Abstract
The endosymbiotic bacterium Wolbachia pipientis infects many species of insects and has been transinfected into the mosquito Aedes aegypti (L.), the primary vector of dengue virus (DENV). Recently, it has been shown that Wolbachia blocks the replication and transmission of RNA viruses, such as DENV, in a number of mosquito species including Ae. aegypti and Aedes albopictus (Skuse), which is naturally infected with Wolbachia and considered a secondary vector for DENV. The mosquito species Aedes notoscriptus (Skuse) is highly prevalent in Australia, including in areas where DENV outbreaks have been recorded. The mosquito has been implicated in the transmission of Ross River and Barmah Forest viruses, but not DENV. We investigated whether Wolbachia naturally infects this mosquito species and whether it has an impact on the ability of Ae. notoscriptus to transmit DENV. We show, for the first time, that Ae. notoscriptus is naturally infected with a strain of Wolbachia that belongs to supergroup B and is localized only in the ovaries. However, Wolbachia infection in Ae. notoscriptus did not induce resistance to DENV and had no effect on overall DENV infection rate or titer. The presence of a native Wolbachia in Ae. notoscriptus cannot explain why this mosquito is an ineffective vector of DENV.
Keywords: Wolbachia pipientis, dengue virus, Aedes notoscriptus, vector competence, tissue tropism
Wolbachia pipientis is an intracellular alpha-Proteobacterium that naturally infects numerous insect species, including mosquitoes ( Jeyaprakash and Hoy 2000 , Hilgenboecker et al. 2008 , Zug and Hammerstein 2012 ). Wolbachia infections can induce a number of different phenotypes in their hosts, including interfering with host reproduction to promote their own transmission ( Hurst et al. 1999 , Rousset et al. 1992 , Stouthamer et al. 1999 , O’Neill and Karr 1990 , Riegler et al. 2005 , Hoffmann et al. 2011 ). Recent studies have found that several Wolbachia strains can block either the replication or the pathogenicity of RNA viruses, including dengue viruses (DENVs), in insect hosts ( Hedges et al. 2008 , Teixeira et al. 2008 , Moreira et al. 2009 , Osborne et al. 2009 , Glaser and Meola 2010 , Bian et al. 2010 , Frentiu et al. 2010 , Walker et al. 2011 , Mousson et al. 2012 ). However, not all Wolbachia strains show antiviral effects. In their natural host, Drosophila simulans , neither the w Ha nor the w No strain shows viral protection against Drosophila C virus (DCV), whereas the transinfected strains w Mel, w MelPop, w Ri and w Au protect their fly hosts from infection ( Osborne et al. 2009 , 2012 ). The variation in protection among strains is thought to correlate with Wolbachia density and tissue distribution, with strains offering viral protection usually present at high densities in reproductive and somatic host tissues ( Lu et al. 2012 , Osborne et al. 2012 , Bian et al. 2013 , Micieli and Glaser 2014 ).
A number of mosquito species are naturally infected with Wolbachia ( Kittayapong et al. 2000 , Dobson et al. 2002 , Osei-Poku et al. 2012 ), but little is known about the role native Wolbachia strains play in pathogen interference in these hosts. Glaser and Meola (2010) showed increased rates of West Nile virus transmission and dissemination as well as amplified virus titers in Wolbachia -uninfected Culex quinquefasciatus compared with naturally Wolbachia -infected mosquitoes, although the magnitude of the effect was small. The mosquito Aedes albopictus (Skuse) is naturally infected with two strains of Wolbachia , w AlbA and w AlbB ( Sinkins et al. 1995 , Zhou et al. 1998 ), and is considered a secondary vector of DENV ( Lambrechts et al. 2010 ), whereas Aedes aegypti (L.), the primary DENV vector, is not naturally infected with Wolbachia. The native Wolbachia strains in Ae. albopictus have been demonstrated to interfere with the ability of the mosquito to transmit DENV by reducing viral infection in salivary glands ( Mousson et al. 2012 ).
The mosquito species Aedes notoscriptus (Skuse) is widespread across Australia and considered a major domestic pest. Aedesnotoscriptus breeds in both natural and domestic environments and readily feeds on humans, as well as flying foxes and brushtail possums ( Kay et al. 2007 ). A survey in the city of Brisbane (Queensland) recorded Ae. notoscriptus larvae as the most common species populating domestic habitats, including plant pots, rubbish, and discarded household items ( Kay et al. 2008 ). Despite being a competent vector of canine heartworm and implicated as a possible vector of Ross River and Barmah Forest viruses ( Watson and Kay 1998 ), Ae. notoscriptus has not been implicated in DENV transmission, and lab-based vector competence studies have demonstrated that it has low susceptibility to oral infection by all four serotypes of DENV ( Watson and Kay 1999 ). To elucidate if the low susceptibility of Ae. notoscriptus to DENV is owing to the presence of Wolbachia , we first determined whether the bacterium is present in the mosquito and its prevalence in natural Ae. notoscriptus populations. We then tested whether Wolbachia modulates the vector competence of Ae. notoscriptus for DENV.
Materials and Methods
Mosquito Rearing and Antibiotic Treatment
Eggs of an Ae. notoscriptus line (denoted Noto) were obtained from a laboratory colony established at The University of Queensland, originating from adult mosquitoes captured in Brisbane in 2007. Adult mosquitoes were kept in a controlled insectary at 25°C, approximately 65% RH, and a photoperiod of 12:12 (L:D) h and fed 10% sucrose solution ad libitum. Eggs were hatched under vacuum and larvae reared in plastic containers at a density of 150 larvae per 3 liters of distilled water. Larvae were fed fish food consisting of one half of a TetraMin (Tetra, Germany) tablet per day until pupation. Pupae were transferred into cages to emerge at a density of 250 adults per cage. Five-day-old adult females were blood-fed for oviposition with human blood from volunteers (The University of Queensland Medical Research Ethics Approval number 2007001379). Oviposition cups, containing scored brown paper and larval water, were placed in the cage 2 days after blood feeding. Eggs were collected at approximately 5 days post-feeding.
Ae des notoscriptus was cleared of Wolbachia infection by feeding adult mosquitoes for four generations on one of three antibiotic treatments: 1) 1 mg/ml tetracycline dissolved in 10% sucrose solution; 2) 1.25 g/liter rifampicin dissolved in 10% sucrose solution; or 3) alternating between both antibiotics. Mosquitoes produced from all three treatment groups were pooled to establish the Wolbachia -free line, denoted Noto.tet. Resident gut microflora was re-introduced into Noto.tet by adding 100 ml of larval water from Noto mosquitoes into the larval water for two generations post-antibiotic treatment ( McMeniman et al. 2009 ). Removal of Wolbachia was confirmed by polymerase chain reaction (PCR).
Wolbachia Prevalence in Laboratory and Field Ae. notoscriptus
Third-instar larvae of Ae. notoscriptus were captured from ovibuckets in four suburbs in Brisbane, Queensland, Australia. In total, 248 larvae were captured and stored at −80°C before DNA extraction. DNA was extracted from 5-day-old adult laboratory females and field-captured larvae using DNeasy® spin columns (Qiagen, Hilden, Germany) as per manufacturer’s protocol. Two microliters of sample was PCR-amplified in 4 µl 5xPhire Buffer (Finnzymes, Woburn, MA), 4 µl 1 mM dNTPs, 0.4 µl Phire Taq polymerase (Invitrogen, Carlsbad, CA), 2 µl each of 5 µM primers specific for Wolbachia surface protein ( wsp ; Braig et al. 1998 ), and insect ribosomal locus 28 S ( Table 1 ) in a total volume of 20 µl. Samples were denatured for 30 s at 98°C, cycled 35 times at 98°C for 5 s, 55°C for 5 s, and 72°C for 15 s, followed by a 1 min extension at 72°C. Wolbachia -infected Ae. aegypti (PGYP1 line) adults and their tetracycline-treated counterparts (PGYP1.tet; McMeniman et al. 2009 ) served as positive and negative controls for the Wolbachia PCR, respectively. PCR products were visualized with ethidium bromide using a 1.5% agarose gel.
Table 1.
Primer sequences used for Wolbachia w Noto PCR ( wsp , 28 S, and 16S), DENV-2 NS5 gene cDNA synthesis and qPCR, and 16S probe sequences for FISH of w Noto
| Target Gene | Primer/Probe Sequence (5′-3′) |
|---|---|
| wsp | 81F: TGG TCC AAT AAG TGA TGA AGA AAC |
| 691R: AAA AAT TAA ACG CTA ACT CCA | |
| 28S | FWD: TAC CGT GAG GGA AAG TTG AAA |
| REV: AGA CTC CTT GGT CCG TGT TT | |
| 16S | FWD: CAT ACC TAT TCG AAG GGA TAG |
| REV: AGC TTC GAG TGA AAC CAA TTC | |
| DENV-2 NS5 | FWD: ACA AGT CGA ACA ACCTGG TCC AT |
| REV: GCC GCA CCA TTG GTC TTC TC | |
| 16S probes | W2: CTT CTG TGA GTA CCG TCA TTA TC |
| W3: AAC CGA CCC TAT CCC TTC GAA TA |
Phylogenetic Grouping of Ae. notoscriptus Wolbachia
To determine the phylogenetic grouping of the Wolbachia strain infecting Ae. notoscriptus , denoted w Noto, DNA was extracted from female mosquitoes, PCR amplified as previously described, and purified using a MinElute kit (Qiagen). Twelve independent PCR amplicons underwent ligation into pGEM®T-Easy vector (Promega, Madison, WI), followed by transformation of the plasmid into E. coli overnight at 37°C. PCR was performed directly on bacterial colonies using T7 and M13R primers (T7: 5′-TAA TAC GAC TCA CTA TAG GG-3′; M13R: 5′-CAG GAA ACA GCT ATG AC-3′). Colonies positive for the plasmid underwent plasmid purification with a MiniPrep DNA kit (Qiagen) and subsequent sequencing at the Australian Genome Research Facility. We used the w Noto wsp sequence as a BLASTn query against the NCBI NT database to identify closely related orthologous sequences from other Wolbachia strains. The sequences were aligned using T-Coffee ( Notredame et al. 2000 ), and the alignment was then manually edited to remove known hypervariable regions. We then used SplitsTree ( Huson and Bryant 2006 ) to construct a Neighbor-Net network ( Bryant and Moulton 2004 ) based on the alignment to investigate the phylogenetic placement of w Noto.
Fluorescence In-Situ Hybridization
We used fluorescence in situ hybridization (FISH) to determine the tissue tropism of w Noto in Ae. notoscriptus. Legs and wings were removed from 7-d-old females prior to bodies being fixed overnight at 4°C in 4% paraformaldehyde in PBS containing 0.5% Triton X-100. Mosquito bodies were dehydrated in an inverse ethanol series followed by two toluene treatments before being embedded in paraffin wax. Embedded mosquitoes were cut into 8-µm sections using a rotary microtome, placed on Superfrost Plus slides (Lomb), and allowed to dry overnight. Slides were then xylene-treated twice; incubated in 100, 90, and 70% ethanol; and allowed to air dry. Slides were incubated overnight at 37°C in hybridization buffer containing two fluorescent probes, 16 S-W2 and 16 S-W3 ( Table 1 ) labeled with rhodamine ( Moreira et al. 2009 ). The w Noto16S gene was cloned and sequenced to ensure that FISH probes bound accurately to the 16 S gene target. Slides were then washed in 1 × SSC (+10 mM DTT) at room temperature, twice in 1 × SSC (+10 mM DTT) and twice in 0.5 × SSC (+10 mM DTT) at 55°C for 15 min each, and followed by a wash in 0.5 × SSC (+10 mM DTT) containing DAPI for 10 min at room temperature. After washing with 0.5 × SSC (+10 mM DTT) for 10 min, antifading reagent (ProLong, Invitrogen) was pipetted onto slides, which were then sealed with a coverslip. Slides were viewed using an Axioscope fluorescent microscope.
Dengue Virus Preparation
The Aedes albopictus C6/36 cell line was used to prepare DENV stock. Cells were infected with DENV serotype 2 strain 92 T (DENV2-92 T; Moreira et al. 2009 ) and then incubated for 7 d at 26°C in RPMI 1640 media (Gibco, Invitrogen) supplemented with 1 × Glutamax (Invitrogen), 25 mM HEPES buffer (Sigma-Aldrich, Australia), and 2% fetal bovine serum. After 7 d, cells and media were centrifuged at 4,000 g at 4°C for 15 min and the supernatant containing the virus was harvested. The supernatant was aliquoted into single-use 1-ml lots and frozen at −80°C.
Exposure of Mosquitoes to DENV-2
Seven-day-old female Noto and Noto.tet mosquitoes were placed in gauze-covered, plastic feeding containers, at a density of 50 mosquitoes per container, and starved overnight before blood feeding occurred. Females were allowed to feed for 1 h on a 1:1 mix of sheep blood and DENV-2 at a titer of 1 × 10 7 pfu/ml, using a glass feeder covered with sausage casing and warmed with circulating 37°C water. Mosquitoes were left overnight in containers with 10% sucrose solution, then anesthetized the following morning using CO 2 and placed on ice for sorting. Non-engorged mosquitoes were discarded, whereas blood-fed mosquitoes were transferred to cups at a density of approximately 10 per cup and maintained on 10% sucrose solution. Mosquito heads, thoraces (including wings and legs), and abdomens were dissected on dry ice at 14 d post-feeding, homogenized in 200 µl TRIzol® (Invitrogen), and stored separately at −80°C until RNA extraction. Wild-type Ae. aegypti mosquitoes (third generation in the laboratory post field collection in Cairns, Australia) were also fed DENV-2 under the same conditions and, at the same time, as a positive control for Ae. notoscriptus virus challenge.
RNA Extraction and cDNA Synthesis
RNA was extracted from heads and abdomens following the manufacturer’s protocol for use of TRIzol® (Invitrogen). Forty microliters of chloroform was added to the sample, followed by centrifugation at 12,000 g for 15 min at 4°C. After centrifugation, the supernatants were collected, and for abdomen samples, chloroform extraction was repeated a second time. Following collection of supernatant, 100 µl of isopropanol was added and the mix incubated overnight at −20°C. The following day, samples were centrifuged for 10 min at 12,000 g at 4°C to form a pellet. The pellet was washed with 75% ethanol and samples left to air-dry before adding 12 µl RNAse-free water and incubating at 4°C for 30 min. RNA concentration and quality was checked on NanoDrop ND-1000 (NanoDrop Technologies). Complementary DNA (cDNA) was synthesized with the DENV-2 NS5 gene reverse primer ( Richardson et al. 2006 ) to detect the presence of the virus ( Table 1 ). The volume of RNA needed for a final quantity of 0.3 µg for head samples or 1 µg for abdomens was added to H 2 O, up to a final volume of 13.85 µl. RNA was incubated with the primer and 10 mM dNTPS at 86°C for 15 min, followed by 10 min at 2°C. cDNA was synthesized using Superscript III (Invitrogen), in reaction mixtures containing 4 µl of reverse transcriptase buffer and 0.5 µl reverse transcriptase in a total volume of 20 µl. Cycling conditions were as follows: 25°C for 10 min, 42°C for 50 min, and 95°C for 10 min, followed by storage at −20°C. Abdomen cDNAs were diluted 1:5 with MilliQ water, but head samples were not diluted owing to low initial concentration of RNA in the extracts.
Quantitative PCR of Mosquitoes Challenged With DENV-2
Two microliters of cDNA was amplified in a quantitative PCR (qPCR) reaction consisting of 5 µl SYBR Green mix (Invitrogen) and 1 µl of 10 µM forward and reverse primers targeting the DENV-2 NS5 gene ( Richardson et al. 2006 ; Table 1 ) in a total volume of 10 µl. Reactions were performed in duplicate using a LightCycler®480 (Roche Applied Biosciences) with the following cycling conditions: 50°C for 2 min, 95°C for 2 min, and then 45 cycles of 95°C for 5 s, 60°C for 5 s, and, finally, 72°C for 10 s, followed by a melting curve to confirm specificity of amplified product. Each sample was compared with a DENV-2 standard curve at known concentrations to calculate the number of DENV-2 copies. The standard curve had been generated previously by cloning the DENV-2 NS5 fragment into a plasmid, followed by serial dilution at known concentrations ( Moreira et al. 2009 ). Samples that recorded zero amplification across both replicates were considered to be negative for DENV-2. DENV infection rate was expressed as the percentage of DENV-2 infected samples out of the total sample number. C t values and DENV-2 copies were averaged for each replicate and the number of DENV-2 copies/1 µg RNA was calculated. Data were standardized per µg RNA to compare abdomen and head samples. Fisher’s exact tests were used to test for significant differences in DENV-2 infection rates between Ae. notoscriptus lines Noto and Noto.tet, and between each line and A e . aegypti. The number of DENV-2 RNA copies was compared for significant differences between the lines using Mann–Whitney U tests.
Results
High Prevalence of Wolbachia Infection in Ae. notoscriptus Ovaries
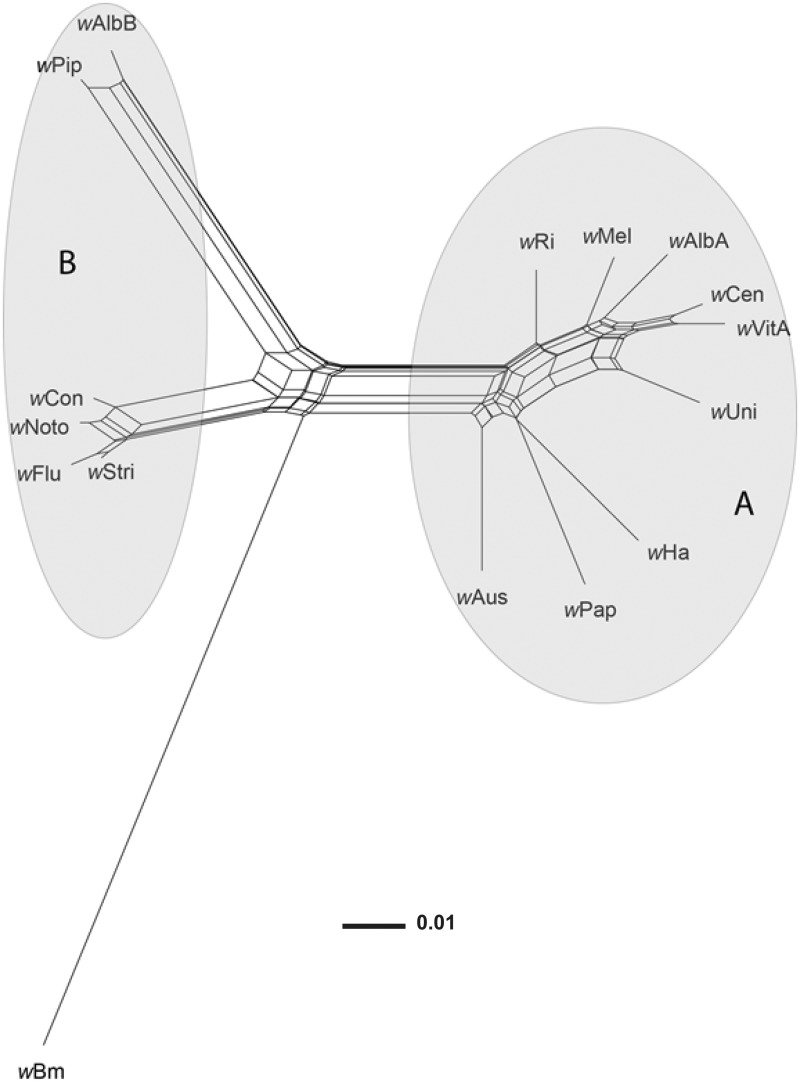
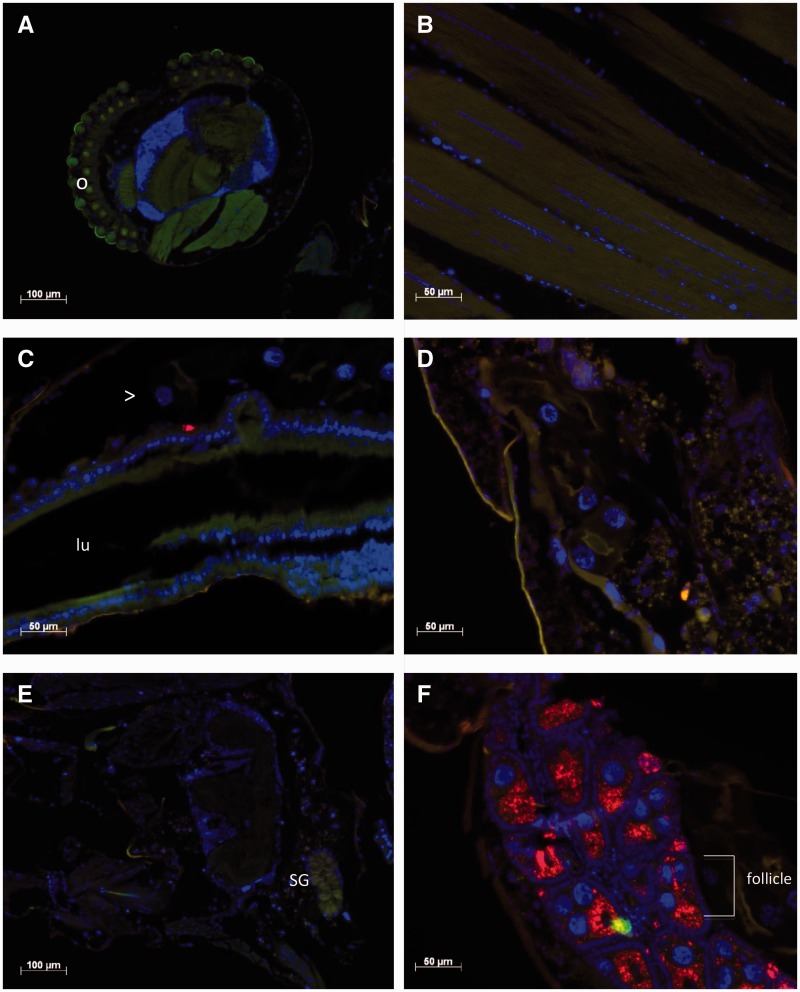
The Ae. notoscriptus laboratory population was 100% infected with Wolbachia , and field-captured Ae. notoscriptus larvae showed a high level of infection with 87.9% of 248 samples positive for Wolbachia. In a phylogenetic network based on the wsp gene, the strain infecting Ae. notoscriptus , w Noto (GenBank record KT962260), clustered closely with the B-group strains w Flu, w Stri, and w Con, which infect the mosquito Aedes fluviatilis , the planthopper Laodelphax striatellus, and the flour beetle Tribolium confusum , respectively ( Fig. 1 ). There is some evidence of conflicting phylogenetic signal in these sequence data, as indicated by boxes in the network. This is likely to be due at least in part to recombination, which occurs frequently within Wolbachia supergroups ( Klasson et al. 2008 , Duplouy et al. 2013 ). Despite the presence of recombination, however, w Noto clearly clusters with B group strains to the exclusion of A group strains. The tissue distribution of w Noto was verified by FISH using Wolbachia -specific probes. Wolbachia was present in the ovaries of Ae. notoscriptus , but absent from all other tissues examined ( Fig. 2 ).
Fig. 1.
Neighbour-Net phylogenetic network based on wsp sequences of Wolbachia strains from A, B, and D supergroups. The Wolbachia strain infecting Ae. notoscriptus ( w Noto) is closely related to B-group strains w Flu, w Con, and w Stri. Conflicting phylogenetic signals (owing to recombination and/or homoplasy) are represented as boxes or parallelograms in the network. Supergroups A and B are indicated by gray ellipses. Supergroup D is represented by w Bm.
Fig. 2.
Localization of Wolbachia in Ae. notoscriptus. Fluorescence in situ hybridization (FISH) of paraffin sections showing the localization of Wolbachia (red) in different tissues of 7-d-old female Ae. notoscriptus. Wolbachia are labeled using two 16 S rRNA rhodamine-labeled probes. DNA is stained with DAPI (blue) and a green GFP filter is used to provide contrast. ( A ) Head. “o” indicates the ommatidia that form the compound eye of the mosquito. ( B ) Thoracic muscle. ( C ) Midgut. The red spot is an artifact and not Wolbachia. “Lu” indicates the midgut lumen and “>“ the malpighian tubule running parallel to the midgut. ( D ) Malpighian tubule. ( E ) Salivary gland, labeled “SG”. ( F ) Ovary showing the presence of Wolbachia (red).
DENV-2 Infection in Noto, Noto.tet, and Ae. aegypti
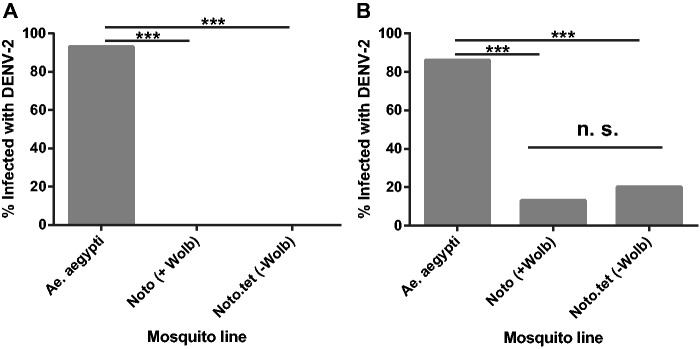
Thirteen percent of abdomen samples (4/30 samples) from Noto mosquitoes were positive for DENV-2, compared with 20% (6/30 samples) for Noto.tet ( Fig. 3 ). The difference in infection rates of the lines was not statistically significant ( P = 0.73 by Fisher’s exact test). cDNA could not be synthesized from some head samples owing to the presence of insufficient RNA, resulting in a decreased total sample size ( N = 20 per Ae. notoscriptus line) in comparison with abdomen samples. We did not find any DENV-positive heads in either of the Ae. notoscriptus lines. In Wolbachia -uninfected Ae. aegypti , a positive control for the virus challenge, DENV-2 was present in 93% of head samples (14/15) and in 86% of abdomens (26/30; Fig. 3 ). Infection rates were significantly higher in Ae. aegypti versus the two Ae. notoscriptus lines ( P < 0.0001 by Fisher’s exact tests).
Fig. 3.
DENV-2 infection rate in A .notoscriptus and Ae. aegypti. Wild-type Ae. aegypti (Wolbachia -), Wolbachia- infected Ae. notoscriptus and their antibiotic-treated counterparts Noto.tet were blood-fed with DENV-2, and viral genomic RNA was then detected by qPCR at 14 d post-infection. Infection rates are expressed as the percentage of DENV-2 infected samples out of the total sample number. ( A ) Head samples (Noto and Noto.tet total N = 20, Ae. aegypti total N = 15). ( B ) Abdomen samples (total N = 30 for all groups). There was no significant difference (ns; P > 0.05) in proportions infected between Noto and Noto.tet abdomen samples, but there were highly significant differences ( P < 0.0001) between Ae. aegypti and other mosquitoes.
The number of DENV-2 copies in abdomen samples was not significantly different between Noto and Noto.tet lines ( P = 0.25 by Mann–Whitney test; Fig. 4 ). Interestingly, the average number of copies for the positive abdomen samples was similar in the two Ae. notoscriptus lines compared with Ae. aegypti.
Fig. 4.
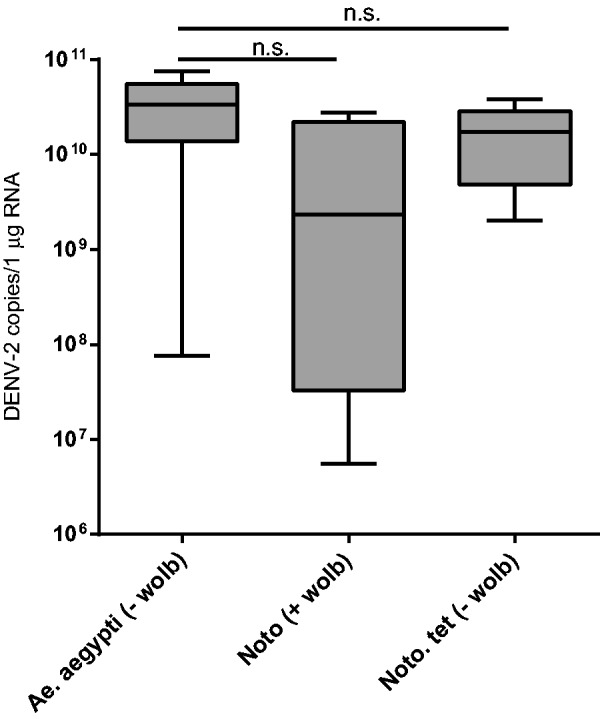
DENV-2 replication in mosquito abdomen samples. Wild-type Ae. aegypti ( ‐ Wolbachia ), Wolbachia- infected Ae. notoscriptus line Noto, and antibiotic-treated Noto.tet were blood-fed with DENV-2, and viral genomic RNA was detected by qPCR. Four Noto, six Noto.tet, and 26 Ae. aegypti abdomen samples were confirmed DENV-2-positive ( N = 30 tested per line). The number of DENV-2 copies/1 µg RNA was calculated from the average number of DENV-2 copies detected by qPCR based on a standard DENV-2 curve multiplied by dilution factors. There was no significant difference (ns) between Noto and Noto.tet for abdomen samples or either line and Ae. aegypti ( P > 0.05 by Mann–Whitney U test).
Discussion
We found no evidence that the Wolbachia strain w Noto interferes with DENV replication in its natural host. Aedesnotoscriptus mosquitoes infected with w Noto did not show lower DENV-2 infection rates, dissemination rates, or titers than the w Noto.tet line, from which Wolbachia had been removed. These observations suggest that the low competence of this vector for DENV-2 is owing to factors other than the presence of Wolbachia.
Phylogenetic network analysis placed wNoto in Wolbachia supergroup B ( Fig. 1 ). Another supergroup B strain, w No from D. simulans , has also been shown not to protect its natural host against two RNA viruses, DCV and Flock house virus ( Osborne et al. 2009 ). Other B-group strains, however, have been reported to confer viral resistance in mosquitoes: w AlbB and w Pip increase resistance to DENV and West Nile virus in their native hosts Ae. albopictus and C. quinquefasciatus, respectively ( Mousson et al. 2012 , Glaser and Meola 2010 ).
Two factors potentially underlying Wolbachia -mediated viral interference are the density and tissue tropism of Wolbachia infection. Wolbachia strains that replicate to high densities within D. simulans confer viral protection ( Osborne et al. 2009 ), and reducing the density of w Au in this host through antibiotic treatment results in the loss of antiviral protection against DCV ( Osborne et al. 2012 ). A strong positive correlation between Wolbachia density and interference with DENVs has also been demonstrated in cell lines ( Frentiu et al. 2010 , Lu et al. 2012 ).
In addition to high Wolbachia density, the presence of the bacterium within somatic tissues, especially the midgut and salivary glands, appears to be essential to conferring resistance to viral infection. The strains w Mel and w MelPop, for example, establish high-density infections of numerous somatic tissues when introduced into Ae. aegypti , and both confer resistance to DENVs ( Moreira et al. 2009 , Walker et al. 2011 ). Similarly, the strong viral resistance to DENV-2 observed in the MTB strain of Aedespolynesiensis has been linked to increased Wolbachia density within somatic tissues of the mosquito ( Bian et al. 2013 ). In contrast, low Wolbachia density within somatic tissues of Ae. albopictus could explain why the mosquito is still able to transmit DENV despite being naturally infected with Wolbachia ( Lu et al. 2012 ). In this study, FISH imaging indicated the presence of Wolbachia in Ae. notoscriptus ovaries but not in somatic tissues ( Fig. 2 ), showing that this strain has a restricted tissue tropism in its native host, which may contribute to the absence of Wolbachia -dependent viral interference.
There was no significant difference between DENV-2 titers in the abdomens of Noto and Noto.tet mosquitoes ( Fig. 4 ). Interestingly, abdominal titers in both mosquito lines were comparable with those in Ae. aegypti ( Fig. 4 ), indicating that Ae. notoscriptus individuals can support levels of DENV-2 infection that could lead to successful dissemination and transmission to another host. However, the rate of DENV-2 infection was significantly lower in Noto and Noto.tet than in Ae. aegypti ( Fig. 3 ), suggesting a low potential vectorial capacity for Ae. notoscriptus compared with the primary dengue vector. These results support previous findings by Watson and Kay (1999) that Ae. notoscriptus is susceptible to oral infection with DENV-2, although less so than Ae. aegypti. We saw no evidence of DENV dissemination to the head in the Noto or Noto.tet lines, however, even for those mosquitoes with high abdominal titer. As Wolbachia does not infect the head or salivary glands of Ae. notoscriptus ( Fig. 2 ), and removal of Wolbachia has no effect on dissemination, this is not the result of Wolbachia limiting virus replication in these tissues.
The precise factors that inhibit DENV infection and dissemination in Ae. notoscriptus remain to be identified. The factors may be genetic and include a lack of compatible cellular receptors compared with Ae. aegypti ( Mercado-Curiel et al. 2008 ), or the presence of naturally resistant variants in the RNAi ( Lambrechts et al. 2013 ) and the Toll, IMD, and JAK-STAT pathways ( Souza-Neto et al. 2009 , Behura et al. 2011 ) that control virus infections in insects. Alternatively, these factors may include a gut microbiome and virome present in Ae. notoscriptus that could induce refractoriness ( Jupatanakul et al. 2014 ). Additional experiments are required to determine if the difference observed between A e . notoscriptus and A e . aegypti holds true for all arboviruses or is DENV-2-specific . Aedesnotoscriptus was identified as a possible vector during a 1999 outbreak of Ross River virus in Brisbane, with laboratory experiments showing that it is able to transmit the virus to suckling mice ( Watson and Kay1998 ). Future studies are needed to identify the effect of Wolbachia on the vector competence of A e . notoscriptus for Ross River virus.
In summary, our results demonstrate that factors other than infection with the native Wolbachia strain w Noto reduce the vector competence of Ae. notoscriptus for DENV, by limiting infection of this mosquito species and preventing dissemination of the virus into the head if an infection is established. The low density and restricted tissue distribution of the w Noto infection may underlie the absence of Wolbachia -mediated viral interference in this system.
Acknowledgments
We would like to thank the members of the O’Neill and McGraw laboratories for their assistance, in particular Nichola Kenny and Jenny Gough for their help with mosquito rearing and blood feeding. We also thank Vincent van Uitreght for supplying Ae. notoscriptus eggs. Funding for this project came from the Foundation for the National Institutes of Health through the Grand Challenges in Global Health Initiative of the Bill and Melinda Gates Foundation.
References Cited
- Behura S. K., Gomez-Machorro C., Harker B. W., deBruyn B., Lovin D. D., Hemme R. R., Mori A., Romero-Severson J., Severson D. W. . 2011. . Global cross-talk of genes of the mosquito Aedes aegypti in response to dengue virus infection . PLoS Negl. Trop. Dis. 5 : e1385 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian G., Xu Y., Lu P., Xie Y., Xi Z. . 2010. . The endosymbiotic bacterium Wolbachia induces resistance to Dengue virus in Aedes aegypti . PLoS Pathog. 4 : 833 – 842 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian G., Zhou G., Lu P., Xi Z. . 2013. . Replacing a native Wolbachia with a novel strain results in an increase in endosymbiont load and resistance to dengue virus in a mosquito vector . PLoS Negl. Trop. Dis. 7 : e.2250 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braig H. R., Zhou W., Dobson S. L., O’Neill S. L. . 1998. . Cloning and characterization of a gene encoding the major surface protein of the bacterial endosymbiont Wolbachia pipientis . J. Bacteriol. 180 : 2373 – 2378 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D., Moulton V. . 2004. . Neighbor-net: An agglomerative method for the construction of phylogenetic networks . Mol. Biol. Evol. 21 : 255 – 265 . [DOI] [PubMed] [Google Scholar]
- Dobson S. L., Marsland E. J., Rattanadechakul W. . 2002. . Mutualistic Wolbachia infection in Aedes albopictus: Accelerating cytoplasmic drive . Genetics 160 : 1087 – 1094 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplouy A., Iturbe-Ormaetxe I., Beatson S. A., Szubert J. M., Brownlie J. C., McMeniman C. J., McGraw E. A., Hurst G. D., Charlat S., O’Neill S. L., et al. . 2013. . Draft genome sequence of the male-killing Wolbachia strain w Bol1 reveals recent horizontal gene transfers from diverse sources . BMC Genomics 14 : 20 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frentiu F. D., Robinson J., Young P. R., McGraw E. A., O’Neill S. L. . 2010. . Wolbachia -mediated resistance to dengue virus infection and death at the cellular level . PLoS ONE 5 : e13398 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser R. L., Meola M. A. . 2010. . The native Wolbachia endosymbionts of Drosophila melanogaster and Culex quinquefasciatus increase host resistance to West Nile Virus infection . PLoS ONE 5 : 977 – 987 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges L. M., Brownlie J. C., O’Neill S. L., Johnson K. N. . 2008. . Wolbachia and virus protection in insects . Science 322 : 702 . [DOI] [PubMed] [Google Scholar]
- Hilgenboecker K., Hammerstein P., Schlattmann P., Telschow A., Werren J. H. . 2008. . How many species are infected with Wolbachia ?–A statistical analysis of current data . FEMS Microbiol. Lett. 281 : 215 – 220 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A. A., Montgomery B. L., Popovici J., Iturbe-Ormaetxe I., Johnson P. H., Muzzi F., Greenfiled M., Durkan M., Leong Y. S., Dong Y., et al. . 2011. . Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission . Nature 476 : 454 – 457 . [DOI] [PubMed] [Google Scholar]
- Hurst G. D., Jiggins F. M., Garf von der Schulenburg J., Bertrand D., West S. A., Goriacheva I. I., Zakharov I. A., Werren J. H., Stouthamer R., et al. . 1999. . Male-killing Wolbachia in two species of insect . Proc. Biol. Sci. 266 : 735 – 740 . [Google Scholar]
- Huson D. H., Bryant D. . 2006. . Application of phylogenetic networks in evolutionary studies . Mol. Biol. Evol. 23 : 254 – 267 . [DOI] [PubMed] [Google Scholar]
- Jeyaprakash A., Hoy M. A. . 2000. . Long PCR improves Wolbachia DNA amplification: wsp sequences found in 76% of sixty-three arthropod species . Insect Mol. Biol. 9 : 393 – 405 . [DOI] [PubMed] [Google Scholar]
- Jupatanakul N., Sim S., Dimopoulos G. . 2014. . The insect microbiome modulates vector competence for arboviruses . Viruses. 6 : 4294 – 4313 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay B. H., Boyd A. M., Ryan P. A., Hall R. A. . 2007. . Mosquito feeding patterns and natural infection of vertebrates with Ross River and Barmah Forest viruses in Brisbane, Australia . Am. J. Trop. Med. Hyg. 76 : 417 – 423 . [PubMed] [Google Scholar]
- Kay B. H., Watson T. M., Ryan P. A. . 2008. . Definition of productive Aedes notoscriptus (Diptera: Culicidae) habitats in western Brisbane, and a strategy for their control . Aust. J. Entomol. 47 : 142 – 148 . [Google Scholar]
- Kittayapong P., Baisley K. J., Baimai V., O’Neill S. L. . 2000. . Distribution and diversity of Wolbachia infections in Southeast Asian mosquitoes (Diptera: Culicidae) . J. Med. Entomol. 37 : 340 – 345 . [DOI] [PubMed] [Google Scholar]
- Klasson L., Walker T., Sebaihia M., Sanders M. J., Quail M. A., Lord A., Sanders S., Earl J., O’Neill S. L., Thomson N., et al. . 2008. . Genome evolution of Wolbachia strain w Pip from the Culex pipiens group . Mol. Biol. Evol. 25 : 1877 – 1887 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts L., Scott T. W., Gubler D. J. . 2010. . Consequences of the expanding global distribution of Aedes albopictus for dengue virus transmission . PLoS Negl. Trop. Dis. 4 : e646 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts L., Quillery E., Noel V., Richardson J. H., Jarman R. G., Scott T. W., Chevillon C. . 2013. . Specificity of resistance to dengue virus isolates is associated with genotypes of the mosquito antiviral gene Dicer-2 . Proc. Biol. Sci. 280 : 20122437 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P., Bian G., Pan X., Xi Z. . 2012. . Wolbachia induces density-dependent inhibition to dengue virus in mosquito cells . PLoS Negl. Trop. Dis. 6 : e1754 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMeniman C. J., Lane R. V., Cass B. N., Fong A. W. C., Sidhu M., Wang Y. F., O’Neill S. L. . 2009. . Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti . Science 323 : 141 – 144 . [DOI] [PubMed] [Google Scholar]
- Mercado-Curiel R. F., Black W. C., Munoz M de L. . 2008. . A dengue receptor as possible genetic marker of vector competence in Aedes aegypti . BMC Microbiol. 8 : 118 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micieli M. V., Glaser R. L. . 2014. . Somatic Wolbachia (Rickettsiales: Rickettsiaceae) levels in Culex quinquefasciatus and Culex pipiens (Diptera: Culicidae) and resistance to West Nile virus infection . J. Med. Entomol. 51 : 189 – 199 . [DOI] [PubMed] [Google Scholar]
- Moreira L. A., Iturbe-Ormaetxe I., Jeffery J. A., Lu G. J., Pyke A. T., Hedges L. M., Rocha B. C., Hall-Mendelin S., Day A., Riegler M., et al. . 2009. . A Wolbachia symbiont in Aedes aegypti limits infection with Dengue, Chikungunya, and Plasmodium . Cell 139 : 1268 – 1278 . [DOI] [PubMed] [Google Scholar]
- Mousson L., Zouache K., Arias-Goeta C., Raquin V., Mavingui P., Failloux A. B. . 2012. . The native Wolbachia symbionts limit transmission of dengue virus in Aedes albopictus . PLoS Negl. Trop. Dis. 6 : e1989 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C., Higgins D. G., Heringa J. . 2000. . T-Coffee: A novel method for fast and accurate multiple sequence alignment . J. Mol. Biol. 302 : 205 – 217 . [DOI] [PubMed] [Google Scholar]
- O’Neill S. L., Karr T. L. . 1990. . Bidirectional incompatibility between conspecific populations of Drosophila simulans . Nature 348 : 178 – 180 . [DOI] [PubMed] [Google Scholar]
- Osborne S. E., Leong Y. S., O’Neill S. L., Johnson K. N. . 2009. . Variation in antiviral protection mediated by different Wolbachia strains in Drosophila simulans . PLoS Pathog. 5 : 656 – 664 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne S. E., Iturbe-Ormaetxe I., Brownlie J. C., O’Neill S. L., Johnson K. N. . 2012. . Antivrial protection and the importance of Wolbachia density and tissue tropism in Drosophila simulans . Appl. Environ. Microbiol. 78 : 69222 – 69229 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osei-Poku J., Mbogo C. M., Palmer W. J., Jiggins F. M. . 2012. . Deep sequencing reveals extensive variation in the gut microbiota of wild mosquitoes from Kenya . Mol. Ecol. 21 : 5138 – 5150 . [DOI] [PubMed] [Google Scholar]
- Richardson J., Molina-Cruz A., Salazar M. I., Black W. . 2006. . Quantitative analysis of dengue-2 virus RNA during the extrinsic incubation period in individual Aedes Aegypti . Am. J. Trop. Med. Hyg. 74 : 132 – 141 . [PubMed] [Google Scholar]
- Riegler M., Sidhu M., Miller W. J., O’Neill S. L. . 2005. . Evidence for a global Wolbachia replacement in Drosophila melanogaster . Curr. Biol. 15 : 1428 – 1433 . [DOI] [PubMed] [Google Scholar]
- Rousset F., Bouchon D., Pintureau B., Jachault P., Solignac M. . 1992. . Wolbachia endosymbionts responsible for various alterations of sexuality in arthropods . Proc. Biol. Sci. 250 : 91 – 98 . [DOI] [PubMed] [Google Scholar]
- Sinkins S. P., Braig H. R., O’Neill S. L. . 1995. . Wolbachia superinfections and the expression of cytoplasmic incompatibility . Proc. Biol. Sci. 261 : 325 – 330 . [DOI] [PubMed] [Google Scholar]
- Souza-Neto J. A., Sim S., Dimopoulos G. . 2009. . An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense . Proc. Natl. Acad. Sci. 106 : 17841 – 17846 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouthamer R., Breeuwer J. A., Hurst G. D. . 1999. . Wolbachia pipientis: microbial manipulator of arthropod reproduction . Annu. Rev. Microbiol. 53 : 71 – 102 . [DOI] [PubMed] [Google Scholar]
- Teixeira L., Ferreira A., Ashburner M. . 2008. . The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster . PLoS Biol. 6 : 2753 – 2763 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker T., Johnson P. H., Moreira L. A., Iturbe-Ormaetxe I., Frentiu F. D., McMeniman C. J., Leong Y. S., Dong Y., Axford J., Kriesner P., et al. . 2011. . The w Mel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations . Nature 476 : 450 – 453 . [DOI] [PubMed] [Google Scholar]
- Watson T. M., Kay B. H. . 1998. . Vector competence of Aedes notoscriptus (Diptera: Culicidae) for Ross River virus in Queensland, Australia . J. Med. Entomol. 35 : 104 – 106 . [DOI] [PubMed] [Google Scholar]
- Watson T. M., Kay B. H. . 1999. . Vector competence of Aedes notoscriptus (Diptera: Culicidae) for Barmah Forest virus and of this species and Aedes aegypti (Diptera: Culicidae) for dengue 1–4 viruses in Queensland, Australia . J. Med. Entomol. 36 : 508 – 514 . [DOI] [PubMed] [Google Scholar]
- Zhou W., Rousset F., O’Neill S. L. . 1998. . Phylogeny and PCR-based classification of Wolbachia strains using WSP gene sequences . Proc. Biol. Sci. 265 : 509 – 515 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zug R., Hammerstein P. . 2012. . Still a host of hosts for Wolbachia : analysis of recent data suggests that 40% of terrestrial arthropod species are infected . PLoS ONE 7 : e38544 . [DOI] [PMC free article] [PubMed] [Google Scholar]