GS et al. Geospatial distribution of intestinal parasitic infections in Rio de Janeiro (Brazil) and its association with social determinants. PLoS Negl Trop Dis 2017; 11:e0005445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu J, Platts-Mills JA, Juma J et al. Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: a reanalysis of the GEMS case-control study. Lancet 2016; 388:1291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gilchrist CA, Petri SE, Schneider BN et al. Role of the gut microbiota of children in diarrhea due to the protozoan parasite Entamoeba histolytica. J Infect Dis 2016; 213:1579–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Morton ER, Lynch J, Froment A et al. Variation in rural african gut microbiota is strongly correlated with colonization by Entamoeba and subsistence. PLoS Genet 2015; 11:e1005658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Groome MJ, Page N, Cortese MM et al. Effectiveness of monovalent human rotavirus vaccine against admission to hospital for acute rotavirus diarrhoea in South African children: a case-control study. Lancet Infect Dis 2014; 14:1096–104. [DOI] [PubMed] [Google Scholar]
- 12. Statistics Council 2013 to the South African Government (STATS SA). Statistics South Africa http://www.statssa.gov.za/. Accessed 3 August 2017.
- 13. Riegler G, Esposito I. Bristol scale stool form. A still valid help in medical practice and clinical research. Tech Coloproctol 2001; 5:163–4. [DOI] [PubMed] [Google Scholar]
- 14. Liu J, Kabir F, Manneh J et al. Development and assessment of molecular diagnostic tests for 15 enteropathogens causing childhood diarrhoea: a multicentre study. Lancet Infect Dis 2014; 14:716–24. [DOI] [PubMed] [Google Scholar]
- 15. Silberman JD, Clark CG, Diamond LS, Sogin ML. Phylogeny of the genera Entamoeba and Endolimax as deduced from small-subunit ribosomal RNA sequences. Mol Biol Evol 1999; 16:1740–51. [DOI] [PubMed] [Google Scholar]
- 16. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28:2731–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Taniuchi M, Sobuz SU, Begum S et al. Etiology of diarrhea in Bangladeshi infants in the first year of life analyzed using molecular methods. J Infect Dis 2013; 208:1794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Scher JU, Sczesnak A, Longman RS et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife 2013; 2:e01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Barman M, Unold D, Shifley K et al. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun 2008; 76:907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mabunda TE, Ramalivhana NJ, Dambisya YM. Mortality associated with tuberculosis/HIV co-infection among patients on TB treatment in the Limpopo province, South Africa. Afr Health Sci 2014; 14:849–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Samie A, Barrett LJ, Bessong PO et al. Seroprevalence of Entamoeba histolytica in the context of HIV and AIDS: the case of Vhembe district, in South Africa’s Limpopo province. Ann Trop Med Parasitol 2010; 104:55–63. [DOI] [PubMed] [Google Scholar]
- 22. Lau YL, Anthony C, Fakhrurrazi SA, Ibrahim J, Ithoi I, Mahmud R. Real-time PCR assay in differentiating Entamoeba histolytica, Entamoeba dispar, and Entamoeba moshkovskii infections in Orang Asli settlements in Malaysia. Parasit Vectors 2013; 6:250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stensvold CR, Lebbad M, Verweij JJ et al. Identification and delineation of members of the Entamoeba complex by pyrosequencing. Mol Cell Probes 2010; 24:403–6. [DOI] [PubMed] [Google Scholar]
- 24. Clark CG, Kaffashian F, Tawari B et al. New insights into the phylogeny of Entamoeba species provided by analysis of four new small-subunit rRNA genes. Int J Syst Evol Microbiol 2006; 56:2235–9. [DOI] [PubMed] [Google Scholar]
- 25. Nxasana N, Baba K, Bhat V, Vasaikar S. Prevalence of intestinal parasites in primary school children of mthatha, eastern cape province, South Africa. Ann Med Health Sci Res 2013; 3:511–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heckendorn F, N’Goran EK, Felger I et al. Species-specific field testing of Entamoeba spp. in an area of high endemicity. Trans R Soc Trop Med Hyg 2002; 96:521–8. [DOI] [PubMed] [Google Scholar]
- 27. Mbae CK, Nokes DJ, Mulinge E, Nyambura J, Waruru A, Kariuki S. Intestinal parasitic infections in children presenting with diarrhoea in outpatient and inpatient settings in an informal settlement of Nairobi, Kenya. BMC Infect Dis 2013; 13:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sylvain PN, Kaur U, Goyal K, Sehgal R, Paul MF. Molecular differentiation of Entamoeba Spp. isolated from Cameroonian human immunodeficiency virus (HIV) infected and uninfected patient. J Parasitol Vector Biol 2015; 7:139–50. [Google Scholar]
- 29. Rastew E, Vicente JB, Singh U. Oxidative stress resistance genes contribute to the pathogenic potential of the anaerobic protozoan parasite, Entamoeba histolytica. Int J Parasitol 2012; 42:1007–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Marie C, Petri WA Jr. Regulation of virulence of Entamoeba histolytica. Annu Rev Microbiol 2014; 68:493–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Galván-Moroyoqui JM, Del Carmen Domínguez-Robles M, Franco E, Meza I. The interplay between Entamoeba and enteropathogenic bacteria modulates epithelial cell damage. PLoS Negl Trop Dis 2008; 2:e266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Abhyankar MM, Shrimal S, Gilchrist CA, Bhattacharya A, Petri WA Jr. The Entamoeba histolytica serum-inducible transmembrane kinase EhTMKB1-9 is involved in intestinal amebiasis. Int J Parasitol Drugs Drug Resist 2012; 2:243–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Burgess SL, Buonomo E, Carey M et al. Bone marrow dendritic cells from mice with an altered microbiota provide interleukin 17A-dependent protection against Entamoeba histolytica colitis. MBio 2014; 5:e01817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Burgess SL, Petri WA Jr. The intestinal bacterial microbiome and E. histolytica infection. Curr Trop Med Rep 2016; 3:71–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Burgess SL, Saleh M, Cowardin CA et al. Role of serum amyloid a, granulocyte-macrophage colony-stimulating factor, and bone marrow granulocyte-monocyte precursor expansion in segmented filamentous bacterium-mediated protection from Entamoeba histolytica. Infect Immun 2016; 84:2824–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol 2016; 14:20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Larsen JM. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology 2017; 151:363–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Haque R, Mondal D, Shu J et al. Correlation of interferon-gamma production by peripheral blood mononuclear cells with childhood malnutrition and susceptibility to amebiasis. Am J Trop Med Hyg 2007; 76:340–4. [PubMed] [Google Scholar]
- 39. Sharma M, Vohra H, Bhasin D. Enhanced pro-inflammatory chemokine/cytokine response triggered by pathogenic Entamoeba histolytica: basis of invasive disease. Parasitology 2005; 131:783–96. [DOI] [PubMed] [Google Scholar]
- 40. Pianta A, Arvikar S, Strle K et al. Evidence of the immune relevance of Prevotella copri, a gut microbe, in patients with Rheumatoid Arthritis. Arthritis Rheumatol 2017; 69:964–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sievers F, Wilm A, Dineen D et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 2011; 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 2010; 59:307–21. [DOI] [PubMed] [Google Scholar]
- 43. Kumar R, Singh SK, Koshkin AA, Rajwanshi VK, Meldgaard M, Wengel J. The first analogues of LNA (locked nucleic acids): phosphorothioate-LNA and 2’-thio-LNA. Bioorg Med Chem Lett 1998; 8:2219–22. [DOI] [PubMed] [Google Scholar]
- 44. Verweij JJ, Blangé RA, Templeton K et al. Simultaneous detection of Entamoeba histolytica, Giardia lamblia, and Cryptosporidium parvum in fecal samples by using multiplex real-time PCR. J Clin Microbiol 2004; 42:1220–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.