MdSnRK1.1 regulates anthocyanin and proanthocyanidin accumulation in apple by phosphorylation and modulation of MdJAZ18 stability, explaining how jasmonic acid acts together with sucrose to regulate anthocyanin biosynthesis.
Keywords: Anthocyanins, JA, JAZ protein, MBW complex, proanthocyanins, SnRK1 protein kinase, sucrose
Abstract
Sugars induce anthocyanin biosynthesis in plants. As a conserved energy sensor, SnRK1 (SNF1-related kinase 1) is involved in sucrose-induced anthocyanin accumulation. However, the exact molecular mechanism by which SnRK1 regulates the biosynthesis of anthocyanins and proanthocyanidins (PAs) in response to sucrose in plants is not clear. In this study, it was found that MdSnRK1.1 interacted with MdJAZ18 protein which acts as a repressor in the jasmonate (JA) signaling pathway. MdSnRK1.1 then phosphorylated MdJAZ18 to facilitate its 26S proteasome-mediated degradation, which released MdbHLH3 thereby activating the expression of the regulatory and structural genes, thus finally promoting the biosynthesis of anthocyanins and PAs. Taken together, these results demonstrate the involvement of MdSnRK1.1 in sucrose-induced accumulation of anthocyanins and PAs. For the first time, our findings shed light on the molecular mechanism by which the crosstalk of sucrose and JA signaling regulates flavonoid biosynthesis.
Introduction
Flavonoids are a class of important secondary metabolites in plants. They mainly include flavonols, anthocyanins, flavones, and proanthocyanins (PAs). Anthocyanins contribute to the colors of flowers and fruit, ranging from blue to red with the purpose of attracting pollinators and seed distributors (Koes et al., 2006). The presence of anthocyanins in fruit mainly determines fruit exterior quality, which is an important consideration in consumer choice (Allan et al., 2008; Li et al., 2012). PAs are present in various organs, particularly in leaf, bark, root, fruit, and seed (Barbehenn and Constabel, 2011). These flavonoids generally exist in the form of colorless polymers, and are oxidized into brown complexes under various stress stimuli (Pourcel et al., 2005). PAs are considered as one of the most effective natural antioxidants responsible for the removal of free radicals from the human body. In addition, anthocyanins and PAs play important roles in resistance to insect attacks and pathogen infection in plants, and are beneficial for human health (Dixon et al., 2005; Scalbert et al., 2005).
Anthocyanins and PAs, also known as condensed tannins, are derived from phenylpropanoids and malonyl-CoA in the flavonoid biosynthetic pathway which contains a series of enzymes (Yoshida et al., 2015). These enzymes are encoded by biosynthetic structural genes which include early biosynthetic genes (EBGs) such as phenylalanine ammonia-lyase (PAL), chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), and flavonoid 3'-hydroxylase (F3'H), as well as late biosynthetic genes (LBGs), including anthocyanidin synthase (ANS), dihydroflavonol reductase (DFR), leucoanthocyanidin oxidase (LDOX), UDP-Glc:flavonoid 3-O-glucosyltransferase (UF3GT), and anthocyanidin reductase (ANR) (Solfanelli et al., 2006; Shan et al., 2009; Jeong et al., 2010). The branch of the anthocyanin and PA synthetic pathway exists in the late stage of the flavonoid biosynthetic pathway, mainly involving ANS, DFR, LDOX, and UF3GT genes whose products are responsible for the synthesis of anthocyanins, and ANR and leucoanthocyanidin reductase (LAR) whose products are responsible for PA synthesis (Koes et al., 2006). The structural genes are regulated by the WD-repeat/bHLH/MYB complex (MBW), which consists of WD-repeat protein, basic helix–loop–helix (bHLH), and MYB transcription factors (TFs). The conserved MBW regulatory mechanism works in various plant species (Espley et al., 2006; Qi et al., 2011; Albert et al., 2014; An et al., 2015; Yoshida et al., 2015). In apple, an increasing number of MBW members has been proven to be involved in the control of anthocyanin and PA biosynthesis. The bHLH TFs such as MdbHLH3 and MdbHLH33 recruit WD-repeat protein MdTTG1 and the R2R3-MYB TF, MdMYB1, to mediate anthocyanin biosynthesis (An et al., 2012). MdbHLH3 interacts with MdMYB9 and MdMYB11 to promote anthocyanin and PA accumulation by binding to the promoter elements of MdANS, MdUFGT, MdANR, and MdDFR genes to activate their expression in apple (Xie et al., 2012; An et al., 2015).
Anthocyanin biosynthesis is affected by multiple environmental stimuli such as intense light, UV irradiation, temperature, wounding, pathogen infection, nutrient deficiency, and drought, as well as by many endogenous developmental signals, such as sugar and plant hormones (Takos et al., 2006; Lillo et al., 2008; Jeong et al., 2010; Li et al., 2012; Sperdouli and Moustakas, 2012; Xie et al., 2012; Zhang et al., 2013; An et al., 2015). Plant hormones such as auxins, jasmonic acid (JA), gibberellins (GAs), cytokinin (CKT), abscisic acid (ABA), and ethylene are involved in the modulation of anthocyanin biosynthesis (Mori et al., 1994; Loreti et al., 2008; Shan et al., 2009; Jeong et al., 2010). In poplar leaves, a variety of biotic and abiotic stresses such as nutrient deficiency, insect herbivory, pathogen attack, mechanical wounding, and intense light stimulate PA synthesis (Mellway et al., 2009). JA also promotes PA accumulation by up-regulating the expression of the MYB genes MdMYB9 and MdMYB11 in apple (An et al., 2015).
Sugar is very important throughout the entire plant life cycle because it acts as an energy source, a structural component, and an important regulatory molecule (Jang et al., 1997). Sugar signaling generally regulates the TFs and the metabolic enzymes associated with pathogenesis, photosynthesis, nutrient mobilization and allocation, senescence, and anthocyanin biosynthesis at both the transcriptional and post-transcriptional levels (Koch, 1996; Buchanan-Wollaston et al., 2005; Teng et al., 2005; Rolland et al., 2006). Sugars induce anthocyanin biosynthesis in Arabidopsis. The effect of sucrose and maltose is the most obvious in terms of elevating anthocyanin levels, followed by glucose, fructose, and turanose (a sucrose isomer). Other sugars such as galactose, lactose, and trehalose do not induce anthocyanin accumulation (Teng et al., 2005; Góraj-Koniarska and Saniewski, 2015). The MYB75/PAP1 gene is essential for sucrose-specific regulation of anthocyanin accumulation in Arabidopsis, which then enhances the transcriptional level of the anthocyanin biosynthetic genes (Solfanelli et al., 2006). In addition, DELLA proteins are also found to be involved in sucrose-induced anthocyanin biosynthesis (Li et al., 2014). However, the exact mechanism through which sucrose signaling controls the biosynthesis of anthocyanins is not yet clear.
Sugar-responsive pathways are highly complex processes involved in plant growth and development, and are integrated with other signaling pathways such as those for light, stresses, nitrogen, and plant hormones (Dijkwel et al., 1997; Roitsch, 1999; Sun et al., 2013; Ljung et al., 2015). In higher plants, several components of sugar-responsive pathways have been identified by their conservation among eukaryotic cells (Jang et al., 1997; Rolland et al., 2006). HEXOKINASE1 (HXK1) and Sucrose-Nonfermenting1 (SNF1)-related protein kinases 1 (SnRK1) are the main players among them (Rolland et al., 2006; Hanson and Smeekens, 2009). As the first plant sugar sensor, HXK1 senses glucose (Jang et al., 1997; Moore et al., 2003). In Arabidopsis, AtHXK1 has dual functions as a glycolytic enzyme and a sugar-responsive regulator which regulates gene expression and several plant hormone signaling transduction processes (Moore et al., 2003). Similarly, two isoforms of the catalytic Arabidopsis SnRK1 α subunit, AtSnRK1.1 and AtSnRK1.2, are proposed to function as central integrators of transcription networks in response to stresses and sugar signaling (Baena-González et al., 2007). They are structurally and functionally homologous to the yeast SNF1 and mammalian AMP-activated protein kinase (AMPK) (Baena-González et al., 2007; Hardie, 2007). SnRK1 in plants is considered as a metabolic sensor that perceives the status of cellular carbohydrates and energy (Baena-González and Sheen, 2008; Ghillebert et al., 2011).
SnRK1 is a heterotrimeric protein complex which is composed of a catalytic subunit α and two regulatory subunits, β and βγ within plants (Ramon et al., 2013; Emanuelle et al., 2016). The α subunit includes two functional domains: an N-terminal kinase domain (KD) that contains a conserved activation loop, and a C-terminal regulatory domain (RD) required for the interaction with β and βγ subunits (Crozet et al., 2014; Emanuelle et al., 2016). The β subunit acts as a scaffold to bridge α and βγ subunits (Crozet et al., 2014). The βγ subunit, which functions as the canonical γ subunit, is required for SnRK1 complex formation in plants (Ramon et al., 2013; Emanuelle et al., 2016). In Arabidopsis, each subunit is encoded by multiple genes. As a result, there is a large variety of possible heterotrimeric combinations for SnRK1 kinase (Polge and Thomas, 2007; Emanuelle et al., 2016).
SnRK1 is a Ser/Thr protein kinase, and it has been proved to phosphorylate and inactivate some important enzymes such as trehalose phosphate synthase 5 (TPS5), sucrose phosphate synthase (SPS), and nitrate reductase (NR) to modulate nutrient balance (Sugden et al., 1999; Harthill et al., 2006; Polge and Thomas, 2007). In addition to metabolic regulation, SnRK1 plays a crucial role in co-ordinating various stresses (Baena-González et al., 2007). In plants, SnRK1 is important for seed filling, plant flowering, senescence, and maturation, as well as affecting both embryo and pollen development (Zhang et al., 2001; Baena-González et al., 2007; Radchuk et al., 2010). It also responds to several phytohormones such as auxin, CKT, and ABA, and thus is involved in the crosstalk between sugar and hormone signaling pathways (Radchuk et al., 2010). However, the exact mechanism underlying the potential link between SnRK1 and hormone signals is poorly understood.
Overexpression of AtSnRK1.1 reduces anthocyanin accumulation under a sucrose concentration of 3% and represses the expression of MYB75/PAP1 in Arabidopsis (Baena-González et al., 2007). However, the exact molecular mechanism by which SnRK1 regulates anthocyanin biosynthesis remains to be elucidated. In this study, it was found that MdSnRK1.1, which is closely related to AtSnRK1.1, promoted the biosynthesis of anthocyanins and PAs in apple. Subsequently, its function in control of anthocyanin and PA biosynthesis by interacting with MdJAZ18 was characterized. Finally, the crosstalk between JA and sugar signaling was discussed.
Materials and methods
Plant materials and growth conditions
The calli of apple cultivar ‘Orin’ were subcultured at 20 d intervals on Murashige and Skoog (MS) medium containing 0.4 mg l−1 6-benzylaminopurine (6-BA) and 1.5 mg l−1 2,4-dichlorophenoxy acetic acid (2,4-D) at 25 °C in the dark. Shoot cultures of apple (Malus domestica ‘Royal Gala’) were subcultured at monthly intervals on MS medium supplemented with 0.5 mg l−1 6-BA and 0.2 mg l−1 naphthylacetic acid (NAA) at 25 °C under long-day conditions.
Arabidopsis thaliana ecotype ‘Columbia’ was used as the background for genetic transformation and control. After vernalization, seeds were sown on MS medium without sucrose for 4 d, and then treated with different sucrose concentrations (0, 1, 3, 6, and 9%) under long-day conditions for 1 week.
To analyze the accumulation of anthocyanin and PA, apple calli in good condition were transferred to the calli culture medium with low nitrogen (0.5 mM) and different concentrations of sucrose (1, 3, 6, 9, and 12%) and mannitol (1, 3, and 6%) in the dark for 2 d, and then were exposed to 17 °C with continuous UVB (280–320 nm) light for 1 week. The in vitro apple shoot cultures were treated at 17 °C under continuous white light (30 mmol m−2 s −1) for 2 weeks after starvation.
Anthocyanin measurement
Anthocyanins were extracted from the samples with a HCl–methanol method, and the content was calculated following the protocol described by An et al. (2015). The experiment was repeated at least three times for each sample.
PA extraction and determination
The DMACA (p-dimethylaminocinnamaldehyde)–methanol method was used to stain PAs in apple calli and leaves. PAs were extracted from the samples by the method described previously (An et al., 2015). (+)-Catechin hydrate was used to check the quantity. The experiment was repeated at least three times for each sample.
RNA extraction and quantitative real-time PCR (qRT-PCR) analysis
The in vitro apple shoot cultures and calli were used for RNA extraction. RNAs were extracted from the in vitro apple shoot cultures and calli using RNAplant Plus Reagent (Tiangen, Beijing, China), and then reverse transcribed using a PrimeScript first-strand cDNA synthesis kit (Takara, Dalian, China), following the manufacturer’s instructions. qRT-PCR assays were performed with the UltraSYBR Mixture (SYBR Green I) (Takara) using an ABI7500 qRT-PCR system. The concentration of cDNA was diluted to 1–10 ng μl−1. A 1 μl aliquot of diluted cDNA was used for qRT-PCR. The calculation method for qRT-PCR is 2–ΔΔCT. The results were normalized by 18S. At least three replicates for each sample were used for qRT-PCR. The primers used are listed in Supplementary Table S1 at JXB online or in An et al. (2015).
Vector constructs and plant transformation
To construct the expression vectors, the full-length cDNA of MdJAZ18 and MdSnRK1.1 genes, and an antisense fragment (asMdSnRK1.1) specific to the MdSnRK1.1 gene were cloned from apple ‘Royal Gala’.
MdSnRK1.1 cDNA was linked with PXSN-MYC (Chen et al., 2009) and PRI–green fluorescent protein (GFP), respectively; the antisense fragment asMdSnRK1.1 was linked with PXSN and PRI, respectively; while MdJAZ18 cDNA was linked with PXSN-FLAG and PRI–β-glucosidase (GUS), respectively. These vectors were driven by the Cauliflower mosaic virus (CaMV) 35S promoter. The primers used are shown in Supplementary Table S1.
The recombinant plasmids were transferred into Agrobacterium tumefaciens strain LBA4404, and then introduced into apple calli using the method described by An et al. (2012). For Arabidopsis, the ecotype ‘Columbia’ was used for transformation with an Agrobacterium tumefaciens GV3101-mediated floral dip method. Transgenic lines were screened with kanamycin monosulfate. Homozygous transgenic lines were used in the experiment.
Yeast two-hybrid (Y2H) screening and Y2H assays
Full-length cDNA of the MdSnRK1.1 gene was cloned into the bait vector pGBT9. Y2H screening of the apple cDNA library was performed according to the yeast transformation system (Clontech, Dalian, China), and screened on yeast dropout medium lacking Trp, Leu, His, and Ade (-T/-L/-H/-A).
Y2H assays were performed as described in the manufacturer’s instructions (Clontech). The domain fragments of the MdSnRK1.1 gene were each cloned into vector pGBT9, while those of the full-length MdJAZ18 gene were cloned into vector pGAD424. To confirm the general interaction in plants, AtSnRK1.1 was cloned into pGBT9, and AtJAZ3, MdJAZ1, MdJAZ8, MdJAZ9, MdJAZ10, MdJAZ12, MdJAZ14, MdJAZ18, and MdJAZ19 were cloned into pGAD424. Primers used for vector construction are shown in Supplementary Table S1.
Pull-down assays
The coding regions of MdSnRK1.1 and MdJAZ18 were introduced into the PET-32a and PGEX-4T-1 vector, respectively, and then recombinant vector was transformed into Escherichia coli BL21(DE3) to express HIS-MdSnRK1.1 or GST–MdJAZ18 protein. The pull-down assay was performed according to the instructions of the Pierce GST Spin Purification Kit (Thermo, MA, USA). Then samples were detected by immunoblotting with anti-GST and anti-HIS antibodies, respectively.
In vitro kinase assays
In vitro kinase assays were implemented according to the method described by Lin et al. (2009). For in vitro kinase assays, the E. coli strain BL21-induced HIS-MdSnRK1.1, HIS-MdJAZ18, and HIS-MdJAZ12 fused proteins were purified and co-incubated in kinase buffer (20 mM Tris, pH 7.5, 20 mM DTT, 50 mM MgCl2, 100 μM ATP, and 100 mM MnCl2) containing 10 μCi of [γ-32P]ATP at 30 °C for 30 min. Proteins were separated by SDS–PAGE, and phosphorylated protein was detected by exposing the dried gels to X-ray films.
Protein extraction and protein degradation in vitro
Dry apple calli were harvested and powdered in liquid nitrogen. The residues were mixed with degradation buffer [25 mM Tris–HCl, pH 7.5, 10 mM NaCl, 10 mM MgCl2, 4 mM phenylmethylsulfonylfluoride (PMSF), 5 mM DTT, and 10 mM ATP]. The mixture was kept for 15 min on ice, and then centrifuged for 15 min at 12 000 g at 4 °C. The supernatant was collected, and the Bradford assay was used to detecte its protein concentration. MdACTIN was used as internal non-degraded control. A concentration of 100 μM MG132 (carbobenzoxy-l-leucyl-l-leucyl-l-leucinal), which is proteasome inhibitor, was added to the degradation system as indicated. For protein degradation in vitro, 100 ng of E. coli strain BL21-induced recombinant HIS-MdJAZ18 protein was incubated in 600 μl extracts (containing 500 µg of total proteins) for each reaction system at 22 °C for the indicated times. The HIS-MdJAZ18 protein abundance was detected by immunoblotting with an anti-HIS antibody, and quantified with Bio-Rad’s QuantityOne software.
Transient expression and GUS analysis assays
The transient expression assays were performed in apple calli. The coding sequence of MdSnRK1.1 was cloned into the virus plasmid vector pIR, and IL-60-BS functioned as an auxiliary construct for the normal replication, movement, and expression of pIR (Peretz et al., 2007). About 2 μg ml−1 of plasmid was introduced into wild-type (WT) and MdJAZ18–GUS calli, while JA and calf intestinal alkaline phosphatase (CIP) were selectively added to various reaction systems as indicated. Thereafter, the calli were placed under normal conditions for 4 d. For GUS histochemical staining, the same quality of dry calli was immersed in GUS staining buffer {0.5 μg μl−1 X-Gluc, 0.075 M sodium phosphate buffer pH 7.2, 0.05 mM K4[Fe(CN)]6·3(H2O), 0.05 mM K3[Fe(CN)6], 10 mM EDTA, 20% methanol, 0.1% Triton X-100}. The samples were incubated overnight at 37 °C.
To detect the GUS activity, 500 mg of calli were ground and extracted with 1 ml of GUS extraction buffer (50 mM sodium phosphate buffer pH 7.0, 10 mM EDTA, 0.1% Triton X-100, 0.1% N-lauroylsarcosine). The GUS activity assay was performed as described by An et al. (2015). The analysis was repeated at least three times.
Results
Sucrose induces anthocyanin and PA accumulation in apple
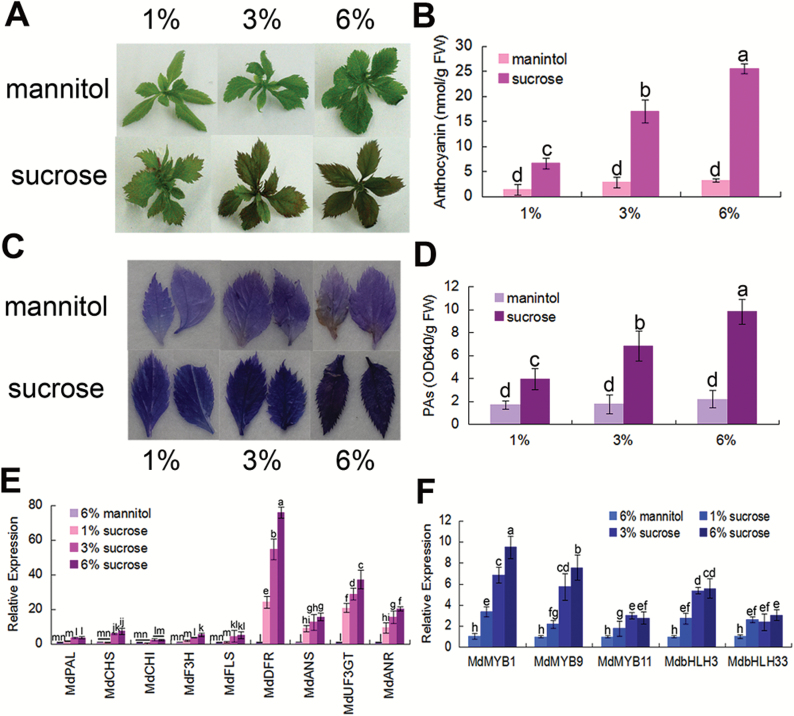
To examine if sucrose promotes anthocyanin and PA accumulation in apple, in vitro shoot cultures of apple grown on MS medium containing sucrose were used. The results showed that anthocyanin levels increased with sucrose, but not with the osmotic control mannitol (Fig. 1A, B). Similarly, sucrose also promoted the accumulation of PAs in the apple leaves (Fig. 1C, D). Furthermore, the expression levels of genes involved in anthocyanin and PA biosynthesis were examined by qRT-PCR assays. When compared with mannitol, the results indicated that sucrose noticeably promoted the transcript levels of anthocyanin and PA structural genes such as MdUF3GT, MdANR, MdANS, and MdDFR, while the transcript levels of MdPAL, MdCHS, MdF3H, MdCHI, and MdFLS increased a little (Fig. 1E). The expression levels of the regulatory MYB and bHLH genes were also assayed. The results indicated that the mRNA levels of MdMYB1, MdMYB9, MdMYB11, MdbHLH3, and MdbHLH33 were also noticeably enhanced in response to sucrose treatment (Fig. 1F).
Fig. 1.
Sucrose induces anthocyanin and PA accumulation by promoting the expression of the regulatory and biosynthetic genes. (A, B) Anthocyanin pigmentation phenotype (A) and anthocyanin contents (B) of 20-day-old in vitro apple shoot cultures treated with different concentrations (1, 3, and 6%) of sucrose and mannitol. (C, D) PA staining and content determination in apple leaves of the shoot cultures in (A). In (B) and (D), FW, fresh weight. Error bars represent the SDs, which were analyzed based on >9 replicates. (E, F) Expression analysis of anthocyanin- and PA-related genes with qRT-PCR in the plants shown in (A). 18S was used as the internal control. Error bars represent the SD based on three independent replicates. In (B), (D), (E), and (F), statistical significance was calculated by the LSD test with DPS software, P<0.05.
MdSnRK1.1 is involved in sucrose-induced accumulation of anthocyanins and PAs
SnRK1 plays a crucial role in sugar and metabolic signaling pathways in plants (Baena-González et al., 2007; Mohannath et al., 2014). To characterize the function of SnRK1 in anthocyanin and PA accumulation in response to sucrose, three AtSnRK1-like genes, MDP0000191788 (MdSnRK1.1), MDP0000173500 (MdSnRK1.2), and MDP0000320932 (MdSnRK1.3) were found in apple. They are similar to each other (Supplementary Fig. S1A). The Neighbor–Joining phylogenetic tree based on the amino acid sequences of AtSnRK1s and MdSnRK1s showed that MdSnRK1.1 had the highest similarity to AtSnRK1.1 (Supplementary Fig. S2). The MdSnRK1.1 gene was cloned from apple (Li et al., 2010). The alignment analysis of the amino acid sequences demonstrated that the predicted MdSnRK1.1 protein shares 82.16% similarity with the AtSnRK1.1 protein in Arabidopsis. It contains a conserved KD and a C-terminal RD (Supplementary Fig. S1B).
To characterize the function of the MdSnRK1.1 gene, the calli of apple cultivar ‘Orin’ were used for genetic transformation. The full-length cDNAs of the MdSnRK1.1 gene were used to construct the overexpression vector 35S::MdSnRK1.1-GFP, while its antisense cDNA fragment was used to construct the suppression vector 35S::asMdSnRK1.1. As a result, two transgenic calli, 35S::MdSnRK1.1-GFP and 35S::asMdSnRK1.1, were obtained. The expression analysis demonstrated that 35S::MdSnRK1.1-GFP transgenic calli produced much more MdSnRK1.1 transcript, but 35S::asMdSnRK1.1 calli produced less compared with the WT control (Suupplementary Fig. S3A), thereby indicating that the expression of the MdSnRK1.1 gene was successfully overexpressed or suppressed in the transgenic calli. Moreover, the expression of MdSnRK1.2 and MdSnRK1.3 was also reduced in 35S::asMdSnRK1.1 transgenic calli, perhaps due to the sequence similarity of their products to MdSnRK1.1 (Supplementary Fig. S3B).
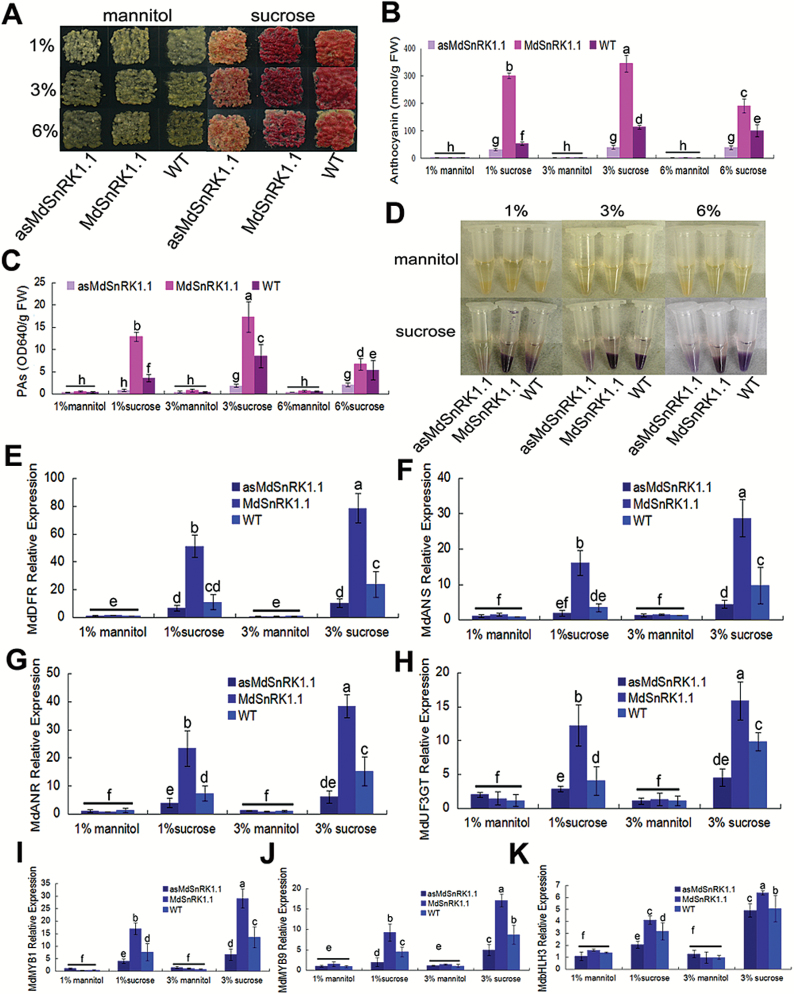
Subsequently, two kinds of transgenic calli were used to examine whether MdSnRK1.1 influences the accumulation of anthocyanins and PAs in response to sucrose, and WT calli were used as the control. In the absence of sucrose, three kinds of calli failed to produce anthocyanins on the medium with mannitol, thus indicating that sugar is necessary for their accumulation. When exposed to different concentrations of sucrose, the WT control produced more anthocyanins in response to 3% sucrose than to 1% and 6% sucrose (Fig. 2A, B).
Fig. 2.
MdSnRK1.1 enhances anthocyanin and PA accumulation. (A) Coloration of the WT apple calli and the transgenic apple calli (MdSnRK1.1 and asMdSnRK1.1) treated with sucrose (1, 3, and 6%) or mannitol (1, 3, and 6%). (B–D) Anthocyanin contents (B), PA contents (C), and staining (D) in the corresponding calli shown in (A). In (B) and (C), FW, fresh weight. (E–K) The expression levels of the regulatory MdbHLH3, MdMYB1, and MdMYB9 genes, as well as the structural MdANS, MdDFR, MdUF3GT, and MdANR genes in anthocyanin and PA biosynthetic pathways in the calli shown in (A), as analyzed with qRT-PCR. 18S was used as the internal control. In (B), (C). and (E–K), error bars represent the SD based on three independent replicates. Statistical significance was calculated by the LSD test with DPS software, P<0.05.
For the transgenic calli, the overexpression of the MdSnRK1.1 gene noticeably increased the accumulation of anthocyanins under 1% sucrose. The 35S::MdSnRK1.1-GFP transgenic calli accumulated more anthocyanins under 1% sucrose, even more than the WT control under the most appropriate sucrose (3%) condition (Fig. 2A, B). However, high sucrose (>3% in the case of apple calli) reduced the accumulation of anthocyanins, compared with 1% sucrose (Supplementary Fig. S4A, B). In contrast, the suppression of MdSnRK1.1 neutralized sucrose-induced anthocyanin accumulation. The 35S::asMdSnRK1.1 transgenic calli produced anthocyanins at similar levels under the different tested sucrose concentrations (Fig. 2A, B), indicating that MdSnRK1.1 is necessary for the sucrose-induced anthocyanin accumulation. In addition, the accumulation of PAs was also detected. The result showed that PAs accumulated in the calli in a pattern highly similar to that of anthocyanins in response to the tested sucrose concentrations (Fig. 2C, D).
Furthermore, the expression of flavonoid biosynthetic genes associated with anthocyanin and PA accumulation was analyzed with qRT-PCRs in WT control as well as 35S::MdSnRK1.1-GFP and 35S::asMdSnRK1.1 transgenic calli treated with different concentrations of sucrose. Corresponding to the anthocyanin and PA phenotypes, the expression levels of LBGs such as MdUF3GT, MdDFR, MdANS, and MdANR were significantly up-regulated in 35S::MdSnRK1.1-GFP transgenic calli (Fig. 2E–H). Besides these structural genes, MdSnRK1.1 also positively regulated the expression of anthocyanin-related regulatory upstream genes such as MdMYB1 and MdMYB9, but only slightly influenced the expression of the MdbHLH3 gene (Fig. 2I–K). In contrast, the suppression of the MdSnRK1.1 gene neutralized the sucrose-induced expression in 35S::asMdSnRK1.1 transgenic calli as compared with the WT control (Fig. 2E–K).
In addition, the expression vector 35S::MdSnRK1.1-GFP was genetically transformed into Arabidopsis to verify the involvement of MdSnRK1.1 in the regulation of anthocyanin biosynthesis (Supplementary Fig. S3C). It was found that MdSnRK1.1 transgenic lines exhibited sucrose-hypersensitive phenotypes. In these MdSnRK1.1 transgenic lines, 1% sucrose promoted root growth and anthocyanin accumulation, while 6% sucrose inhibited them (Supplementary Figs S4C, D, S5).
MdSnRK1.1 interacts with MdJAZ18 protein
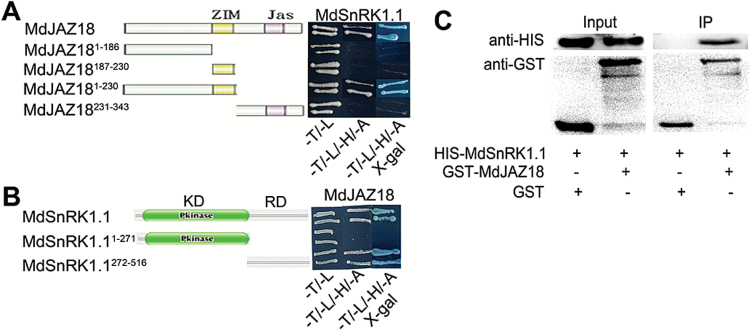
To elucidate how MdSnRK1.1 modulates the accumulation of anthocyanins and PAs, Y2H screening through a cDNA library was performed. The full-length cDNA of the MdSnRK1.1 gene was inserted into the pGBT9 vector. The resultant vector BD-MdSnRK1.1 was used as a bait to screen the library for MdSnRK1.1-interacting proteins. The result showed that a positive colony contained a cDNA fragment which is a part of the MdJAZ18 gene (Li et al., 2015). The evidence suggests that MdJAZ18 plays an important role in the regulation of anthocyanin and PA biosynthesis (An et al., 2015). Therefore, the Y2H assay was conducted to verify the interaction. The full-length cDNA of the MdJAZ18 gene was inserted into the pGAD424 vector as prey (AD-MdJAZ18). The different combinations of bait and prey vectors as well as empty control were transformed into yeast for Y2H assays. The result showed that MdSnRK1.1 interacted with MdJAZ18 protein (Fig. 3A). Furthermore, the interactions between MdSnRK1.1 and other MdJAZs such as MdJAZ1, MdJAZ8, MdJAZ9, MdJAZ10, MdJAZ12, MdJAZ14, and MdJAZ19, were also assayed (Supplementary Fig. S6). The result indicated that MdSnRK1.1 interacted only with MdJAZ18, but not with the other proteins (Supplementary Fig. S7A). In addition, Arabidopsis AtJAZ3 is the orthologous protein of MdJAZ18. The Y2H assay demonstrated that AtJAZ3 also interacted with both MdSnRK1.1 and AtSnRK1.1 proteins (Supplementary Fig. S7A, B).
Fig. 3.
MdSnRK1.1 interacts with MdJAZ18. (A, B) MdSnRK1.1 interacts with MdJAZ18 in Y2H assays. According to the domains of MdSnRK1.1 (KD and RD domain) and MdJAZ18 (ZIM and Jas domain), the full length or derivatives thereof were used to assess their interactions. Yeast dropout medium lacking Leu and Trp (-T/-L) was used as transformation control, and that lacking Leu, Trp, His, and Ade (-T/-L/-H/-A) acted as a screen. (C) In vitro pull-down assay to verify the interaction between MdSnRK1.1 and MdJAZ18. HIS-MdSnRK1.1 was co-incubated with GST or GST–MdJAZ18 protein and then purified using a GST purification kit. The resultant protein samples were immunoblotted with anti-HIS and anti-GST antibodies, respectively.
To map the domains in MdSnRK1.1 and MdJAZ18 proteins necessary for their interaction, MdSnRK1.1 was divided into an N-terminal kinase domain MdSnRK1.11–271 and a C-terminal regulatory domain MdSnRK1.1272–516, while MdJAZ18 was divided into MdJAZ181–186, MdJAZ18187–230, MdJAZ181–230, and MdJAZ18231–343. They were then inserted into pGBT9 and pGAD424 vectors, respectively. As a result, six vectors BD-MdSnRK1.11–271, BD-MdSnRK1.1272–516, AD-MdJAZ181–186, AD-MdJAZ18187–230, AD-MdJAZ181–230, and AD-MdJAZ18230–343 were obtained and used for Y2H assays. The results showed that C-terminal regulatory domain (272–516) of MdSnRK1.1 protein and the N-terminal ZIM domain (1–230) of MdJAZ18 protein were responsible for their interaction (Fig. 3A, B).
To verify further the interaction between MdSnRK1.1 and MdJAZ18, an in vitro pull-down assay was conducted. Purified recombinant HIS-MdSnRK1.1 and GST–MdJAZ18 proteins were expressed and purified from E. colli BL21. Subsequently, HIS-MdSnRK1.1 proteins were incubated with GST–MdJAZ18 and GST, respectively, and then separated by SDS–PAGE for western blotting with an anti-HIS antibody. The HIS-MdSnRK1.1 proteins were enriched by GST–MdJAZ18, but not by the GST control (Fig. 3C), indicating that MdSnRK1.1 physically interacted with MdJAZ18.
MdSnRK1.1 phosphorylates and destabilizes MdJAZ18 protein
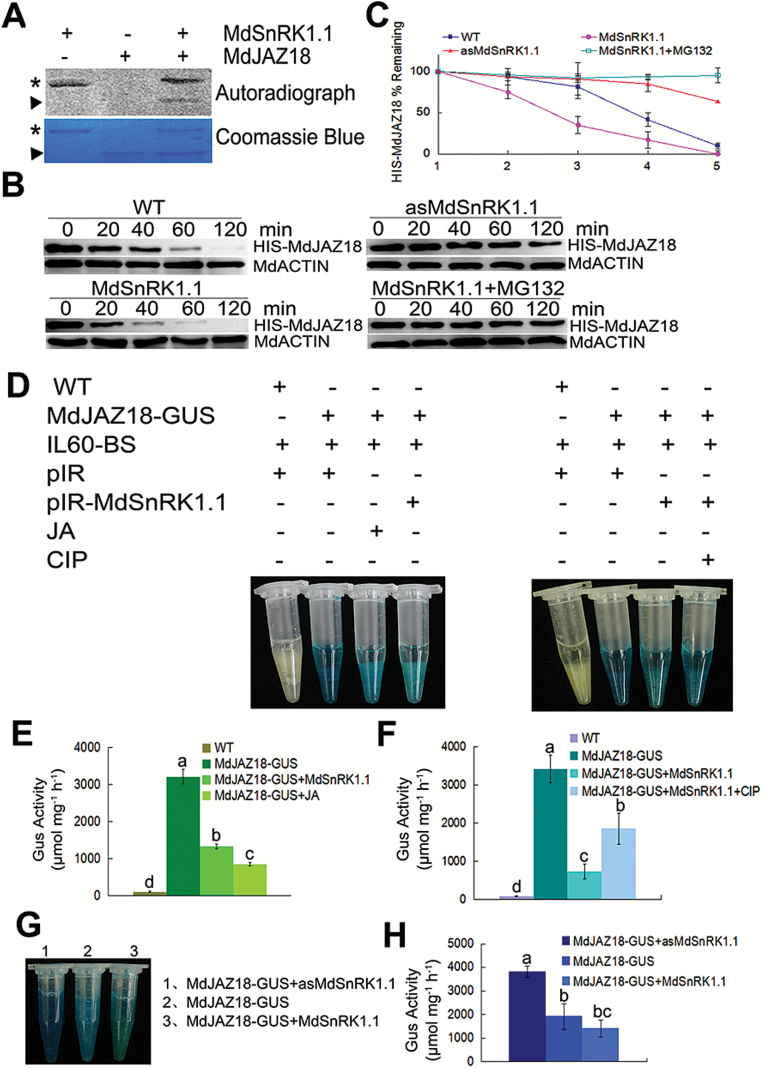
MdSnRK1.1 is a Ser/Thr protein kinase. To examine whether MdSnRK1.1 phosphorylates MdJAZ18 protein, the purified HIS-MdSnRK1.1, HIS-MdJAZ18, and HIS-MdJAZ12 proteins were used for an in vitro phosphorylation experiment. The result showed that MdSnRK1.1 efficiently autophosphorylated itself, indicating its kinase activity. The phosphorylated MdJAZ18 protein was detected in the MdSnRK1.1 and MdJAZ18 co-incubated sample, but not in the MdJAZ18 sample alone, indicating that MdSnRK1.1 directly phosphorylated MdJAZ18 protein in vitro (Fig. 4A). In addition, MdSnRK1.1 failed to phosphorylate MdJAZ12 protein (Supplementary Fig. S8).
Fig. 4.
MdSnRK1.1 phosphorylates and degrades MdJAZ18 protein. (A) MdSnRK1.1 phosphorylates MdJAZ18 in vitro. The asterisk indicates the autophosphorylation of the purified HIS-MdSnRK1.1, while the triangle indicates the phosphorylation of the purified HIS-MdJAZ18 by HIS-MdSnRK1.1 in the autoradiogram. The asterisk and the triangle refer to protein loading of HIS-MdJAZ18 and HIS-MdSnRK1.1, respectively, in Coomassie blue staining. (B) MdJAZ18 stability is reduced by MdSnRK1.1 and the effect is inhibited by MG132. Total proteins extracted from WT, MdSnRK1.1, and asMdSnRK1.1 apple calli were incubated with the purified HIS-MdJAZ18 protein treated with or without MG132. The samples were harvested at the indicated time. MdACTIN was used as internal reference. (C) The half-life plot for in vitro HIS-MdJAZ18 degradation was analyzed from protein bands with Bio-Rad QuantityOne software. (D) MdSnRK1.1 degrades MdJAZ18, and CIP inhibits MdSnRK1.1-mediated degradation. GUS staining images of transiently expressed pIR-MdSnRK1.1 or pIR in MdJAZ18-GUS calli treated with or without JA or CIP. WT calli were used as a negative control. (E, F) Quantitative analysis of GUS activity in (D). (G) MdSnRK1.1 promoted the degradation of MdJAZ18 in vivo. GUS staining images of MdJAZ18–GUS+MdSnRK1.1, MdJAZ18–GUS+asMdSnRK1.1, and MdJAZ18–GUS calli. (H) Quantitative analysis of GUS activity in (G). In (C), (E), (F), and (H), error bars represent the SD. Statistical significance was calculated by the LSD test with DPS software, P<0.05.
JAZs are typical proteins which interact with the SCFCOI1 complex and are degraded via the ubiquitination–26S proteasome pathway (Thines et al., 2007). To examine if MdSnRK1.1-mediated phosphorylation influences the stability of MdJAZ18 protein, a cell-free degradation assay was carried out. The prokaryon-expressed and purified HIS-MdJAZ18 protein was incubated with the plant total proteins that were extracted from WT control, 35S::MdSnRK1.1-GFP, and 35S::asMdSnRK1.1 transgenic apple calli. Subsequently, immunoblottings were performed with an anti-HIS antibody to detect the protein abundance. The results showed that the abundance of HIS-MdJAZ18 protein was less in the protein extracts of 35S::MdSnRK1.1-GFP transgenic calli than in those of the WT control (Fig. 4B, C), whereas it was more stable in the protein extracts of 35S::asMdSnRK1.1 transgenic calli than in those of the WT control (Fig. 4B, C). In addition, MdSnRK1.1-mediated HIS-MdJAZ18 degradation was more rapid in protein samples of 35S::MdSnRK1.1-GFP transgenic calli treated with 1% sucrose than those treated with 3% and 6% sucrose (Supplementary Fig. S9). These results suggest that MdSnRK1.1-mediated phosphorylation of MdJAZ18 protein promotes its degradation in response to sucrose. Furthermore, the MG132 treatment inhibited MdSnRK1.1-mediated degradation of MdJAZ18 protein (Fig. 4B, C), indicating that the degradation may be through a 26S proteasome pathway.
To detect the degradation of MdJAZ18 in vivo, the 35S::MdJAZ18-GUS construct was genetically transformed into apple calli. The GUS staining assay demonstrated that 35S::MdJAZ18-GUS transgenic calli exhibited GUS activity, indicating that MdJAZ18–GUS fusion protein was successfully expressed in the transgenic calli. After treatment with 100 μM JA for 2 h, the transgenic calli showed a reduced GUS activity, indicating that MdJAZ18 proteins were degraded in response to JA (Supplementary Fig. S10).
To check rapidly if MdSnRK1.1 influences the degradation of MdJAZ18, the viral vectors pIR and pIR-MdSnRK1.1 were used to transiently transform 35S::MdJAZ18-GUS transgenic calli. The GUS staining assay demonstrated that 35S::MdJAZ18-GUS+pIR-MdSnRK1.1 double transgenic calli showed a lower GUS activity than the 35S::MdJAZ18-GUS+pIR control, indicating that MdSnRK1.1 promoted the degradation of MdJAZ18 protein (Fig. 4D, E). However, CIP treatment partially neutralized MdSnRK1.1-mediated degradation of MdJAZ18–GUS protein (Fig. 4D, F), suggesting that phosphorylation modification was involved in the degradation process.
To verify further that MdSnRK1.1 promotes MdJAZ18 degradation, double transgenic calli 35S::MdJAZ18-GUS+35S:: MYC-MdSnRK1.1 and 35S::MdJAZ18-GUS+35S::asMdSn RK1.1 were obtained (Supplementary Fig. S11A, B). Subsequently, they were used to detect the abundance of MdJAZ18–GUS protein with a GUS staining assay, while 35S::MdJAZ18-GUS transgenic calli were used as the control. The results showed that 35S::MdJAZ18-GUS+35S:: MYC-MdSnRK1.1 double transgenic calli exhibited much lower GUS activity, while that in 35S::MdJAZ18-GUS+35S:: asMdSnRK1.1 calli was higher than in the 35S::MdJAZ18-GUS control (Fig. 4G, H).
Taken together, these findings indicate that MdSnRK1.1 phosphorylates MdJAZ18 to facilitate its 26S proteasome-mediated degradation.
MdJAZ18 is involved in MdSnRK1.1-induced anthocyanin and PA accumulation
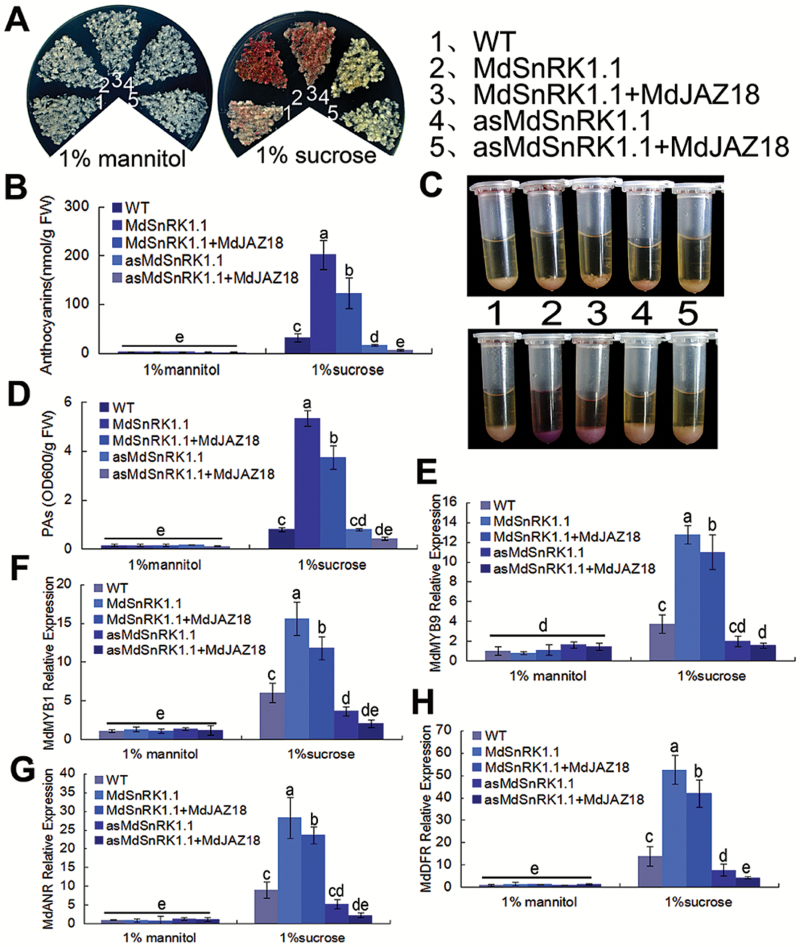
In plants, JAZ proteins degrade in response to JA signal, which release bHLH and MYB TFs to promote anthocyanin and PA biosynthesis (Qi et al., 2011; An et al., 2015). To examine if MdJAZ18 functions in MdSnRK1.1-mediated anthocyanin and PA accumulation, the fused Flag-MdJAZ18 driven by a 35S promoter was genetically transformed into WT calli, as well as into 35S::MdSnRK1.1-GFP and 35S::asMdSnRK1.1 transgenic calli. The resultant 35S::MdSnRK1.1-GFP, 35S::asMdSnRK1.1, 35S::MdSnRK1.1-GFP+35S::Flag-MdJAZ18, and 35S::asMdSnRK1.1 + 35S::Flag-MdJAZ18 transgenic calli were obtained and used for anthocyanin and PA analysis (Supplementaty Fig. S11A), while WT calli were applied as the control. The results showed that sucrose is necessary for anthocyanin and PA biosynthesis in all tested calli, and that MdSnRK1.1 promoted anthocyanin and PA accumulation in response to sucrose (Fig. 5A–D).
Fig. 5.
MdJAZ18 is involved in MdSnRK1.1-induced anthocyanin and PA accumulation. (A, B) Coloration (A) and anthocyanin contents (B) of WT, 35S::MdSnRK1.1-GFP, 35S::MdSnRK1.1-GFP+35S::Flag-MdJAZ18, 35S::asMdSnRK1.1, and 35S::asMdSnRK1.1 + 35S::Flag-MdJAZ18 treated with 1% sucrose or mannitol. (C, D) PA staining (C) and contents (D) in the corresponding calli shown in (A). In (B) and (D), FW, fresh weight. (E–H) The expression levels of regulatory genes (MdMYB1 and MdMYB9) and structural genes (MdDFR and MdANR) in anthocyanin and PA biosynthetic pathways by qRT-PCR analysis in the calli shown in (A). 18S acted as the internal control. In (B) and (D–H), error bars represent the SD based on three independent replicates. Statistical significance was calculated by the LSD test with DPS software, P<0.05.
When MdJAZ18 was overexpressed in 35S::MdSnRK1.1-GFP transgenic calli, 35S::MdSnRK1.1-GFP+35S::Flag-MdJAZ18 double transgenic calli produced less anthocyanins and PAs than 35S::MdSnRK1.1-GFP, indicating that Flag-MdJAZ18 oversupply neutralized MdSnRK1.1-mediated MdJAZ18 degradation, thereby inhibiting MdSnRK1.1-promoted anthocyanin and PA accumulation (Fig. 5A–D). In addition, it was found that the suppression of the MdSnRK1.1 gene inhibited sucrose-induced anthocyanin and PA accumulation in 35S::asMdSnRK1.1 transgenic calli, and that the MdJAZ18 overexpression exacerbated this inhibition in 35S::antiMdSnRK1.1 + 35S::Flag-MdJAZ18 calli (Fig. 5A–D). These findings indicated that MdJAZ18 was involved in MdSnRK1.1-mediated anthocyanin and PA accumulation in response to sucrose.
Consistent with the anthocyanin and PA phenotypes, the expression levels of the regulatory MYB genes MdMYB1 and MdMYB9, as well as the structural genes MdDFR, MdANS, MdANR, and MdUF3GT, were lower in 35S::MdSnRK1.1-GFP+35S::Flag-MdJAZ18 and 35S::asMdSnRK1.1 + 35S::Flag-MdJAZ18 transgenic calli than in the corresponding 35S::MdSnRK1.1-GFP and 35S::asMdSnRK1.1 calli (Fig. 5E–H: Supplementary Fig. S12A, B). These findings suggested that MdJAZ18 was involved in MdSnRK1.1-induced anthocyanin and PA accumulation by modulating the expression of the regulatory and structural genes.
Discussion
The stimulatory effects of sucrose on anthocyanin biosynthesis have been reported in different plant species such as grapevine, petunia, mulberry, and Arabidopsis (Pirie and Mullins, 1976; Weiss, 2000; Teng et al., 2005; Tsai et al., 2005). The sucrose-induced anthocyanin biosynthesis depends on the MYB75/PAP1 TF, but not on the glucose sensor AtHXK1 in Arabidopsis (Teng et al., 2005). Signaling intermediates such as Ca2+, protein kinases, and protein phosphatases participate in sucrose-induced anthocyanin accumulation (Vitrac et al., 2000). In parallel, various stresses and phytohormones induce the accumulation of PAs by modulating the expression of their biosynthetic genes (Mellway et al., 2009; An et al., 2015; Yoshida et al., 2015). In this study, it was found that sucrose induced the biosynthesis of anthocyanins and PAs in apple, and that MdSnRK1.1 played a crucial role by interacting with and phosphorylating MdJAZ18 protein in this process (Figs 1A–D, 2A–D, 3A–C, 4A).
SnRK1 is an important regulator of plant growth and development (Baena-González et al., 2007; Lu et al., 2007; Cho et al., 2012). It positively regulates the accumulation of starch in potato tubers (Lin et al., 2014). It also inhibits seed germination and seedling growth by positively regulating the expression of MYBS1 and aAmy3 genes in rice (Lu et al., 2007). In addition, sugar is indispensible for anthocyanin accumulation. Both sugar starvation and oversupply inhibit anthocyanin biosynthesis (Sivitz et al., 2008). SnRK1 is proposed to function as a central component of kinase cascades in the sugar signaling pathway (Baena-González et al., 2007; Jossier et al., 2009). Its activity is regulated by sugar (Baena-González and Sheen, 2008).
Correspondingly, SnRK1.1 overexpression increases the sensitivity to sucrose both in Arabidopsis and in apple calli (Jossier et al., 2009; Fig. 2A; Supplementary Fig. S5). In this case, 3% sucrose is appropriate for the WT Arabidopsis plants, but is too high for the transgenic plants. Therefore, 3% sucrose inhibited anthocyanin accumulation in AtSnRK1.1 transgenic Arabidopsis (Baena-González et al., 2007). In this study, ectopic expression of the MdSnRK1.1 gene in Arabidopsis promoted anthocyanin biosynthesis under 1% sucrose, but inhibited this process under 9% sucrose (Supplementary Figs S4C, D, S5A). Similarly, compared with 1% sucrose, 12% sucrose inhibited anthocyanin biosynthesis in MdSnRK1.1 transgenic apple calli (Fig. 2A, B; Supplementary S4A, B). Therefore, there is a threshold sucrose concentration for the induction of anthocyanin biosynthesis in apple calli and Arabidopsis. Sucrose at concentrations higher and lower than this threshold inhibits the accumulation of anthocyanins.
The demand for and utilization of sucrose vary with plant species, developmental stages, and environment cues, which may affect the SnRK1 activity (Rolland et al., 2006; Baena-González and Sheen, 2008). Compared with zero sucrose, 9% sucrose inhibits anthocyanin biosynthesis in MdSnRK1.1 transgenic Arabidopsis but promotes this process in transgenic apple calli (Supplementary Fig. S4), suggesting that apple calli have a higher sugar tolerance than Arabidopsis. This also explains the discrepancy between our data concerning MdSnRK1.1 ectopic transgenic Arabdidopsis and those of a previous study about AtSnRK1.1 overexpression in transgenic Arabidopsis (Baena-González et al., 2007; Supplementary Fig. S5).
In Arabidopsis, SnRK1 is involved in various life processes by phosphorylating its specific substrates (Zhang et al., 2008). The substrates not only include the key metabolic enzymes such as sucrose phosphate synthase, nitrate reductase, and HMG-CoA reductase, but also the TFs such as FUS3 (Sugden et al., 1999; Harthill et al., 2006; Tsai and Gazzarrini, 2012). The tomato SlSnRK1 phosphorylates the βC1 protein that is the pathogenicity determinant and a suppressor of RNA silencing (Shen et al., 2011). SnRK1 is also involved in the innate antiviral defense by interacting with AL2 and L2, which are the geminivirus pathogenicity proteins (Hao et al., 2003). SnRK1 interacts with and phosphorylates adenosine kinase (ADK) to maintain energy homeostasis and to regulate the responses to biotic and abiotic stresses (Mohannath et al., 2014). In this study, it was found that MdSnRK1.1 interacted with and phosphorylated the MdJAZ18 protein in apple (Figs 3A–C, 4A), which further promoted its degradation (Fig. 4B–H). In mammals, AMPK is structurally and functionally analogous to SnRK1 in higher plants. It phosphorylates TXNIP (thioredoxin-interacting protein) and accelerates its degradation upon energy stress (Wu et al., 2013). In our study, MG132 treatment eliminated the effect of MdSnRK1.1-induced MdJAZ18 phosphorylation on the degradation of MdJAZ18 protein, suggesting that the degradation depends on the 26S proteasome (Fig. 4B, C).
The JAZ proteins act as repressors in the JA signaling pathway. They are recruited by the F-box protein (COI1) for ubiquitination and subsequent degradation through the 26S proteasome in response to JA signal, thereby releasing the downstream JA-responsive factors (Qi et al., 2011; Wasternack and Hause, 2013). The JAZ proteins contain two domains, which are the N-terminal ZIM domain and the C-terminal Jas domain (Wasternack and Hause, 2013). The conserved ZIM (TIFY) domain of JAZ proteins is responsible for JAZ dimerization and interaction with the co-repressors NINJA (Pauwels et al., 2010). In this study, it was found that the N-terminal ZIM domain of MdJAZ18 protein is essential for its interaction with MdSnRK1.1 (Fig. 3A). Hence, it is possible that MdSnRK1.1 competes with NINJA to interact with MdJAZ18, which consequently results in JA responses and therefore links sugar and JA signaling pathways.
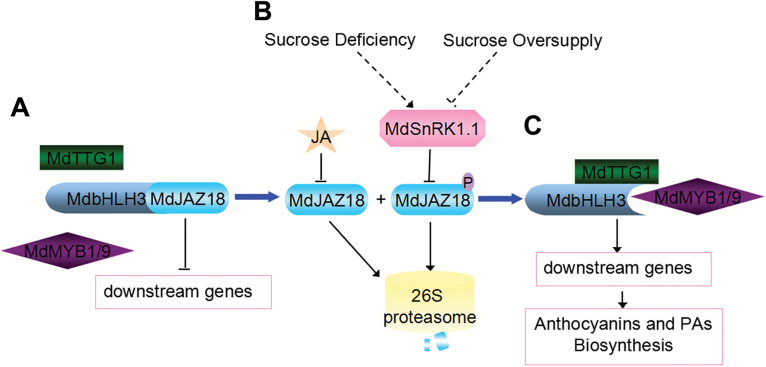
In Arabidopsis, JAZ proteins directly interact with bHLH TFs (GL3, EGL3, and TT8) and MYB TFs (MYB75 and GL1). These bHLH and MYB TFs are essential components of the MBW complex to mediate anthocyanin accumulation (Qi et al., 2011). In apple, MdJAZ18, MdJAZ1, and MdJAZ19 interact with the MdbHLH3 protein. MdbHLH3 not only improves the transcription level of MdMYB1, MdMYB9, and MdMYB11, but also interacts with three MdMYB proteins to activate the expression of MdANS, MdANR, and MdLAR to regulate anthocyanin and PA biosynthesis (Xie et al., 2012; An et al., 2015). Therefore, JAs stimulate anthocyanin and PA accumulation by up-regulating the expression of these flavonoid biosynthetic genes (Qi et al., 2011; Wasternack and Hause, 2013; An et al., 2015). Therefore, our findings support a model in which MdSnRK1.1 promotes the accumulation of anthocyanins and PAs by phosphorylating and degrading MdJAZ18 to release MdbHLH3 in response to sucrose deficiency, which subsequently activates the expression of the regulatory and structural genes (Fig. 6). This mechanism explains the long known fact that JA acts together with sucrose to improve anthocyanin biosynthesis (Loreti et al., 2008). The integration of sugar and JA signals was verified by the interaction between MdSnRK1.1 and MdJAZ18 proteins, which controls anthocyanin and PA accumulation through the modulation of the function of the MBW complex in plants.
Fig. 6.
Proposed model for the involvement of MdSnRK1.1 in sucrose- and JA-regulated anthocyanin and PA biosynthesis in apple. (A) The interaction of MdJAZ18 with the bHLH transcription factor MdbHLH3 represses the transcriptional function of the MBW complex to activate downstream genes. (B) Under sucrose deficiency, MdSnRK1.1 phosphorylates MdJAZ18, and then accelerates its degradation through the 26S proteasome. However, under sucrose oversupply, this process is inhibited. Sucrose and JA have a synergistic effect on the degradation of MdJAZ18 protein. (C) Once the degradation of MdJAZ18 protein is completed, MdbHLH3 is released and then interacts with MdMYBs (MdMYB1 and MdMYB9) and MdTTG1 to form an active MBW complex, or bind to the promoters of MdMYB (MdMYB1 and MdMYB9) genes to regulate the biosynthesis of anthocyanins and PAs.
Besides anthocyanin biosynthesis, JAs modulate male and female fertility, root growth, and trichome formation (Stintzi, 2000; Pauwels et al., 2010; Qi et al., 2011). Furthermore, they also regulate a wide range of defense processes, such as pathogen infection, insect attack, UV damage, wounding, and many other abiotic stresses (Nibbe et al., 2002; Xiao et al., 2004; Schilmiller and Howe, 2005; Wasternack and Hause, 2013). JAs activate the defense responses against herbivorous insects and necrotrophic pathogens through regulating the expression of VSP2 and PDF1.2 (Wasternack and Hause, 2013). Similarly, sugars regulate stress-inducible and pathogenesis-related genes (Sadka et al., 1994; Loreti et al., 2008). In plants, elevated levels of cellular sugar increase the expression level of genes associated with defense responses (Price et al., 2004). The sensitivity to virus attack is increased in the transgenic tobacco plants which contain an antisense sequence of the Arabidopsis SnRK1 gene (Hao et al., 2003; Polge and Thomas, 2007). For the first time, our findings and the model concerning the interaction between MdSnRK1.1 and MdJAZ18 shed light on the molecular mechanism by which the crosstalk of sugar and JA signaling regulates defense against various abiotic and biotic stresses.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Protein sequence comparison of MdSnRK1s or MdSnRK1.1 with AtSnRK1.1.
Fig. S2. Phylogenic tree of Arabidopsis and apple SnRK1 protein.
Fig. S3. Generation of MdSnRK1.1 and asMdSnRK1.1 transgenic plants.
Fig. S4. MdSnRK1.1 mediates anthocyanin accumulation in response to sucrose.
Fig. S5. Ectopic expression of the MdSnRK1.1 gene enhances sucrose sensitivity and promotes anthocyanin accumulation in Arabidopsis under 1% sucrose.
Fig. S6. Phylogenic tree of Arabidopsis and apple JAZ proteins.
Fig. S7. Y2H assay to test interactions of SnRK1.1 with the MdJAZs proteins in apple and AtJAZ3 in Arabidopsis.
Fig. S8. MdSnRK1.1 fails to phosphorylate MdJAZ12 in vitro.
Fig. S9. Sucrose influences MdSnRK1.1-mediated MdJAZ18 degradation.
Fig. S10 GUS activity of MdJAZ18–GUS transgenic calli in response to JA.
Fig. S11. Generation of transgenic calli that transform MdJAZ18 in the MdSnRK1.1 and asMdSnRK1.1 background, and which express MdSnRK1.1 and asMdSnRK1.1 in MdJAZ18–GUS transgenic calli.
Fig. S12. The expression levels of MdUF3GT and MdANS genes in the WT and transgenic calli in Fig. 5A.
Table S1. Primers used for qRT-PCR and vector construction.
Supplementary Material
Acknowledgements
This work was supported by the NSFC (31325024, 31471854), the Ministry of Education of China (IRT15R42), and the Ministry of Agriculture of China (CARS-28). We thank Professor Yan Guo and Dr Shuang-Shuang Zhao of China Agricultural University for technical assistance with the in vitro phosphorylation assay.
References
- Albert NW, Davies KM, Lewis DH, Zhang H, Montefiori M, Brendolise C, Boase MR, Ngo H, Jameson PE, Schwinn KE. 2014. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. The Plant Cell 26, 962–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan AC, Hellens RP, Laing WA. 2008. MYB transcription factors that colour our fruit. Trends in Plant Science 13, 99–102. [DOI] [PubMed] [Google Scholar]
- An XH, Tian Y, Chen KQ, Liu XJ, Liu DD, Xie XB, Cheng CG, Cong PH, Hao YJ. 2015. MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant and Cell Physiology 56, 650–662. [DOI] [PubMed] [Google Scholar]
- An XH, Tian Y, Chen KQ, Wang XF, Hao YJ. 2012. The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. Journal of Plant Physiology 169, 710–717. [DOI] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J. 2007. A central integrator of transcription networks in plant stress and energy signalling. Nature 448, 938–942. [DOI] [PubMed] [Google Scholar]
- Baena-González E, Sheen J. 2008. Convergent energy and stress signaling. Trends in Plant Science 13, 474–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbehenn RV, Constabel CP. 2011. Tannins in plant–herbivore interactions. Phytochemistry 72, 1551–1565. [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, et al. 2005. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. The Plant Journal 42, 567–585. [DOI] [PubMed] [Google Scholar]
- Chen S, Songkumarn P, Liu J, Wang GL. 2009. A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiology 150, 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YH, Hong JW, Kim EC, Yoo SD. 2012. Regulatory functions of SnRK1 in stress-responsive gene expression and in plant growth and development. Plant Physiology 158, 1955–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozet P, Margalha L, Confraria A, Rodrigues A, Martinho C, Adamo M, Elias CA, Baena-González E. 2014. Mechanisms of regulation of SNF1/AMPK/SnRK1 protein kinases. Frontiers in Plant Science 5, 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkwel PP, Huijser C, Weisbeek PJ, Chua NH, Smeekens SC. 1997. Sucrose control of phytochrome A signaling in Arabidopsis. The Plant Cell 9, 583–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Xie DY, Sharma SB. 2005. Proanthocyanidins—a final frontier in flavonoid research?New Phytologist 165, 9–28. [DOI] [PubMed] [Google Scholar]
- Emanuelle S, Doblin MS, Stapleton DI, Bacic A, Gooley PR. 2016. Molecular insights into the enigmatic metabolic regulator, SnRK1. Trends in Plant Science 21, 341–353. [DOI] [PubMed] [Google Scholar]
- Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC. 2006. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. The Plant Journal 49, 414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghillebert R, Swinnen E, Wen J, Vandesteene L, Ramon M, Norga K, Rolland F, Winderickx J. 2011. The AMPK/SNF1/SnRK1 fuel gauge and energy regulator: structure, function and regulation. FEBS Journal 278, 3978–3990. [DOI] [PubMed] [Google Scholar]
- Góraj-Koniarska J, Saniewski M. 2015. The effect of sugars in relation to methyl jasmonate on anthocyanin formation in the roots of Kalanchoe blossfeldiana (Poelln.). Acta Agrobotanica 68, 173–178. [Google Scholar]
- Hanson J, Smeekens S. 2009. Sugar perception and signaling—an update. Current Opinion in Plant Biology 12, 562–567. [DOI] [PubMed] [Google Scholar]
- Hao L, Wang H, Sunter G, Bisaro DM. 2003. Geminivirus AL2 and L2 proteins interact with and inactivate SNF1 kinase. The Plant Cell 15, 1034–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG. 2007. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nature Reviews Molecular Cell Biology 8, 774–785. [DOI] [PubMed] [Google Scholar]
- Harthill JE, Meek SE, Morrice N, Peggie MW, Borch J, Wong BH, Mackintosh C. 2006. Phosphorylation and 14-3-3 binding of Arabidopsis trehalose-phosphate synthase 5 in response to 2-deoxyglucose. The Plant Journal 47, 211–223. [DOI] [PubMed] [Google Scholar]
- Jang JC, León P, Zhou L, Sheen J. 1997. Hexokinase as a sugar sensor in higher plants. The Plant Cell 9, 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SW, Das PK, Jeoung SC, et al. 2010. Ethylene suppression of sugar-induced anthocyanin pigmentation in Arabidopsis. Plant Physiology 154, 1514–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossier M, Bouly JP, Meimoun P, Arjmand A, Lessard P, Hawley S, Grahame Hardie D, Thomas M. 2009. SnRK1 (SNF1-related kinase 1) has a central role in sugar and ABA signalling in Arabidopsis thaliana. The Plant Journal 59, 316–328. [DOI] [PubMed] [Google Scholar]
- Koch KE. 1996. Carbohydrate-modulated gene expression in plants. Annual Review of Plant Physiology and Plant Molecular Biology 47, 509–540. [DOI] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F. 2006. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends in Plant Science 10, 1360–1385. [DOI] [PubMed] [Google Scholar]
- Li G, Peng F, Zhang L, Shi X, Wang Z. 2010. Cloning and characterization of a SnRK1-encoding gene from Malus hupehensis Rehd. and heterologous expression in tomato. Molecular Biology Reports 37, 947–954. [DOI] [PubMed] [Google Scholar]
- Li XQ, Yin XJ, Wang H, Li J, Guo CL, Gao H, Zheng Y, Fan CH, Wang XP. 2015. Genome-wide identification and analysis of the apple (Malus×domestica Borkh.) TIFY gene family. Tree Genetics and Genomes 11, 1–13. [Google Scholar]
- Li Y, Van den Ende W, Rolland F. 2014. Sucrose induction of anthocyanin biosynthesis is mediated by DELLA. Molecular Plant 7, 570–572. [DOI] [PubMed] [Google Scholar]
- Li YY, Mao K, Zhao C, Zhao XY, Zhang HL, Shu HR, Hao YJ. 2012. MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiology 160, 1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillo C, Lea US, Ruoff P. 2008. Nutrient depletion as a key factor for manipulating gene expression and product formation in different branches of the flavonoid pathway. Plant, Cell and Environment 31, 587–601. [DOI] [PubMed] [Google Scholar]
- Lin CR, Lee KW, Chen CY, Hong YF, Chen JL, Lu CA, Chen KT, Ho TH, Yu SM. 2014. SnRK1A-interacting negative regulators modulate the nutrient starvation signaling sensor SnRK1 in source–0sink communication in cereal seedlings under abiotic stress. The Plant Cell 26, 808–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Yang Y, Quan R, Mendoza I, Wu Y, Du W, Zhao S, Schumaker KS, Pardo JM, Guo Y. 2009. Phosphorylation of SOS3-LIKE CALCIUM BINDING PROTEIN8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. The Plant Cell 21, 1607–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung K, Nemhauser JL, Perata P. 2015. New mechanistic links between sugar and hormone signalling networks. Current Opinion in Plant Biology 25, 130–137. [DOI] [PubMed] [Google Scholar]
- Loreti E, Povero G, Novi G, Solfanelli C, Alpi A, Perata P. 2008. Gibberellins, jasmonate and abscisic acid modulate the sucrose-induced expression of anthocyanin biosynthetic genes in Arabidopsis. New Phytologist 179, 1004–1016. [DOI] [PubMed] [Google Scholar]
- Lu CA, Lin CC, Lee KW, Chen JL, Huang LF, Ho SL, Liu HJ, Hsing YI, Yu SM. 2007. The SnRK1A protein kinase plays a key role in sugar signaling during germination and seedling growth of rice. The Plant Cell 19, 2484–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellway RD, Tran LT, Prouse MB, Campbell MM, Constabel CP. 2009. The wound-, pathogen-, and ultraviolet B-responsive MYB134 gene encodes an R2R3 MYB transcription factor that regulates proanthocyanidin synthesis in poplar. Plant Physiology 150, 924–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohannath G, Jackel JN, Lee YH, Buchmann RC, Wang H, Patil V, Adams AK, Bisaro DM. 2014. A complex containing SNF1-related kinase (SnRK1) and adenosine kinase in Arabidopsis. PLoS One 9, e87592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J. 2003. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300, 332–336. [DOI] [PubMed] [Google Scholar]
- Mori T, Sakurai M, Seki M, Furusaki S. 1994. Use of auxin and cytokinin to regulate anthocyanin production and composition in suspension cultures of strawberry cell. Journal of the Science of Food and Agriculture 65, 271–276. [Google Scholar]
- Nibbe M, Hilpert B, Wasternack C, Miersch O, Apel K. 2002. Cell death and salicylate- and jasmonate-dependent stress responses in Arabidopsis are controlled by single cet genes. Planta 216, 120–128. [DOI] [PubMed] [Google Scholar]
- Pauwels L, Barbero GF, Geerinck J, et al. 2010. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretz Y, Mozes-Koch R, Akad F, Tanne E, Czosnek H, Sela I. 2007. A universal expression/silencing vector in plants. Plant Physiology 145, 1251–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirie A, Mullins MG. 1976. Changes in anthocyanin and phenolics content of grapevine leaf and fruit tissues treated with sucrose, nitrate, and abscisic acid. Plant Physiology 58, 468–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polge C, Thomas M. 2007. SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control?Trends in Plant Science 12, 20–28. [DOI] [PubMed] [Google Scholar]
- Pourcel L, Routaboul JM, Kerhoas L, Caboche M, Lepiniec L, Debeaujon I. 2005. TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. The Plant Cell 17, 2966–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J, Laxmi A, St Martin SK, Jang JC. 2004. Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. The Plant Cell 16, 2128–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D. 2011. The jasmonate-ZIM-domain proteins interact with the WD-repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. The Plant Cell 23, 1795–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radchuk R, Emery RJ, Weier D, Vigeolas H, Geigenberger P, Lunn JE, Feil R, Weschke W, Weber H. 2010. Sucrose non-fermenting kinase 1 (SnRK1) coordinates metabolic and hormonal signals during pea cotyledon growth and differentiation. The Plant Journal 61, 324–338. [DOI] [PubMed] [Google Scholar]
- Ramon M, Ruelens P, Li Y, Sheen J, Geuten K, Rolland F. 2013. The hybrid Four-CBS-Domain KINβγ subunit functions as the canonical γ subunit of the plant energy sensor SnRK1. The Plant Journal 75, 11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitsch T. 1999. Source–sink regulation by sugar and stress. Current Opinion in Plant Biology 2, 198–206. [DOI] [PubMed] [Google Scholar]
- Rolland F, Baenagonzalez E, Sheen J. 2006. Sugar sensing and signaling in plants: conserved and novel mechanisms. Plant Biology 57, 675–709. [DOI] [PubMed] [Google Scholar]
- Sadka A, DeWald DB, May GD, Park WD, Mullet JE. 1994. Phosphate modulates transcription of soybean VspB and other sugar-inducible genes. The Plant Cell 6, 737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalbert A, Manach C, Morand C, Rémésy C, Jiménez L. 2005. Dietary polyphenols and the prevention of diseases. Critical Reviews in Food Science and Nutrition 45, 287–306. [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Howe GA. 2005. Systemic signaling in the wound response. Current Opinion in Plant Biology 8, 369–377. [DOI] [PubMed] [Google Scholar]
- Shan X, Zhang Y, Peng W, Wang Z, Xie D. 2009. Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. Journal of Experimental Botany 60, 3849–3860. [DOI] [PubMed] [Google Scholar]
- Shen Q, Liu Z, Song F, Xie Q, Hanley-Bowdoin L, Zhou X. 2011. Tomato SlSnRK1 protein interacts with and phosphorylates βC1, a pathogenesis protein encoded by a geminivirus β-satellite. Plant Physiology 157, 1394–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivitz AB, Reinders A, Ward JM. 2008. Arabidopsis sucrose transporter AtSUC1 is important for pollen germination and sucrose-induced anthocyanin accumulation. Plant Physiology 147, 92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P. 2006. Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiology 140, 637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperdouli I, Moustakas M. 2012. Interaction of proline, sugars, and anthocyanins during photosynthetic acclimation of Arabidopsis thaliana to drought stress. Journal of Plant Physiology 169, 577–585. [DOI] [PubMed] [Google Scholar]
- Stintzi A. 2000. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proceedings of the National Academy of Sciences, USA 97, 10625–10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden C, Donaghy PG, Halford NG, Hardie DG. 1999. Two SNF1-related protein kinases from spinach leaf phosphorylate and inactivate 3-hydroxy-3-methylglutaryl-coenzyme A reductase, nitrate reductase, and sucrose phosphate synthase in vitro. Plant Physiology 120, 257–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Huang A, Sang Y, Fu Y, Yang Z. 2013. Carbon–nitrogen interaction modulates plant growth and expression of metabolic genes in rice. Journal of Plant Growth Regulation 32, 575–584. [Google Scholar]
- Takos AM, Jaffé FW, Jacob SR, Bogs J, Robinson SP, Walker AR. 2006. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiology 142, 1216–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S. 2005. Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiology 139, 1840–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. 2007. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665. [DOI] [PubMed] [Google Scholar]
- Tsai AY, Gazzarrini S. 2012. AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. The Plant Journal 69, 809–821. [DOI] [PubMed] [Google Scholar]
- Tsai PJ, Delva L, Yu TY, Huang YT, Dufosse L. 2005. Effect of sucrose on the anthocyanin and antioxidant capacity of mulberry extract during high temperature heating. Food Research International 38, 1059–1065. [Google Scholar]
- Vitrac X, Larronde F, Krisa S, Decendit A, Deffieux G, Mérillon JM. 2000. Sugar sensing and Ca2+-calmodulin requirement in Vitis vinifera cells producing anthocyanins. Phytochemistry 53, 659–665. [DOI] [PubMed] [Google Scholar]
- Wasternack C, Hause B. 2013. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Annals of Botany 111, 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D. 2000. Regulation of flower pigmentation and growth: multiple signaling pathways control anthocyanin synthesis in expanding petals. Physiologia Plantarum 110, 152–157. [Google Scholar]
- Wu N, Zheng B, Shaywitz A, et al. 2013. AMPK-dependent degradation of TXNIP upon energy stress leads to enhanced glucose uptake via GLUT1. Molecular Cell 49, 1167–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Dai L, Liu F, Wang Z, Peng W, Xie D. 2004. COS1: an Arabidopsis coronatine insensitive1 suppressor essential for regulation of jasmonate-mediated plant defense and senescence. The Plant Cell 16, 1132–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie XB, Li S, Zhang RF, Zhao J, Chen YC, Zhao Q, Yao YX, You CX, Zhang XS, Hao YJ. 2012. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant, Cell and Environment 35, 1884–1897. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Ma D, Constabel CP. 2015. The MYB182 protein down-regulates proanthocyanidin and anthocyanin biosynthesis in poplar by repressing both structural and regulatory flavonoid genes. Plant Physiology 167, 693–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Andralojc PJ, Hey SJ, Primavesi LF, Specht M, Koehler J, Parry MAJ, Halford NG. 2008. Arabidopsis sucrose non-fermenting-1-related protein kinase-1 and calcium-dependent protein kinase phosphorylate conserved target sites in ABA response element binding proteins. Annals of Applied Biology 153, 401–409. [Google Scholar]
- Zhang Y, Butelli E, De Stefano R, et al. 2013. Anthocyanins double the shelf life of tomatoes by delaying overripening and reducing susceptibility to gray mold. Current Biology 23, 1094–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Shewry PR, Jones H, Barcelo P, Lazzeri PA, Halford NG. 2001. Expression of antisense SnRK1 protein kinase sequence causes abnormal pollen development and male sterility in transgenic barley. The Plant Journal 28, 431–441. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.