A correlation between the conformation of the gating residues and the size and shape of the bound compound suggests a metabolite-driven mechanism for transport across the carboxysome shell.
Keywords: BMC domain, carboxysome, β-cyanobacteria, gated transport, microcompartment, shell protein
Abstract
Cyanobacterial CO2 fixation is promoted by encapsulating and co-localizing the CO2-fixing enzymes within a protein shell, the carboxysome. A key feature of the carboxysome is its ability to control selectively the flux of metabolites in and out of the shell. The β-carboxysome shell protein CcmP has been shown to form a double layer of pseudohexamers with a relatively large central pore (~13 Å diameter), which may allow passage of larger metabolites such as the substrate for CO2 fixation, ribulose 1,5-bisphosphate, through the shell. Here we describe two crystal structures, at 1.45 Å and 1.65 Å resolution, of CcmP from Synechococcus elongatus PCC7942 (SeCcmP). The central pore of CcmP is open or closed at its ends, depending on the conformation of two conserved residues, Glu69 and Arg70. The presence of glycerol resulted in a pore that is open at one end and closed at the opposite end. When glycerol was omitted, both ends of the barrel became closed. A binding pocket at the interior of the barrel featured residual density with distinct differences in size and shape depending on the conformation, open or closed, of the central pore of SeCcmP, suggestive of a metabolite-driven mechanism for the gating of the pore.
Introduction
Carboxysomes are bacterial protein-bound microcompartments with the function to increase the concentration of carbon dioxide (CO2) close to the carbon-fixing enzyme Rubisco (ribulose-1,5-bisphosphate carboxylase/oxygenase). CO2 is assimilated by the Rubisco-catalysed carboxylation of ribulose 1,5-bisphosphate (RuBP) to produce two molecules of 3-phosphoglycerate (3PGA). Rubisco is characterized by a low carboxylation turnover rate and is further slowed by a competing oxygenase activity. Cyanobacteria are highly productive organisms on a global scale (Field et al., 1998), but their efficiency is impeded by slow CO2 diffusion and a low CO2 concentration in aqueous environments. To overcome this crippling condition, cyanobacteria and some chemoautotrophic bacteria have evolved carboxysomes, which encapsulate Rubisco and carbonic anhydrase in a semi-permeable icosahedral protein shell. HCO3− is actively pumped into the cell and enters the carboxysome where it is converted to CO2 by the activity of carbonic anhydrase (reviewed in Price et al., 2008). Positively charged channels in the protein shell are believed to allow entry of the charged HCO3− molecule into the carboxysome but to prevent diffusion of CO2 back into the cytosol, thereby creating an environment with an elevated CO2 concentration near Rubisco (Dou et al., 2008).
Two different types of carboxysomes occur, named α and β, based on gene arrangement and the form of encapsulated Rubisco (reviewed in Rae et al., 2013). In α-carboxysomes, the cso operon codes for Form 1A Rubisco, shell proteins, β-carbonic anhydrase, and a protein suggested to have an essential role in α-carboxysome assembly (CsoS2; Cai et al., 2015). β-Carboxysomes instead contain Form1B Rubisco and a γ-carbonic anhydrase analogue, and their shell proteins are encoded by the more scattered ccm genes (Tanaka et al., 2008). The shell proteins in the two types of carboxysomes are orthologues and share a common fold. In the present models of bacterial microcompartments (Tanaka et al., 2008), the facets are built up by several paralogues containing one or two copies of the bacterial microcompartment (BMC) domain (PF00936; Punta et al., 2012) forming hexamers, or pseudohexamers in the case of the tandem BMC domains (Cai et al., 2013). The less abundant pentamer-forming proteins (PF03319) are believed to close the vertices of the icosahedral compartment (Tanaka et al., 2008; Wheatley et al., 2013) and prevent diffusion of CO2 from the carboxysome (Cai et al., 2009). Analysis of knockouts of the ccm shell proteins in Synechococcus elongatus PCC 7942 show that inactivation of the genes coding for the most abundant shell proteins, CcmK2, the tandem BMC domain protein CcmO, or the pentameric vertex protein CcmL, generate high-CO2-requiring mutants (Rae et al., 2012). The double inactivation mutation of ccmK3 and ccmK4 resulted in normal appearance of carboxysomes but functional deficiency under low CO2 conditions (Rae et al., 2012).
CcmP is a less abundant and recently identified shell protein (Cai et al., 2012, 2013) that comprises two BMC domains. The overall fold of CcmP is similar to that of the other facet shell protein, CcmK, but the connectivity between the secondary structure elements varies due to rearrangements in the genome that result in a circularly permuted protein (Crowley et al., 2008). The two-domain CcmP forms a trimer resembling the hexamer of the CcmK proteins. In addition, two trimeric CcmP rings appear to be stacked on top of each other and form a barrel (Cai et al., 2013). Three-dimensional crystal structures of CcmP (Cai et al., 2013) and its orthologue in α-carboxysomes, CsoS1D (Klein et al., 2009), show that the pores at the three-fold symmetry axis may be either open or closed. The open central pore (~13 Å diameter) of these permuted tandem shell proteins is wider than those of the CcmK proteins (reviewed in Yeates et al., 2011, 2013; Kinney et al., 2011) and may potentially allow larger metabolites, such as RuBP and 3PGA, to pass through. The alternative conformations were suggested to facilitate the selective transport of metabolites through the protein shell (Klein et al., 2009; Cai et al., 2013). However, specific details on the transport process, in terms of the identity of the metabolites transported or the interactions that might trigger the conformational changes leading to the shuttling of metabolites, are presently lacking.
Here we describe two high-resolution structures of CcmP from S. elongatus PCC 7942 (named SeCcmP_P213 and SeCcmP_I213) co-crystallized with RuBP, the substrate of Rubisco. SeCcmP_P213 crystals were obtained from a protein solution containing glycerol, whereas SeCcmP_I213 crystals were obtained from protein produced free of glycerol throughout, including protein expression, purification, and crystallization. Both structures consist of a barrel formed by two layers of a SeCcmP trimer with a central pore, but the two structures show distinct conformational states. In SeCcmP_P213, the pore is open at one end and closed at the opposite end, whereas in SeCcmP_I213, the pore is closed at both ends. Electron density for a bound ligand is found in a binding pocket which is located between the two BMC domains within the CcmP monomer.
Materials and methods
DNA cloning
The sequence of the gene (Synpcc7942_0520; UniProt id Q31QW7) coding for CcmP was amplified from S. elongatus PCC 7942 chromosomal DNA using Phusion High-Fidelity DNA PCR Master Mix (Thermo Scientific) with forward primer 5'- AGCGGCTCTTCAATGGGCGTTGAGCTGCGCAGT-3' and reverse primer 5'- AGCGGCTCTTCTCCCCTCCCGCGAGCGA TCGC-3'. The gene was cloned into pENTRY-IBA51 using a StarGate® Combi Entry Cloning Set (IBA Life Sciences) and, after confirming the correct sequence, subsequently cloned into pPSG-IBA33 using Stargate® Transfer Reagent Set (IBA Life Sciences), resulting in plasmid pPSG-IBA33_0520.
Protein expression
pPSG-IBA33_0520 was transformed into Escherichia coli strain BL21-AI (Life Technologies) for heterologous protein expression. Escherichia coli was grown in 2× YT medium (1.6% tryptone, 1% yeast extract, 0.5% NaCl) containing 100 μg ml−1 ampicillin until OD600=0.7 was reached. Cells were cooled in an ice bath, and protein expression was induced by adding 0.02% l-arabinose. Cells were harvested after further cultivation for 16 h at 291 K.
Protein purification
The SeCcmP protein was purified using a three-step protocol comprising affinity, ion exchange, and size exclusion chromatographies at room temperature using an NGC purifier system (BioRad). Cells were resuspended in binding buffer (20 mM Bis-Tris propane pH 8, 500 mM NaCl, 20 mM imidazole, 10% glycerol) and lysed by sonication. After clearing the lysate using centrifugation at 40 000 g (JA-25.50, Beckman Coulter), the supernatant was applied to a HisTrap™ HP column (5 ml, GE Healthcare), washed thoroughly with washing buffer (20 mM Bis-Tris propane pH 8, 500 mM NaCl, 65 mM imidazole, 10% glycerol), and eluted with a gradient of 65–500 mM imidazole in the same buffer. After lowering the salt concentration by diluting the eluate in buffer A (20 mM Bis-Tris propane pH 9.5, 10% glycerol), the sample was applied to a MonoQ column (GE Healthcare) and eluted with a 0–50% gradient of buffer B (20 mM Bis-Tris propane pH 9.5, 1 M NaCl, 10% glycerol). Size exclusion chromatography was carried out using a HiLoad 26/60 Superdex 200 column (GE Healthcare), equilibrated with buffer C (20 mM Bis-Tris propane pH 8, 100 mM NaCl, 10% glycerol) and a flow rate of 2 ml min−1. Glycerol, which was initially present in all buffers as a stabilizing agent, was found to bind to the protein (see below). In subsequent rounds of purification, glycerol was therefore excluded from all steps in the purification protocol. The purified SeCcmP was concentrated to a final concentration of ~3 mg ml−1 (Vivaspin, Sartorius Stedim Biotech).
Protein crystallization and data collection
SeCcmP (3 mg ml−1 in 20 mM Bis-Tris propane, 50 mM NaCl, 10 mM glycerol) was co-crystallized with 50 mM RuBP at room temperature (293 K). Crystallization conditions were screened using commercial screens and a mosquito® Crystal robot (TTP Labtech) with 0.2 μl drop size. The best crystals were obtained in 0.2 M Li sulphate, 0.1 mM Tris–HCl pH 7.5, and 5% (w/v) polyethylene glycol (PEG) 4000 (Proplex Screen, Molecular Dimensions). These crystals belong to space group P213 with unit cell parameters a, b, c=107.3 Å, α, β, γ=90°, and two molecules per asymmetric unit, giving a VM of 2.1 Å3 Da−1 and 42% solvent content. Glycerol-free SeCcmP (3 mg ml−1 in 20 mM Bis-Tris propane, 50 mM NaCl) was crystallized at 293 K using the hanging drop vapour diffusion method. Crystals were obtained using a reservoir solution consisting of 0.1 mM Tris–HCl pH 7.0, 0.2 mM potassium thiocyanate, and 6% poly-γ-glutamic acid (Molecular Dimensions), with drops formed from 4 μl of protein solution, 4 μl of reservoir solution, and 1 μl of 1 M RuBP. Crystals appeared after a few days and belong to space group I213 with unit cell parameters a, b, c=178.7 Å, α, β, γ=90°, and three molecules per asymmetric unit. Prior to data collection, crystals were soaked in a reservoir solution containing cryoprotectant (20% glycerol), transferred into a nylon loop (Hampton Research), flash-cooled in liquid nitrogen, and maintained at 100 K for data collection. When glycerol was omitted, 25% ethylene glycol was used as cryoprotectant.
X-ray diffraction data were collected at 100 K on a Pilatus 6M-F detector at beam line ID29 of the European Synchrotron Radiation Facility (ESRF) in Grenoble, France.
Structure determination and refinement
Diffraction data were processed using XDS (Kabsch, 2010) and scaled with AIMLESS (Evans, 2006). Phases were obtained by molecular replacement in PHASER (McCoy et al., 2007) using one monomer of the SeCcmP X-ray crystal structure (PDB code 4HT5) as a search model for the SeCcmP_P213 and a single BMC domain for the SeCcmP_I213 structure. ZANUDA in the CCP4 suite (Winn et al., 2011) was used to verify the space group and to improve the model. Refinement was performed with BUSTER (Bricogne et al., 2011) alternated with manual rebuilding in O (Jones et al., 1991). Solvent molecules were added using the water insertion command in BUSTER. Assignment of water molecules was made based on peak heights of residual electron density, hydrogen-bonding patterns, and B-factors. Analysis of ligand occupancy was performed with Phenix (Adams et al., 2010). Data collection and refinement statistics are presented in Table 1.
Table 1.
Data collection and refinement statistics for SeCcmP
Values in parentheses are for the outer resolution shell.
| SeCcmP_P2 1 3 | SeCcmP_I2 1 3 | |
|---|---|---|
| Protein Data Bank id | 5LT5 | 5LSR |
| Data collection | ||
| Beam line | ID29 | ID29 |
| Wavelength (Å) | 0.97239 | 0.97625 |
| Space group | P213 | I213 |
| Unit cell parameters (Å) | a, b, c=107.2; α, β, γ=90° | a, b, c=178.7; α, β, γ=90° |
| V M (Å3 Da–1) | 2.1 | 3.4 |
| Solvent content (%) | 42 | 64 |
| Resolution (Å) | 47.9–1.45 (1.47–1.45) | 44.7–1.65 (1.68–1.65) |
| No. of observations | 721 918 (34 433) | 2 272 675 (98 426) |
| No. of unique reflections | 72 625 (3579) | 113 271 (5592) |
| R meas a | 0.061 (0.943) | 0.095 (1.445) |
| <I/(σI)> | 21.7 (2.6) | 18.9 (2.3) |
| Completeness (%) | 100.0 (100.0) | 100.0 (100.0) |
| Multiplicity | 9.9 (9.6) | 20.1 (17.6) |
| Refinement | ||
| Resolution range (Å) | 47.9–1.45 (1.49–1.45) | 44.7–1.65 (1.69–1.65) |
| No. of reflections | 72 565 | 113 268 |
| R work b | 0.165 | 0.181 |
| R free c | 0.184 | 0.191 |
| No. of atoms | ||
| Protein | 3246 | 4774 |
| Ligands | 13 | 6 |
| Waters | 325 | 357 |
| Average B-values (Å2) | ||
| Estimated from Wilson plot | 19.2 | 29.2 |
| Protein | 20.1 | 32.1 |
| Ligands | 26.1 | 37.2 |
| Waters | 33.1 | 39.8 |
| Rms deviations from ideal values | ||
| Bond lengths (Å) | 0.010 | 0.010 |
| Bond angles (°) | 1.05 | 1.03 |
| Ramachandran analysisd | ||
| Outliers (%) | 0 | 0 |
a R meas=∑h ∑l (nh/nh−1)1/2|Ihl−<Ih>|/∑h ∑l<Ih> (Evans, 2006; Evans and Murshudov, 2013).
b R work=∑hkl||Fo|−|Fc||/∑hkl |Fo| where Fo and Fc are the observed and calculated structure factor amplitudes, respectively.
c R free calculated from a randomly chosen 5% of all unique reflections.
dFrom MolProbity (Chen et al., 2010).
Structure analysis
The structures were evaluated using wwPDB Validation Server (Berman et al., 2003). Electron density fit was analysed using Coot (Emsley et al., 2010). The DALI server (Holm and Rosenström, 2010) was used to identify similar structures in the PDB, and the lsq commands in O (Jones et al., 1991) were used for comparison of SeCcmP_P213 chain B with hits in the DALI server.
The figures were created with PyMOL (Version 1.6.0.0, Schrödinger, LLC).
Results
Structure solution
SeCcmP was expressed in E. coli with a C-terminal 6×histidine-tag. Crystallization could only be achieved at a relatively low protein concentration (3 mg ml−1) and using freshly purified SeCcmP because protein precipitation occurred at higher concentration and lower (cold room) temperatures. Crystals appeared after incubation at 293 K for a few days and belong to the cubic space group P213 with two monomers per asymmetric unit. The phases were obtained using the molecular replacement method. Data to 1.45 Å resolution were included in the processing and refinement of the final model, SeCcmP_P213. When glycerol was omitted from the purification and crystallization protocols, SeCcmP crystallized in space group I213 with three molecules per asymmetric unit. The resulting model was refined to 1.65 Å resolution and is referred to as SeCcmP_I213. Statistics for data collection and refinement are presented in Table 1.
Overall structures
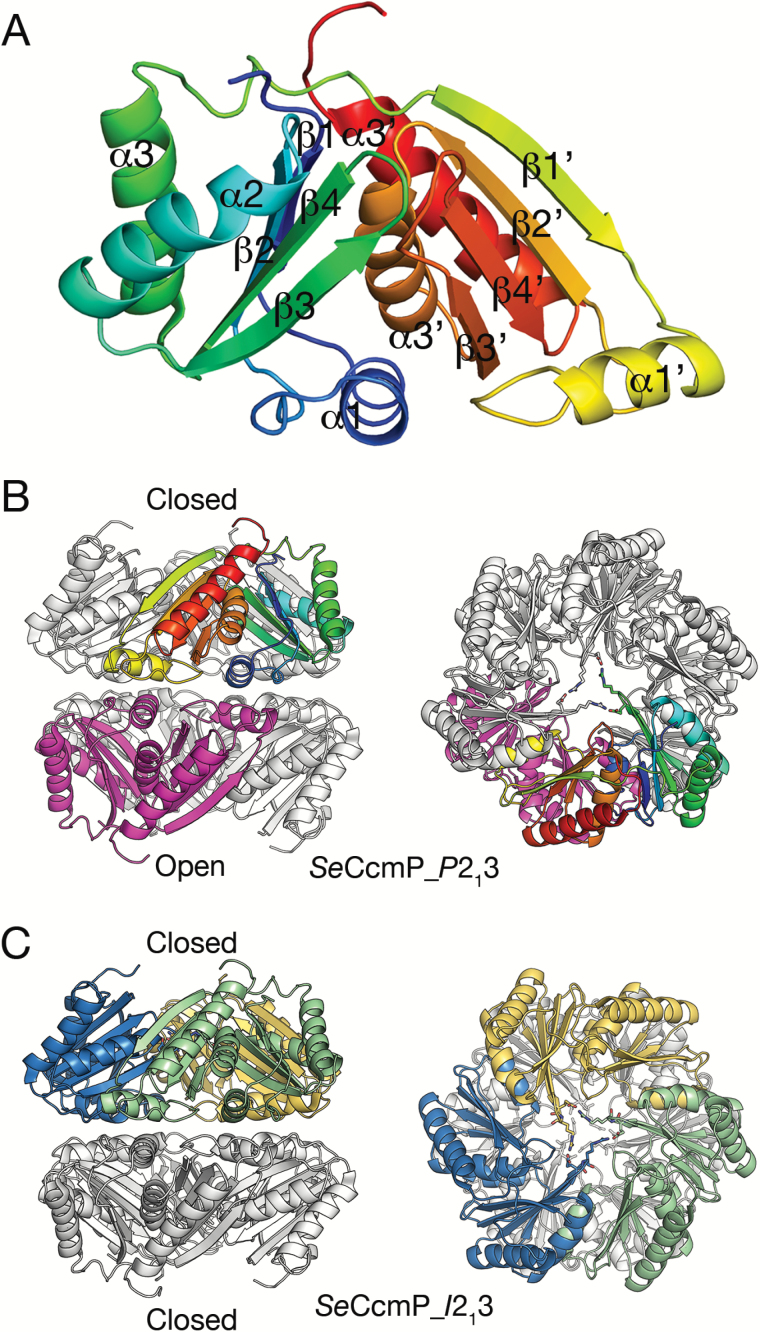
The SeCcmP monomer contains two BMC domains in tandem, the N- and C-BMC domains, each consisting of an α+β fold with three α-helices and a single β-sheet formed by four antiparallel β-strands (Fig. 1A). The SeCcmP_P213 structure contains two polypeptide chains in the asymmetric unit, chain A with 204 and chain B with 205 amino acids out of a total of 213 (residues Val3 or Gly2, respectively, to His206). In addition we identified 325 water molecules, two glycerol molecules, and a chloride ion, which is located between two hexamers in the crystal lattice. Seven amino acids in the C-terminus of SeCcmP, preceding the histidine-tag in the construct, were disordered and could not be modelled in the structure because of lack of interpretable electron density. The SeCcmP_I213 structure contains three monomers per asymmetric unit consisting of amino acids Val3 (chains A and C) or Gly2 (chain B) to Pro207. The stereochemistry of SeCcmP_P213 and SeCcmP_I213 is excellent, with no Ramachandran outliers and 99% and 98%, respectively, of the residues in the most favoured region. Two residues (Gly69 and Tyr72) in SeCcmP_P213 adopted more unusual conformations (identified as non-rotamer side chains in the wwPDB validation report; Berman et al., 2003). Tyr72 in chain A is located in a flexible region and displays weak density. Glu69 in chain B has well-defined density and is one of the two principal pore-closing residues (see below). In the SeCcmP_I213 structure, Glu69 and Arg129 adopt unusual (non-rotamer) side chain conformations in all three monomers of the asymmetric unit.
Fig. 1.
Overall structure of SeCcmP and arrangement of the six monomers in the asymmetric unit. (A) The SeCcmP monomer, consisting of two BMC domains, rainbow coloured from blue at the N-terminus to red at the C-terminus. (B) The SeCcmP_P213 structure with two monomers per asymmetric unit. One monomer (rainbow coloured) has the gating residues in a closed conformation, the second monomer (magenta) has the gating residues disordered. The biologically relevant hexamer is generated by adding symmetry-related monomers (grey). (C) The SeCcmP_I213 structure with three molecules per asymmetric unit (blue, yellow, and green) and the gating residues in a closed conformation. Crystal symmetry-related monomers in the biologically relevant hexamer are coloured grey. The two views in (B) and (C) are related by a 90 ° rotation with respect to the horizontal axis.
The overall SeCcmP quaternary structure is a barrel made up by six SeCcmP monomers in two trimeric rings stacked on top of each other, with the same faces interacting (Fig. 1B). The two ends of the barrel feature pores that may be either open or closed depending on the conformations of residues lining the apertures at the three-fold axis. In particular, two highly conserved residues, Glu69 and Arg70, adopt different conformations in the two monomers in the asymmetric unit of the SeCcmP_P213 structure (Fig. 1B), resulting in a shift of the polypeptide backbone around these gating residues. In the SeCcmP_P213 structure, one monomer (chain A) forms a trimeric ring with an open central pore on the three-fold symmetry axis, whereas the second monomer (chain B) forms a ring with a closed central pore. This arrangement of the two rings leads to formation of a barrel with one end open and the other closed. The three monomers in the SeCcmP_I213 structure form a ring with a closed end. Addition of a second trimeric ring related by crystal symmetry results in a barrel with both ends closed (Fig. 1C).
In several places, additional electron density was observed that could not be assigned to protein. These densities occurred reproducibly in multiple structures, but unambiguous interpretation was not possible in most cases. The difficulties in interpreting the extra electron density might be explained by unspecific binding and the propensity of CcmP to interact with other shell proteins.
Open and closed conformations
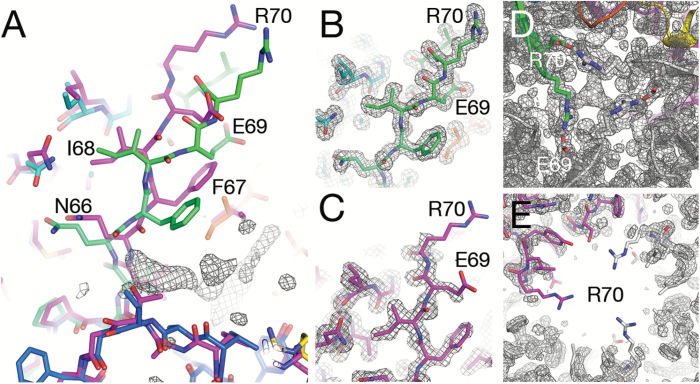
The two molecules in the SeCcmP_P213 structure that represent the open and closed conformations superimpose well overall (rms deviations of 0.739 Å for 203 equivalent Cα atoms out of a total of 204 amino acids). This superposition uses the default cut-off limit for equivalent Cα atoms of 3.8 Å; as a result, Arg70 is the only Cα atom that does not fall within the cut-off limit. In fact, the largest differences between the open and closed conformations of SeCcmP_P213 are observed for residues 66–72 in a β-hairpin structure comprising the gating residue Arg70 (Gln66 1.4 Å, Phe67 1.5 Å, Ile68 1.1 Å, Glu69 3.4 Å, Arg70 5.4 Å, Leu71 3.0 Å, and Tyr72 1.4 Å). In the closed state, there is an interaction between Glu69 and Arg70 from two adjacent protein molecules (Fig. 1B), which effectively closes the pore. In the transition from the closed to open state, the side chain of Arg70 changes its conformation and interacts with Glu69 in the monomer on the opposite side. Despite the large movement of Arg70 (>12 Å movement of its guanidinum group), the structure of the β-hairpin is essentially intact and shifted such that the hydrogen bonds between β-strands β3 and β4 including the interactions between Glu69 and Tyr72 and between Phe67 and Leu74 are kept intact. Besides the changes in position of the gating residues Glu69 and Arg70, the side chain of Phe67 displays a large positional shift (Fig. 2A). Weaker electron density is observed for residues 67–72 in the open conformation compared with the corresponding residues in the closed conformation, indicating a higher flexibility of this region for the open state (Fig. 2B–E). Most notable is the lack of density for the carboxyl group of Glu69 and nearly all of the side chain of Arg70 in monomer A, which forms the open pore. The same tendency, but with only slightly lower density fit for the open state, is observed in loop A171–A175 of the C-BMC domain, which corresponds to the N-BMC loop containing Glu69 and Arg70. The side chain of Ile172 assumes different conformations in the two states; this shift appears to be coupled to the large displacement of Phe67, with which it forms hydrophobic interactions.
Fig. 2.
Gating residues Glu69 and Arg70 in open and closed conformations in the two monomers of SeCcmP_P213. (A) Superposition of the two monomers of SeCcmP_P213. Monomer A (magenta) in an open conformation and monomer B (rainbow coloured) in a closed conformation. The electron density of the ligand extends from the binding pocket of monomer B toward the carbonyl oxygen of Gln66. (B) Well-defined electron density for the gating residues in the closed conformation (monomer B). (C) Electron density for the gating residues in the open conformation (monomer A). (D) The closed end of the pore seen down the three-fold symmetry axis. (E) The open end of the pore seen down the three-fold symmetry axis. Electron densities are from 2Fo–Fc maps contoured at the 1 σ level.
The three monomers in SeCcmP_I213 form a pseudohexameric ring with a closed pore (Fig. 1C). Crystallographic symmetry-related monomers make up the second ring, resulting in a barrel that is closed at both ends. The three molecules in the structure superimpose with low rms deviations (0.122–0.140 Å) when including 205 Cα atoms. Superposition of the two different structures shows that SeCcmP_I213 (chain A) is most similar to the closed conformation chain of SeCcmP_P213 (chain B, rms deviation 0.333 Å). The open conformation SeCcmP_P213 chain A superimposes on SeCcmP_I213 (chain A) with an rms deviation of 0.701 Å.
Binding pocket at the interface of the N- and C-BMC domains
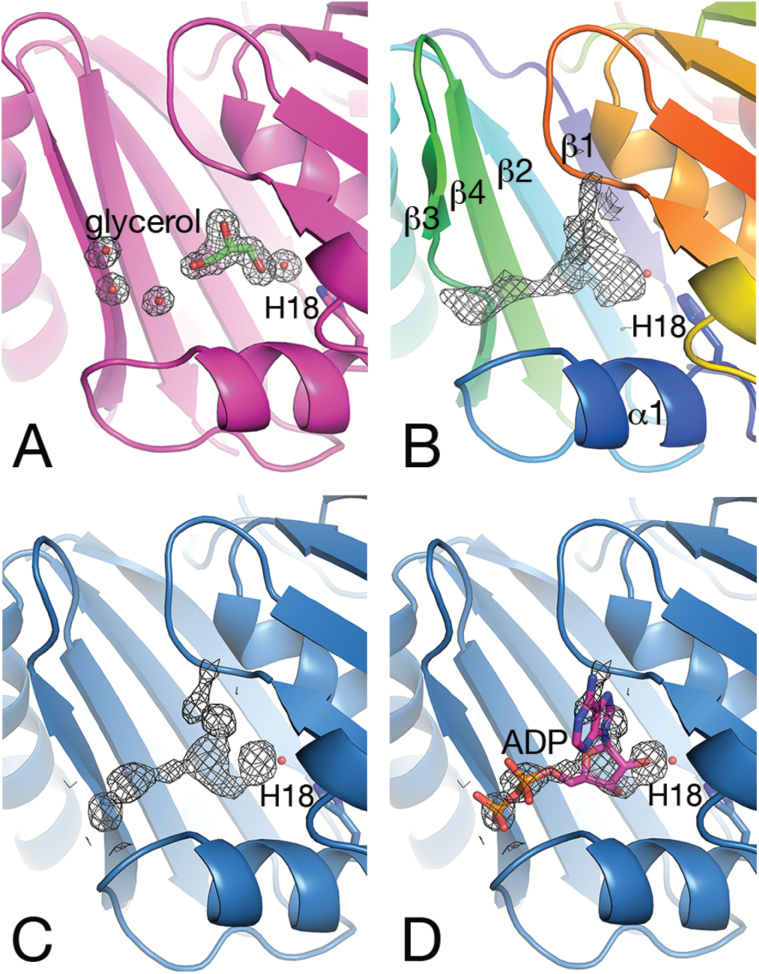
SeCcmP contains a binding pocket which is located between the N- and C-BMC domains (Cai et al., 2013). The hexameric structure of SeCcmP thus potentially gives rise to six binding pockets that line the interior of the barrel. The pocket is confined by residues from helix α1 and strand β3 from the N-BMC domain and strand β3' of the C-BMC domain. In particular, a conserved histidine residue (His18) is strategically located in the innermost part of the binding pocket. After careful refinement of the protein, residual electron density was apparent in the pockets. The shape of the density showed distinct differences depending on the conformation, open or closed, of the SeCcmP molecule. Well-defined electron density for solvent molecules (glycerol and a water molecule) was observed in the pocket of the SeCcmP_P213 monomer forming the open pore (Fig. 3A). The water molecule is at hydrogen-bonding distance to the ε2-nitrogen atom of His18 and one of the hydroxyl oxygen atoms of glycerol. In the pocket of the second molecule in the asymmetric unit, with the gating loop in a closed conformation, the interpretation of the electron density was more ambiguous (Fig. 3B). Electron density potentially originating from a water molecule is situated 2.8 Å from the ε2-nitrogen of the His18 imidazole ring in a position corresponding to the location of the solvent molecule in the pocket of the open pore molecule. Adjacent to this solvent site, an elongated electron density was observed that expands in two directions (Fig. 3B). The density is bounded on one side by the backbone carbonyl group of Val169 and the backbone amide nitrogen atoms of Ala171 and Ile172. Extending toward the N-BMC domain, a strong patch of electron density is positioned 3.6 Å from the backbone amide nitrogen of Gln66 in the β-strand preceding the loop that determines the open or closed state of the pore (see below; Fig. 2A). The peak is located 10 Å away from the water molecule that interacts with His18. This is a reasonable distance between the two phosphate groups of the substrate RuBP (Duff et al., 2000) which was present during crystallization. Considering that the density is strongest at the centre, the binding of a mixture of compounds in the closed monomers B of the CcmP_P213 crystal may be considered, which manifests itself as a complex and smeared density in the electron density maps.
Fig. 3.
Electron density 2Fo–Fc maps in the binding pocket contoured at the 1 σ level. The side chain of His18 and the adjacent water molecule are shown as a stick representation and a red sphere, respectively. (A) Monomer A of SeCcmP_P213 (magenta) with the gating residues in an open conformation binds glycerol and a water molecule in the binding pocket between the N- and C-BMC domains. (B) Monomer B of SeCcmP_P213 (rainbow coloured from blue in the N-terminus to red in the C-terminus) with an extended electron density and the gating residues in a closed conformation. (C) Monomer A of SeCcmP_I213 (blue) with the gating residues in a closed conformation displaying a similar density as in (B). (D) ADP modelled into the density in (C).
The discovery of well-defined density for a glycerol molecule in the binding pocket of the open-pore monomer prompted a revision of the purification and crystallization protocols omitting glycerol. The resulting protein crystallized in space group I213 in the presence of thiocyanate and assumed a closed conformation only. Elongated electron densities of a similar shape were also observed in all three binding pockets of the closed conformation SeCcmP_I213 structure (Fig. 3C). In monomer A, the density displays two strong peaks 2.8 Å apart, with the more distant peak 3.3 Å away from the backbone amide nitrogen of Gln66. The electron density in the cleft is not a chance observation; similarly shaped densities were consistently observed in all data sets analysed in this project (unpublished results). Co-crystallization and soaking experiments in order to elucidate the identity of the bound ligands have so far been inconclusive. Bicarbonate and 3PGA used in co-crystallization experiments were not detected and may have been outcompeted by the cryoprotectant.
Model building of RuBP into the difference density did not give a conclusive result. Given the shape of the density (resembling a boomerang), we identified ADP as a possible candidate to fit the density, with the ribose ring at hydrogen-bonding distance to the water molecule, the two phosphate groups pointing toward the N-BMC domain, and the adenine ring contacting strand β3 of the C-BMC domain. Refinement of the structure with ADP present was not convincing, and the estimated occupancy of ADP was low (0.3–0.4). Weak density was present for the adenine ring, and the positioning of ADP resulted in too short contact distances to the protein in several places. However, some of the residues close to the ligand have dual conformations, indicating the presence of two subpopulations. Co-crystallization of ADP or ATP increased the occupancy slightly, but did not result in well-defined density for the ligand (unpublished results). Considering these uncertainties, we have opted not to include a ligand in the final model and conclude that the binding of a larger molecule, extending toward Thr27 and the backbone nitrogen of Gln66, coincides with the gating residues in a closed conformation, whereas the binding of a smaller molecule, corresponding to glycerol present in one of our crystallization conditions, coincides with an open pore.
Structural comparison with other BMC shell proteins
Two structures of SeCcmP are available in the PDB—a 2.51 Å resolution structure in space group P21212 and a low-resolution (3.3 Å) structure in space group P212121 (PDB codes 4HT5 and 4HT7, respectively; Cai et al., 2013). In the former structure, a solvent content of 70.7% and empty space between layers of well-defined SeCcmP hexamers indicate that not all molecules in the asymmetric unit had been located. The difficulties in placing molecules in the missing layer were suggested (Cai et al., 2013) to be caused by lattice-translocation disorder, as previously detected in CsoS1C (Tsai et al., 2009), but no analysis of native Patterson peaks or local intensity differences were presented to justify this proposal and no corrections were applied. The high R and Rfree factors for the P21212 structure may thus be a result of parts missing from the model, causing problems during refinement. The 3.3 Å resolution SeCcmP structure provides limited information due to the low resolution. In addition, two structures of the related CsoS1D from the α-carboxysome Prochlorococcus marinus subsp. pastoris str. CCMP1986 (Klein et al., 2009) are available in PDB; a well-refined SeMet-substituted structure to 2.3 Å resolution (PDB code 3F56) and a native structure to 2.2 Å resolution, which was solved from merohedrally twinned data with a twin factor of 0.45 and a very low fit (23.2% RSRZ outliers) to the density (PDB code 3FCH).
When comparing a single BMC domain (residues 3–103 or 104–206) of SeCcmP with other structures in the PDB using DALI, the more similar structure unexpectedly appears to be the SeMet-substituted structure of CsoS1D (Klein et al., 2009), and not the previously deposited SeCcmP structures (Cai et al., 2013). The same result is obtained using either the N-BMC or C-BMC domains as search models. The higher Z-score may be a result of both CsoS1D and SeCcmP_P213 being well-refined structures and may be less influenced by the lower sequence identity (48% compared with identical) in this case.
If the complete two-domain SeCcmP_P213 structure is used in the DALI search, SeCcmP structures are given the highest similarity scores followed by the P. marinus CsoS1D structures (Table 2). Surprisingly, the next structures in the list are PduT from Citrobacter freundii and a PduT analogue from Desulfitbacterium hafniense. PduT is a shell protein with tandem repeats. The two domains are not circularly permuted in this case, but feature a duplication of a canonical domain. A lower similarity score is obtained for PduT compared with the other proteins if single domains are used for the comparision. Slightly lower Z-scores were observed for PduU and EutS (9.2 and 10.2, respectively); these are shell proteins that contain a single permuted BMC domain. Despite higher sequence identity, shell proteins with permuted tandem repeats get a lower Z-score, when the complete structures are compared, than the permuted single domain proteins (Table 2). The higher similarity to PduT when using the complete SeCcmP_P213 molecule as a search model indicates that the two domains of SeCcmP_P213 are arranged in a way more similar to PduT than to the other non-carboxysomal permuted tandem proteins.
Table 2.
Structure comparison of SeCcmP_P213 chain B (closed) with BMC shell proteins (permuted and/or duplicated) in the PDB
| Protein | Organism | PDB | No. of aaa | Z-score | Rmsd | No.b | No. idc |
|---|---|---|---|---|---|---|---|
| CcmP | Synechococcus elongatus PCC 7942 | 4HT5-Ad | 204 | 39.1 | 0.252 | 204 | 204 |
| CsoS1D | Prochlorococcus marinus MED4 | 3F56-Ae | 207 | 34.8 | 0.667 | 201 | 91 |
| PduT | Citrobacter freundii | 3PAC-Af | 183 | 11.1 | 1.608 | 90 | 19 |
| EutS | Escherichia coli K-12 | 3IA0-Eg | 111 | 10.2 | 1.236 | 88 | 17 |
| PduU | Salmonella enterica | 3CGI-Ch | 119 | 9.2 | 1.271 | 82 | 17 |
| EutL | Clostridium perfringens | 4U6I-Ci | 216 | 8.5 | 1.673 | 157 | 32 |
| EtuB | Clostridium kluyveri DSM 555 | 3IO0-Aj | 229 | 8.1 | 1.624 | 82 | 13 |
| GrpU | Clostridiales bacterium 1_7_47_FAA | 4OLO-Bk | 84 | 6.9 | 1.380 | 74 | 12 |
| PduB | Lactobacillus reuteri SD2112 | 4I61-Cl | 227 | 6.4 | 1.536 | 76 | 12 |
aNo. of aa, number of amino acids in the chain.
bNo., number of amino acids within the 3.8 Å cut-off.
cNo. id, number of identical residues within the cut-off.
dCai et al.(2013).
eKlein et al. (2009).
gTanaka et al. (2010).
hCrowley et al. (2008).
iThompson et al. (2014a).
jHeldt et al. (2009).
kThompson et al. (2014b).
lNot published.
Communication between binding sites
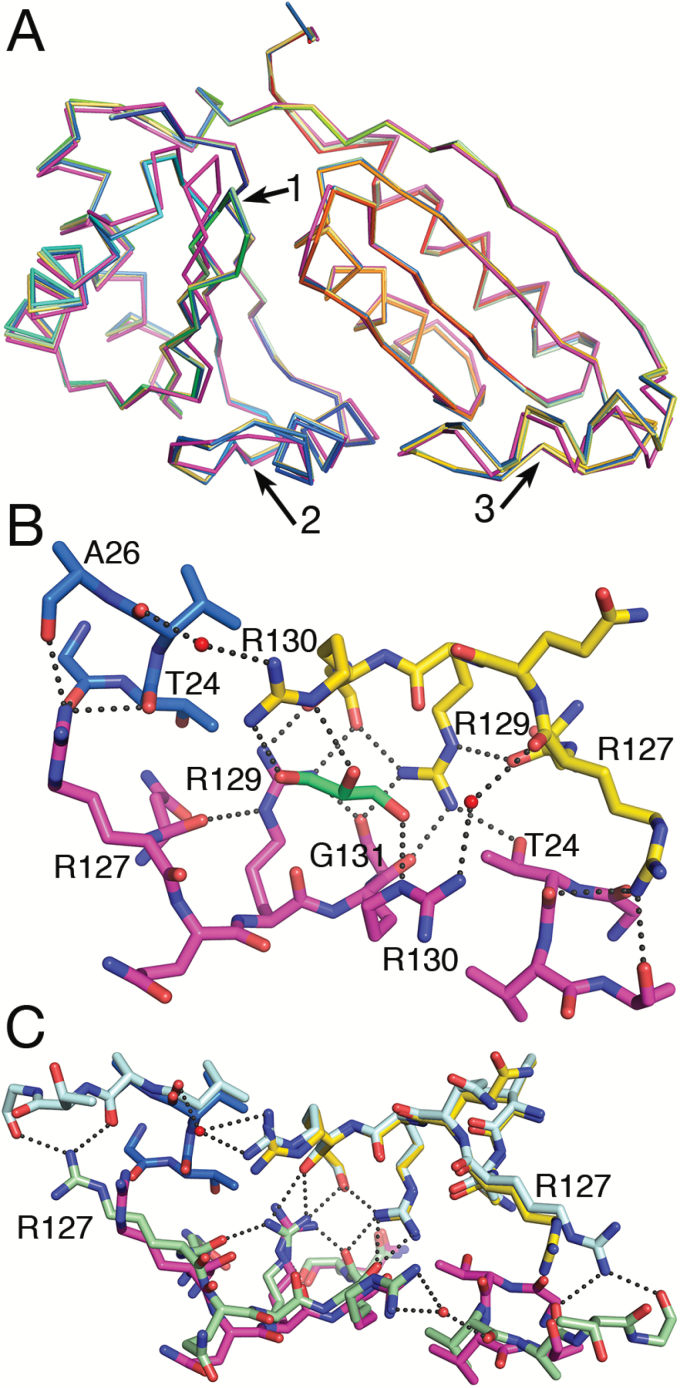
To date, three out of four available structures of the tandem BMC domain proteins CcmP and CsoS1D feature open and closed pores in the same barrel (Klein et al., 2009; Cai et al., 2013). This may suggest communication between the open and closed states such that one end of the barrel is closed (gating residues organized), while the other end is open (gating residues flexible). Such gating may be controlled by the binding of an extended molecule in one site making the binding in the other site less likely. To elucidate such interactions, we superposed SeCcmP_P213 chains A and B and SeCcmP_I213 chains B and C on SeCcmP_I213 chain A (Fig. 4A). The strongest electron density peaks of the potential ligand are positioned within contact distance to the main chain carbonyl group of Gln66 in strand β3 and the side chain of Thr27 in the loop following helix α1 in the N-BMC domain. The largest structural differences between the monomers that make up the open and closed conformers are localized to the gating loop containing Glu69 and Arg70, but we also observe distinct differences in helix α1 in the N-BMC domain and in the corresponding helix α1'-loop region of the C-BMC domain. The helix α1-loop region of the N-BMC domain of monomer B, featuring a closed central pore, interacts with Arg127 of the helix α1'-loop of the C-BMC domain in the trimeric ring with an open central pore (monomer A) via three hydrogen bonds to the backbone carbonyl groups of Gly23, Thr24, and Ala26. Arg127 in SeCcmP_I213 adopts a more extended conformation, compared with in SeCcmP_P213, and forms hydrogen bonds to the carbonyl groups of Ala26 and Gly28 (Fig. 4C). The two C-terminal helix α1-loop regions from the open and closed monomers are interacting symmetrically with each other via several hydrogen bond interactions between Arg129 in chains A/B and the backbone carbonyl groups of Arg130 and Gly131 in chains B/A (Fig. 4B). Arg129 also forms a hydrogen bond to the carbonyl group of Gly131 in the same polypeptide chain; this contact further stabilizes the interactions between the two helix–loop regions of the C-BMC domains. Thus Arg127 of the closed pore trimer may affect the helix–loop region of the open pore trimer through this network of hydrogen bonds.
Fig. 4.
Interactions between monomers in the CcmP hexamer. (A) Differences in the SeCcmP main chain backbone in the open and closed conformation shown by superimposing SeCcmP_P213 monomer A (magenta) and monomer B (rainbow) and SeCcmP_I213 monomer B (yellow) and monomer C (green) on SeCcmP_I213 monomer A (blue). The only monomer with the gating residues in an open conformation is SeCcmP_P213 chain A (magenta). Structural deviations of the backbone in open and closed conformations are observed in the gating loops region (arrow 1), in helix α1 and the following loop region of the N-BMC domain (arrow 2) and in the corresponding helix α1'-loop region in the C-BMC domain (arrow 3). (B) Interactions in the helix–loop region between the two molecules in the asymmetric unit of SeCcmP_P213 (monomer A magenta and monomer B rainbow coloured). Salt bridges between Arg127 and Arg129 to backbone carbonyl groups and solvent form a rigid network of interactions that may mediate changes in the binding site of one trimeric ring to the binding site in the second ring. (C) SeCcmP_I213 monomer A (light blue) superimposed on SeCcmP_P213 chain B (rainbow coloured), and SeCcmP_I213 monomer C (green) superimposed on SeCcmP_P213 monomer A (magenta). Note the differences in salt bridges formed by the side chains of Arg127 in the two structures.
In the CsoS1D and CcmP structures (Klein et al., 2009; Cai et al., 2013), which display open and closed conformations in the same barrel, Arg127, or the corresponding Arg178 in CsoS1D, is in an extended conformation in the open pore trimer and in a bent conformation in the closed pore trimer. The conflicting result seen for SeCcmP_P213 and SeCcmP_I213, where Arg127 is in an extended conformation in the closed/closed structure and in a bent conformation for the open/closed structure, may be explained by high concentration of molecules that bind to the protein during crystallization (Fig. 4C). In SeCcmP_P213, density corresponding to a glycerol molecule is present at the interface forming hydrogen bonds to Arg130 in both monomers A and B (Fig. 4B). SeCcmP_I213 crystallizes in the presence of thiocyanate. Densities interpreted as thiocyanate were observed close to Phe29 in monomers A and C. The binding of thiocyanate may impair Phe29 from assuming the two different conformations observed in the open and closed conformations of SeCcmP_P213.
Discussion
Structure determination of BMC shell proteins has proved to be a challenging task (Klein et al., 2009; Tsai et al., 2009; Cai et al., 2013; Thompson and Yeates, 2014). Non-crystallographic symmetry may give rise to twinning, and preference for packing in layers can result in disorder of entire layers and the inability to trace all the molecules present in the asymmetric unit. This has resulted in difficulties in refining the structures, yielding high R factors. In this context, our well-refined high-resolution SeCcmP structures add important information and provide models for further analysis.
Speculation that some carboxysomal shell proteins function as selective channels with gated pores to allow the passage of substrate or cofactors through the microcompartment shell was inspired by the three-dimensional structure of CsoS1D—the CcmP orthologue in α-carboxysomes (Klein et al., 2009). In the crystal structure of CsoS1D, one trimer features a pore that is closed by the formation of salt bridges between conserved glutamate and arginine residues, whereas the second trimer of the hexamer has a relatively large open pore. Both these two-domain BMC proteins feature circularly permuted secondary structure elements not observed in the more abundant single-domain shell proteins in both types of carboxysomes.
Recently, molecular transport through the protein shell of another type of microcompartment, the metabolosome, was suggested. Chowdhury et al. (2015) described how PduA may selectively transport the substrate 1,2-propanediol into the Pdu microcompartment while the outflow of the toxic intermediate propionaldehyde is restricted. The trimeric PduB is another pseudohexameric shell protein suggested to be a selective transporter of glycerol (Pang et al., 2012), a second substrate metabolized in the Pdu microcompartment. In the PduB crystal structure (PDB code 4FAY) glycerol molecules are bound to conserved residues in the three channels formed between the two BMC domains within each PduB subunit (Pang et al., 2012). For the related protein EutL in the ethanolamine utilization (Eut) microcompartment, structures are available with a closed pore (PDB code 3I82), as in the PduB structures, but also with an open 10–12 Å wide central pore (PDB code 3I87) on the three-fold symmetry axis of the trimer (Tanaka et al., 2010). The binding of the substrate, ethanolamine, in the three channels within each subunit was suggested to be a negative allosteric control that prevents the open conformation of the pore (Thompson et al., 2015).
The binding pocket between the two BMC domains of SeCcmP contains a conserved His18. Conservation is not strict between α- and β-carboxysomes; in CsoS1D, the corresponding residue is a leucine. The difference electron density map for the CsoS1D structure showed additional density near the modelled water molecules in the regions corresponding to the binding pocket. The additional density extends from SeMet73 toward the opening of the cleft (selenomethionine replaces methionine in CsoS1D to facilitate phasing; Klein et al., 2009) and indicates that a molecule larger than water is bound in the pocket. Alternatively the density might just be an artefact due to the incorporation of a selenium atom instead of a sulphur atom in residue 73.
No residue with a similar function to His18 in SeCcmP could be identified in the corresponding cleft of Csos1D. However, on the other side of strand β3 and the gating loop in the CsoS1D structure, His100—conserved in α-carboxysomes from both cyanobacteria and chemoautotrophs (Cai et al., 2013)—is located in a pocket where a molecule may bind and stabilize the closing of the pore. Additional density was observed in SeCcmP_I213 and SeCcmP_P213 monomer B (closed conformation) in the cleft that corresponds to the His100 cleft in CsoS1D, but not in SeCcmP_P213 monomer A (open conformation). Differences in the binding pockets of the α- and β-carboxysomes may be a result of distinct preferences for the molecule bound. Alternatively, and perhaps more likely, the gating control has developed independently from a common ancestor.
The SeCcmP_P213 structure to 1.45 Å resolution presented herein reveals that glycerol was bound in the binding pocket between the N- and C-BMC domains close to the gating loop when the pore is open, whereas a density for a larger molecule was observed in the binding pocket when the pore is closed. In earlier structures of SeCcmP, Cai et al. (2013) observed density corresponding to a 2–4-carbon molecule in all six binding pockets, and interpreted this as 3PGA included in the crystallization solution (Cai et al., 2013). Based on this scattered information, an attractive model emerges for the gated transport through the CcmP shell, in which the binding of the shorter product 3PGA promotes the opening of the pore while the binding of the larger substrate RuBP promotes the closing. This model is only partly supported by our structural data, which show the binding of glycerol in the pocket of the open pore monomer, whereas a larger, as yet unidentified, molecule binds in the pocket of the closed pore monomer of SeCcmP. Our co-crystallization and soaking experiments aimed at elucidating the identity of the bound compounds have so far been inconclusive. The smaller HCO3− and 3PGA were not observed and either may have been outcompeted by cryoprotectants or may not have bound. The co-crystallization or the soaking into pre-formed crystals of the larger compounds, RuBP and ADP/ATP, did not result in well-defined density. There may be several reasons for this. Binding may be hindered by the crystal lattice packing. Alternatively, the binding of a metabolite in the pocket during protein expression and purification may have blocked the binding of other compounds during crystallization. However, the high concentration of RuBP (50 mM) used during crystallization makes this unlikely, at least if this site is poised to bind RuBP. It is also possible that the crystal lattice may have forced the protein into a slightly altered conformation that does not permit the binding of the compound. The low occupancy may also be a result of the protein allowing the binding of the ligand in only one out of the three pockets, which would then be localized randomly within the trimer in the crystal. Given the proposed nature of CcmP as a carrier of metabolites in and out of the carboxysome shell, weak binding and low occupancy of the ligand is to be expected. Finally, we note that there may be an advantage of using a metabolite related to the energy content of the cell as a gating signal.
The answer to the nature of the conformational switch clearly awaits further research, but we note the following: β-carboxysomes have not yet been purified to homogeneity and it is not known if ADP is able to enter the carboxysome. However, the occurrence in some organisms (albeit not in S. elongatus) of genes for the ATP-dependent Rubisco activases, Rca, CbbX, and CbbQ, with their carboxysome operon (Zarzycki et al., 2013; Sutter et al., 2015) hints at the targeting of these proteins to the carboxysome and indicates that the possible binding of ADP to CcmP may not be as far-fetched as may at first seem. According to the taxonomy classification by Axen et al. (2014), some metabolosome genetic loci with unknown function are described to have an incomplete core, with enzymes responsible for cofactor recycling missing. Since one of the main functions of the characterized metabolosomes is believed to be the maintenance of a pool of recycled cofactors, the loss of the recycling machinery raises the question of the actual function of these types of metabolosome loci. For the type of metabolosomes where the machinery for recycling is not present, alternative solutions may be proposed; one of the suggestions is the selective transport of cofactors through the shells (Axen et al., 2014). In the PDU microcompartment, the iron-dependent alcohol dehydrogenase (PduQ) is responsible for recycling of NAD+ (Cheng et al., 2012), and a coenzyme A phosphotransacylase (PduL) recycles coenzyme A (Liu et al., 2015). In the two loci of the type Rhodococcus and Mycobacterium Microcompartment (RMM), RMM1 and RMM2, the genes coding for the two enzymes responsible for recycling are missing from a set of core enzymes otherwise similar to the PDU loci (Axen et al., 2014). Instead, the RMM loci contain genes coding for two CcmP homologues with the glutamate–arginine pair responsible for the pore closing conserved. The Metabolosomes with an Incomplete Core 1 (MIC1) locus has two putative alcohol dehydrogenase genes but no phosphotransacylase for the recycling of coenzyme A. A single gene for a CcmP homologue is also present in the MIC1 locus. It is tempting to speculate that these CcmP homologues function as gated pores for the selective transport of the cofactors through the BMC shell and that this may also apply to some carboxysomal shell proteins such as SeCcmP.
Acknowledgements
We wish to acknowledge the European Synchrotron Radiation Facility, Grenoble for providing beam time and data collection facilities, and thank the staff at beamline ID29 for excellent support. This work was supported by the Swedish Research Council, the Knut and Alice Wallenberg Foundation, and the Röntgen-Ångström Cluster.
Glossary
Abbreviations:
- BMC
bacterial microcompartment
- PDB
Protein Data Bank
- 3PGA
3-phosphoglycerate
- Rubisco
ribulose-1,5-bisphosphate carboxylase/oxygenase
- RuBP
ribulose 1,5-bisphosphate
- SeCcmP_I213
Synechococcus elongatus PCC7942 crystallized in space group I213
- SeCcmP_P213
Synechococcus elongatus PCC7942 crystallized in space group P213.
References
- Adams PD, Afonine PV, Bunkóczi G, et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica. Section D, Biological Crystallography 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axen SD, Erbilgin O, Kerfeld CA. 2014. A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLoS Computational Biology 10, e1003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H, Henrick K, Nakamura H. 2003. Announcing the worldwide Protein Data Bank. Nature Structural Biology 10, 980. [DOI] [PubMed] [Google Scholar]
- Bricogne G, Blanc E, Brandl M, et al. 2011. BUSTER version 2.10.0. Cambridge: Global Phasing Ltd. [Google Scholar]
- Cai F, Dou Z, Bernstein SL, Leverenz R, Williams EB, Heinhorst S, Shively J, Cannon GC, Kerfeld CA. 2015. Advances in understanding carboxysome assembly in Prochlorococcus and Synechococcus implicate CsoS2 as a critical component. Life 5, 1141–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai F, Kerfeld CA, Sandh G. 2012. Bioinformatic identification and structural characterization of a new carboxysome shell protein. In: Burnap R, Vermaas W, eds. Functional genomics and evolution of photosynthetic systems. Dordrecht: Springer, 345–356. [Google Scholar]
- Cai F, Menon BB, Cannon GC, Curry KJ, Shively JM, Heinhorst S. 2009. The pentameric vertex proteins are necessary for the icosahedral carboxysome shell to function as a CO2 leakage barrier. PLoS One 4, e7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai F, Sutter M, Cameron JC, Stanley DN, Kinney JN, Kerfeld CA. 2013. The structure of CcmP, a tandem bacterial microcompartment domain protein from the β-carboxysome, forms a subcompartment within a microcompartment. Journal of Biological Chemistry 288, 16055–16063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallographica. Section D, Biological Crystallography 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Fan C, Sinha S, Bobik TA. 2012. The PduQ enzyme is an alcohol dehydrogenase used to recycle NAD+ internally within the Pdu microcompartment of Salmonella enterica. PLoS One 7, e47144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury C, Chun S, Pang A, Sawaya MR, Sinha S, Yeates TO, Bobik TA. 2015. Selective molecular transport through the protein shell of a bacterial microcompartment organelle. Proceedings of the National Academy of Sciences, USA 112, 2990–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley CS, Sawaya MR, Bobik TA, Yeates TO. 2008. Structure of the PduU shell protein from the Pdu microcompartment of Salmonella. Structure 16, 1324–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Z, Heinhorst S, Williams EB, Murin CD, Shively JM, Cannon GC. 2008. CO2 fixation kinetics of Halothiobacillus neapolitanus mutant carboxysomes lacking carbonic anhydrase suggest the shell acts as a diffusional barrier for CO2. Journal of Biological Chemistry 283, 10377–10384. [DOI] [PubMed] [Google Scholar]
- Duff AP, Andrews TJ, Curmi PM. 2000. The transition between the open and closed states of rubisco is triggered by the inter-phosphate distance of the bound bisphosphate. Journal of Molecular Biology 298, 903–916. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallographica. Section D, Biological Crystallography 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. 2006. Scaling and assessment of data quality. Acta Crystallographica. Section D, Biological Crystallography 62, 72–82. [DOI] [PubMed] [Google Scholar]
- Evans PR, Murshudov GN. 2013 How good are my data and what is the resolution?Acta Crystallographica. Section D, Biological Crystallography 69, 1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. 1998. Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281, 237–240. [DOI] [PubMed] [Google Scholar]
- Heldt D, Frank S, Seyedarabi A, Ladikis D, Parsons JB, Warren MJ, Pickersgill RW. 2009. Structure of a trimeric bacterial microcompartment shell protein, EtuB, associated with ethanol utilization in Clostridium kluyveri. Biochemical Journal 423, 199–207. [DOI] [PubMed] [Google Scholar]
- Holm L, Rosenström P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Research 38, W545–W549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallographica. Section A, Foundations of Crystallography 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kabsch W. 2010. XDS. Acta Crystallographica. Section D, Biological Crystallography 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney JN, Axen SD, Kerfeld CA. 2011. Comparative analysis of carboxysome shell proteins. Photosynthesis Research 109, 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein MG, Zwart P, Bagby SC, Cai F, Chisholm SW, Heinhorst S, Cannon GC, Kerfeld CA. 2009. Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. Journal of Molecular Biology 392, 319–333. [DOI] [PubMed] [Google Scholar]
- Liu Y, Jorda J, Yeates TO, Bobik TA. 2015. The PduL phosphotransacylase is used to recycle coenzyme A within the Pdu microcompartment. Journal of Bacteriology 197, 2392–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. Journal of Applied Crystallography 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang A, Liang M, Prentice MB, Pickersgill RW. 2012. Substrate channels revealed in the trimeric Lactobacillus reuteri bacterial microcompartment shell protein PduB. Acta Crystallographica. Section D, Biological Crystallography 68, 1642–1652. [DOI] [PubMed] [Google Scholar]
- Pang A, Warren MJ, Pickersgill RW. 2011. Structure of PduT, a trimeric bacterial microcompartment protein with a 4Fe–4S cluster-binding site. Acta Crystallographica. Section D, Biological Crystallography 67, 91–96. [DOI] [PubMed] [Google Scholar]
- Price GD, Badger MR, Woodger FJ, Long BM. 2008. Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. Journal of Experimental Botany 59, 1441–1461. [DOI] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, et al. 2012. The Pfam protein families database. Nucleic Acids Research 40, D290–D301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae BD, Long BM, Badger MR, Price GD. 2012. Structural determinants of the outer shell of β-carboxysomes in Synechococcus elongatus PCC 7942: roles for CcmK2, K3-K4, CcmO, and CcmL. PLoS One 7, e43871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae BD, Long BM, Badger MR, Price GD. 2013. Functions, compositions, and evolution of the two types of carboxysomes: polyhedral microcompartments that facilitate CO2 fixation in cyanobacteria and some proteobacteria. Microbiology and Molecular Biology Reviews 77, 357–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter M, Roberts EW, Gonzalez RC, Bates C, Dawoud S, Landry K, Cannon GC, Heinhorst S, Kerfeld CA. 2015. Structural characterization of a newly identified component of α-carboxysomes: the AAA+ domain protein CsoCbbQ. Scientific Reports 5, 16243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, Cannon GC, Yeates TO. 2008. Atomic-level models of the bacterial carboxysome shell. Science 319, 1083–1086. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Sawaya MR, Yeates TO. 2010. Structure and mechanisms of a protein-based organelle in Escherichia coli. Science 327, 81–84. [DOI] [PubMed] [Google Scholar]
- Thompson MC, Cascio D, Leibly DJ, Yeates TO. 2015. An allosteric model for control of pore opening by substrate binding in the EutL microcompartment shell protein. Protein Science 24, 956–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MC, Crowley CS, Kopstein J, Bobik TA, Yeates TO. 2014a Structure of a bacterial microcompartment shell protein bound to a cobalamin cofactor. Acta Crystallographica. Section D, Biological Crystallography 70, 1584–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MC, Wheatley NM, Jorda J, Sawaya MR, Gidaniyan SD, Ahmed H, Yang Z, McCarty KN, Whitelegge JP, Yeates TO. 2014b Identification of a unique Fe–S cluster binding site in a glycyl-radical type microcompartment shell protein. Journal of Molecular Biology 426, 3287–3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MC, Yeates TO. 2014. A challenging interpretation of a hexagonally layered protein structure. Acta Crystallographica. Section D, Biological Crystallography 70, 203–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y, Sawaya MR, Yeates TO. 2009. Analysis of lattice-translocation disorder in the layered hexagonal structure of carboxysome shell protein CsoS1C. Acta Crystallographica. Section D, Biological Crystallography 65, 980–988. [DOI] [PubMed] [Google Scholar]
- Wheatley NM, Gidaniyan SD, Liu Y, Cascio D, Yeates TO. 2013. Bacterial microcompartment shells of diverse functional types possess pentameric vertex proteins. Protein Science 22, 660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, et al. 2011. Overview of the CCP4 suite and current developments. Acta Crystallographica. Section D, Biological Crystallography 67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeates TO, Jorda J, Bobik TA. 2013. The shells of BMC-type microcompartment organelles in bacteria. Journal of Molecular Microbiology and Biotechnology 23, 290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeates TO, Thompson MC, Bobik TA. 2011. The protein shells of bacterial microcompartment organelles. Current Opinion in Structural Biology 21, 223–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarzycki J, Axen SD, Kinney JN, Kerfeld CA. 2013. Cyanobacterial-based approaches to improving photosynthesis in plants. Journal of Experimental Botany 64, 787–798. [DOI] [PubMed] [Google Scholar]