Endogenous unsaturated fatty acids identified from salt-treated Arabidopsis by a bioassay-guided fractionation method activate plasma membrane H+-ATPase by binding to its C-terminus.
Keywords: Arabidopsis, bioassay-guided fractionation, plasma membrane H+-ATPase, unsaturated fatty acids
Abstract
Plasma membrane (PM) H+-ATPase is essential for plant growth and development. Various environmental stimuli regulate its activity, a process that involves many protein cofactors. However, whether endogenous small molecules play a role in this regulation remains unknown. Here, we describe a bio-guided isolation method to identify endogenous small molecules that regulate PM H+-ATPase activity. We obtained crude extracts from Arabidopsis seedlings with or without salt treatment and then purified them into fractions based on polarity and molecular mass by repeated column chromatography. By evaluating the effect of each fraction on PM H+-ATPase activity, we found that fractions containing the endogenous, free unsaturated fatty acids oleic acid (C18:1), linoleic acid (C18:2), and linolenic acid (C18:3) extracted from salt-treated seedlings stimulate PM H+-ATPase activity. These results were further confirmed by the addition of exogenous C18:1, C18:2, or C18:3 in the activity assay. The ssi2 mutant, with reduced levels of C18:1, C18:2, and C18:3, displayed reduced PM H+-ATPase activity. Furthermore, C18:1, C18:2, and C18:3 directly bound to the C-terminus of the PM H+-ATPase AHA2. Collectively, our results demonstrate that the binding of free unsaturated fatty acids to the C-terminus of PM H+-ATPase is required for its activation under salt stress. The bio-guided isolation model described in this study could enable the identification of new endogenous small molecules that modulate essential protein functions, as well as signal transduction, in plants.
Introduction
Plasma membrane (PM) H+-ATPase is a P-type H+-ATPase found in plants and fungi (Serrano, 1989). It hydrolyses ATP and pumps protons from the cytoplasm to the apoplast to establish electrochemical gradients across the plasma membrane, which are used to drive secondary transport processes including ion, lipid, sugar, and nutrient transport. PM H+-ATPase functions in many cellular and developmental processes in plants such as cell expansion, turgor regulation, cellular pH changes, tip growth, stomatal movement, and stress responses (Michelet and Boutry, 1995; Bibikova et al., 1998; Palmgren, 1998, 2001; Morsomme and Boutry, 2000; Duby and Boutry, 2009; Lang et al., 2014; Yan et al., 2015).
PM H+-ATPase activity is tightly regulated during plant growth and development and in response to environmental changes. It is regulated by a number of endogenous and exogenous factors, such as hormones (auxin and abscisic acid), calcium, blue light, fungal elicitors, and stress stimuli (Johansson et al., 1993; Niu et al., 1993; Frías et al., 1996; Kinoshita and Shimazaki, 1999; Janicka-Russak and Kłobus, 2007; Hayashi et al., 2014; Spartz et al., 2014). The C-terminal region of PM H+-ATPase is an auto-inhibitory domain, and PM H+-ATPase is constitutively activated when this region is truncated (Palmgren et al., 1991). Phosphorylation of several Thr and Ser residues in the C-terminal region is crucial for the regulation of PM H+-ATPase activity (Rudashevskaya et al., 2012; Haruta et al., 2015). 14-3-3 proteins interact directly with the C-terminal region of the Thr-947 phosphorylation site to activate PM H+-ATPase (Jahn et al., 1997; Svennelid et al., 1999). Two phosphorylated residues in this region, Ser-931 and Ser-899, negatively regulate the activity (Duby et al., 2009; Haruta et al., 2014). The Arabidopsis Ser/Thr protein kinase PKS5 negatively regulates PM H+-ATPase activity by phosphorylating its Ser-931 residue, preventing its interaction with the 14-3-3 protein (Fuglsang et al., 2007; Yang et al., 2010). Another phosphorylation site, Ser-899, mediates RALF-FERONIA signal transduction (Haruta et al., 2014).
Lipids play important roles in the regulation of PM H+-ATPase activity. In vitro studies suggest that PM H+-ATPase activity is affected by exogenous-added lipids in isolated vesicles, including fatty acids (FAs), phosphatidylserine (PS), phosphatidylinositol (PI), phosphatidylcholine (PC), phosphatidylglycerol (PG) and phosphatidic acid (PA) (Dufour and Goffeau, 1980; Palmgren et al., 1988; Kasamo, 1990; Sánchez-Nieto et al., 2011). Lysophospholipids stimulate PM H+-ATPase activity (Palmgren et al., 1988; Palmgren and Sommarin, 1989; Kasamo, 1990; Pedchenko et al., 1990; Gomès et al., 1996; Wielandt et al., 2015), and the activation depends on autoinhibitory N- and C-terminal domains of PM H+-ATPase (Wielandt et al., 2015). These studies suggest that lipids are part of the regulatory machinery governing plant PM H+-ATPase activity. However, it is still unclear whether and how these lipids regulate activity in plants.
The levels of thousands of metabolites dynamically fluctuate throughout plant development and in response to various environmental stimuli. Some of these metabolites function as small, bioactive molecules that regulate plant growth and development, such as phytohormones (Lu and Xu, 2015). Many studies have been conducted on the pharmacological effects of plant-derived metabolites on human health; however, less is known about how these molecules function in plants and which targets they work on. In this study, based on the fact that salt stress rapidly activates PM H+-ATPase, we used a bio-guided isolation strategy to identify the bioactive endogenous compounds that activate PM H+-ATPase activity in Arabidopsis under salt stress. We isolated the free, unsaturated fatty acids C18:1, C18:2, and C18:3 from NaCl-treated cell fractions, evaluated them by spectrometric parameters, and found that they directly bind to and activate PM H+-ATPase.
Materials and methods
Plant materials
Arabidopsis thaliana Columbia (Col-0) wild-type, Nössen (NӦ) wild-type, ssi2 mutant, and complemented lines ProSSI2::SSI2-1 and ProSSI2::SSI2-2 were used in this study. The NӦ and ssi2 have been described previously (Kachroo et al., 2001). The ProSSI2::SSI2-1 and ProSSI2::SSI2-2 were generated as follows: the 4421 bp SSI2 genomic sequence (including 1262 bp upstream of the ATG start codon and 770 bp downstream of the TAA stop codon) was amplified from NӦ genomic DNA using primers 5′-GCGCGAATTCCTACATA TTTCTTCTGTGTGCTAAC-3′ containing an EcoRI restriction site and 5′-GCGTCGACTCAACTTTGCTCTCACAATGATTC-3′ containing a SalI restriction site. Then the ProSSI2::SSI2 was cloned into the EcoRI and SalI sites in the pCAMBIA1300 binary vector and the resulting construct was transformed into the ssi2.
Extraction of endogenous small molecules
Arabidopsis Col-0 seeds were sterilized and sown in liquid 1/2 Murashige and Skoog (MS) medium with 10 g l–1 sucrose, grown under dark conditions and maintained shaking in a shaking incubator for 20 d, and treated with 150 mM NaCl or 1/2 MS for 6 h. The seedlings were gently squeezed to remove extra water and ground under liquid nitrogen. The material (1040 g treated with NaCl; 1170 g without salt treatment) was extracted in 5 l of methanol for 30 min under agitation, respectively. After filtration through a Buchner funnel, the material was re-extracted in 5 l of methanol. The two liquid extracts were combined and concentrated under reduced pressure using a rotary evaporator, resulting in 2.07 g of total crude extract (with salt treatment) and 2.20 g of total crude extract (without salt treatment).
Isolation of plasma membrane vesicles and PM H+-ATPase activity assays
Plasma membrane-enriched vesicles were obtained from Arabidopsis seedlings with or without salt treatment using aqueous two-phase (Dextron-PEG3350) partitioning as described previously (Qiu et al., 2002). Arabidopsis Col-0 seeds were sterilized, sown on solid MS medium with 25 g l–1 sucrose, and grown under continuous white light (light intensity of 50 μmol m−2 s−1) at 23 °C for 7 d. The seedlings were then transferred to soil for 4 weeks under a 16 h light (22 °C)–8 h dark (20 °C) cycle. For the salt treatment, NӦ, ssi2, ProSSI2::SSI2-1, and ProSSI2::SSI2-2 seedlings were treated with 250 mM NaCl for 3 d prior to being collected for plasma membrane vesicles isolation. The isolation of plasma membrane vesicles was performed at 4 °C or on ice. Plants were homogenized in buffer (2 ml buffer per gram plant tissue) containing 10% (w/v) glycerol, 0.33 M sucrose, 0.2% (w/v) BSA, 5 mM dithiothreitol (DTT), 5 mM EDTA, 0.2% (w/v) casein, 5 mM ascorbate, 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.6% (w/v) polyvinylpyrrolidone, 1× protease inhibitor, and 50 mM HEPES–KOH, pH 7.5. The homogenate was filtered through two layers of Miracloth and centrifuged at 12 000 g for 10 min. The supernatant was centrifuged for 1 h at 100 000 g to obtain a microsomal pellet, which was resuspended in a buffer containing 3 mM KCl, 0.33 M sucrose, 1 mM DTT, 1 mM PMSF, 0.1 mM EDTA, 1× protease inhibitor, and 5 mM K2HPO4–KH2PO4, pH 7.8. The suspension was added to a two-phase mixture to obtain a phase consisting of 6.2% (w/w) dextran T-500, and 6.2% (w/w) polyethylene glycol 3350 in 5 mM K2HPO4–KH2PO4 buffer, pH 7.8, containing 3 mM KCl and 0.33 M sucrose. The final upper phases were collected, diluted with resuspension buffer containing 10% (w/v) glycerol, 0.33 M sucrose, 0.1% (w/v) BSA, 0.1 mM EDTA, 1× protease inhibitor, 2 mM DTT and 20 mM HEPES–KOH, pH 7.5, and centrifuged for 1 h at 100 000 g. The pellet was collected and resuspended in the above-mentioned resuspension buffer plus 1 mM EDTA. Inside-out vesicles were produced by adding 0.05% (w/v) Brij58 to the medium as described previously (Johansson et al., 1995). PM H+-ATPase activity was determined as described previously (Qiu et al., 2002). H+-ATPase activity was evaluated based on the quenching (decrease) in fluorescence of quinacrine (a pH-sensitive fluorescent probe). The assays contained 5 mM quinacrine, 100 mM KCl, 3 mM MgSO4, 250 mM mannitol, 25 mM 1,3-bis[tris(hydroxylmethyl)methylamino]propane–HEPES, pH 6.5, and 50 μg ml–1 of vesicle protein. The reactions were mixed several times by inversion, incubated at 25 °C, and placed in a dark chamber in a fluorescence spectrophotometer (Hitachi F-7000). The assay was initiated by the addition of ATP (final concentration of 3 mM), and H+-ATPase activity was evaluated at excitation and emission wavelengths of 430 and 500 nm, respectively. At the end of the reaction, the protonophore carbonyl cyanide m-chlorophenyl hydrazone (CCCP; final concentration of 10 mM) was added to the assay solution to dissipate any remaining pH gradient.
PM H+-ATPase hydrolytic activity was determined by measuring the release of Pi from ATP (Sandstrom et al., 1987). The resuspended plasma membrane vesicles (1 μg of protein) were added to 490 μl of assay buffer containing 30 mM MES/Tris (pH 6.5), 125 mM sucrose, 50 mM KCl, 5 mm MgSO4, 5 mM Na2ATP (pH 6.5), 0.1 mM (NH4)6Mo7O24, 50 mM KNO3, 1 mM NaN3, and 0.0125% (w/v) Triton X-100. The reaction proceeded for 20 min at 37 °C and was stopped by the addition of 1 ml of a stopping solution containing 5% (w/v) sodium dodecyl sulfate (SDS), 2% (w/v) H2SO4, and 0.5% (w/v) (NH4)6Mo7O24. Total Pi from ATP hydrolysis was measured spectrophotometrically by adding 50 μl of 10% (w/v) ascorbic acid and determining the absorbance at 660 nm after incubating at 45 °C for 20 min. Hydrolytic activity was evaluated in terms of μM Pi liberated per mg protein per min.
H+ flux measurement
Net H+ fluxes were measured in the meristematic zone (approximately 120 μm from the root tip) of Arabidopsis roots using non-invasive micro-test technology (NMT; NMT100 Series, Younger USA LLC, Amherst, MA, USA; Xuyue (Beijing) Sci & Tech Co., Ltd, Beijing, China). The H+ concentration was evaluated by moving the H+-selective microelectrode between two positions close to the root at a programmable frequency in the range of 0.3–0.5 Hz. The micropipettes were front-filled with 15 μm columns of H+-selective liquid exchange cocktails (LIXs) (Fluka 95293). An Ag/AgCl wire electrode holder was inserted into the back of the electrode to make contact with the electrolyte buffer. The reference electrode was an Ag/AgCl half-cell connected to the assay solution. H+ fluxes were calculated by MageFlux, which is based on Fick’s law of diffusion. The samples were prepared and treated as described previously with some modifications (Fuglsang et al., 2007). Arabidopsis NӦ, ssi2, ProSSI2::SSI2-1, and ProSSI2::SSI2-2 seeds were sterilized and sown in solid MS medium plus 25 g l–1 sucrose and grown under continuous white light (light intensity of 50 μmol m−2 s−1) at 23 °C for 5 d. To observe the effect of exogenous addition of C18:1, C18:2, or C18:3 on the activation of PM H+-ATPase activity, the NӦ, ssi2, ProSSI2::SSI2-1, and ProSSI2::SSI2-2 seedlings were pre-incubated in a buffer containing 0.5 mM KCl, 0.1 mM CaCl2, and 0.3 mM MES, pH 6.0, with the addition of 100 μM of C18:0, C18:1, C18:2, C18:3, or 0.1% dimethyl sulfoxide (DMSO) (v/v) for 20 min and assayed.
Bio-guided isolation
The total methanol extract of seedlings with salt treatment and extract without salt treatment were separated simultaneously by normal-phase column chromatography on silica gel using a gradient elution of methanol–dichloromethane (DCM). The crude extract was loaded onto a silica gel column (200 ml) and fractioned by gradient elution with DCM as the starting elution solvent, followed by an increasing gradient of the polar solvent methanol from 5% to 100%. Every gradient elution needs 5× column volume of solvent. All subfractions were tested for their effect on PM H+-ATPase activity. The fractions that affected PM H+-ATPase activity were combined based on their thin layer chromatography (TLC) characteristics and further fractioned by repeated column chromatography, as shown in Supplementary Figs S1 (positive effect) and S2 (negative effect). Finally, the combined positive fraction in seedlings with salt treatment was fractioned by preparative high performance liquid chromatography–ultraviolet mass spectrometry (prep-HPLC-UV-MS); low resolution electrospray ionization mass spectrometry (LRESIMS) information about the compounds in active fractions is shown in Supplementary Fig. S3A–C.
Structural elucidation of active compounds
Structural elucidation of the compounds in positive fractions was carried out using 1H-NMR (Varian, 400 MHz) and high resolution electrospray ionization mass spectrometry (HRESIMS; Thermo Fisher, Q-Exactive). Further verification was carried out via comparison with standards after derivation using gas chromatography–mass spectrometry (GC-MS) (see the method below).
LC-MS analysis and preparation
The samples were analysed by HPLC–mass spectrometry on a Waters Auto Purification LC/MS system (3100 mass detector, 2545 binary gradient module, 2767 sample manager, 2998 photodiode array (PDA) detector, and system fluidics organizer). The system was equipped with a Waters C18 5 μm Xbridge analytical column (4.6mm×150 mm) or a preparative column (19mm×150 mm). The mobile phase consisted of acetonitrile (phase A) and H2O (phase B) at a flow rate of 1 ml min–1 in analytical mode and 15 ml min–1 in preparative mode. The preparative HPLC gradient elution conditions were as follows: 0 min, 50% A:50% B; 55 min, 73% A:27% B; 65 min, 82% A:18% B; and 75 min, 100% A:0% B. The fractions were collected in 15 ml glass tubes, with 10 ml elution sample in each glass tube, and column-wash elution solvent (100% A) was collected in a 500 ml round-bottomed flask. All fractions were solvent-evaporated, redissolved in methanol, and a PM H+-ATPase activity test performed.
Fatty acid analysis
Direct transesterification of total fatty acids was performed as described previously (Miquel and Browse, 1992). Briefly, seedlings were ground under liquid nitrogen and lyophilized. Each lyophilized sample (30 mg) was combined with 3 ml of 3% H2SO4 in methanol in a screw-capped glass tube and incubated at 75 °C for 2.5 h. When the reaction was completed, 1 ml of 0.9% NaCl and 3 ml hexane were added, and the sample was mixed well. After standing for 1 h, the upper hexane layer was transferred to a new tube for analysis. GC-MS analysis of the methylated fatty acids was performed using an Agilent 7890 GC with a DB-23 column and 5975 mass selective detector. The oven conditions were as follows: 45 °C for 3 min, followed by 15 °C min–1 to 140 °C for 5 min, 1.5 °C min–1 to 144 °C for 12 min, 1.5 °C min–1 to 164 °C for 6 min, 1.5 °C min–1 to 170 °C for 0 min, and 20 °C min–1 to 230 °C for 5 min.
Lipid-protein overlay assay
The fatty acids C18:0, C18:1, C18:2, and C18:3 were dissolved in DCM:methanol 1:1, and fatty acid test strips were prepared by spotting the indicated amounts of fatty acids onto a polyvinylidene difluoride membrane and allowing them to dry for 1 h at room temperature. The strips were incubated at room temperature for 2 h with 1 μg ml–1 of His fusion proteins in 5% skim milk–20 mM Tris–HCl (pH 8.0) plus 150 mM NaCl for blocking. After four washes, the bound proteins were detected using anti-His antibodies as previously described (Sun et al., 2013). Bromophenol blue reagent was prepared by dissolving 0.04 g bromophenol blue in 100 ml ethanol.
Fatty acid agarose affinity chromatography
Fatty acid sodium salts (C18:0, C18:1, C18:2, or C18:3, from J&K Scientific Ltd) were coupled to the Sepharose matrix (EAH Sepharose from GE Healthcare) as described previously (Peters et al., 1973; Kim et al., 2005; Mandal et al., 2012). Briefly, Na-C18:1, Na-C18:2, and Na-C18:3 were dissolved in ddH2O, and Na-C18:0 was dissolved in 25% ethanol–ddH2O at a concentration of 100 μM. EAH-Sepharose, 1.5 volumes of 100 μM fatty acid sodium salts, and 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (50 mg ml–1 of Sepharose) were mixed by inversion and incubated at 37 °C for 3 d. The Sepharose matrix was washed with 50% ethanol, followed by ethanol–75 mM NaH2PO4 (1:1), ethanol–50 mM NaOH (1:1), and phosphate buffered saline (PBS) at pH 7.0. The binding assay was performed as described previously (Kim et al., 2005). Briefly, 2 μM His fusion protein in 200 μl PBS was incubated with 200 μl fatty acid–Sepharose at 37 °C for 1 h. The Sepharose was then washed with PBS and eluted with 50% ethanol. Binding proteins were analysed by immunoblot using anti-His antibodies.
Membrane conductivity detection
The membrane conductivity assay was performed as previously described (Eich et al., 2000). Briefly, ten Arabidopsis leaves (5-week-old plants) were pre-treated with 100 μM C18:0, C18:1, C18:2, or C18:3 or 150 mM NaCl for 12 h. A hole-puncher was used to obtain leaf tissue samples of the same size for each treatment. The leaf discs were immersed in ddH2O under vacuum for 10 min until the leaf discs were submerged, and electrolyte loss was measured after 3 h. The total electrolyte content of the leaves was determined by boiling the leaves for 10 min at 95 °C. Relative conductivity is the percentage of electrolyte loss versus total electrolyte content.
Membrane fluidity detection
Five-day-old seedlings were treated with 100 μM C18:0, C18:1, C18:2, or C18:3 or 500 mM NaCl for 10 min, washed three times with ddH2O, and stained with 4 μM N-(3-triethylammoniumpropyl)-4-(6-[4-(diethylamino)phenyl]-hexatrienyl) pyridinium dibromide (FM 4-64, from Molecular Probes) for 10 min. The fluorescence recovery after photobleaching (FRAP) assay was conducted under a microscope, using Andor IQ software according to the manufacturer’s instructions. FRAP data analysis was performed as described previously (Goral et al., 2010). Briefly, a plasma membrane region was selected and the fluorescence intensity of the region was measured in pre- and post-bleach images. The fluorescence intensity was analysed using ImageJ software. The fluorescence recovery rate is the percentage of post-bleach fluorescence intensity versus pre-bleach fluorescence intensity.
Antibody and protein-blot analyses
The AHA2 centerloop and C-terminus coding sequences were amplified by RT-PCR using primer pairs 5′-CGGGATCCGCAGG AATGGATGTCCTGTGC-3′ and 5′-CGGAATTCTTAGA GCACGATATCTGAAGCACC-3′, and 5′-CGGGATCCGCGT GGCTCAACTTGTTTGAGAAC-3′ and 5′-CGGAATTCCTACA CAGTGTAGTGACTG-3′, respectively. The PCR products were cloned into the pET-28a-SMT3 vector to generate His-SMT3-AHA2 centerloop or His-SMT3-AHA2 C-terminus fusion protein. All plasmids were verified by sequencing. The recombinant proteins were expressed purified in E. coli bacterial cells as described in the manufacturer’s instructions.
Polyclonal antibody was raised against the AHA2 centerloop region (321–620 amino acid residue) from an antiserum of mouse. The AHA2 centerloop recombination protein can be recognized by the anti-AHA2 antibody; as a negative control the AHA2 C-terminus recombination protein cannot be recognized by the antibody (Supplementary Fig. S4A). The anti-H+-ATPase AHA2 antibody recognized not only AHA2 but also other plasma membrane H+-ATPase isoforms in Arabidopsis (Supplementary Fig. S4B). Protein samples (plasma membrane vesicles) were mixed with 5× SDS loading buffer (250 mM Tris–HCl pH 6.8, 30% glycerol, 10% SDS, 0.01% bromophenol blue and 200 mM dithiothreitol) followed by boiling for 5 min. Samples were loaded onto 12% SDS-PAGE gels and subjected to electrophoresis in denaturing SDS running buffer (3.03 g l−1 Tris, 1.0 g l−1 SDS, 14.4 g l−1 glycine). Proteins were transferred to nitrocellulose membranes. Membranes were probed with anti-AHA2 antisera at a 1:1000 dilution. Secondary antibody was horseradish peroxidase-conjugated goat anti-mouse IgG, and diluted at a 1:5000.
Results
Identification of fractions that activate PM H+-ATPase activity using a bio-guided isolation protocol
To investigate the endogenous small molecules involved in activating/deactivating PM H+-ATPase activity, we first utilized a bio-guided isolation procedure to isolate bioactive compounds. PM H+-ATPase has relatively low activity under normal growth conditions; however, this activity increases significantly in response to salt stress (Yang et al., 2010). To determine whether small molecules are involved in this increased activation, we used our newly developed bio-guided isolation technique to fractionate crude extracts of Arabidopsis Col-0 seedlings with or without NaCl treatment. In total, 25 fractions were collected (based on polarity; Supplementary Figs S1 and S2), solvent-evaporated, and dissolved in methanol.
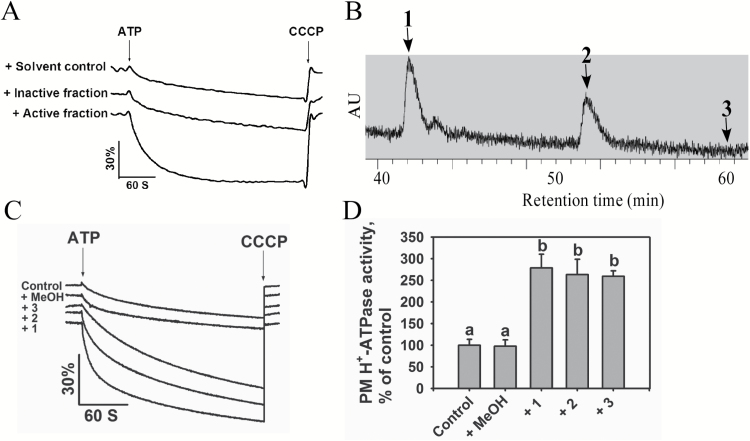
To perform the PM H+-ATPase activity assay, we prepared plasma membrane vesicles from 5-week-old soil-grown Col-0 plants without NaCl treatment (Yang et al., 2010). We evaluated the purity and H+-transport competency of the isolated vesicles and found them to be transport-competent (Supplementary Table S1 and Supplementary Fig. S5). We then pre-incubated the plasma membrane vesicles with each small-molecule fraction at a concentration of 10 μg ml–1 and evaluated the effect of each fraction on PM H+-ATPase activity (Fig. 1A). The fractions A2 and A3 isolated from the seedlings with salt treatment enhanced PM H+-ATPase activity, whereas the A2 and A3 fractions from the seedlings without salt treatment had no such effect (Supplementary Figs S1 and S2). Active fractions A2 and A3 were combined and further separated into four new fractions, B1–B4, using Sephadex LH-20 column chromatography, in which, fraction B2 was shown to activate PM H+-ATPase activity. We also separated the A2 and A3 fractions from the seedlings without salt treatment into four new fractions and none of them had an effect on the activity (Supplementary Fig. S2). We then separated the active fraction B2 from seedlings with salt treatment into five fractions by gel filtration on silica gel eluted with PE (petroleum ether)–acetone (10:1), in which fractions C2, C3, and C4 were shown to activate PM H+-ATPase activity. We then combined fractions C2, C3, and C4 and subjected them to further separation using Sephadex LH-20 column chromatography eluted with methanol. The resulting subfractions, D2 and D3, increased PM H+-ATPase activity. We separated fractions D2 and D3 by normal-phase column chromatography on silica gel using PE–DCM (1:1) as the elution solvent and found that the resulting fraction, E8, activated PM H+-ATPase activity (Supplementary Fig. S1). Finally, we separated fraction E8 by prep-HPLC-UV-MS and identified three active fractions, 1, 2, and 3 (Fig. 1B), all of which increased PM H+-ATPase activity (Fig. 1C, D). Active compounds in fractions 1 and 2 were further elucidated by 1H-NMR spectra, HRESIMS and GC-MS. An active compound in fraction 3 had a weak signal in 1H-NMR, and was further elucidated by HRESIMS and GC-MS.
Fig. 1.
Identification of fractions that induce PM H+-ATPase activity using a bio-guided isolation protocol. (A) PM H+-ATPase activity measured in the vesicles in the presence of fractions. Plasma membrane vesicles were obtained from 5-week-old Arabidopsis (ecotype Col-0) seedlings. The reaction (proton-pumping resulting in intravesicular acidification and a pH gradient) was initiated by the addition of 3 mM ATP, and the pH gradient collapsed from the addition of 10 μM carbonyl cyanide m-chlorophenyl hydrazone (CCCP). (B) HPLC-UV profiling of active fractions 1, 2, and 3 in the process of preparation. (C) PM H+-ATPase activities measured in vesicles in the presence of fractions 1, 2, and 3. (D) Comparison of PM H+-ATPase activities in (C). Fraction-dependent change values in proton-pumping activity were calculated by setting the activity of the control to 100%. The data in (D) represent means±standard deviation (SD) of five replicates. Student’s t-test was used to analyse the statistical significance; significant differences (P≤0.05) in (D) are indicated by different lower-case letters.
Structural elucidation of compounds in active fractions 1, 2 and 3
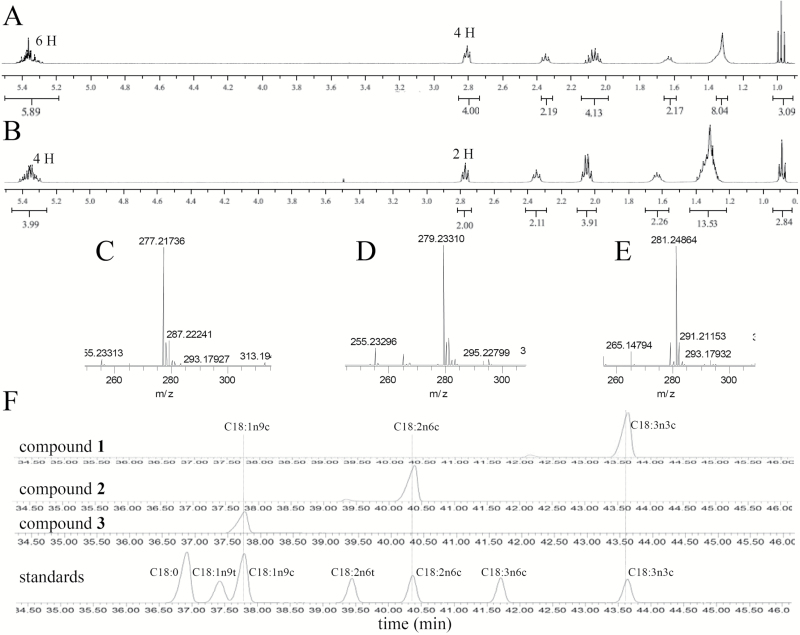
LRESIMS (electrospray ionization in negative ion mode; ESI−) revealed values of m/z of 277 for a compound in fraction 1, 279 for a compound in fraction 2, and 281 for compound in fraction 3, suggesting that these molecules have similar structures (Supplementary Fig. S3A–C). The molecular formula of the compound in fraction 1 was determined to be C18H29O2 by HRESIMS at m/z 277.21736 [M–H]− (calculated 277.21676, Fig. 2C), which indicates three degrees of unsaturation. The 1H-NMR spectra of the active compound in fraction 1 (Fig. 2A and Supplementary Table S2) displayed six hydrogens (Hs) at δ 5.36 ppm, which correlated with three degrees of unsaturation based on its HRESIMS data. Four protons at δ 2.05 ppm indicated the presence of two α-H groups next to one double bond, and four protons at δ 2.81 ppm indicated the presence of two α-H groups between two double bonds. A carbonyl moiety was present, with two α-H protons at δ 2.35 ppm and two β-H protons at δ 1.65 ppm next to the carbonyl moiety. This information, combined with the finding that methyl Hs at δ 0.98 ppm and high-abundance alkyl Hs at δ 1.31 ppm were observed in the 1H-NMR spectrum, indicated that the compound in active fraction 1 might be C18:3. GC-MS analysis of the methyl derivative of the compound in fraction 1 showed the same retention time at 43.6 min (Fig. 2F) and the same characteristic mass fragment ions (data not shown) as the commercially available methyl-linolenate standard. On this basis, the compound in active fraction 1 was identified as C18:3 (C18:3n3c).
Fig. 2.
Structural elucidation of compounds in fractions 1, 2, and 3. (A) 1H-NMR spectrum of compound in fraction 1 (CDCl3, 400 MHz). (B) 1H-NMR spectrum of compound in fraction 2 (CDCl3, 400 MHz). (C) HRESIMS of compound in fraction 1 (ESI−). (D) HRESIMS of compound in fraction 2 (ESI−). (E) HRESIMS of compound in fraction 3 (ESI−). (F) Comparison of methyl derivatives of compounds in fractions 1, 2, and 3 with a standard fatty acid methyl ester mix.
The molecular formula of the compound in fraction 2 was determined to be C18H31O2 by HRESIMS at m/z 279.23310 [M–H]− (calculated 279.23241, Fig. 2D), indicating two degrees of unsaturation. The 1H-NMR data for the compound in fraction 2 (Fig. 2B and Supplementary Table S2) were similar to those of the compound in fraction 1 except for the presence of four double-bond Hs at δ 5.36 ppm and two α-H protons between the two double bonds at δ 2.81 ppm, indicating that two double bonds exist in the structure. Based on structural analysis of the compound in fraction 2, we postulated that it might be C18:2. To further verify the structure of the compound in fraction 2, we compared its methylated derivative with standard methyl-C18:2 via GC-MS. Both showed the same characteristic mass fragment ions and the same retention time at 40.35 min (Fig. 2F). Thus, the compound in active fraction 2 was identified as C18:2 (C18:2n6c).
The molecular formula for the compound in fraction 3 was determined to be C18H33O2 based on the HRESIMS peak at m/z 281.24864 [M–H]− (calculated 281.24806, Fig. 2E), with two more Hs than the compound in fraction 2, suggesting that compound 3 might be C18:1. We compared its methylated derivative with standard methyl-C18:1, both of which showed the same retention time at 37.8 min (Fig. 2F). Thus, we confirmed that the compound in active fraction 3 is C18:1 (C18:1n9c).
The effect of exogenous C18:1, C18:2, or C18:3 addition on PM H+-ATPase activity
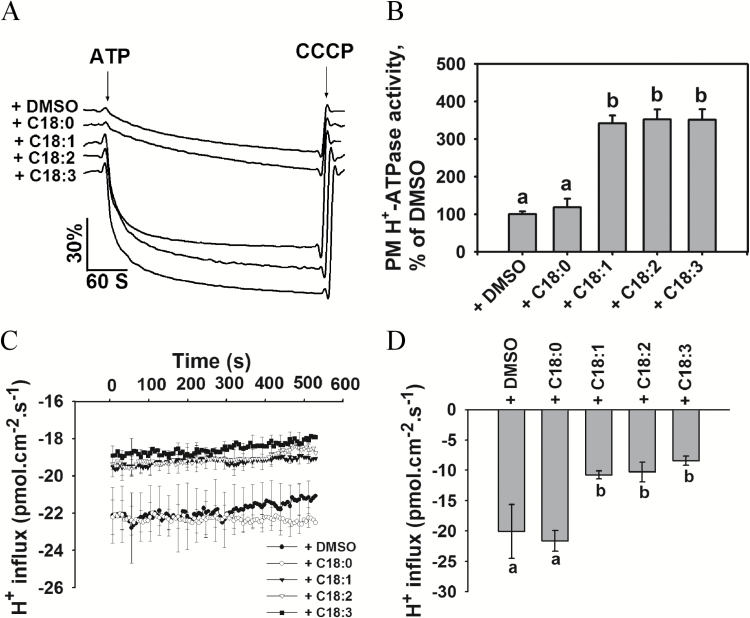
To exclude the possibility that endogenous C18:1, C18:2, and C18:3 isolated from Arabidopsis may contain other substances involved in activating PM H+-ATPase activity, we performed a verification experiment using commercially available standard samples. We pre-incubated the PM vesicles with 100 μM C18:0, C18:1, C18:2, or C18:3, or 0.1% DMSO (v/v) and measured the PM H+-ATPase activity. As shown in Fig. 3A, B, exogenous addition of C18:1, C18:2, or C18:3 to the vesicles caused a significant increase in H+-transport activity compared with the solvent control (0.1% DMSO, v/v). However no obvious effect on PM H+-ATPase activity was observed in the presence of C18:0. When we evaluated the effect of unsaturated fatty acids on hydrolytic activity, the presence of C18:1, C18:2, or C18:3 in the assay solution resulted in an increase in hydrolytic activity, but incubation with C18:0 had no obvious effect on this activity (Supplementary Table S3).
Fig. 3.
Exogenous addition of fatty acids increases H+ extrusion. (A) PM H+-ATPase activities measured in vesicles in the presence of the compounds. Plasma membrane vesicles were isolated from 5-week-old Arabidopsis (ecotype Col-0) seedlings. (B) Comparison of PM H+-ATPase activities in (A). (C) Net H+ fluxes in root tips of Col-0 plants in the presence of the compounds. The non-invasive microsensing system technique was used to monitor ion flux. After the roots were incubated in buffer (0.5 mM KCl, 0.1 mM CaCl2, and 0.3 mM MES, pH 6.0) in the presence of a DMSO control or 100 μM of one of the compounds for 20 min, the transient net H+ fluxes were recorded in the buffer mentioned above. (D) Calculated net H+ fluxes from (C). The data in (B–D) represent means±SD of five replicates. The three biological replicates displayed similar results. Student’s t-test was used to analyse the statistical significance; significant differences (P≤0.05) in (B, D) are indicated by different lower-case letters.
We further verified the effects of unsaturated fatty acids on H+ fluxes in Arabidopsis roots using the non-invasive microsensing system (NMS), which can be used to monitor transport of various ions and molecules in intact samples (Xu et al., 2006). Net H+ fluxes were measured in the root apex of seedlings pre-incubated with 100 μM C18:0, C18:1, C18:2, or C18:3, or 0.1% DMSO (v/v). The H+ influxes were reduced after treatment with C18:1, C18:2, or C18:3 (Fig. 3C, D), but C18:0 did not affect H+ fluxes in the root compared with the DMSO control. Taken together, these results suggest that C18:1, C18:2, and C18:3 exert a positive regulatory effect on PM H+-ATPase activity.
C18:1, C18:2, and C18:3 bind to the C-terminus of AHA2
To understand how the unsaturated fatty acids regulate PM H+-ATPase activity and to identify which part of AHA2 is involved in this regulation, we performed binding assays between C18:1, C18:2, or C18:3 and AHA2 peptides. Specifically, DNA fragments encoding the centerloop (amino acids 321–620) and C-terminus (amino acids 849–948) of AHA2 were cloned into the pET-28a-SMT3 vector and the resulting plasmids were transformed into E. coli strain BL21. The recombinant proteins were then purified.
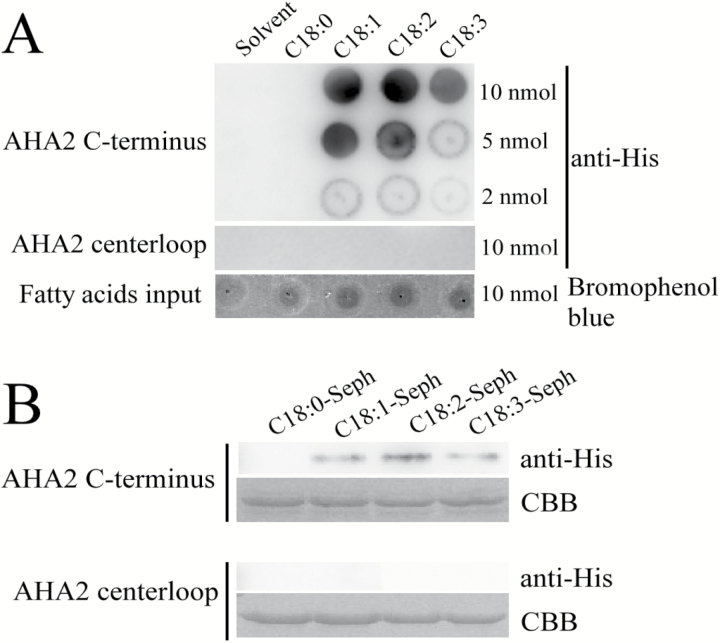
We used a lipid–protein overlay experiment to investigate the interaction between C18:0, C18:1, C18:2, or C18:3 and the peptides. The unsaturated fatty acids C18:1, C18:2, and C18:3 physically interacted with the C-terminus of AHA2, but not its centerloop (Fig. 4A). By contrast, C18:0 did not interact with the centerloop or the C-terminus of AHA2.
Fig. 4.
C18:1, C18:2, and C18:3 bind to the C-terminus of PM H+-ATPase AHA2. (A) Lipid–protein overlay experiment between AHA2 peptides (C-terminus and centerloop) as His fusion proteins extracted from E. coli and C18:1, C18:2, or C18:3. The solvent (DCM:methanol, 1:1) and C18:0 were used as controls. The amount that each lipid spot contained is shown on the right side. The upper lane shows the interaction between the AHA2 C-terminus and the fatty acids. The middle lane shows the interaction between the AHA2 centerloop and the fatty acids. The lower lane shows the fatty acid input control, which was stained by bromophenol blue. The experiment was repeated three times with similar results. (B) C18:0, C18:1, C18:2, and C18:3 agarose affinity chromatography experiment performed with the AHA2 peptides (C-terminus and centerloop) as His fusion proteins extracted from E. coli. The fatty acid–Sepharose matrixes were incubated with the peptides (C-terminus and centerloop) and washed with PBS, and the bound peptides were eluted and detected by immunoblot using anti-His antibodies. The upper lanes show detection using the C-terminus and the lower lanes show detection using the centerloop. The input control of each experiment was detected with Coomassie brilliant blue staining (CBB). The experiment was repeated three times with similar results.
To further verify the interaction between C18:1, C18:2, or C18:3 and the C-terminus of PM H+-ATPase, we prepared C18:0–, C18:1–, C18:2–, and C18:3–Sepharose columns, which we loaded with the peptides of the C-terminus or centerloop of AHA2. The C-terminus was pulled down by the C18:1, C18:2, and C18:3 Sepharose matrix, but the centerloop was not (Fig. 4B). Consistently, none of the AHA2 peptides were pulled down by the C18:0 matrix (Fig. 4B). These results suggest that C18:1, C18:2, and C18:3 activate PM H+-ATPase activity by directly interacting with its C-terminus.
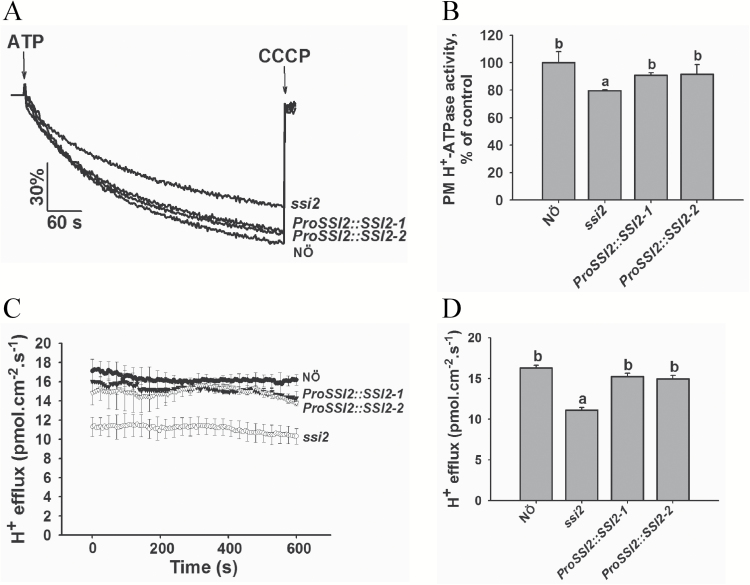
PM H+-ATPase activity is decreased in an Arabidopsis mutant with low levels of unsaturated fatty acids
In Arabidopsis, SSI2 encodes a fatty acid desaturase involved in the desaturation of fatty acid C18:0 to C18:1. To determine the genetic linkage between unsaturated fatty acids and PM H+-ATPase activity, we obtained the ssi2 mutant (in the NӦ background) (Kachroo et al., 2001) and generated ProSSI2:SSI2 transgenic rescue lines (ProSSI2::SSI2-1 and ProSSI2::SSI2-2) in this mutant background. The levels of C18:1, C18:2, and C18:3 were significantly reduced in the ssi2 mutant compared with wild-type, whereas the ProSSI2::SSI2-1 and ProSSI2::SSI2-2 transgenic plants had levels of unsaturated fatty acids similar to those of wild-type (Supplementary Fig. S6). We isolated plasma membrane vesicles from leaves of wild-type NӦ, ssi2, ProSSI2::SSI2-1, and ProSSI2::SSI2-2 plants treated with 250 mM NaCl for 3 d and measured PM H+-ATPase H+-transport and hydrolytic activity. These activities were lower in vesicles isolated from the ssi2 mutant than those of NӦ, whereas in the ProSSI2::SSI2-1 and ProSSI2::SSI2-2 genotypes, PM H+-ATPase H+-transport and hydrolytic activity were restored to wild-type levels (Fig. 5A, B and Supplementary Table S4). We monitored the protein level of PM H+-ATPase via immunoblot analysis using H+-ATPase AHA2 antibody and found no significant difference in AHA2 levels among NӦ, ssi2, ProSSI2::SSI2-1, and ProSSI2::SSI2-2 plants (Supplementary Fig. S7).
Fig. 5.
PM H+-ATPase activity is reduced in the ssi2 mutant. (A) PM H+-ATPase activity was measured in the vesicles of NӦ, ssi2, ProSSI2::SSI2-1, and ProSSI2::SSI2-2 seedlings. Plasma membrane vesicles were isolated from 5-week-old Arabidopsis (ecotype Col-0) seedlings treated with 250 mM NaCl for 3 d. (B) Comparison of PM H+-ATPase activity in (A). (C) The net H+ effluxes in root tips of NӦ, ssi2, ProSSI2::SSI2-1, and ProSSI2::SSI2-2 seedlings. The non-invasive microsensing system technique was used to monitor ion flux. After the roots were incubated under alkaline conditions (0.5 mM KCl, 0.1 mM CaCl2, 0.3 mM MES, and 75 mM NaCl, pH 8.1) for 20 min, the transient net H+ fluxes were recorded in the same buffer. (D) Calculated net H+ effluxes from (C). The data in (B–D) represent means±SD of five replicates. The three biological replicates displayed similar results. Student’s t-test was used to analyse the statistical significance; significant differences (P≤0.05) in (B, D) are indicated by different lower-case letters.
We measured net H+ fluxes in the root apices of 7-day-old NӦ, ssi2, ProSSI2::SSI2-1, and ProSSI2::SSI2-2 seedlings using the NMS method. Specifically, we pretreated the seedlings under salt–alkaline conditions (0.5 mM KCl, 0.1 mM CaCl2, 0.3 mM MES, 75 mM NaCl, pH 8.1) to activate PM H+-ATPase activity and evaluated the difference in net H+ efflux. The rate of transmembrane H+ efflux was significantly lower in the ssi2 mutant than in wild-type NӦ, whereas no difference in net H+ efflux was detected among NӦ, ProSSI2::SSI2-1, and ProSSI2::SSI2-2 plants (Fig. 5C, D). These results further support the notion that PM H+-ATPase activity is activated by unsaturated fatty acids C18:1, C18:2, and C18:3.
Exogenous addition of C18:1, C18:2, or C18:3 does not influence membrane conductivity or fluidity
To exclude the possibility that C18:1, C18:2, and C18:3 influence membrane properties, we tested their effects on membrane conductivity and membrane fluidity. In our membrane conductivity assay, samples treated with 100 μM C18:0, C18:1, C18:2, or C18:3 showed similar relative conductivity values to that of the solvent control (0.1% DMSO, v/v); however, treatment with 150 mM NaCl led to a significantly larger change in relative conductivity compared with unsaturated fatty acid treatments (Supplementary Fig. S8A). In the membrane fluidity assay, we used fluorescence recovery after photobleaching (FRAP) to investigate the effect of C18:0, C18:1, C18:2, or C18:3 on membrane fluidity in Arabidopsis seedlings. Treatment with 100 μM C18:0, C18:1, C18:2, or C18:3 had similar effects on membrane fluidity to that of the solvent control (0.1% DMSO, v/v) (Supplementary Fig. S8B–D). However, NaCl treatment reduced membrane fluidity. This result is consistent with the previous finding that C18:2 does not influence membrane fluidity (Lin et al., 2011).
Discussion
Many metabolites are generated in plants during metabolism, and their levels are altered throughout development and in response to environmental stimuli. Metabolites function in many physiological processes and can also serve as cofactors with stimulatory or inhibitory effects on enzymes. In plants, the biological functions of many metabolites have not yet been discovered.
In this study, we fractionated small molecules from NaCl-treated or untreated Arabidopsis seedlings according to their polarity and molecular mass and used a PM H+-ATPase activity-guided assay to identify fractions that activate or deactivate PM H+-ATPase activity. Using this strategy, we obtained three small molecules, unsaturated fatty acids C18:1, C18:2, and C18:3. We performed a direct binding assay that showed that unsaturated fatty acids C18:1, C18:2, and C18:3 bound to the C-terminus of PM H+-ATPase and stimulated its activity. We also performed a PM H+-ATPase activity assay in a mutant with low levels of unsaturated fatty acids, as well as transgenic rescued lines, which gave the genetic evidence that the unsaturated fatty acids play a role in activating PM H+-ATPase activity. Salt stress triggers many plant responses, including osmotic, ion and ROS signals, and also causes many physiological changes, including membrane properties. It is believed that Na+ causes membrane destabilization by reducing the hydration of fatty acids and displacing the Ca2+ in the membranes, which in turn change membrane properties (Cramer et al., 1985; Lynch et al., 1987). However, the unsaturated fatty acids do not affect membrane properties, indicating that they function in plant salt response by a specific pathway. It is still unclear how these unsaturated fatty acids activate H+-ATPase. It is possible that the binding of the unsaturated fatty acids to the C-terminus changes its conformation and in turn alters the inhibition activity of PM H+-ATPase, or that the binding changes the phosphorylation status of H+-ATPase, repressing Ser-931 phosphorylation by PKS5 and increasing Thr-947 phosphorylation to activate PM H+-ATPase. Further studies are needed to investigate these possibilities.
Our bio-guided isolation result is in agreement with previous studies that exogenous addition of free fatty acids enhances PM H+-ATPase activity (Kasamo, 1990; Palmgren et al., 1988), which supports the reliability of our bio-guided method. It is also reported that various lysophospholipids can also enhance PM H+-ATPase activity (Kasamo, 1990; Palmgren et al., 1988). Lysophosphatidylcholine (LPC) has been studied in yeast for its possible function in regulating PM H+-ATPase activity through both the N- and C-terminus of PM H+-ATPase (Wielandt et al., 2015). However, genetic and biochemical information is lacking on whether these small molecules directly bind to PM H+-ATPase and affect its activity in plants.
Phospholipids, one of the major classes of metabolites in plants, play critical roles in plant development and stress responses (Xue et al., 2007). Using our bio-guided method, we also found the existence of phosphatidylcholine (PC) in active fractions (A22) from both of the salt-treated and untreated seedlings (Supplementary Fig. S9A–D). PC (particularly PC with unsaturated acyl chains) was reported to activate PM H+-ATPase (Kasamo, 1990), and the commercially standard PC sample can activate PM H+-ATPase (Supplementary Fig. S9E). The PC is enriched in both salt-treated and untreated seedlings and the free unsaturated fatty acids were enriched only in the samples of the salt-treated seedlings, but in different fractions from those that activate PM H+-ATPase activity. Based on our small-molecule fractionation method, the free fatty acids are unlikely to result from lipolysis during the fractionation process; however, they might be generated from some phospholipids via phospholipases A (PLAs) in plants.
There is a complex network of different phospholipid signaling pathways, and phospholipids exert their physiological function by dynamic conversion of different phospholipids. Although various lysophospholipids and free unsaturated fatty acids appear to have a similar ability to activate plant PM H+-ATPase activity, their specificity is yet to be determined. In plants, lysophospholipids and free unsaturated fatty acids are generated from phospholipids simultaneously, and either differences in binding affinity or local changes in small molecule concentrations may result in this binding specificity and guide plant responses. It is possible that different lipids play a specific role to fine tune PM H+-ATPase activity in a timing- and locality-dependent manner under different stress stimuli. Further studies to determine when and where lysophospholipids and free unsaturated fatty acids are produced may help us to understand the difference in the regulation.
Several studies have investigated total fatty acid contents in plants (Kodama et al., 1995; Murakami et al., 2000; Kachroo et al., 2001; Devaiah et al., 2007; Zhang et al., 2009; Zhang et al., 2012). In the current study, the content of total C18 unsaturated fatty acids did not change under salt treatment (Supplementary Fig. S10). The free unsaturated fatty acids were only identified in the fractions isolated from more than 1 kg of seedlings treated with NaCl, but not from the same amount of seedlings without NaCl treatment, suggesting that the free unsaturated fatty acid content is very low and induced by salt stress. We attempted to measure the content of free unsaturated fatty acids in Arabidopsis with 0.5 g of material using two reported methods: GC-MS analysis after conversion to Weinreb amides and direct high-performance thin-layer chromatography analysis, which are used in animal and yeast cells (Maeda et al., 2013, 2014; Ubhayasekera et al., 2013); however, we failed to quantify the content of free unsaturated fatty acids. Perhaps the content of free unsaturated fatty acids in small samples is too low to reach the quantification limit of these two methods.
PLAs play important roles in the generation of unsaturated free fatty acids. They consist of a superfamily of enzymes that catalyse the hydrolysis of the ester bonds of phospholipids to generate lysophospholipids and free fatty acids (Ryu, 2004). PLAs function in many physiological processes, including cell elongation, shoot gravitropism and stomatal opening (Lee et al., 2003; Seo et al., 2008), which require the stimulation of PM H+-ATPase activity. These results suggest that PLAs may also be involved in the regulation of PM H+-ATPase activity. The degree of unsaturation of free fatty acids depends on desaturase in plants. In our study, SSI2, a plastid-located desaturase (Wu et al., 2009), is involved in the regulation of PM H+-ATPase activity. PLA1 (class I, II, and III), hydrolysing fatty acids at the sn-1 position, is predicted to localize in chloroplasts, cytosol, and mitochondria based on sequence analysis (Ishiguro et al., 2001; Ryu, 2004; Wang et al., 2012); sPLA2 in Arabidopsis (AtsPLA2α, AtsPLA2β, AtsPLA2γ, and AtsPLA2δ), hydrolysing fatty acids at the sn-2 position, is reported to localize in apoplasts, Golgi bodies and/or endoplasmic reticulum (Seo et al., 2008; Froidure et al., 2010; Kim et al., 2011; Jung et al., 2012). It is reported that PLAs in plant cells can be relocalized (Froidure et al., 2010; Jung et al., 2012). PLA2α is translocated from cytoplasmic vesicles to nucleus mediated by AtMYB30 (Froidure et al., 2010). It can also be translocated from Golgi bodies to apoplasts during development and bacterial infection in Arabidopsis (Jung et al., 2012). Phospholipids and free fatty acids could be translocated in the cell through membrane contact sites and vesicle traffic (Li et al., 2016). Although many transport proteins are involved in the lipid translocation, no exact protein is reported to transport free fatty acids to plasma membrane (Li et al., 2016). It is possible that either unknown proteins are involved in this process or relocated PLAs produce free fatty acids in plasma membrane to activate PM H+-ATPase.
The levels of LPC and lysophosphatidic acid (LPA) increased markedly after hyperosmotic shock in Dunaliella salina (Einspahr et al., 1988), and LPA accumulated in a dose- and time-dependent manner after NaCl treatment (Meijer et al., 2001) in Chlamydomonas. Because free unsaturated fatty acids are generated with LPA and LPC simultaneously, it is possible that free fatty acid levels are also elevated under osmotic and salt stress. The expression of SSI2 was induced after salt treatment (Supplementary Fig. S11). These results suggest that the content of free unsaturated fatty acids increased under salt stress.
The expression of genes involved in lipid metabolism is regulated by various environmental stimuli. In Arabidopsis, salt stress induces the expression of genes encoding fatty acid desaturase-2 (FAD2), fatty acid desaturase-6 (FAD6), and phospholipase D, which hydrolyses phospholipids to produce phosphatidic acid (Zhang et al., 2009, 2012; Yu et al., 2010), and low temperature induces the expression of fatty acid desaturase-8 (FAD8) (Gibson et al., 1994). In tomato, the expression of the fatty acid desaturase-7 gene (LeFAD7) is induced by chilling stress but reduced by high temperature (Liu et al., 2006). These findings suggest that different lipid products may play different roles in the regulation of physiological function in facing multiple environments. The technique developed in this study can be used as a starting point to investigate the roles of these and other small molecules that activate PM H+-ATPase in more detail.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. The bio-guided isolation procedure for seedlings with salt treatment.
Fig. S2. The bio-guided isolation procedure for seedlings without salt treatment.
Fig. S3. LRESIMS spectrum of compounds in fractions 1, 2, and 3.
Fig. S4. Availability of the AHA2 antibody.
Fig. S5. Proton transport competency of the isolated vesicles.
Fig. S6. Fatty acid analyses of NӦ, ssi2, ProSSI2::SSI2-1, and ProSSI2::SSI2-2 seedlings.
Fig. S7. PM H+-ATPase AHA2 protein levels in NӦ, ssi2, ProSSI2::SSI2-1, and ProSSI2::SSI2-2 seedlings.
Fig. S8. Exogenous application of C18:1, C18:2, or C18:3 does not influence membrane conductivity and membrane fluidity.
Fig. S9. The fraction containing PC activates PM H+-ATPase activity.
Fig. S10. Total fatty acid analyses of col-0 under salt stress.
Fig. S11. Expression of SSI2 after salt treatment.
Table S1. Plasma membrane purity determination.
Table S2. 1H NMR spectral data for compounds in fractions 1 and 2 (CDCl3, 400 MHz).
Table S3. Effect of fatty acids on plasma membrane H+-ATPase hydrolytic activity.
Table S4. Plasma membrane H+-ATPase hydrolytic activity of NӦ, ssi2, ProSSI2::SSI2-1, and ProSSI2::SSI2-2 seedlings.
Supplementary Material
Acknowledgements
We thank Pradeep Kachroo for NӦ and ssi2 seeds. This work was supported by the National Natural Science Foundation of China (Grant 31430012, Grant 31670260), Foundation for Innovative Research Group of the National Natural Science Foundation of China (Grant 31421062), and National Genetically Modified Organisms Breeding Major Projects (2016ZX08009002).
References
- Bibikova TN, Jacob T, Dahse I, Gilroy S. 1998. Localized changes in apoplastic and cytoplasmic pH are associated with root hair development in Arabidopsis thaliana. Development 125, 2925–2934. [DOI] [PubMed] [Google Scholar]
- Cramer GR, Läuchli A, Polito VS. 1985. Displacement of Ca2+ by Na+ from the plasmalemma of root cells: a primary response to salt stress?Plant Physiology 79, 207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah SP, Pan X, Hong Y, Roth M, Welti R, Wang X. 2007. Enhancing seed quality and viability by suppressing phospholipase D in Arabidopsis. The Plant Journal 50, 950–957. [DOI] [PubMed] [Google Scholar]
- Duby G, Boutry M. 2009. The plant plasma membrane proton pump ATPase: a highly regulated P-type ATPase with multiple physiological roles. Pflugers Archiv 457, 645–655. [DOI] [PubMed] [Google Scholar]
- Duby G, Poreba W, Piotrowiak D, Bobik K, Derua R, Waelkens E, Boutry M. 2009. Activation of plant plasma membrane H+-ATPase by 14-3-3 proteins is negatively controlled by two phosphorylation sites within the H+-ATPase C-terminal region. The Journal of Biological Chemistry 284, 4213–4221. [DOI] [PubMed] [Google Scholar]
- Dufour JP, Goffeau A. 1980. Phospholipid reactivation of the purified plasma membrane ATPase of yeast. The Journal of Biological Chemistry 255, 10591–10598. [PubMed] [Google Scholar]
- Eich J, Dürholt H, Steger-Hartmann T, Wagner E. 2000. Specific detection of membrane-toxic substances with a conductivity assay. Ecotoxicology and Environmental Safety 45, 228–235. [DOI] [PubMed] [Google Scholar]
- Einspahr KJ, Maeda M, Thompson GA., Jr 1988. Concurrent changes in Dunaliella salina ultrastructure and membrane phospholipid metabolism after hyperosmotic shock. The Journal of Cell Biology 107, 529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frías I, Caldeira MT, Pérez-Castiñeira JR, Navarro-Aviñó JP, Culiañez-Maciá FA, Kuppinger O, Stransky H, Pagés M, Hager A, Serrano R. 1996. A major isoform of the maize plasma membrane H+-ATPase: characterization and induction by auxin in coleoptiles. The Plant Cell 8, 1533–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froidure S, Canonne J, Daniel X, Jauneau A, Briere C, Roby D, Rivas S. 2010. AtsPLA2-α nuclear relocalization by the Arabidopsis transcription factor AtMYB30 leads to repression of the plant defense response. Proceedings of the National Academy of Sciences, USA 107, 15281–15286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglsang AT, Guo Y, Cuin TA, et al. 2007. Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. The Plant Cell 19, 1617–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson S, Arondel V, Iba K, Somerville C. 1994. Cloning of a temperature-regulated gene encoding a chloroplast omega-3 desaturase from Arabidopsis thaliana. Plant Physiology 106, 1615–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomès E, Venema K, Simon-Plas F, Milat ML, Palmgren MG, Blein JP. 1996. Activation of the plant plasma membrane H+-ATPase. Is there a direct interaction between lysophosphatidylcholine and the C-terminal part of the enzyme?FEBS Letters 398, 48–52. [DOI] [PubMed] [Google Scholar]
- Goral TK, Johnson MP, Brain AP, Kirchhoff H, Ruban AV, Mullineaux CW. 2010. Visualizing the mobility and distribution of chlorophyll proteins in higher plant thylakoid membranes: effects of photoinhibition and protein phosphorylation. The Plant Journal 62, 948–959. [DOI] [PubMed] [Google Scholar]
- Haruta M, Gray WM, Sussman MR. 2015. Regulation of the plasma membrane proton pump (H+-ATPase) by phosphorylation. Current Opinion in Plant Biology 28, 68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR. 2014. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343, 408–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Takahashi K, Inoue S, Kinoshita T. 2014. Abscisic acid suppresses hypocotyl elongation by dephosphorylating plasma membrane H+-ATPase in Arabidopsis thaliana. Plant & Cell Physiology 55, 845–853. [DOI] [PubMed] [Google Scholar]
- Ishiguro S, Kawai-Oda A, Ueda J, Nishida I, Okada K. 2001. The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. The Plant Cell 13, 2191–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn T, Fuglsang AT, Olsson A, Brüntrup IM, Collinge DB, Volkmann D, Sommarin M, Palmgren MG, Larsson C. 1997. The 14-3-3 protein interacts directly with the C-terminal region of the plant plasma membrane H+-ATPase. The Plant Cell 9, 1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicka-Russak M, Kłobus G. 2007. Modification of plasma membrane and vacuolar H+-ATPases in response to NaCl and ABA. Journal of Plant Physiology 164, 295–302. [DOI] [PubMed] [Google Scholar]
- Johansson F, Olbe M, Sommarin M, Larsson C. 1995. Brij 58, a polyoxyethylene acyl ether, creates membrane vesicles of uniform sidedness. A new tool to obtain inside-out (cytoplasmic side-out) plasma membrane vesicles. The Plant Journal 7, 165–173. [DOI] [PubMed] [Google Scholar]
- Johansson F, Sommarin M, Larsson C. 1993. Fusicoccin activates the plasma membrane H+-ATPase by a mechanism involving the C-terminal inhibitory domain. The Plant Cell 5, 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J, Kumar K, Lee HY, Park YI, Cho HT, Ryu SB. 2012. Translocation of phospholipase A2α to apoplasts is modulated by developmental stages and bacterial infection in Arabidopsis. Frontiers in Plant Science 3, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo P, Shanklin J, Shah J, Whittle EJ, Klessig DF. 2001. A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proceedings of the National Academy of Sciences, USA 98, 9448–9453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasamo K. 1990. Mechanism for the activation of plasma membrane H+-ATPase from rice (Oryza sativa L.) culture cells by molecular species of a phospholipid. Plant Physiology 93, 1049–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Ok SH, Bahn SC, Jang J, Oh SA, Park SK, Twell D, Ryu SB, Shin JS. 2011. Endoplasmic reticulum- and Golgi-localized phospholipase A2 plays critical roles in Arabidopsis pollen development and germination. The Plant Cell 23, 94–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Nakatomi R, Akagi T, Hashikawa T, Takahashi R. 2005. Unsaturated fatty acids induce cytotoxic aggregate formation of amyotrophic lateral sclerosis-linked superoxide dismutase 1 mutants. The Journal of Biological Chemistry 280, 21515–21521. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Shimazaki Ki. 1999. Blue light activates the plasma membrane H+-ATPase by phosphorylation of the C-terminus in stomatal guard cells. The EMBO Journal 18, 5548–5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama H, Horiguchi G, Nishiuchi T, Nishimura M, Iba K. 1995. Fatty acid desaturation during chilling acclimation is one of the factors involved in conferring low-temperature tolerance to young tobacco leaves. Plant Physiology 107, 1177–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang V, Pertl-Obermeyer H, Safiarian MJ, Obermeyer G. 2014. Pump up the volume—a central role for the plasma membrane H+ pump in pollen germination and tube growth. Protoplasma 251, 477–488. [DOI] [PubMed] [Google Scholar]
- Lee HY, Bahn SC, Kang YM, Lee KH, Kim HJ, Noh EK, Palta JP, Shin JS, Ryu SB. 2003. Secretory low molecular weight phospholipase A2 plays important roles in cell elongation and shoot gravitropism in Arabidopsis. The Plant Cell 15, 1990–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Xu C, Li-Beisson Y, Philippar K. 2016. Fatty acid and lipid transport in plant cells. Trends in Plant Science 21, 145–158. [DOI] [PubMed] [Google Scholar]
- Lin ZF, Liu N, Lin GZ, Peng CL. 2011. Factors altering the membrane fluidity of spinach thylakoid as determined by fluorescence polarization. Acta Physiologiae Plantarum 33, 1019–1024. [Google Scholar]
- Liu XY, Yang JH, Li B, Yang XM, Meng QW. 2006. Antisense-mediated depletion of tomato chloroplast omega-3 fatty acid desaturase enhances thermal tolerance. Journal of Integrative Plant Biology 48, 1096–1107. [DOI] [PubMed] [Google Scholar]
- Lu Y, Xu J. 2015. Phytohormones in microalgae: a new opportunity for microalgal biotechnology?Trends in Plant Science 20, 273–282. [DOI] [PubMed] [Google Scholar]
- Lynch J, Cramer GR, Läuchli A. 1987. Salinity reduces membrane-associated calcium in corn root protoplasts. Plant Physiology 83, 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K, Anand K, Chiapparino A, Kumar A, Poletto M, Kaksonen M, Gavin AC. 2013. Interactome map uncovers phosphatidylserine transport by oxysterol-binding proteins. Nature 501, 257–261. [DOI] [PubMed] [Google Scholar]
- Maeda K, Poletto M, Chiapparino A, Gavin AC. 2014. A generic protocol for the purification and characterization of water-soluble complexes of affinity-tagged proteins and lipids. Nature Protocols 9, 2256–2266. [DOI] [PubMed] [Google Scholar]
- Mandal MK, Chandra-Shekara AC, Jeong RD, Yu K, Zhu S, Chanda B, Navarre D, Kachroo A, Kachroo P. 2012. Oleic acid-dependent modulation of NITRIC OXIDE ASSOCIATED1 protein levels regulates nitric oxide-mediated defense signaling in Arabidopsis. The Plant Cell 24, 1654–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer HJ, Arisz SA, Van Himbergen JA, Musgrave A, Munnik T. 2001. Hyperosmotic stress rapidly generates lyso-phosphatidic acid in Chlamydomonas. The Plant Journal 25, 541–548. [DOI] [PubMed] [Google Scholar]
- Michelet B, Boutry M. 1995. The Plasma membrane H+-ATPase (a highly regulated enzyme with multiple physiological functions). Plant Physiology 108, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel M, Browse J. 1992. Arabidopsis mutants deficient in polyunsaturated fatty acid synthesis. Biochemical and genetic characterization of a plant oleoyl-phosphatidylcholine desaturase. The Journal of Biological Chemistry 267, 1502–1509. [PubMed] [Google Scholar]
- Morsomme P, Boutry M. 2000. The plant plasma membrane H+-ATPase: structure, function and regulation. Biochimica et Biophysica Acta 1465, 1–16. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Tsuyama M, Kobayashi Y, Kodama H, Iba K. 2000. Trienoic fatty acids and plant tolerance of high temperature. Science 287, 476–479. [DOI] [PubMed] [Google Scholar]
- Niu X, Narasimhan ML, Salzman RA, Bressan RA, Hasegawa PM. 1993. NaCl regulation of plasma membrane H+-ATPase gene expression in a glycophyte and a halophyte. Plant Physiology 103, 713–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren MG. 1998. Proton gradients and plant growth: Role of the plasma membrane H+-ATPase. Advances in Botanical Research 28, 1–70. [Google Scholar]
- Palmgren MG. 2001. Plant plasma membrane H+-ATPases: powerhouses for nutrient uptake. Annual Review of Plant Physiology and Plant Molecular Biology 52, 817–845. [DOI] [PubMed] [Google Scholar]
- Palmgren MG, Sommarin M. 1989. Lysophosphatidylcholine stimulates ATP dependent proton accumulation in isolated oat root plasma membrane vesicles. Plant Physiology 90, 1009–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren MG, Sommarin M, Serrano R, Larsson C. 1991. Identification of an autoinhibitory domain in the C-terminal region of the plant plasma membrane H+-ATPase. The Journal of Biological Chemistry 266, 20470–20475. [PubMed] [Google Scholar]
- Palmgren MG, Sommarin M, Ulvskov P, Jorgensen PL. 1988. Modulation of plasma membrane H+-ATPase from oat roots by lysophosphatidylcholine, free fatty acids and phospholipase A2. Physiologia Plantarum 74, 11–19. [Google Scholar]
- Pedchenko VK, Nasirova GF, Palladina TA. 1990. Lysophosphatidylcholine specifically stimulates plasma membrane H+-ATPase from corn roots. FEBS Letters 275, 205–208. [DOI] [PubMed] [Google Scholar]
- Peters T, Jr, Taniuchi H, Anfinsen CB., Jr 1973. Affinity chromatography of serum albumin with fatty acids immobilized on agarose. The Journal of Biological Chemistry 248, 2447–2451. [PubMed] [Google Scholar]
- Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK. 2002. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proceedings of the National Academy of Sciences, USA 99, 8436–8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudashevskaya EL, Ye J, Jensen ON, Fuglsang AT, Palmgren MG. 2012. Phosphosite mapping of P-type plasma membrane H+-ATPase in homologous and heterologous environments. The Journal of Biological Chemistry 287, 4904–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu SB. 2004. Phospholipid-derived signaling mediated by phospholipase A in plants. Trends in Plant Science 9, 229–235. [DOI] [PubMed] [Google Scholar]
- Sánchez-Nieto S, Enríquez-Arredondo C, Guzmán-Chávez F, Hernández-Muñoz R, Ramírez J, Gavilanes-Ruíz M. 2011. Kinetics of the H+-ATPase from dry and 5-hours-imbibed maize embryos in its native, solubilized, and reconstituted forms. Molecular Plant 4, 505–515. [DOI] [PubMed] [Google Scholar]
- Sandstrom RP, Deboer AH, Lomax TL, Cleland RE. 1987. Latency of plasma membrane H-ATPase in vesicles isolated by aqueous phase partitioning: increased substrate accessibility or enzyme activation. Plant Physiology 85, 693–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano R. 1989. Structure and function of plasma membrane ATPase. Annual Review of Plant Physiology and Plant Molecular Biology 40, 61–94. [Google Scholar]
- Seo J, Lee HY, Choi H, Choi Y, Lee Y, Kim YW, Ryu SB, Lee Y. 2008. Phospholipase A2β mediates light-induced stomatal opening in Arabidopsis. Journal of Experimental Botany 59, 3587–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz AK, Ren H, Park MY, Grandt KN, Lee SH, Murphy AS, Sussman MR, Overvoorde PJ, Gray WM. 2014. SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. The Plant Cell 26, 2129–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Kale SD, Azurmendi HF, Li D, Tyler BM, Capelluto DG. 2013. Structural basis for interactions of the Phytophthora sojae RxLR effector Avh5 with phosphatidylinositol 3-phosphate and for host cell entry. Molecular Plant-Microbe Interactions 26, 330–344. [DOI] [PubMed] [Google Scholar]
- Svennelid F, Olsson A, Piotrowski M, Rosenquist M, Ottman C, Larsson C, Oecking C, Sommarin M. 1999. Phosphorylation of Thr-948 at the C terminus of the plasma membrane H+-ATPase creates a binding site for the regulatory 14-3-3 protein. The Plant Cell 11, 2379–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubhayasekera SJ, Staaf J, Forslund A, Bergsten P, Bergquist J. 2013. Free fatty acid determination in plasma by GC-MS after conversion to Weinreb amides. Analytical and Bioanalytical Chemistry 405, 1929–1935. [DOI] [PubMed] [Google Scholar]
- Wang G, Ryu S, Wang X. 2012. Plant phospholipases: an overview. Methods in Molecular Biology 861, 123–137. [DOI] [PubMed] [Google Scholar]
- Wielandt AG, Pedersen JT, Falhof J, Kemmer GC, Lund A, Ekberg K, Fuglsang AT, Pomorski TG, Buch-Pedersen MJ, Palmgren M. 2015. Specific activation of the plant P-type plasma membrane H+-ATPase by lysophospholipids depends on the autoinhibitory N- and C-terminal domains. The Journal of Biological Chemistry 290, 16281–16291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Liu T, Liu H, Zheng GC. 2009. Unsaturated fatty acid: metabolism, synthesis and gene regulation. African Journal of Biotechnology 8, 1782–1785. [Google Scholar]
- Xu Y, Sun T, Yin LP. 2006. Application of non-invasive microsensing system to simultaneously measure both H+ and O2 fluxes around the pollen tube. Journal of Integrative Plant Biology 48, 823–831. [Google Scholar]
- Xue H, Chen X, Li G. 2007. Involvement of phospholipid signaling in plant growth and hormone effects. Current Opinion in Plant Biology 10, 483–489. [DOI] [PubMed] [Google Scholar]
- Yan S, McLamore ES, Dong S, Gao H, Taguchi M, Wang N, Zhang T, Su X, Shen Y. 2015. The role of plasma membrane H+-ATPase in jasmonate-induced ion fluxes and stomatal closure in Arabidopsis thaliana. The Plant Journal 83, 638–649. [DOI] [PubMed] [Google Scholar]
- Yang Y, Qin Y, Xie C, et al. 2010. The Arabidopsis chaperone J3 regulates the plasma membrane H+-ATPase through interaction with the PKS5 kinase. The Plant Cell 22, 1313–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Nie J, Cao C, Jin Y, Yan M, Wang F, Liu J, Xiao Y, Liang Y, Zhang W. 2010. Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytologist 188, 762–773. [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu H, Sun J, Li B, Zhu Q, Chen S, Zhang H. 2012. Arabidopsis fatty acid desaturase FAD2 is required for salt tolerance during seed germination and early seedling growth. PLoS ONE 7, e30355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JT, Zhu JQ, Zhu Q, Liu H, Gao XS, Zhang HX. 2009. Fatty acid desaturase-6 (Fad6) is required for salt tolerance in Arabidopsis thaliana. Biochemical and Biophysical Research Communications 390, 469–474. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.