Protein persulfidation in the Arabidopsis proteome is widespread and is the molecular mechanism by which hydrogen sulfide performs its signaling role.
Keywords: Cysteine, hydrogen sulfide, mass spectrometry, persulfidation, post-translational modification, proteomics
Abstract
Hydrogen sulfide-mediated signaling pathways regulate many physiological and pathophysiological processes in mammalian and plant systems. The molecular mechanism by which hydrogen sulfide exerts its action involves the post-translational modification of cysteine residues to form a persulfidated thiol motif, a process called protein persulfidation. We have developed a comparative and quantitative proteomic analysis approach for the detection of endogenous persulfidated proteins in wild-type Arabidopsis and L-CYSTEINE DESULFHYDRASE 1 mutant leaves using the tag-switch method. The 2015 identified persulfidated proteins were isolated from plants grown under controlled conditions, and therefore, at least 5% of the entire Arabidopsis proteome may undergo persulfidation under baseline conditions. Bioinformatic analysis revealed that persulfidated cysteines participate in a wide range of biological functions, regulating important processes such as carbon metabolism, plant responses to abiotic and biotic stresses, plant growth and development, and RNA translation. Quantitative analysis in both genetic backgrounds reveals that protein persulfidation is mainly involved in primary metabolic pathways such as the tricarboxylic acid cycle, glycolysis, and the Calvin cycle, suggesting that this protein modification is a new regulatory component in these pathways.
Introduction
Hydrogen sulfide (H2S) has been referred to as the third gasotransmitter in animal and plant cells and is as important as nitric oxide (NO), carbon monoxide (CO), and hydrogen peroxide (H2O2) (García-Mata and Lamattina, 2010; Vandiver and Snyder, 2012; Kimura, 2014). These small molecules possess high permeability that allows them to cross biological membranes and to act as signaling molecules. H2S dissociates easily under physiological conditions. It is therefore assumed that hydrogen sulfide pools include H2S, HS-, and S2-, but the active form of hydrogen sulfide in cells has not been fully clarified. H2S is involved in many physiological and pathological processes in animals, including cell proliferation, apoptosis, inflammatory processes, hypoxia protection, neuromodulation, and cardioprotection, as previously described (Wang, 2014; Olas, 2015; Paul and Snyder, 2015b).
More recently, regulatory properties of H2S have emerged in plants, with protective effects reported against oxidative and metal stresses (Zhang et al., 2008; Wang et al., 2010; Zhang et al., 2010; Li et al., 2012a; Shen et al., 2013; Sun et al., 2013; Fang et al., 2016;), drought and heat tolerance (Li et al., 2012b; Shen et al., 2013), and osmotic and saline stresses (Shi et al., 2013). H2S is involved in regulating important physiological processes in plants, such as stomatal closure/aperture (García-Mata and Lamattina, 2010; Lisjak et al., 2010; Jin et al., 2013; Scuffi et al., 2014; Papanatsiou et al., 2015), the modulation of photosynthesis (Chen et al., 2011), and autophagy regulation (Alvarez et al., 2012b; Gotor et al., 2013; Romero et al., 2014; Laureano-Marin et al., 2016).
In mammals, endogenous H2S is produced by the action of two pyridoxal phosphate (PLP)–dependent enzymes, cystathionine γ-lyase (CSE) and cystathionine β-synthase (CBS), and the PLP-independent enzyme 3-mercaptopyruvate sulfurtransferase (3MST), as steps in the cysteine catabolic pathway (Kimura, 2015). In plant systems, the bulk of sulfide production occurs in chloroplasts through the photosynthetic sulfate-assimilation pathway, in which sulfate is reduced to sulfide and assimilated into cysteine (Takahashi et al., 2011; Garcia et al., 2015). In mitochondria, the enzyme CYANOALANINE SYNTHASE C1 (CAS-C1) also generates sulfide through the catalysis of cysteine and cyanide to form β-cyanoalanine (Yamaguchi et al., 2000; Alvarez et al., 2012c). However, the sulfide produced in these organelles is dissociated into its ionized forms and therefore is unable to be transported across the membrane into the cytosol. Although several enzymatic reactions have been described but have not been characterized in detail (Riemenschneider et al., 2005; Van Hoewyk et al., 2008; Jin et al., 2013), the production of endogenous cytosolic H2S has been demonstrated by L-CYSTEINE DESULFHYDRASE 1 (DES1). This enzyme catalyzes the desulfuration of cysteine to sulfide, ammonia, and pyruvate (Alvarez et al., 2010; Gotor et al., 2010; Alvarez et al., 2012b). In plants, DES1 is primarily responsible for sulfide production in the cytosol and this sulfide and other sulfurating species such as sulfane sulfur or polysulfides are essential for signaling (Romero et al., 2013b; Gotor et al., 2015).
Despite numerous reports highlighting the importance of H2S as a signaling molecule, its mechanism of action is not yet fully understood. However, the primary signaling mechanism of H2S occurs through the persulfidation of reactive cysteine residues on target proteins via conversion of the thiol group (-SH) into a persulfide group (-SSH). This posttranslational modification of target proteins results in functional changes in enzymatic structures and activities, including those of Arabidopsis ascorbate peroxidase, glyceraldehyde-3-phosphate dehydrogenase, and glutamine synthetase (Aroca et al., 2015), and in subcellular localizations (Mustafa et al., 2009; Kimura, 2015; Paul and Snyder, 2015c; Aroca et al., 2017). Persulfidation, previously known as S-sulfhydration, typically increases the reactivity of the modified cysteine due to the increased nucleophilicity of persulfide compared with the thiol group (Paul and Snyder, 2012; Cuevasanta et al., 2015). Furthermore, quantitative data have indicated the widespread nature of persulfides in animal and plant cells (Mustafa et al., 2009; Ida et al., 2014). This reinforces the importance of persulfidation as a signaling process and explains the increasing interest in understanding its underlying mechanism.
A growing number of persulfidated proteins have been described using different tagging methods and proteomics approaches, including the elution of alkylated persulfides by reducing agent methods (Gao et al., 2015; Longen et al., 2016). Using the first described method, the modified biotin switch assay, a large number of persulfidated proteins were identified in animal and plant systems (Mustafa et al., 2009; Aroca et al., 2015). Nevertheless, the specificity of the blocking reagent S-methyl-methanothiosulfonate (MMTS) has been questioned by several authors. Thus, a new approach to detect persulfidated proteins was recently described, the tag-switch method (Zhang et al., 2014). This method employs methylsulfonylbenzothiazole (MSBT) to block both thiols and persulfide groups in the first step; then, the disulfide bonds in persulfide adducts possess enhanced reactivity to nucleophilic attack by the cyanoacetate-based reagent CN-biotin, while thiol adducts are thioethers that do not react with nucleophiles.
Due to the growing importance of sulfide as a signaling molecule and considering our scarce knowledge regarding the functions and targets of persulfidation in plants, in this paper we performed a large-scale proteomics study of endogenously persulfidated proteins in wild-type Arabidopsis and DES1-defective (des1) mutant plants using the tag-switch method to shed light on the role of persulfidation and provide targets for further studies of this post-translational modification in plants.
Materials and methods
Plant material and growth conditions
Arabidopsis (Arabidopsis thaliana) wild-type ecotype Col-0 and the des1 T-DNA insertion mutant (des1-1; SALK_103855) were grown in soil under a photoperiod of 16 h of white light (120 μE m-2 s-1) at 20°C and 8 h of dark at 18°C (Bermudez et al., 2012).
Tag-switch method
The biotinylation of endogenously persulfidated proteins from 30-day-old Arabidopsis plants was performed by the tag-switch method as previously described (Zhang et al., 2014).
Plant leaf material weighing 1 g was ground in a mortar under liquid nitrogen and an enriched cytosolic extract was obtained following a previously described adapted method (Giavalisco et al., 2003). Ground material was homogenized in a solution containing 0.125 parts (v/w) of buffer I [50 mM Tris, pH 7.1, amended with 100 mM KCl, 20% glycerol and protease inhibitor cocktail (25x) (Roche)] and 0.05 parts (v/w) of buffer II (1 mM pepstatin and 1.4 µM PMSF dissolved in ethanol) and then centrifuged at 50000 rpm for 1 h at 4ºC.
The enriched cytosolic extract was incubated with 50 mM MSBT in 50 mM Tris-HCl at pH 8, amended with 2.5% SDS, at 37°C for 30 min, followed by the addition of 20 mM CN-biotin at 37°C for 4 h. Proteins were then precipitated and pellets were washed with ice-cold acetone and resuspended in 50 mM Tris-HCl at pH 8. To purify the labeled proteins, the solution was incubated with streptavidin beads for 1 h at room temperature with frequent vortexing. The beads were intensively washed with Tris-HCl at pH 8, 600 mM NaCl, 1 mM EDTA, and 0.5% Triton X-100 and centrifuged at 3000 rpm for 5 s at room temperature. Bound proteins were eluted with a solution of 2% SDS, 30 mM biotin, 50 mM phosphate, 100 mM NaCl, 6 M urea, and 2 M thiourea for 15 min at room temperature, followed by 15 min at 96°C as previously described (Rybak et al., 2004).
To determine the specificity of the method, a cytosol-enriched protein extract was obtained from the 30-day-old wild-type and des1 Arabidopsis plants. One aliquot was incubated directly with 20 mM CN-biotin as described for tag-switch labeling, skipping the MSBT blocking step. Another aliquot was incubated with 100 mM DTT for 1 h at 37°C prior to tag-switch labeling. The proteins were then precipitated and washed, followed by incubation with MSBT and CN-biotin as described above. Samples were purified with streptavidin beads and immunoblotted using anti-HPDP-biotin (Abcam) as previously described (Aroca et al., 2015).
Identification of persulfidated proteins by LC-MS/MS analysis
To identify persulfidated proteins, tryptic digestion was performed in solution and peptide analysis was carried out by liquid chromatography and mass spectrometry analysis at the Proteomics Facility of the Centro Nacional de Biotecnología, Spain.
Protein concentration from eluted streptavidin beads was determined using a Pierce 660-nm protein assay (Thermo). An aliquot of 40 µg of protein from each treatment group was digested with sequencing-grade modified trypsin (Sigma-Aldrich) at 37°C overnight on a shaker. Three biological replicates for each condition, wild type and des1, were analyzed in this study.
A 1 µg aliquot of each sample was subjected to 1D-nano LC ESI-MSMS analysis using a nano liquid chromatography system (Eksigent Technologies nanoLC Ultra 1D plus, AB SCIEX) coupled to a high-speed TripleTOF 5600 mass spectrometer (AB SCIEX) with a Nanospray III source. The analytical column was a silica-based reversed-phase column C18 ChromXP, 75 µm × 15 cm, 3-µm particle size, with 120-Å pore size (Eksigent Technologies, AB SCIEX). The trap column was a C18 ChromXP (Eksigent Technologies, AB SCIEX). Peptides were separated using a 250 min gradient ranging from 2% to 90% mobile phase B (mobile phase A, 2% acetonitrile, 0.1% formic acid; mobile phase B, 100% acetonitrile, 0.1% formic acid). The injection volume was 5 µL.
Data acquisition was performed with a TripleTOF 5600 System (AB SCIEX). For IDA parameters, a 0.25 s MS survey scan in the mass range of 350–1250 m/z followed by 35 MS/MS scans of 100 ms in the mass range of 100–1800, with a total cycle time of 4 s, was performed. Switching criteria were set to ions with a mass-to-charge ratio (m/z) greater than 350 and smaller than 1250 with a charge state of 2–5 and an abundance threshold of more than 90 counts (cps). Former target ions were excluded for 20 s. The IDA rolling collision energy (CE) parameters script was used to automatically control for the CE.
MS and MS/MS data obtained for individual samples were processed using Analyst® TF 1.5.1 Software (AB SCIEX). Searches were done with an A. thaliana protein database from UniProt, which contains 66814 protein-coding genes and their corresponding reversed entries using the Mascot Server v. 2.5.1 (Matrix Science, London, UK). Search parameters were set as follows: acetyl (Protein N-term), CN-Biotin-Na-Sulfide (Cysteine), CN-Biotin-Sulfide (Cysteine), Methylthio (Cysteine), MSBT (Cysteine), Oxidation (Methionine), and Sulfide (Cysteine) as variable modifications. The peptide mass tolerance was set to 25 ppm and 0.05 Da for fragment masses, and two missed cleavages were allowed. False discovery rates (FDR≤1% at the PSM level) for peptide identification were manually calculated.
The mass spectrometry data have been deposited with the ProteomeXchange Consortium via the PRIDE (Vizcaino et al., 2016) partner repository with the dataset identifiers PXD005168 and 10.6019/PXD005168.
Protein functional analysis and classification were performed with MapMan (Thimm et al., 2004; Klie and Nikoloski, 2012). Singular enrichment analysis (SEA) of Gene Ontology (GO) was performed using the web-based tool and database AgriGO (Du et al., 2010).
Identification and quantification of persulfidated proteins by Tandem Mass Tag (TMT) SixplexTM
The identification and quantification of persulfidated proteins using TMTsixplexTM were performed at the same facility described above and the tryptic digestion was performed as described in the previous section.
The resulting peptides were subsequently labeled using a TMTsixplex Isobaric Mass Tagging Kit (Thermo Scientific) according to the manufacturer’s instructions, as follows: 126, wt-1; 127, des1-1; 128, wt-2; 129, des1-2; 130, wt-3; 131, des1-3. Three biological replicates for both conditions, wild-type and des1, were analyzed in this study.
A 1 µg aliquot of the labeled mixture was subjected to 1D-nano LC-electrospray ionization (ESI)-MS/MS analysis using the same methodology and equipment as in the previous section for protein identification, but in this case a 200 min gradient was used for peptide separation. Data acquisition was performed with a TripleTOF 5600 System (AB SCIEX) under the same parameters as described in the previous section. For IDA, the procedure was the same as above, except that the 0.25 s MS survey scan was followed by 30 MS/MS scans of 150 ms. Switching criteria were set the same as in the previous section, except that an abundance threshold of more than 70 counts (cps) was used. Other parameters were the same.
The mass spectrometry data have been deposited with the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD006140.
Statistical analysis
Label-based quantification was performed using Proteobotics S.L. (Madrid, Spain). MS/MS spectra were exported to mgf format using Peak View v1.2.0.3 and searched using Mascot Server 2.5.1, OMSSA 2.1.9, X! Tandem 2013.02.01.1, X! Tandem with k-score plug-in and Myrimatch 2.2.140 against a composite target/decoy database built from the 31 551 sequences in the A. thaliana reference proteome at UniProt Knowledgebase, together with commonly occurring contaminants. Search engines were configured to match potential peptide candidates with a mass error tolerance of 25 ppm and fragment ion tolerance of 0.02 Da, allowing for up to two missed tryptic cleavage sites and a maximum isotope error (13C) of 1, setting Methylthio (C) as fixed modification, and TMTsixplex (N-term, K, Y), acetyl (Protein N-term), Oxidation (M), Gln->pyro-Glu (N-term Q), and Glu->pyro-Glu (N-term E) as variable modifications. An additional OMSSA search allowed for no-enzyme cleavage. CN-Biotin-Na-Sulfide (C), CN-Biotin-Sulfide (C), Sulfide (C), Methylthio (C), and MSBT (C) were incorporated as variable modifications in additional MASCOT and X! Tandem searches, in which Methylthio (C) was also considered a variable modification. Score distribution models were used to compute peptide-spectrum match p-values (Ramos-Fernandez et al., 2008) and spectra recovered by a peptide-level FDR<0.01 filter were selected for quantitative analysis. Approximately 1% of the signals with the lowest quality were removed prior to further analysis. Peptides modified with CN-Biotin-Na-Sulfide (C), CN-Biotin-Sulfide (C) or Sulfide (C) were quantified separately from their parent proteins. Differential regulation was measured using linear models (Lopez-Serra et al., 2014) and statistical significance was measured using q-value<0.05.
Results
Identification and functional classification of persulfidated proteins in wild-type Arabidopsis plants
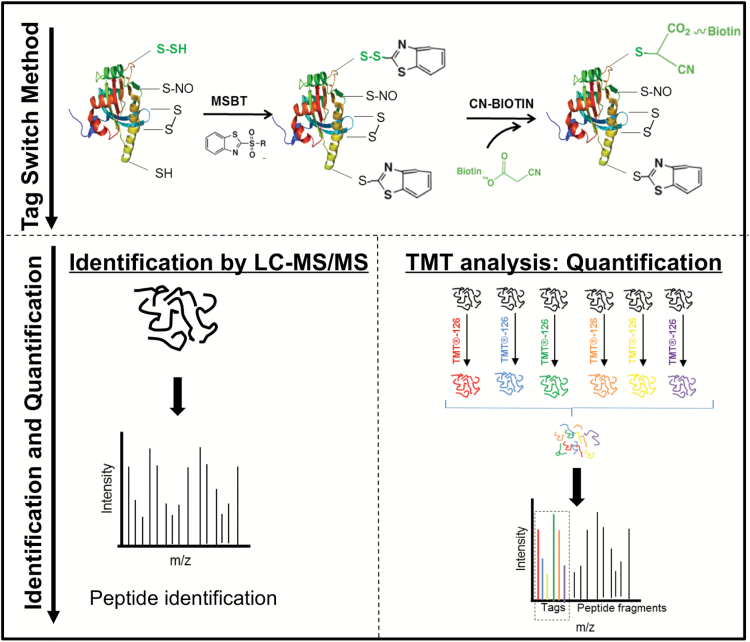
We recently reported the presence of persulfidation-modified cysteine residues in 106 proteins from Arabidopsis leaf extracts by using the modified biotin switch method (Aroca et al., 2015). Recent articles have questioned the validity of this method because thiols and persulfides demonstrate similar reactivity towards MMTS (Filipovic et al., 2012; Pan and Carroll, 2013). The number of Arabidopsis proteins previously reported may therefore be significantly underestimated. The publication of new methods for selective identification of persulfides on cysteine residues has motivated us to undertake a new study. The detection of endogenous persulfidated proteins in Arabidopsis leaves was performed using the tag-switch method, which labels persulfidated proteins with CN-biotin (Zhang et al., 2014) (Fig. 1).
Fig. 1.
Schematic illustration of the procedure to identify and quantify persulfidated proteins in Arabidopsis leaves using the tag-switch method.
Cytosol-enriched leaf extracts from 30-day-old wild-type and des1 mutant Arabidopsis plants grown under physiological conditions were subjected to the tag-switch method (Fig. 1). To validate the method in our plant system, protein labeling was first analyzed by performing immunoblotting using anti-biotin antibodies. Several abundant protein bands were detected in whole protein extracts labeled with the tag-switch method prior to streptavidin bead purification (see Supplementary Fig. S1A at JXB online). When protein extracts were treated with DTT to reduce persulfide residues prior to blocking with MSBT and CN-biotin and streptavidin-bead purification, we were unable to detect biotin-labeled proteins in the immunoblots in any wild-type or des1 extract (Supplementary Fig. S1B). Similarly, when protein extracts were treated directly with CN-biotin without the MSBT blocking step, no signals were detected in the streptavidin-purified samples. Analogously, in the absence of the streptavidin purification step, only one biotinylated protein band was immunodetected in both wild-type and des1 extracts previously treated with DTT (Supplementary Fig. S1B). The method was therefore considered suitable and specific for the study of persulfidation patterns in Arabidopsis in combination with a proteomics approach.
Initially, three biological replicates of CN-biotinylated proteins from Arabidopsis wild-type cytosol-enriched leaf extracts were streptavidin-purified and analyzed by LC-MS/MS. In the three biologically independent samples, we identified a total of 3147 persulfidated proteins with FDR≤1% (Supplementary Dataset S1; Comparative WT SAMPLES). Although for further analysis we have considered persulfidated proteins found in all three biological samples to be statistically significant, we cannot exclude that other proteins identified in one or two of the replicates may also be susceptible to persulfidation. Based on this criterion, we report in this work a list of 2015 persulfidated proteins in wild-type leaf extracts (Supplementary Dataset S1; WT proteins and locus). The peptides used for protein identification are provided as supplementary information (Supplementary Dataset S1). Because the protein samples analyzed in this work were isolated from the leaves of plants grown under controlled conditions with no apparent symptoms of stress, at least 5% of the Arabidopsis proteome may undergo persulfidation under baseline conditions. As a control experiment, samples with the same protein concentrations were pre-treated with DTT and processed under the same conditions as described. Following elution of the streptavidin beads, we were unable to detect persulfidated proteins via LC-MS/MS analysis in these controls.
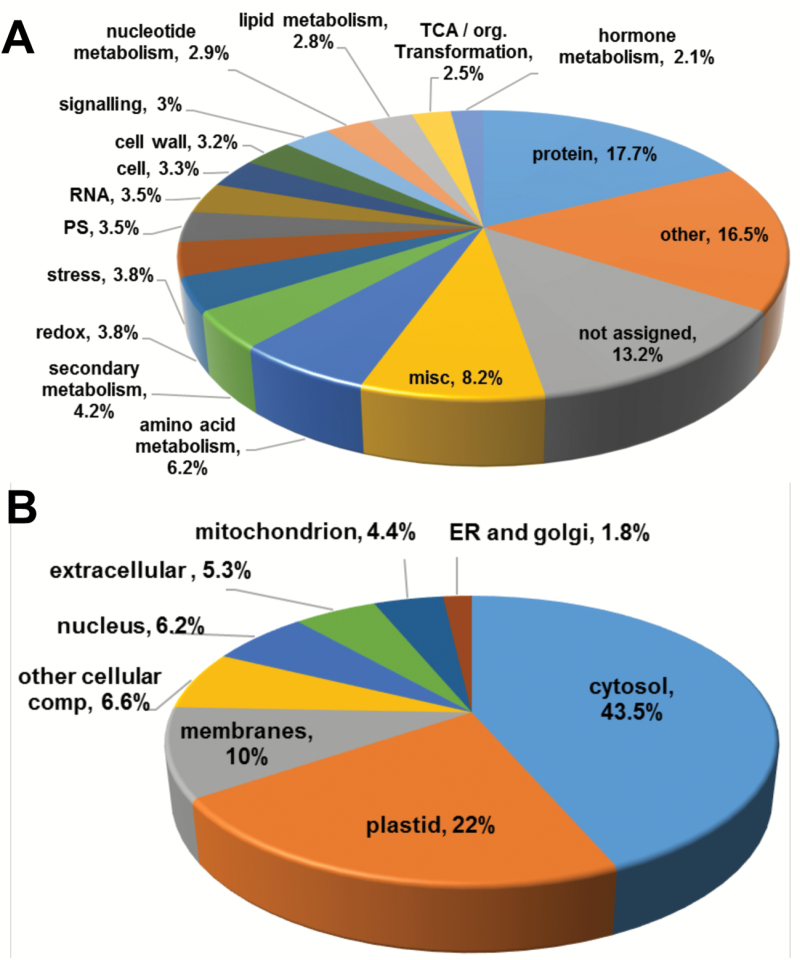
The 2015 identified proteins were analyzed based on their assigned functions and classified into 34 functional groups using the MapMan nomenclature developed for plant-specific pathways and processes (Thimm et al., 2004; Klie and Nikoloski, 2012) (Fig. 2A and Supplementary Table S1). The most abundant set corresponded to the general protein group, which included 17.7% of total identified proteins with 367 elements involved in protein synthesis (79 elements), subcellular targeting (25 elements), post-translational modification (38 elements), degradation (144 elements) and folding, glycosylation and assembly (38 elements). Two other important groups contained proteins involved in amino acid metabolism and miscellaneous enzyme families, representing 6.2% and 8.2% of the total, respectively (Fig. 2A), many of which are primarily involved in the regulation of primary metabolism, glycolysis, the Calvin cycle, and the tricarboxylic acid cycle. The subcellular localization of the identified proteins primarily consisted of the cytoplasm (43.5%) and plastids (22%), although proteins from the membrane and mitochondria were also highly represented (Fig. 2B).
Fig. 2.
Functional classification of persulfidated proteins identified by LC-MS/MS in leaf extracts of 30-day-old Arabidopsis wild-type plants. (A) Functional classification of gene ontology (GO) terms categorized by biological processes. (B) Functional classification by subcellular localization.
In a previous persulfidation proteomics approach involving the biotin-switch method, 106 proteins were identified in Arabidopsis under physiological conditions (Romero et al., 2013a; Aroca et al., 2015), constituting 5% of the total newly identified persulfidated proteins. These results demonstrate a significant improvement in the method used to identify persulfidated proteins.
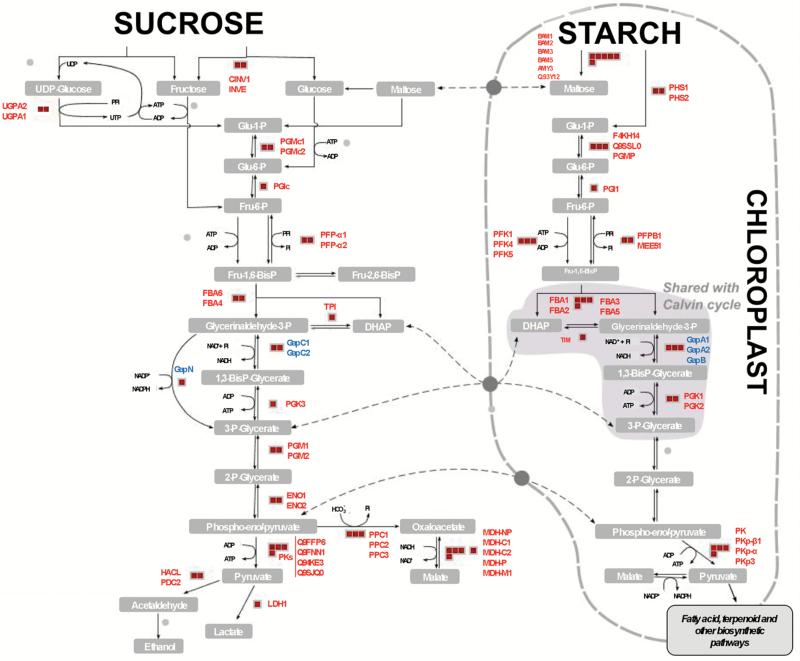
To study the regulatory roles and the putative functions of persulfidation in specific biological processes and to identify over-represented functional categories, we performed a singular enrichment analysis (SEA) of Gene Ontology (GO) terms using AgriGO (Du et al., 2010). SEA compares each annotated gene to all annotated expressed genes. In total, 634 GO terms demonstrated significant over-representation (FDR≤0.001) (Supplementary Fig. S2). The GO enrichment analysis revealed a significant number of persulfidated proteins involved in various aspects of carbon metabolism and glycolysis; for example, 57 proteins annotated in the reference A. thaliana database related to the GO category ‘glycolysis’ (GO:0006096), of which 30 proteins (52.6%) were identified as persulfidated proteins, including the two cytosolic glyceraldehyde-3-phosphate dehydrogenases, GapC1 and GapC2, previously identified in Arabidopsis (Aroca et al., 2015). Moreover, the regulatory function of sulfide in the nuclear localization of these proteins has been very recently demonstrated and the persulfidated cysteine residue on nuclear GapC1 identified (Aroca et al., 2017). The three chloroplastic glyceraldehyde-3-phosphate dehydrogenase forms, GapA1, GapA2, and GapB, and the NADP-dependent isoforms, GapN, were also identified in the GO category. Other prominent proteins involved in glycolysis were identified as persulfidated, including several phosphofructokinases and enolases (Fig. 3 and Supplementary Table S2). An enriched subset of proteins involved in ‘tRNA aminoacylation’ (GO:0006418) was also identified, highlighting the importance of persulfidation in protein translation. Thus, almost half, 41 out of 84, of the total tRNA aminoacylation proteins represented in the reference database were identified as persulfidated proteins (Supplementary Table S3). Two other enriched categories included proteins involved in ‘jasmonic acid biosynthesis’ (GO:0009695), with 11 proteins (37.9%) out of the 29 annotated in Arabidopsis, including 12-oxophytodienoate reductases 1 and 3 and lipoxygenase 2 (Supplementary Table S4), and ‘abiotic stress responses’ (GO:0009628), comprising 214 proteins (14.5%) out of 1471 (Supplementary Table S5). Thus, the GO categorization of persulfidated proteins revealed the involvement of a large number of these proteins in regulating important biological processes, such as carbon metabolism, plant responses to abiotic stresses, plant growth and development, and RNA translation.
Fig. 3.
Persulfidated proteins in the plant glycolysis pathway. Red squares represent persulfidated proteins. Cytosolic, GapC1 and GapC2, chloroplastic, GapA1, GapA2, and GapB, and NADP-dependent isoforms of glyceraldehyde-3-phosphate dehydrogenase are highlighted in blue.
Identification of persulfidated proteins in the des1 mutant
Although the Arabidopsis genome contains several genes coding for putative D- and L-cysteine desulfhydrases that produce sulfide (Jin and Pei, 2015), only the protein DES1 has been extensively characterized in vitro and in vivo. The loss of protein function and reduced sulfide production in the mutant des1 are related to plant physiological processes, such as senescence, autophagy, stomatal closure, and immunity (Alvarez et al., 2010; Alvarez et al., 2012a; Alvarez et al., 2012b; Scuffi et al., 2014; Laureano-Marin et al., 2016). To determine whether the synthesis of sulfide catalyzed by DES1 acts on cells through the protein persulfidation mechanism, we analyzed the basal levels of persulfidation in cytosol-enriched des1 leaf extracts following the same methodology previously utilized with wild-type plants (Fig. 1). Although we used des1 leaves from 30-day-old plants for comparison purposes, the mutation in DES1 causes accelerated plant growth, premature leaf senescence, and altered transcriptional levels compared with wild-type plants (Alvarez et al., 2012b).
Three biological replicates of CN-biotinylated proteins were analyzed under the same conditions and protocols as those employed for wild-type plants. A total of 3242 proteins were identified in the three samples (Supplementary Dataset S2; Comparative DES SAMPLES); 85% of these proteins were also identified in wild-type plants. Following the same criteria implemented for wild-type plants, 2130 proteins were identified in all three samples and, therefore, considered to be statistically significant as persulfidated proteins in the des1 mutant (Supplementary Dataset S2; DES1 Proteins and locus). Functional classification of the 2130 proteins based on MapMan ontology showed no significant differences compared with wild-type (Supplementary Fig. S3).
A qualitative comparison between the lists of identified persulfidated proteins in the wild-type and des1 lines was performed based on our criterion of being present in all three biological replicates. The comparison showed that 184 proteins were predominantly found in wild-type plants and 299 proteins in the des1 mutant, suggesting the ability of sulfurating species generated in the cells in each genetic background to modify specific targets (Supplementary Table S6 and Datasets S1 and S2).
Quantitative comparison of persulfidation patterns in wild-type and des1 plants
To better understand the role of DES1 in sulfide-mediated persulfidation, a quantitative approach was performed using 6-plex tandem mass tag (TMT) isobaric peptide labeling. For this purpose, the same samples used for wild-type (three replicate samples) and des1 (three replicate samples) LC-MS/MS analysis were labeled with a set of six isobaric compounds containing different numbers of heavy isotopes in the mass reporter region, which result in unique reporter masses during tandem MS/MS for sample identification and relative quantitation (Fig. 1).
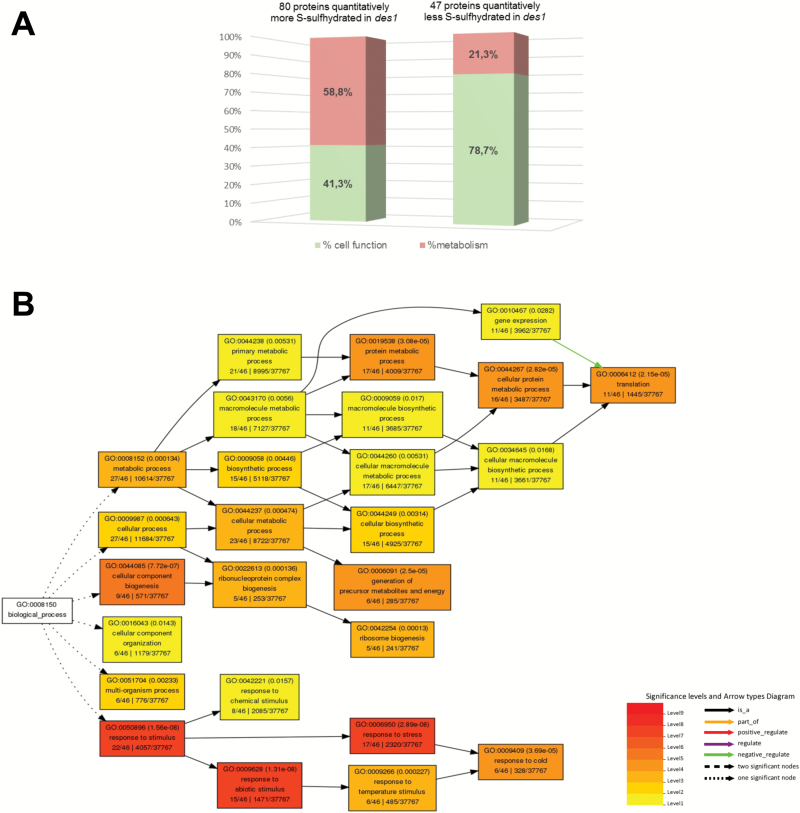
TMTsixplex identified 1937 proteins based on 41213 unique peptides with a FDR≤1% (Supplementary Dataset S3). The quantification of proteins found in the des1 samples revealed a significantly different representation of 127 persulfidated proteins compared with wild-type, with 80 proteins being more abundant and 47 proteins being less abundant, with a q-value≤0.05 (Supplementary Table S7 and Dataset S3). MapMan ontology indicated the involvement of 58.8% of these over-represented proteins in primary metabolism routes, such as the tricarboxylic acid cycle, glycolysis, and the Calvin cycle. However, 78.7% of the under-represented proteins in des1 also took part in different cell functions, such as protein synthesis or degradation, biotic and abiotic stress responses, the redox response, and calcium signaling, while only 21.3% of these proteins participated in primary metabolism pathways (Fig. 4A). A singular enrichment analysis of gene ontology was performed using the agriGO web-based toolkit to analyze the 47 proteins associated with higher levels of persulfidation in wild-type plants than in the des1 mutant. A large number of these proteins were identified in translation, 11 proteins out of 46 (GO:0006412), and response to stress, 17 proteins out of 46 (GO:0006950) (Fig. 4B).
Fig. 4.
Functional characterization of proteins quantitatively regulated by persulfidation. (A) Functional distribution of proteins differentially regulated by persulfidation in des1 and wild-type plants. (B) Singular enrichment analysis performed with AgriGO to identify enriched gene ontologies associated with proteins negatively regulated by persulfidation in des1 mutants in comparison with persulfidation patterns in wild-type plants. Box colors indicate levels of statistical significance: yellow, 0.05; orange, e−5; red, e−9.
Comparison analysis of persulfidation and nitrosylation proteomes
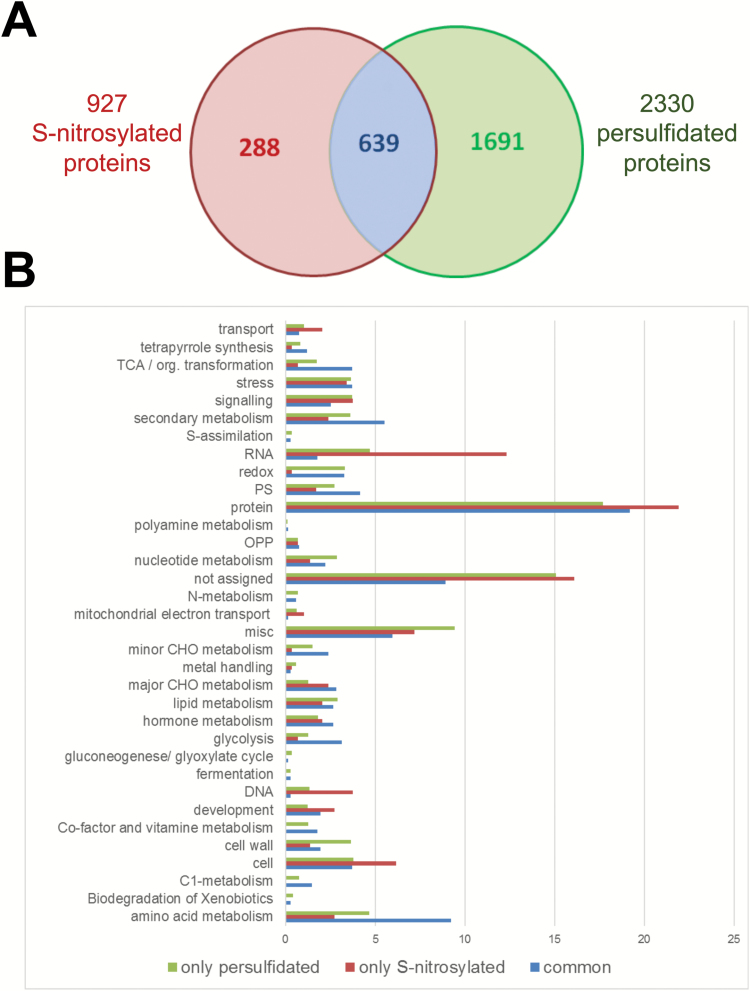
Although the sulfhydryl groups in protein Cys residues undergo an array of oxidative reactions and modifications, many studies have established NO-dependent S-nitrosylation as fundamental for the regulation of diverse protein activities. Since protein persulfidation and nitrosylation are similar in terms of their chemical and biological determinants, we have compared our persulfidation results with the S-nitrosylation proteome analysis previously reported. The latest and largest nitrosylation proteome of Arabidopsis has been reported by the Zuo group (Hu et al., 2015), identifying 927 endogenously S-nitrosylated proteins in the gsnor1-3 mutant that show increased levels of NO compared with wild-type. A comparison of the persulfidated and nitrosylated Arabidopsis proteomes shows that 639 proteins are susceptible to being both persulfidated and nitrosylated (Fig. 5A and Supplementary Table S8). Among others, the cytosolic glyceraldehyde-3-phosphate dehydrogenase (GAPC1) may be nitrosylated at residue Cys156 (Vescovi et al., 2013) and persulfidated at residue Cys160 (Aroca et al., 2017), and the functional consequences differ between modifications; persulfidation determines its nuclear localization and nitrosylation does not. However, the same GAPDH protein sites reported to undergo nitrosylation have also been found to undergo persulfidation in mammalian tissue (Hara et al., 2005; Mustafa et al., 2009).
Fig. 5.
Comparison of S-nitrosylated proteins and persulfidated proteins identified in Arabidopsis plants. (A) Venn diagram of total S-nitrosylated identified proteins in wild-type and gsnor1-3 mutant (Hu et al., 2015) and of total persulfidated identified proteins in wild-type and des1 mutant in this work. (B) Functional classification of gene ontology (GO) terms categorized by biological processes.
Functional classification of the GO terms categorized by biological processes shows that proteins that are only nitrosylated are predominantly enriched in the RNA and DNA categories despite the number of identified proteins being significantly lower (Fig. 5B).
Discussion
Protein persulfidation of cysteine residues is an important mechanism involved in diverse biological processes
Many physiological and pathophysiological processes are mediated by signaling pathways involving H2S in mammalian and plant systems (Gotor et al., 2013; Romero et al., 2014; Jin and Pei, 2015; Kimura, 2015; Paul and Snyder, 2015a). The molecular mechanism by which H2S exerts its action has been identified and involves the defined oxidative post-translational modification of cysteine residues to form a persulfidated thiol motif, a process called protein persulfidation (Mustafa et al., 2009; Mishanina et al., 2015; Paul and Snyder, 2015c). One of the major challenges in the last few years has been the development of detection methods specific to protein persulfidation because one critical step is to differentiate cysteine persulfide residues from cysteine thiols. The tag-switch method, which directly labels proteins with persulfidated residues by forming stable thioethers, has allowed us to significantly increase the number of identified proteins that undergo this modification in Arabidopsis leaf extracts from 106, identified using the modified biotin-switch method (Aroca et al., 2015), to more than 2000 proteins present in all the three biological replicates in this work. In the traditional method, the SH-blocking reagent MMTS does not adequately distinguish between cysteine persulfide (-SSH) and cysteine thiols (-SH) as reported (Mustafa et al., 2009) because both demonstrate similar reactivity (Pan and Carroll, 2013). The reaction with biotin-HPDP to form biotin-labeled proteins is therefore under-represented. The tag-switch method detected almost 4000 proteins in samples extracted from mature leaves; however, for further protein analysis, we took a more restrictive approach in which we considered only those proteins detected in all three biological samples to be validated. This method was previously developed in mammalian systems to investigate recombinant proteins, such as bovine serum albumin (BSA) and glyceraldehyde-3-phophate dehydrogenase (GAPDH), followed by testing to detect intracellular protein persulfides in cell extracts, protein immunoprecipitation, and fluorescence microscopy (Zhang et al., 2014; Park et al., 2015; Wedmann et al., 2016). We demonstrated the specificity of the tag-switch method on Arabidopsis protein extracts by treating the protein samples with DTT to chemically reduce cysteine persulfide residues to thiols prior to the procedure. Under these conditions, we were not able to detect labeled proteins by protein blotting or even by LC-MS/MS after streptavidin-bead purification.
The 2015 persulfidated proteins identified in wild-type plants represent at least 5% of the entire proteome of 35386 proteins encoded by the A. thaliana genome. However, this number may be even higher because although the tag-switch method involves enrichment for protein identification, very low abundance proteins may be lost during the procedure. In addition, we have used the stringent criterion of being identified in all three replicates but we cannot exclude that those proteins identified in one or two replicates are also persulfidated at a lower abundance and thus went undetected. Our data suggest the widespread distribution of this post-translational modification in the plant proteome and that persulfidation may have the same level of impact as phosphorylation or glycosylation modifications. Moreover, our proteomics analysis was performed on plants grown under physiological conditions and the magnitude of this modification may be even higher under conditions where sulfide has been demonstrated to play a signaling role, such as plant responses to a variety of plant stresses (Zhang et al., 2008; Wang et al., 2010; Li et al., 2012b; Shen et al., 2013; Shi et al., 2013), autophagy (Alvarez et al., 2012b; Gotor et al., 2013; Romero et al., 2014; Laureano-Marin et al., 2016), stomatal movement (Garcia-Mata and Lamattina, 2010; Lisjak et al., 2010; Jin et al., 2013; Scuffi et al., 2014; Papanatsiou et al., 2015) and photosynthesis (Chen et al., 2011). Another factor that we are unable to exclude is changes in persulfidation patterns under specific conditions; thus, performing the described method on protein extracts from Arabidopsis plants subjected to any of these specific conditions is of major interest.
The GO classification of proteins with persulfidation-modified cysteine residues indicates their involvement in a large range of biological processes. The majority of the identified proteins are located in the cytosol (46%) and the chloroplast (25%), which are compartments with sulfide concentrations calculated to be approximately 55 µM and 125 µM, respectively, in Arabidopsis (Krueger et al., 2009). It has been estimated that chloroplasts contain between 2000 and 3500 proteins, approximately 10% of all predicted protein-encoded genes (van Wijk and Baginsky, 2011); thus, protein persulfidation may be highly significant in this compartment. Indeed, hydrogen sulfide has been described to enhance photosynthesis in spinach seedlings by, among other effects, increasing the activity of the ribulose-1,5-bisphosphate carboxylase (Rubisco) enzyme (Chen et al., 2011). One of the largest groups identified in this work consists of proteins involved in carbon assimilation, energy metabolism, and glycolytic flux. Several enzymes from the Calvin-Benson cycle are also present in our analysis, such as the Rubisco large and small subunits, the Rubisco activase, the chloroplastic glyceraldehyde-3-phosphate dehydrogenase, phosphoribulokinase (PRK), fructose-1,6-bisphosphatase (FBPase), sedoheptulose-1,7-bisphosphatase (SBPase), and transketolase. Regulation of Calvin–Benson cycle enzymes based on redox post-translational modifications was recognized very early in plants and involves different states of cysteine residue oxidation (Buchanan and Balmer, 2005), now also including persulfidation. These enzymes exhibit low activity in the dark, with oxidized cysteine residues forming disulfides, and are activated in the light, with reduced cysteine residues. All of these enzymes are regulated by the ferredoxin/thioredoxin (Fd/TRX) system, which plays a crucial role in redox- and light-dependent reactions in chloroplasts through the reduction of regulatory disulfides (Buchanan and Balmer, 2005).). Clearly, there is a substantial interconnection between photosynthesis and the source of hydrogen sulfide in chloroplasts because the primary sulfate assimilation pathway in photosynthetic organisms occurs in the chloroplast, where sulfate ions are reduced to sulfide in the light (Schmidt and Trebst, 1969). Sulfide per se is not the sulfurating species but rather polysulfides generated via the mild oxidation of sulfide (Kimura, 2015; Mishanina et al., 2015). Post-translational modification via persulfidation may be an intermediate regulatory step for the enzymes of the Calvin–Benson cycle between inhibition in the dark, via the oxidation of cysteines to disulfides, and activated enzyme activity under illumination via reduction to thiol. The persulfidation of cysteine residues is reverted in vitro by thioredoxins and glutaredoxin/glutathione reductase/glutathione systems as demonstrated for persulfidated mammalian PROTEIN TYROSINE PHOSPHATASE 1B (PTP1B) and BSA (Krishnan et al., 2011; Doka et al., 2016; Wedmann et al., 2016). Persulfidated proteins in the chloroplast could therefore also be reduced and regulated by the chloroplastic thioredoxin machinery.
Sulfate and nitrate assimilation are dependent upon and interact with carbon assimilation because they are regulated by the supply of carbon skeletons produced during CO2 assimilation (Oaks, 1994; Kopriva et al., 2002). Furthermore, the regulatory interaction between the sulfate and nitrate assimilation pathways is well established as a deficiency in one element represses the other pathway (Takahashi et al., 2011). Levels of O-acetylserine, the carbon-nitrogen molecule into which sulfide is incorporated to form cysteine, connect these pathways. Curiously, the major enzymes involved in nitrogen assimilation are susceptible to persulfidation, such as nitrate reductase, NIA1 and NIA2, nitrite reductase, NIR1, glutamine synthetase, GS1, and ferredoxin-dependent glutamate synthase, GLU1(this work and Aroca et al., 2015). We previously characterized the regulation of glutamine synthetase by hydrogen sulfide via the inhibition of its activity; this process is reversed by reduction with dithiothreitol (Aroca et al., 2015). Enzyme regulation by persulfidation in the nitrogen-assimilation pathway therefore represents another step in the molecular mechanism underlying the coordination of both routes.
In addition to playing a regulatory role in primary carbon and nitrogen assimilation, H2S is also essential for the degradation and recycling of cellular components in plant cells induced by carbon or nitrogen starvation, regulating the mechanism of autophagy and acting as a repressor of this process under normal growth conditions and sufficient nutrient availability (Alvarez et al., 2012b; Laureano-Marin et al., 2016). In des1 mutants exhibiting a reduced cytosolic H2S repressor signal, autophagy is therefore induced under sufficient nutrient availability via an unknown signaling mechanism. Proteomics analysis performed on wild-type samples revealed the susceptibility of the autophagy (ATG)-related proteins, ATG18a, ATG3, ATG5, and ATG7, to persulfidation, although the three last proteins were identified in only one replicate. In the proteomics analysis performed in des1 mutants, we detected ATG18a, but no other ATG members, in the full protein set, suggesting that persulfidation may be the molecular mechanism through which sulfide regulates autophagy in plant cells. However, the identification of autophagy-related molecular protein targets requires further specific studies.
Similar to the results obtained in this work, proteomics analysis of pancreatic ß cells recently revealed a subset of proteins with induced persulfidation involved in aminoacyl-tRNA biosynthesis (Gao et al., 2015). Nearly half of the proteins involved in the synthesis of aminoacyl-tRNA have been identified as persulfidated in wild-type Arabidopsis samples. Efficient protein translation depends on the availability of aminoacyl-tRNA and the ratio of correctly acylated to misacylated tRNA. Although there is little information regarding the redox regulation of these enzymes, the sulfenylation of cysteine residues in aminoacyl-tRNA synthases in Escherichia coli during oxidative stress increases misacylation and leads to error-prone translation (Ling and Soll, 2010). As hydropersulfides do not form in a direct reaction between H2S and protein –SH, the persulfidation reaction occurs when one of the cysteine residues in the target protein, found in the oxidized form, is subsequently modified to a persulfide moiety. Additional control of translation by thiolation was reported in yeast, in which intracellular methionine and cysteine availability directly control the thiolation status of wobble-uridine (U34) nucleotides present on lysine, glutamine, and glutamate tRNAs to regulate cellular translational capacity and metabolic homeostasis (Laxman et al., 2013). Cellular sulfur content therefore plays a fundamental role in the translational machinery of cells.
Sulfide generated by DES1 has specific regulatory roles
A quantitative comparison between wild-type and des1 plants revealed small but significant differences in protein persulfidation profiles. Although the majority of proteins carrying persulfidated residues are the same in wild-type and des1 plants, some of these proteins slightly but significantly change their persulfidation levels as determined by quantitative proteomics analysis. In addition, wild-type and des1 plants also demonstrated specific persulfidated protein targets under our experimental conditions. Although the des1 mutant is defective for the cytosolic H2S-generating enzyme DES1, DES1 is not the only cysteine-dependent sulfide producer present in the cell and therefore des1 mutants retain the capacity to generate sulfide via both photosynthetic sulfate reduction and the actions of other cysteine desulfhydrase enzymes. The des1 mutant demonstrates two clearly distinct characteristics compared with wild-type plants that affect protein persulfidation in the mutant plants. First, the des1 mutant contains 30% less sulfur under steady-state growth conditions, primarily in the cytosol; second, it contains higher cysteine levels that may locally promote the production of oxidative species by driving the Fenton reaction (Park and Imlay, 2003; Alvarez et al., 2010).
Several proteins whose persulfidation levels are altered in des1 plants are involved in intracellular signaling processes, including many protein kinases, phosphatases, defense response molecules, and components of the proteasome complex. Among the kinase targets, we detected SNF1-RELATED PROTEIN KINASE 2.2 (SNRK2.2) and 2.6 (OST1), which play essential roles in the abscisic acid (ABA)-dependent regulation of stomatal movement. In addition, we also detected persulfidation of the abscisic acid receptors PYRABACTIN RESISTANCE 1 (PYR1) and PYR1-LIKE PROTEIN 1 (PYL1) in both in wild-type and des1 samples (Miyakawa et al., 2013). The participation of H2S in stomatal closure has been widely reported (Garcia-Mata and Lamattina, 2010; Papanatsiou et al., 2015) and ABA does not induce stomatal closure in isolated epidermal strips of des1 mutants; however, this effect is restored via the application of exogenous H2S. Furthermore, H2S acts upstream of NO to regulate stomatal closure because NO depletion blocks H2S-dependent effects (Scuffi et al., 2014). ABA binds to PYR/PYL/RCAR receptors and this complex promotes the phosphorylation and activation of SNRK2, which phosphorylates numerous downstream protein targets involved in the ABA response. OST1/SNRK2 is S-nitrosylated by NO at Cys137, a residue adjacent to the kinase catalytic site, provoking the inhibition of its phosphorylating activity (Wang et al., 2015). As persulfidation and nitrosylation influence protein function in an opposing manner, similar to ascorbate peroxidase and GAPDH (Aroca et al., 2015), OST1 is a candidate protein to modulate the cross-talk of these two gasotransmitters.
Comparison of the persulfidated proteome with a reported nitrosylated proteome shows that many of the identified proteins are susceptible to both modifications. It is interesting that in wild-type plants, with a significantly lower level of NO content, the differences in protein nitrosylation compared with the gsnor1-3 mutant were not important, like in wild-type and des1 plants at the level of persulfidation. These observations suggest that the production of intracellular NO and H2S is not enough to induce protein nitrosylation or persulfidation, respectively; therefore, additional events are necessary to generate and regulate protein modifications.
In conclusion, the tag-switch method, which is more specific and selective than the methods previously published, has allowed us to significantly increase the number of identified proteins that may be susceptible to persulfidation. Functional analysis of these proteins reveals the impact that regulation by hydrogen sulfide can exert on metabolism and cellular regulation in plants. The fact that a high number of proteins are persulfidated in both wild-type and des1 mutant lines indicates that the basal level of persulfidation is high under normal growth conditions and that in addition to DES1, there are other enzymes or processes that can generate sulfurating species as they exist in mammalian systems. Further proteomic analyses under conditions of biotic or abiotic environmental stress and at different stages of development will shed more light on the regulatory role of sulfide in plant cells.
Supplementary Data
Supplementary data are available at JXB online.
Fig. S1. Validation of the tag-switch method in wild-type and des1 mutant Arabidopsis leaf extracts by immunoblotting with anti-biotin antibodies.
Fig. S2. Enriched Gene Ontology analysis of the 2015 loci corresponding to the persulfidated proteins identified by the tag-switch method in wild-type plants.
Fig. S3. Functional classification of persulfidated proteins identified by LC-MS/MS in the leaf extracts of 30-day-old Arabidopsis thaliana des1 mutant plants.
Table S1. Ontology classification of persulfidated proteins in wild-type samples according to MapMan.
Table S2. Singular enrichment analysis of Gene Ontology (GO) terms related to the category “glycolysis”.
Table S3. Singular enrichment analysis of Gene Ontology (GO) terms related to the category “tRNA aminoacylation for protein translation”.
Table S4. Singular enrichment analysis of Gene Ontology (GO) terms related to the category “jasmonic acid biosynthesis”.
Table S5. Singular enrichment analysis of Gene Ontology (GO) terms related to the category “abiotic stress responses”.
Table S6. Proteins differentially labeled persulfidated in wild-type and des1 samples.
Table S7. Quantitative comparison of persulfidation patterns in wild-type and des1 plants.
Table S8. Commonly identified proteins in the persulfidation and nitrosylation proteome analysis.
Dataset S1. List of identified proteins in wild-type (WT) samples (FDR<1%).
Dataset S2. List of identified proteins in des1 samples (FDR<1%).
Dataset S3. Quantitative comparison of persulfidation patterns in wild-type and des1 plants (FDR<1%).
Data Deposition
Mass spectrometry data. PRIDE Archive, ProteomeXchange Consortium. Dataset identifiers PXD005168, 10.6019/PXD005168, and PXD006140.
Supplementary Material
Acknowledgements
This work was supported in part by the European Regional Development Fund through the Ministerio de Economia y Competitividad (grant no. BIO2013-44648-P) and the Agencia Estatal de Investigación (grant no. BIO2016-76633-P).
Glossary
Abbreviations:
- ABA
abscisic acid
- DES1
L-CYSTEINE DESULFHYDRASE 1
- FDR
false discovery rate
- MMTS
S-methyl-methanothiosulfonate
- MSBT
methylsulfonylbenzothiazole.
References
- Álvarez C, Bermúdez MÁ, Romero LC, Gotor C, García I. 2012a. Cysteine homeostasis plays an essential role in plant immunity. New Phytologist 193, 165–177. [DOI] [PubMed] [Google Scholar]
- Alvarez C, Calo L, Romero LC, García I, Gotor C. 2010. An O-acetylserine(thiol)lyase homolog with L-cysteine desulfhydrase activity regulates cysteine homeostasis in Arabidopsis. Plant Physiology 152, 656–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez C, García I, Moreno I, Pérez-Pérez ME, Crespo JL, Romero LC, Gotor C. 2012b. Cysteine-generated sulfide in the cytosol negatively regulates autophagy and modulates the transcriptional profile in Arabidopsis. The Plant Cell 24, 4621–4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez C, García I, Romero LC, Gotor C. 2012c. Mitochondrial sulfide detoxification requires a functional isoform O-acetylserine(thiol)lyase C in Arabidopsis thaliana. Molecular Plant 5, 1217–1226. [DOI] [PubMed] [Google Scholar]
- Aroca A, Benito JM, Gotor C, Romero LC. PRIDE Archive, ProteomeXchange Consortium; 2017. https://www.ebi.ac.uk/pride/archive/projects/PXD005168 PXD006140https://www.ebi.ac.uk/pride/archive/projects/ Data from: Persulfidation proteome reveals the regulation of protein function by hydrogen sulfide in diverse biological processes in Arabidopsis. [Google Scholar]
- Aroca A, Schneider M, Scheibe R, Gotor C, Romero LC. 2017. Hydrogen sulfide regulates the cytosolic/nuclear partitioning of glyceraldehyde-3-phosphate dehydrogenase by enhancing its nuclear localization. Plant & Cell Physiology 58, 983–992. [DOI] [PubMed] [Google Scholar]
- Aroca Á, Serna A, Gotor C, Romero LC. 2015. S-sulfhydration: a cysteine posttranslational modification in plant systems. Plant Physiology 168, 334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermúdez MÁ, Galmés J, Moreno I, Mullineaux PM, Gotor C, Romero LC. 2012. Photosynthetic adaptation to length of day is dependent on S-sulfocysteine synthase activity in the thylakoid lumen. Plant Physiology 160, 274–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan BB, Balmer Y. 2005. Redox regulation: a broadening horizon. Annual Review of Plant Biology 56, 187–220. [DOI] [PubMed] [Google Scholar]
- Chen J, Wu FH, Wang WH, Zheng CJ, Lin GH, Dong XJ, He JX, Pei ZM, Zheng HL. 2011. Hydrogen sulphide enhances photosynthesis through promoting chloroplast biogenesis, photosynthetic enzyme expression, and thiol redox modification in Spinacia oleracea seedlings. Journal of Experimental Botany 62, 4481–4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevasanta E, Lange M, Bonanata J, Coitiño EL, Ferrer-Sueta G, Filipovic MR, Alvarez B. 2015. Reaction of Hydrogen Sulfide with Disulfide and Sulfenic Acid to Form the Strongly Nucleophilic Persulfide. The Journal of Biological Chemistry 290, 26866–26880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dóka É, Pader I, Bíró A et al. . 2016. A novel persulfide detection method reveals protein persulfide- and polysulfide-reducing functions of thioredoxin and glutathione systems. Science Advances 2, e1500968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. 2010. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Research 38, W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H, Liu Z, Jin Z, Zhang L, Liu D, Pei Y. 2016. An emphasis of hydrogen sulfide-cysteine cycle on enhancing the tolerance to chromium stress in Arabidopsis. Environmental Pollution 213, 870–877. [DOI] [PubMed] [Google Scholar]
- Filipovic MR, Miljkovic JLj, Nauser T, Royzen M, Klos K, Shubina T, Koppenol WH, Lippard SJ, Ivanović-Burmazović I. 2012. Chemical characterization of the smallest S-nitrosothiol, HSNO; cellular cross-talk of H2S and S-nitrosothiols. Journal of the American Chemical Society 134, 12016–12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XH, Krokowski D, Guan BJ et al. . 2015. Quantitative H2S-mediated protein sulfhydration reveals metabolic reprogramming during the integrated stress response. Elife 4, e10067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia I, Gotor C, Romero LC. 2015. Cysteine homeostasis. In: D’Mello JPF, ed. Amino Acids in Higher Plants. Wallingford, United Kingdom: CABI Publishing, 219–233. [Google Scholar]
- García-Mata C, Lamattina L. 2010. Hydrogen sulphide, a novel gasotransmitter involved in guard cell signalling. New Phytologist 188, 977–984. [DOI] [PubMed] [Google Scholar]
- Giavalisco P, Nordhoff E, Lehrach H, Gobom J, Klose J. 2003. Extraction of proteins from plant tissues for two-dimensional electrophoresis analysis. Electrophoresis 24, 207–216. [DOI] [PubMed] [Google Scholar]
- Gotor C, Alvarez C, Bermúdez MA, Moreno I, García I, Romero LC. 2010. Low abundance does not mean less importance in cysteine metabolism. Plant Signaling & Behavior 5, 1028–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotor C, García I, Crespo JL, Romero LC. 2013. Sulfide as a signaling molecule in autophagy. Autophagy 9, 609–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotor C, Laureano-Marín AM, Moreno I, Aroca Á, García I, Romero LC. 2015. Signaling in the plant cytosol: cysteine or sulfide?Amino Acids 47, 2155–2164. [DOI] [PubMed] [Google Scholar]
- Hara MR, Agrawal N, Kim SF et al. . 2005. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nature Cell Biology 7, 665–674. [DOI] [PubMed] [Google Scholar]
- Hu J, Huang X, Chen L, Sun X, Lu C, Zhang L, Wang Y, Zuo J. 2015. Site-specific nitrosoproteomic identification of endogenously S-nitrosylated proteins in Arabidopsis. Plant Physiology 167, 1731–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ida T, Sawa T, Ihara H et al. . 2014. Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proceedings of the National Academy of Sciences USA 111, 7606–7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Pei Y. 2015. Physiological implications of hydrogen sulfide in plants: pleasant exploration behind its unpleasant odour. Oxidative Medicine and Cellular Longevity 2015, 397502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Xue S, Luo Y, Tian B, Fang H, Li H, Pei Y. 2013. Hydrogen sulfide interacting with abscisic acid in stomatal regulation responses to drought stress in Arabidopsis. Plant Physiology and Biochemistry 62, 41–46. [DOI] [PubMed] [Google Scholar]
- Kimura H. 2014. The physiological role of hydrogen sulfide and beyond. Nitric Oxide: Biology and Chemistry 41, 4–10. [DOI] [PubMed] [Google Scholar]
- Kimura H. 2015. Signaling of hydrogen sulfide and polysulfides. Antioxidants & Redox Signaling 22, 347–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klie S, Nikoloski Z. 2012. The choice between mapman and gene ontology for automated gene function prediction in plant science. Frontiers in Genetics 3, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva S, Suter M, von Ballmoos P, Hesse H, Krähenbühl U, Rennenberg H, Brunold C. 2002. Interaction of sulfate assimilation with carbon and nitrogen metabolism in Lemna minor. Plant Physiology 130, 1406–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan N, Fu C, Pappin DJ, Tonks NK. 2011. H2S-Induced sulfhydration of the phosphatase PTP1B and its role in the endoplasmic reticulum stress response. Science Signaling 4, ra86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger S, Niehl A, Lopez Martin MC et al. . 2009. Analysis of cytosolic and plastidic serine acetyltransferase mutants and subcellular metabolite distributions suggests interplay of the cellular compartments for cysteine biosynthesis in Arabidopsis. Plant, Cell & Environment 32, 349–367. [DOI] [PubMed] [Google Scholar]
- Laureano-Marín AM, Moreno I, Romero LC, Gotor C. 2016. Negative regulation of autophagy by sulfide is independent of reactive oxygen species. Plant Physiology 171, 1378–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxman S, Sutter BM, Wu X, Kumar S, Guo X, Trudgian DC, Mirzaei H, Tu BP. 2013. Sulfur amino acids regulate translational capacity and metabolic homeostasis through modulation of tRNA thiolation. Cell 154, 416–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wang Y, Shen W. 2012. Roles of hydrogen sulfide and nitric oxide in the alleviation of cadmium-induced oxidative damage in alfalfa seedling roots. BioMetals 25, 617–631. [DOI] [PubMed] [Google Scholar]
- Li ZG, Gong M, Xie H, Yang L, Li J. 2012. Hydrogen sulfide donor sodium hydrosulfide-induced heat tolerance in tobacco (Nicotiana tabacum L) suspension cultured cells and involvement of Ca(2+) and calmodulin. Plant Science 185-186, 185–189. [DOI] [PubMed] [Google Scholar]
- Ling J, Soll D. 2010. Severe oxidative stress induces protein mistranslation through impairment of an aminoacyl-tRNA synthetase editing site. Proceedings of the National Academy of Sciences USA 107, 4028–4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisjak M, Srivastava N, Teklic T, Civale L, Lewandowski K, Wilson I, Wood ME, Whiteman M, Hancock JT. 2010. A novel hydrogen sulfide donor causes stomatal opening and reduces nitric oxide accumulation. Plant Physiology and Biochemistry 48, 931–935. [DOI] [PubMed] [Google Scholar]
- Longen S, Richter F, Köhler Y, Wittig I, Beck KF, Pfeilschifter J. 2016. Quantitative Persulfide Site Identification (qPerS-SID) reveals protein targets of h2s releasing donors in mammalian cells. Scientific Reports 6, 29808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Serra P, Marcilla M, Villanueva A et al. . 2014. A DERL3-associated defect in the degradation of SLC2A1 mediates the Warburg effect. Nature Communications 5, 3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishanina TV, Libiad M, Banerjee R. 2015. Biogenesis of reactive sulfur species for signaling by hydrogen sulfide oxidation pathways. Nature Chemical Biology 11, 457–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa T, Fujita Y, Yamaguchi-Shinozaki K, Tanokura M. 2013. Structure and function of abscisic acid receptors. Trends in Plant Science 18, 259–266. [DOI] [PubMed] [Google Scholar]
- Mustafa AK, Gadalla MM, Sen N, Kim S, Mu W, Gazi SK, Barrow RK, Yang G, Wang R, Snyder SH. 2009. H2S signals through protein S-sulfhydration. Science Signaling 2, ra72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oaks A. 1994. Efficiency of nitrogen utilization in C3 and C4 cereals. Plant Physiology 106, 407–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olas B. 2015. Hydrogen sulfide in signaling pathways. Clinica Chimica Acta 439, 212–218. [DOI] [PubMed] [Google Scholar]
- Pan J, Carroll KS. 2013. Persulfide reactivity in the detection of protein s-sulfhydration. ACS Chemical Biology 8, 1110–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanatsiou M, Scuffi D, Blatt MR, García-Mata C. 2015. Hydrogen sulfide regulates inward-rectifying K+ channels in conjunction with stomatal closure. Plant Physiology 168, 29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CM, Macinkovic I, Filipovic MR, Xian M. 2015. Use of the “tag-switch” method for the detection of protein S-sulfhydration. Methods in Enzymology 555, 39–56. [DOI] [PubMed] [Google Scholar]
- Park S, Imlay JA. 2003. High levels of intracellular cysteine promote oxidative DNA damage by driving the fenton reaction. Journal of Bacteriology 185, 1942–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul BD, Snyder SH. 2012. H₂S signalling through protein sulfhydration and beyond. Nature Reviews. Molecular Cell Biology 13, 499–507. [DOI] [PubMed] [Google Scholar]
- Paul BD, Snyder SH. 2015a. H2S: A novel gasotransmitter that signals by sulfhydration. Trends in Biochemical Sciences 40, 687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul BD, Snyder SH. 2015b. Modes of physiologic H2S signaling in the brain and peripheral tissues. Antioxidants & Redox Signaling 22, 411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul BD, Snyder SH. 2015c. Protein sulfhydration. Methods in Enzymology 555, 79–90. [DOI] [PubMed] [Google Scholar]
- Ramos-Fernández A, Paradela A, Navajas R, Albar JP. 2008. Generalized method for probability-based peptide and protein identification from tandem mass spectrometry data and sequence database searching. Molecular & Cellular Proteomics 7, 1748–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemenschneider A, Riedel K, Hoefgen R, Papenbrock J, Hesse H. 2005. Impact of reduced O-acetylserine(thiol)lyase isoform contents on potato plant metabolism. Plant Physiology 137, 892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero LC, Aroca MÁ, Laureano-Marín AM, Moreno I, García I, Gotor C. 2014. Cysteine and cysteine-related signaling pathways in Arabidopsis thaliana. Molecular Plant 7, 264–276. [DOI] [PubMed] [Google Scholar]
- Romero LC, Aroca MÁ, Serna A, Gotor C. 2013a. Proteomic analysis of endogenous S-sulfhydration in Arabidopsis thaliana. Nitric Oxide 31, S23. [Google Scholar]
- Romero LC, García I, Gotor C. 2013b. L-Cysteine Desulfhydrase 1 modulates the generation of the signaling molecule sulfide in plant cytosol. Plant Signaling & Behavior 8, e24007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak JN, Scheurer SB, Neri D, Elia G. 2004. Purification of biotinylated proteins on streptavidin resin: a protocol for quantitative elution. Proteomics 4, 2296–2299. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Trebst A. 1969. The mechanism of photosynthetic sulfate reduction by isolated chloroplasts. Biochimica et Biophysica Acta 180, 529–535. [DOI] [PubMed] [Google Scholar]
- Scuffi D, Álvarez C, Laspina N, Gotor C, Lamattina L, García-Mata C. 2014. Hydrogen sulfide generated by L-cysteine desulfhydrase acts upstream of nitric oxide to modulate abscisic acid-dependent stomatal closure. Plant Physiology 166, 2065–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Xing T, Yuan H, Liu Z, Jin Z, Zhang L, Pei Y. 2013. Hydrogen sulfide improves drought tolerance in Arabidopsis thaliana by microRNA expressions. PLoS ONE 8, e77047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Ye T, Chan Z. 2013. Exogenous application of hydrogen sulfide donor sodium hydrosulfide enhanced multiple abiotic stress tolerance in bermudagrass (Cynodon dactylon (L). Pers.). Plant Physiology and Biochemistry 71, 226–234. [DOI] [PubMed] [Google Scholar]
- Sun J, Wang R, Zhang X, Yu Y, Zhao R, Li Z, Chen S. 2013. Hydrogen sulfide alleviates cadmium toxicity through regulations of cadmium transport across the plasma and vacuolar membranes in Populus euphratica cells. Plant Physiology and Biochemistry 65, 67–74. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Kopriva S, Giordano M, Saito K, Hell R. 2011. Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annual Review of Plant Biology 62, 157–184. [DOI] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y et al. . 2004. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. The Plant Journal 37, 914–939. [DOI] [PubMed] [Google Scholar]
- Van Hoewyk D, Pilon M, Pilon-Smits EAH. 2008. The functions of NifS-like proteins in plant sulfur and selenium metabolism. Plant Science 174, 117–123. [Google Scholar]
- van Wijk KJ, Baginsky S. 2011. Plastid proteomics in higher plants: current state and future goals. Plant Physiology 155, 1578–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandiver M, Snyder S. 2012. Hydrogen sulfide: a gasotransmitter of clinical relevance. Journal of Molecular Medicine 90, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vescovi M, Zaffagnini M, Festa M, Trost P, Lo Schiavo F, Costa A. 2013. Nuclear accumulation of cytosolic glyceraldehyde-3-phosphate dehydrogenase in cadmium-stressed Arabidopsis roots. Plant Physiology 162, 333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno JA, Csordas A, del-Toro N et al. . 2016. 2016 update of the PRIDE database and its related tools. Nucleic Acids Research 44, D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BL, Shi L, Li YX, Zhang WH. 2010. Boron toxicity is alleviated by hydrogen sulfide in cucumber (Cucumis sativus L.) seedlings. Planta 231, 1301–1309. [DOI] [PubMed] [Google Scholar]
- Wang P, Du Y, Hou YJ, Zhao Y, Hsu CC, Yuan F, Zhu X, Tao WA, Song CP, Zhu JK. 2015. Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1. Proceedings of the National Academy of Sciences USA 112, 613–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. 2014. Gasotransmitters: growing pains and joys. Trends in Biochemical Sciences 39, 227–232. [DOI] [PubMed] [Google Scholar]
- Wedmann R, Onderka C, Wei S et al. . 2016. Improved tag-switch method reveals that thioredoxin acts as depersulfidase and controls the intracellular levels of protein persulfidation. Chemical Science 7, 3414–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Nakamura T, Kusano T, Sano H. 2000. Three Arabidopsis genes encoding proteins with differential activities for cysteine synthase and beta-cyanoalanine synthase. Plant & Cell Physiology 41, 465–476. [DOI] [PubMed] [Google Scholar]
- Zhang D, Macinkovic I, Devarie-Baez NO, Pan J, Park CM, Carroll KS, Filipovic MR, Xian M. 2014. Detection of protein S-sulfhydration by a tag-switch technique. Angewandte Chemie 53, 575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Hu LY, Hu KD, He YD, Wang SH, Luo JP. 2008. Hydrogen sulfide promotes wheat seed germination and alleviates oxidative damage against copper stress. Journal of Integrative Plant Biology 50, 1518–1529. [DOI] [PubMed] [Google Scholar]
- Zhang H, Tan ZQ, Hu LY, Wang SH, Luo JP, Jones RL. 2010. Hydrogen sulfide alleviates aluminum toxicity in germinating wheat seedlings. Journal of Integrative Plant Biology 52, 556–567. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.