FUS3 phosphorylation at the SnRK1 target site regulates embryo growth rate, seed yield, and plant growth at control and elevated temperature.
Keywords: AKIN10, embryogenesis, FUS3, heat stress, high temperature, phosphorylation, seed development, SnRK1, transcriptional regulation
Abstract
The transcription factor FUSCA3 (FUS3) acts as a major regulator of seed maturation in Arabidopsis. FUS3 is phosphorylated by the SnRK1 catalytic subunit AKIN10/SnRK1α1, which belongs to a conserved eukaryotic kinase complex involved in energy homeostasis. Here we show that AKIN10 and FUS3 share overlapping expression patterns during embryogenesis, and that FUS3 is phosphorylated by AKIN10 in embryo cell extracts. To understand the role of FUS3 phosphorylation, we generated fus3-3 plants carrying FUS3 phosphorylation-null (FUS3S>A) and phosphorylation-mimic (FUS3S>D) variants. While FUS3S>A and FUS3S>D rescued all the fus3-3 seed maturation defects, FUS3S>A showed reduced transcriptional activity and enhanced fus3-3 previously uncharacterized phenotypes. FUS3S>A embryos displayed increased seed abortion due to maternal FUS3S>A and delayed embryo development, which correlated with a strong decrease in seed yield (~50%). Accordingly, the akin10 and akin11 mutants displayed a frequency of seed abortion similar to fus3-3. When plants were grown at elevated temperature, most phenotypes were exaggerated in FUS3S>A plants, and progeny seedlings overall grew poorly, suggesting that phosphorylation of FUS3 plays an important role during early embryogenesis and under heat stress. Collectively, these results suggest that FUS3 phosphorylation and SnRK1 are required for embryogenesis and integration of environmental cues to ensure the survival of the progeny.
Introduction
Seed production is vital to the success of higher plants and integrally important to human diet and animal feed. Seed development is a highly regulated and complex process. Arabidopsis thaliana follows a classical pattern of double fertilization forming the zygote and endosperm. Cell division dominates early embryogenesis up until the globular stage; afterwards cell differentiation overtakes division in mid to late embryogenesis. In conjunction, seed storage reserves accumulate, dormancy is established, and desiccation tolerance is acquired, resulting in the final mature seed (Capron et al., 2009).
The LAFL proteins, LEAFY COTYLEDON 2 (LEC2), ABSCISIC ACID INSENSTIVE 3 (ABI3), and FUSCA3 (FUS3) B3 domain transcription factors, together with LEC1 and LEC1-LIKE (L1L), homologs of the NF-YB subunit of the CCAAT-binding complex, are considered master regulators of seed development. Mutations in these genes cause reduced levels of seed storage compounds, desiccation intolerance, and/or enhanced precocious germination of immature embryos, indicating their critical role in seed maturation (Jia et al., 2013; Nonogaki, 2014; Fatihi et al., 2016). Mutant analyses suggest that each gene acts during different developmental stages of seed maturation to arrest embryo growth, with fus3-3 mutants acting earliest and showing premature growth of excised embryos, from the torpedo stage onwards (Raz et al., 2001). Almost complete repression of somatic embryogenic potential is seen in lec1/2 and fus3 mutants, further demonstrating the requirement of these genes for the embryonic program (Gaj et al., 2005). In accordance, in ChIP using somatic embryo tissue, FUS3 was bound to seed-specific promoters, with the majority containing the FUS3/LEC2/ABI3 target RY cis-motif (Reidt et al., 2000; Wang and Perry, 2013). Transcription of some of these seed-specific genes, such as those encoding storage proteins, CRUCIFERIN 3 (CRU3), LATE EMBRYOGENESIS ABUNDANT (LEA), and SEED STORAGE ALBUMIN 3 (2S3), have been demonstrated to be regulated by both FUS3 and ABI3 (Parcy et al., 1997; Nambara et al., 2000; Kagaya et al., 2005; Yamamoto et al., 2010; Roscoe et al., 2015).
Restriction of AFL function to seed development has been shown to be essential for the transition to vegetative growth and to be under epigenetic regulation (Jia et al., 2013; Fatihi et al., 2016). Ectopic expression of FUS3 during vegetative growth up-regulates seed-specific gene transcription, delays germination, growth, and flowering, and causes embryonic-like leaf development (Gazzarrini et al., 2004; Kagaya et al., 2005; Tsai and Gazzarrini, 2012b). Embryonic phenotypes are also shown in plants overexpressing the LEC genes (Lotan et al., 1998; Stone et al., 2001). Accordingly, in higher order viviparous1/abi3-like- (val), curly leaf- (clf), and pickle- (pkl) related mutants involved in chromatin remodeling, failure to suppress FUS3/LEC2/ABI3 expression during germination results in expression of embryonic traits and developmental arrest of seedlings (Jia et al., 2013; Fatihi et al., 2016).
Genetic, transcriptomic, and ChIP studies have shown that spatio-temporal expression of FUS3/LEC2/ABI3 is transcriptionally cross-regulated during seed development (Parcy et al., 1997; To et al., 2006; Mönke et al., 2012; Wang and Perry, 2013). Although extensive work has shown the importance of these genes in transcriptional regulation, little is known about how these proteins are themselves regulated during seed development. LAFL genes interact with hormone signaling and synthesis pathways at various levels to regulate seed development and inhibit germination (Jia et al., 2013). FUS3 in particular is a node in hormone crosstalk and acts as a molecular switch in the dormancy to germination transition (Gazzarrini and Tsai, 2015). FUS3 inhibits the synthesis of gibberellic acid (GA) and ethylene, and promotes that of abscisic acid (ABA) to induce dormancy while inhibiting germination and vegetative growth (Curaba et al., 2004; Gazzarrini et al., 2004; Lumba et al., 2012). ABA and GA positively and negatively regulate the FUS3 protein through a PEST degron (sequence rich in proline, glutamic acid, serine, and threonine), respectively (Gazzarrini et al., 2004; Lu et al., 2010). The FUS3 protein, which is not detected during germination at optimal temperature, accumulates during seed imbibition at high temperature due to an increase in the ABA/GA ratio, which inhibits FUS3 degradation through inhibition of proteasome activity. Seeds overexpressing FUS3 are hypersensitive to high temperature and inhibit germination through de novo ABA synthesis (Chiu et al., 2012, 2016a, b). This indicates that FUS3 accumulation is under tight hormonal control and regulates both primary and secondary dormancy.
Snf1-related kinase1 (SnRK1) is a conserved eukaryotic kinase complex involved in cellular energy homeostasis. In Arabidopsis, there are three homologs of the kinase α-subunit, of which AKIN10/SnRK1α1 and AKIN11/SnRK1α2 are functional (Crozet et al., 2014). Single mutants of SnRK1α show a slight delay in flowering and lower transcript levels of darkness-induced genes, while a virally induced akin10/akin11 RNAi double mutant undergoes premature senescence and is non-viable. SnRK1 overexpression elicits transcriptome shifts to inhibit growth and promote survival (Baena-González et al., 2007; Mair et al., 2015; Nukarinen et al., 2016). In peas (Pisum sativum), an antisense SnRK1α construct driven by a seed-specific promoter results in enhancement of vivipary, reduced desiccation tolerance, and lower levels of seed storage compounds and ABA, similar to fus3/lec1/lec2/abi3 mutants, and correspondingly causes reduced transcript levels of FUS3 and LEC1 orthologs. SnRK1 knockdown lines also displayed seed abortion and delayed embryo development, suggesting that SnRK1 plays an important role during seed development (Radchuk et al., 2006, 2010).
FUS3 is phosphorylated by AKIN10/SnRK1α1 (Tsai and Gazzarrini, 2012a). Overexpression of AKIN10 delays germination, senescence, and flowering (Baena-González et al., 2007; Tsai and Gazzarrini 2012a, b; Jeong et al., 2015), which can be rescued by fus3-3, indicating that FUS3 acts downstream of AKIN10 (Tsai and Gazzarrini, 2012a). Although overexpression of AKIN10 delays FUS3 degradation, the role of FUS3 phosphorylation by AKIN10 in seeds is unknown (Tsai and Gazzarrini, 2012a). In this study, we examined the function of the SnRK1 phosphorylation sites of FUS3 (S55, S56, S57) through analysis of FUS3 phosphorylation-null (FUS3S>A) and phosphorylation-mimic (FUS3S>D) mutants. While the FUS3S>D variant rescued fus3-3 seed maturation defects, FUS3S>A showed increased seed abortion, an increased number of seedling showing polycotyledons [previously observed in fus3-3 (Tsai and Gazzarrini, 2012a)], delayed embryo development, and reduced seed yield. Interestingly, akin10 and akin11 mutants displayed a frequency of seed abortion similar to fus3-3. Furthermore, when fus3-3 and FUS3S>A complemented lines were grown under elevated temperature, they displayed reduced plant vigor, seed yield, and seedling growth of the progeny. These results uncover a new function for FUS3 phosphorylation during embryogenesis and under heat stress.
Materials and methods
Plant material, growth conditions, and phenotypic analysis
The AKIN10–green fluorescent protein (GFP) transgenic line, and akin10 and akin11 mutants were previously described (Bitrián et al., 2011; Tsai and Gazzarrini, 2012a; Mair et al., 2015). FUS3 phosphomutants were generated by site-directed mutagenesis of a previously described pFUS3:FUS3-eGFP/pBI construct (Gazzarrini et al., 2004) using QuikChange® (Agilent). The non-phosphorylatable or phospho-null (pFUS3:FUS3S>A-GFP) and phosphorylation-mimic or phospho-mimic (pFUS3:FUS3S>D-GFP) transgenes were then introduced into the Arabidopsis Columbia fus3-3 mutant (Keith et al., 1994) by Agrobacterium-mediated plant transformation. Several transgenic plants were selected, and experiments were conducted using two independent homozygous lines. Arabidopsis seeds were germinated on half-strength Murashige and Skoog (MS) medium, transferred to soil at 7 d after germination, and grown under long days at 16 h light 21 °C/8 h dark 18 °C (optimal temperature regime) or 16 h light 27 °C/8 h dark 24 °C (high temperature regime).
For germination and seedling establishment assays, three sets of 50 seeds were cold stratified and germinated for 7 d. Bolting time was counted when the primary inflorescence reached 2 cm in length. An average of 15–20 plants per genotype was used. Experiments were repeated twice, and one is shown. Total seed production per silique and seed abortion were determined by counting the number of funiculi and seeds in eight siliques per genotype. Embryo development was scored by the morphological shape of dissected embryos from four siliques per genotype. Growth arrest was scored by growth of dissected embryos on 0.4% water agar after 5 d under constant light at 22 °C. Seed yield was measured from the total weight of dry seeds of eight plants per genotype grown in two pots.
Microscopy
Light microscopy images were taken using an SMZ1500 dissecting microscope (Nikon). Confocal images were taken using an LSM510 (Zeiss) with a 488 nm excitation laser with a 515–535 nm bandpass filter for GFP emission (green channel) and a 595 nm high-pass filter (red channel). Plant tissue was directly mounted on glass slides in 10% glycerol.
Protein expression analysis
Proteins were extracted by grinding frozen plant material in 2× Laemmli buffer (120 mM Tris pH 6.8, 20% glycerol, 4% SDS, 10% β-mercaptoethanol, 0.004% bromophenol blue) followed by 10 min of boiling and separation of debris by centrifugation. AKIN10 protein was detected using an AKIN10 antibody (Agrisera).
In-gel kinase activity assay
The in-gel kinase activity assay was performed as previously described (Tsai and Gazzarrini, 2012a). Six to ten frozen siliques were homogenized in extraction buffer (100 mM HEPES pH 7.5, 5 mM EDTA, 5 mM EGTA, 10 mM Na3VO4, 10 mM NaF,50 mM β-glycerophosphate, 10 mM DTT, 1 mM phenylmethylsulfonyl fluoride,1× protease inhibitor, 5% glycerol), and centrifuged twice. A 15 µg aliquot of cell lysate was separated on a 10% SDS–polyacrylamide gel containing 0.1 mg ml–1 glutathione S-transferase (GST)–FUS3. SDS was removed by washing the gel three times for 30 min at room temperature in washing buffer (25 mM Tris–HCl pH 7.5,0.5 mM DTT, 0.1 mM Na3VO4, 5 mM NaF, 0.5 mg ml–1 BSA, 0.1% Triton X-100). Proteins were renatured by three washes with renaturation buffer overnight (25 mM Tris–HCl, pH 7.5,1 mM DTT, 0.1 mM Na3VO4, 5 mM NaF). The gel was incubated in reaction buffer for 30 min (25 mM HEPES pH 7.5 2 mM EGTA, 12 mM MgCl2, 1 mM DTT,0.1 mM Na3VO4), then in 30 ml of reaction buffer with 200 nM ATP and 50 μCi of [γ-32P]ATP. The reaction was quenched in 5% trichloroacetic acid (w/v) and 1% sodium pyrophosphate (w/v) solution, washed five times to remove any unincorporated radioactivity, and then subjected to autoradiography.
Gene expression analysis
RNA was extracted from ground frozen plant material using the RNeasy Plant Mini Kit (Qiagen), followed by reverse transcription using GoScript™ (Promega). Quantative PCR was performed using SsoFast™ EvaGreen® (BioRad) with the CFX Connect™ real-time PCR detection system (BioRad). Primers used are listed in Supplementary Table S1 at JXB online.
Results
AKIN10 is expressed during embryogenesis and phosphorylates FUS3
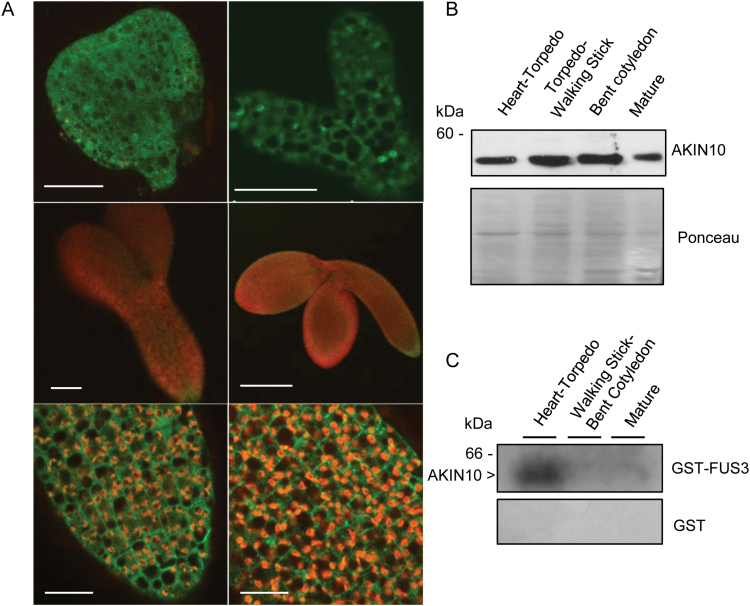
It has been previously shown that FUS3 is phosphorylated by AKIN10 on serine residues 55, 56, and 57 in an in-gel kinase assay using cell extract from seedlings (Tsai and Gazzarrini, 2012a). However, the FUS3 protein has only been detected during embryogenesis using a FUS3–GFP reporter, although mRNA is also found post-embryonically (Gazzarrini et al., 2004; Lu et al., 2010; Tsai and Gazzarrini, 2012a). To test whether AKIN10 may phosphorylate FUS3 in vivo, we first examined the expression pattern of AKIN10 during seed development. In transcriptomic studies, FUS3 and AKIN10 transcripts have been detected throughout embryo development, and as early as the elongated zygote stage (Le et al., 2010; Xiang et al., 2011). Using the AKIN10–GFP line previously described (Bitrián et al., 2011), the AKIN10 protein was found to be expressed in all cells from the heart to mature embryo stages, which was also confirmed by western blots (Fig. 1A, B; Supplementary Fig. S1). Thus, the AKIN10–GFP expression pattern during embryogenesis overlaps with that of FUS3–GFP previously published (Gazzarrini et al., 2004). Furthermore, in-gel kinase assays show that AKIN10 from cell extracts of siliques corresponding to early embryo stages can phosphorylate FUS3 (Fig. 1C; Supplementary Fig. S2), suggesting that AKIN10 interaction with and phosphorylation of FUS3 is biologically relevant and may play a role during early to mid embryogenesis.
Fig. 1.
AKIN10 is expressed during embryogenesis and phosphorylates FUS3. (A) Expression pattern of AKIN10 during seed development in Arabidopsis grown under long days (21 °C/18 °C). Confocal images showing AKIN10–GFP expression predominantly in the cytoplasm and occasionally in the nuclei of the embryo proper in the triangular (top, left), heart (top, right), torpedo (center and bottom, left), and bent cotyledon (center and bottom, right) stage embryos. Bottom images are magnifications of torpedo (left) and bent cotyledon (right) embryos shown in the center panels. Scale bars from left: top, 20 µm; middle, 50 µm, 200 µm; bottom, 20 µm. (B) AKIN10 protein was detected throughout seed development from total soluble protein of isolated seeds using anti-AKIN10. Ponceau stain is shown as the loading control. (C) In-gel kinase assay using purified GST–FUS3 (top) or GST (bottom) as the substrates and cell extracts from siliques showing a phosphorylation band at the expected size of AKIN10 at the heart/torpedo stages. Purified GST was used as the negative control. (This figure is available in colour at JXB online.)
FUS3 phosphorylation at SnRK1 sites positively regulates expression of FUS3 target genes
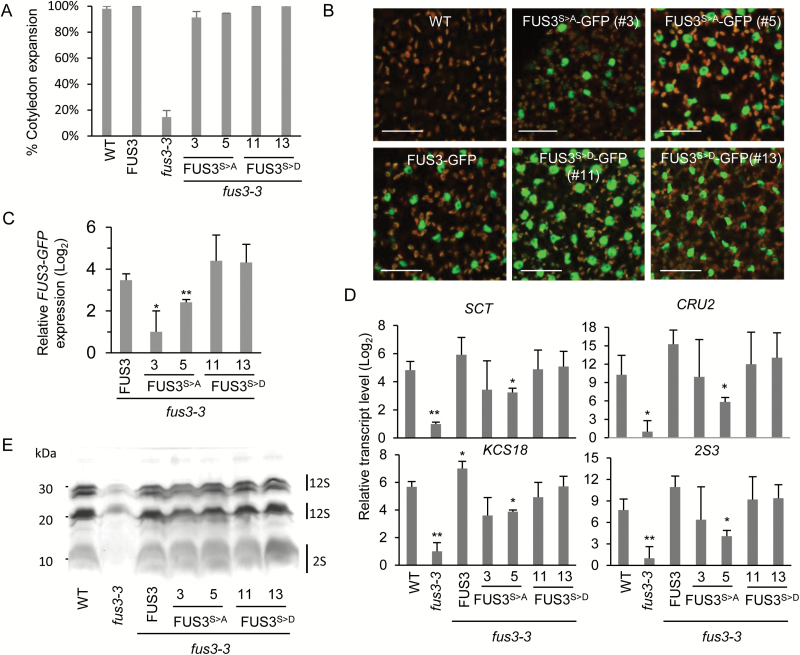
To elucidate the function of FUS3 phosphorylation by SnRK1, we mutated serine residues 55, 56, and 57 of the pFUS3:FUS3-GFP construct (Gazzarrini et al., 2004) to alanine and aspartic acid to generate phosphorylation-null (FUS3S>A) and potential phosphorylation-mimic (FUS3S>D) constructs driven by the FUS3 promoter, respectively. Substitution of serine with aspartic acid has been shown to mimic phosphorylation in several proteins including transcription factors (Maréchal et al., 1999; Kawamoto et al., 2015; Zorzatto et al., 2015). Both constructs were introduced into the fus3-3 background and rescued the desiccation intolerance phenotype of fus3-3 seeds (Fig. 2A). In the embryo, FUS3S>A–GFP and FUS3S>D–GFP were predominately localized to the nucleus, similar to wild-type FUS3–GFP, indicating that phosphorylation was not required for FUS3 subcellular localization and both mutant proteins were functional (Fig. 2B).
Fig. 2.
Expression levels of FUS3 and FUS3 target genes in FUS3 phosphomutants. (A) Seedling establishment (cotyledon expansion) assay of 3-month-old seeds showing rescue of fus3-3 desiccation intolerance by FUS3:FUS3-GFP, FUS3:FUS3S>A-GFP (lines #3 and #5), and FUS3:FUS3S>D-GFP (lines #11 and #13) phosphomutant lines. Seeds were cold stratified for 3 d and grown under constant light at 22 °C. The averages of three plates of 50 seeds ±SD are shown. (B) FUS3S>A–GFP and FUS3S>D–GFP were detected in the nuclei of almost all embryo proper cells in the cotyledon of walking stick/bent embryos. Scale bar=20 µm. (C) qPCR showing decreased FUS3S>A-GFP and slightly increased FUS3S>D-GFP transcript levels in transgenic lines. All plants were grown under long days (21 °C/18 °C) and siliques were collected at ~10 DAP (walking stick/bent cotyledon stages). (D) Transcript levels of FUS3 target genes measured by qPCR. 12S storage protein CRU2 (At1G03880), 2S storage protein 2S3 (At4G27160), fatty acid elongase FAE1/KCS18 (AT4G34520), and scorpion toxin proteinase/trypsin inhibitor SCT (At1g47540). FUS3S>A lines show lower transcript levels of most genes. Averages of three biological replicates ±SD are shown. Statistical significance against the wild type (WT) calculated by Welch’s t-test (*P<0.05, **P<0.01). All plants were grown under long days (21 °C/18 °C) and siliques were collected at ~10 DAP (walking stick/bent cotyledon stages). (E) Seed storage proteins extracted from 1 mg of dry seed visualized by Coomassie staining on a 15% SDS–polyacrylamide gel electrophoresis. (This figure is available in colour at JXB online.)
Interestingly, fewer FUS3S>A lines were recovered compared with FUS3S>D, suggesting that phospho-null mutations provide weaker rescue. This may be due to lower mRNA levels of FUS3S>A compared with FUS3 and FUS3S>D transgenes (Fig. 2C). Given that FUS3 binds to its own promoter and increases its own expression (Parcy et al., 1997; To et al., 2006), these data suggest that FUS3 transcriptional activity may be increased by phosphorylation. To test this hypothesis, we investigated FUS3 transcriptional target gene expression. ChIP studies have shown that FUS3 binds to the promoters of genes that are highly expressed during seed maturation (Wang and Perry 2013), including those encoding 12S storage protein CRU2 (At1G03880), 2S storage protein 2S3 (At4G27160), fatty acid elongase FAE1/KCS18 (AT4G34520), and scorpion toxin proteinase/trypsin inhibitor SCT (At1g47540). These genes are expressed throughout embryogenesis (Toufighi et al., 2005) and their transcript levels were strongly reduced in fus3-3; thus, they were good markers for FUS3 transcriptional activity (Fig. 2D; Yamamoto et al., 2010). Transcript measurements taken at the bent cotyledon stage [10 days after pollination (DAP)] show that the FUS3S>A line has lower transcript levels of these genes compared with the FUS3 and FUS3S>D lines (Fig. 2D). However, the levels of storage proteins in dry seeds did not appear to be affected in FUS3S>A lines (Fig. 2E). Thus, although phosphorylation modulates FUS3 transcriptional activity, it did not appear to be required for seed maturation processes such as desiccation tolerance and storage protein accumulation under normal growth conditions.
FUS3 phosphorylation is required for early embryo development and positively impacts seed yield
Previous work has shown that silencing and overexpression of AKIN10 cause dramatic phenotypes during vegetative and reproductive development, including alteration of growth rate, senescence, and fertility (Baena-González et al., 2007; Tsai and Gazzarrini, 2012a, b). This suggests that phosphorylation of FUS3 by SnRK1 could affect vegetative and reproductive development. When grown under long days at 21 °C/18 °C, FUS3S>D lines consistently displayed a delay in flowering (Fig. 3A), similar to plants overexpressing FUS3 and AKIN10 (Gazzarrini et al., 2004; Baena-González et al., 2007; Lu et al., 2010; Tsai and Gazzarrini, 2012b). Given that neither FUS3 nor FUS3-phosphomutant proteins were expressed during vegetative development (Supplementary Fig. S3), embryonic FUS3S>D could affect the regulation of genes that control flowering time and are also expressed in the embryo, such as the floral repressor FLOWERING LOCUS C or other regulators of flowering that are expressed in the embryo (Sheldon et al., 2008).
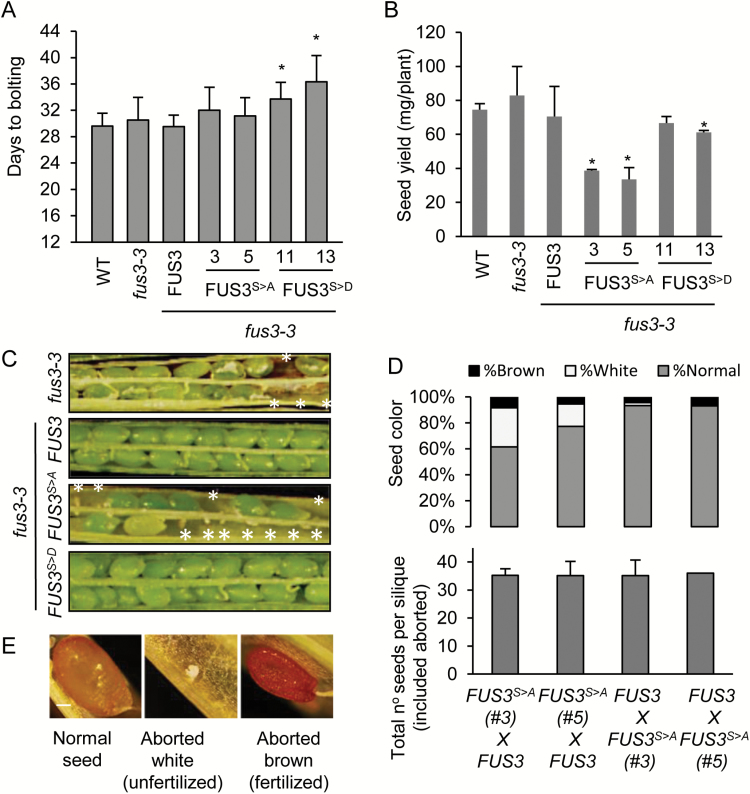
Fig. 3.
Increased seed abortion and reduced seed yield in FUS3 phospho-null mutants. (A) Bolting time of wild-type, fus3-3, FUS3S>A, and FUS3S>D plants. Averages of 15–20 plants ±SEM (P<0.01) per genotype are shown. Arabidopsis plants were grown under long days at 21 °C/18 °C. (B) Seed yield of FUS3S>A and FUS3S>D plants grown under long days at 21 °C/18 °C. Averages ±SD of total dry seed yield from eight plants grown in two pots is shown (P<0.01). (C) Representative picture showing seed morphology in yellowing siliques. fus3-3 and FUS3S>A show aborted seeds (empty spaces indicated by*). (D) Frequency of seed abortion in F1 progeny from reciprocal crosses (♀ × ♂) between FUS3 and FUS3S>A. The total number of seeds per silique (bottom) and frequency of aborted white or brown seeds are shown (top). (E) Representative picture showing normal, aborted white (unfertilized), and brown (fertilized) seeds. (This figure is available in colour at JXB online.)
In contrast, FUS3S>A plants appeared less vigorous and their seed yield was reduced by ~50% compared with the wild type and fus3-3 (Fig. 3B). To understand the cause of decreased seed yield of the FUS3S>A plants, the seed set per silique and quality of seeds produced were examined. When grown under long days, 25–35% of the FUS3S>A seeds were aborted (Fig. 3C; Table 1). Aborted seeds were randomly distributed within a silique and among siliques. Surprisingly, seed abortion was also displayed by fus3-3, but at a much lower frequency, ~8% (Table 1). To test if the higher number of aborted seeds of FUS3S>A was due to a zygotic or gametic effect, we crossed FUS3S>A with FUS3 and analyzed the F1 progeny of reciprocal crosses. When FUS3S>A was the pollen donor, seed abortion was comparable with fus3-3, at 7%, indicating that maternal FUS3 and paternal FUS3S>A did not rescue the fus3-3 seed abortion phenotype. However, in the reciprocal cross with maternal FUS3 S>A and paternal FUS3, seed abortion was increased to 23–39%, with a higher frequency of white aborted seeds due to unfertilized ovules (Fig. 3D, E; Table 2). These results show that the lower seed yield of FUS3S>A is due in part to an increase in seed abortion due to a maternal effect of FUS3S>A. Lastly, akin10 and akin11 mutants showed a rate of seed abortion similar to that of fus3-3 (Table 1), suggesting that FUS3 and AKIN10 may interact to regulate early embryo development processes.
Table 1.
Frequency of aborted seeds in various genotypes grown at optimal temperature
| Genotype | Total no. of seeds | Total no. of aborted seeds | % Aborted seeds |
|---|---|---|---|
| Wild type | 398 | 8 | 2% |
| fus3-3 | 342 | 26 | 8% |
| fus3-3, FUS3 | 381 | 7 | 2% |
| fus3-3, FUS3 S>A (3) | 284 | 100 | 35% |
| fus3-3, FUS3 S>A (5) | 327 | 81 | 25% |
| fus3-3, FUS3 S>D (11) | 383 | 23 | 6% |
| fus3-3, FUS3 S>D (13) | 396 | 8 | 2% |
| akin10 | 381 | 44 | 12% |
| akin11 | 402 | 39 | 10% |
Total seeds produced from eight siliques including aborted seeds were counted.
Seed abortion included both fertilized and unfertilized ovules.
Plants were grown under long days at 21 °C/18 °C.
Table 2.
Frequency of aborted seeds in progeny from F1 crosses
| Crosses (♀×♂) | Total no. of seeds | White aborted seeds | Brown aborted seeds | Total no. of aborted seeds |
|---|---|---|---|---|
| FUS3 S>A (3)×FUS3 | 179 | 54 (30%) | 15 (8%) | 69 (39%) |
| FUS3 S>A (5)×FUS3 | 314 | 54 (17%) | 17 (5%) | 61 (23%) |
| FUS3×FUS3 S>A (3) | 210 | 5 (2%) | 9 (4%) | 14 (7%) |
| FUS3×FUS3 S>A (5) | 72 | 0 (0%) | 5 (7%) | 5 (7%) |
Total seeds produced including aborted seeds from crossed plants were counted after mature seeds began browning.
Aborted seeds were classified as unfertilized ovules (white) and aborted embryos (brown) based on coat color and morphology.
Plants were grown under long days at 21 °C /18 °C.
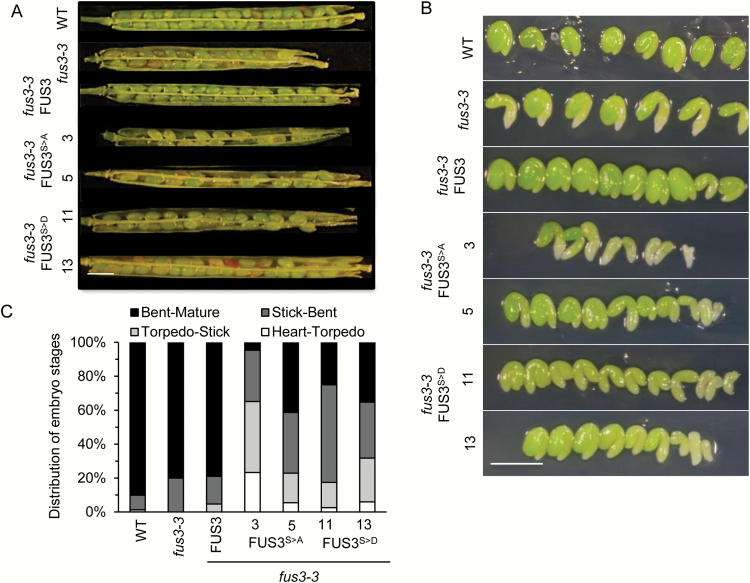
FUS3 phosphorylation positively regulates embryo growth rate
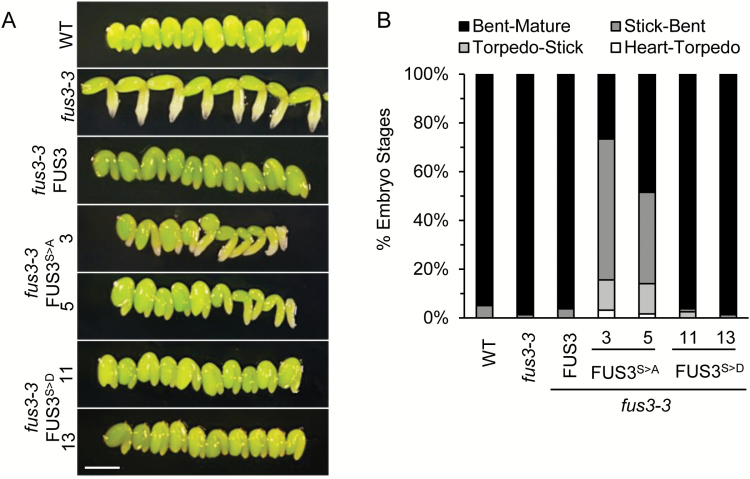
Upon closer inspection, many FUS3S>A siliques also showed delayed embryo development. While almost all wild-type embryos reached the late bent cotyledon stage at 11 DAP, <50% of the FUS3S>A embryos were at a comparable stage (Fig. 4). Most FUS3S>A embryos were closer to the walking stick stage and 10–15% were still at the torpedo or earlier stages. In contrast, the developmental progression of FUS3S>D embryos was not significantly different from that of the wild type. Unlike FUS3S>D, FUS3S>A did not rescue and instead enhanced the fus3-3 cotyledon defects, such as polycotyledons (1.5%), previously reported (Table 3; Tsai and Gazzarrini, 2012a). Together, these results indicate that FUS3 phosphorylation promotes embryo growth and is required for proper cotyledon development.
Fig. 4.
Delayed seed development of FUS3 phospho-null mutants at control temperature. Embryo development of wild-type, fus3-3, FUS3S>A, and FUS3S>D plants grown under long days at 21 °C/18 °C. (A) Representative images showing the morphology of FUS3 phosphomutant embryos excised from 11 DAP siliques, corresponding to the bent cotyledon/mature stage in the wild type. fus3-3 embryos skip dormancy and enter vegetative growth precociously, as seen from root growth and embryo size. FUS3S>A embryo growth is delayed, as seen from the presence of early embryo stages (torpedo and walking stick). Scale bar=0.5 mm. (B) Quantification of embryo stages shown in (A). Embryos dissected from four siliques per genotype were scored for morphological stages (n=60–80). (This figure is available in colour at JXB online.)
Table 3.
Frequency of cotyledon defects in FUS3 phosphomutants
| Genotype | Total no. of seedlings | Defective cotyledonsa | % Defective cotyledons |
|---|---|---|---|
| Wild type | 1050 | 0 | 0% |
| fus3-3, FUS3 | 1050 | 2 | 0.2% |
| fus3-3, FUS3 S>A (3) | 1050 | 60 | 5.7% |
| fus3-3, FUS3 S>A (5) | 1050 | 46 | 4.4% |
| fus3-3, FUS3 S>D (11) | 1050 | 3 | 0.3% |
| fus3-3, FUS3 S>D (13) | 1050 | 2 | 0.2% |
a Cotyledon defects include altered cotyledon number (polycotyledons or single cotyledon), fused cotyledons, and twin embryos.
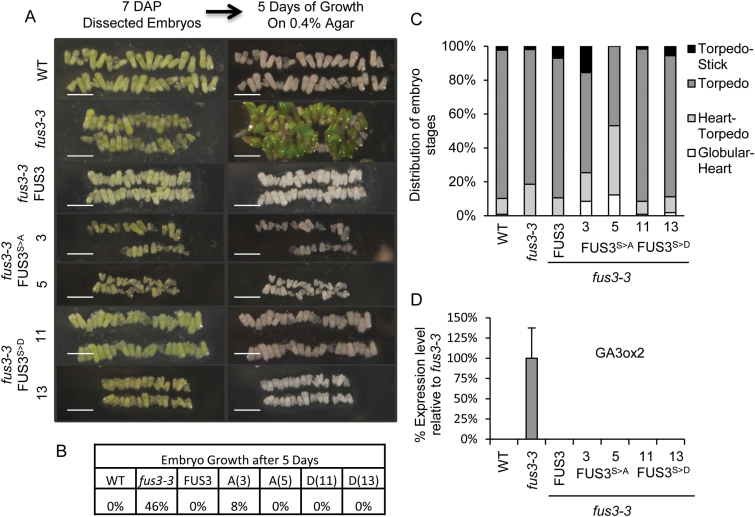
In agreement with the literature, fus3-3 embryos showed precocious vegetative growth, as shown by growth of the radicle (Keith et al., 1994), and all FUS3 phosphomutants rescued this phenotype (Fig. 4A). Previous work has shown that fus3-3 mutants fail to suppress growth of excised embryos (Raz et al., 2001). Therefore, we asked if the same growth suppression mechanism caused the difference in embryo development arrest or delay. Using the same growth assay on torpedo stage embryos (7 DAP), we observed no significant difference in the ability to complement the growth suppression defect of fus3-3 by both FUS3 variants (Fig. 5A, B). Accordingly, expression of GA3ox2, a GA biosynthetic gene, is derepressed in fus3-3 but not in the FUS3 phospho-mutant embryos (Fig. 5D; Curaba et al., 2004; Gazzarrini et al., 2004; Wang and Perry, 2013). A lower level of asynchronous embryo development was also observed in the early embryo stage of fus3-3 (Fig. 5C). Overall, these data indicate that FUS3 phosphorylation positively regulates the embryo growth rate, but is not required for suppression of precocious germination.
Fig. 5.
Suppression of growth in excised FUS3 phosphomutant embryos. (A) Representative images showing growth (fus3-3) or suppression of growth of embryos on 0.4% agar for 5 d. Arabidopsis plants were grown under long days at 21 °C/18 °C, and embryos at 7 DAP were dissected from four siliques (n=100–200). Scale bar=1 mm. (B) Quantification of excised embryo growth. (C) Distribution of total developing seeds excised from four siliques (7 DAP). (D) Transcript levels of GA3ox2 measured by qPCR. GA3ox2 expression was only detected in fus3-3. All plants were grown under long days at 21 °C/18 °C and siliques were collected at ~10 DAP (walking stick/bent cotyledon stages). (This figure is available in colour at JXB online.)
FUS3 phosphorylation is essential for plant growth and seed development at high temperature
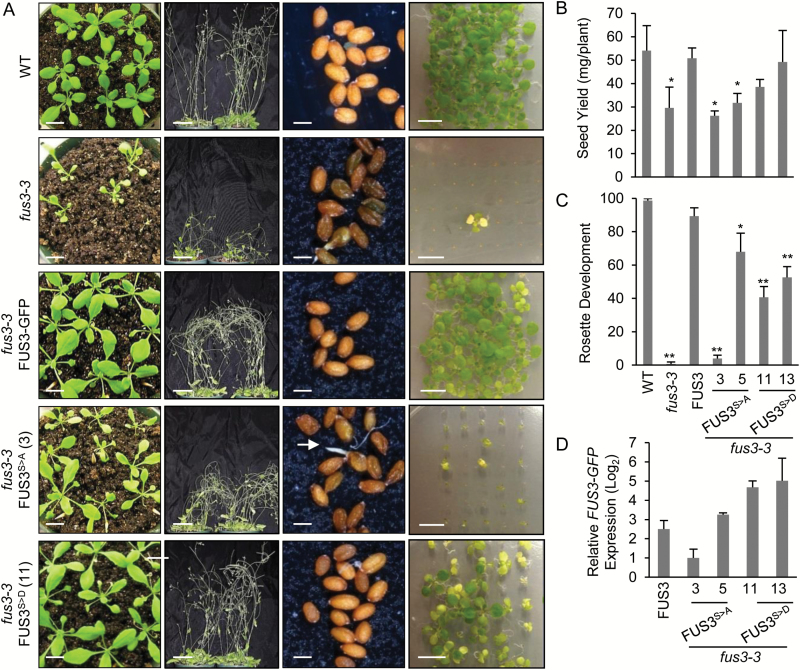
The FUS3 protein is not detected post-embryonically under optimal growth conditions; however, it accumulates in seeds imbibed at supraoptimal temperature to inhibit germination (Chiu et al., 2012). We examined whether FUS3 phosphorylation by SnRK1, a major regulator of the stress response, plays a role during growth at elevated temperature. When grown under long days at 27/24 °C, the overall growth was negatively impacted in fus3-3 and FUS3S>A plants, which showed smaller rosettes, weaker inflorescences that were unable to stand upright, and reduced seed yield compared with the wild type (Fig. 6A, B). FUS3S>A line 3 also produced shriveled seeds with a low germination rate, poor progeny seedling establishment, and yellowing of the cotyledons at 27 °C, similar to fus3-3 (Fig. 6A, C). The mRNA level of FUS3S>A of line 3 was strongly reduced, explaining the lack of fus3-3 rescue of this line at 27 °C (Fig. 6D). When plants were grown at elevated temperature, both FUS3S>A and FUS3S>D variants caused delayed embryo development and increased seed abortion compared with the wild type and fus3-3 (Fig. 7; Table 4). This suggests that while aspartic acid may mimic phosphorylation under control conditions, effective similarity is reduced by elevated temperature. Alternatively, constitutive phosphorylation (S>D) may not be as effective as regulated phosphorylation. Altogether, these data indicate an essential role for FUS3 phosphorylation during growth at high temperature. Given that neither FUS3 nor its phosphomutant variants were detected during vegetative growth at high temperature (Supplementary Fig. S4), these results also suggest that FUS3 phosphorylation in the mother plant integrates temperature cues to regulate growth and survival of the progeny indirectly.
Fig. 6.
Vegetative and reproductive growth of FUS3 phosphomutants is greatly affected at elevated temperatures. Phenotypic analysis of FUS3 phosphomutants germinated for 7 d on agar plates and transferred to soil under long days at elevated temperatures (27 °C/24 °C). (A) Vegetative growth of fus3-3 and FUS3S>A mutants is greatly affected at high temperature, as can be seen from reduced rosette size (first column), less erect inflorescences (second column), and increased numbers of shriveled seeds with occasional vivipary (white root shown by arrow) (third column) and lowered seedling establishment of the progeny (fourth column). Scale bars=(left to right) 1, 5, 0.1, 0.9 cm. (B) Seed yield is greatly reduced in fus3-3 and FUS3S>A plants compared with the wild type. n=8 plants; P<0.05 (Student’s t-test). (C) The progeny of FUS3 phosphomutants show lower seedling establishment (emergence of leaf primordia) 15 d after imbibition. The averages of three plates of 50 seeds ±SD are shown. (D) Relative transcript levels were measured by qPCR. Siliques were collected approximately at the bent cotyledon stage (9 DAP). The FUS3S>D transgene is expressed at higher levels compared with FUS3, while the FUS3S>A transgene (line 3) shows a lower expression level. (This figure is available in colour at JXB online.)
Fig. 7.
Development of FUS3 phosphomutant embryos at elevated temperature. Embryo development was examined in FUS3 phosphomutants grown under long days at elevated temperature (27 °C/24 °C). (A) Representative images showing aborted seeds in siliques at 14+ DAP. Scale bar=1 mm. (B) Representative images showing embryo morphology of FUS3 phosphomutants dissected from siliques at 9 DAP corresponding to the bent cotyledon/mature stage in the wild type. Scale bar=0.5 mm. (C) Quantification of embryo developmental stages in siliques at 9 DAP; four siliques per genotype (n=60–80). (This figure is available in colour at JXB online.)
Table 4.
Frequency of aborted seeds in various genotypes grown at high temperature
| Genotype | Total no. of seeds | Total no. of aborted seeds | % Aborted seeds |
|---|---|---|---|
| Wild type | 283 | 29 | 10% |
| fus3-3 | 202 | 33 | 16% |
| fus3-3, FUS3 | 254 | 20 | 8% |
| fus3-3, FUS3 S>A (3) | 220 | 71 | 32% |
| fus3-3, FUS3 S>A (5) | 263 | 85 | 32% |
| fus3-3, FUS3 S>D (11) | 280 | 75 | 27% |
| fus3-3, FUS3 S>D (13) | 283 | 61 | 22% |
Total seeds produced from eight siliques including aborted seeds were counted.
Seed abortion included both fertilized and unfertilized ovules.
Plants were grown under long days at 27 °C/24 °C.
Discussion
The LAFL transcription factors are master regulators of the seed maturation program. Through alteration of the transcriptome, they co-ordinate the accumulation of seed storage compounds, the establishment of dormancy, and acquisition of desiccation tolerance, while suppressing germination (Jia et al., 2013; Sreenivasulu and Wobus 2013; Fatihi et al., 2016). Compared with their role in transcription, there is less known about upstream regulators and modulators of their functions. Previously, we have shown that FUS3 interacts with and is phosphorylated by the SnRK1α1/AKIN10 kinase on three serine residues in the conserved B2 domain (Tsai and Gazzarrini, 2012a). However, the biological role of this post-translational modification was not clear. Here we found that AKIN10 is expressed in embryos, from the triangular to mature stages, and phosphorylates FUS3 during early to mid stages of embryogenesis. Using FUS3 phosphomutants at the SnRK1 target sites, we show that FUS3 phosphorylation plays a crucial role during embryogenesis, by regulating embryo growth rate and seed development under optimal and stress conditions. Impairment of FUS3 phosphorylation negatively affects FUS3 transcriptional activity, increases seed abortion, delays embryo development, and ultimately results in a large decrease in seed yield. The importance of FUS3 phosphorylation is more evident when plants are grown at high temperature, as even germination and vegetative growth of the next generation is greatly impacted. This suggests that SnRK1 and FUS3 may integrate endogenous signals (energy level) and environmental cues to regulate seed development and ensure survival of the next generation.
Previously, we showed that SnRK1 phosphorylation positively regulates FUS3, and that FUS3 acts downstream of AKIN10 in Arabidopsis (Tsai and Gazzarrini, 2012a). However, AKIN10 was previously found to be expressed only in the ovule, but not in the embryo where FUS3 mainly functions (Bitrián et al., 2011). Here we show that AKIN10 is indeed expressed in the embryo, and that FUS3 and AKIN10 protein expression patterns partially overlap during embryogenesis. This is in agreement with the expression of the regulatory subunit SnRK1βγ in embryos (Bitrián et al., 2011). Furthermore, FUS3 is phosphorylated by AKIN10 in cell extracts of early to mid embryos, thus their interaction is biologically relevant. In pea seeds, SnRK1α knockdown lines are impaired in maturation and show phenotypes similar to those displayed by fus3 and abi3 mutants, including precocious germination, reduced ABA and seed storage levels, and reduced desiccation tolerance (Radchuk et al., 2006, 2010). This suggests that SnRK1 plays a prominent role during seed development and maturation, and probably acts through regulation of AFL functions.
To understand the role of FUS3 phosphorylation by SnRK1, we transformed fus3-3 with FUS3:FUS3S>D-GFP phospho-mimic and FUS3:FUS3S>A-GFP phospho-null constructs. Our expression studies indicate that down-regulated gene transcription in fus3-3 mutants is fully restored in FUS3S>D, but only partially restored in FUS3S>A embryos, suggesting that FUS3 transcriptional activity is increased by phosphorylation. Furthermore, FUS3S>A transgene expression was reduced while that of FUS3S>D was increased compared with FUS3. This may be due to reduced FUS3S>A and slightly increased FUS3S>D ability, respectively, to regulate the FUS3 promoter positively, as previously shown for FUS3 (Parcy et al., 1997; To et al., 2006; Wang and Perry, 2013). However, FUS3 phosphorylation by SnRK1 did not appear to be required for seed maturation at normal temperature. The dramatic fus3-3 phenotypes, including reduced dormancy, low levels of seed storage compounds, desiccation intolerance, and ectopic trichomes on the cotyledons (Bäumlein et al., 1994; Keith et al., 1994; Meinke et al., 1994), were rescued by expressing either FUS3S>A or FUS3S>D transgenes. Altogether, these data suggest that SnRK1 phosphorylation positively regulates FUS3 expression and transcriptional activity, but does not play a prominent role during seed maturation. It is possible that the full rescue of fus3-3 seed maturation phenotypes in FUS3S>A lines may be due to the action of other LAFL genes. Expression of FUS3 and its target genes is induced by other B3 domain proteins through binding of the RY motif, which may compensate for reduced FUS3S>A transcriptional activity (Jia et al., 2013).
Upon closer inspection of siliques from plants grown under optimal temperatures, we found that ~8% of the fus3-3 seeds were aborted and, surprisingly, FUS3S>A lines showed an increase in the frequency of aborted seeds (~30%) due to a maternal FUS3S>A effect. Furthermore, FUS3S>A lines also showed delayed embryo development, with 10–15% of the seeds still at the heart to torpedo stages when all wild-type embryos already reached the mature stage. This resulted in a reduction of seed yield by ~50% in FUS3S>A plamts. Thus, it appears that in FUS3S>A siliques only half of the seeds were able to develop fully and reach the mature stage, and these embryos did not show maturation defects. Notably, akin10 and to a lesser extent akin11 mutants showed frequencies of seed abortion close to that of fus3-3, and similar phenotypes were observed during seed development of antisense SnRK1α lines (with 30–50% reduction in SnRK1 activity) in pea, where 10% seed abortion and delayed embryo development was also reported (Radchuk et al., 2006). Interestingly, FUS3 phosphorylation affects the vegetative development and flowering time of the progeny, despite the lack of FUS3 protein expression post-embryonically, suggesting a link between seed development and flowering time. While FUS3S>D rescued all fus3-3 embryonic phenotypes, the FUS3S>D progeny flowered later than the wild type at control temperature, similarly to ML1:FUS3 plants that overexpress FUS3 post-embryonically (Gazzarrini et al., 2004). During seed development, FUS3S>D may alter the regulation of flowering-related genes such as FLC, whose expression in the embryo was shown to be required for its repression of flowering (Sheldon et al., 2008), or DELAY OF GERMINATION1 (DOG1), which controls dormancy and flowering time through miRNA 156/172 (Huo et al., 2016). DOG1 and miRNA 156/172 are FUS3 targets based on ChIP studies (Wang and Perry, 2013).
When plants were grown at high temperature, plant growth was accelerated and seed yield was reduced in all genotypes. All phenotypes displayed by FUS3S>A lines were enhanced when plants were grown at high temperature, in particular, both fus3-3 and FUS3S>A plants were less vigorous and unable to stand upright, and the progeny seedlings grew poorly compared with the wild type. This indicates that phosphorylation of FUS3 by SnRK1 is critical during heat stress and affects the survival of the next generation. Possibly, specific transcriptional activities may be altered if phosphorylation of FUS3 changes the nature of ternary transcription complexes with LEC1/NF-YB, which increase the transcriptional activity of ABI3/LEC2 on the OLE1 promoter (Baud et al., 2016). Alternatively, this may arise from novel protein–protein interactions. Thus, FUS3 phosphorylation in the mother plant integrates temperature cues to regulate growth and survival of the progeny. Interestingly, progeny dormancy is regulated by maternal temperature through activation of the florigen Flowering Locus T in siliques (Chen et al., 2014; Penfield and MacGregor, 2017). It will be interesting to determine if maternal temperature affects both progeny dormancy and flowering time through similar pathways, thus passing environmental memory across generations to optimize plant growth.
The mechanism of how prevention of FUS3 phosphorylation causes seed abortion and delayed embryo development is not known. However, one possibility is that FUS3-mediated regulation of cell division is dependent on energy levels, which fluctuate in response to growth conditions and are signaled through SnRK1. FUS3 was previously shown to regulate cell division negatively; ectopic cell division was observed in the fus3-3 embryo, while FUS3 overexpression represses cell cycling (Raz et al., 2001; Gazzarrini et al., 2004). FUS3 directly binds to the promoter of cycD3;1, and cycD activity suppressors KIP-RELATED PROTEIN (KRP) 4 and 6, which have reduced expression in the fus3-3 mutant (Yamamoto et al., 2010; Wang and Perry, 2013). Higher order mutants of the cyclin-D3 (cycD3) cell cycle regulator subfamily also display seed abortion and delayed embryo development phenotypes shown by FUS3S>A lines (Collins et al., 2012). Thus, FUS3 phosphorylation may control the rate of cell cycling to modulate the rate of embryo development and transition to maturation. Recently, a mutant affected in a novel nuclear-localized protein, red1, also displayed seed abortion, delayed embryo development, and desiccation intolerance phenotypes described in this study (Du and Wang, 2016). However, the function of this gene is currently unknown.
In conclusion, this work uncovered a novel and essential role for FUS3 phosphorylation by SnRK1 during early embryogenesis at optimal and especially at high temperatures. Lack of phosphorylation impairs FUS3 function and results in unfertilized ovules, aborted seeds, and delayed embryo growth, suggesting metabolic and signaling impairment. Seed abortion, reduced seed yield, and overall plant growth were further impacted when plants were grown at elevated temperature, suggesting that phosphorylation is essential for FUS3 function under stress, possibly integrating energy and stress levels through SnRK1 phosphorylation to ensure the survival of the next generation. Future work will focus on elucidating the molecular mechanisms underlying these processes.
Supplementary data
Supplementary data are available at JXB online.
Table S1. List of primers used in this research.
Fig. S1. AKIN10 protein expression levels during seed development.
Fig. S2. In-gel kinase assays.
Fig. S3. FUS3–GFP and its phospho-variants are undetectable during early vegetative development at control temperature.
Fig. S4. FUS3–GFP and its phospho-variants are undetectable during early vegetative development under elevated temperature.
Supplementary Material
Acknowledgements
This study was supported by a National Science and Engineering Council of Canada (NSERC) grant to SG. The authors have no conflict of interest to declare.
References
- Baena-González E, Rolland F, Thevelein JM, Sheen J. 2007. A central integrator of transcription networks in plant stress and energy signalling. Nature 448, 938–942. [DOI] [PubMed] [Google Scholar]
- Baud S, Kelemen Z, Thévenin J et al. 2016. Deciphering the molecular mechanisms underpinning the transcriptional control of gene expression by master transcriptional regulators in Arabidopsis seed. Plant Physiology 171, 1099–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäumlein H, Miséra S, Luerßen H, Kölle K, Horstmann C, Wobus U, Müller A. 1994. The FUS3 gene of Arabidopsis thaliana is a regulator of gene expression during late embryogenesis. The Plant Journal 6, 379–387. [Google Scholar]
- Bitrián M, Roodbarkelari F, Horváth M, Koncz C. 2011. BAC-recombineering for studying plant gene regulation: developmental control and cellular localization of SnRK1 kinase subunits. The Plant Journal 65, 829–842. [DOI] [PubMed] [Google Scholar]
- Capron A, Chatfield S, Provart N, Berleth T. 2009. Embryogenesis: pattern formation from a single cell. The Arabidopsis Book 7, e0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, MacGregor DR, Dave A, Florance H, Moore K, Paszkiewicz K, Smirnoff N, Graham IA, Penfield S. 2014. Maternal temperature history activates Flowering Locus T in fruits to control progeny dormancy according to time of year. Proceedings of the National Academy of Sciences, USA 111, 18787–18792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu RS, Nahal H, Provart NJ, Gazzarrini S. 2012. The role of the Arabidopsis FUSCA3 transcription factor during inhibition of seed germination at high temperature. BMC Plant Biology 12, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu RS, Pan S, Zhao R, Gazzarrini S. 2016a. ABA-dependent inhibition of the ubiquitin proteasome system during germination at high temperature in Arabidopsis. The Plant Journal 88, 749–761. [DOI] [PubMed] [Google Scholar]
- Chiu RS, Saleh Y, Gazzarrini S. 2016b. Inhibition of FUSCA3 degradation at high temperature is dependent on ABA signaling and is regulated by the ABA/GA ratio. Plant Signaling and Behavior 11, e1247137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C, Dewitte W, Murray JA. 2012. D-type cyclins control cell division and developmental rate during Arabidopsis seed development. Journal of Experimental Botany 63, 3571–3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozet P, Margalha L, Confraria A, Rodrigues A, Martinho C, Adamo M, Elias CA, Baena-González E. 2014. Mechanisms of regulation of SNF1/AMPK/SnRK1 protein kinases. Frontiers in Plant Science 5, 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curaba J, Moritz T, Blervaque R, Parcy F, Raz V, Herzog M, Vachon G. 2004. AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiology 136, 3660–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q, Wang H. 2016. Retarded Embryo Development 1 (RED1) regulates embryo development, seed maturation and plant growth in Arabidopsis. Journal of Genetics and Genomics 43, 439–449. [DOI] [PubMed] [Google Scholar]
- Fatihi A, Boulard C, Bouyer D, Baud S, Dubreucq B, Lepiniec L. 2016. Deciphering and modifying LAFL transcriptional regulatory network in seed for improving yield and quality of storage compounds. Plant Science 250, 198–204. [DOI] [PubMed] [Google Scholar]
- Gaj MD, Zhang S, Harada JJ, Lemaux PG. 2005. Leafy cotyledon genes are essential for induction of somatic embryogenesis of Arabidopsis. Planta 222, 977–988. [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, Tsai AY. 2015. Hormone cross-talk during seed germination. Essays in Biochemistry 58, 151–164. [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, Tsuchiya Y, Lumba S, Okamoto M, McCourt P. 2004. The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Developmental Cell 7, 373–385. [DOI] [PubMed] [Google Scholar]
- Huo H, Wei S, Bradford KJ. 2016. DELAY OF GERMINATION1 (DOG1) regulates both seed dormancy and flowering time through microRNA pathways. Proceedings of the National Academy of Sciences, USA 113, E2199–E2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong EY, Seo PJ, Woo JC, Park CM. 2015. AKIN10 delays flowering by inactivating IDD8 transcription factor through protein phosphorylation in Arabidopsis. BMC Plant Biology 15, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, McCarty DR, Suzuki M. 2013. Distinct roles of LAFL network genes in promoting the embryonic seedling fate in the absence of VAL repression. Plant Physiology 163, 1293–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y, Okuda R, Ban A, Toyoshima R, Tsutsumida K, Usui H, Yamamoto A, Hattori T. 2005. Indirect ABA-dependent regulation of seed storage protein genes by FUSCA3 transcription factor in Arabidopsis. Plant and Cell Physiology 46, 300–311. [DOI] [PubMed] [Google Scholar]
- Kawamoto N, Sasabe M, Endo M, Machida Y, Araki T. 2015. Calcium-dependent protein kinases responsible for the phosphorylation of a bZIP transcription factor FD crucial for the florigen complex formation. Scientific Reports 5, 8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith K, Kraml M, Dengler NG, McCourt P. 1994. fusca3: a heterochronic mutation affecting late embryo development in Arabidopsis. The Plant Cell 6, 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le BH, Cheng C, Bui AQ et al. 2010. Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proceedings of the National Academy of Sciences, USA 107, 8063–8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ. 1998. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93, 1195–1205. [DOI] [PubMed] [Google Scholar]
- Lu QS, dela Paz J, Pathmanathan A, Chiu RS, Tsai AY, Gazzarrini S. 2010. The C-terminal domain of FUSCA3 negatively regulates mRNA and protein levels and mediates sensitivity to the hormones abscisic acid and gibberellic acid in Arabidopsis. The Plant Journal 64, 100–113. [DOI] [PubMed] [Google Scholar]
- Lumba S, Tsuchiya Y, Delmas F, Hezky J, Provart NJ, Shi Lu Q, McCourt P, Gazzarrini S. 2012. The embryonic leaf identity gene FUSCA3 regulates vegetative phase transitions by negatively modulating ethylene-regulated gene expression in Arabidopsis. BMC Biology 10, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal E, Hiratsuka K, Delgado J, Nairn A, Qin J, Chait BT, Chua NH. 1999. Modulation of GT-1 DNA-binding activity by calcium-dependent phosphorylation. Plant Molecular Biology 40, 373–386. [DOI] [PubMed] [Google Scholar]
- Mair A, Pedrotti L, Wurzinger B et al. 2015. SnRK1-triggered switch of bZIP63 dimerization mediates the low-energy response in plants. Elife 4, e05828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke DW, Franzmann LH, Nickle TC, Yeung EC. 1994. Leafy cotyledon mutants of Arabidopsis. The Plant Cell 6, 1049–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mönke G, Seifert M, Keilwagen J et al. 2012. Toward the identification and regulation of the Arabidopsis thaliana ABI3 regulon. Nucleic Acids Research 40, 8240–8254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Hayama R, Tsuchiya Y, Nishimura M, Kawaide H, Kamiya Y, Naito S. 2000. The role of ABI3 and FUS3 loci in Arabidopsis thaliana on phase transition from late embryo development to germination. Developmental Biology 220, 412–423. [DOI] [PubMed] [Google Scholar]
- Nambara E, Keith K, McCourt P, Naito S. 1994. Isolation of an internal deletion mutant of the Arabidopsis thaliana ABI3 gene. Plant and Cell Physiology 35, 509–513. [PubMed] [Google Scholar]
- Nonogaki H. 2014. Seed dormancy and germination—emerging mechanisms and new hypotheses. Frontiers in Plant Science 5, 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nukarinen E, Nägele T, Pedrotti L et al. 2016. Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation. Scientific Reports 6, 31697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F, Valon C, Kohara A, Miséra S, Giraudat J. 1997. The ABSCISIC ACID-INSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 loci act in concert to control multiple aspects of Arabidopsis seed development. The Plant Cell 9, 1265–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, MacGregor DR. 2017. Effects of environmental variation during seed production on seed dormancy and germination. Journal of Experimental Botany 68, 819–825. [DOI] [PubMed] [Google Scholar]
- Radchuk R, Emery RJ, Weier D, Vigeolas H, Geigenberger P, Lunn JE, Feil R, Weschke W, Weber H. 2010. Sucrose non-fermenting kinase 1 (SnRK1) coordinates metabolic and hormonal signals during pea cotyledon growth and differentiation. The Plant Journal 61, 324–338. [DOI] [PubMed] [Google Scholar]
- Radchuk R, Radchuk V, Weschke W, Borisjuk L, Weber H. 2006. Repressing the expression of the SUCROSE NONFERMENTING-1-RELATED PROTEIN KINASE gene in pea embryo causes pleiotropic defects of maturation similar to an abscisic acid-insensitive phenotype. Plant Physiology 140, 263–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz V, Bergervoet JH, Koornneef M. 2001. Sequential steps for developmental arrest in Arabidopsis seeds. Development 128, 243–252. [DOI] [PubMed] [Google Scholar]
- Reidt W, Wohlfarth T, Ellerström M, Czihal A, Tewes A, Ezcurra I, Rask L, Bäumlein H. 2000. Gene regulation during late embryogenesis: the RY motif of maturation-specific gene promoters is a direct target of the FUS3 gene product. The Plant Journal 21, 401–408. [DOI] [PubMed] [Google Scholar]
- Roscoe TT, Guilleminot J, Bessoule JJ, Berger F, Devic M. 2015. Complementation of seed maturation phenotypes by ectopic expression of ABSCISIC ACID INSENSITIVE3, FUSCA3 and LEAFY COTYLEDON2 in Arabidopsis. Plant and Cell Physiology 56, 1215–1228. [DOI] [PubMed] [Google Scholar]
- Sheldon CC, Hills MJ, Lister C, Dean C, Dennis ES, Peacock WJ. 2008. Resetting of FLOWERING LOCUS C expression after epigenetic repression by vernalization. Proceedings of the National Academy of Sciences, USA 105, 2214–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasulu N, Wobus U. 2013. Seed-development programs: a systems biology-based comparison between dicots and monocots. Annual Review of Plant Biology 64, 189–217. [DOI] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ. 2001. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proceedings of the National Academy of Sciences, USA 98, 11806–11811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To A, Valon C, Savino G, Guilleminot J, Devic M, Giraudat J, Parcy F. 2006. A network of local and redundant gene regulation governs Arabidopsis seed maturation. The Plant Cell 18, 1642–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. 2005. The botany array resource: e-northerns, expression angling, and promoter analyses. The Plant Journal 43, 153–163. [DOI] [PubMed] [Google Scholar]
- Tsai AY, Gazzarrini S. 2012a. AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. The Plant Journal 69, 809–821. [DOI] [PubMed] [Google Scholar]
- Tsai AY, Gazzarrini S. 2012b. Overlapping and distinct roles of AKIN10 and FUSCA3 in ABA and sugar signaling during seed germination. Plant Signaling and Behavior 7, 1238–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Perry SE. 2013. Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiology 161, 1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang D, Venglat P, Tibiche C et al. 2011. Genome-wide analysis reveals gene expression and metabolic network dynamics during embryo development in Arabidopsis. Plant Physiology 156, 346–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Kagaya Y, Usui H, Hobo T, Takeda S, Hattori T. 2010. Diverse roles and mechanisms of gene regulation by the Arabidopsis seed maturation master regulator FUS3 revealed by microarray analysis. Plant and Cell Physiology 51, 2031–2046. [DOI] [PubMed] [Google Scholar]
- Zorzatto C, Machado JP, Lopes KV et al. 2015. NIK1-mediated translation suppression functions as a plant antiviral immunity mechanism. Nature 520, 679–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.