Abstract
Background
In a recent trial of intermittent preventive treatment in pregnancy (IPTp) in Uganda, dihydroartemisinin-piperaquine (DP) was superior to sulfadoxine-pyrimethamine (SP) in preventing maternal and placental malaria.
Methods
We compared genotypes using sequencing, fluorescent microsphere, and quantitative polymerase chain reaction assays at loci associated with drug resistance in Plasmodium falciparum isolated from subjects receiving DP or SP.
Results
Considering aminoquinoline resistance, DP was associated with increased prevalences of mutations at pfmdr1 N86Y, pfmdr1 Y184F, and pfcrt K76T compared to SP (64.6% vs 27.4%, P < .001; 93.9% vs 59.2%, P < .001; and 87.7% vs 75.4%, P = .03, respectively). Increasing plasma piperaquine concentration at the time of parasitemia was associated with increasing pfmdr1 86Y prevalence; no infections with the N86 genotype occurred with piperaquine >2.75 ng/mL. pfkelch13 propeller domain polymorphisms previously associated with artemisinin resistance were not identified. Recently identified markers of piperaquine resistance were uncommon and not associated with DP. Considering antifolate resistance, SP was associated with increased prevalence of a 5-mutation haplotype (pfdhfr 51I, 59R, and 108N; pfdhps 437G and 581G) compared to DP (90.8% vs 60.0%, P = .001).
Conclusions
IPTp selected for genotypes associated with decreased sensitivity to treatment regimens, but genotypes associated with clinically relevant DP resistance in Asia have not emerged in Uganda.
Keywords: P. falciparum, drug resistance, dihydroartemisinin-piperaquine, sulfadoxine-pyrimethamine, IPT
Intermittent preventive treatment during pregnancy (IPTp) with sulfadoxine-pyrimethamine (SP) is recommended by the World Health Organization in areas of Africa with moderate to high malaria transmission [1]. However, resistance to SP is widespread in eastern and southern Africa [2], and recent studies have suggested a loss of effectiveness of IPTp with SP [3–6]. In a recent randomized controlled trial among 300 pregnant women in Tororo, Uganda, an area in which malaria transmission intensity and the prevalence of SP resistance are high [7], IPTp with the artemisinin-based combination therapy (ACT) dihydroartemisinin-piperaquine (DP) every 4 weeks (DP4w) or every 8 weeks (DP8w) was superior to IPTp with SP every 8 weeks in decreasing parasitemia, symptomatic malaria, and placental malaria. Similarly, in western Kenya, IPTp with DP was associated with decreased parasitemia and symptomatic malaria compared to IPTp with SP [8]. However, the emergence and spread of Plasmodium falciparum resistance to available antimalarial drugs is a serious threat, and there is concern that IPTp will select for drug resistance.
Mediators of decreased drug sensitivity are characterized for a number of antimalarial drugs. Resistance to antifolates is conferred by polymorphisms in genes encoding 2 enzymes in the folic acid pathway, pfdhps and pfdhfr, the targets of sulfadoxine and pyrimethamine, respectively [9]. A quintuple mutant haplotype, comprised of pfdhfr 51I, 59R, and 108N and pfdhps 437G and 540E, has been associated with SP treatment failure and is highly prevalent in Uganda [10, 11]. Additional mutations at pfdhfr 164 and pfdhps 581 and 613 are associated with high-level SP resistance in South America and Asia, but are uncommon in Africa [9]. Resistance to the aminoquinolines chloroquine and amodiaquine is mediated largely by polymorphisms in putative drug transporters encoded by pfcrt and pfmdr1 [12, 13]. Chloroquine resistance–associated mutations in pfcrt (76T) and pfmdr1 (86Y and 1246Y) have been common in Africa, but with discontinuation of chloroquine and wide use of the ACT artemether-lumefantrine (AL), the prevalence of these mutations has been decreasing in Uganda [10, 14] and other countries [15–17]. Piperaquine (PQ) is a bisaminoquinoline related to chloroquine and amodiaquine. Use of DP has selected inconsistently for mutations associated with resistance to chloroquine and amodiaquine [14, 18–22]. Recently, frequent failures have been seen after treatment for uncomplicated malaria with DP in Cambodia [23, 24]. These failures were associated with mutations in the propeller domain of the pfkelch13 gene (PF3D7_1343700) linked to artemisinin resistance [25, 26], and with 2 polymorphisms (increased copy number of plasmepsin genes [27, 28] and an exonuclease mutation, exo-E415G [28]) linked to decreased PQ sensitivity. In Uganda, the efficacy of DP for the treatment of malaria remains excellent [29, 30], pfkelch13 polymorphisms are uncommon and not associated with resistance [31, 32], and it is unknown if newly identified polymorphisms associated with decreased PQ activity are prevalent.
With few ACT failures in Africa, insights into the emergence and spread of drug resistance can be gained by considering selection by antimalarial drugs of resistance-associated polymorphisms in parasites that emerge after therapy. In Uganda, recent use of DP for both treatment [31] and chemoprevention [19] selected for pfmdr1 mutations associated with decreased sensitivity to these drugs. Interestingly, lumefantrine, a component of the Ugandan first-line ACT AL, exerts the opposite selective pressure. Specifically, parasites that emerged after therapy with AL showed selection of the wild-type polymorphisms pfcrt K76 and pfmdr1 N86 and D1246 [14, 18]. As DP showed clear efficacy advantages, it is under consideration to replace SP for IPTp. To gain insights into effects of different IPT regimens on drug resistance, we compared the prevalences of P. falciparum resistance–associated polymorphisms in samples collected from women enrolled in a trial comparing DP and SP for IPTp.
MATERIALS AND METHODS
Clinical Trial
We assessed polymorphisms of interest in samples collected as part of a double-blind, randomized trial comparing the efficacies of different IPTp regimens in Tororo District, Uganda [7]. In brief, human immunodeficiency virus–uninfected pregnant women between 12 and 20 weeks of gestation were enrolled and randomized to receive (i) SP (500 mg sulfadoxine and 25 mg pyrimethamine) every 8 weeks (20, 28, and 36 weeks’ gestational age), (ii) DP (40 mg dihydroartemisinin and 320 mg PQ) every 8 weeks (20, 28, and 36 weeks’ gestational age [DP8w]), or (iii) DP every 4 weeks (starting at 16 weeks’ gestational age [DP4w]). Participants were encouraged to visit the study clinic for all illnesses. Febrile participants with thick blood smears showing parasites were diagnosed with malaria and treated with AL. Capillary or venous blood samples were obtained from participants at enrollment, every 4 weeks during pregnancy, and when malaria was diagnosed. Blood spots were stored on filter paper, and plasma was stored at –80°C for PQ quantitation. DNA was extracted from dried blood spots using Chelex-100 [33] and tested for the presence of Plasmodium DNA by loop-mediated isothermal amplification (LAMP), as previously described [34]. The study was approved by the Makerere University Research and Ethics Committee, the Uganda National Council for Science and Technology, and the University of California, San Francisco Committee for Human Research.
Characterization of Parasite Polymorphisms
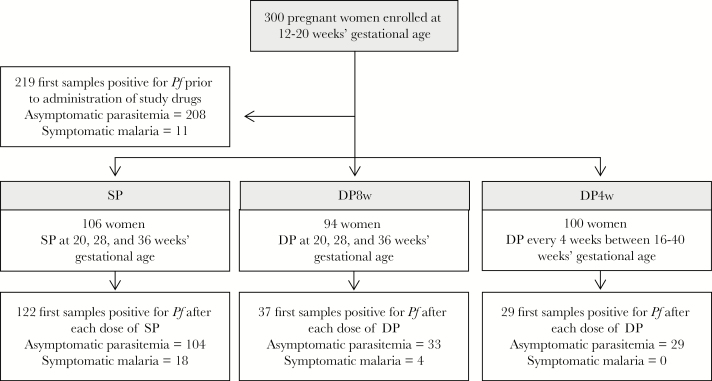
Samples were tested for polymorphisms in pfcrt, pfmdr1, pfdhfr, and pfdhps if they were positive for P. falciparum and either the first positive sample (if any) identified for each study participant prior to administration of study drugs (219 samples) or the first positive sample identified following administration of each dose of study drug (122, 37, and 29 samples for the SP, DP8w, and DP4w arms, respectively) (Figure 1). Genotyping was performed using a ligase detection reaction–fluorescent microsphere assay as previously described [35], with minor modifications including nested polymerase chain reaction (PCR) amplification of templates, as described previously [14, 36]. The pfkelch13 propeller domain was amplified and sequenced as previously described [32]. The exonuclease gene (PF3D7_1362500) was amplified using nested PCR as described in Supplementary Table 1. PCR products were sequenced in both directions (Eurofins Genomics) using second-round PCR primers. Sequencing results were assembled, visually examined, and aligned with the reference 3d7 sequence using CodonCode version 6.0.2 (CodonCode Corporation). The pfkelch13 and exonuclease gene sequences were submitted to GenBank under accession numbers MF285352–MF285413 and MF285277–MF285277351, respectively.
Figure 1.
Selection of samples for inclusion in the study. Abbreviations: DP, dihydroartemisinin-piperaquine; DP4w, dihydroartemisinin-piperaquine every 4 weeks; DP8w, dihydroartemisinin-piperaquine every 8 weeks; Pf, Plasmodium falciparum parasitemia; SP, sulfadoxine-pyrimethamine.
Plasmepsin-2 (pfpm2) copy number was determined using a TaqMan quantitative PCR assay. Pfpm2 primers and probes were designed with Primer Express 3.0 (Supplementary Table 2). The pfpm2 probe was FAM (6-carboxyfluorescein) labeled at the 5ʹ-end and had a minor groove binder quencher at the 3ʹ-end. The β-tubulin primers and probe were as described elsewhere [37], and were VIC labeled at the 5ʹ-end, with a TAMRA (6-carboxytetramethylrhodamine) quencher at the 3ʹ-end (Thermo Fisher Scientific). Multiplex PCR reactions (20 µL) contained TaqMan Universal PCR Master Mix (Thermo Fisher Scientific), 300 nM of each forward and reverse primer, 100 nM of each probe, and 2 µL of template DNA. Thermocycler conditions were as follows: 2 minutes at 50°C, then 10 minutes at 95°C, followed by 50 cycles of 95°C for 15 seconds and 60°C for 1 minute. All reactions were performed in quadruplicate and contained a single pfpm2 copy control, a multicopy control (generated using pfpm2-containing plasmids with 3–4 copies of the gene and kindly provided by Didier Menard), and a negative control. The pfpm2 and β-tubulin reactions had similar relative amplification efficiencies and were analyzed using the comparative cycle threshold (Ct) method (2-ΔΔCt) as applied in the SDS Software package associated with the Applied Biosystems 7500 real-time PCR instrument on which the reactions were run. Pfmdr1 copy number was quantified using TaqMan real-time PCR as previously described [14]. Data were rejected if they did not conform to exponential kinetics. Assays were repeated if (i) the ΔCt standard error was >0.25; (ii) Ct values were >35; or (iii) the relative quantification (RQ) value was <0.70 or >1.3. Data from assays with standard errors >0.25, with Ct values were >35, or RQ values <0.70 were rejected. RQ values were averaged between repeated assays when quality controls were met.
Plasma Piperaquine Quantitation
Venous plasma samples were collected at enrollment, at 20, 28, and 36 weeks’ gestation, and at delivery. Capillary samples were obtained at 16, 24, 32, and 40 weeks’ gestation, or when malaria was diagnosed. Blood samples were centrifuged within 60 minutes of collection at 2000g for 10 minutes, and plasma was transferred to cryovials and stored at –80°C. Concentrations of PQ were determined using high-performance liquid chromatography–tandem mass spectrometry, as previously described [38], with the modification that the calibration range was lowered to 0.500–50.0 ng/mL. The lower limit of quantification was 0.500 ng/mL, and the coefficient of variation was <10% for quality control measurements. A correlation equation, InCCapillary = 0.673*lnCvenous+1.574, was previously determined for this population [39], and was used to convert capillary PQ values to venous concentration estimates. Venous and converted capillary PQ concentrations were used for analyses.
Statistical Methods
Data analysis was performed with Stata version 14.0 (StataCorp) and RStudio version 0.99.903 (RStudio, Inc) software. Outcomes of interest were the prevalence of mutant alleles (including mixed infections for transporter loci and excluding mixed infections for folate loci) for each locus of interest, the prevalence of pfmdr1 and pfcrt haplotypes, and the prevalence of the quintuple folate mutant haplotype (pfdhfr 51I, 59R, and 108N and pfdhps 437G and 540E). For pfmdr1 and pfcrt haplotypes, mixed-genotype samples were excluded. The quintuple mutant haplotype was considered present if a sample had only mutant genotypes at all 5 loci, and absent if there were wild-type or mixed genotypes at any of the loci. Samples with missing data were excluded. Exposure variables of interest were the IPTp arm to which the patient was randomized, the duration of time since IPTp dosing, and the plasma PQ concentration. Associations between outcomes and categorical exposure variables were measured in Stata using log-binomial regression with generalized estimating equations to account for repeated measures in the same patient. Associations between prevalences of mutant alleles and PQ concentrations were measured using generalized linear models in the R package “ggplot2,” implementing binomial variance and logit link functions. A P value <.05 was considered significant.
RESULTS
Impact of DP on Transporter Polymorphisms
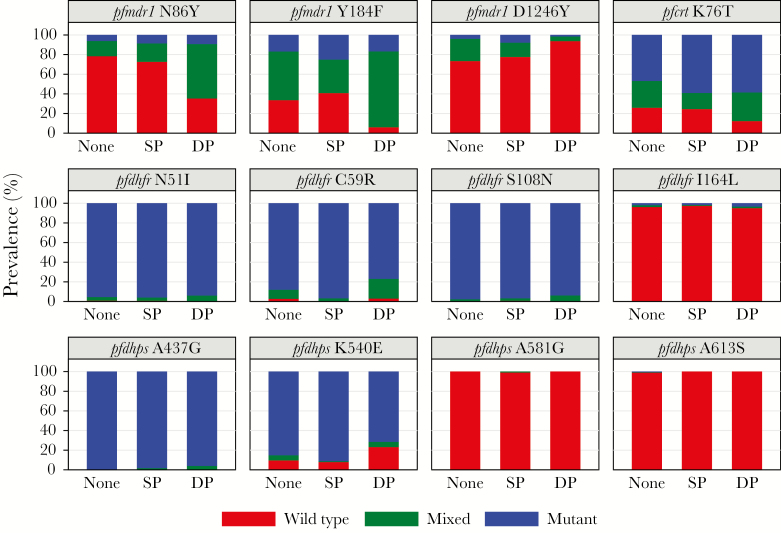
We compared the prevalence of mutant alleles (mixed genotype and pure mutant infections) at pfmdr1 N86Y, pfmdr1 Y184F, pfmdr1 D1246Y, and pfcrt K76T in P. falciparum–infected samples before and after the initiation of study drugs and between patients assigned to receive DP or SP. Treatment with DP was associated with increased prevalences of mutant alleles at pfmdr1 86Y, pfmdr1 184F, and pfcrt 76T compared to no treatment (samples collected before treatment was initiated; 64.6% vs 21.6%, P < .001; 93.9% vs 59.2%, P < .001; and 87.7% vs 74.1%, P = .004, respectively) or treatment with SP (64.6% vs 27.4%, P < .001; 93.9% vs 59.2%, P < .001; and 87.7% vs 75.4%, P = .03, respectively; Table 1 and Figure 2). Treatment with DP was associated with decreased prevalence for mutant pfmdr1 1246Y compared to no treatment (6.3% vs 26.6%, P = .02) or treatment with SP (6.3% vs 22.4%, P = .03). We found no differences in the prevalence of transporter polymorphisms between the samples collected before treatment and from individuals treated with SP. Considering impact of DP on haplotypes, we found that receipt of DP was selected for the pfmdr1 N86-184F, pfmdr1 86Y-D1246, pfmdr1 N86-184F-1246D, and pfcrt 76T-mdr1 N86-184F-1246D haplotypes (Supplementary Tables 3–6).
Table 1.
Associations Between Exposure to Intermittent Preventive Treatment in Pregnancy Drugs and Transporter Gene Polymorphisms
| No Exposure vs IPTp | SP vs DP | |||||
|---|---|---|---|---|---|---|
| Locus | IPTp Drug Exposure | Prevalence of Mutant and Mixed Infections | Prevalence Ratio (95% CI) |
P Value | Prevalence Ratio (95% CI) |
P Value |
| pfmdr1 N86Y | None | 46/213 (21.6%) | reference | … | … | … |
| SP | 32/117 (27.4%) | 1.26 (.83–1.93) | .28 | reference | … | |
| DP | 42/65 (64.6%) | 2.99 (2.13–4.21) | <.001 | 2.42 (1.64–3.56) | <.001 | |
| pfmdr1 Y184F | None | 142/214 (66.4%) | reference | … | … | … |
| SP | 71/120 (59.2%) | 0.89 (.73–1.08) | .25 | reference | … | |
| DP | 62/66 (93.9%) | 1.42 (1.26–1.60) | <.001 | 1.58 (1.33–1.87) | <.001 | |
| pfmdr1 D1246Y | None | 55/207 (26.6%) | reference | … | … | … |
| SP | 26/116 (22.4%) | 0.85 (.56–1.29) | .44 | reference | … | |
| DP | 4/64 (6.3%) | 0.22 (.06–.74) | .02 | 0.25 (.08–.84) | .03 | |
| pfcrt K76T | None | 160/216 (74.1%) | reference | … | … | … |
| SP | 92/122 (75.4%) | 1.02 (.89–116) | .80 | reference | … | |
| DP | 57/65 (87.7%) | 1.19 (1.05–1.33) | .004 | 1.16 (1.02–1.33) | .03 | |
Abbreviations: CI, confidence interval; DP, dihydroartemisinin-piperaquine; IPTp, intermittent preventive treatment in pregnancy; SP, sulfadoxine-pyrimethamine.
Figure 2.
Prevalence of wild-type, mixed, or mutant alleles at Plasmodium falciparum loci of interest in parasitemic samples collected from patients before intermittent preventive treatment in pregnancy (IPTp) treatment was initiated (None), in patients receiving sulfadoxine-pyrimethamine (SP) as IPTp, and in patients receiving dihydroartemisinin-piperaquine (DP) as IPTp.
We considered recent use of AL as a potential confounder. The proportion of samples collected within 60 days of prior AL treatment did not differ statistically between the treatment arms (11% in the SP arm and 5% in the DP arms, P = .18), and, when analyzed as a covariate, was not associated with differences for any of the considered loci.
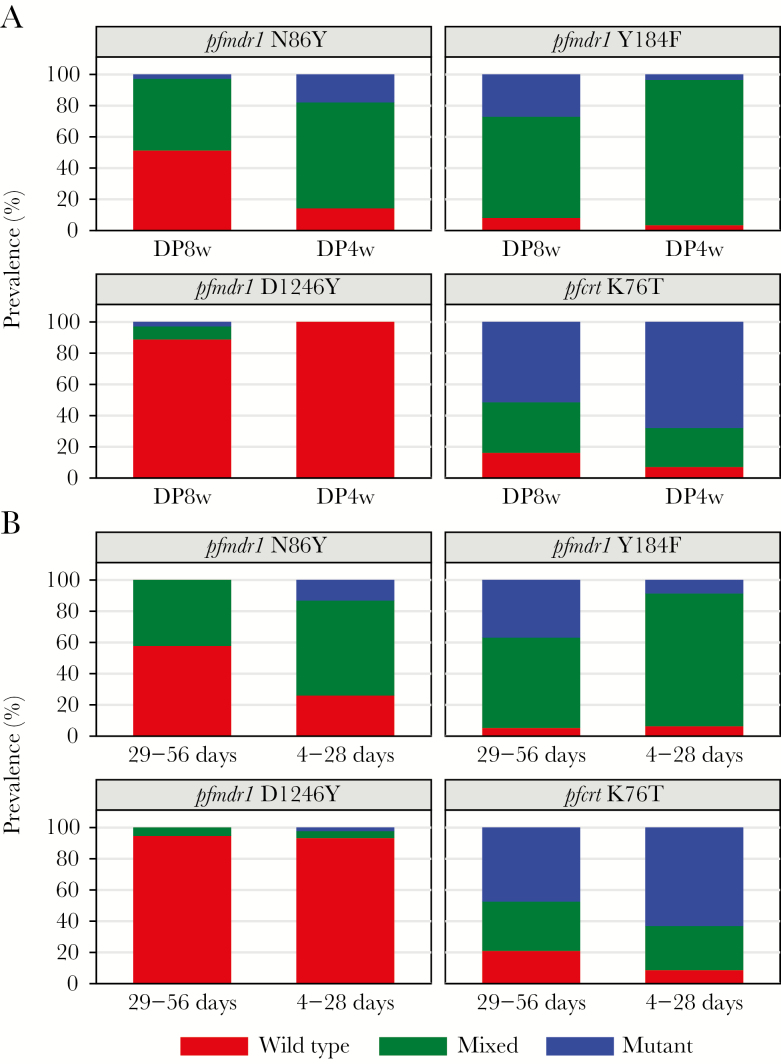
Considering DP dosing frequency, all 4 transporter loci demonstrated at least a modest dose effect (Figure 3A), with more frequent dosing associated with increased differences in allele prevalences between treatment arms, although the difference between the DP8w and DP4w arms was significant only for pfmdr1 86 (48.7% vs 85.7%, P = .003; Table 2). Considering time since last DP dosing, recent (within 4–28 days) DP dosing was associated with an increase in the prevalence of pfmdr1 86Y mutant infections compared with dosing 29–56 days before sample collection (73.9% vs 42.1%, P = .04; Table 2 and Figure 3B).
Figure 3.
Impact of dosing frequency and recent use of dihydroartemisinin-piperaquine (DP) on the prevalence of wild-type, mixed, or mutant alleles at Plasmodium falciparum loci of interest in parasitemic samples. A, Allele prevalences in parasites from patients receiving DP every 8 weeks (DP8w) or every 4 weeks (DP4w). B, Allele prevalences in parasites from patients who received DP 29–56 days or 4–28 days before parasitemic episodes.
Table 2.
Associations Between Extent of Dihydroartemisinin-Piperaquine Exposure and Transporter Gene Polymorphisms
| Locus | Level of DP Exposure | Prevalence of Mutant and Mixed Infections | Prevalence Ratio (95% CI) |
P Value | |
|---|---|---|---|---|---|
| pfmdr1 N86Y | Frequency of DP dosing | Every 8 weeks | 18/37 (48.7%) | reference | … |
| Every 4 weeks | 24/28 (85.7%) | 1.76 (1.21–2.55) | .003 | ||
| No. of days since last dose of DP | 29–56 days | 8/19 (42.1%) | reference | … | |
| 4–28 days | 34/46 (73.9%) | 1.76 (1.01–3.07) | .04 | ||
| pfmdr1 Y184F | Frequency of DP dosing | Every 8 weeks | 34/37 (91.9%) | reference | … |
| Every 4 weeks | 28/29 (96.6%) | 1.05 (.94–1.18) | .40 | ||
| No. of days since last dose of DP | 29–56 days | 18/19 (94.7%) | reference | … | |
| 4–28 days | 44/47 (93.6%) | 0.99 (.87–1.12) | .87 | ||
| pfmdr1 D1246Y | Frequency of DP dosing | Every 8 weeks | 4/36 (11.1 %) | reference | … |
| Every 4 weeks | 0/28 (0%) | NA | NA | ||
| No. of days since last dose of DP | 29–56 days | 1/19 (5.3%) | reference | … | |
| 4–28 days | 3/45 (6.7%) | NA | NA | ||
| pfcrt K76T | Frequency of DP dosing | Every 8 weeks | 31/37 (83.8%) | reference | … |
| Every 4 weeks | 26/28 (92.9%) | 1.11 (.94–1.32) | .23 | ||
| No. of days since last dose of DP | 29–56 days | 15/19 (79.0%) | reference | … | |
| 4–28 days | 42/46 (91.3%) | 1.19 (.93–1.51) | .17 | ||
Abbreviations: CI, confidence interval; DP, dihydroartemisinin-piperaquine; NA, not applicable.
Association Between Piperaquine Plasma Concentrations and Loci Associated With Drug Resistance
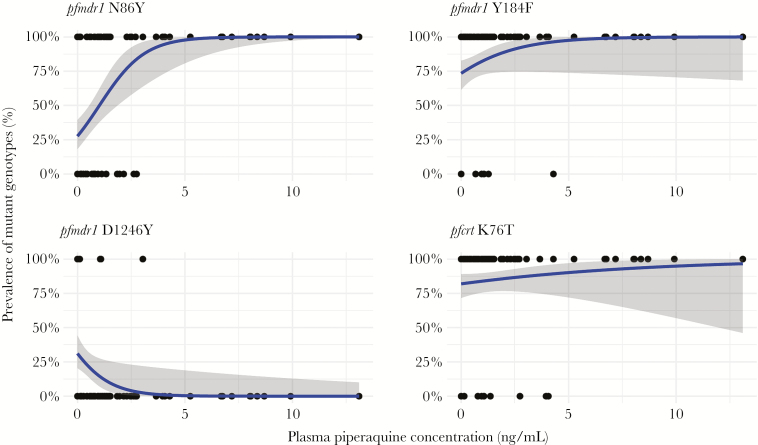
A PQ concentration was available for 56 of the 66 (85%) genotyped samples with prior exposure to DP. We used generalized linear models to determine the relationship between the prevalence of infections with mutant transporter genotypes and the plasma concentration of PQ. Increasing PQ concentration was associated with increasing prevalence of the mutant pfmdr1 86Y allele, consistent with the selection observed in the DP treatment arms (Figure 4). In samples with PQ concentrations >2.70 ng/mL, the prevalence of mutant infections was 94.1% (16/17); in contrast, in samples with PQ concentrations ≤2.70 ng/mL, the prevalence of mutant infections was 44.4% (20/45) (prevalence ratio, 2.16 [95% confidence interval, 1.56–2.99], P < .001). Only parasites with the pfmdr1 86Y mutant genotype were seen in samples with PQ levels >2.75 ng/mL.
Figure 4.
Generalized linear models representing the predicted prevalence of mutant infections with increasing plasma piperaquine concentrations. Each dot represents the genotype of a single sample (top: mutant and mixed infections; bottom: pure wild-type infections). Blue curves represent estimated prevalence of mutant alleles. Gray-shaded areas indicate 95% confidence intervals.
Markers of Artemisinin and Piperaquine Resistance
Sequencing of 61 randomly selected samples detected pfkelch mutations in 2 of 19 samples collected from patients receiving DP (A569T and G638R), 1 of 22 samples from patients receiving SP (N594K), and 0 of 20 samples collected before the initiation of chemoprevention. None of these mutations have been reported to be associated with decreased sensitivity to artemisinins [26, 40], and the prevalences of pfkelch13 mutations did not differ significantly between treatment groups. We detected no clear evidence of increased pfmdr1 copy number, although average RQ values between 1.3 and 1.5 were detected for 7 of 46 samples. The prevalence of these modest increases did not vary between treatment arms.
Considering recently identified polymorphisms associated with decreased PQ sensitivity, we measured pfpm2 copy number variation and sequenced a 395-bp amplicon surrounding the exo-E415G locus. We detected modest increases in pfpm2 copy number in 1 of 18 samples from patients receiving DP (RQ = 1.3), 1 of 21 samples from patients receiving SP (RQ = 1.3), and 1 of 27 samples collected before the initiation of chemoprevention (RQ = 2.1). We did not detect the exo-E415G mutation, but we detected other nonsynonymous exo mutations in 1 of 19 samples collected from patients receiving DP (R346T), 2 of 28 samples from patients receiving SP (L341F and Y384F), and 2 of 26 samples collected before the initiation of chemoprevention (D380H and N436I); prevalences of mutations did not differ significantly between treatment groups.
Impact of SP on Folate Polymorphisms
We compared the prevalences of mutant alleles at pfdhfr N51I, C59R, S108N/T, and I164L and pfdhps A437G, K540E, A581G, and A613S before and after the initiation of study drugs, and between patients assigned to receive DP or SP. Although the population had high prevalences of mutant alleles at pfdhfr 51, 59, and 108 and pfdhps 437 and 540 (>85% for all loci in samples collected before the initiation of study drugs), treatment with SP was associated with increased prevalences for mutant alleles at pfdhfr 59, pfdhps 540, and the quintuple mutant haplotype compared to samples collected prior to the administration of study drugs (96.7% vs 87.9%, P = .001; 91.2% vs 85.1%, P = .11; and 90.8% vs 77.8%, P = .002, respectively) or samples collected from the DP arm (96.7% vs 76.9%, P = .002; 91.2% vs 71.4%, P = .009; and 90.8% vs 60.0%, P = .001, respectively) (Table 3 and Figure 2). We found no differences in allele prevalences between samples from patients dosed with SP 29–76 or 2–28 days prior to sample collection.
Table 3.
Associations Between Exposure to Intermittent Preventive Treatment in Pregnancy Drugs and Folate Enzyme Polymorphisms
| Pre vs Post-IPTp | SP vs DP | |||||
|---|---|---|---|---|---|---|
| Locus | IPTp Drug Exposure | Prevalence of Mutant Infections | Prevalence Ratio (95% CI) |
P Value | Prevalence Ratio (95% CI) |
P Value |
| pfdhfr N51I | None | 208/218 (95.4%) | reference | … | … | … |
| DP | 61/65 (93.9%) | 0.98 (.92–1.05) | .61 | reference | … | |
| SP | 115/120 (95.8%) | 1.01 (.96–1.05) | .83 | 1.02 (.95–1.09) | .61 | |
| pfdhfr C59R | None | 189/215 (87.9%) | reference | … | … | … |
| DP | 50/65 (76.9%) | 0.88 (.75–1.02) | .09 | reference | … | |
| SP | 117/121 (96.7%) | 1.10 (1.04–1.17) | .001 | 1.26 (1.09–1.46) | .002 | |
| pfdhfr S108N | None | 207/212 (97.6%) | reference | … | … | … |
| DP | 60/64 (93.8%) | 0.96 (.90–1.03) | .24 | reference | … | |
| SP | 117/121 (96.7%) | 0.99 (.95–1.03) | .61 | 1.03 (.96–1.11) | .40 | |
| pfdhfr I164L | None | 4/213 (1.9%) | reference | … | … | … |
| DP | 2/61 (3.3%) | 1.76 (.33–9.50) | .51 | reference | … | |
| SP | 2/118 (1.7%) | 0.89 (.16–4.88) | .89 | 0.52 (.07–3.62) | .51 | |
| pfdhps A437G | None | 203/204 (99.5%) | reference | … | … | … |
| SP | 48/50 (96.0%) | 0.97 (.91–1.02) | .23 | reference | … | |
| DP | 109/111 (98.2%) | 0.99 (.96–1.01) | .32 | 1.02 (.96–1.09) | .48 | |
| pfdhps K540E | None | 177/208 (85.1%) | reference | … | … | … |
| DP | 40/56 (71.4%) | 0.84 (.70–1.01) | .06 | reference | … | |
| SP | 103/113 (91.2%) | 1.08 (.98–1.18) | .11 | 1.29 (1.07–1.56) | .009 | |
| pfdhps A581G | None | 0/209 (0.0%) | reference | … | … | … |
| DP | 0/56 (0.0%) | NA | … | reference | … | |
| SP | 0/115 (0.0%) | NA | … | NA | … | |
| Pfdhps A613S | None | 1/205 (0.5%) | reference | … | … | … |
| DP | 0/51 (0.0%) | NA | … | reference | … | |
| SP | 0/113 (0.0%) | NA | … | NA | … | |
| Quintuple mutant | None | 154/198 (77.8%) | reference | … | … | … |
| DP | 30/50 (60.0%) | 0.77 (.61–.98) | .04 | reference | … | |
| SP | 99/109 (90.8%) | 1.18 (1.06–1.31) | .002 | 1.54 (1.20–1.98) | .001 | |
Abbreviations: CI, confidence interval; DP, dihydroartemisinin-piperaquine; IPTp, intermittent preventive treatment in pregnancy; NA, not applicable; SP, sulfadoxine-pyrimethamine.
DISCUSSION
We surveyed established and recently described P. falciparum drug resistance markers in samples from subjects randomized to receive DP or SP as IPTp. DP selected for mutant alleles at pfmdr1 86, pfmdr1 184, and pfcrt 76, and against mutant alleles at pfmdr1 1246, and this selection increased with increasing PQ exposure. Recently described markers of artemisinin resistance and PQ resistance were uncommon. SP selected for mutant alleles at pfdhfr 59 and pfdhps 540, and for the quintuple pfdhfr/pfdhps mutant haplotype, all of which confer resistance to antifolate drugs. Our findings indicate that IPTp with DP or SP selects parasite genotypes previously associated with decreased sensitivity to these regimens, but polymorphisms associated with clinically relevant DP resistance in Southeast Asia appear not to have emerged in Uganda.
The transporter polymorphisms pfmdr1 N86Y, pfmdr1 D1246Y, and pfcrt K76T modulate parasite sensitivity to numerous antimalarials. Mutant pfmdr1 86Y, pfmdr1 1246Y, and pfcrt 76T are selected by prior use of the aminoquinolines chloroquine and amodiaquine and associated with reduced sensitivity to these drugs [18, 30, 41–44]. In contrast, wild-type sequences at these same alleles are selected by prior use of AL and associated with reduced sensitivity to lumefantrine [14, 30, 45–47]. Another allele, pfmdr1 Y184F, is also highly polymorphic, but its relationship to antimalarial sensitivity is not straightforward [13] and may be haplotype dependent [48].
Piperaquine is similar in structure to chloroquine and amodiaquine, but perhaps due to its larger size, its selective pressure on transporter mutations and the impacts of these mutations on drug sensitivity differ [19]. Our new results, considering impacts of DP when used for IPTp, are generally consistent with those seen in Uganda for DP used for treatment [14] or prevention [18, 19] of malaria in children. Most clearly, prior use of DP selects for parasites with increased prevalence of the pfmdr1 86Y mutation. This selection is the same as that of other aminoquinolines, and opposite that of lumefantrine. Results have been more complex for other alleles. Notably, for pfmdr1 D1246Y, our new results show selection by DP of wild-type D1246 parasites, opposite the selection seen with amodiaquine-containing regimens [43] and also opposite that with DP in some [14, 18], but not other [19], studies. For pfcrt, DP selected for the 76T mutation associated with resistance to other aminoquinolines, the same selection seen in one other recent study [19] but not others [14, 18]. Taken together, our results suggest that DP selects in Uganda for the pfmdr1 86Y mutation and other polymorphisms associated with resistance to aminoquinolines, but results for alleles other than 86Y have been inconsistent, and specific selection appears to depend on background parasite sequences that vary geographically.
Our study benefited from the availability of plasma PQ levels at the time of presentation with parasitemia or symptomatic malaria. The selective effects of DP were clearly dose dependent, with greater selection of the pfmdr1 86Y mutation and other alleles with increased exposure. However, selection for mutant pfmdr1 86Y occurred at low concentrations, and only parasites with the mutant 86Y allele were detected at PQ concentrations >2.75 ng/mL. In a recent study in Uganda, mean PQ levels 21 days after treatment were 11.8 ng/mL in pregnant women and 14.5 ng/mL in nonpregnant women [39]. Recent surveys have shown that the prevalence of parasites with the wild-type pfmdr1 N86 allele has increased to >90% in multiple sites across Uganda [10]. Taken together, our results suggest that, in Uganda, with nearly all parasites now harboring the pfmdr1 N86 genotype, monthly DP will be highly effective, with minimal incidence of breakthrough parasitemia, but that continued drug pressure might select for pfmdr1 86Y mutant parasites more likely to cause breakthroughs. It should be noted that although more frequent DP dosing was associated with stronger selection of resistance mutations, it was also more protective against parasitemia during pregnancy. Only 26 of 496 (5.2%) samples collected from the DP4w arm during our trial demonstrated parasitemia with a highly sensitive LAMP assay, compared to 74 of 445 (16.6%) samples from the DP8w arm. Determining the ideal dosing regimen to minimize exposure of parasites to PQ concentrations that are high enough to select for resistance but too low to eliminate parasitemia will be of importance to ensure the continued efficacy of DP as IPTp.
Mutations at 5 loci in the folate enzyme genes associated with resistance to SP have long been highly prevalent in Uganda [10, 49]. We detected significant selection by SP of the common mutations most clearly associated with SP resistance, pfdhfr 59R and pfdhps 540E [49]. These results reinforce concerns about the continued protective efficacy of SP as IPTp and the consequences of its continued use. Reassuringly, additional mutations associated with higher-level resistance to SP, notably pfdhfr 164L and pfdhps 581G, which have shown modest increases in prevalence in some regions of Uganda [10], were not selected by SP when used for IPTp.
Our results have important implications for the choice of drug for IPTp. We have previously demonstrated the superiority of DP over SP for young children [50], schoolchildren [19], and pregnant women [7]. Our current results show that, when used as IPTp, both DP and SP select for P. falciparum mutations previously associated with decreased sensitivity to the drugs. In the case of DP, low nanomolar PQ levels that are maintained for weeks after standard dosing were protective against drug-sensitive parasites currently circulating in Uganda, but associated with selection of parasites harboring mutations previously associated with decreased susceptibility. Importantly, for the most part, the selective pressure of DP on transporter polymorphisms is opposite that of AL, the national malaria treatment regimen in Uganda. As a result, AL selects for parasites that are highly sensitive to DP. Thus, a rational strategy to effectively treat and control malaria while limiting resistance selection may be to continue to use AL to treat malaria while instituting DP for chemoprevention in high-risk groups.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Financial support. This work was supported by the National Institutes of Health (NIH) (grant numbers R01AI075045 to P. J. R.; R01AI117001 to P. J. R. and F. A.; U19AI089674 to G. D.; and P01DH059454 to D. V. H. and M. R. K.). M. C. was supported by NIH Research Training Grant (number R25 TW009343) funded by the Fogarty International Center.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Footnotes
Presented in part: 65th Annual Meeting of the American Society of Tropical Medicine and Hygiene, Atlanta, Georgia, November 2016. Abstract 1491.
References
- 1. World Health Organization. Updated WHO policy recommendation: intermittent preventive treatment of malaria in pregnancy using sulfadoxine-pyrimethamine (IPTp-SP). Geneva, Switzerland: WHO, 2014. [Google Scholar]
- 2. Naidoo I, Roper C. Drug resistance maps to guide intermittent preventive treatment of malaria in African infants. Parasitology 2011; 138:1469–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gutman J, Mwandama D, Wiegand RE, Ali D, Mathanga DP, Skarbinski J. Effectiveness of intermittent preventive treatment with sulfadoxine-pyrimethamine during pregnancy on maternal and birth outcomes in Machinga district, Malawi. J Infect Dis 2013; 208:907–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Moussiliou A, De Tove YS, Doritchamou J et al. . High rates of parasite recrudescence following intermittent preventive treatment with sulphadoxine-pyrimethamine during pregnancy in Benin. Malar J 2013; 12:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arinaitwe E, Ades V, Walakira A et al. . Intermittent preventive therapy with sulfadoxine-pyrimethamine for malaria in pregnancy: a cross-sectional study from Tororo, Uganda. PLoS One 2013; 8:e73073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harrington WE, Mutabingwa TK, Kabyemela E, Fried M, Duffy PE. Intermittent treatment to prevent pregnancy malaria does not confer benefit in an area of widespread drug resistance. Clin Infect Dis 2011; 53:224–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kakuru A, Jagannathan P, Muhindo MK et al. . Dihydroartemisinin-piperaquine for the prevention of malaria in pregnancy. N Engl J Med 2016; 374:928–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Desai M, Gutman J, L’lanziva A et al. . Intermittent screening and treatment or intermittent preventive treatment with dihydroartemisinin-piperaquine versus intermittent preventive treatment with sulfadoxine-pyrimethamine for the control of malaria during pregnancy in western Kenya: an open-label, three-group, randomised controlled superiority trial. Lancet 2015; 386:2507–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Naidoo I, Roper C. Mapping ‘partially resistant’, ‘fully resistant’, and ‘super resistant’ malaria. Trends Parasitol 2013; 29:505–15. [DOI] [PubMed] [Google Scholar]
- 10. Tumwebaze P, Tukwasibwe S, Taylor A et al. . Changing antimalarial drug resistance patterns identified by surveillance at three sites in Uganda. J Infect Dis 2017; 215:631–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gregson A, Plowe CV. Mechanisms of resistance of malaria parasites to antifolates. Pharmacol Rev 2005; 57:117–45. [DOI] [PubMed] [Google Scholar]
- 12. Ecker A, Lehane AM, Clain J, Fidock DA. PfCRT and its role in antimalarial drug resistance. Trends Parasitol 2012; 28:504–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rosenthal PJ. The interplay between drug resistance and fitness in malaria parasites. Mol Microbiol 2013; 89:1025–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Conrad MD, LeClair N, Arinaitwe E et al. . Comparative impacts over 5 years of artemisinin-based combination therapies on Plasmodium falciparum polymorphisms that modulate drug sensitivity in Ugandan children. J Infect Dis 2014; 210:344–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Malmberg M, Ngasala B, Ferreira PE et al. . Temporal trends of molecular markers associated with artemether-lumefantrine tolerance/resistance in Bagamoyo district, Tanzania. Malar J 2013; 12:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Okombo J, Kamau AW, Marsh K, Sutherland CJ, Ochola-Oyier LI. Temporal trends in prevalence of Plasmodium falciparum drug resistance alleles over two decades of changing antimalarial policy in coastal Kenya. Int J Parasitol Drugs Drug Resist 2014; 4:152–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kublin JG, Cortese JF, Njunju EM et al. . Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect Dis 2003; 187:1870–5. [DOI] [PubMed] [Google Scholar]
- 18. Tumwebaze P, Conrad MD, Walakira A et al. . Impact of antimalarial treatment and chemoprevention on the drug sensitivity of malaria parasites isolated from Ugandan children. Antimicrob Agents Chemother 2015; 59:3018–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nankabirwa JI, Conrad MD, Legac J et al. . Intermittent preventive treatment with dihydroartemisinin-piperaquine in Ugandan schoolchildren selects for Plasmodium falciparum transporter polymorphisms that modify drug sensitivity. Antimicrob Agents Chemother 2016; 60:5649–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Somé AF, Zongo I, Compaoré YD et al. . Selection of drug resistance-mediating Plasmodium falciparum genetic polymorphisms by seasonal malaria chemoprevention in Burkina Faso. Antimicrob Agents Chemother 2014; 58:3660–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Somé AF, Séré YY, Dokomajilar C et al. . Selection of known Plasmodium falciparum resistance-mediating polymorphisms by artemether-lumefantrine and amodiaquine-sulfadoxine-pyrimethamine but not dihydroartemisinin-piperaquine in Burkina Faso. Antimicrob Agents Chemother 2010; 54:1949–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baraka V, Tinto H, Valea I et al. . In vivo selection of Plasmodium falciparum Pfcrt and Pfmdr1 variants by artemether-lumefantrine and dihydroartemisinin-piperaquine in Burkina Faso. Antimicrob Agents Chemother 2015; 59:734–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fairhurst RM. Understanding artemisinin-resistant malaria: what a difference a year makes. Curr Opin Infect Dis 2015; 28:417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Amaratunga C, Lim P, Suon S et al. . Dihydroartemisinin-piperaquine resistance in Plasmodium falciparum malaria in Cambodia: a multisite prospective cohort study. Lancet Infect Dis 2016; 16:357–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ariey F, Witkowski B, Amaratunga C et al. . A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 2014; 505:50–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ashley EA, Dhorda M, Fairhurst RM et al. ; Tracking Resistance to Artemisinin Collaboration (TRAC) Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 2014; 371:411–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Witkowski B, Duru V, Khim N et al. . A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: a phenotype-genotype association study. Lancet Infect Dis 2017; 17:174–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Amato R, Lim P, Miotto O et al. . Genetic markers associated with dihydroartemisinin-piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype-phenotype association study. Lancet Infect Dis 2017; 17:164–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kamya MR, Yeka A, Bukirwa H et al. . Artemether-lumefantrine versus dihydroartemisinin-piperaquine for treatment of malaria: a randomized trial. PLoS Clin Trials 2007; 2:e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yeka A, Kigozi R, Conrad MD et al. . Artesunate/amodiaquine versus artemether/lumefantrine for the treatment of uncomplicated malaria in Uganda: a randomized trial. J Infect Dis 2016; 213:1134–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Conrad MD, Bigira V, Kapisi J et al. . Polymorphisms in K13 and falcipain-2 associated with artemisinin resistance are not prevalent in Plasmodium falciparum isolated from Ugandan children. PLoS One 2014; 9:e105690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cooper RA, Conrad MD, Watson QD et al. . Lack of artemisinin resistance in Plasmodium falciparum in Uganda based on parasitological and molecular assays. Antimicrob Agents Chemother 2015; 59:5061–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Plowe CV, Djimde A, Bouare M, Doumbo O, Wellems TE. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am J Trop Med Hyg 1995; 52:565–8. [DOI] [PubMed] [Google Scholar]
- 34. Hopkins H, González IJ, Polley SD et al. . Highly sensitive detection of malaria parasitemia in a malaria-endemic setting: performance of a new loop-mediated isothermal amplification kit in a remote clinic in Uganda. J Infect Dis 2013; 208:645–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. LeClair NP, Conrad MD, Baliraine FN, Nsanzabana C, Nsobya SL, Rosenthal PJ. Optimization of a ligase detection reaction-fluorescent microsphere assay for characterization of resistance-mediating polymorphisms in African samples of Plasmodium falciparum. J Clin Microbiol 2013; 51:2564–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Conrad MD, Mota D, Musiime A et al. . Comparative prevalence of Plasmodium falciparum resistance-associated genetic polymorphisms in parasites infecting humans and mosquitoes in Uganda. Am J Trop Med Hyg 2017. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Price RN, Uhlemann AC, Brockman A et al. . Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet 2004; 364:438–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kjellin LL, Dorsey G, Rosenthal PJ, Aweeka F, Huang L. Determination of the antimalarial drug piperaquine in small volume pediatric plasma samples by LC-MS/MS. Bioanalysis 2014; 6:3081–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kajubi R, Huang L, Jagannathan P et al. . Antiretroviral therapy with efavirenz accentuates pregnancy-associated reduction of dihydroartemisinin-piperaquine exposure during malaria chemoprevention. Clin Pharmacol Ther 2017; 102:520–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ménard D, Khim N, Beghain J et al. ; KARMA Consortium A worldwide map of Plasmodium falciparum K13-propeller polymorphisms. N Engl J Med 2016; 374:2453–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Holmgren G, Hamrin J, Svärd J, Mårtensson A, Gil JP, Björkman A. Selection of pfmdr1 mutations after amodiaquine monotherapy and amodiaquine plus artemisinin combination therapy in East Africa. Infect Genet Evol 2007; 7:562–9. [DOI] [PubMed] [Google Scholar]
- 42. Eyase FL, Akala HM, Ingasia L et al. . The role of Pfmdr1 and Pfcrt in changing chloroquine, amodiaquine, mefloquine and lumefantrine susceptibility in western-Kenya P. falciparum samples during 2008–2011. PLoS One 2013; 8:e64299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nsobya SL, Dokomajilar C, Joloba M, Dorsey G, Rosenthal PJ. Resistance-mediating Plasmodium falciparum pfcrt and pfmdr1 alleles after treatment with artesunate-amodiaquine in Uganda. Antimicrob Agents Chemother 2007; 51:3023–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Humphreys GS, Merinopoulos I, Ahmed J et al. . Amodiaquine and artemether-lumefantrine select distinct alleles of the Plasmodium falciparum mdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrob Agents Chemother 2007; 51:991–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sisowath C, Strömberg J, Mårtensson A et al. . In vivo selection of Plasmodium falciparum pfmdr1 86N coding alleles by artemether-lumefantrine (Coartem). J Infect Dis 2005; 191:1014–7. [DOI] [PubMed] [Google Scholar]
- 46. Zongo I, Dorsey G, Rouamba N et al. . Artemether-lumefantrine versus amodiaquine plus sulfadoxine-pyrimethamine for uncomplicated falciparum malaria in Burkina Faso: a randomised non-inferiority trial. Lancet 2007; 369:491–8. [DOI] [PubMed] [Google Scholar]
- 47. Baliraine FN, Rosenthal PJ. Prolonged selection of pfmdr1 polymorphisms after treatment of falciparum malaria with artemether-lumefantrine in Uganda. J Infect Dis 2011; 204:1120–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Veiga MI, Dhingra SK, Henrich PP et al. . Globally prevalent PfMDR1 mutations modulate Plasmodium falciparum susceptibility to artemisinin-based combination therapies. Nat Commun 2016; 7:11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dorsey G, Dokomajilar C, Kiggundu M, Staedke SG, Kamya MR, Rosenthal PJ. Principal role of dihydropteroate synthase mutations in mediating resistance to sulfadoxine-pyrimethamine in single-drug and combination therapy of uncomplicated malaria in Uganda. Am J Trop Med Hyg 2004; 71:758–63. [PubMed] [Google Scholar]
- 50. Bigira V, Kapisi J, Clark TD et al. . Protective efficacy and safety of three antimalarial regimens for the prevention of malaria in young Ugandan children: a randomized controlled trial. PLoS Med 2014; 11:e1001689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.