Clustering of PAMP-induced transcripts regulated by Arabidopsis MAP kinase phosphatase 1 delineates signaling pathways that are MKP1 dependent and MPK6 dependent and independent, providing a validated set of molecular pathway markers.
Keywords: Arabidopsis, mitogen-activated protein kinase, MKP1, MPK6, pathogen-associated molecular pattern (PAMP), phosphatase, RNAseq, transcriptome
Abstract
Plant immunity is initiated by extracellular detection of pathogen-associated molecular patterns (PAMPs) through surface-localized pattern recognition receptors (PRRs). PRR activation induces many responses including the activation of mitogen-activated protein kinases (MAPKs) that ultimately limit bacterial growth. Previous work identified Arabidopsis MAP kinase phosphatase 1 (MKP1) as a negative regulator of signaling pathways required for some, but not all, of PAMP-initiated responses. Specifically, loss of MAPK MPK6 in an mkp1 background suppressed a subset of the mkp1-dependent biological phenotypes, indicating the requirement for MPK6 in MKP1-dependent signaling. To further genetically separate the outputs of PAMP-responsive signaling pathways, we performed a transcriptome analysis in Arabidopsis wild type, mkp1 and mkp1 mpk6 seedlings treated with the bacterially derived PAMP elf26 for 0, 30, and 90 min. Using differential genetic and temporal clustering analyses between and within genotypes, we identified and separated 6963 elf26-responsive transcripts based on both genetic requirements of MKP1 (with or without a requirement for MPK6) and temporal transcriptional accumulation patterns, and some of these novel response markers were validated by qRT-PCR over a more extended time course. Taken together, our transcriptome analysis provides novel information for delineating PAMP signaling pathways.
Introduction
To protect themselves against potential bacterial pathogens, plants need to activate immune responses rapidly and effectively when invading microbes are detected. Bacteria initially are perceived through the recognition of pathogen-associated molecular patterns (PAMPs) such as the flagellin protein required for motility (Felix et al., 1999), lipopolysaccharides from the bacterial cell wall (Desaki et al., 2006), and the bacterial translation elongation factor EF-Tu (Zipfel et al., 2006). This non-self recognition is mediated by specific plasma membrane-localized pattern recognition receptors (PRRs) that initiate a diverse array of signaling events including reactive oxygen species (ROS) production, cellular Ca2+ influx, reversible protein phosphorylation, and transcriptional reprogramming (Boller and Felix, 2009). Some or all of these immune responses ultimately restrict the growth of the bacterial invader, contributing to the basal defense program called pattern-triggered immunity (PTI).
Transcriptional profiling of plant tissue treated with different single elicitors has identified sets of transcripts that change in response to diverse elicitors in Arabidopsis cell culture or seedlings (Navarro et al., 2004; Zipfel et al., 2004, 2006; Ramonell et al., 2005; Bae et al., 2006; Qutob et al., 2006; Denoux et al., 2008). These studies demonstrated considerable overlap in response to multiple elicitors, indicating that different elicitors activate conserved basal defense responses (Jones and Dangl, 2006). In addition, they also found that the most abundant categories of gene products encoded by PAMP-responsive genes included signaling components such as transcription factors, protein kinases and/or phosphatases, and proteins that regulate protein turnover (Navarro et al., 2004; Zipfel et al., 2004, 2006; Denoux et al., 2008). Recently, a high-resolution time course study investigated genome-wide expression changes following challenge with the virulent pathogen Pseudomonas syringae pv tomato DC3000 (DC3000) and the non-pathogenic mutant strain DC3000 hrpA−. This work captured gene expression dynamics of PTI induced by DC3000 hrpA− and how these expression profiles were modulated by DC3000 (Lewis et al., 2015). However, time course studies that only involve treatment of wild type plants do not provide any immediate information on the specific pathways connecting perception from the plasma membrane localized receptor(s) to specific downstream transcriptional changes.
Mitogen-activated protein kinases (MAPKs) and calcium-dependent protein kinases (CDPKs) are signaling components that link the perception from PRR complexes to downstream defense responses (Tena et al., 2011; Li et al., 2016). Studies of innate immunity in Arabidopsis mainly focus on three MAPKs: MPK3, MPK6, and MPK4. MPK3 and MPK6 appear to be partially redundant and are activated by the upstream MAPK kinases MKK4/MKK5, whereas MPK4 is regulated by MKK1/MKK2 (Nühse et al., 2000; Asai et al., 2002; Suarez-Rodriguez et al., 2007; Qiu et al., 2008). A comparative transcriptome analysis of mpk3, mpk4, and mpk6 mutants revealed that 36% of the flg22-up-regulated genes and 68% of the flg22-down-regulated genes were affected in at least one of the mpk mutants, pointing out the essential role of these MAPKs in regulating the transcriptional reprogramming during PAMP signaling (Frei dit Frey et al., 2014). A separate study using constitutively active forms of two CDPKs, CPK5 and CPK11, transiently expressed in the protoplasts identified target genes downstream of these kinases (Boudsocq et al., 2010). Adding a layer of complexity, MAPKs and CDPKs may act either alone or synergistically in controlling PAMP-induced transcript accumulation. By transiently expressing constitutively active CPK5 and/or MKK4 in protoplasts, several marker genes for CDPK-specific, MAPK-specific and CDPK–MAPK synergistic pathways were identified (Boudsocq et al., 2010), and these few markers have been widely used to begin delineating the PAMP signaling pathways by placing newly discovered putative signaling components into different pathways (Desclos-Theveniau et al., 2012; Lozano-Durán et al., 2013; Prince et al., 2014; Smith et al., 2014; Domínguez-Ferreras et al., 2015). The utility of just this small number of pathway markers clearly demonstrates how identifying additional transcripts regulated by separate pathways can assist the field in better molecular characterization of genetic mutants with altered defense responses. Therefore, the goal of this study was to make use of a novel set of genetic mutants to define a new set of molecular pathways.
Protein phosphatases are important components required for the proper regulation of PAMP signaling. Phosphatases often act as negative regulators that function by dephosphorylating and inactivating kinases, including MAPKs, to prevent overactive stimulation of defense responses. In a screen for phosphatases involved in the proper regulation of PAMP responses, we uncovered MAPK phosphatase 1 (MKP1) as an important negative regulator as evidenced by heightened early responses to PAMP elicitation in the mkp1 null mutant such as enhanced MAPK activation, ROS production, and transcript accumulation of some but not all PAMP-regulated genes (Anderson et al., 2011). In addition, later PAMP responses such as seedling growth inhibition and resistance against normally virulent DC3000 were also enhanced in the mkp1 mutant. Interestingly, these responses were suppressed in mkp1 mpk6 double mutants but not in mkp1 mpk3 mutants, indicating that enhanced biological responses (i.e. resistance and growth inhibition) in mkp1 specifically require MPK6 but not MPK3 (Anderson et al., 2011). The results were consistent with the fact that although MKP1 could interact at least somewhat with MPK3 and MPK4, it showed a much stronger preference for MPK6 (Ulm et al., 2002). More recently, we found that the enhanced resistance against DC3000 in mkp1 can be explained by the decreased abundance of specific extracellular metabolites from the plant that DC3000 uses as signals to activate its virulence program (Anderson et al., 2014). Interestingly, during this study we found that while all the bioactive metabolites were restored to wild type (WT) levels in mkp1 mpk6 plants, some of the non-bioactive metabolite levels altered in mkp1 were not affected by the loss of MPK6 (Anderson et al., 2014). These results provided the first evidence that not all mkp1-dependent changes involve MPK6, indicating the existence of MKP1-dependent, MPK6-independent pathways. Thus, it seemed likely that these mutants that affect early signaling responses would be useful tools to genetically separate different signaling pathways leading to specific changes in transcript accumulation.
In this study, we compared transcript accumulation profiles of Arabidopsis seedlings of wild type (Ws), mkp1 (Ws), and mkp1 mpk6 (Ws) treated with elf26 (a 26-amino acid peptide corresponding to a conserved domain of bacterial elongation factor EF-Tu) at 0, 30, and 90 min post-treatment. By differential expression analysis, we separated the PAMP-responsive transcripts into defined genetic pathways. In addition, by clustering transcripts within discrete temporal accumulation profiles, we also predicted genes that are likely targets of different transcriptional regulation within these genetic pathways. Gene Ontology (GO) analysis indicated distinct biological processes that are specifically associated with MKP1-dependent genes. Finally, the expression patterns of selected marker genes were confirmed by quantitative real-time PCR over a more extended time course following elicitation, providing a validated set of novel molecular pathway markers for plant defense studies.
Materials and methods
Plant material
Arabidopsis ecotype Wassilewskija (Ws) was used in this study. The mutants mkp1-1 (Ws) and mkp1-1 mpk6-1 (Ws) have been described previously (Ulm et al., 2001; Liu and Zhang, 2004; Bartels et al., 2009; Anderson et al., 2011, 2014) and also have been confirmed by genotyping (Supplementary Fig. S1 at JXB online). Seeds were sterilized with 1% sodium hypochlorite and 0.01% Tween-20 for 20 min, rinsed with water, and plated aseptically on 0.5% agar containing 2.1 g l−1 Murashige and Skoog (MS) salts (PhytoTechnology Laboratories, http://www.phytotechlab.com/), pH 5.7, 1% sucrose and 6.4 g ml−1 MS salts vitamin powder (PhytoTechnology Laboratories). After stratification for 2 d at 4 °C, seeds were grown at 22 °C with a 16/8 h light–dark cycle and 70% humidity. Seedlings were maintained in the same growth chamber under the same conditions during elf26 treatments.
Transcriptome studies
Prior to elicitation, 12-day-old seedlings of Ws, mkp1, mkp1 mpk6 were transferred from MS agar plates to incubate in sterile water overnight for 16–20 h. For elicitor treatments, water from the overnight incubation was removed and replaced with sterile water containing 1 µM elf26. Seedlings were harvested at 0, 30, and 90 min post-treatment. Three independent experiments that included each genotype and time point were performed. Total RNA was isolated with the RNeasy Plant Mini Kit (Qiagen, http://www.qiagen.com/). RNAseq libraries were prepared using the TruSeq RNA sample preparation kit (Illumina) following the manufacturer’s protocol (Illumina, http://www.illumina.com). All samples were sequenced by an Illumina HiSeq 2500 at DNA core in Bond Life Sciences Center, University of Missouri–Columbia. Raw reads were filtered using the NGS QC tool kit (Patel and Jain, 2012). The first 13 bp from the 3′ end of the reads, which showed unstable base composition as determined by the percentage of four different nucleotides (A, T, C, and G) and low quality bases [PHRED-like score (Q score)<20], were trimmed. The reads containing primer/adaptor sequences and low quality reads (the percentage of low quality bases ≥30%) were removed. All high-quality reads from all biological experiments were mapped to Arabidopsis TAIR 10 genome with TopHat (http://tophat.cbcb.umd.edu/), and only the uniquely mapped reads were chosen for differential gene analysis. Transcript abundance and differential gene expression were calculated with the Cufflinks package (Trapnell et al., 2012). Specifically, the mapped reads were assembled into transcripts by Cufflinks (http://cufflinks.cbcb.umd.edu/). The resulting assemblies were merged using Cuffmerge. Aligned reads and merged assemblies were used to calculate expression levels and to determine statistical significance using Cuffdiff. Transcript abundance of each gene was estimated by fragments per kilo base of transcript per million fragments mapped (FPKM). The raw Illumina reads generated from RNAseq experiments were deposited at NCBI Sequence Read Archive (SRP101277).
Differential gene expression analysis
Transcripts with significant changes (absolute value of log2-fold change ≥1, q≤0.01) in Ws or mkp1 post-30 min and/or 90 min elf26 elicitation compared with 0 min were considered to be elf26-responsive transcripts. Transcripts with at least 1.5-fold difference (q≤0.01) between Ws and mkp1 in the same treatment condition were considered to be MKP1 dependent. The MKP1-independent transcripts were defined as the set of total elf26-responsive transcripts with MKP1-dependent transcripts subtracted. Within the MKP1-dependent category, transcripts with a significant (q≤0.01) difference between mkp1 and mkp1 mpk6 but no significant difference (q>0.01) between Ws and mkp1 mpk6 post-elf26 elicitation were considered to be both MKP1 and MPK6 dependent. Transcripts with no significant difference (q>0.01) between mkp1 and mkp1 mpk6 but significant difference (q≤0.01) between Ws and mkp1 mpk6 were MKP1 dependent/MPK6 independent. MKP1-dependent transcripts that do not meet either criterion were considered to be partially MPK6 dependent.
Clustering analysis
The dataset for the co-expression analysis was built from the results of the differential transcript abundance analyses. Transcripts that differentially accumulated at either 30 min or 90 min post-elf26 elicitation were included for the analysis; and the clustering was performed with STEM (Short Time-series Expression Miner) using the temporal profiles in Ws (Ernst and Bar-Joseph, 2006). Default values were selected for two variable parameters: 50 as maximum number of model profiles and 2 as maximum unit change in model profiles between time points and combined clusters with similar general trends. Cluster profiles were graphed with the R package and represented as line graphs in three groups based on the different scale ranges (low, medium, high).
Validation using quantitative real-time PCR
Elicitation treatment was performed using the same conditions as described for the transcriptome analysis. For each time point, seedlings were blotted dry and frozen in liquid nitrogen. Total RNA was isolated using TRI reagent (Sigma-Aldrich) and treated with DNase I (Thermo Fisher Scientific) before 1 µg of RNA was reverse-transcribed in 25 µl reactions containing 5 µM DTT, 0.5 µl RnaseOUT (Thermo Fisher Scientific), 2 µM oligo(dT), 1 mM each of dNTPs and 0.5 µl M-MLV reverse transcriptase (Promega, http://www.promega.com/) for 1 h at 42 °C, followed by 5 min at 85 °C. Reverse transcription reactions were diluted to 100 µl using diethylpyrocabonate-treated water. Real-time PCR reactions were performed using primers listed in Supplementary Table S1. Briefly, 10 µl real-time PCR reactions containing 5 µl SYBR Green PCR master mix (Thermo Fisher Scientific), 1 µl cDNA and 0.2 µM of each primer were performed using an ABI7500 real-time thermal cycler (Thermo Fisher Scientific). Three independent experiments were performed. Expression levels were calculated using the equation: expression level=(PCR efficiency)−ΔCt, where ΔCt=Ct(sample)−Ct(control), and the cycle threshold Ct and PCR efficiency for each reaction were obtained using the software program LINREGPCR (Ramakers et al., 2003). At2g28390 (SAND family protein) was used as the reference gene for normalizing Ct values (Czechowski et al., 2005). Statistical analyses were performed using Student’s two-sample unpaired t tests.
Gene Ontology analysis
The web-based agriGO software (http://bioinfo.cau.edu.cn/agriGO/index.php) was used to assign GO functional categories. Singular enrichment analysis was used to compute over-represented categories in the sets of MKP1-dependent genes by comparing them with GO terms in the set of elf26-up-regulated genes or elf26-down-regulated genes using Fisher’s exact test (Du et al., 2010). Over-represented GO terms in the MKP1-dependent genes, elf26-responsive genes as compared with all expressed genes in the whole Arabidopsis genome were also calculated. The cut-off (q≤0.05) was used for significantly over-represented GO terms.
Results and discussion
Identification of transcriptomes of Ws, mkp1, and mkp1 mpk6 in response to elf26
MKP1 is a negative regulator of multiple PAMP-induced defense responses in Arabidopsis (Anderson et al., 2011). In a limited characterization of a few early defense related genes, only six out of eight PAMP-induced transcript profiles were altered in mkp1, indicating that MKP1 regulates some, but not all, transcriptional pathways (Anderson et al., 2011). Loss of function mutations in MPK6 but not in MPK3 suppress a number of the molecular and biological phenotypes of the mkp1 mutant, including the enhanced resistance against DC3000 and PAMP-induced growth inhibition, indicating that MPK6 but not MPK3 acts within MKP1-dependent signaling pathways leading to enhanced resistance (Anderson et al., 2011). A subsequent study showed that all biologically active metabolites involved in the enhanced resistance in the mkp1 mutant are restored to wild type levels in the mkp1 mpk6 double mutant (Anderson et al., 2014). However, some of the non-bioactive extracellular metabolites with altered abundance in mkp1 were not restored in the mkp1 mpk6 double mutant (Anderson et al., 2014). These observations indicated two important findings: (i) not all MKP1-regulated phenotypes are MPK6 dependent; and (ii) The use of the mkp1 mpk6 double mutant can be used to eliminate molecular responses that are not directly correlated with resistance in mkp1. Therefore, it was likely that not all mkp1-dependent changes in PAMP-regulated transcript accumulation patterns would require MPK6; and the identification of mkp1-dependent changes that also required MPK6 would define transcript changes most associated with the enhanced resistance.
To genetically separate PAMP-responsive transcripts into MKP1-dependent and MKP1-independent categories, as well as to further subdivide the MKP1-dependent transcripts into MPK6-dependent and MPK6-independent pathways, we used RNAseq to perform a whole transcriptome analysis of Ws, mkp1, and mkp1 mpk6 seedlings treated for 0, 30, or 90 min with 1 μM elf26. For transcriptome sequencing, a total of 737 407 322 reads from all biological experiments passed the quality filter and were mapped to the Arabidopsis TAIR10 genome. Approximately 89.29% of high-quality reads mapped uniquely to a single annotated TAIR10 gene (Supplementary Table S2). These high-quality data were then analysed by different comparisons as described below.
Identification of MKP1-dependent transcripts in elf26-triggered transcriptional reprogramming
First, transcriptomes of Ws and the mkp1 mutant in response to 30 and 90 min treatment with elf26 were compared; and transcripts that showed significant (q≤0.01) changes in response to elf26 in Ws and mkp1 mutant were considered to be elf26-responsive. At 30 min, 2895 genes had altered transcript levels (≥2-fold) after elf26 treatment. By 90 min, the response to elf26 increased to include 5989 transcripts with altered levels. Among all elf26-responsive transcripts at either time point, 5109 were induced by elf26, whereas 3775 were repressed (Supplementary Table S3; for a complete gene list including normalized expression values, log2-fold change (Fc), q-values and annotations, see Supplementary Table S4).
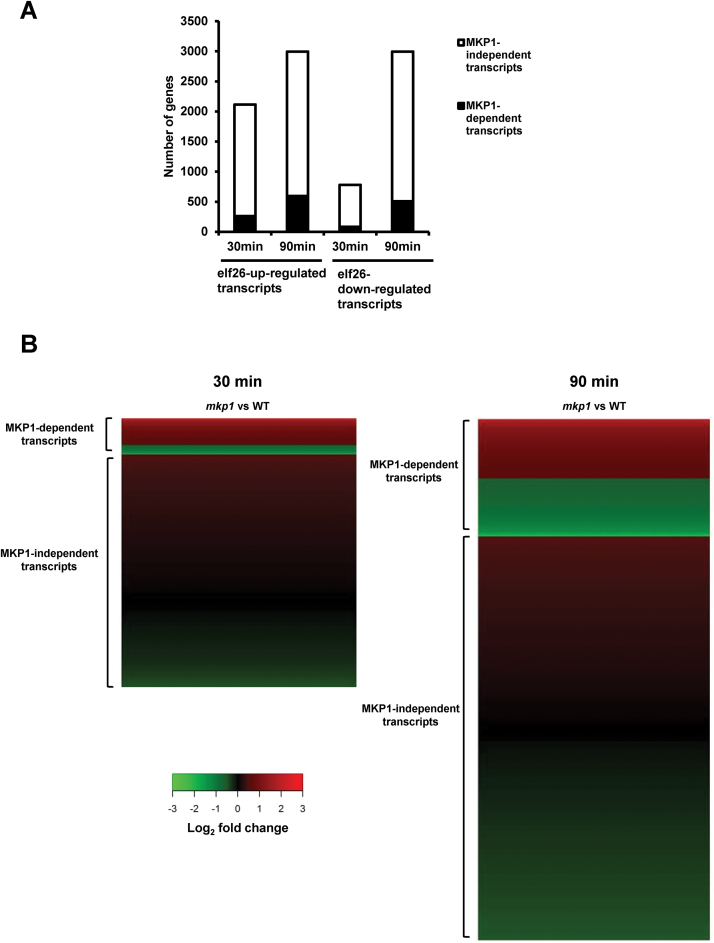
Transcripts with at least 1.5-fold difference between the mkp1 mutant and Ws were considered to be differentially accumulating in mkp1 and, therefore, MKP1 dependent. At 30 min post-elicitation, we observed that 12% (262/2114) of the elf26-induced transcripts and 11% (85/785) of elf26-repressed transcripts were affected in the mkp1 mutant (Fig. 1; for a complete gene list see Supplementary Table S5). At 90 min, the proportion of MKP1-dependent transcripts increased to 20% (594/2995) of elf26 up-regulated and 17% (508/2994) of elf26-down-regulated transcripts. Altogether, these results were consistent with the previous limited study (Anderson et al., 2011) indicating that the abundance of only a subset of PAMP-responsive transcripts is regulated by MKP1. We should note that the comparison of untreated samples indicated that some of the MKP1-dependent differences exist between transcripts at 0 min, and these are summarized in Supplementary Table S5. However, these small changes did not significantly contribute to the quantitative differences between genotypes following elf26 treatment; so all elf26-induced differences result from the stimulus rather than any pre-existing differences within the genotypes.
Fig. 1.
Only a subset of PAMP-regulated transcripts is MKP1 dependent at 30 and 90 min post-elicitation. (A) The total PAMP-regulated (elf26) transcripts are divided into transcripts differentially accumulating in mkp1 mutant (black bars) and transcripts not affected by MKP1 mutation (white bars). (B) These are also are shown represented by a heat map.
Gene Ontology characterization of MKP1- dependent genes
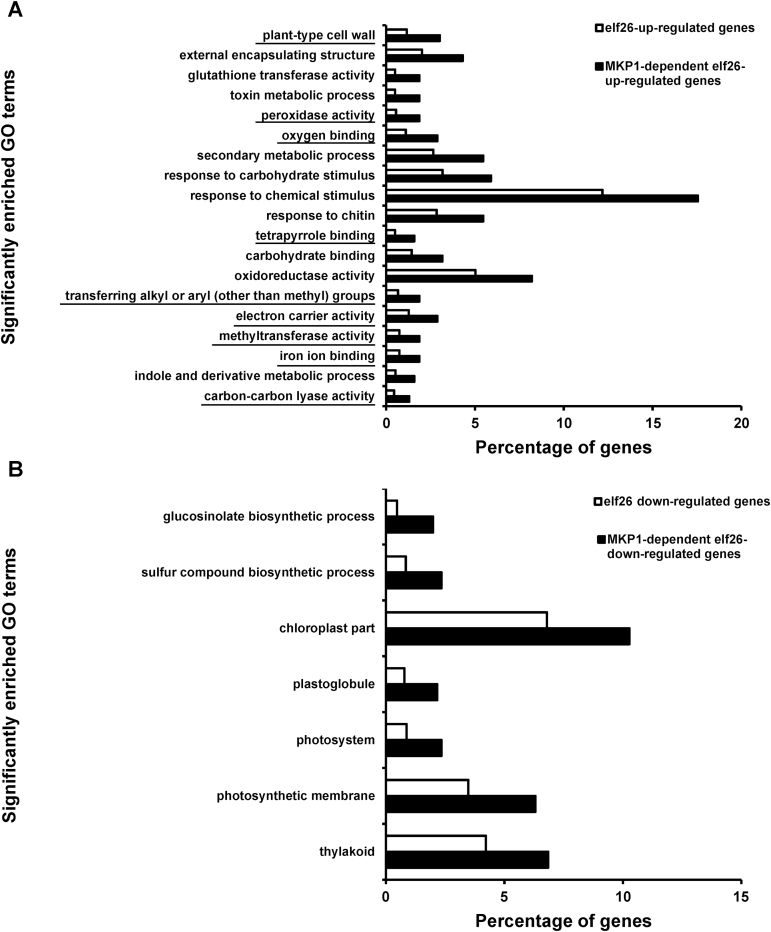
To investigate the biological processes that are over-represented in MKP1-dependent genes, Gene Ontology (GO) analysis with agriGO was performed (Du et al., 2010). First, over-represented GO categories within MKP1-dependent genes in comparison with all elf26-regulated genes were found by singular enrichment analysis. We observed that 19 GO categories were significantly (q≤0.05) over-represented in the MKP1-dependent elf26 up-regulated genes, whereas seven GO terms were enriched in the MKP1-dependent elf26 down-regulated genes (Fig. 2, Supplementary Table S6). In addition, GO terms significantly (q≤0.05) enriched in MKP1-dependent genes and in all elf26-regulated genes were also compared with the whole Arabidopsis genome (Supplementary Table S6). Most of the GO enriched terms in the mkp1 mutant were similarly enriched in the set of elf26-responsive genes. However, by comparing the GO terms enriched in elf26-regulated genes with those in MKP1-dependent genes, we observed that nine GO terms were uniquely enriched in mkp1 as compared with the whole genome (underlined in Fig. 2). Within the categories uniquely enriched in mkp1, we found GO terms related to multiple defense responses such as cell wall modification, redox reactions, and iron binding. Previous studies found that rapidly PAMP-induced genes were functionally enriched for enzymes involved in antimicrobial compound biosynthesis and for proteins involved in signal perception and transduction (Navarro et al., 2004; Zipfel et al., 2004, 2006; Denoux et al., 2008; Frei dit Frey et al., 2014). By contrast, genes associated with photosynthesis-related process, fatty acid metabolism and glucosinolate biosynthesis were significantly repressed by PAMPs, suggesting that plants may redirect secondary metabolism and reduce the production of photosynthates to restrict the resources required for pathogen growth (Lewis et al., 2015). By comparison, our GO analysis of MKP1-dependent genes indicates that some biological processes enriched in MKP1-dependent genes are unique (i.e. not enriched in wild type elf26-responsive genes), and that some MKP1-dependent overlap with ones normally occur during PTI signaling, with the representation of these processes being further amplified. These distinct patterns of transcript accumulation suggest that loss of MKP1 may lead to aberrant signaling outputs that are not typically observed in wild type seedlings, as well as the hyper-response of a small subset of genes that are normally PAMP responsive.
Fig. 2.
MKP1-dependent genes show significant enrichment in discrete GO categories. Significantly enriched GO terms (q≤0.05) of MKP1-regulated genes (black bars) compared with all elf26-regulated genes (white bars) for both up-regulated genes (A) and down-regulated genes (B). The underlined GO terms are uniquely enriched in the MKP1-dependent genes (not in elf26-regulated genes) compared with the whole Arabidopsis genome.
Subdivision of MKP1-dependent transcripts into MPK6-dependent and MPK6-independent categories
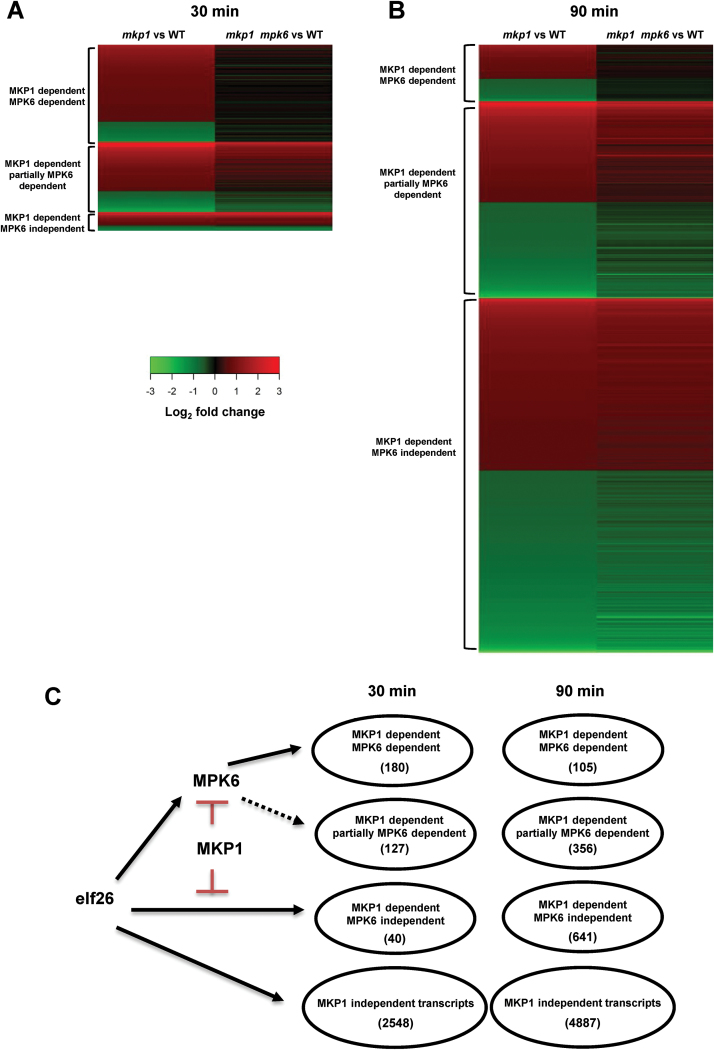
We next investigated if MKP1-dependent transcripts could be further subdivided into MPK6-dependent or MPK6-independent pathways using comparisons with responses in the mkp1 mpk6 double mutant. MKP1-dependent transcripts were considered MPK6 dependent if the changes in the mkp1 mutant were completely reversed to being indistinguishable from WT levels in the mkp1 mpk6 double mutant. Conversely, transcripts were considered MPK6 independent if the transcript levels in the mkp1 mpk6 double mutant were not significantly (q>0.01) different from those in the mkp1 mutant but were significantly (q≤0.01) different from those in Ws. Heat map analyses show the log2-fold change ratio in gene expression between different genotypes (Fig. 3A, B). Transcripts not discreetly meeting either of these criteria were considered to be partially MPK6 dependent and not included in the heat map (Supplementary Table S7 compiles all data including the transcripts in different genetic categories at different time points post-elicitation).
Fig. 3.
MKP1 plays a complex role in the regulation of PAMP-responsive transcripts. (A, B) Heat maps show the log2-fold change in transcript accumulation level between indicated comparison of genotypes. (C) Numbers of transcripts in different genetic categories.
Using these criteria, we found that at 30 min post-elf26 treatment, 180 (82%) of the MKP1-dependent transcripts were also dependent on MPK6, whereas only 40 (18%) of the MKP1-dependent transcripts were independent of MPK6. However, at 90 min the pattern flipped, with 86% (641) of the MKP1-dependent transcripts being MPK6 independent with only 14% (105) being MPK6 dependent (Fig. 3C). These observations indicate that MPK6 plays a more significant role in MKP1-dependent transcript accumulation during early PAMP responses.
Previous quantitative RT-PCR results showed that six out of eight PAMP-responsive transcripts accumulated to higher levels but none to lower levels (the other two were unchanged) in the mkp1 mutant compared with WT, indicating that MKP1 acts primarily as a negative regulator in controlling elf26-responsive transcript accumulation (Anderson et al., 2011). The results of the current comprehensive study are consistent with these initial observations, with 80% (143/180) of the transcripts in the MKP1-dependent/MPK6-dependent category showing hyper-induction in the mkp1 mutant compared with WT at 30 min elf26 treatment (Fig. 3A, Supplementary Table S7). However, this study also identified a subset of PAMP-responsive transcripts with lower accumulation in mkp1 (Fig. 3). Moreover, the proportion of transcripts with either increased or decreased level in the mkp1 mutant was close to equivalent at 90 min, indicating a more complex role of MKP1 in regulating PAMP-responsive transcription as responses progress (Fig. 3B, Supplementary Table S7).
Because there are only five MAPK phosphatases in Arabidopsis but 20 MAPKs (Colcombet and Hirt, 2008), it is likely that MKP1 regulates pathways involving other MAPKs. MKP1 was previously shown to interact with both MPK3 and MPK4 (Ulm et al., 2002). In addition, MPK3 was also hyperactivated in mkp1 mutants along with MPK6 during elf26 treatment (Anderson et al., 2011). Therefore, it is quite likely that at least a portion of the MKP1-dependent but MPK6-independent transcript changes are regulated by MPK3. However, we cannot rule out that additional MAPKs such as MPK1, MPK11, or MPK13 (Nitta et al., 2014) may also contribute to these MKP1-dependent changes in transcript accumulation.
Temporal clustering analysis to predict putative co-regulated genes
The differential analysis performed on the transcriptome data between genotypes separated transcripts based on statistically significant differential accumulation at specific time points but did not consider the accumulation pattern across different time points. Genes with similar accumulation profiles may share a regulatory protein (e.g. transcription factor) or mechanism, possibly placing the transcripts in the same signaling pathways. To identify transcripts that behave similarly during elicitation, we performed a co-expression analysis using the STEM (Short Time-series Expression Miner) tool because of its effectiveness in clustering short time-series data (Ernst and Bar-Joseph, 2006). We performed this analysis with PAMP-responsive transcripts and separated them based on their temporal profiles in the wild type, because only the magnitude of the PAMP-responsive transcripts and not the temporal accumulation pattern was altered in the mkp1 mutant (i.e. the patterns were the same as in wild type).
According to the clustering algorithm from STEM, 96% (6709/6964) of the PAMP-responsive transcripts could be assigned to a single model profile (the remaining could not be confidently assigned to a single category). This analysis yielded eight major group clusters (the three different graphs are different scales to allow visualization of all genes with similar patterns): (i) early induced and transient (cluster 1); (ii) late induced (cluster 2); (iii) early induced and sustained (cluster 3); (iv) early induced and amplified (cluster 4); (v) early repressed and transient (cluster 5); (vi) early repressed and sustained (cluster 6); (vii) late repressed (cluster 7); (viii) early repressed and amplified (cluster 8) (Fig. 4; for a complete list of genes associated with each cluster, see Supplementary Table S8). To investigate the possible correlation between accumulation kinetics and genetically distinct pathways, we determined the representation of different clusters within the genotype-specific response categories. In general, we did not observe a unique correlation between a type of transcript accumulation pattern with a specific genetic category, indicating that the regulation of MKP1 and or MPK6 is not restricted to transcripts within a certain type of accumulation pattern but widely covers transcripts displaying different types of temporal profiles. However, we observed that MKP1-dependent transcripts showed a higher representation in several clusters. For instance, in cluster 7 (late repressed) at 30 min post-elf26 treatment, the percentage of MKP1-dependent transcripts was 75 times that of MKP1-independent transcripts. At 90 min in clusters 5 (early repressed and transient) and 8 (early repressed and amplified), the proportion of MKP1-dependent transcripts was almost five times that of MKP1-independent transcripts. Because clusters 5, 7 and 8 were all associated with elf26-repressed transcripts, these results indicate an important role of MKP1 in regulating the abundance of elf26-repressed transcripts as well as elf-induced transcripts (Table 1).
Fig. 4.
PAMP-responsive genes can be clustered based on temporal expression patterns. PAMP-responsive genes were clustered by STEM (Short Time-series Expression Miner) according to their expression kinetics in wild type plants. Eight major clusters were obtained. The temporal expression profiles of genes in each cluster are represented by line graphs (the top, middle, and bottom graphs of each cluster represent transcripts separated by high, medium, and low expression levels, respectively). The y-axis of each graph is the normalized expression value of each gene in artificial units, and the x-axis is the corresponding time points after elf26 treatment. The numbers in parentheses are the number of genes within each cluster. (This figure is available in color at JXB online.)
Table 1.
Percentage of cluster in each genetic category
| Cluster | Description | 30 min | 90 min | ||||
|---|---|---|---|---|---|---|---|
| MKP1- independent transcripts | MKP1-dependent MPK6-dependent transcripts | MKP1-dependent MPK6-independent transcripts |
MKP1- independent transcripts | MKP1-dependent MPK6-dependent transcripts | MKP1-dependent MPK6-independent transcripts | ||
| 1 |
|
44.58% (1136/2548) |
46.67% (84/180) |
30.00% (12/40) |
9.94% (486/4887) |
12.38% (13/105) |
20.44% (131/641) |
| 2 |
|
6.75% (172/2548) |
10.00% (18/180) |
10.00% (4/40) |
22.88% (1118/4887) |
21.90% (23/105) |
14.51% (93/641) |
| 3 |
|
7.73% (197/2548) |
12.22% (22/180) |
17.50% (7/40) |
3.56% (174/4887) |
3.81% (4/105) |
5.77% (37/641) |
| 4 |

|
13.23% (337/2548) |
8.33% (15/180) |
25.00% (10/40) |
12.11% (592/4887) |
13.33% (14/105) |
12.48% (80/641) |
| 5 |
|
9.50% (242/2548) |
9.44% (17/180) |
2.50% (1/40) |
0.90% (44/4887) |
2.86% (3/105) |
1.87% (12/641) |
| 6 |
|
12.72% (324/2548) |
8.89% (16/180) |
5.00% (2/40) |
14.22% (695/4887) |
12.38% (13/105) |
11.23% (72/641) |
| 7 |
|
0.04% (1/2548) |
0.56% (1/180) |
2.50% (1/40) |
28.14% (1375/4887) |
20.95% (22/105) |
20.28% (130/641) |
| 8 |

|
2.63% (67/2548) |
2.22% (4/180) |
2.50% (1/40) |
4.01% (196/4887) |
11.43% (12/105) |
12.32% (79/641) |
| Total | 97% (2476/2548) |
98% (177/180) |
95% (38/40) |
95% (4680/4887) |
99% (104/105) |
99% (634/641) |
|
Validation of unique transcripts that are representative of different regulatory pathways during PTI signaling
By comparing the transcriptional changes in three different genotypes (Ws, mkp1, and mkp1 mpk6 mutants), we were able to genetically separate the elf26-responsive transcripts into MKP1-independent, MKP1-dependent/MPK6-dependent, and MKP1-dependent/MPK6-independent categories. Temporal profile clustering analysis further separated the transcripts into different clusters with similar transcriptional accumulation patterns. Based on both genetic separation and expression kinetics, we have generated a novel set of candidates that can serve as marker genes representative of different regulatory pathways during PTI signaling. For validation, we selected nine MKP1-dependent/MPK6-dependent (Fig. 5), six MKP1-dependent/MPK6-independent (Fig. 6), and four MKP1-independent (Fig. 7) candidates representative of different termporal accumulation patterns. qRT-PCR was used to measure the expression of 19 genes in response to elf26 in the same genotypes treated for RNAseq. We also extended the time course from 0.5 to 2 h following elf26 treatment to potentially clarify patterns of transcript accumulation that might have been missed in the RNAseq analysis at only 30 and 90 min post-elicitation.
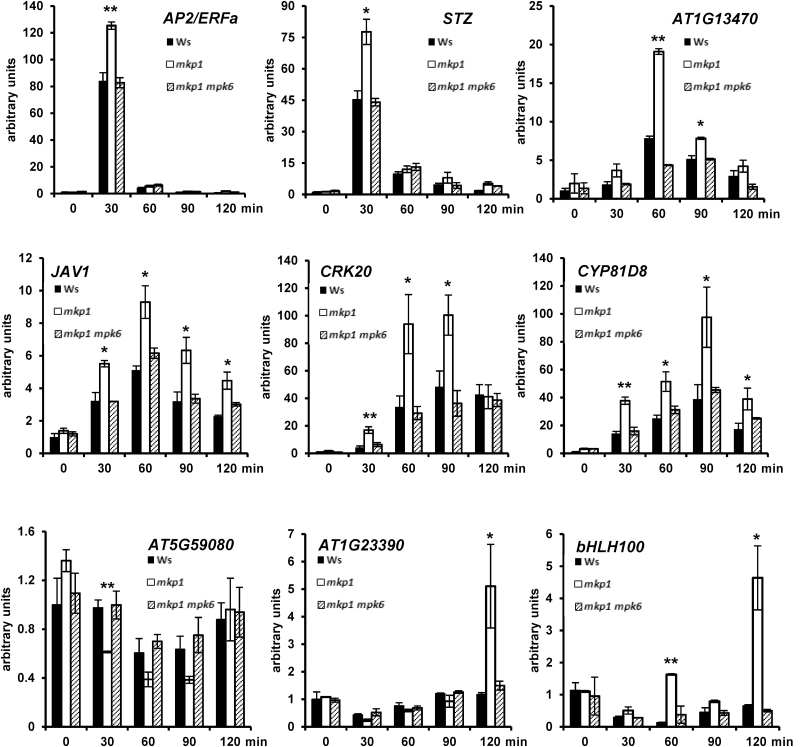
Fig. 5.
Independent qRT-PCR verification of transcripts that are both MKP1 dependent and MPK6 dependent. Transcript levels of candidate genes measured by quantitative RT-PCR from 12-day-old seedlings treated with or without 1 μM elf26 for indicated time points. Transcript levels were normalized to At2g28390 in each sample, then to transcript level at time 0 in Ws. Values are means±SE, n=3. Asterisks indicate significant differences between Ws and mkp1 (*P<0.05, **P<0.01). Ws and mkp1 mpk6 are not significantly different in any result. Data are pooled from three biological replicates.
Fig. 6.
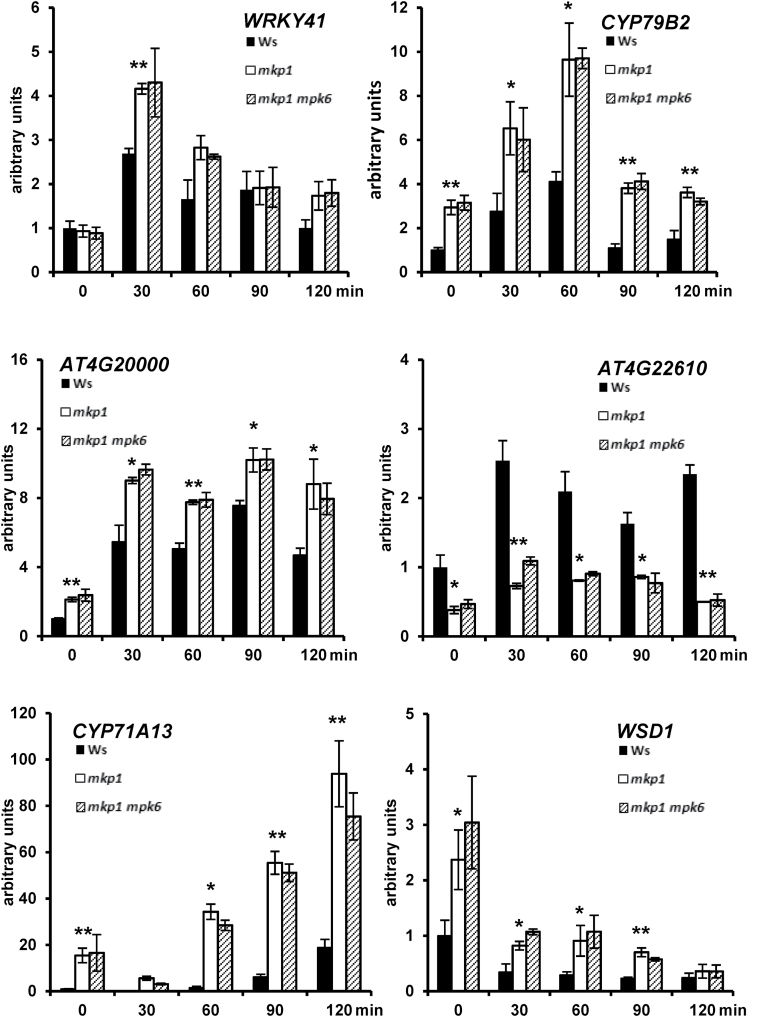
Independent qRT-PCR verification of transcripts that are MKP1 dependent but MPK6 independent. Transcript levels of candidate genes measured by quantitative RT-PCR from 12-day-old seedlings treated with or without 1 μM elf26 for indicated time points. Transcript levels were normalized to At2g28390 in each sample, then to transcript level at time 0 in Ws. Values are means±SE, n=3. Asterisks indicate significant differences between Ws and mkp1 (*P<0.05, **P<0.01). Results from mkp1 and mkp1 mpk6 are not significantly different in any experiment. Data are pooled from three biological replicates.
Fig. 7.
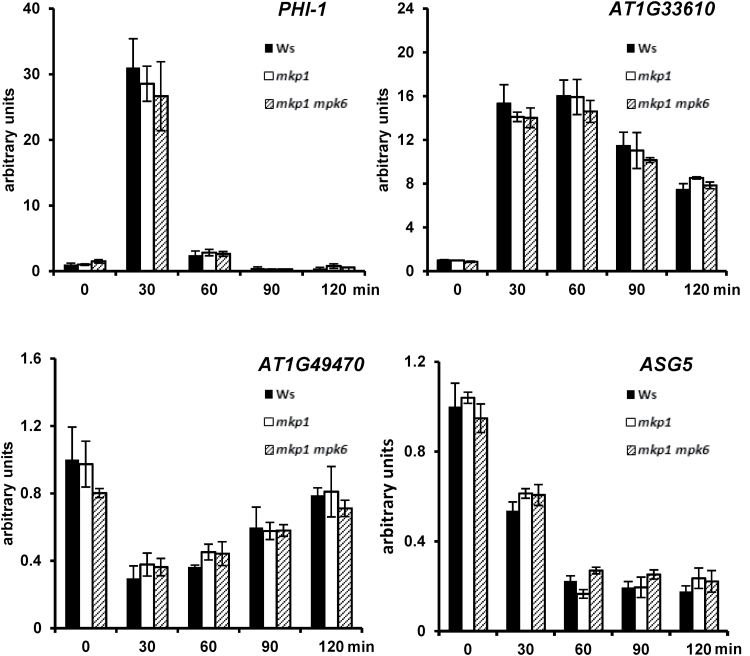
Independent qRT-PCR verification of transcripts that are MKP1 independent. Transcript levels of candidate genes measured by quantitative RT-PCR from 12-day-old seedlings treated with or without 1 μM elf26 for indicated time points. Transcript levels were normalized to At2g28390 in each sample, then to transcript level at time 0 in Ws. No significant difference was detected between any genotype. Values are means±SE, n=3. Data are pooled from three biological replicates.
Overall, the results from qRT-PCR analyses were consistent with that from RNAseq, thereby validating the large-scale analysis. Two conclusions arise from the results of qRT-PCR that are consistent with our conclusions from the RNAseq dataset. First, there are diverse accumulation patterns for elf26-responsive transcripts within all different genotype-specific response categories (Figs 5–7). Second, transcripts with similar accumulation patterns have different genetic requirements. For instance, both AP2/ERFa and PHI-1 (PHOSPHATE-INDUCED 1) were rapidly and strongly induced at 30 min but rapidly returned to basal levels by 90 min. However, AP2/ERFa belongs to the MKP1-dependent/MPK6-dependent category (Fig. 5), whereas PHI-1 was MKP1 independent (Fig. 7).
In addition to validating our RNAseq results, the extended qRT-PCR time course provided additional information that helped to refine gene expression patterns. For example, AT1G13470 and JAV1 (JASMONATE-ASSOCIATED VQ MOTIF GENE 1), marker genes for the MKP1-dependent/MPK6-dependent pathway, were initially identified from the RNAseq data as being rapidly induced at 30 min with the induction either sustained or amplified at 90 min. However, when assayed over an extended time course by qRT-PCR, we observed that both AT1G13470 and JAV1 showed a peak expression at 60 min that actually declined by 90 min (Fig. 5). Similarly, CYP79B2 in the MKP1-dependent/MPK6-independent pathway appeared to have a peak expression at 30 min from RNAseq analysis, but actually peak expression was observed at 60 min by qRT-PCR (Fig. 6). We also observed two marker genes (AT1G23390 and bHLH100) within the MKP1-dependent/MPK6-dependent category that strongly accumulated in mkp1 at 2 h post-elicitation, whereas no accumulation compared with the 0 min was observed in wild type or mkp1 mpk6 double mutant (Fig. 5). This last result indicates that over longer times, the mkp1 mutation results in the expression of genes that are not normally induced by elf26.
A next step will be to identify the transcription factors responsible for regulating these transcripts during PAMP responses. Initial candidates for some of the MKP1-dependent transcripts include WRKY18, WRKY33, and WRKY40, which were shown by ChIP analyses (Birkenbihl et al., 2017) to interact with promoters from 15–32% of the genes (data and individual genes are summarized in Supplementary Table S9). Future genetic experiments introducing mutations lacking these WRKY factors into the mkp1 mutant will be necessary to begin to dissect these early signaling pathways.
Separation of previously identified PTI signaling components downstream MPK6
MAPK cascades are important components for integrating responses to extracellular stimuli, and a number of signaling components or modules downstream of MAPK cascades have been described (Li et al., 2016). WRKY22 and WRKY29 are two transcription factors induced by flg22 treatment that are downstream of a flg22-activated MEKK1/MTK–MKK4/MKK5–MPK3/MPK6 cascade (Asai et al., 2002). WRKY33 is a direct substrate for MPK3 and MPK6, and plays an essential role in immunity against the necrotrophic fungus Botrytis (Mao et al., 2011). PAMP-activated MPK6 also phosphorylates BES1, a transcription factor involved in brassinosteroid (BR) signaling pathway, which positively regulates the resistance to bacteria and PTI gene expression (Kang et al., 2015).
Multiple signaling components involved in ethylene signaling are also downstream of MPK3 and MPK6. Transcripts encoding ACS2 and ACS6, two isoforms of the rate-limiting enzyme in ethylene biosynthesis, are induced upon pathogen attack in an MPK3- and MPK6-dependent manner through phosphorylation of WRKY33 (Li et al., 2012). The ethylene response factor ERF104, a regulator of basal immunity, is also a substrate of PAMP-activated MPK6 (Bethke et al., 2009). Similarly, ERF6 is phosphorylated by MPK3 and MPK6 upon Botrytis infection (Meng et al., 2013). Both the protein stability and transcript encoded by ERF6 are regulated by MPK3/MPK6 activation (Meng et al., 2013).
Although previous studies placed all these signaling components downstream of MPK6, our transcriptome analysis indicates that they can be separated by their requirement for MKP1, indicating that they are within different sub-pathways of MAP kinase signaling. For instance, the transcripts of WRKY22, WRKY29, and BES1 are all MKP1 independent at either 30 or 90 min post-elicitation (Table 2). However, WRKY33 switches from the MKP1-independent category at 30 min to MKP1-dependent category at 90 min (Table 2). For the components involving in ethylene signaling, ACS2 transcript accumulation remains independent of MKP1 at both 30 and 90 min, whereas ACS6 shifts from MKP1 independent at 30 min to MKP dependent at 90 min (Table 2). ERF104 and ERF6 are also distinct, with ERF104 being MKP1 dependent while ERF6 is MKP1 independent (Table 2). Therefore, our transcriptome analysis provides important additional information that further distinguishes components that had previously been clustered together as merely downstream of MPK6 into those that are or are not MKP1 dependent.
Table 2.
Comparison of previously identified MPK6 substrates and/or downstream signaling components based on requirement of MKP1-regulation in response to elf26
| Gene | Gene identifier | 30 min | 90 min |
|---|---|---|---|
| WRKY22 | AT4G01250 | MKP1 independent | MKP1 independent |
| WRKY29 | AT4G23550 | MKP1 independent | MKP1 independent |
| WRKY33 | AT2G38470 | MKP1 independent | MKP1 dependent MPK6 independent |
| BES1 | AT4G18890 | MKP1 independent | MKP1 independent |
| ACS2 | AT1G01480 | MKP1 independent | MKP1 independent |
| ACS6 | AT4G11280 | MKP1 independent | MKP1 dependent partially MPK6 dependent |
| ERF104 | AT5G61600 | MKP1 dependent MPK6 dependent |
Not PAMP responsive |
| ERF6 | AT4G17490 | MKP1 independent | Not PAMP responsive |
Identification of new potential signaling components downstream of MPK6
Our transcriptome analysis has identified a novel set of transcripts that require the regulation of MKP1, with or without the contribution of MPK6. This set of transcripts can contribute to understanding PAMP-induced signaling by delineating the positions of putative downstream components of MPK6 in the context of MKP1-dependent signaling. That is, if MPK6 is required for the correct transcript accumulation pattern, MPK6 substrates or downstream signaling components must also be required to connect MPK6 to the transcript changes. The use of molecular phenotypes (e.g. transcript accumulation) is likely to be more efficient than using biological phenotypes such as resistance or growth inhibition. In this regard, if the output(s) of MPK6 kinase activity is integrated through multiple signaling pathways, analysis of knockout mutants for individual substrates may not result in easily detectable biological phenotypes as these outputs tend to be more variable. However, screens based on molecular phenotypes are likely to be more sensitive to quantitative contributions of individual substrates or downstream signaling components. Therefore, the MPK6-dependent gene markers identified from our studies are likely to be more refined readouts for separating substrates/signaling components downstream of MPK6 within the MKP1-dependent pathway. By the same logic, the improved list of potential pathway markers for MKP1-independent and MKP1-dependent/MPK6-independent pathways will provide more refined tools for phenotyping mutants in diverse PAMP-induced signaling pathways.
Supplementary data
Supplementary data are available at JXB online
Fig. S1. Genotyping of mkp1 and mkp1 mpk6 mutants.
Table S1. qRT-PCR primers used in this study.
Table S2. RNAseq reads and mapping statistics.
Table S3. Numbers of PAMP-responsive transcripts at 30 and 90 min post-elf26 elicitation.
Table S4. Comprehensive list of elf26-responsive genes in Ws and mkp1 at 30 and 90 min post-elicitation including normalized expression values, log2-fold change (Fc), q values and annotations.
Table S5. Comprehensive list of MKP1-dependent and MKP1-independent genes at 30 and 90 min post-elicitation including normalized expression values, log2-fold change (Fc), q values and annotations.
Table S6. Analysis of Gene Ontology (GO) enrichment in MKP1-dependent genes and elf26-regulated genes.
Table S7. Comprehensive list of MKP1-dependent, MPK6-dependent, and MKP1-dependent MPK6-independent genes at 30 and 90 min post-elicitation including normalized expression values, log2-fold change (Fc), q values and annotations.
Table S8. Comprehensive list of genes in each clusters including the normalized expression values at different time points in different genotypes and annotations.
Supplementary Material
Acknowledgements
This work was supported by National Science Foundation Grants IOS-1051286 and IOS-1456256 (to SCP) and IOS-1545780 and DBI-1149224 (to JC).
References
- Anderson JC, Bartels S, Gonzalez Besterio MA, Shahollari B, Ulm R, Peck SC. 2011. Arabidopsis MAP kinase phosphatase 1 (AtMKP1) negatively regulates MPK6-mediated PAMP responses and resistance against bacteria. The Plant Journal 67, 258–268. [DOI] [PubMed] [Google Scholar]
- Anderson JC, Wan Y, Kim YM, Pasa-Tolic L, Metz TO, Peck SC. 2014. Decreased abundance of type III secretion system-inducing signals in Arabidopsis mkp1 enhances resistance against Pseudomonas syringae. Proceedings of the National Academy of Sciences, USA 111, 6846–6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. 2002. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983. [DOI] [PubMed] [Google Scholar]
- Bae H, Kim MS, Sicher RC, Bae HJ, Bailey BA. 2006. Necrosis- and ethylene-inducing peptide from Fusarium oxysporum induces a complex cascade of transcripts associated with signal transduction and cell death in Arabidopsis. Plant Physiology 141, 1056–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels S, Anderson JC, González Besteiro MA, Carreri A, Hirt H, Buchala A, Métraux JP, Peck SC, Ulm R. 2009. MAP KINASE PHOSPHATASE 1 and PROTEIN TYROSINE PHOSPHATASE 1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. The Plant Cell 21, 2884–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke G, Unthan T, Uhrig JF, Pöschl Y, Gust AA, Scheel D, Lee J. 2009. Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proceeding of the National Academy of Sciences, USA 106, 8067–8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl RP, Kracher B, Somssich IE. 2017. Induced genome-wide binding of three Arabidopsis WRKY transcription factors during early MAMP-triggered immunity. The Plant cell 29, 20–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G. 2009. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annual Review of Plant Biology 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng SH, Sheen J. 2010. Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464, 418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colcombet J, Hirt H. 2008. Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. The Biochemical Journal 413, 217–226. [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology 139, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoux C, Galletti R, Mammarella N, Gopalan S, Werck D, De Lorenzo G, Ferrari S, Ausubel FM, Dewdney J. 2008. Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Molecular Plant 1, 423–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desaki Y, Miya A, Venkatesh B, Tsuyumu S, Yamane H, Kaku H, Minami E, Shibuya N. 2006. Bacterial lipopolysaccharides induce defense responses associated with programmed cell death in rice cells. Plant & Cell Physiology 47, 1530–1540. [DOI] [PubMed] [Google Scholar]
- Desclos-Theveniau M, Arnaud D, Huang TY, Lin GJ, Chen WY, Lin YC, Zimmerli L. 2012. The Arabidopsis lectin receptor kinase LecRK-V.5 represses stomatal immunity induced by Pseudomonas syringae pv. tomato DC3000. PLoS Pathogens 8, e1002513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Ferreras A, Kiss-Papp M, Jehle AK, Felix G, Chinchilla D. 2015. An Overdose of the Arabidopsis coreceptor BRASSINOSTEROID INSENSITIVE1-ASSOCIATED RECEPTOR KINASE1 or its ectodomain causes autoimmunity in a SUPPRESSOR OF BIR1-1-dependent manner. Plant Physiology 168, 1106–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. 2010. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Research 38, W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Bar-Joseph Z. 2006. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics 7, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T. 1999. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. The Plant Journal 18, 265–276. [DOI] [PubMed] [Google Scholar]
- Frei dit Frey N, Garcia AV, Bigeard J et al. 2014. Functional analysis of Arabidopsis immune-related MAPKs uncovers a role for MPK3 as negative regulator of inducible defences. Genome Biology 15, R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. 2006. The plant immune system. Nature 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kang S, Yang F, Li L, Chen H, Chen S, Zhang J. 2015. The Arabidopsis transcription factor BRASSINOSTEROID INSENSITIVE1-ETHYL METHANESULFONATE-SUPPRESSOR1 is a direct substrate of MITOGEN-ACTIVATED PROTEIN KINASE6 and regulates immunity. Plant Physiology 167, 1076–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis LA, Polanski K, de Torres-Zabala M et al. 2015. Transcriptional dynamics driving MAMP-triggered immunity and pathogen effector-mediated immunosuppression in arabidopsis leaves following infection with Pseudomonas syringae pv tomato DC3000. The Plant Cell 27, 3038–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Meng X, Shan L, He P. 2016. Transcriptional regulation of pattern-triggered immunity in plants. Cell Host & Microbe 19, 641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Meng X, Wang R, Mao G, Han L, Liu Y, Zhang S. 2012. Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genetics 8, e1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang S. 2004. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. The Plant Cell 16, 3386–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Durán R, Macho AP, Boutrot F, Segonzac C, Somssich IE, Zipfel C, Nürnberger T. 2013. The transcriptional regulator BZR1 mediates trade-off between plant innate immunity and growth. eLife 2, e00983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao G, Meng X, Liu Y, Zheng Z, Chen Z, Zhang S. 2011. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. The Plant Cell 23, 1639–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Xu J, He Y, Yang KY, Mordorski B, Liu Y, Zhang S. 2013. Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. The Plant Cell 25, 1126–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Zipfel C, Rowland O, Keller I, Robatzek S, Boller T, Jones JD. 2004. The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiology 135, 1113–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta Y, Ding P, Zhang Y. 2014. Identification of additional MAP kinases activated upon PAMP treatment. Plant Signaling & Behavior 9, e976155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nühse TS, Peck SC, Hirt H, Boller T. 2000. Microbial elicitors induce activation and dual phosphorylation of the Arabidopsis thaliana MAPK 6. The Journal of Biological Chemistry 275, 7521–7526. [DOI] [PubMed] [Google Scholar]
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE 7, e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince DC, Drurey C, Zipfel C, Hogenhout SA. 2014. The leucine-rich repeat receptor-like kinase BRASSINOSTEROID INSENSITIVE1-ASSOCIATED KINASE1 and the cytochrome P450 PHYTOALEXIN DEFICIENT3 contribute to innate immunity to aphids in Arabidopsis. Plant Physiology 164, 2207–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JL, Zhou L, Yun BW, Nielsen HB, Fiil BK, Petersen K, Mackinlay J, Loake GJ, Mundy J, Morris PC. 2008. Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiology 148, 212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qutob D, Kemmerling B, Brunner F et al. 2006. Phytotoxicity and innate immune responses induced by Nep1-like proteins. The Plant Cell 18, 3721–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF. 2003. Assumption-free analysis of quantitative real-time PCR data. Neuroscience Letters 339, 62–66. [DOI] [PubMed] [Google Scholar]
- Ramonell K, Berrocal-Lobo M, Koh S, Wan J, Edwards H, Stacey G, Somerville S. 2005. Loss-of-function mutations in chitin responsive genes show increased susceptibility to the powdery mildew pathogen Erysiphe cichoracearum. Plant Physiology 138, 1027–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM, Leslie ME, Robinson SJ, Korasick DA, Zhang T, Backues SK, Cornish PV, Koo AJ, Bednarek SY, Heese A. 2014. Loss of Arabidopsis thaliana Dynamin-Related Protein 2B reveals separation of innate immune signaling pathways. PLoS Pathogens 10, e1004578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Rodriguez MC, Adams-Phillips L, Liu Y, Wang H, Su SH, Jester PJ, Zhang S, Bent AF, Krysan PJ. 2007. MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiology 143, 661–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena G, Boudsocq M, Sheen J. 2011. Protein kinase signaling networks in plant immunity. Current Opinion in Plant Biology 14, 519–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R, Ichimura K, Mizoguchi T, Peck SC, Zhu T, Wang X, Shinozaki K, Paszkowski J. 2002. Distinct regulation of salinity and genotoxic stress responses by Arabidopsis MAP kinase phosphatase 1. The EMBO Journal 21, 6483–6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R, Revenkova E, di Sansebastiano GP, Bechtold N, Paszkowski J. 2001. Mitogen-activated protein kinase phosphatase is required for genotoxic stress relief in Arabidopsis. Genes & Development 15, 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. 2006. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125, 749–760. [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T. 2004. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428, 764–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.