Antagonistic growth effects of two T-DNA alleles in THESEUS1 correlate with the expression of a predicted functional THE1 protein lacking the kinase domain and antisense transcripts, respectively.
Keywords: Cell elongation, cellulose synthesis, cell wall integrity signaling, CrRLK1L receptor, gene silencing, isoxaben
Abstract
Perturbation of cellulose synthesis in plants triggers stress responses, including growth retardation, mediated by the cell wall integrity-sensing receptor-like kinase (RLK) THESEUS1 (THE1). The analysis of two alleles carrying T-DNA insertions at comparable positions has led to conflicting conclusions concerning the impact of THE1 signaling on growth. Here we confirm that, unlike the1-3 and other the1 alleles in which cellular responses to genetic or pharmacological inhibition of cellulose synthesis are attenuated, the1-4 showed enhanced responses, including growth inhibition, ectopic lignification, and stress gene expression. Both the1-3 and the1-4 express a transcript encoding a predicted membrane-associated truncated protein lacking the kinase domain. However, the1-3, in contrast to the1-4, strongly expresses antisense transcripts, which are expected to prevent the expression of the truncated protein as suggested by the genetic interactions between the two alleles. Seedlings overexpressing such a truncated protein react to isoxaben treatment similarly to the1-4 and the full-length THE overexpressor. We conclude that the1-4 is a hypermorphic allele; that THE1 signaling upon cell wall damage has a negative impact on cell expansion; and that caution is required when interpreting the phenotypic effects of T-DNA insertions in RLK genes.
Introduction
The inhibition of cellulose synthesis in plants causes stress responses, including growth retardation. The latter is not only the result of the structural changes in the cell wall linked to a reduced cellulose content but involves active growth inhibition mediated by a receptor kinase that appears to act as a wall integrity sensor (Hématy et al., 2007). This receptor kinase was identified in a screen for suppressors of the short hypocotyl and ectopic lignin phenotype of the cellulose synthase mutant cesa6prc1-1. Three mutant alleles were identified in the THESEUS1 (THE1) gene, which all partially suppressed this phenotype in cesa6prc1-1 and other cellulose-deficient mutants. Alleles the1-1 and the1-2 carry amino acid changes in the predicted ectodomain. This domain comprises two copies of a malectin-like domain, which in animal cells binds to carbohydrate epitopes and also in plants may bind to cell wall carbohydrates (Schallus et al., 2008; Nissen et al., 2016; Voxeur and Höfte, 2016). Allele the1-3 carries a T-DNA insertion behind the cytosolic and transmembrane domain-encoding portion of the intron-less gene (Fig. 1). The suppressor phenotype of the three independent alleles suggested that THE1 negatively regulates growth as part of a more general stress response including the accumulation of reactive oxygen species (ROS) and ectopic lignin (Denness et al., 2011), and the up-regulation of jasmonic acid (JA)-regulated genes. The latter include genes involved in ROS detoxification and the synthesis of defense compounds such as indole-glucosinolates (Hématy et al., 2007).
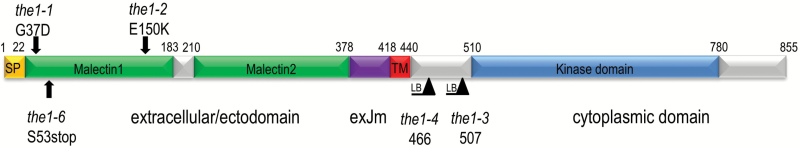
Fig. 1.
Overview of the domain structure and mutant alleles of THESEUS1. SP, TM, and exJM correspond to signal peptide, transmembrane domain, and extracellular juxtamembrane region, respectively. Numbers indicate amino acid residues. Arrows indicate the position and amino acid changes of the mutant alleles. LB indicates the left border of the T-DNA insertion. G, D, E, K, and S denote glycine, aspartic acid, glutamic acid, lysine, and serine, respectively.
THE1 belongs to the 17-member Catharanthus roseus receptor-like kinase 1-like (CrRLK1L) gene family (Li et al., 2016; Nissen et al., 2016), the most intensively studied member of which is FERONIA (FER). FER has been implicated in numerous biological processes, such as mechanosensing (Shih et al., 2014), pollen tube recognition at the micropyle (Escobar-Restrepo et al., 2007), bacterial pathogen interaction (Keinath et al., 2010), brassinosteroid responsiveness (Deslauriers and Larsen, 2010), and root hair development (Duan et al., 2010). In addition, FER is a receptor for secreted peptides of the RALF (rapid alkalinization factor) family (Haruta et al., 2014; Stegmann et al., 2017), which trigger alkalinization of the cell surface and inhibition of cell elongation (Haruta et al., 2014).
Roles in cell elongation have also been demonstrated for other family members CURVY (CVY1), HERKULES1 (HERK), HERK2, and ANXUR1 and 2 (ANX1/2). CVY1 regulates trichome and pavement cell morphogenesis and cvy1 mutants develop faster and have a higher fecundity (Gachomo et al., 2014). Double mutants of ANX1/2 are required to maintain elongation in pollen tubes (Boisson-Dernier et al., 2009; Miyazaki et al., 2009) while overexpression of ANX1/2 inhibits pollen tube elongation (Boisson-Dernier et al., 2013). Mutants for HERK1 or 2 did not show growth defects, but developed, in combination with the THE1 mutant allele the1-4, smaller leaves and petioles. This suggested a partially redundant growth-promoting role for HERK1, HERK2, and THE1 even in the absence of cell wall damage (Guo et al., 2009a, b). The the1-4 allele in combination with a AtKINESIN-13A mutant triggered a similar reduction of cell expansion in petals (Fujikura et al., 2014). These findings were contradictory to the growth-inhibiting rather than the growth-promoting role for THE1 inferred from the cesa6prc1-1 suppressor mutants (Hématy et al., 2007).
Here we show that the1-4, in contrast to the other four alleles, enhances rather than suppresses the growth defects of cesa6prc1-1 and cesa3je5, and thus mimics the effect of THE1 overexpression. We next demonstrate that the1-4 is a hypermorphic gain-of-function allele, which expresses a transcript encoding a predicted membrane-associated truncated protein, lacking the kinase domain. This truncated protein appears to trigger a stress response more efficiently than the wild-type protein upon inhibition of cellulose synthesis, perhaps through the interaction with other membrane receptor kinases. In contrast, the1-3, which carries a T-DNA insertion at a position comparable with that of the1-4, strongly expresses antisense transcripts, which presumably interfere with the expression of the truncated THE1 protein from the sense transcripts and hence can be considered a partial or complete loss-of-function allele like the1-1, the1-2, and the new knock out allele the1-6. These observations confirm that THE1 signaling negatively, not positively, affects cell expansion upon inhibition of cellulose synthesis. The availability of both loss-of-function and gain-of-function alleles will be useful for the study of cell wall integrity signaling in Arabidopsis and, more generally, underscore that caution is required when interpreting the phenotypic effects of RLK and T-DNA insertion mutants.
Materials and methods
Plant materials
The the1-1 (G37D) and the1-2 (E150K) mutants were discovered in a suppressor screen with the cellulose-deficient mutant cesa6prc1-1 (Hématy et al., 2007) and the novel ethyl methanesulfonate (EMS)-induced allele the1-6 (S53Stop) in a suppressor screen of ctl1-1/pom1. the1-3 (FLAG_201C06) and the1-4 (SAIL_683_H03) are both T-DNA mutants in either the Wassilewskija (WS-0) or Columbia (Col-0) background (Hématy et al., 2007; Guo et al., 2009a). Both T-DNA lines were crossed into the cellulose-deficient CELLULOSE SYNTHASE A6 and A3 mutants: cesa6prc1-1, cesa6prc1-8, cesa3je5, and cesa3eli1-1. Homozygous plants were selected after PCR genotyping and through sequencing of the cesa3je5 gene.
An overexpressing green fluorescent protein (GFP) reporter fusion (THE1:GFP) controlled by a double Cauliflower mosaic virus (CaMV) 35S promoter in the Col-0 accession (Hématy et al., 2007) was crossed with cesa3je5 and with double mutants of cesa3je5the1-3 and cesa3je5the1-4. Homozygous F3 generations were selected based upon their growth phenotype and PCR genotyping.
Generating ECD–TMTHE1–YFP-overexpressing plants
A 1510 bp fragment of THE1 coding for the extracellular domain (ECD), the transmembrane (TM) domain, and the juxtamembrane sequence up to the kinase domain (amino acid 510) was PCR amplified with primers THE1-Start and TM-JTM-Lo, and cloned into the reconstituted SmaI site of pSmile-YFP. pSmile-YFP was generated by inserting the yellow fluorescent protein (YFP) coding sequence (CDS) into the SmaI site of the pBIB-Hygro-derived plasmid pMagic (Nesi et al., 2002) containing a double CaMV 35S promoter. Wild-type Arabidopsis thaliana accession Col-0 was transformed with the Agrobacterium tumefaciens strain C58 pMP90 with the floral-dip method (Clough and Bent, 1998). Transformants were selected on Murashige and Skoog (MS) medium containing 50 mg l–1 hygromycin (Duchefa, France).
Growth conditions
Seeds were surface-sterilized and placed on sterile MS agar plates supplemented with 4.5% sucrose (w/v) as described previously (Hauser et al., 1995). After 2 d (1 week for the WS background) of imbibition at 4 °C in darkness, the plates were placed in a 22 °C growth chamber with constant white light (80 µmol m–2 s–1).
For the production of etiolated seedlings, seeds were sterilized and placed on MS plates without sucrose or supplemented with 2.5% sucrose. After imbibition and before wrapping the plates in aluminum foil, they were exposed for 4–5 h to white light and subsequently incubated in a vertical position at 22 °C.
Isoxaben treatment was done with 4-day-old etiolated seedlings by cultivating them on a 75 µm nylon mesh for easy transfer to MS medium containing 100 nM isoxaben or the same amount of methanol as solvent control. After 6 h further growth in the dark, the seedlings were harvested, shock frozen in liquid nitrogen, and RNA was isolated for expression analyses.
Plant growth on soil (1:1 mix of perlite and soil) was initiated from in vitro cultured, 10- to 14-day-old seedlings. Rosette and inflorescence phenotypes were photographed after 4 or 5 weeks, respectively.
Growth analyses and lignin staining
Hypocotyl lengths were measured from pictures of either 5- or 7-day-old etiolated plants of the1 mutants in the cesa6prc1-1 and cesa3je5 background, respectively, with the ImageJ freehand tracking and the measuring tool from photographs.
For lignin staining, seedlings were fixed and cleared in methanol/acetate (3:1) for 1 h, washed with water, stained with 1% phloroglucinol in 6 N HCl/46% ethanol at room temperature for 15 min, and mounted in either water or chloralhydrate solution (16 g of chloralhydrate dissolved in 5 ml of phosphate buffer with 17.5% glycerol).
Expression analyses by semi-quantitative and quantitative real-time PCR
Total RNA was isolated with TRI REAGENT (MRC, Cincinnati) according to the manufacturers’ protocols. First-strand cDNA was synthesized from 2.5 µg of RNA with the Superscript III Moloney murine leukemia virus (MMLV) reverse transcriptase (Invitrogen) after DNase I digestion (Roche) as described by Karsai et al. (2002). For sense cDNA synthesis, oligo(dT)18 primer, and for antisense cDNA synthesis, the gene-specific primer THE1_midF or the T-DNA-specific primers LB3 and LB4, were used (Supplementary Table S1 at JXB online).
For semi-quantitative determination of sense and antisense expression primers downstream (THE1_3endF/THE1_endR) and upstream (THE1_5endF/THE1_5endR), the T-DNA insertions were used. For antisense fragments originating from the T-DNA of the1-3, the primers THE1_midF and LB4 were used and for those originating from the T-DNA of the1-4 the primers THE1_midF and LB3 were used and compared with the expression of the housekeeping gene BETA-TUBULIN9 (TUB9). Duplicate PCRs from 3–4 independent cDNAs (biological replicates) were evaluated and shown by representative, ethidium bromide-stained gel images.
Reverse transcription–quantative PCR (RT–qPCR) expression analyses were carried out using the Hot FirePol EvaGreen qPCR Mastermix (Solis Biodyne) with the Rotorgene 3000 (Qiagen). Primers for THE1 expression analyses were the same as for the semi-quantitative approach. Primers of the THE1-dependent genes, 5g19110/EDGP and 2g26530/AR781, and the reference gene UBIQUTIN EXTENSION PROTEIN 5 (UBQ) are listed in Supplementary Table S1. Absolute and relative expression was calculated with a dilution series of purified PCR fragments of known molar concentrations in each RT–qPCR run. Each sample was measured in triplicate and from 3–4 independent cDNAs. The identities of amplicons were verified with melting curve analyses.
Results
THE1 alleles have opposing effects on plant growth, ectopic lignification, and gene expression
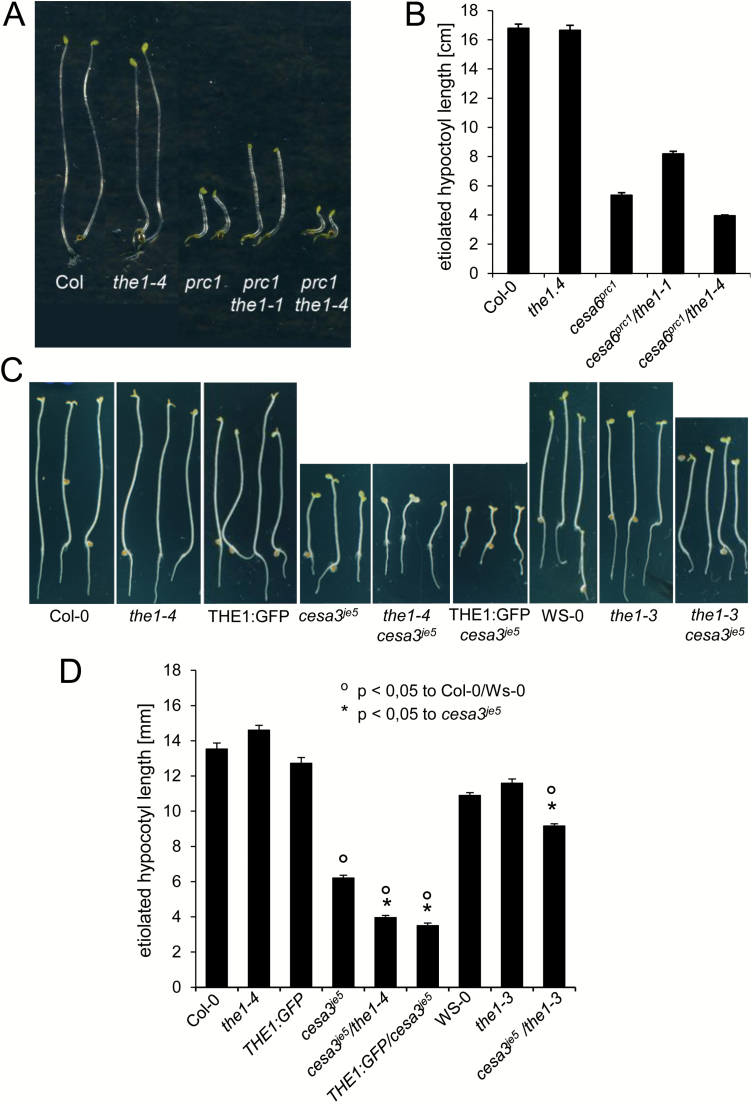
We studied four previously described THE1 alleles (the1-1 to the1-4) and a novel allele (the1-6) isolated from a suppressor screen of the cellulose-deficient mutant ctl1-1/pom1. the1-6 carries a point mutation causing a (TCA to TAA) change of S53 into a stop codon (Fig. 1). This truncates the protein within the first malectin domain and hence can be considered as a complete loss-of-function allele. The the1-6 allele was separated from the ctl1-1 mutation before further analysis. The positions of the respective mutations in the THE1 sequence are summarized in Fig. 1. We showed previously that the1-1, the1-2, and the1-3 all partially suppress the dark-grown hypocotyl growth defect of the cesa6prc1-1 mutant (Hématy et al., 2007). Interestingly, the the1-4 allele had the opposite effect: it enhanced the phenotype of cesa6prc1-1, while it did not show a hypocotyl growth phenotype in a wild-type background (Fig. 2A, B). To confirm these observations for another cellulose-deficient mutant, we combined the1-3, the1-4, and a 35S::THE1:GFP overexpression line (named THE1:GFP), respectively, with the weak allele of CELLULOSE SYNTHASEA3, cesa3je5. This allele is identical to multiple response expansion1 (mre1) (Pysh et al., 2012) and carries a point mutation at position 916 causing an amino acid change from a conserved glycine to glutamate. Based on in silico analysis, this mutation causes a shift in the location of transmembrane domain (TMD) 5 from amino acids 913–929 in the wild type to amino acids 919–935 in cesa3je5. This shift leads to an enlargement of the cytoplasmic loop between TMD 4 and 5 and a reduction of the cell wall-facing loop between TMD 5 and 6 of six amino acids. Another possibility is that TMD 5 is a cytosolic interfacial helix similar to the bacterial catalytic unit of cellulose synthase (BcsA) and involved in the regulation of UDP-glucose access to the nearby catalytic pocket (Slabaugh et al., 2014). This scenario and topological change, however, are incompatible with the recent finding of cysteine acylation at the C-terminus (Kumar et al., 2016) unless TMD 6 behaves as a re-entrant segment that enters the membrane but does not cross it. Again, the hypocotyl growth defect of cesa3je5 was suppressed in the1-3 and enhanced in the1-4 and THE1:GFP, whereas neither the1-3, the1-4, nor THE1:GFP showed a detectable dark-grown phenotype in a wild-type background (Fig. 2C, D). Similar observations were made for seedlings grown in the light on 4.5% sucrose, a condition that enhances growth phenotypes of many cell wall mutants (Hauser et al., 1995). While cesa3je5 seedlings develop short radially swollen roots, they are even shorter in the1-4/cesa3je5 and cesa3je5/THE1:GFP, and longer and less swollen in the1-3/cesa3je5 double mutants (Fig. 7B; Supplementary Fig. S1). It has been shown previously that ectopic lignin accumulation in cesa6prc1-1 and cesa3eli1-1 depends on functional THE1 (Hématy et al., 2007). Also cesa3je5 accumulates ectopic lignin in etiolated hypocotyls. While this phenotype is reduced in cesa3je5/the1-3, it is strongly enhanced in cesa3je5/the1-4 and cesa3je5/THE1:GFP (Supplementary Fig. S2).
Fig. 2.
Seedling development of loss- and gain-of-function alleles of THE1 in combination with cellulose-deficient mutants. (A) Five-day-old dark-grown seedlings of the wild type (Col-0), the1-4, prc1, and the double mutants prc1/the1-1 and prc1/the1-4. (B) Hypocotyl length quantification of etiolated seedlings in (A). (C) Seven-day-old etiolated seedlings of the wild type (Col-0, WS), the1-4, THE1:GFP, cesa3je5, the1-3, and the double mutants the1-4/cesa3je5, THE1:GFP/cesa3je5, and the1-3/cesa3je5. (D) The quantification of hypocotyl length of (C). The graphs show the average ±SE of 37–40 seedlings in (B) and 27–152 seedlings in (D). * and °P<0.05 according to Student’s t-test compared with the wild type or cesa3je5. (This figure is available in colour at JXB online.)
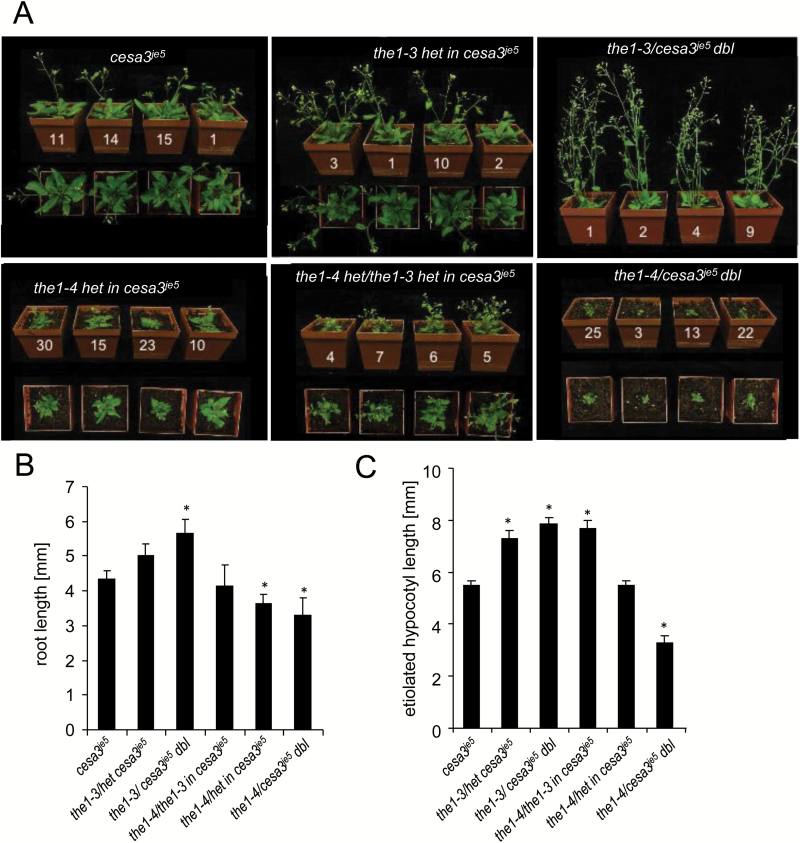
Fig. 7.
Semi-dominant phenotypes of the1-3 and the1-4 in cellulose-deficient backgrounds. (A) Four-week-old plants. All plants are homozygous for cesa3je5 and either wild type, heterozygous for the1-3 and/or the1-4, and homozygous for the1-3 or the1-4. (B) Root length 7 d after germination and (C) etiolated hypocotyl length of 5-day-old seedlings of the above-mentioned genotypes. Graphs represent the mean ±SE of 5–35 seedlings in (B) and 9–24 seedlings in (C). Significant differences from cesa3je5 are indicated according to Student’s t-tests with *P<0.05.
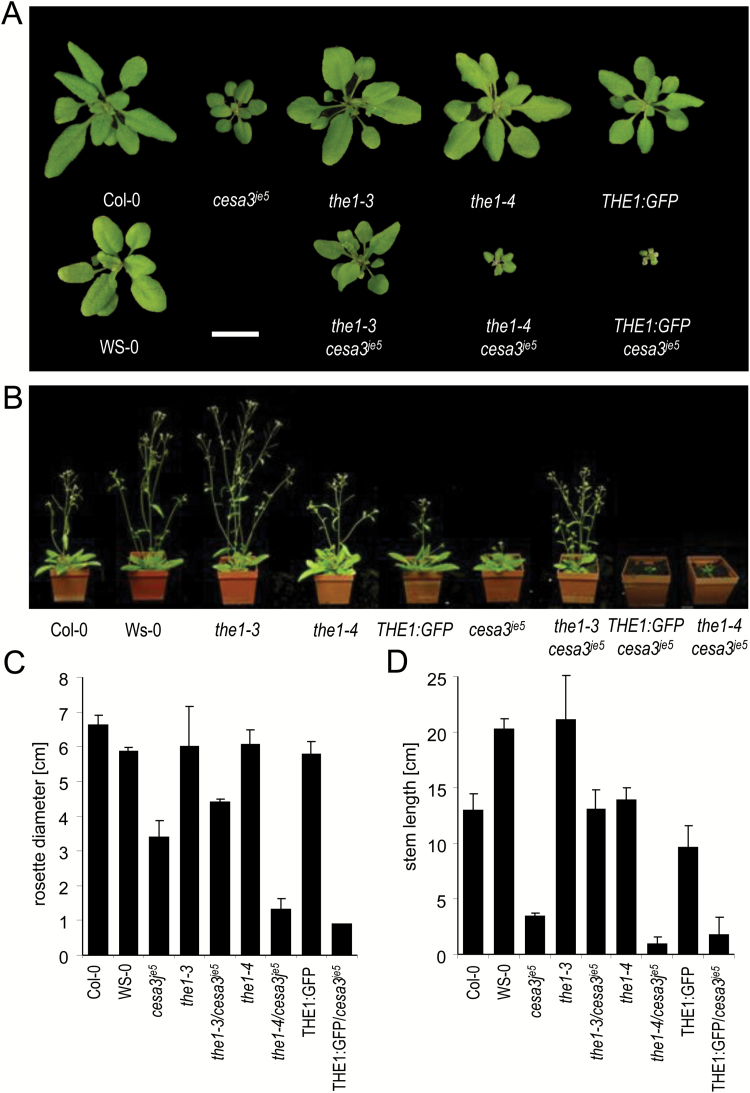
The opposite effects of the1-3 and the1-4 alleles were also observed in soil-grown adult plants of cesa3je5 (Fig. 3; Supplementary Fig. S3). The rosette diameter was not significantly different among wild types, the1-3, the1-4, and THE1:GFP, while cesa3je5 showed a significantly smaller rosette diameter (Fig. 3A, C). This phenotype was also reverted in cesa3je5/the1-3 and enhanced in cesa3je5/the1-4 or cesa3je5/THE1:GFP. Plant heights followed the same trend of opposite effects, with cesa3je5/the1-4 or cesa3je5/THE1:GFP hardly surviving on soil (Fig 3B, D; Supplementary Fig. S3). In conclusion, alleles the1-1, the1-2, and the1-3 partially or completely suppressed the dark-grown hypocotyl phenotypes of cesa6prc1-1 or cesa3je5 as well as the adult plant phenotypes of cesa3je5 and cesa3eli1-1, whereas the1-4, like THE1:GFP, enhanced these phenotypes.
Fig. 3.
Phenotypes of loss- and gain-of-function alleles in combination with cellulose-deficient mutants on soil. (A) Rosette phenotypes of 4-week-old plants of the wild type (Col-0, WS), the1-4, THE1:GFP, cesa3je5, the1-3, and the double mutants the1-4/cesa3je5, THE1:GFP/cesa3je5, and the1-3/cesa3je5, (B) The same genotypes 2 weeks later. (C) Quantification of the rosette diameters and (D) the stem lengths. The graphs show the average ±SE for at least four plants.
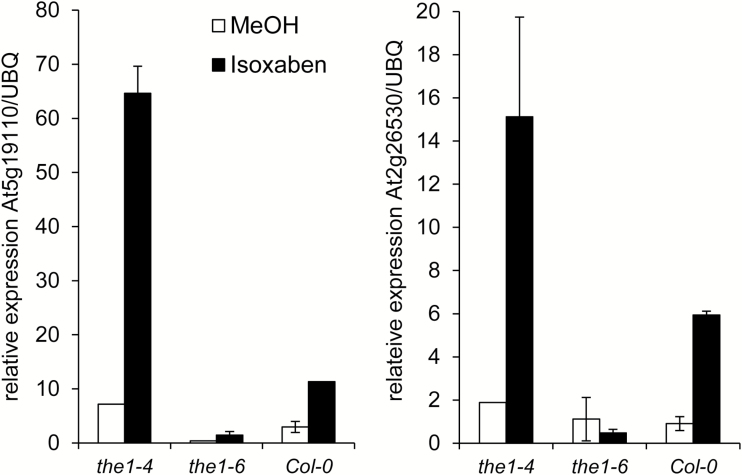
To avoid long-term pleiotropic effects typically observed in cellulose-deficient mutants, we used the cellulose synthesis inhibitor isoxaben to challenge the THE1 pathway in a more controlled way (Heim et al., 1989; Scheible et al., 2001; Desprez et al., 2002, 2007; Wormit et al., 2012). As a read out, we quantified the transcript levels of two genes that previously had been shown to be up-regulated in cesa6prc1-1 but not in cesa6prc1-1/the1-1. As expected, 6 h isoxaben treatment of 4-day-old dark-grown seedlings caused a 3.8-fold and 6.5-fold increase of At5g19110 and At2g26530 transcript levels in wild-type seedlings compared with the solvent control (Fig. 4). Interestingly, in the complete loss-of-function background the1-6, the expression induction was abolished, whereas in the1-4 the induction was strongly enhanced in a way similar to that observed in THE1:GFP (Hématy et al., 2007). Together, these results demonstrate that the the1-4 allele has an effect on growth and gene expression opposite to that of loss-of-function the1 alleles.
Fig. 4.
Opposite effects of loss- and gain-of-function alleles of THE1 on gene expression. Relative expression of THE1-dependent genes At5g19110 and At2g26530 normalized with the reference gene UBQ5 of etiolated seedlings after 6 h of isoxaben or mock treatment in the dark. The graph represents the mean ±SE of three biological replicates with each of three technical replicates.
the1-3 and the1-4 are loss-of-function and gain-of-function alleles, respectively
The opposite effect of the two alleles is surprising given the fact that they are both the result of T-DNA insertions at positions very close to each other (amino acids 507 and 466 for the1-3 and the1-4, respectively), corresponding to the cytoplasmic region just downstream of the membrane-spanning domain (Fig. 1). If expressed, the resulting truncated proteins would comprise the N-terminal extracellular domain, the membrane-spanning domain, and a short stretch of amino acids derived from the intracellular domain, but without the kinase domain.
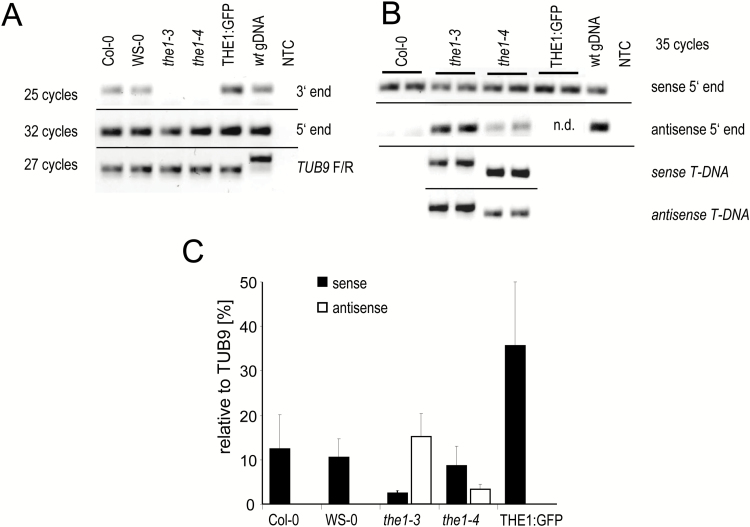
To understand the opposing phenotypes, we investigated the transcripts produced at the THE1 locus in the two mutants. RT–qPCR showed that in both mutants, THE1 transcripts downstream of the T-DNA insertion were absent (Fig. 5A), whereas the transcript levels corresponding to the 5' end of THE1 were unaltered in the wild type, the1-3, and the1-4 (Fig. 5A). Although the two alleles were produced with different T-DNA vectors [the1-3, pGKB5 (Bouchez et al., 1993; Samson et al., 2002); the1-4, pDAP101 (Sessions et al., 2002)], both contain promoters facing towards the left border (LB). While in the1-3 the 35S promoter of the T-DNA was driving a BASTA resistance gene and is ~2000 bp from the LB, in the1-4, pDAP101 contains the bidirectional MAS (mannopine synthase) promoter (Velten et al., 1984) 96 bp from the LB. To test the possibility that T-DNA-driven antisense transcripts were present, we quantified these as well. Indeed, in both alleles, we detected antisense transcripts upstream of the T-DNA insertion (Fig. 5B) but the1-3 was present in significantly higher amounts than the1-4 (Fig. 5C). The ratio between antisense and sense transcripts was ~7 for the1-3 and 0.7 for the1-4 (Fig. 5C).
Fig. 5.
THE1 sense and antisense expression in the1-3 and the1-4. (A) Semi-quantitative expression of THE1 5' and 3' of the T-DNA insertion. (B) Semi-quantitative expression of sense and antisense transcripts of THE1 5' of the T-DNA and with one primer located on the T-DNA. Horizontal lines signify images from the same analysis and gel. (C) RT–qPCR expression analyses of the wild type (Col-0, WS), the1 mutant alleles, and the THE1:GFP overexpressor normalized to the reference gene TUB9. The graph represents the mean ±SE of four biological with each of three technical replicates.
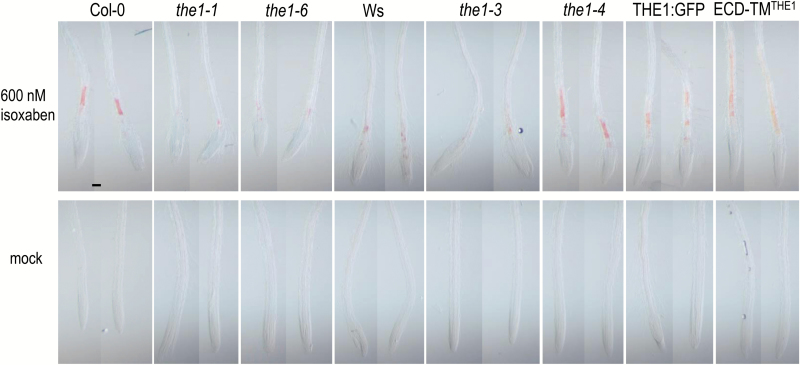
We hypothesize that the1-4 sense transcripts produce a truncated protein, whereas in the1-3, the accumulation of the encoded protein is prevented by the presence of antisense transcripts, which presumably act post-transcriptionally. Unfortunately, despite repeated attempts, we have not been able to detect such truncated proteins with an antiserum directed against the THE1 ectodomain. To examine this hypothesis, we overexpressed the truncated THE1 protein without the kinase domain (ECD–TMTHE1) in Col-0 and studied the response to isoxaben. While ectopic lignification in roots of all the knock out mutants, the1-1, the1-3, and the1-6 was weaker as in their corresponding wild types, the full-length THE:GFP overexpressor, the1-4, and the transgenes overexpressing the truncated ECD–TMTHE1 accumulated lignin to a much higher extent than the wild type upon isoxaben treatment (Fig. 6). Thus the kinase domain is not required for THE1-induced responses upon cellulose deficiency.
Fig. 6.
Ectopic lignification of loss- and gain-of-function alleles upon chemical inhibition of cellulose biosynthesis. Shown are roots from 6-day-old seedlings cultivated on half-strength MS with 1% sucrose and incubated for 16 h with 600 nM isoxaben or the mock control. Seedlings were stained with 1% phloroglucinol for 15 min. Pictures were taken immediately after mounting the seedlings in water. The scale bar corresponds to 100 µm.
Both the1-3 and the1-4 alleles are semi-dominant
According to our hypothesis, both the increased and decreased sensitivity to cell wall perturbation in the1-4 and the1-3, respectively, would involve mechanisms with dominant effects. Indeed, the accumulation of a truncated membrane-bound ectodomain in the1-4 is also expected to enhance the signaling strength in a heterozygote, whereas the accumulation of an inhibitory antisense transcript in the1-3 might also have dominant effects, by inhibiting the accumulation of the endogenous THE1 protein. To investigate this, we analyzed the growth phenotype of heterozygotes for one or the other allele in a homozygous cesa3je5 background. As expected, heterozygote seedlings for the1-3 and the1-4 developed primary roots and etiolated hypocotyls of a length intermediate between those of cesa3je5 and their homozygous counterparts (Fig. 7B, C). The semi-dominance of the hypocotyl elongation was also seen in the the1-3/cesa6prc1-8 combinations (Supplementary Fig. S4). In addition, consistent with our hypothesis, the1-3 suppressed the enhancing effects of the1-4 in the transheterozygote.
For soil-grown plants, the intermediate phenotypes of heterozygous rosettes and inflorescences and the epistasis of the1-3 over the1-4 were even more evident (Fig. 7A).
Discussion
Here we showed that different alleles of THE1 have opposite phenotypic effects: the1-4 enhances whereas the1-1, the1-2, the1-3, and the novel allele the1-6 suppress the reduced growth and (at least for the1-1, -2, and -3) ectopic lignification phenotypes in a cellulose-deficient background. In addition, the expression of two genes induced by the cellulose synthesis inhibitor isoxaben was enhanced and suppressed in the1-4 and the1-6, respectively. The same two genes were also up-regulated in a cellulose-deficient cesa6prc1-1 background relative to the wild type, and this effect was further enhanced by THE1:GFP and suppressed by the1-1 and the1-3 (Hématy et al., 2007).
The T-DNA insertions of the1-3 and the1-4 are only 120 bp apart in a region between the transmembrane and kinase domain. The enhancing effect of the1-4 can be explained by the presence of a truncated THE1 protein lacking the kinase domain that is able to trigger cell wall damage-induced stress responses more efficiently than the wild-type protein. The kinase domain therefore is not required for THE1 activation, like for FER, for which a kinase-dead version still complements the defective pollen tube reception phenotype (Kessler et al., 2015). The hypersensitive the1-4 phenotype suggests that the THE1 kinase domain is required for turning down THE1 activity. RLKs often occur in heteromeric complexes, which can also contain kinase-defective RLKs or receptor-like proteins (RLPs) that influence RLK-mediated signaling. Therefore, it is possible that THE1 can act as co-receptor in such a complex (Castells and Casacuberta, 2007; Gust and Felix, 2014; Liebrand et al., 2014) (Fig. 8). Potential candidates for such signaling partners are other CrRLK1L family members including HERK1/2 or FER, some of which are co-expressed with THE1 (Lindner et al., 2012). The predicted truncated THE1 protein in the1-4 contains the ectodomain, the TMD, and ~26 amino acids of the cytoplasmic domain. Within the ectodomain, the extracellular juxtamembrane region (exJM) is highly conserved in all CrRLKs. In FER, this region interacts with the glycosylphophatidylinositol (GPI)-anchored co-receptors LORELEI (LRE) and LRE-LIKE (LLG1-3) (Li et al., 2015). Furthermore, TMDs can also serve as interaction platforms (Cymer et al., 2012), for instance through the ‘glycine zipper’ motif in the TMD of the RLK SOBIR1/EVR (SUPPRESSOR OF BIR1-1/EVERSHED), which mediates interaction with several RLPs (Bi et al., 2016). Confirmation of these hypotheses awaits the identification of TMD and/or ectodomain interaction partners of THE1 and functional studies with constitutively kinase-active and kinase-dead versions.
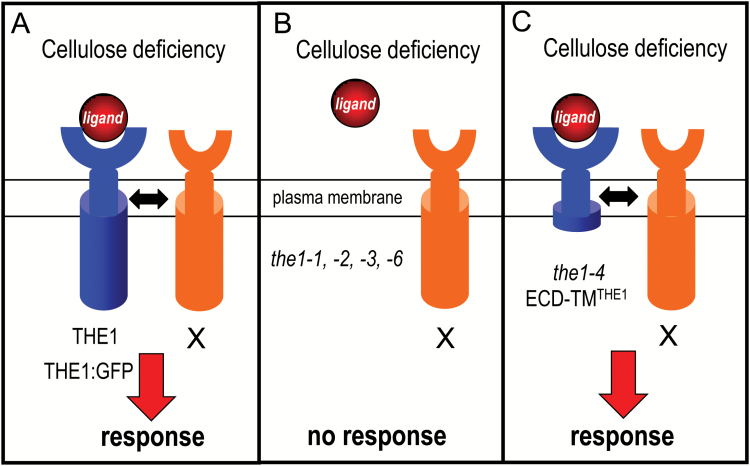
Fig. 8.
Model for the action of the loss- and gain-of-function alleles of THESEUS1. (A) Cellulose deficiency generates a ligand (red) which binds to the extracellular domain and activates THE1 or THE1:GFP (blue). THE1 either signals directly or associates with one or multiple partners (orange). The activation of this pathway suppresses growth and cell elongation and induces transcriptional changes. (B) In loss-of-function plants (the1-1, -2, -3, and -6), plants express no or reduced levels of THE1 which cannot trigger growth responses to or sufficiently efficient associations with signal transmitting partners. (C) In gain-of-function plants (the1-4), the truncated THE1 protein without the cytoplasmic kinase domain is able to associate with the signal transmitting partners and induce strong growth effects. The truncated the1-4 and the overexpressed THE1:GFP proteins are less sensitive to feedback inhibition mediated by the cytoplasmic domain and cause semi-dominant phenotypes.
The expression of the 5' end of THE1 is reduced in the1-3 and correlates with the presence of an antisense RNA initiated from the T-DNA. Interestingly, both T-DNAs contain promoters at the LB, which, given their orientation, would generate THE1 antisense transcripts. Sense or antisense (McElver et al., 2001) read-through transcription of flanking genes from the same type of T-DNA-associated promoters has been reported (Ülker et al., 2008). For instance, the terminator used for pGKB5 (Bouchez et al., 1993) of the FLAG lines is weak, and in conjunction with the 35S promoter can lead to read-through transcription (Meierhoff et al., 2003; Yoo et al., 2005; Ülker et al., 2008). The 10-fold difference in the antisense transcript abundance in the1-3 versus the1-4 is probably due to the strong activity of the bidirectional MAS promoter (the1-3) and the weak read-through of the 35S promoter in the FLAG line, the1-4. The high levels of antisense transcripts in the1-3 might act similarly to natural antisense transcripts (NATs) at different levels including mRNA processing, cellular transport, and translation (Faghihi and Wahlestedt, 2009; Britto-Kido et al., 2013).
The findings of Guo et al. (2009a, b) that herk1 and herk2 only in combination with the hypermorphic the1-4 allele inhibit cell elongation might indicate cell wall integrity defects in these mutants. Another possibility would be that HERK1 and HERK2 act antagonistically to THE1. Seemingly opposite functions of CrRLK family members have been observed for FER and ANX1/2 during pollination (Escobar-Restrepo et al., 2007; Boisson-Dernier et al., 2009; Miyazaki et al., 2009). Kessler et al. 2015) found that FER and ANX most probably do not compete for the same ligand and proposed that an as yet unknown mechanism leads to antagonistic functions. Moreover, it is not formally excluded that the two HERK mutants used by Guo et al. (2009a, b) are hypermorphic alleles, since the authors did not report on expression analyses of the 5' end of these mutants, and the insertion of the T-DNA would allow full ECD expression in both of them.
Our results illustrate one of the pitfalls of using T-DNA insertion lines to infer gene function from quickly characterized mutants and in particular mutants for RLKs. Finally, the availability of both gain-of-function and loss-of-function alleles for THE1 will be helpful for the dissection of cell wall integrity signaling networks in the future.
Supplementary data
Supplementary data are available at JXB online.
Table S1. List of primers used.
Fig. S1. Root growth of the1-3, the1-4, and THE1:GFP seedlings in cellulose-deficient genetic backgrounds
Fig. S2. Ectopic lignification of etiolated seedlings of the1 alleles in cellulose-deficient genetic backgrounds
Fig. S3. Inflorescence phenotypes of loss- and gain-of-function alleles in combination with cellulose-deficient backgrounds
Fig. S4. Semi-dominance of the1-3 in combination with cesa6prc1-8.
Supplementary Material
Acknowledgements
The authors are grateful to Susanne Neubert for her help in generating the crosses, Viktoria Kantner and Martin Frauenlob for genotyping, and Tobias Seethaler and Catarina Fernandes for plant propagation and seed harvesting. The project was supported by grants of the Austrian Science Fund and the French National Research Agency projects FWF-SFB F3707-B22 and ANR-FWF I 1725-B16.
Glossary
Abbreviations:
- CaMV
Cauliflower mosaic virus
- EMS
ethyl methanesulfonate
- GFP
green fluorescent protein
- MAS
mannopine synthase
- MMLV
Moloney murine leukemia virus
- qPCR
quantitative PCR
- RLK
receptor-like-kinases
- ROS
reactive oxygen species
- RT–qPCR
reverse transcription–qPCR.
References
- Bi G, Liebrand TW, Bye RR, Postma J, van der Burgh AM, Robatzek S, Xu X, Joosten MH. 2016. SOBIR1 requires the GxxxG dimerization motif in its transmembrane domain to form constitutive complexes with receptor-like proteins. Molecular Plant Pathology 17, 96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A, Lituiev DS, Nestorova A, Franck CM, Thirugnanarajah S, Grossniklaus U. 2013. ANXUR receptor-like kinases coordinate cell wall integrity with growth at the pollen tube tip via NADPH oxidases. PLoS Biology 11, e1001719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A, Roy S, Kritsas K, Grobei MA, Jaciubek M, Schroeder JI, Grossniklaus U. 2009. Disruption of the pollen-expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development 136, 3279–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchez D, Camilleri C, Caboche M. 1993. A binary vector based on Basta resistance for in planta transformation of Arabidopsis thaliana. Comptes Rendus de l’Academie des Sciences, Serie 3. Sciences de la Vie 316, 1188–1193. [Google Scholar]
- Britto-Kido SdA, Ferreira Neto J, Costa R, Pandolfi V, Marcelino-Guimaraes CF, Nepomuceno AL, Vilela Abdelnoor R, Benko-Iseppon AM, Kido EA. 2013. Natural antisense transcripts in plants: a review and identification in soybean infected with Phakopsora pachyrhizi SuperSAGE library. ScientificWorldJournal 2013, 219798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castells E, Casacuberta JM. 2007. Signalling through kinase-defective domains: the prevalence of atypical receptor-like kinases in plants. Journal of Experimental Botany 58, 3503–3511. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cymer F, Veerappan A, Schneider D. 2012. Transmembrane helix–helix interactions are modulated by the sequence context and by lipid bilayer properties. Biochimica et Biophysica Acta 1818, 963–973. [DOI] [PubMed] [Google Scholar]
- Denness L, McKenna JF, Segonzac C, Wormit A, Madhou P, Bennett M, Mansfield J, Zipfel C, Hamann T. 2011. Cell wall damage-induced lignin biosynthesis is regulated by a reactive oxygen species- and jasmonic acid-dependent process in Arabidopsis. Plant Physiology 156, 1364–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslauriers SD, Larsen PB. 2010. FERONIA is a key modulator of brassinosteroid and ethylene responsiveness in Arabidopsis hypocotyls. Molecular Plant 3, 626–640. [DOI] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Höfte H, Gonneau M, Vernhettes S. 2007. Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 104, 15572–15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T, Vernhettes S, Fagard M, Refrégier G, Desnos T, Aletti E, Py N, Pelletier S, Höfte H. 2002. Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiology 128, 482–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Q, Kita D, Li C, Cheung AY, Wu H-M. 2010. FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proceedings of the National Academy of Sciences, USA 107, 17821–17826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Restrepo JM, Huck N, Kessler S, Gagliardini V, Gheyselinck J, Yang WC, Grossniklaus U. 2007. The FERONIA receptor-like kinase mediates male–female interactions during pollen tube reception. Science 317, 656–660. [DOI] [PubMed] [Google Scholar]
- Faghihi MA, Wahlestedt C. 2009. Regulatory roles of natural antisense transcripts. Nature Reviews. Molecular Cell Biology 10, 637–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikura U, Elsaesser L, Breuninger H, Sánchez-Rodríguez C, Ivakov A, Laux T, Findlay K, Persson S, Lenhard M. 2014. Atkinesin-13A modulates cell-wall synthesis and cell expansion in Arabidopsis thaliana via the THESEUS1 pathway. PLoS Genetics 10, e1004627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachomo EW, Jno Baptiste L, Kefela T, Saidel WM, Kotchoni SO. 2014. The Arabidopsis CURVY1 (CVY1) gene encoding a novel receptor-like protein kinase regulates cell morphogenesis, flowering time and seed production. BMC Plant Biology 14, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Li L, Ye H, Yu X, Algreen A, Yin Y. 2009a. Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 106, 7648–7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ye H, Li L, Yin Y. 2009. A family of receptor-like kinases are regulated by BES1 and involved in plant growth in Arabidopsis thaliana. Plant Signaling and Behavior 4, 784–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust AA, Felix G. 2014. Receptor like proteins associate with SOBIR1-type of adaptors to form bimolecular receptor kinases. Current Opinion in Plant Biology 21, 104–111. [DOI] [PubMed] [Google Scholar]
- Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR. 2014. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343, 408–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser MT, Morikami A, Benfey PN. 1995. Conditional root expansion mutants of Arabidopsis. Development 121, 1237–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim DR, Roberts JL, Pike PD, Larrinua IM. 1989. Mutation of a locus of Arabidopsis thaliana confers resistance to the herbicide isoxaben. Plant Physiology 90, 146–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hématy K, Sado PE, Van Tuinen A, Rochange S, Desnos T, Balzergue S, Pelletier S, Renou JP, Höfte H. 2007. A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Current Biology 17, 922–931. [DOI] [PubMed] [Google Scholar]
- Karsai A, Müller S, Platz S, Hauser MT. 2002. Evaluation of a homemade SYBR green I reaction mixture for real-time PCR quantification of gene expression. Biotechniques 32, 790–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinath NF, Kierszniowska S, Lorek J, Bourdais G, Kessler SA, Shimosato-Asano H, Grossniklaus U, Schulze WX, Robatzek S, Panstruga R. 2010. PAMP (pathogen-associated molecular pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. Journal of Biological Chemistry 285, 39140–39149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler SA, Lindner H, Jones DS, Grossniklaus U. 2015. Functional analysis of related CrRLK1L receptor-like kinases in pollen tube reception. EMBO Reports 16, 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Wightman R, Atanassov I, Gupta A, Hurst CH, Hemsley PA, Turner S. 2016. S-Acylation of the cellulose synthase complex is essential for its plasma membrane localization. Science 353, 166–169. [DOI] [PubMed] [Google Scholar]
- Li C, Wu HM, Cheung AY. 2016. FERONIA and her pals: functions and mechanisms. Plant Physiology 171, 2379–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Yeh F-L, Cheung AY et al. . 2015. Glycosylphosphatidylinositol-anchored proteins as chaperones and co-receptors for FERONIA receptor kinase signaling in Arabidopsis. eLife 4, e06587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebrand TW, van den Burg HA, Joosten MH. 2014. Two for all: receptor-associated kinases SOBIR1 and BAK1. Trends in Plant Science 19, 123–132. [DOI] [PubMed] [Google Scholar]
- Lindner H, Müller LM, Boisson-Dernier A, Grossniklaus U. 2012. CrRLK1L receptor-like kinases: not just another brick in the wall. Current Opinion in Plant Biology 15, 659–669. [DOI] [PubMed] [Google Scholar]
- McElver J, Tzafrir I, Aux G et al. . 2001. Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159, 1751–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meierhoff K, Felder S, Nakamura T, Bechtold N, Schuster G. 2003. HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB–psbT–psbH–petB–petD RNAs. The Plant Cell 15, 1480–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki S, Murata T, Sakurai-Ozato N, Kubo M, Demura T, Fukuda H, Hasebe M. 2009. ANXUR1 and 2, sister genes to FERONIA/SIRENE, are male factors for coordinated fertilization. Current Biology 19, 1327–1331. [DOI] [PubMed] [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Stewart AJ, Jenkins GI, Caboche M, Lepiniec L. 2002. The TRANSPARENT TESTA16 locus encodes the ARABIDOPSIS BSISTER MADS domain protein and is required for proper development and pigmentation of the seed coat. The Plant Cell 14, 2463–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen KS, Willats WG, Malinovsky FG. 2016. Understanding CrRLK1L function: cell walls and growth control. Trends in Plant Science 21, 516–527. [DOI] [PubMed] [Google Scholar]
- Pysh L, Alexander N, Swatzyna L, Harbert R. 2012. Four alleles of AtCESA3 form an allelic series with respect to root phenotype in Arabidopsis thaliana. Physiologia Plantarum 144, 369–381. [DOI] [PubMed] [Google Scholar]
- Samson F, Brunaud V, Balzergue S, Dubreucq B, Lepiniec L, Pelletier G, Caboche M, Lecharny A. 2002. FLAGdb/FST: a database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Research 30, 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schallus T, Jaeckh C, Fehér K et al. . 2008. Malectin: a novel carbohydrate-binding protein of the endoplasmic reticulum and a candidate player in the early steps of protein N-glycosylation. Molecular Biology of the Cell 19, 3404–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible W-R, Eshed R, Richmond T, Delmer D, Somerville C. 2001. Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants. Proceedings of the National Academy of Sciences, USA 98, 10079–10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G et al. . 2002. A high-throughput Arabidopsis reverse genetics system. The Plant Cell 14, 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih HW, Miller ND, Dai C, Spalding EP, Monshausen GB. 2014. The receptor-like kinase FERONIA is required for mechanical signal transduction in Arabidopsis seedlings. Current Biology 24, 1887–1892. [DOI] [PubMed] [Google Scholar]
- Slabaugh E, Davis JK, Haigler CH, Yingling YG, Zimmer J. 2014. Cellulose synthases: new insights from crystallography and modeling. Trends in Plant Science 19, 99–106. [DOI] [PubMed] [Google Scholar]
- Stegmann M, Monaghan J, Smakowska-Luzan E, Rovenich H, Lehner A, Holton N, Belkhadir Y, Zipfel C. 2017. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355, 287–289. [DOI] [PubMed] [Google Scholar]
- Velten J, Velten L, Hain R, Schell J. 1984. Isolation of a dual plant promoter fragment from the Ti plasmid of Agrobacterium tumefaciens. EMBO Journal 3, 2723–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voxeur A, Höfte H. 2016. Cell wall integrity signaling in plants: ‘to grow or not to grow that’s the question’. Glycobiology 26, 950–960. [DOI] [PubMed] [Google Scholar]
- Wormit A, Butt SM, Chairam I et al. . 2012. Osmosensitive changes of carbohydrate metabolism in response to cellulose biosynthesis inhibition. Plant Physiology 159, 105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SY, Bomblies K, Yoo SK, Yang JW, Choi MS, Lee JS, Weigel D, Ahn JH. 2005. The 35S promoter used in a selectable marker gene of a plant transformation vector affects the expression of the transgene. Planta 221, 523–530. [DOI] [PubMed] [Google Scholar]
- Ulker B, Peiter E, Dixon DP, Moffat C, Capper R, Bouché N, Edwards R, Sanders D, Knight H, Knight MR. 2008. Getting the most out of publicly available T-DNA insertion lines. The Plant Journal 56, 665–677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.