Genetic evidence is provided for the role of potato PM H+-ATPases in tuberization, demonstrating that the isoform PHA1 promotes stolon elongation and tuber growth, and is involved in sugar–starch metabolism.
Keywords: Branching, PHA1, plant growth, PM H+-ATPase, potato, stolon elongation, tuber growth, tuberization
Abstract
This study presents the characterization of the plasma membrane (PM) H+-ATPases in potato, focusing on their role in stolon and tuber development. Seven PM H+-ATPase genes were identified in the Solanum tuberosum genome, designated PHA1–PHA7. PHA genes show distinct expression patterns in different plant tissues and under different stress treatments. Application of PM H+-ATPase inhibitors arrests stolon growth, promotes tuber induction, and reduces tuber size, indicating that PM H+-ATPases are involved in tuberization, acting at different stages of the process. Transgenic potato plants overexpressing PHA1 were generated (PHA1-OE). At early developmental stages, PHA1-OE stolons elongate faster and show longer epidermal cells than wild-type stolons; this accelerated growth is accompanied by higher cell wall invertase activity, lower starch content, and higher expression of the sucrose–H+ symporter gene StSUT1. PHA1-OE stolons display an increased branching phenotype and develop larger tubers. PHA1-OE plants are taller and also present a highly branched phenotype. These results reveal a prominent role for PHA1 in plant growth and development. Regarding tuberization, PHA1 promotes stolon elongation at early stages, and tuber growth later on. PHA1 is involved in the sucrose–starch metabolism in stolons, possibly providing the driving force for sugar transporters to maintain the apoplastic sucrose transport during elongation.
Introduction
Plasma membrane (PM) H+-ATPases are integral membrane proteins that pump protons out of the cell, generating an electrochemical gradient of protons across the plasmalemma. As a result, PM H+-ATPases provide the driving force for the transport of ions and metabolites through channels and transporters (Palmgren, 2001). This process is required for several physiological responses, such as cell expansion, phloem loading/unloading, stress adaptation, and plant growth and development (Michelet and Boutry, 1995; Palmgren, 2001; Gaxiola et al., 2007; Duby and Boutry, 2009; Schumacher and Krebs, 2010). PM H+-ATPases have been described in many plant species, such as Arabidopsis thaliana (Gaxiola et al., 2007), Nicotiana plumbaginifolia (Morsomme and Boutry, 2000), Oryza sativa (Baxter et al., 2003), Solanum lycopersicum (Kalampanayil and Wimmers, 2001), Zea mays (Santi et al., 2003), and Cucumis sativus (Wdowikowska and Klobus, 2016).
The plant PM H+-ATPases are regulated at different levels. With respect to post-translational regulation, the mechanism involves the autoinhibitory action of the C-terminal domain that can be displaced by phosphorylation of the penultimate residue, a threonine, and the subsequent binding of 14-3-3 proteins, resulting in pump activation (Palmgren et al., 1991; Svennelid et al., 1999; Maudoux et al., 2000). More recently, it has been described that phosphorylation of other residues can also affect PM H+-ATPase activity (Duby et al., 2009; Piette et al., 2011), suggesting that these proton pumps are controlled by a complex regulatory mechanism. Other factors can regulate PM H+-ATPase activity. A novel interaction partner of PM H+-ATPase named PPI (proton pump interactor) was identified in A. thaliana (PPI1; Morandini et al., 2002) and Solanum tuberosum (StPPI1; Muñiz García et al., 2011); this protein increases the activity of the proton pump in vitro once the C-terminus has been displaced.
Several studies have indicated that regulation of PM H+-ATPase also occurs at the genetic level. PM H+-ATPases are encoded by five distinct gene subfamilies. The members of subfamilies I and II show ubiquitous expression profiles, while the expression of genes belonging to subfamilies III, IV, and V is restricted to specific tissues (Arango et al., 2003; Gaxiola et al., 2007). PM H+-ATPase transcript levels are differentially regulated by environmental and hormonal signals, such as salt stress (Kalampanayil and Wimmers, 2001; Janicka-Russak and Kłobus, 2007; Sahu and Shaw, 2009), dehydration (Surowy and Boyer, 1991), low temperature (Ahn et al., 1999), and exogenous application of hormones (Frías et al., 1996; Janicka-Russak and Kłobus, 2007).
In potato (Solanum tuberosum cv. Désirée), two genes encoding PM H+-ATPases (PHA1 and PHA2) were isolated long ago by Harms et al. (1994). However, their physiological functions have not yet been elucidated. The economic importance of potato plants resides in the capacity to produce tubers. Potato tubers develop from stolons, which are underground stems. All aspects of tuber development are co-ordinated by a complex interaction of phytohormones and environmental signals (Sarkar, 2008). Gibberellic acid (GA) is a central negative regulator of tuberization that stimulates stolon elongation and prevents tuber induction (Xu et al., 1998). Abscisic acid (ABA) and auxin also regulate tuberization, acting as promoting factors (Xu et al., 1998; Roumeliotis et al., 2012; Kolachevskaya et al., 2015).
At early stages of tuber formation, stolons display a cessation of elongation and the initiation of subapical radial growth, which is followed by a deposition of starch and storage proteins (Visser et al., 1994). During the transition from stolon to tuber, a switch from apoplastic to symplastic sucrose phloem unloading occurs (Viola et al., 2001). Phloem loading of sucrose in source leaves requires the activity of sucrose–H+ symporters, and apoplastic sucrose unloading in sink tissues involves sucrose–H+ and hexose–H+ symporters (Lemoine et al., 2013). These sugar transporters depend on the proton motive force generated by PM H+-ATPases; therefore, these proton pumps might play an important role in stolon and tuber development.
This study presents a comprehensive overview of the PM H+-ATPase gene family (PHA) in potato, including the identification of PHA genes and expression profiles to explore which PHA genes are possibly involved in tuberization. Based on the results of the expression analysis, PHA1 was chosen for further characterization by the development of transgenic plants.
Materials and methods
Plant material
Soil-grown potato plants (Solanum tuberosum cv. Spunta) were cultivated in a growth chamber at 19 °C or 22 °C, under a 12 h or 16 h light photoperiod (4000 lux light intensity), or in a greenhouse maintained between 22 °C and 24 °C, under a 16 h light photoperiod. In vitro plants were obtained by micropropagation of virus-free single-node cuttings in Murashige and Skoog medium (MS medium; Prod no. M519, PhytoTechnology Laboratories, Shawnee Mission, KS, USA) containing 20 g l–1 sucrose solidified with 0.7% (w/v) agar, under a 16 h light photoperiod (4000 lux light intensity) at 22 °C.
Abiotic stress treatments
The first two fully expanded leaves detached from plants grown in soil, in a greenhouse, for 4 weeks were used for salt stress and drought treatments. Prior to stress treatment, leaves were placed in individual containers with water in a growth chamber at 22 °C, under a 16 h light photoperiod, for 48 h, to allow the wound response components induced by excision to be restored to basal levels. Only the cut end of the petioles was immersed in the solution. For salt stress, leaves were treated with 250 mM NaCl. For drought treatment, water was removed from the containers and leaves were kept in air. For cold treatment, in vitro grown potato plants were exposed to 4 °C, while control plants remained at 22 °C, under a 16 h light photoperiod.
Stolon growth conditions and in vitro tuberization
Tuberization can be studied in vitro, reproducing the process occurring in vivo with the advantages of generating tubers in a controlled environment (Xu et al., 1998; Roumeliotis et al., 2012; Muñiz García et al., 2016). Single-node cuttings obtained from potato plants do not induce tubers when cultured in darkness on standard propagation media (MS medium plus 2% sucrose); however, increasing sucrose concentration (8%) increases the frequency of tuberization.
Shoot apices derived from in vitro plants were cultured on solid MS media containing 20 g l–1 sucrose for 2 weeks, in a growth chamber at 22 °C, under a 16 h light photoperiod, prior to harvesting single-node explants. Nodal segments were grown on MS medium containing 20 g l–1 or 80 g l–1 sucrose (non-inducing and tuber-inducing conditions, respectively) solidified with 0.7% (w/v) agar, in a growth chamber in darkness at 19 °C. In some experiments, the media was supplemented with hormones (5 μM GA3, 25 μM 3-indoleacetic acid, or 5 μM ABA; purchased from Sigma, St Louis, MO, USA) or the inhibitors of H+-ATPase (1 mM sodium orthovanadate or 50 μM erythrosine B; purchased from Sigma). The single-node cuttings formed etiolated shoots/stolons that, under tuber-inducing conditions, developed tubers. For tuber-inducing conditions, the process was studied for 9–10 weeks. After this time of culture, most of the tubers were fully developed, and the rate of tuber growth decreased significantly.
All the experiments were performed at least three times independently, using 10–20 stolons per condition. A tuberizing stolon was defined as a stolon presenting visible subapical swelling or tubers. Stolons presenting more than one subapical swelling/tuber were considered as one tuberizing stolon. A branched stolon was defined as a stolon presenting at least one branch ≥5 mm. The total stolon length was determined as the sum of the primary and secondary stolon lengths. Tuber weight was calculated as the sum of the weight of all the tubers divided by number of tubers obtained. If the stolon presented more than one tuber, only the fully developed tubers (minor diameter ≥4 mm) were used for weight determination, since, in most cases, the additional tubers were very small or undeveloped.
Tuberization in soil-grown plants
Yield determination was carried out on plants transferred to soil ex vitro and cultivated in a growth chamber at 19 °C, under a 12 h light photoperiod, for 10 weeks, in 0.5 liter pots with commercial soil mixture (Grow Mix Multipro, Terrafertil Argentina). Stolon length determination was carried out on plants obtained from seed tubers, grown in 0.8 liter pots with commercial soil mixture, in greenhouse, for 4 weeks.
Semi-quantitative reverse transcription–PCR (RT–PCR)
Semi-quantitative RT–PCR to determine PHA gene expression was performed as described in País et al. (2010) using the primers and reaction conditions shown in Supplementary Table S1 at JXB online. RT–PCR bands were quantified relative to the elongation factor 1-α gene (EF1-α) using ImageJ software (http://rsb.info.nih.gov/ij/).
Real-time quantitative RT–PCR (RT–qPCR) analysis
Relative expression of PHA1-3 and StSUT1 was determined by RT–qPCR. RNA was isolated and cDNA synthesis was performed as described in País et al. (2010). The cDNA was used as template for PCR amplification. The potato EF1-α gene was used as reference gene using the primers forward, ATTGGAAACGGATATGCTCCA; and reverse, TCCTTACCTGAACGCCTGTCA. The primer sequences for PHA1–PHA3 were PHA1 5' UTR forward, GGAAGAGAGGAAATTGAGAAAGATG; and PHA1 5' UTR reverse, CTCCTCTAGTTTGTTGTACCC (PCR efficiency: 2.04); PHA2 forward, AGAAAAGAAGAGACACACAAGC; and PHA2 reverse, GACACAATCCCTTTCAATGG (PCR efficiency: 1.97); PHA3 3' UTR forward, GTTGGTGTTGTGATGAGAGCG; and PHA3 3' UTR reverse, GAAGGCTCCAGGAAACAGC (PCR efficiency: 2.02). The primer sequences for StSUT1 were obtained from Krügel et al. (2012) (LC-SUT1 fw, TTCCATAGCTGCTGGTGTTC; and LC-SUT1 rev, TACCAGAAATGGGTCCACAA). Reactions were performed in a final volume of 20 μl containing 4 μl of 5XHOT FIREPol EvaGreen qPCR Mix Plus (Solis-BioDyne, Tartu, Estonia). The amount of cDNA used in each reaction was derived from 1 ng of total RNA for EF1-α, 10 ng for PHA1–PH3, and 5 ng for StSUT1 (each cDNA sample was diluted accordingly). Reactions for EF1-α amplification were carried out under the following conditions: 50 °C/2 min (1 cycle); 95 °C/15 min (1 cycle); 95 °C/15 s; 60 °C/1 min; 72 °C/30 s (35–40 cycles). For PHA1–PHA3, reactions were carried out as follows: 95 °C/15 min (1 cycle); 94 °C/2 min (1 cycle); 58 °C/1 min (1 cycle); 72 °C/1 min (1 cycle); 94 °C/30 s, 58 °C/30 s; 72 °C/45 s (35–40 cycles). For StSUT1, reactions were performed with the following program: 95 °C/15 min (1 cycle); 95 °C/30 s, 61 °C/30 s, 72 °C/30 s (35–45 cycles). Amplification of a single product of the correct size for each gene was confirmed by agarose gel electrophoresis. The relative expression level was calculated using the 2–ΔΔCt (cycle threshold) method (Livak and Schmittgen, 2001).
Cloning of the PHA1 gene from Solanum tuberosum cv. Spunta
The PHA1 coding sequence was obtained by RT–PCR from RNA isolated from potato (S. tuberosum cv. Spunta) flower bud using the primers PHA1 5'UTR Fw, GGAAGAGAGGAAATTGAGAAAGATG; and PHA1 3'UTR Rv, GCCGATAATGAATGCTGTTATAG. The amplified product was cloned into the pCR-Blunt II-TOPO vector (Thermo Fisher Scientific, Waltham, MA, USA), for sequencing.
Yeast complementation assay
The complementation assay was carried out using the Saccharomyces cerevisiae strain YAK2 (MATα, ade 2-101, leu2Δ1, his3-Δ200, ura3-52, trp1Δ63, lys2-801 pma1Δ::HIS3, pma2-Δ::TRP1) lacking the genomic copies of PMA1 and PMA2, the two endogenous H+-ATPase genes. Survival is possible by the expression of the PMA1 gene under the GAL1 promoter on a URA3-bearing centromeric plasmid (de Kerchove d’Exaerde et al., 1995). The YEplac181 plasmid (bearing the 2μ origin of replication and the LEU2 marker) containing the promoter region of the yeast PMA1 gene was used to express the different plant PM H+-ATPase genes. Colony PCR was performed to detect the presence of the corresponding plasmids in the YAK2 transformants. The transformed YAK2 strains were grown at 30 °C in a synthetic medium containing 0.7% yeast nitrogen base without amino acids, supplemented with all of the amino acids except those used for selection (His, Leu, Ura and Trp), and 2% glucose (MGlu-His, Leu, Ura, Trp) or 2% galactose/1% raffinose (MGal-His, Leu, Ura, Trp). Solid media contained 2% agar. The media were supplemented with 20 mM K2HPO4 and buffered to a pH of 6.5. In addition, the YAK2 transformants were grown on MGlu-His, Leu, Trp medium at pH 6.5, containing 0.1% 5-fluoro-orotic acid (5-FOA) to counter-select the plasmid bearing the yeast PM H+-ATPase gene.
Generation of PHA1-OE transgenic potato plants
The PHA1 sequence was subcloned into the pBI121 binary vector downstream of the Cauliflower mosaic virus 35S promoter (35S CaMV). Transformation of potato discs was performed using Agrobacterium tumefaciens strain LBA4404 as described in Muñiz García et al. (2014). Regenerated plants carrying no plasmid but obtained from the same explants and by the same regeneration method were used as controls (wild type). All plants were obtained from the same tuber; therefore, regeneration controls and transgenic lines have the same genetic background. Three independent transgenic lines (L1, L2, and L3) were selected and characterized in this study.
PCR analysis was performed to confirm the presence of the transgene using genomic DNA as template and the primers PHA1 5' UTR forward (GGAAGAGAGGAAATTGAGAAAGATG) and tNOS reverse (TGATAATCATCGCAAGACCG) to amplify the transgene but not the endogenous PHA1 gene. PCR was also performed using primers to detect the nptII gene: nptII Fw, ATGATTGAAGAAGATGGATTG; and nptII Rv, GAAGAACTCGTCAAGAAGGCG. The 18S rRNA gene was used as a positive control utilizing the primers 18S forward, GGGCATTCGTATTTCATAGTCAGAG, and 18S reverse, CGGTTCTTGATTAATGAAAACATCCT.
PM H+-ATPase activity
PM H+-ATPase activity was determined in purified membranes isolated by a two-step aqueous two-phase partitioning system as described in Olivari et al. (2000) from leaves of potato plants grown in soil for 60 d or stolons cultured in vitro on MS medium plus 8% sucrose for 3 weeks. The PM H+-ATPase activity was determined in 5 mM ATP, 5 mM MgCl2, and 10 mM PIPES pH 7.3, with the addition of 5 mM NaN3, 0.1 mM sodium molybdate, and 100 mM KNO3, in the absence or presence of 100 μM Na3VO4. The assay was performed in the presence of 0.01 mg ml–1 lysophosphatidylcholine (Papini and De Michelis, 1997). The assay was carried out using the plasma membrane samples (2.5 μg of protein) in a final reaction volume of 150 µl for 30 min at 30 °C. Released Pi was measured using the malachite green method (Hohenwallner and Wimmer, 1973). The PM H+-ATPase activity was determined as the difference between the activity measured in the absence and presence of vanadate.
Observation of stolon epidermal cell imprints
A thick layer of clear nail polish was brushed onto the epidermis of each stolon. Once dried, the nail polish was peeled off, placed on a clear glass, and observed using a light microscope (Olympus BX41, Olympus Optical Co. Ltd, Tokyo, Japan). Photomicrographs of the imprints were obtained at ×200 magnification in the microscope.
Starch content
The samples (50 mg) were ground to a fine powder in liquid nitrogen and extracted three times with 10 ml of 80% ethanol, by boiling the samples in a 90 °C water bath for 20 min to dissolve sugars, dextrins, and tannins, and centrifuged at 1500 g for 5 min. The residues were dried, resuspended in 15 ml of 1 M HCl, and incubated at 99 °C for 45 min for starch hydrolysis. The contents were filtered through Whatman No. 40 filter paper. The filtrates were adjusted to pH 7.0 with NaOH and analyzed for glucose by the Somogyi–Nelson method (Nelson, 1994). Starch content was estimated by multiplying the glucose content by the glucose equivalent of 0.9. Results were expressed as g starch 100 g–1 FW.
Cell wall invertase activity
The pellet mix procedure was used to assess the activities of cell wall invertase (Albertson et al., 2001). About 50 mg of stolon tissue was homogenized in ice-cold extraction buffer at a 1:5 (w/v) ratio containing 50 mM HEPES (pH 7.5), 10 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM benzamidine, 1 mM phenylmethylsulfonyl fluoride (PMSF), 2 μg ml–1 trypsin inhibitor, 5 mM aminocaproic acid, 5 mM DTT, 0.1% (v/v) Triton X-100, and 2% (w/v) polyvinylpyrrolidone (PVP). The extract was centrifuged at 12 000 g for 10 min at 4 °C; the pellet was washed three times with extraction buffer without PVP and used for activity assay after a final resuspension in a 1:4 (w/v) ratio with the same buffer. The activities were assayed in a final volume of 1 ml, containing 125 mM Na-acetate buffer (pH 4.5), 100 mM sucrose, and 80 μl of the pellet mix. The reactions were incubated at 37 °C for 30 min. Reactions were stopped in boiling water for 1 min, centrifuged at 12 000 g for 5 min, and glucose was quantified in the supernatant according to the Nelson–Somogyi method (Nelson, 1994).
Statistical analysis
Statistical analysis was carried out using the Student’s t-test. A P-value <0.05 was considered statistically significantly.
Results
Identification of PHA genes in potato
A search in the Potato Genome Sequencing Consortium database, (http://solanaceae.plantbiology.msu.edu/pgsc_download.shtml) revealed the existence of seven sequences presenting a high degree of homology with PM H+-ATPases in the S. tuberosum Phureja genome, designated PHA1–PHA7 (Supplementary Fig. S1; Supplementary Table S2). Analysis of the protein primary structure revealed that the potato PM H+-ATPases present the characteristic features of PM proton pumps: the C-terminal autoinhibitory domain, 10 transmembrane segments, the small cytosolic loop, between the second and third transmembrane segments, and the large cytosolic loop, between the fourth and fifth transmembrane segments (Supplementary Fig. S1). PHA6 is truncated at its C-terminus and presents a unique region of 88 amino acids, absent in the other members of the PHA family. PHA3 also presents a 34 amino acid sequence which is missing in the other PHAs. Scanning the PM H+-ATPase protein sequences for motif analysis confirmed the presence of typical motifs for plant P-ATPases: TGES, crucial for ATPase activity, DKTGTLT, in which the aspartate is reversibly phosphorylated during catalysis, KGAP, essential for ATP binding, and the GDGVNDA motif, involved the hydrolysis of the acyl-phosphate intermediate (Thever and Saier, 2009). The last three motifs are not completely conserved in PHA6. The 14-3-3-binding site, located at the extreme C-terminus, is absent in PHA5 and PHA6 (Supplementary Fig. S1).
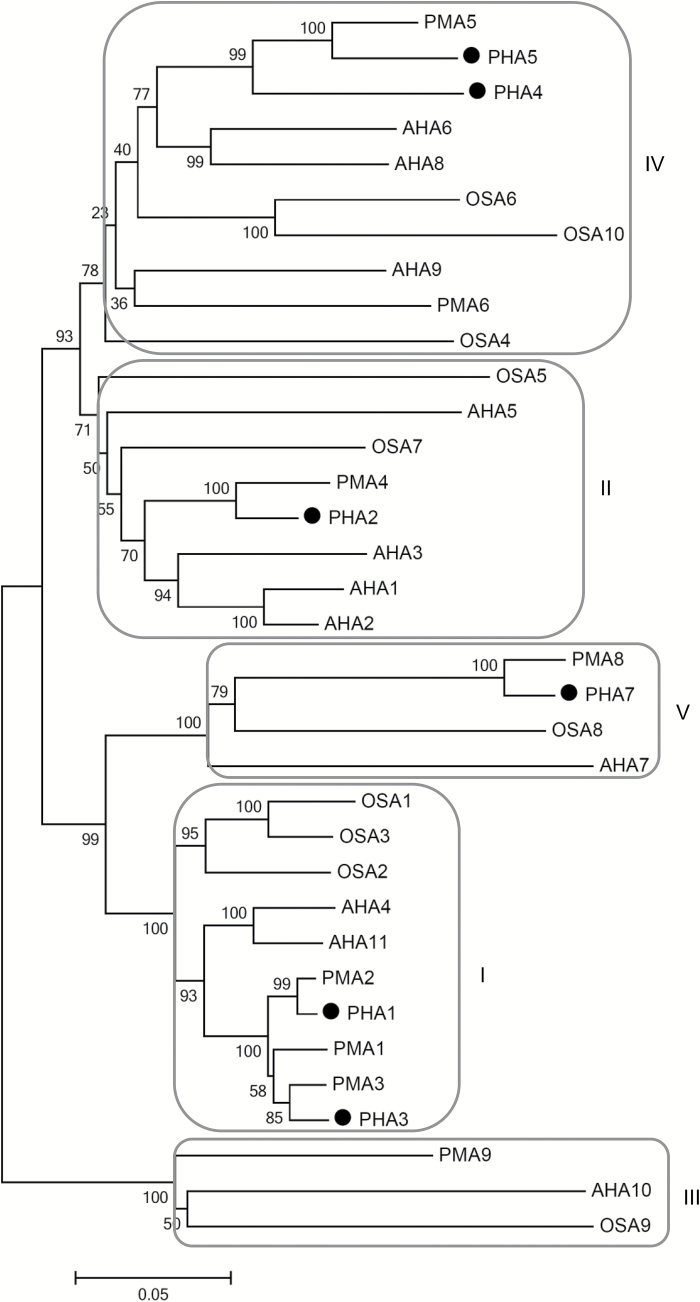
Multiple alignment of the protein sequences of A. thaliana (AHA), N. plumbaginifolia (PMA), O. sativa (OSA), and S. tuberosum Phureja (PHA) PM H+-ATPases was carried out to generate a phylogenetic tree (Fig. 1). The potato proteins were grouped into four of the five well-established subfamilies (Arango et al., 2003), except for PHA6, which does not fall into any of the subfamilies defined, and was excluded from the phylogenetic analysis. PHA1–PHA5 and PHA7 were clustered into the subfamilies I, II, IV, and V, while no members of PHAs were represented in subfamily III.
Fig. 1.
Phylogenetic analysis of potato, Arabidopsis, N. plunbaginifolia, and rice PM H+-ATPases. The amino acid sequences of PM H+-ATPases from S. tuberosum Phureja (PHA1–PHA5 and PHA7), A. thaliana (AHA1–AHA11), N. plunbaginifolia (PMA1–PMA6, PMA8, and PMA9), and O. sativa (OSA1–OSA10) were compared to generate the phylogenetic tree using the Neighbor–Joining method of MEGA5.05 (http://www.megasoftware.net/). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is indicated next to the branches. GenBank accession numbers are: A. thaliana AHA1, P20649; AHA2, P19456; AHA3, P20431; AHA4, Q9SU58; AHA5, Q9SJB3; AHA6, Q9SH76; AHA7, Q9LY32; AHA8, Q9M2A0; AHA9, Q42556; AHA10, Q43128; AHA11, Q9LV11; N. plumbaginifolia PMA1, Q08435; PMA2, Q42932; PMA3, Q08436; PMA4, Q03194; PMA6, Q9SWH2; PMA8, Q9SWH1; PMA9, Q9SWH0); O. sativa OSA1, Q43001; OSA2, Q43002; OSA3, AF110268; OSA4, AJ440002; OSA5, AJ440216; OSA6, AJ440217; OSA7, AJ440218; OSA8, AJ440219; OSA9, AJ440220; and OSA10, AJ440221. Accession numbers for S. tuberosum Phureja (Potato Genome Sequencing Consortium Public Data Release): PHA1, PGSC0003DMP400055772; PHA2, PGSC0003DMP400007331; PHA3, PGSC0003DMP400043938; PHA4, PGSC0003DMP400021001; PHA5, PGSC0003DMP400013900; and PHA7, PGSC0003DMP400060260).
Expression profile of PHA genes
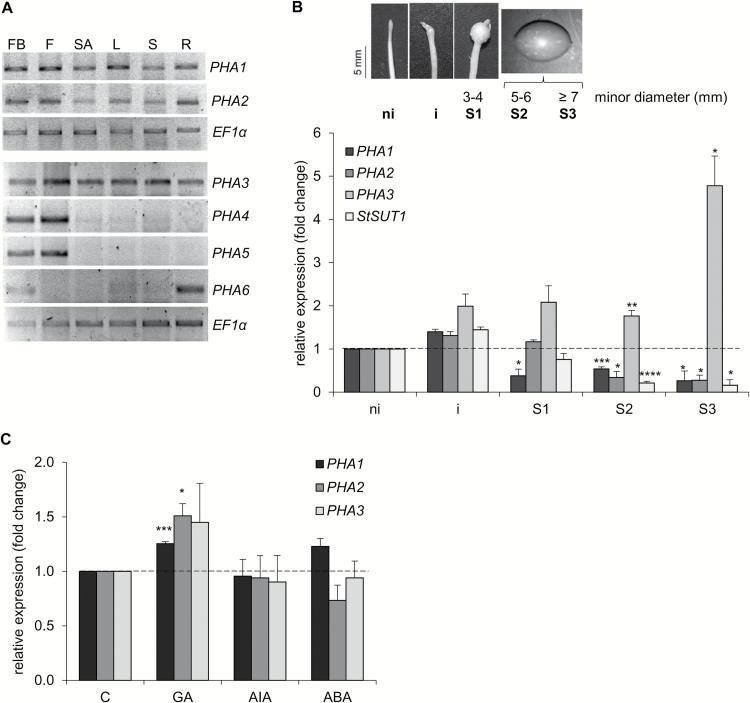
The expression profile of PHA genes in different organs was determined (Fig. 2A; Supplementary Fig. S2). PHA1, PHA2, and PHA3 were expressed in all organs, while expression of PHA4 and PHA5 was restricted to flower buds and flowers. PHA6 was expressed predominantly in flower buds and roots. PHA7 transcripts were not detected in any of the organs analyzed (not shown).
Fig. 2.
Expression profile of PHA genes. (A) Total RNA was isolated from different tissues of soil-grown potato plants, cDNA was synthesized, and semi-quantitative RT–PCR was performed. A representative RT–PCR analysis of three independent experiments is shown. FB, flower bud; F, flower; SA, shoot apex; L, leaf; S, stem; R, root. Quantitative data of RT–PCR bands are shown in Supplementary Fig. S2. (B) RT–qPCR analysis of PHA genes and StSUT1 during tuberization in vitro. Total RNA was obtained from stolons cultured under tuber-inducing conditions (MS medium plus 8% sucrose) at progressive stages of tuber development (ni, non-induced stolons; i, induced stolons with visible subapical swelling; S1–S3, tubers at different stages of growth). Quantitative data (mean ±SE) of three independent experiments, each consisting of four technical replicates, are displayed in the bar graph. The asterisks indicate statistical significance (*P<0.05, **P<0.01, ***P<0.005, ****P<0.001, with respect to non-induced stolons). (C) RT–qPCR analysis of PHA genes in stolons cultured under tuber-inducing conditions for 2 weeks, in the absence or presence of 5 μM GA3 (GA), 25 μM 3-indoleacetic acid (IAA), or 5 μM ABA. Quantitative data (mean ±SE) of three independent experiments, each consisting of four technical replicates, are displayed in the bar graph. The asterisks indicate statistical significance (*P<0.05, ***P<0.005, with respect to control, C).
The mRNA levels of PHA genes were determined in stolons cultured in vitro. PHA1, PHA2, and PHA3 were expressed in stolons cultured under tuber-inducing conditions, while PHA4, PHA5, PHA6, and PHA7 transcripts were not detected (Supplementary Fig. S3). PHA1 and PHA2 were expressed at higher levels in early stages of tuber organogenesis than in the tuber growth stages, while PHA3 displayed an opposite expression pattern (Fig. 2B). The sucrose–H+ symporter StSUT1 showed an expression pattern similar to that of PHA1 and PHA2 (Fig. 2B). GA increased PHA1 and PHA2 expression in stolons, while no significant changes were observed for PHA3 (Fig. 2C); no significant changes in the expression of PHA genes were detected in response to auxin or ABA (Fig. 2C). PHA4, PHA5, PHA6, and PHA7 transcripts were not detected at any stage of tuberization, with or without hormone treatment (not shown).
The expression of PHA genes in response to abiotic stress was determined in leaves. Salt stress increased PHA1, PHA2, and PHA3 mRNA levels, while drought only up-regulated the PHA3 gene (Supplementary Fig. S4). Cold stress increased the expression of PHA2 (Supplementary Fig. S4). PHA4, PHA5, PHA6, and PHA7 transcripts were not detected in leaves in the absence or presence of stress (not shown).
Involvement of PM H+-ATPase in tuberization
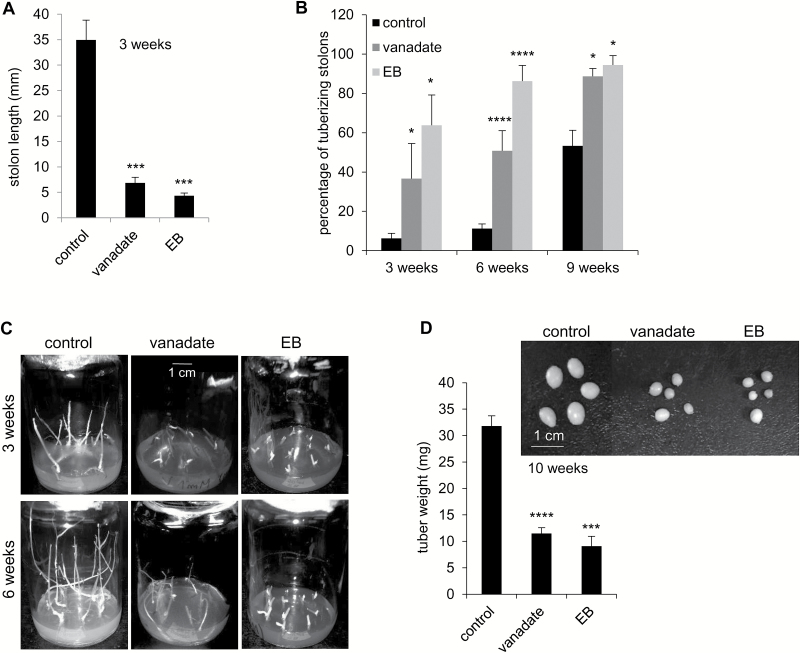
In a first attempt to elucidate the role of PHAs in tuberization, the inhibitors of H+-ATPase, sodium orthovanadate (vanadate) and erythrosine B (Kanczewska et al., 2005; Hayashi et al., 2014; Yuan et al., 2017), were used in tuberization experiments in vitro. Dose–response experiments were carried out to determine the appropriate concentration of inhibitors, which were 1 mM and 50 μM for vanadate and erythrosine B, respectively (not shown). The length of inhibitor-treated stolons was drastically reduced (Fig. 3A, C). Both inhibitors increased the percentage of tuberizing stolons cultured under tuber-inducing conditions (MS medium plus 8% sucrose) (Fig. 3B, C). The tubers obtained from stolons treated with vanadate or erythrosine B were significantly smaller than those obtained from untreated stolons (Fig. 3D). When stolons were cultured under non-inducing conditions (MS medium plus 2% sucrose), inhibition of PM H+-ATPases by vanadate or erythrosine B also inhibited stolon growth and promoted tuber initiation (Supplementary Fig. S5).
Fig. 3.
Effect of vanadate and erythrosine B (EB) on stolons cultured under tuber-inducing conditions. Stolons were cultured under tuber-inducing conditions (MS medium plus 8% sucrose) in the absence (control) or presence of 1 mM vanadate or 50 μM erythrosine B. The stolon length was measured after 3 weeks (A), and the percentage of tuberizing stolons was determined at the indicated times (B). (C) Representative images of stolons. (D) Fresh weight of tubers obtained after 10 weeks of culture under tuber-inducing conditions in the absence (control) or presence of 1 mM vanadate or 50 μM erythrosine B; a representative image of the tubers obtained is shown. Quantitative data of three independent experiments (mean ±SE) are displayed in the bar graphs. The asterisks indicate statistical significance (*P<0.05, ***P<0.005, ****P<0.001, with respect to control).
Cloning of the PHA1 gene from S. tuberosum cv. Spunta
PHA1 was selected for further analysis. The full-length coding sequence of PHA1 was cloned from S. tuberosum cv. Spunta. The nucleotide sequence appears in the GenBank database under the accession number KX827766. This sequence is almost identical to the S. tuberosum Phureja PHA1 sequence (PGSC0003DMP400055772; Potato Genome Sequencing Consortium database), except for one nucleotide difference within the coding region, with no amino acid change. Comparing the sequence of PHA1 from S. tuberosum cv. Spunta with the sequence of PHA1 from S. tuberosum cv. Désirée (Harms et al., 1994; GenBank accession number: X76536.1), 15 nucleotide differences were found within the coding region, one of which causes an amino acid change (R170A, in Spunta).
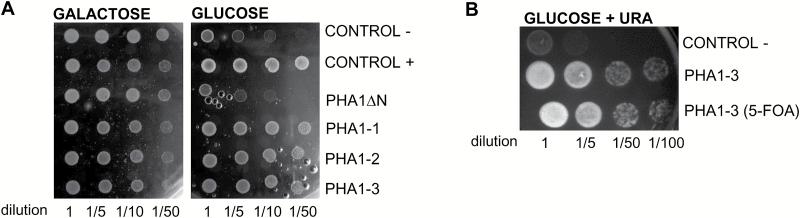
To confirm that the PHA1 gene cloned encodes a protein with PM H+-ATPase activity, a functional complementation assay was carried out. The S. tuberosum cv. Spunta PHA1 coding sequence was subcloned in YEplac181 under the control of the constitutive promoter of the yeast PMA1 gene. This plasmid was introduced into the yeast strain YAK2, which is deleted from its own two PM H+-ATPase genes (PMA1 and PMA2), and is able to survive on galactose medium with a centromeric plasmid carrying the yeast PMA1 controlled by the GAL1 promoter (de Kerchove d’Exaerde et al., 1995). Three independent transformants (PHA1-1, PHA1-2, and PHA1-3) obtained in a galactose medium were able to sustain the growth of YAK2 yeast cells when shifted to a glucose medium (Fig. 4A), demonstrating that the PHA1 gene cloned from S. tuberosum cv. Spunta has ATPase and proton pumping activity. The same result was obtained after eliminating the plasmid bearing the yeast PM H+-ATPase from the YAK2-PHA1-3 strain by 5-FOA (Fig. 4B).
Fig. 4.
Functional complementation of a null mutation of yeast PM H+-ATPase (strain YAK2) by the PHA1 gene from S. tuberosum cv. Spunta. (A) Serial dilutions (starting from an OD600nm=1.7) of the following YAK2 yeast strains were spotted onto solid media containing glucose (MGlu-His, Leu, Trp) or galactose (MGal-His, Leu, Trp), buffered at pH 6.5: Control –, YAK2 transformed with empty YEplac181; Control +, YAK2 transformed with YEplac181-E14D, that expresses the constitutively active form of the PM H+-ATPase PMA2 Q42932 from N. plumbaginifolia under the yeast PMA1 promoter; PHA1∆N, YAK2 transformed with YEplac181-PHA1∆N that expresses a truncated, inactive form of PHA1, lacking the first 553 nucleotides of the N-terminus (devoid of the TGES motif) under the yeast PMA1 promoter; PHA1-1/2/3, YAK2 transformed with YEplac181-PHA1, that expresses PHA1 under the yeast PMA1 promoter (three different transformed yeast clones were used). (B) Serial dilutions (starting from an OD600nm=1.7) of the following YAK2 yeast strains spotted onto solid media containing glucose (MGlu-His, Leu, Ura, Trp): Control –, PHA1–PHA3, and PHA1–PHA3 after 5-FOA treatment to eliminate the plasmid bearing the yeast PM H+-ATPase gene. Yeast strains were grown at 30 °C for 48 h.
Overexpression of PHA1 promotes stolon growth and branching, and increases tuber weight
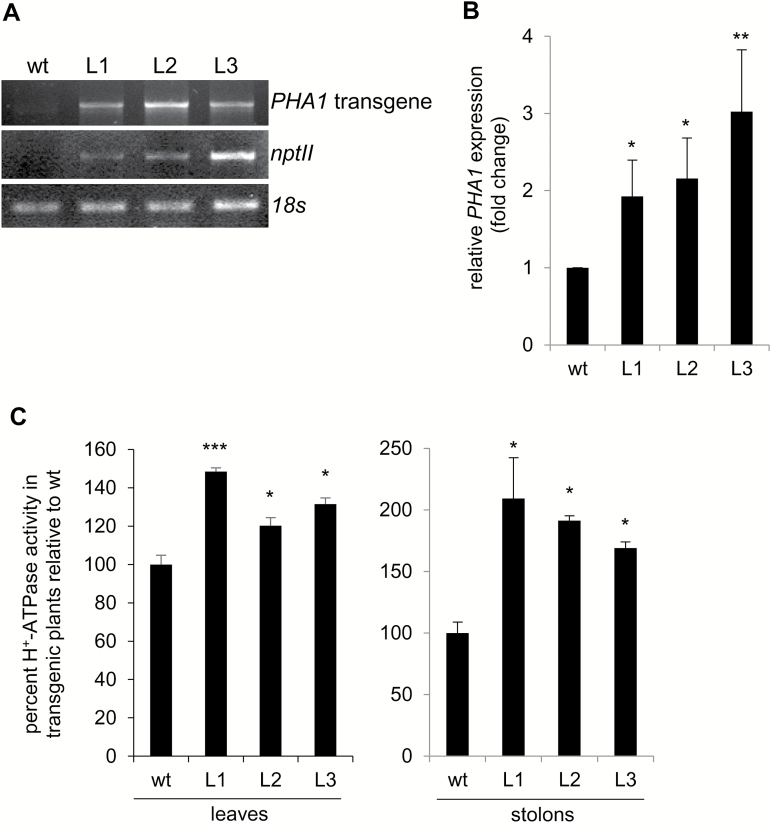
Transgenic plants expressing the PHA1 gene under the control of the 35S CaMV promoter (PHA1-OE) were developed. Three independent PHA1-OE transgenic lines (L1–L3) were selected for detailed characterization. The presence of the transgene was confirmed by PCR amplification (Fig. 5A). RT–qPCR analysis showed that the mRNA abundance of PHA1 was higher in transgenic lines than in wild-type plants (Fig. 5B). Accordingly, transgenic lines exhibited a higher PM H+-ATPase activity in leaves and stolons (Fig. 5C).
Fig. 5.
Analysis of transgenic plants overexpressing PHA1 (PHA1-OE). (A) PCR analysis of genomic DNA isolated from leaves of wild-type (wt) and transgenic plants (L) grown in vitro, to detect the presence of the transgene and the nptII gene (B) RT–qPCR analysis of RNA isolated from leaves of wild-type and transgenic plants grown in vitro, to determine PHA1 expression. Data are presented as the expression level relative to the wt. Quantitative data (mean ±SE) of three independent experiments, each consisting of four technical replicates, are displayed in the bar graph. (C) PM H+-ATPase activity in leaves and stolons, expressed as the percentage increase in proton pump activity of transgenic lines with respect to wild-type plants. As a reference, average PM H+-ATPase activity was 44.7 pmol Pi min–1 µg–1 protein for wild-type leaves and 18.7 pmol Pi min–1 µg–1 protein for wild-type stolons. Means ±SE of two independent experiments each performed in quadruplicate, are shown. The asterisks indicate statistical significance (*P<0.05, **P<0.01, ***P<0.005, with respect to the wt).
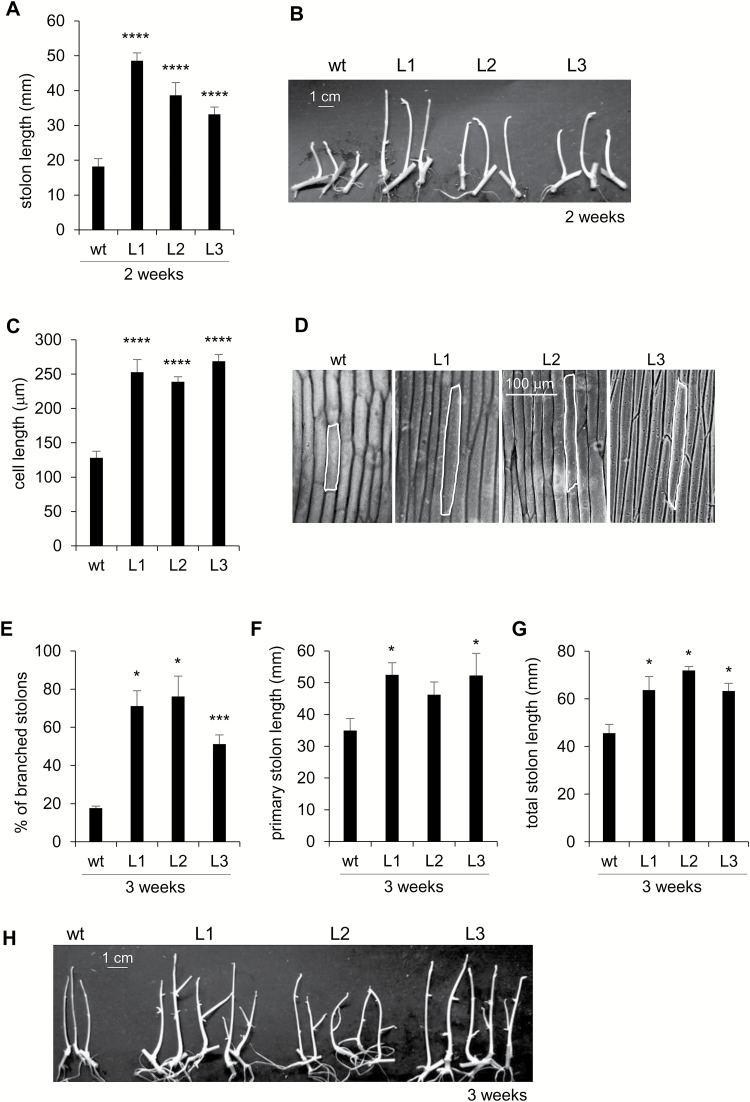
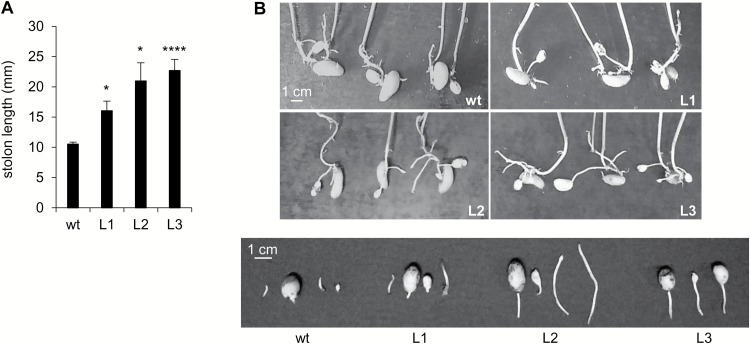
After 2 weeks of culture under tuber-inducing conditions, PHA1-OE stolons were longer than wild-type stolons (Fig. 6A, B). Accordingly, transgenic stolons had longer epidermal cells (Fig. 6C, D). No branching was observed after 2 weeks (Fig. 6B). After 3 weeks of culture, the percentage of branched stolons was significantly higher in PHA1-OE stolons than in wild-type stolons (Fig. 6E, H). The primary stolon length of L1 and L3 was higher, with no statistically significant difference between L2 and wild-type stolons (Fig. 6F), although the total stolon length was increased in all three transgenic lines, due to the increased number of branches (Fig. 6G). PHA1-OE plants grown in soil presented longer stolons than wild-type plants (Fig. 7A, B), confirming the long-stolon phenotype observed in vitro.
Fig. 6.
Phenotypic analysis of PHA1-OE stolons cultured in vitro. Stolons from wild-type (wt) and PHA1-OE plants (L) were cultured under tuber-inducing conditions (MS medium plus 8% sucrose). (A) Stolon length was determined after 2 weeks of culture; quantitative data of three independent experiments (mean ±SE) are displayed in the bar graph; a representative image is shown (B). (C) Longitudinal length of epidermal cells of the medial region of the stolons cultured for 2 weeks; the data represent the means ±SE of five biological replicates; lengths of 30–60 cells per replicate were measured. (D) Imprints of epidermal cells from the medial region of stolons, viewed by light microscopy. After 3 weeks, the percentage of branched stolons (E), primary stolon length (F), and total stolon length (G) were determined; quantitative data of three independent experiments (mean ±SE) are displayed in the bar graphs. (H) Representative images of stolons after 3 weeks of culture. The asterisks indicate statistical significance (*P<0.05, ***P<0.005, ****P<0.001, with respect to the wt).
Fig. 7.
Stolon length of PHA1-OE plants grown in soil. (A) Stolon length of plants obtained from seed tubers, grown in soil, in a greenhouse, for 4 weeks. The data of four independent experiments (mean ±SE) are displayed in the bar graph; each experiment consisted of 3–4 soil-grown plants per condition, with 3–5 stolons per plant. The asterisks indicate statistical significance [*P<0.05, ****P<0.001, with respect to the wild type (wt)]. (B) Representative image of the underground part of the plants (upper panel) and detached stolons (lower panel).
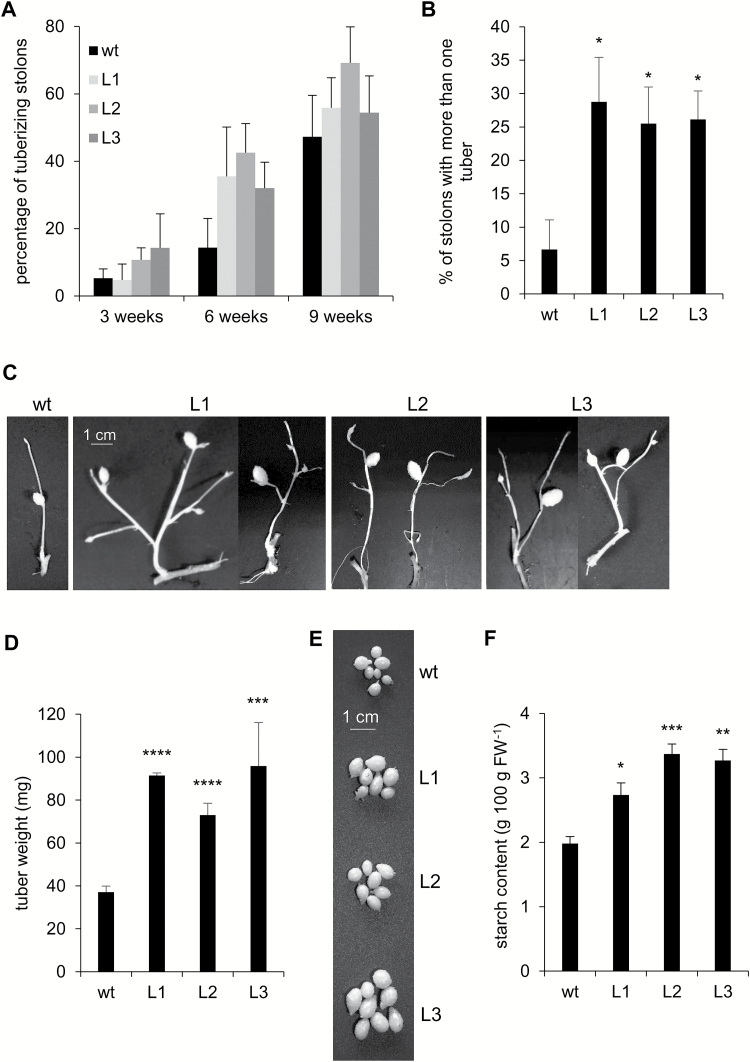
The percentage of tuberizing stolons was not significantly affected in PHA1-OE lines (Fig. 8A); however, after 10 weeks of culture, ~26% of PHA1-OE stolons presented more than one tuber (2–5 tubers per stolon), while only 7% of wild-type stolons developed more than one tuber (Fig 8B, C). In most cases, the additional tubers were very small or undeveloped. The tubers developed from PHA1-OE stolons were larger and presented higher starch content than those obtained from wild-type stolons (Fig. 8D–F).
Fig. 8.
Tuberization of PHA1-OE stolons cultured in vitro. Stolons from wild-type (wt) and PHA1-OE (L) plants were cultured under tuber-inducing conditions (MS medium plus 8% sucrose). (A) Percentage of tuberizing stolons, determined at the indicated times. (B) Percentage of stolons presenting more than one tuber after 10 weeks of culture; a representative image of the stolons is shown (C). (D) Fresh weight of tubers obtained from wild-type and PHA1-OE stolons after 10 weeks of culture; a representative image of the tubers is shown (E). (F) Starch content of tubers obtained from wild-type and PHA1-OE stolons after 10 weeks of culture. Quantitative data of three independent experiments (mean ±SE) are displayed in the bar graphs. The asterisks indicate statistical significance (*P<0.05, **P<0.01, ***P<0.005, ****P<0.001, with respect to the wt).
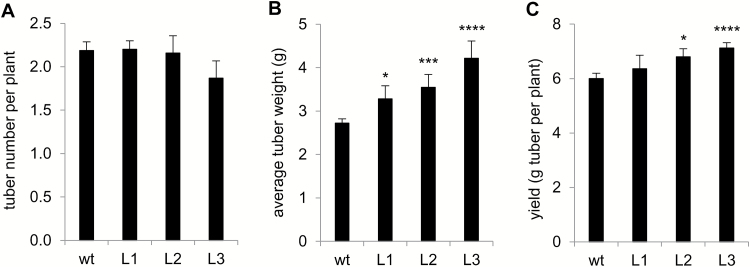
The tuberization capacity of PHA1-OE plants grown in soil was determined. There were no significant differences in the number of tubers obtained per plant between PHA1-OE and the wild type (Fig. 9A). The average weight of tubers obtained from PHA1-OE plants was higher than that of wild-type tubers (Fig. 9B). The tuber yield per plant was higher in transgenic lines with respect to wild-type plants, although this difference was statistically significant only for L2 and L3 (Fig. 9C). It is important to note that the tuber yield was determined in plants transferred to soil ex vitro, which are much smaller than potato plants obtained from seed tubers, and consequently have significantly lower yields.
Fig. 9.
Tuberization of PHA1-OE plants in soil. Wild-type (wt) and PHA1-OE plants (L) transferred to soil ex vitro were cultivated in a growth chamber. After 10 weeks, the tubers were harvested and the number of tubers obtained per plant (A), the average tuber fresh weight (B), and the tuber yield, defined as grams (FW) of tuber obtained per plant (C) were determined. Data are the results (mean ±SE) of 30–60 plants obtained from five different harvests performed over an 18 month period. The asterisks indicate statistical significance (*P<0.05, ***P<0.005, ****P<0.001, with respect to the wt).
Overexpression of PHA1 alters cell wall invertase activity, starch content, and StSUT1 expression in stolons
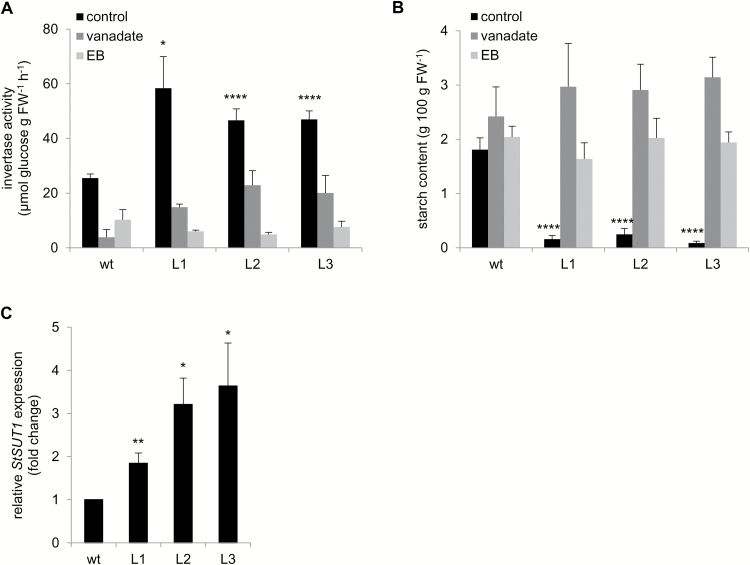
Cell wall invertase activity, starch content, and StSUT1 expression were determined in stolons during the elongation stage. After 2 weeks of culture, PHA1-OE stolons showed higher cell wall invertase activity (Fig. 10A) and accumulated significantly less starch than wild-type stolons, showing very low values of starch content (Fig. 10B). Treatment with H+-ATPase inhibitors inhibited stolon elongation (Supplementary Fig. S6), decreased cell wall invertase activity (Fig. 10A), and increased starch accumulation (Fig. 10B) in PHA1-OE stolons, abolishing the phenotypic differences between transgenic and wild-type stolons. PHA-OE stolons showed higher expression levels of the sucrose–H+ symporter gene StSUT1 than wild-type stolons (Fig. 10C).
Fig. 10.
Cell wall acid invertase activity, starch content, and StSUT1 expression in stolons from wild-type (wt) and PHA1-OE (L) plants. Stolons were cultured under tuber-inducing conditions (MS medium plus 8% sucrose) for 2 weeks; alternatively, 1 mM vanadate or 50 μM erythrosine B (EB) was applied to the medium. (A) Cell wall acid invertase activity was determined in the apical portion (1 cm) of stolons cultured in the absence or presence of PM H+-ATPase inhibitors. (B) Starch content was determined in the apical portion (1 cm) of stolons cultured in the absence or presence of PM H+-ATPase inhibitors. (C) RT–qPCR analysis of StSUT1 in wt and transgenic stolons; data are presented as the expression level relative to the wt. Quantitative data (mean ±SE) of three independent experiments, each consisting of four technical replicates, are displayed in the bar graphs. The asterisks indicate statistical significance (*P<0.05, **P<0.01, ****P<0.001, with respect to the wt).
Overexpression of PHA1 promotes stem growth and branching
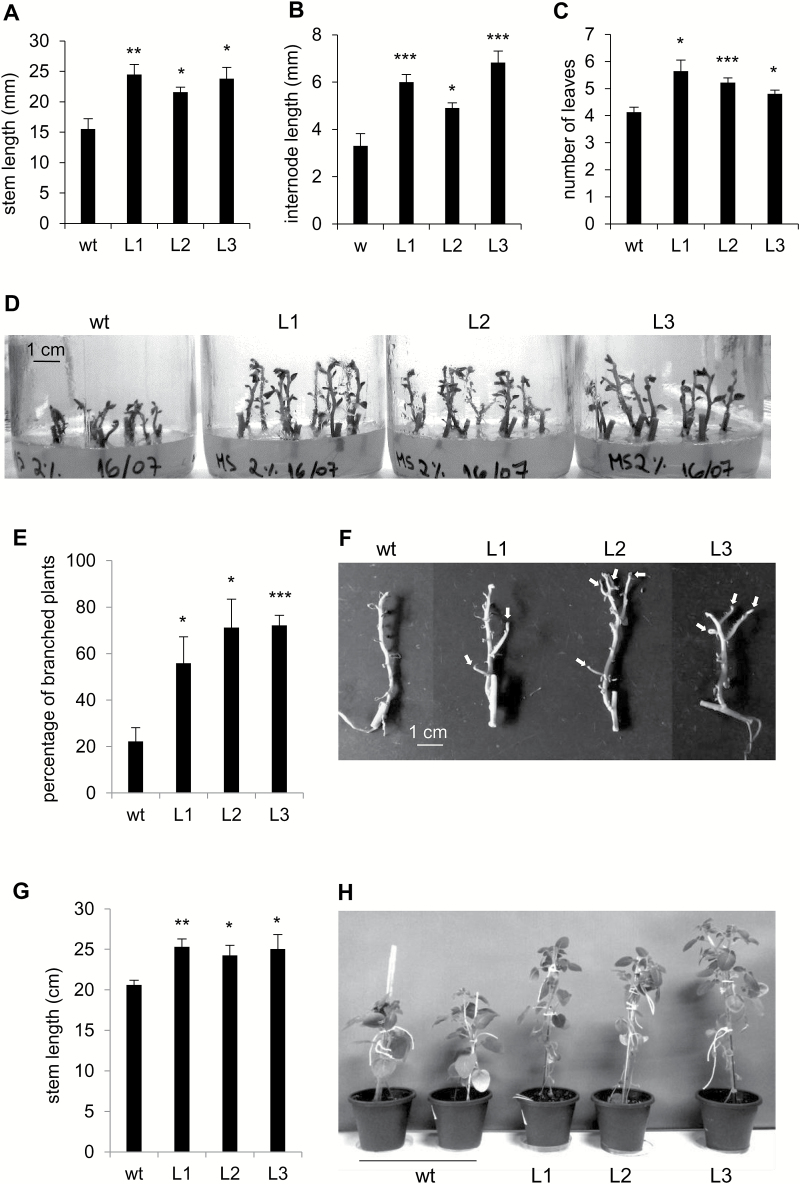
Plant growth parameters were determined in 2-week-old plants generated in vitro from internode cuttings. PHA1-OE plants showed higher stem and first internode lengths, and higher number of leaves than wild-type plants (Fig.11A–D). No significant differences were observed in the number of primary roots or total root length, between transgenic and wild-type plants (Supplementary Fig. S7). Inhibition of PM H+-ATPase activity by vanadate or erythrosine B resulted in shorter plants (Supplementary Fig. S8). No branching was observed in 2-week-old plants (Fig. 11D). After 4 weeks of growth in vitro, the percentage of plants presenting branches was significantly higher in PHA1-OE plants than in wild-type plants (Fig. 11E, F); a similar result was obtained using plants generated from shoot apexes (Supplementary Fig. S9). PHA1-OE plants grown in soil were ~20% taller than wild-type plants (Fig. 11G, H).
Fig. 11.
Phenotypic analysis of PHA1-OE plants grown in vitro. Wild-type (wt) and transgenic (L) plants generated from single-node cuttings were grown on MS medium plus 2% sucrose. After 2 weeks, the stem length (A), first internode length (B), and number of leaves (C) were determined. (D) Representative image of plants after 2 weeks of culture. (E) After 4 weeks, the percentage of branched plants was determined as the number of plants presenting at least one branch ≥5 mm, with respect to the total number of plants. (F) Representative image of wt and PHA1-OE plants grown in vitro for 4 weeks; leaves appear shrunken, since plants were allowed to air-dry for better visualization of branches, which are indicated with arrows. (G) Stem length of plants grown in soil, in a growth chamber, for 4 weeks after ex vitro transfer; a representative image of the plants is shown (H). Quantitative data of four independent experiments (mean ±SE) are displayed in the bar graphs; each experiment consisted of 15–20 in vitro cultured plants or 8–10 soil-grown plants per condition. The asterisks indicate statistical significance (*P<0.05, **P<0.01, ***P<0.005, with respect to the wt).
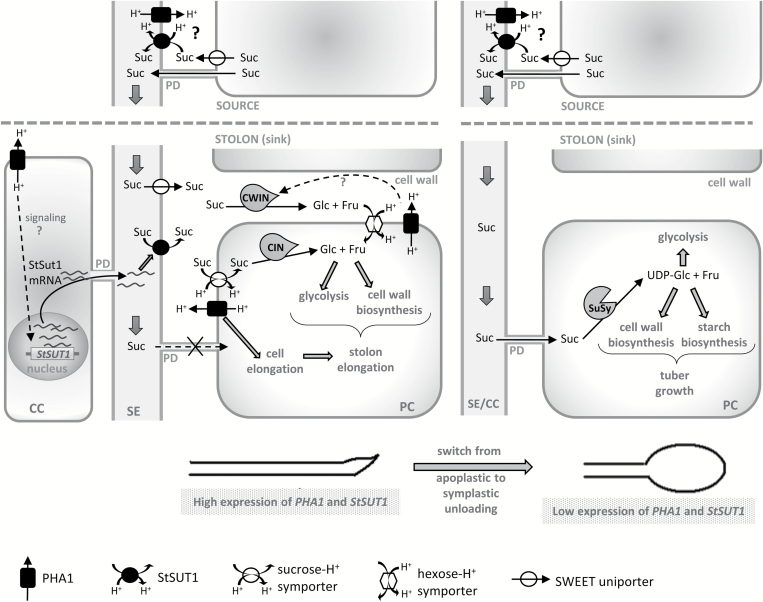
Fig. 12.
Model for the potential function of PHA1 in stolons, based on the results obtained in this study (see text for details). Suc, sucrose; SE, sieve element; CC, companion cell; CWIN, cell wall invertase; CIN, cytoplasmic invertase; SuSy, sucrose synthase; PD, plasmodesmata; PC, parenchymal cell.
Discussion
The potato PM H+-ATPase family
An in silico screening in the S. tuberosum Phureja genome database retrieved seven sequences, designated PHA1–PHA7, with a high degree of homology with PM H+-ATPases (Fig. 1; Supplementary Table S2; Supplementary Fig. S1), including two genes (PHA1 and PHA2) identified earlier in S. tuberosum cv. Désirée (Harms et al., 1994).
The expression patterns of PHAs were determined in S. tuberosum cv. Spunta. PHA1, PHA2, and PHA3 are ubiquitously expressed throughout the plant (Fig. 2A). These results are consistent with previous reports describing the expression patterns in other species. PHA1 and PHA3 belong to subfamily I, and PHA2 to subfamily II; the genes of these subfamilies (A. thaliana AHA1, AHA2, AHA3, AHA4, and AHA11; tobacco PMA1, PMA2, PMA3, and PMA4; maize MHA2; tomato LHA1; and cucumber CsHA2, CsHA3, CsHA4, CsHA8, CsHA9, and CsHA10) have shown a broad expression pattern throughout the plant (Ewing and Bennett, 1994; Frías et al., 1996; Moriau et al., 1999; Oufattole et al., 2000; Santi et al., 2003; Lefebvre et al., 2005; Gaxiola et al., 2007; Wdowikowska and Klobus, 2016). In contrast, PHA4 and PHA5 expression is limited to flower buds and flowers, while PHA6 is only expressed in flower buds and roots (Fig. 2A), suggesting that the pumps encoded by these genes have more specialized functions. PHA4 and PHA5 belong to subfamily IV; it has been shown that the expression of genes of this subfamily is limited to floral organs: Arabidopsis AHA6 and AHA9 transcripts are present only in anthers, AHA8 and the tobacco PMA5 are expressed only in pollen, and cucumber CsHA5, CsHA6, and CsHA7 are exclusively expressed in young flowers (Houlne and Boutry, 1994; Lefebvre et al., 2005; Gaxiola et al., 2007; Wdowikowska and Klobus, 2016). Salt stress significantly increases PHA1, PHA2, and PHA3 expression (Supplementary Fig. S4), suggesting a role for these PM H+-ATPases in salinity tolerance, as described in previous reports (Harms et al., 1994; Kalampanayil and Wimmers, 2001; Janicka-Russak and Klobus, 2007; Sahu and Shaw, 2009).
Only PHA1, PHA2, and PHA3 are expressed in stolons (Supplementary Fig. S3), and their expression levels change during the tuberization process (Fig. 2B), suggesting a role for these PM H+-ATPases in the development of stolons and tubers. Inhibition of PM H+-ATPase activity by vanadate or erythrosine B inhibits stolon elongation, promotes tuber induction, and impairs tuber growth (Fig. 3; Supplementary Fig. S5), confirming that PM H+-ATPases are involved in tuberization acting at different stages of the process.
PHA1 promotes stolon elongation
PHA1 was selected for functional analysis by overexpression in potato plants. PHA1-OE stolons elongate faster and present longer epidermal cell length than wild-type stolons after 2 weeks of culture (Fig. 6A–D), indicating that PHA1 activity contributes to the stolon elongation that occurs prior to tuber initiation by promoting cell elongation. In agreement with this, inhibition of PM H+-ATPase activity with vanadate or erythrosine B results in shorter stolon length (Fig. 3A; Supplementary Fig. S5A). These results are consistent with the role of PM H+-ATPases in cell elongation and expansion described for other tissues such as hypocotyls and roots (Haruta and Sussman, 2012; Inoue et al., 2016), and Nicotiana tabacum BY-2 cells (Niczyj et al., 2016). Acidification of the apoplasm by PM H+-ATPases leads to the wall-loosening process and cell elongation (reviewed in Hager, 2003). GA promotes stolon elongation by stimulating both cell division and cell elongation (Loy, 1977; Xu et al., 1998). PHA1 expression is induced in stolons in response to GA (Fig. 2C), thus it is possible to speculate that PHA1 mediates the GA-induced stolon elongation. However, more studies are needed to establish the mechanistic link between GA and PHA1.
PHA1 regulates sucrose–starch metabolism in stolons
During the stolon elongation phase, apoplastic sucrose unloading predominates (Viola et al., 2001). Although the exact mechanism occurring during this process remains unknown, sucrose can be unloaded from the phloem into the apoplast by the sucrose–H+ symporter StSUT1 in its inverse transport mode (Kühn et al., 2003; Carpaneto et al., 2005), or possibly by recently discovered facilitators of the SWEET family (Chen et al., 2012). The apoplastic sucrose can be converted to hexoses by a cell wall invertase, and taken up by the parenchyma cells through hexose–H+ symporters, not yet characterized in potato. Sucrose uptake from the aploplast might be mediated by sucrose–H+ symporters, possibly different from StSUT1, since it was reported that StSUT1 is localized in sieve elements but not in storage parenchyma in sink tubers (Kühn et al., 2003) (Fig. 12). Apoplastic unloading correlates with high acid invertase activity (Ross et al., 1994; Appeldoorn et al., 1997; Viola et al., 2001), and is associated with rapidly growing vegetative sink tissues (Ehness and Roitsch, 1997; Herbers and Sonnewald, 1998). During the transition from stolon to tuber, a switch from apoplastic to symplastic unloading occurs, accompanied by a switch from an invertase–sucrolytic to a sucrose synthase (SuSy)–sucrolytic pathway, leading to the starch accumulation phase and tuber growth (Viola et al., 2001) (Fig. 12).
During the elongation phase, PHA1-OE stolons grow faster (Fig. 6A), and present higher levels of cell wall invertase activity (Fig. 10A) and significantly lower starch content than wild-type stolons; moreover, starch is almost undetectable in transgenic stolons (Fig. 10B). These metabolic differences are accompanied by higher expression levels of StSUT1 in PHA1-OE stolons (Fig. 10C). In agreement with this, the PHA1-OE stolon phenotype can be reversed by the application of PM H+-ATPase inhibitors, that results in shorter length, decreased cell wall invertase activity, and increased starch content (Fig. 10A, B; Supplementary Fig. S6). Based on these data, it may be hypothesized that, besides promoting stolon growth by mediating cell elongation, PHA1 regulates the sucrose–starch metabolism in elongating stolons (Fig. 12). PHA1 positively regulates cell wall invertase activity (by an unknown mechanism), and its proton pumping activity can sustain the secondary transport of hexose–H+ and sucrose–H+ symporters, promoting the uptake of hexoses and sucrose by parenchyma cells; moreover, PHA1 increases the expression of StSUT1 in stolons, promoting sucrose unloading into the apoplast (Fig. 12). By doing so, PHA1 favors apoplastic unloading, sucrolysis by invertase and stolon elongation, and prevents starch accumulation, which is associated with symplastic unloading. Supporting this hypothesis, PHA1 and StSUT1 expression is high in wild-type stolons during the elongation phase, and decreases during tuber growth (Fig. 2B). In this context, PHA1 seems to play a key role in the molecular mechanism that determines the switch from the apoplastic unloading/invertase–sucrolytic pathway to the symplastic unloading/SuSy–sucrolytic pathway.
PHA1-OE stolons show higher expression levels of StSUT1 (Fig. 10C), suggesting that this PM H+-ATPase may be part of the signaling pathway that leads to the activation of StSUT1 transcription. It is possible that PHA1 induces the expression of StSUT1 in companion cells, and the StSUT1 mRNA is transported via plasmodesmata to the sieve elements as previously described (Kühn et al., 1997) (Fig. 12).
Role of PHA1 in tuber development
PHA1-OE tubers are larger and present higher starch content than wild-type tubers (Figs 8D–F, 9B). StSUT1 is essential for sucrose phloem loading (Kühn et al., 1996); therefore, it is possible to speculate that PHA1 might enhance sucrose phloem loading by energizing this sucrose–H+ symporter (Fig. 12), promoting sucrose translocation to sink tubers and starch accumulation. The mechanism of sucrose phloem loading in whole plants is different from the process occurring in stolons cultured in vitro; however, in both cases sucrose loading requires a proton symport mechanism energized by PM H+-ATPases. In plants, sucrose is loaded into the phloem in source leaves (Lemoine et al., 2013), while in stolons cultured in vitro, sucrose is obtained from the culture medium and loaded into the phloem as either hexoses (after hydrolysis by invertase) or sucrose (De Riek et al. 1997).
PHA1-OE stolons show an increased branching phenotype (Fig. 6E, H), which results in a higher percentage of stolons presenting more than one tuber, although the additional tubers are undeveloped (Fig. 8B, C). Stolon branching increases the number of potential tuber sites; however, developing more than one tuber per stolon leads to a competition for resources that negatively affects tuber size (Celis-Gamboa et al., 2003). Interestingly, the fully developed tubers obtained from PHA1-OE stolons are larger than those from wild-type stolons (Fig. 8D, E), indicating that PHA1 positively regulates tuber growth. This result agrees with the observation that inhibition of PM H+-ATPase activity results in smaller average tuber weight (Fig. 3D). There are no significant differences in the percentage of tuberizing stolons between PHA1-OE and wild-type plants (Fig. 8A); however, treatment with vanadate or erythrosine B enhances tuber induction (Fig. 3B; Supplementary Fig. S5B); this effect might be due to the inhibition of PHA2 or PHA3, which are also expressed in stolons, or to a non-specific action.
As observed in vitro, PHA1-OE plants grown in soil also develop larger tubers and show an increased tuber yield with respect to wild-type plants (Fig. 9B, C), suggesting that the PHA1 gene might be a potential tool to increase potato crop yield.
Branching phenotype of PHA1-OE plants
Stems of PHA1-OE plants grown in vitro present a highly branched phenotype, similar to PHA1-OE stolons (Fig. 11E, F; Supplementary Fig. S9). It has recently been shown that lateral bud growth depends on the amount of sucrose translocated to those buds, with sugar distribution, not auxin, being the initial regulator of apical dominance (Mason et al., 2014). It is now clear that sugar supply to axillary buds is not only essential to trigger outgrowth, but is also required to release bud dormancy (Barbier et al., 2015). Sucrose transporters and PM H+-ATPases have been implicated in this process; in rose bush, the onset of bud outgrowth correlates with increased sugar translocation to axillary buds and the up-regulation of the sucrose–H+ symporter gene RhSUC2 (Girault et al., 2010; Henry et al., 2011); the dormancy break of Prunus persica buds is associated with sucrose uptake and PM H+-ATPase activity (Aue et al., 1999; Maurel et al., 2004). The branched phenotype of PHA1-OE plants could be due to an increased sucrose unloading towards the lateral buds via sucrose–H+ symporters energized by PHA1; however, more studies are required to determine the role of PHA1 in axillary bud outgrowth.
PHA1 promotes plant growth
Like PHA1-OE stolons, the stems of PHA1-OE plants grown in vitro elongate faster, and show longer internodes and more leaves than wild-type plants (Fig. 11A–D). Likewise, PHA1-OE plants grown in soil are taller than wild-type plants (Fig. 11G, H). Thus, PHA1 mediates growth in the aerial part of the plant as well as in underground stolons. Accordingly, inhibition of PM H+-ATPase activity leads to a reduction in stem length (Supplementary Fig. S8). There is much evidence for the role of PM H+-ATPase in plant growth based on studies using proton pump inhibitors; however, strong genetic evidence has so far been sparse. It was reported that overexpression of the unmodified PM H+-ATPase gene PMA4 in tobacco has no effect on plant growth; however, the constitutive expression of a constitutively activated form of PMA4 results in abnormal leaf inclination and twisted stems, suggesting alterations in cell expansion (Gévaudant et al., 2007). Another study has shown that the Arabidopsis aha2 mutant completes its life cycle without any observable growth alteration, and the reduced-growth phenotype only becomes apparent under stress conditions that reduce the transmembrane electrical gradient and/or external proton chemical gradient (Haruta and Sussman, 2012). The present study provides strong genetic evidence for the role of PHA1 as a driver of growth in potato plants, showing a clear enhanced-growth phenotype of PHA1-OE plants.
Supplementary data
Supplementary data are available at JXB online.
Table S1. List of primers used for semi-quantitative RT–PCR.
Table S2. Chromosome localization, coding region length, predicted protein length, and molecular weight of the PHA isoforms.
Fig. S1. Alignment of the protein sequences of PHA isoforms.
Fig. S2. Quantification of RT–PCR bands of Fig. 2A.
Fig. S3. RT–PCR analysis of PHA genes in stolons.
Fig. S4. Expression of PHA genes in response to abiotic stress.
Fig. S5. Effect of vanadate and erythrosine B on stolons cultured under non-inducing conditions.
Fig. S6. Stolon length of PHA1-OE plants cultured in the presence of PM H+-ATPase inhibitors.
Fig. S7. Phenotypic analysis of PHA1-OE plants.
Fig. S8. Effect of vanadate and erythrosine B on stem length.
Fig. S9. Branching phenotype of PHA1-OE plants.
Supplementary Material
Acknowledgements
This work was supported by grants from CONICET, ANPCYT, and the University of Buenos Aires. We thank Dr Mark Boutry for the kind gift of the YAK2 strain and plasmids for the complementation assay. We are grateful to Dra Silvia Fernández Villamil and Dr Guillermo Alonso for their gifts of reagents. We thank Dra Paula Portela for her assistance in the yeast assays.
Glossary
Abbreviations:
- EB
erythrosine B
- GA
gibberellic acid.
References
- Ahn S, Im Y, Chung G, Cho B. 1999. Inducible expression of plasma membrane H+-ATPase in the roots of figleaf gourd plants under chilling root temperature. Physiologia Plantarum 106, 35–40. [Google Scholar]
- Albertson PL, Peters KF, Grof CPL. 2001. An improved method for the measurement of cell wall invertase activity in sugarcane tissue. Functional Plant Biology 28, 323–328. [Google Scholar]
- Appeldoorn NJG, de Bruijn SM, Koot-Gronsveld EAM, Visser RGF, Vreugdenhil D, van der Plas L. 1997. Developmental changes involved in conversion of sucrose to hexose-phosphate during early tuberization of potato. Planta 202, 220–226. [Google Scholar]
- Arango M, Gévaudant F, Oufattole M, Boutry M. 2003. The plasma membrane proton pump ATPase: the significance of gene subfamilies. Planta 216, 355–365. [DOI] [PubMed] [Google Scholar]
- Aue HL, Lecomte I, Gendraud M, Petel G. 1999. Change in plasma membrane ATPase activity during dormancy release of vegetative peach-tree buds. Physiologia Plantarum 106, 41–46. [Google Scholar]
- Barbier FF, Lunn JE, Beveridge CA. 2015. Ready, steady, go! A sugar hit starts the race to shoot branching. Current Opinion in Plant Biology 25, 39–45. [DOI] [PubMed] [Google Scholar]
- Baxter I, Tchieu J, Sussman MR, Boutry M, Palmgren MG, Gribskov M, Harper JF, Axelsen KB. 2003. Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiology 132, 618–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpaneto A, Geiger D, Bambe1rg E, Sauer N, Fromm J, Hedrich R. 2005. Phloem-localized, proton-coupled sucrose carrier ZmSUT1 mediates sucrose efflux under the control of the sucrose gradient and the proton motive force. Journal of Biological Chemistry 280, 21437–21443. [DOI] [PubMed] [Google Scholar]
- Celis-Gamboa C, Struik PC, Jacobsen E, Visser RGF. 2003. Temporal dynamics of tuber formation and related processes in a crossing population of potato (Solanum tuberosum). Annals of Applied Biology 143, 175–186. [Google Scholar]
- Chen LQ, Qu XQ, Hou BH, Sosso D, Osorio S, Fernie AR, Frommer WB. 2012. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335, 207–211. [DOI] [PubMed] [Google Scholar]
- de Kerchove d’Exaerde A, Supply P, Dufour JP, Bogaerts P, Thinés D, Goffeau A, Boutry M. 1995. Functional complementation of a null mutation of the yeast Saccharomyces cerevisiae plasma membrane H+-ATPase by a plant H+-ATPase gene. Journal of Biological Chemistry 270, 23828–23837. [DOI] [PubMed] [Google Scholar]
- De Riek J, Piqueras A, Debergh PC. 1997. Sucrose uptake and metabolism in a double layer system for micropropagation of Rosa multiflora. Plant Cell, Tissue and Organ Culture 47, 269–278. [Google Scholar]
- Duby G, Boutry M. 2009. The plant plasma membrane proton pump ATPase: a highly regulated P-type ATPase with multiple physiological roles. Pflugers Archiv: European Journal of Physiology 457, 645–655. [DOI] [PubMed] [Google Scholar]
- Duby G, Poreba W, Piotrowiak D, Bobik K, Derua R, Waelkens E, Boutry M. 2009. Activation of plant plasma membrane H+-ATPase by 14-3-3 proteins is negatively controlled by two phosphorylation sites within the H+-ATPase C-terminal region. Journal of Biological Chemistry 284, 4213–4221. [DOI] [PubMed] [Google Scholar]
- Ehness R, Roitsch T. 1997. Co-ordinated induction of mRNAs for extracellular invertase and a glucose transporter in Chenopodium rubrum by cytokinins. The Plant Journal 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Ewing NN, Bennett AB. 1994. Assessment of the number and expression of P-type H+-ATPase genes in tomato. Plant Physiology 106, 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frías I, Caldeira MT, Pérez-Castiñeira JR et al. . 1996. A major isoform of the maize plasma membrane H+-ATPase: characterization and induction by auxin in coleoptiles. The Plant Cell 8, 1533–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaxiola RA, Palmgren MG, Schumacher K. 2007. Plant proton pumps. FEBS Letters 581, 2204–2214. [DOI] [PubMed] [Google Scholar]
- Gévaudant F, Duby G, von Stedingk E, Zhao R, Morsomme P, Boutry M. 2007. Expression of a constitutively activated plasma membrane H+-ATPase alters plant development and increases salt tolerance. Plant Physiology 144, 1763–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girault T, Abidi F, Sigogne M, Pelleschi-Travier S, Boumaza R, Sakr S, Leduc N. 2010. Sugars are under light control during bud burst in Rosa sp. Plant, Cell and Environment 33, 1339–1350. [DOI] [PubMed] [Google Scholar]
- Hager A. 2003. Role of the plasma membrane H+-ATPase in auxin-induced elongation growth: historical and new aspects. Journal of Plant Research 116, 483–505. [DOI] [PubMed] [Google Scholar]
- Harms K, Wöhner RV, Schulz B, Frommer WB. 1994. Isolation and characterization of P-type H+-ATPase genes from potato. Plant Molecular Biology 26, 979–988. [DOI] [PubMed] [Google Scholar]
- Haruta M, Sussman MR. 2012. The effect of a genetically reduced plasma membrane protonmotive force on vegetative growth of Arabidopsis. Plant Physiology 158, 1158–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Takahashi K, Inoue S, Kinoshita T. 2014. Abscisic acid suppresses hypocotyl elongation by dephosphorylating plasma membrane H+-ATPase in Arabidopsis thaliana. Plant and Cell Physiology 55, 845–853. [DOI] [PubMed] [Google Scholar]
- Henry C, Rabot A, Laloi M et al. . 2011. Regulation of RhSUC2, a sucrose transporter, is correlated with the light control of bud burst in Rosa sp. Plant, Cell and Environment 34, 1776–1789. [DOI] [PubMed] [Google Scholar]
- Herbers K, Sonnewald U. 1998. Molecular determinants of sink strength. Current Opinion in Plant Biology 1, 207–216. [DOI] [PubMed] [Google Scholar]
- Hohenwallner W, Wimmer E. 1973. The Malachite green micromethod for the determination of inorganic phosphate. Clinica Chimica Acta 45, 169–175. [DOI] [PubMed] [Google Scholar]
- Houlne G, Boutry M. 1994. Identification of an Arabidopsis thaliana gene encoding a plasma membrane H+-ATPase whose expression is restricted to anther tissues. The Plant Journal 5, 311–317. [DOI] [PubMed] [Google Scholar]
- Inoue SI, Takahashi K, Okumura-Noda H, Kinoshita T. 2016. Auxin influx carrier AUX1 confers acid resistance for arabidopsis root elongation through the regulation of plasma membrane H+-ATPase. Plant and Cell Physiology 57, 2194–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicka-Russak M, Kłobus G. 2007. Modification of plasma membrane and vacuolar H+-ATPases in response to NaCl and ABA. Journal of Plant Physiology 164, 295–302. [DOI] [PubMed] [Google Scholar]
- Kalampanayil BD, Wimmers LE. 2001. Identification and characterization of a salt stressed-induced plasma membrane H+-ATPase in tomato. Plant, Cell and Environment 24, 999–1005. [Google Scholar]
- Kanczewska J, Marco S, Vandermeeren S, Maudoux O, Rigaud JL, Boutry M. 2005. Activation of the plant plasma membrane H+-ATPase by phosphorylation and binding of 14-3-3 proteins converts a dimer into a hexamer. Proceedings of the National Academy of Sciences, USA 102, 11675–11680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolachevskaya OO, Alekseeva VV, Sergeeva LI, Rukavtsova EB, Getman IA, Vreugdenhil D, Buryanov YI, Romanov GA. 2015. Expression of auxin synthesis gene tms1 under control of tuber-specific promoter enhances potato tuberization in vitro. Journal of Integrative Plant Biology 57, 734–744. [DOI] [PubMed] [Google Scholar]
- Krügel U, He HX, Gier K, Reins J, Chincinska I, Grimm B, Schulze WX, Kühn C. 2012. The potato sucrose transporter StSUT1 interacts with a DRM-associated protein disulfide isomerase. Molecular Plant 5, 43–62. [DOI] [PubMed] [Google Scholar]
- Kühn C, Franceschi VR, Schulz A, Lemoine R, Frommer WB. 1997. Macromolecular trafficking indicated by localization and turnover of sucrose transporters in enucleate sieve elements. Science 275, 1298–1300. [DOI] [PubMed] [Google Scholar]
- Kühn C, Hajirezaei MR, Fernie AR, Roessner-Tunali U, Czechowski T, Hirner B, Frommer WB. 2003. The sucrose transporter StSUT1 localizes to sieve elements in potato tuber phloem and influences tuber physiology and development. Plant Physiology 131, 102–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn C, Quick WP, Schulz A, Riesmeier JW, Sonnewald U, Frommer WB. 1996. Companion cell-specific inhibition of the potato sucrose transporter SUT1. Plant, Cell and Environment 19, 1115–1123. [Google Scholar]
- Lefebvre B, Arango M, Oufattole M, Crouzet J, Purnelle B, Boutry M. 2005. Identification of a Nicotiana plumbaginifolia plasma membrane H+-ATPase gene expressed in the pollen tube. Plant Molecular Biology 58, 775–787. [DOI] [PubMed] [Google Scholar]
- Lemoine R, La Camera S, Atanassova R et al. . 2013. Source-to-sink transport of sugar and regulation by environmental factors. Frontiers in Plant Science 4, 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Loy JB. 1977. Hormonal regulation of cell division in the primary elongation meristems of shoots. In: Rost TL, Gifford EM Jr, eds. Mechanisms and control of cell division. Stroudsburg, PA: Dowden, Hutchinson, and Ross, Inc, 92–110. [Google Scholar]
- Mason MG, Ross JJ, Babst BA, Wienclaw BN, Beveridge CA. 2014. Sugar demand, not auxin, is the initial regulator of apical dominance. Proceedings of the National Academy of Sciences, USA 111, 6092–6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maudoux O, Batoko H, Oecking C, Gevaert K, Vandekerckhove J, Boutry M, Morsomme P. 2000. A plant plasma membrane H+-ATPase expressed in yeast is activated by phosphorylation at its penultimate residue and binding of 14-3-3 regulatory proteins in the absence of fusicoccin. Journal of Biological Chemistry 275, 17762–17770. [DOI] [PubMed] [Google Scholar]
- Maurel K, Leite GB, Bonhomme M, Guilliot A, Rageau R, Pétel G, Sakr S. 2004. Trophic control of bud break in peach (Prunus persica) trees: a possible role of hexoses. Tree Physiology 24, 579–588. [DOI] [PubMed] [Google Scholar]
- Michelet B, Boutry M. 1995. The plasma membrane H+-ATPase. Plant Physiology 108, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morandini P, Valera M, Albumi C, Bonza MC, Giacometti S, Ravera G, Murgia I, Soave C, De Michelis MI. 2002. A novel interaction partner for the C-terminus of Arabidopsis thaliana plasma membrane H+-ATPase (AHA1 isoform): site and mechanism of action on H+-ATPase activity differ from those of 14-3-3 proteins. The Plant Journal 31, 487–497. [DOI] [PubMed] [Google Scholar]
- Moriau L, Michelet B, Bogaerts P, Lambert L, Michel A, Oufattole M, Boutry M. 1999. Expression analysis of two gene subfamilies encoding the plasma membrane H+-ATPase in Nicotiana plumbaginifolia reveals the major transport functions of this enzyme. The Plant Journal 19, 31–41. [DOI] [PubMed] [Google Scholar]
- Morsomme P, Boutry M. 2000. The plant plasma membrane H+-ATPase: structure, function and regulation. Biomembranes 1465, 1–16. [DOI] [PubMed] [Google Scholar]
- Muñiz García MN, Muro MC, Mazzocchi LC, País SM, Stritzler M, Schlesinger M, Capiati DA. 2016. The protein phosphatase 2A catalytic subunit StPP2Ac2b acts as a positive regulator of tuberization induction in Solanum tuberosum L. Plant Molecular Biology 93, 227–245. [DOI] [PubMed] [Google Scholar]
- Muñiz García MN, País SM, Téllez-Iñón MT, Capiati DA. 2011. Characterization of StPPI1, a proton pump interactor from Solanum tuberosum L. that is up-regulated during tuber development and by abiotic stress. Planta 233, 661–674. [DOI] [PubMed] [Google Scholar]
- Muñiz García MN, Stritzler M, Capiati DA. 2014. Heterologous expression of Arabidopsis ABF4 gene in potato enhances tuberization through ABA–GA crosstalk regulation. Planta 239, 615–631. [DOI] [PubMed] [Google Scholar]
- Nelson N. 1994. A photometric adaptation of the Somogyi method for the determination of glucose. Journal of Biological Chemistry 153, 375–380. [Google Scholar]
- Niczyj M, Champagne A, Alam I, Nader J, Boutry M. 2016. Expression of a constitutively activated plasma membrane H+-ATPase in Nicotiana tabacum BY-2 cells results in cell expansion. Planta 244, 1109–1124. [DOI] [PubMed] [Google Scholar]
- Olivari C, Albumi C, Pugliarello MC, De Michelis MI. 2000. Phenylarsine oxide inhibits the fusicoccin-induced activation of plasma membrane H+-ATPase. Plant Physiology 122, 463–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oufattole M, Arango M, Boutry M. 2000. Identification and expression of three new Nicotiana plumbaginifolia genes which encode isoforms of a plasma-membrane H+-ATPase, and one of which is induced by mechanical stress. Planta 210, 715–722. [DOI] [PubMed] [Google Scholar]
- País SM, García MN, Téllez-Iñón MT, Capiati DA. 2010. Protein phosphatases type 2A mediate tuberization signaling in Solanum tuberosum L. leaves. Planta 232, 37–49. [DOI] [PubMed] [Google Scholar]
- Palmgren MG. 2001. Plant plasma membrane H+-ATPases: powerhouses for nutrient uptake. Annual Review of Plant Physiology and Plant Molecular Biology 52, 817–845. [DOI] [PubMed] [Google Scholar]
- Palmgren MG, Sommarin M, Serrano R, Larsson C. 1991. Identification of an autoinhibitory domain in the C-terminal region of the plant plasma membrane H+-ATPase. Journal of Biological Chemistry 266, 20470–20475. [PubMed] [Google Scholar]
- Papini R, De Michelis MI. 1997. Changes in the level and activation state of the plasma membrane H+-ATPase during aging of red beet slices. Plant Physiology 114, 857–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piette AS, Derua R, Waelkens E, Boutry M, Duby G. 2011. A phosphorylation in the C-terminal auto-inhibitory domain of the plant plasma membrane H+-ATPase activates the enzyme with no requirement for regulatory 14-3-3 proteins. Journal of Biological Chemistry 286, 18474–18482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross HA, Davies HV, Burch LR, Viola R, McRae D. 1994. Developmental changes in carbohydrate content and sucrose degrading enzymes in tuberizing stolons of potato (Solanum tuberosum). Physiologia Plantarum 90, 748–756. [Google Scholar]
- Roumeliotis E, Kloosterman B, Oortwijn M, Kohlen W, Bouwmeester HJ, Visser RG, Bachem CW. 2012. The effects of auxin and strigolactones on tuber initiation and stolon architecture in potato. Journal of Experimental Botany 63, 4539–4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu BB, Shaw BP. 2009. Salt-inducible isoform of plasma membrane H+ATPase gene in rice remains constitutively expressed in natural halophyte, Suaeda maritima. Journal of Plant Physiology 166, 1077–1089. [DOI] [PubMed] [Google Scholar]
- Santi S, Locci G, Monte R, Pinton R, Varanini Z. 2003. Induction of nitrate uptake in maize roots: expression of a putative high-affinity nitrate transporter and plasma membrane H+-ATPase isoforms. Journal of Experimental Botany 54, 1851–1864. [DOI] [PubMed] [Google Scholar]
- Sarkar D. 2008. The signal transduction pathways controlling in planta tuberization in potato: an emerging synthesis. Plant Cell Reports 27, 1–8. [DOI] [PubMed] [Google Scholar]
- Schumacher K, Krebs M. 2010. The V-ATPase: small cargo, large effects. Current Opinion in Plant Biology 13, 724–730. [DOI] [PubMed] [Google Scholar]
- Surowy TK, Boyer JS. 1991. Low water potentials affect expression of genes encoding vegetative storage proteins and plasma membrane proton ATPase in soybean. Plant Molecular Biology 16, 251–262. [DOI] [PubMed] [Google Scholar]
- Svennelid F, Olsson A, Piotrowski M, Rosenquist M, Ottman C, Larsson C, Oecking C, Sommarin M. 1999. Phosphorylation of Thr-948 at the C terminus of the plasma membrane H+-ATPase creates a binding site for the regulatory 14-3-3 protein. The Plant Cell 11, 2379–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thever MD, Saier MH Jr. 2009. Bioinformatic characterization of p-type ATPases encoded within the fully sequenced genomes of 26 eukaryotes. Journal of Membrane Biology 229, 115–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola R, Roberts AG, Haupt S, Gazzani S, Hancock RD, Marmiroli N, Machray GC, Oparka KJ. 2001. Tuberization in potato involves a switch from apoplastic to symplastic phloem unloading. The Plant Cell 13, 385–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser RGF, Vreugdenhil D, Hendriks T, Jacobsen EJ. 1994. Gene expression and carbohydrate content during stolon to tuber transition in potatoes (Solanum tuberosum). Physiologia Plantarum 90, 285–292. [Google Scholar]
- Wdowikowska A, Klobus G. 2016. The plasma membrane proton pump gene family in cucumber. Acta Physiologiae Plantarum 38, 135. [Google Scholar]
- Xu X, van Lammeren AA, Vermeer E, Vreugdenhil D. 1998. The role of gibberellin, abscisic acid, and sucrose in the regulation of potato tuber formation in vitro. Plant Physiology 117, 575–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Zhang D, Song T et al. . 2017. Arabidopsis plasma membrane H+-ATPase genes AHA2 and AHA7 have distinct and overlapping roles in the modulation of root tip H+ efflux in response to low-phosphorus stress. Journal of Experimental Botany 68, 1731–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.