Summary
SdrF, a Staphylococcus epidermidis surface protein, was found to facilitate binding to keratins 1 and 10, human nasal epithelial cells, and neonatal human epithelial keratinocytes. SdrF may facilitate S. epidermidis adherence to skin surfaces.
Keywords: Staphylococcus epidermidis, adhesion, keratin, keratinocyte, skin.
Abstract
Background.
Staphylococcus epidermidis, a major component of skin flora, is an opportunist, often causing prosthetic device infections. A family of structurally related proteins mediates staphylococcal attachment to host tissues, contributing to the success of S. epidermidis as a pathogen. We examined the ability of the surface protein SdrF to adhere to keratin, a major molecule expressed on the skin surface.
Methods.
A heterologous Lactococcus lactis expression system was used to express SdrF and its ligand-binding domains. Adherence to keratin types 1 and 10, human foreskin keratinocytes, and nasal epithelial cells was examined.
Results.
SdrF bound human keratins 1 and 10 and adhered to keratinocytes and epithelial cells. Binding involved both the A and B domains. Anti-SdrF antibodies reduced adherence of S. epidermidis to keratin and keratinocytes. RNA interference reduced keratin synthesis in keratinocytes and, as a result, SdrF adherence. Direct force measurements using atomic force microscopy showed that SdrF mediates bacterial adhesion to keratin 10 through strong and weak bonds involving the A and B regions; strong adhesion was primarily mediated by the A region.
Conclusions.
These studies demonstrate that SdrF mediates adherence to human keratin and suggest that SdrF may facilitate S. epidermidis colonization of the skin.
Staphylococcus epidermidis is a major opportunistic nosocomial pathogen [1]. Coagulase-negative staphylococci, especially S. epidermidis, are the most common cause of prosthetic device infections. This includes catheter-related bloodstream infections, prosthetic valve endocarditis and vascular graft infections, all of which seriously affect patient morbidity and mortality rates [2]. S. epidermidis is also a common colonizer of the skin and mucous membranes [3–5]. This capacity to colonize different skin and mucosal sites is a critical step in the initiation of infection. It allows this species to function as an opportunist, initiating infection when there is a breach of cutaneous barriers. Among the factors that seem to contribute to this colonization potential are its capacity for biofilm formation, its resistance to antimicrobial peptides, and the presence of surface adhesins that facilitate adherence to different host tissue molecules [2, 3, 6].
Many of these staphylococcal adhesins are part of a family of structurally related proteins referred to as S. epidermidis surface (SES) proteins [7]. These cell wall–anchored proteins interact with host matrix molecules, such as fibronectin, collagen, and fibrinogen [7]. These interactions are critical for bacterial adherence to different host tissue surfaces and for initiating infections [8, 9]. Redundancy of function may allow staphylococcal species with varying arrays of these proteins to colonize different tissue surfaces [10].
In earlier studies, we found that SdrF, one of these SES proteins, is present in both colonizing and clinical (eg, bacteremia) isolates (approximately 54%–67%) and that it binds collagen [7, 11]. Collagen was found coating the driveline surfaces of ventricular assist devices; thus, adherence to collagen seemed to play a role in the initiation of these device infections [11]. In a series of preliminary assays designed to identify other potential host tissue receptors for this protein, we found that SdrF adhered to keratin, a major constituent of the outer layer of human skin [12, 13]. This led to our interest in exploring the role of SdrF as a facilitator of S. epidermidis adherence to and colonization of skin. Here, we examine the interaction of SdrF with keratin, human nasal epithelial cells, and keratinocytes.
METHODS
Bacterial Strains, Growth Conditions, Recombinant Proteins, and DNA Constructs
Lactococcus lactis MG1363 was the host strain used for the cell surface expression of recombinant S. epidermidis proteins. Strains were grown in appropriate media as described elsewhere [14]. We used previously generated DNA constructs, as described elsewhere [14] (Supplementary Table 1). These constructs included the ligand-binding A and B domains each expressed in L. lactis. The SdrF-positive (SdrF+) S. epidermidis strain 9491 was used in binding studies [14].
A construct expressing the full-length SdrF protein was created. SdrF was subcloned from a pOri23 construct into pQE-30 into the BamH1 and Pst1 restriction sites. The full-length construct was verified by sequencing (Genewiz). Expression of the full-length SdrF was verified by means of flow cytometry, as described elsewhere [14]. For the expression of recombinant SdrF domains (rASdrF or rBSdrF) and control LukS-PV (rLukS), previously generated recombinant pQE-30 plasmids were used [14].
Polyclonal Antibody Preparation and Use in Adherence Assays
Previously prepared polyclonal antibodies directed against rASdrF or rBSdrF were used [14]. Total immunoglobulin (Ig) G was purified using the ImmunoPure (A) IgG purification kit. Specific IgGs were further purified by affinity chromatography with the MicroLink Protein Coupling kit (Pierce Biotechnology). In the antibody inhibition assays, purified rBSdrF, rASdrF, or the S. epidermidis strain 9491 were preincubated with increasing concentrations of anti-rBSdrF IgG and anti-rASdrF IgG for 1 hour before being added to keratin-coated microtiter wells.
Keratin and Pan-Cytokeratin Antibodies
Keratin 1 derived from human epidermis was purchased from Sigma. The purity of Sigma keratin 1 was validated by mass spectrophotometry. Keratin 10 recombinant protein was purified as described elsewhere, using a complementary DNA construct (kindly provided by Tim Foster, Trinity College) [13]. Pan-cytokeratin antibody (Sigma) was used in inhibition assays.
Bacterial Adherence Assays
Microtiter plates (MaxiSorp; Nalge Nunc International) were coated with 10 µg of keratin 1 in 100 µL of phosphate-buffered saline (PBS) per well overnight at 4°C. Wells were washed with PBS, blocked with 1% (wt/vol) bovine serum albumin (BSA; Sigma) in PBS for 1 hour, and washed. Log-phase S. epidermidis or L. lactis cultures were centrifuged, bacterial cells were resuspended in PBS, adjusted to an optical density (OD) (absorbance at 600 nm [A600]) of 0.5, and added to the microtiter wells. After a 1-hour incubation (37°C), the wells were washed with PBS, and the adherent bacteria were collected using 2 sequential incubations with trypsin/ethylenediaminetetraacetic acid. Bacterial cell suspensions were serially diluted, plated onto agar, and quantified after 24 hours.
Solid-Phase Assays Measuring Protein-Protein Interactions
The interaction between keratin and different truncated forms of SdrF was tested using a protocol described elsewhere, with minor modifications [9]. Microtiter plates were coated in a manner similar to that described for the bacterial adherence assay and blocked with 2% Milk (Bio-Rad) in Tris-buffered saline with Tween 20 for 2 hours, and then purified recombinant proteins (1 µg/mL unless otherwise stated) were added. After a 2-hour incubation (37°C), wells were washed with PBS containing 0.05% (vol/vol) of Tween 20 (PBST) and subsequently incubated for 45 minutes with anti-His horseradish peroxidase (Roche) conjugated in blocking solution (0.1 µg/mL). Wells were then extensively washed with PBST and development was performed using 1-Step Ultra TMB-ELISA (Pierce), according to the manufacturer’s instructions. Absorbance at (at 450 nm [A450]) was measured using a Bio-Rad 680 Microplate Reader (Bio-Rad).
Cell Cultures and Binding Assays
Normal human epidermal keratinocytes (NHEKs), derived from neonatal foreskin keratinocytes, were maintained serum-free in keratinocyte basal medium KBM Gold (supplemented with KGM-SingleQuot; Lonza) and grown in 12-well plates to 90% confluence [15]. S. epidermidis and L. lactis overnight cultures were diluted and grown to mid-log phase and adjusted to an OD (A600) of 0.5, and 600 µL of bacteria were added to washed NHEK cell layers in triplicate. Samples were incubated for 1 hour (37°C), washed 8 times with cell culture medium, and treated with 300 µL of trypsin for 5 minutes (37°C), and bacteria were then lifted. Serial dilutions were plated onto agars and incubated overnight.
Immunostaining of NHEKs
NHEKs were grown to 80% confluence on 35mm glass bottom dishes (MatTek). Samples were blocked with Cnt-07 medium (CELLnTEC ) with 1% BSA for 1 hour at 37°C, incubated with pan-cytokeratin antibody diluted in Cnt-07.S medium with 1% BSA for 1 hour at 37°C, and then incubated with anti-mouse IgG conjugated to fluorescein isothiocyanate (FITC) for 1 hour (at 37°C). CellMask Deep Red Plasma Membrane Stain (ThermoFisher Scientific) was added for the final 10 minutes. Cells were washed and resuspended in Cnt-07.S medium with 1% BSA. A negative control was performed using only FITC and CellMask stain.
Confocal Microscopy
Confocal microscopy was performed with an A1R MP laser scanning confocal attachment on an Eclipse Ti microscope stand (Nikon Instruments) using a 60x/1.49 Apo TIRF objective (Nikon). FITC was excited using a 488-nm laser, with emission detected between 500 and 550 nm; CellMask Deep Red was excited using a 640-nm laser, with emission detected between 660 and 740 nm. Z-stack images were created using a step size of 0.5 μm. ImageJ software (version 2.0.0.0-rc-15/1.51k) was used to merge the 2 channels [16].
Nasal Cell Binding Assays
Adherence to desquamated nasal epithelial cells was determined using a protocol described elsewhere [13]. Nasal epithelial cells were collected from the anterior nares of volunteers using a sterile cotton-tipped swab (Fisher Scientific) rotated in each nostril (protocol approved by the Columbia University institutional review board). Nasal cells were collected in 3 mL of PBS, then centrifuged and washed with PBS. Cell density was adjusted to 1 × 105. Host commensal bacteria were removed from the cells by incubation with 50-µg/mL gentamicin for 1 hour. Residual antibiotic was removed through centrifugation and washing with PBS.
Bacterial strains were brought to log phase, and the density was adjusted to an OD (A600) of 0.5. Clumping was prevented by passing bacterial aliquots through a 25-gauge needle. The bacterial suspension (100 µL) was added to the nasal cell suspension (100 µL), and the mixture was incubated (37°C) with gentle shaking for 1 hour. Bacterial adherence was halted by the addition of 800 µL of PBS. The assay mixture was applied to a 12-µm isopore polycarbonate filter (Millipore) under vacuum conditions and subsequently washed with 2 mL of PBS. Once the filter was dry, cells were fixed with Cytofix/Cytoperm (BD Biosciences), stained with crystal violet, and mounted on a glass slide. Bacteria adherent to 25 desquamated nasal nonnucleated epithelial cells (the most superficial cells) per assay were then enumerated by means of oil immersion microscopy (magnification, ×1000). The observer was blinded to the treatment group. Each assay was performed in duplicate for each of 3 independent experiments.
Small Interfering RNA Transfection of NHEKs
NHEKs were grown in culture to 90% confluency. The keratin 10 small interfering RNA (siRNA) or the control (scrambled sequence) (Supplementary Table 1) and transfection reagent (GE Dharmacon) were incubated with Opti-MEM reduced serum media (Gibco) for 5 minutes, followed by incubation of the 2 combined. This suspension (400 µL) was then added to the NHEKs for 24 or 48 hours, followed by performance of the adherence assay (described above). The final concentration of siRNA in each well was 50 nmol/L.
Atomic Force Microscopy
To prepare keratin-coated substrates for atomic force microscopy (AFM) experiments, glass coverslips coated with a thin layer of gold were immersed overnight in an ethanol solution containing 1 mmol/L of 10% 16-mercaptododecahexanoic acid/90% 1-mercapto-1-undecanol (Sigma), rinsed with ethanol, and dried with nitrogen. Substrates were then immersed for 30 minutes into a solution containing 10 mg/mL N-hydroxysuccinimide and 25 mg/mL 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (Sigma), rinsed 5 times with Ultrapure water (ELGA LabWater), incubated with 0.1 mg/mL keratin 10 for 1 hour, rinsed further with PBS, and then immediately used without dewetting.
Bacterial cell probes were obtained as described elsewhere [17], using triangular tipless cantilevers (NP-O10, Microlevers, Bruker Corporation) with a nominal spring constant of approximately 0.06 N/m, as determined using the thermal noise method (Picoforce; Bruker Corporation). Single bacteria were attached on the center of colloidal probes [17] using a Bioscope Catalyst AFM (Bruker Corporation) equipped with a Zeiss Axio Observer Z1 and a Hamamatsu camera (C10600). The cell probe was positioned over keratin substrates without dewetting, and interaction forces were measured at room temperature (20°C) by recording multiple force curves on 3 spots, using a maximum applied force of 250 pN, contact time of 1 second, and constant approach and retraction speeds of 1000 nm/s.
Statistical Analysis
All data from the protein-protein interaction assays and bacterial adherence assays represent the means and standard errors for 2 or 3 different experiments, using triplicate wells for each condition tested. Statistical analysis was performed using pairwise comparisons with the Student t test.
RESULTS
Mediation of L. lactis Adherence to Keratin 1 and 10 by SdrF and the SdrF A and B Domains
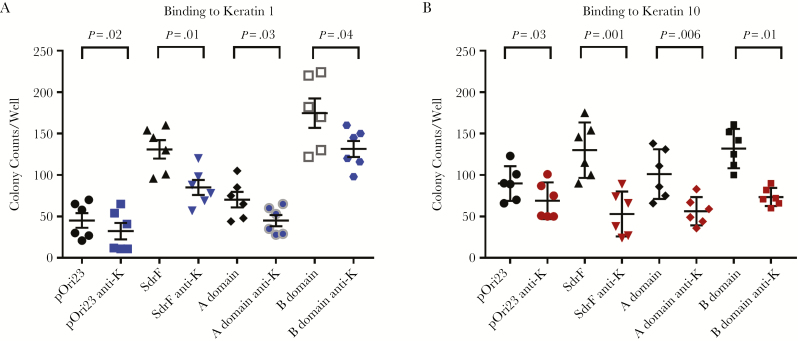
The full-length SdrF expressed in L. lactis increased adherence to both keratin types 1 and 10 when compared with controls (MG1363 cells containing pOri23) (P < .05) (Figure 1). Similarly, L. lactis strains expressing either the SdrF A or B domains bound both keratin types. Binding of the B domain was comparable to that of the full-length SdrF and was significantly higher than that of the A domain (P < .05). Binding of the full-length SdrF construct and the A and B domains to keratins 1 and 10 was significantly reduced (P < .05) in the presence of pan-cytokeratin antibody (Figure 1).
Figure 1.
SdrF-mediated binding to keratins 1 and 10. The binding of Lactococcus lactis surface-expressed SdrF (SdrF on x-axis) and the ligand-binding domains A and B was compared with L. lactis containing the control vector pOri23 (pOri23). Pan-cytokeratin antibody (1 µg/mL) was used in inhibition assays (anti-K). A, Binding of full-length SdrF and the A and B domains to keratin 1. B, Binding to keratin 10. Full-length SdrF, as well as ligand-binding domains A and B, bind keratins 1 and 10. Antibodies directed against SdrF and the A and B domains significantly reduced binding. Data represent the number of bacterial colony counts per well and represent means and standard errors from ≥3 experiments (3 microtiter wells per experiment) for the binding studies and 2 experiments (3 microtiter wells per experiment) for the antibody inhibition assays.
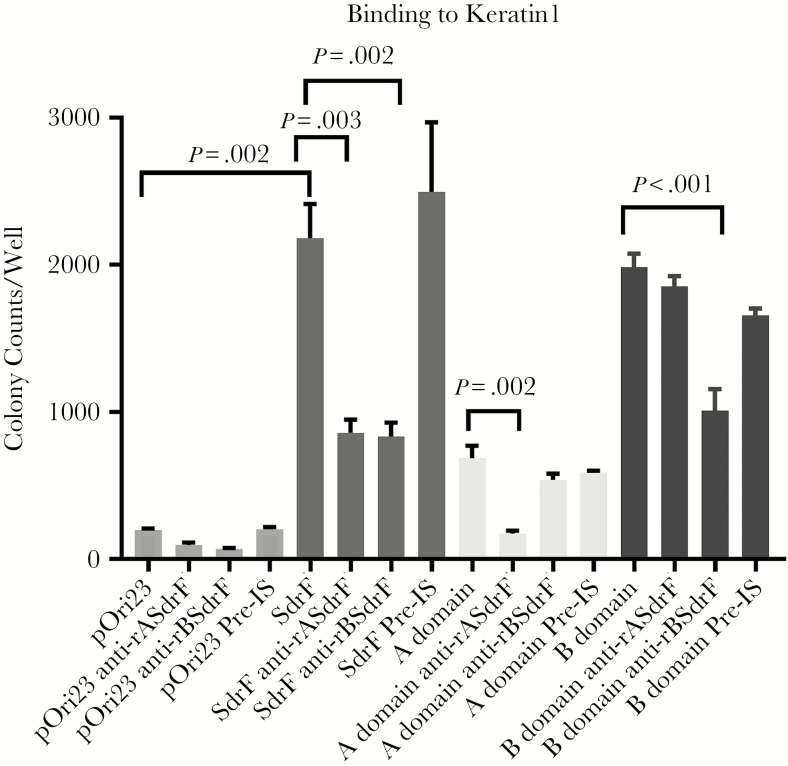
Antibodies directed against either the A or B domains of SdrF reduced adherence of the full-length SdrF to keratin 1 (Figure 2). Antibodies directed against the A or B domains reduced binding of their respective domains but not that of the other domain to keratin 1. Preimmune IgG had no effect on adherence. These results suggest that SdrF binds keratins 1 and 10 and that both the A and B domains contribute to this interaction.
Figure 2.
Effect of anti-SdrF domain antibodies on binding to keratin 1. Antibodies directed against the Lactococcus lactis surface-expressed ligand-binding domains of SdrF (anti-rASdrF and anti-rBSdrF) reduced binding of SdrF to keratin 1 compared with preimmune control serum (pre-IS). Data represent the number of bacterial colonies per well and represent the means and standard errors from ≥2 experiments (3 microtiter wells per experiment).
Binding of the SdrF A and B Domain Recombinant Proteins to Keratin 1 and 10
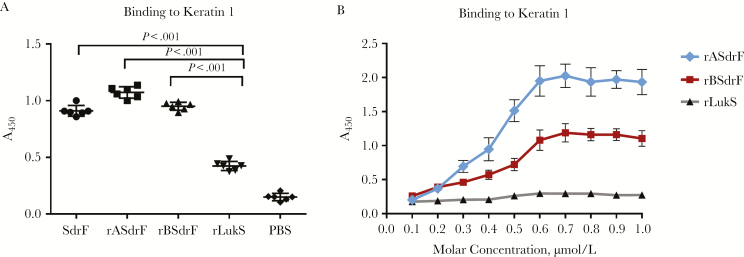
The ability of recombinant proteins from the A and B domains to bind keratin was examined. LukS-PV, a Staphylococcus aureus leukocidin, was chosen as an irrelevant control protein in these solid-phase assays [14]. There was significantly enhanced binding with rBSdrF, rASdrF, and the rSdrF when compared with rLukS (P < .05) (Figure 3A). A dose-response assay showed that binding of both domains to keratin 1 was saturable in a concentration-dependent manner (Figure 3B).
Figure 3.
Binding of the purified recombinant SdrF polypeptide domains A and B to immobilized keratin 1. A, Keratin-coated wells were incubated with purified full-length SdrF (SdrF on x-axis) and the truncated ligand-binding domain proteins A and B (rASdrF and rBSdrF, respectively). An irrelevant protein, rLukS, was used as a control. PBS, phosphate-buffered saline. B, Dose-response curve of the 2 SdrF ligand-binding domains adhering to keratin 1 compared with rLukS. Keratin-coated wells were incubated with increasing concentrations of purified rASdrF, rBSdrF and rLukS. Data represent means and standard errors from ≥2 experiments (3 microtiter wells per experiment).
SdrF Adherence to NHEKs
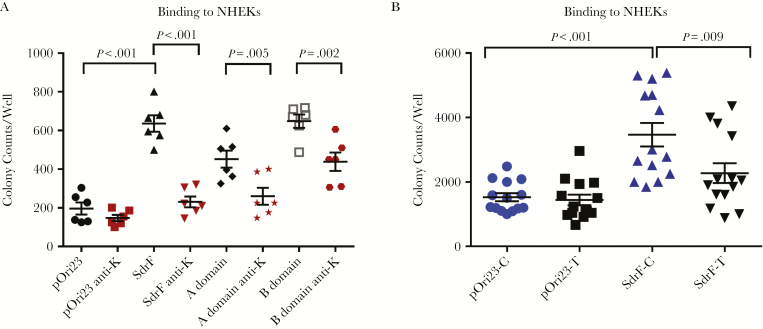
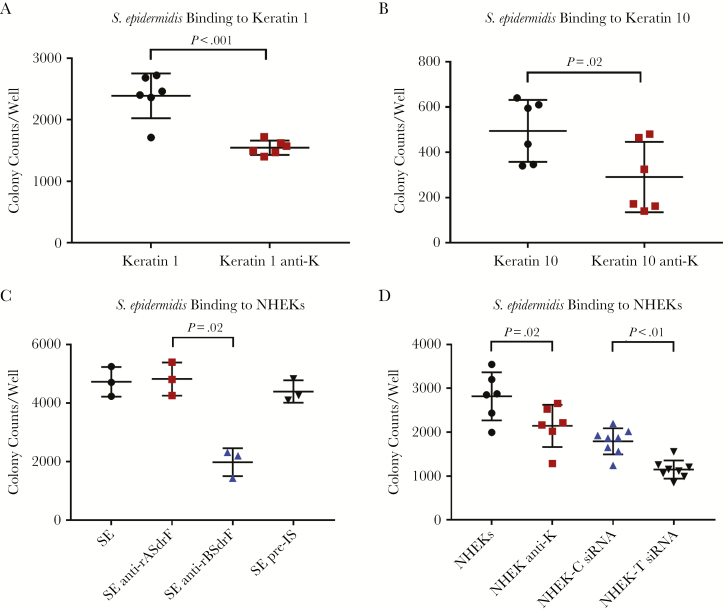
Confocal imaging was used to demonstrate that keratin expression occurred on the NHEK surface (Supplement Figure 1). The full-length SdrF and both the A and B domains significantly increased adherence compared with the L. lactis control. Pan-cytokeratin or anti-A or B domain antibodies reduced adherence (Figure 4A). Adherence of SdrF to NHEKs transfected with keratin 10 siRNA was reduced compared with control NHEKs (P < .03) (Figure 4B). These studies demonstrated SdrF adherence to NHEKs and the contribution of keratin to this binding interaction.
Figure 4.
A, Binding of Lactococcus lactis expressing pOri23 (cloning vector), SdrF, and the A and B domains to normal human epidermal keratinocytes (NHEKs) in cell culture. Pan-cytokeratin antibody (1 µg/mL) (anti-K) was used in the antibody inhibition assays. Results (number of adherent colonies per well) represent means and standard errors from ≥3 experiments (3 microtiter wells per experiment). B, Binding of L. lactis expressing SdrF to NHEKs treated with RNA interference (RNAi) (SdrF-T or pOri-T) to reduce keratin synthesis (SdrF-C and pOri23-C represent RNAi scramble-treated controls). Data (number of bacterial colonies per well) represent means and standard errors from 3 experiments (3 microtiter wells per experiment).
SdrF Adherence to Human Nasal Epithelial Cells
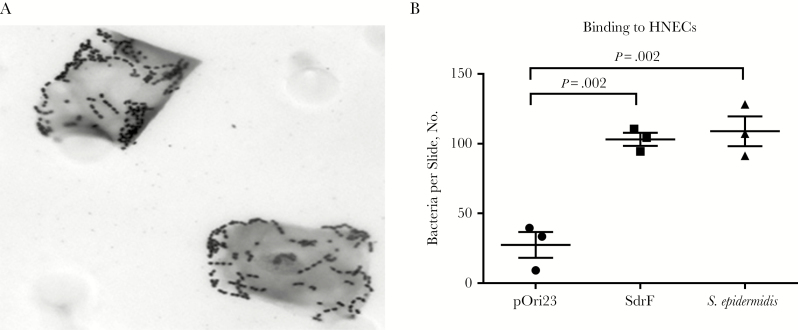
To determine whether SdrF increased adherence to desquamated human nasal epithelial cells, binding studies were performed with the L. lactis constructs (Figure 5A). Binding of the L. lactis SdrF and an SdrF+S. epidermidis strain, 9491, to the nasal epithelial cells was significantly increased compared with the control (Figure 5B).
Figure 5.
Lactococcus lactis expressing SdrF binding to human nasal epithelial cells (HNECs). A, Adherence to the epithelial cells was measured by examining crystal violet stained cells with microscopy. B, Adherence of L. lactis expressing SdrF or an SdrF-positive Staphylococcus epidermidis, strain 9491, to nasal epithelial cells. Numbers of adherent bacteria per nasal epithelial cell were determined under oil immersion. Data represent the mean number of bacteria per epithelial cell per slide.
Binding of an SdrF+S. epidermidis Isolate to Keratin 1 and 10 and Keratinocytes
Adherence of a SdrF+S. epidermidis strain, 9491, was measured in the presence or absence of pan-cytokeratin antibody [14]. Binding to both keratins was reduced in the presence of the antibody (Figure 6A and 6B). Binding of strain 9491 to NHEKs was significantly reduced by anti-rBSdrF but not by anti-rASdrF IgG (Figure 6C). When the NHEKs were treated with pan-cytokeratin antibody or with siRNA, adherence of 9491 was significantly reduced (Figure 6D).
Figure 6.
Staphylococcus epidermidis strain 9491 binding to keratins and to normal human epidermal keratinocytes (NHEKs) in the presence or absence of pan-cytokeratin antibody. A, Adherence to keratin 1. B, Adherence to keratin 10. C, Adherence to NHEKs in the presence or absence of anti-A and B domain antibodies (anti-rASdrF and anti-rBSdrF, respectively) or preimmune control serum (pre-IS). D, Adherence to RNA interference (RNAi) treated (control-scrambled vs keratin-specific experimental) NHEKs. Keratin-coated plates were incubated with S. epidermidis for 1 hour at 37°C, followed by washing and plating. Data represent mean and standard errors from ≥3 experiments (3 microtiter wells per experiment).
AFM Results
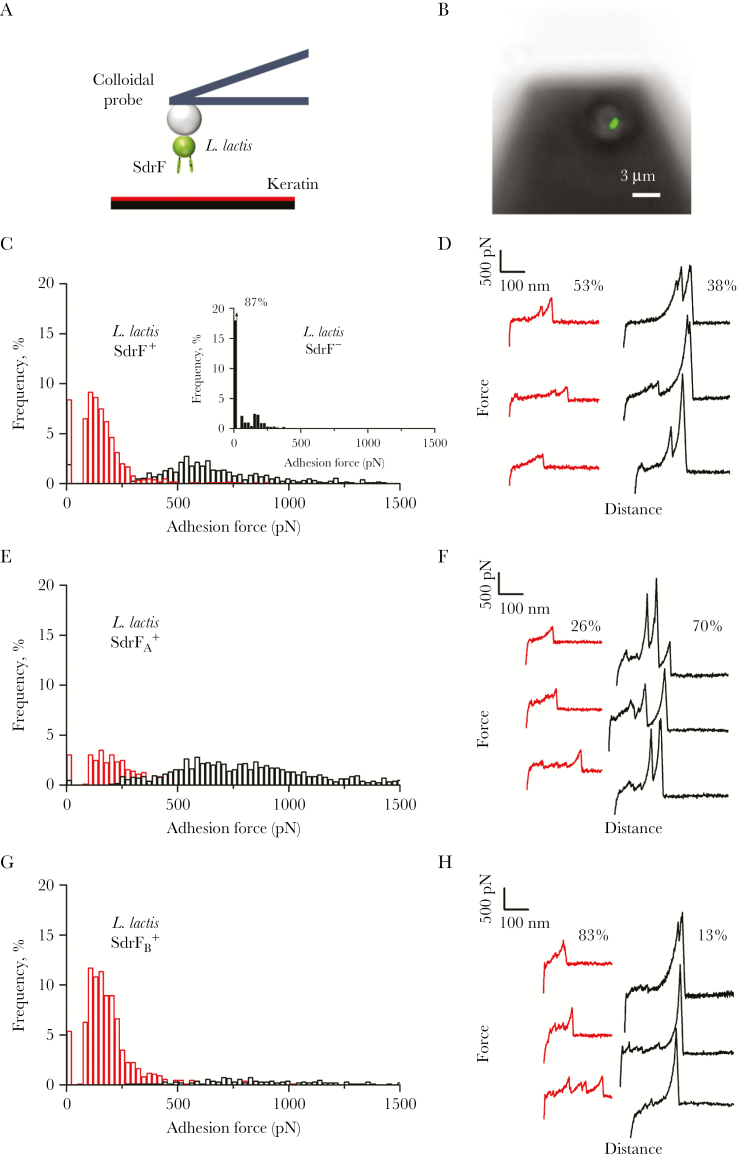
AFM was used to quantify the forces by which SdrF binds to keratin. Single L. lactis bacteria expressing SdrF (SdrF+ cells) were attached onto colloidal cantilevers coated with polydopamine (Figure 7A and 7B), and force-distance curves were recorded between cell probes and keratin 10–coated substrates [17–19]. Shown in Figure 7C and 7D is the adhesion forces and representative force curves obtained for 3 SdrF+ cells. Most curves showed adhesion events ranging from approximately 50 to approximately 1500 pN. Two groups of adhesion signatures were observed, weak forces with a mean (standard error) magnitude of 141 (51) pN (53%), and strong forces of 589 (151) pN (38%) (n = 1327 curves from 3 cells), suggesting that they represent distinct interactions. These forces were not observed on SdrF−L. lactis cells lacking the adhesin (Figure 7C inset), indicating that they represent specific interactions between SdrF and keratin. It is striking to note that the approximately 150-pN and 600-pN forces are very close to the forces measured for single SdrF-collagen bonds [20], suggesting strongly that they reflect individual SdrF-keratin interactions.
Figure 7.
Adhesion forces between SdrF and keratin. A, To probe single-cell adhesion forces, living bacteria were attached on polydopamine-coated colloidal cantilevers and force curves were obtained between cellular probes and keratin-coated substrates. B, Lactococcus lactis cell attached to the colloidal cantilever. The image was obtained from the underside of the cantilever, and the stained cell is attached, because it remained present after washing. Labeling of the cell (green; Baclight LIVE/DEAD stain) showed that the method is nondestructive. C, D, Adhesion force histogram (C) and representative force signatures (D) obtained by recording multiple force-distance curves in phosphate-buffered saline (PBS) between 3 SdrF+ cells and keratin 10. Inset in C shows force data obtained in the same conditions for an SdrF− cell. E–H, Adhesion force histogram (E, G) and representative force signatures (F, H), obtained by recording multiple force-distance curves in PBS between 3 SdrFA+ cells (E, F) or SdrFB+ cells (G, H) and keratin 10. All curves were obtained using a contact time of 1 second, a maximum applied force of 250 pN, and approach and retraction speeds of 1000 nm/s.
L. lactis bacteria expressing only A or B domains (SdrFA+ and SdrFB+ cells) were also probed. Forces recorded for SdrFA+ cells were similar to those for SdrF+ cells, yet with a higher frequency of strong forces (70%) (Figure 7E and 7F). On SdrFB+ cells, however, mostly weak forces were seen, and strong forces were only rarely observed (13%) (Figure 7G and 7H). This suggests that although both the A and B regions bind to keratin, only the A region mediates strong adhesion. In summary, AFM revealed that SdrF binds to keratin 10 via both weak (approximately 150 pN) and strong (approximately 600 pN) bonds involving the A and B regions, with strong adhesion primarily controlled by the A domain.
DISCUSSION
S. epidermidis is a successful opportunistic nosocomial pathogen with a unique capacity to cause prosthetic device infections [2, 3, 21]. This is due in part to its ability to persistently colonize the skin and mucous membranes [2]. Infection is often initiated by a breach of a cutaneous barrier or when bacteria are introduced into surgical wounds at the time of surgery. The capacity to adhere to prosthetic material and the matrix molecules coating these devices, to elaborate biofilm and to resist a diversity of antimicrobials contributes to their unique success. Less well understood are the factors that allow S. epidermidis to persistently colonize cutaneous surfaces. Many factors contributing to the success of S. epidermidis as a pathogen are also likely to contribute to its colonization potential. Included among these virulence determinants is the family of structurally related SES adhesins [7, 22–26].
We examined whether the SES protein SdrF contributes to the ability of S. epidermidis to colonize skin and mucosal surfaces. Relatively little is known about the factors that contribute to staphylococcal colonization of skin and mucosal surfaces. We investigated SdrF adherence to keratin and to cells that express this ligand on their surface. Keratins are a family of related structural proteins that are found in abundance on epithelial surfaces [12]. They are subdivided into 2 sequence types, types I and II, that are typically coexpressed as specific pairs with complex expression patterns [12]. Bibel et al [27] demonstrated increased S. aureus adherence to more heavily keratinized skin and nasal epithelial cells. Human-type cytokeratin 10 has been shown to be a receptor for clumping factor B and, as a result, facilitates S. aureus colonization of the nares [13]. Other studies have examined the interaction of clumping factor with cytokeratin 8 [28]. Comparable studies have not been performed with S. epidermidis.
Several complementary approaches were used to define the role of SdrF as a potential mediator of skin colonization. We used keratin types 1 (type II) and 10 (type I), which, as noted, are coexpressed in the spinous layer of human skin [12, 29], as well as NHEKs and desquamated nasal epithelial cells to determine whether SdrF contributed to colonization of skin and mucosal surfaces. The results show that SdrF facilitates adherence to both keratin types, NHEKs, and desquamated human nasal epithelial cells, suggesting that SdrF may contribute to S. epidermidis colonization of these surfaces.
We further investigated the contribution of the A and B ligand-binding domains to adherence. Our previous studies identified a role for the B domain in adherence to collagen (14). Both domains, expressed in L. lactis, bound keratins 1 and 10. The B domain showed significantly higher adherence than A. Antibodies directed against either the A or B domains significantly reduced full-length SdrF adherence; however, the domain-specific antibodies only reduced binding of their respective domains. This difference may reflect conformational changes resulting from the different manner of protein presentation in the 2 assays.
Few studies have investigated the mechanism of SES protein binding to matrix molecules. Studies with S. aureus microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) have defined one binding interaction that involves a “dock, lock, and latch” mechanism. SdrF is most similar to the SdrC, SdrD, and SdrE S. aureus MSCRAMMs, wherein both the A and B domains seem to contribute to binding interactions [30]. Findings of studies using AFM suggest that SdrF adheres to collagen via a dual-binding activity that involves both weak and strong bonds [20]. These studies suggest that the A and B domains contribute in different ways to SdrF adherence to keratin. It is tempting to speculate on the molecular origin of the measured forces. The dual binding activity measured in this study is reminiscent of that demonstrated for the SdrF-collagen interaction [20]. We suggest that some of the N1, N2, and N3 subdomains of the A region promote strong binding to keratin through a new mechanism different from the dock, lock, and latch mechanism, because the latter involves stronger forces in the range of approximately 2 nN [31]. Weaker binding would be primarily mediated by the B repeats that form a rod projecting the A domain away from the cell surface.
Limitations of this study include the lack of a S. epidermidis knockout to compare with the SdrF+ strain. Preparation of these knockout strains in S. epidermidis has been unsuccessful, for our group and for others [32]. To compensate for this, we used several different strategies, including binding to multiple different cells and surfaces, antibody inhibition, and siRNA, to demonstrate the role of SdrF. In lieu of testing SdrF+ and SdrF− strains, we elected to examine the binding of an SdrF+ strain after siRNA treatment of NHEKs. Use of the different strains would have required careful evaluation of the clonal background of candidate strains to account for potentially confounding factors including the potential of other SES proteins binding keratin. It seems that both domains A and B contribute to the binding interaction. However, the results differed somewhat between in the protein interaction studies and the L. lactis binding studies. As noted above, this difference may reflect differences in presentation of the molecule in the different assays, but it does warrant further investigation. Selecting the appropriate cell type to mimic skin is problematic. We used 2 types of cells to address this concern, foreskin and nasal epithelial cells.
In summary, this study demonstrates that SdrF, a member of the Sdr family of surface proteins, binds keratin and facilitates adherence to human epithelial cells. Although both domains seem to contribute to this binding interaction, the A domain is responsible for the strongest binding. This interaction may facilitate colonization of the skin and mucosal membranes, and, as a result, the invasive potential of this opportunistic pathogen.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Acknowledgments. We thank the Florence and Herbert Irving Center for Dermatology and Skin Cancer Research for providing the human epidermal keratinocytes (NHEKs).
Financial support This work was supported by the National Cancer Institute, National Institutes of Health (grants P30 CA013696 and S10 RR025686 to the Confocal and Specialized Microscopy Shared Resource of the Herbert Irving Comprehensive Cancer Center at Columbia University), the European Research Council under the European Union’s Horizon 2020 research and innovation program (grant 693630 to Université Catholique de Louvain), the National Fund for Scientific Research (FNRS), the FNRS-WELBIO (grant WELBIO-CR-2015A-05), the Belgian Federal Office for Scientific, Technical, and Cultural Affairs (Interuniversity Poles of Attraction Program), and the Research Department of the Communauté Française de Belgique (Concerted Research Action).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 2004; 39:309–17. [DOI] [PubMed] [Google Scholar]
- 2. Becker K, Heilmann C, Peters G. Coagulase-negative staphylococci. Clin Microbiol Rev 2014; 27:870–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Otto M. Staphylococcus epidermidis—the ‘accidental’ pathogen. Nat Rev Microbiol 2009; 7:555–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. von Eiff C, Peters G, Heilmann C. Pathogenesis of infections due to coagulase-negative staphylococci. Lancet Infect Dis 2002; 2:677–85. [DOI] [PubMed] [Google Scholar]
- 5. Grice EA, Kong HH, Renaud G et al. A diversity profile of the human skin microbiota. Genome Res 2008; 18:1043–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Otto M, Peschel A, Götz F. Producer self-protection against the lantibiotic epidermin by the ABC transporter EpiFEG of Staphylococcus epidermidis Tü3298. FEMS Microbiol Lett 1998; 166:203–11. [DOI] [PubMed] [Google Scholar]
- 7. Bowden MG, Chen W, Singvall J et al. Identification and preliminary characterization of cell-wall-anchored proteins of Staphylococcus epidermidis. Microbiology 2005; 151:1453–64. [DOI] [PubMed] [Google Scholar]
- 8. Heilmann C, Götz F. Further characterization of Staphylococcus epidermidis transposon mutants deficient in primary attachment or intercellular adhesion. Zentralbl Bakteriol 1998; 287:69–83. [DOI] [PubMed] [Google Scholar]
- 9. Bowden MG, Visai L, Longshaw CM, Holland KT, Speziale P, Hook M. Is the GehD lipase from Staphylococcus epidermidis a collagen binding adhesin? J Biol Chem 2002; 277:43017–23. [DOI] [PubMed] [Google Scholar]
- 10. Foster TJ, Höök M. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol 1998; 6:484–8. [DOI] [PubMed] [Google Scholar]
- 11. Arrecubieta C, Toba FA, von Bayern M et al. SdrF, a Staphylococcus epidermidis surface protein, contributes to the initiation of ventricular assist device driveline-related infections. PLoS Pathog 2009; 5:e1000411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fuchs E. Keratins and the skin. Annu Rev Cell Dev Biol 1995; 11:123–53. [DOI] [PubMed] [Google Scholar]
- 13. O’Brien LM, Walsh EJ, Massey RC, Peacock SJ, Foster TJ. Staphylococcus aureus clumping factor B (ClfB) promotes adherence to human type I cytokeratin 10: implications for nasal colonization. Cell Microbiol 2002; 4:759–70. [DOI] [PubMed] [Google Scholar]
- 14. Arrecubieta C, Lee MH, Macey A, Foster TJ, Lowy FD. SdrF, a Staphylococcus epidermidis surface protein, binds type I collagen. J Biol Chem 2007; 282:18767–76. [DOI] [PubMed] [Google Scholar]
- 15. Luchinskaya D, Du R, Owens DM, Tarnow D, Bittner N. Various surface treatments to implant provisional restorations and their effect on epithelial cell adhesion: a comparative in vitro study. Implant Dent 2017; 26:12–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rasband WS. ImageJ. Bethesda, MD: National Institutes of Health, 1997–2016. [Google Scholar]
- 17. Beaussart A, El-Kirat-Chatel S, Herman P et al. Single-cell force spectroscopy of probiotic bacteria. Biophys J 2013; 104:1886–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dufrêne YF. Atomic force microscopy in microbiology: new structural and functional insights into the microbial cell surface. MBio 2014; 5:e01363–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dufrêne YF. Sticky microbes: forces in microbial cell adhesion. Trends Microbiol 2015; 23:376–82. [DOI] [PubMed] [Google Scholar]
- 20. Herman-Bausier P, Dufrêne YF. Atomic force microscopy reveals a dual collagen-binding activity for the staphylococcal surface protein SdrF. Mol Microbiol 2016; 99:611–21. [DOI] [PubMed] [Google Scholar]
- 21. Otto M. Staphylococcal biofilms. Curr Top Microbiol Immunol 2008; 322:207–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McCrea KW, Hartford O, Davis S et al. The serine-aspartate repeat (Sdr) protein family in Staphylococcus epidermidis. Microbiology 2000; 146(pt 7):1535–46. [DOI] [PubMed] [Google Scholar]
- 23. Patti JM, Allen BL, McGavin MJ, Höök M. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol 1994; 48:585–617. [DOI] [PubMed] [Google Scholar]
- 24. Potter A, Ceotto H, Giambiagi-Demarval M, dos Santos KR, Nes IF, Bastos Mdo C. The gene bap, involved in biofilm production, is present in Staphylococcus spp. strains from nosocomial infections. J Microbiol 2009; 47:319–26. [DOI] [PubMed] [Google Scholar]
- 25. Cheung GY, Rigby K, Wang R et al. Staphylococcus epidermidis strategies to avoid killing by human neutrophils. PLoS Pathog 2010; 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Otto M. Staphylococcus colonization of the skin and antimicrobial peptides. Expert Rev Dermatol 2010; 5:183–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bibel DJ, Aly R, Shinefield HR, Maibach HI, Strauss WG. Importance of the keratinized epithelial cell in bacterial adherence. J Invest Dermatol 1982; 79:250–3. [DOI] [PubMed] [Google Scholar]
- 28. Haim M, Trost A, Maier CJ et al. Cytokeratin 8 interacts with clumping factor B: a new possible virulence factor target. Microbiology 2010; 156:3710–21. [DOI] [PubMed] [Google Scholar]
- 29. Collin C, Moll R, Kubicka S, Ouhayoun JP, Franke WW. Characterization of human cytokeratin 2, an epidermal cytoskeletal protein synthesized late during differentiation. Exp Cell Res 1992; 202:132–41. [DOI] [PubMed] [Google Scholar]
- 30. Foster TJ, Geoghegan JA, Ganesh VK, Höök M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol 2014; 12:49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Herman P, El-Kirat-Chatel S, Beaussart A, Geoghegan JA, Foster TJ, Dufrêne YF. The binding force of the staphylococcal adhesin SdrG is remarkably strong. Mol Microbiol 2014; 93:356–68. [DOI] [PubMed] [Google Scholar]
- 32. Otto M, . Staphylococcus epidermidis pathogenesis in: Staphylococcus epidermidis: methods and protocols. Methods in molecular biology. Clifton, NJ: Springer protocols (Series), Humana Press, 2014; 1106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.