OsABF1 mediates drought tolerance by directly up-regulating the expression of drought-associated genes, including COR413-TM1, and forms a complex feedback circuit in the drought/abscisic acid signaling pathway.
Keywords: bZIP, COR413-TM1, drought tolerance, OsABF1, rice, transcription factor
Abstract
Water deprivation causes substantial losses in crop yields around the world. In this study, we show that when overexpressed in transgenic rice (Oryza sativa), the bZIP transcription factor OsABF1 confers distinctly different drought-tolerance phenotypes when tethered to the transcriptional activator VP16 versus the transcriptional repressor EAR. We performed chromatin immunoprecipitation sequencing (ChIP-seq) and RNA sequencing (RNA-seq) assays on transgenic rice lines and determined that OsABF1 binds to DNA sequences containing an ACGT core motif. Analysis of the overlap between the RNA-sequencing and chromatin immunoprecipitation-sequencing data identified 242 OsABF1 target genes involved in multiple aspects of the drought response. Overexpression of one of these genes, COR413-TM1, which encodes a putative thylakoid membrane protein, resulted in a drought-tolerance phenotype without obvious side effects. In addition, OsABF1 directly regulates the expression of the protein phosphatase 2C (OsPP48 and OsPP108) and bZIP (OsbZIP23, OsbZIP46, and OsbZIP72) genes, thus forming a complex feedback circuit in the drought/abscisic acid signaling pathway.
Introduction
Globally, drought is the primary abiotic stress that limits crop yields. Breeders have capitalized on natural occurring genetic variation to improve crop yields under drought (Eisenstein, 2013). However, in some crops natural variations that can maximize productivity under drought stress may not exist. A better understanding of physiological and molecular responses of plant under drought stress enables us to improve plant performance by breeding and genetic engineering approaches.
A hierarchy of multiple transcription factors (TFs) mediates the plant response to water deprivation (Song et al., 2016). Among them, the basic leucine zipper (bZIP) TFs, containing a basic region for DNA binding and a leucine zipper motif for dimerization, belong to one of the largest TF families in eukaryotes. Plant bZIP TFs have been classified into three groups based on their DNA-binding specificity to G-box (CACGTG) or C-box (GACGTC) elements (Izawa et al., 1993). To date, at least 75 and 89 bZIP TFs have been identified in the Arabidopsis thaliana and rice (Oryza sativa) genomes, respectively, and these TFs have been further classified into 10 and 11 groups, respectively, according to sequence similarities of their basic DNA-binding region or their predicted DNA-binding preferences (Jakoby et al., 2002; Nijhawan et al., 2008). The bZIP TFs of the different groups play diverse and critical roles in abiotic stress responses, light signaling, flower development, pathogen defense, and seed maturation. The group A bZIP TFs in A. thaliana and group VI bZIP TFs in rice preferentially interact with abscisic acid (ABA)-responsive elements (ABREs), which contain the (T/G/C)ACGT(G/T)GC consensus sequence and are present in the promoter regions of ABA-inducible genes. Overexpression of these bZIPs, such as ABRE-binding Factor 3 (ABF3), OsbZIP23, or OsbZIP72, enhances abiotic stress tolerance in rice (Kang et al., 2002; Oh et al., 2005; Xiang et al., 2008; Lu et al., 2009), and mutations in OsABF1/OsbZIP12 or OsABF2/OsbZIP46 cause a hypersensitive response to drought and/or salinity stress (Amir Hossain et al., 2010a, 2010b). Interestingly, overexpression of a truncated form of OsbZIP46 with a deletion of an intrinsic transcriptional repression domain, but not the intact OsbZIP46, increase the tolerance of rice to drought/osmotic stress (Tang et al., 2012). However, it remains unclear how these ABRE-binding bZIP TFs target specialized downstream genes and function differentially or co-operatively in conferring tolerance to abiotic stresses. Extensive identification of the direct target genes of each bZIP TF will help elucidate their regulatory networks and how they bind to their specific targets and fine-tune gene expression in a spatio-temporal manner.
Previously, we described the approach of using hybrid transcription factors (HTFs), which cover 1500 rice transcription factors fused with the transcription activation module VP64 (a tetrameric repeat of VP16) or the repression module 4EAR (a tetrameric repeat of EAR) (Sadowski et al., 1988; Beerli et al., 1998; Wysocka and Herr, 2003; Imaizumi et al., 2005; Song et al., 2005; Song and Galbraith, 2006; Szemenyei et al., 2008; Pauwels et al., 2010; Kagale and Rozwadowski, 2011), to investigate the roles of different TFs in plant growth and development (Zhao et al., 2015). By screening more than 50 000 independent HTF transgenic events, we identified a bZIP transcription factor, named OsABF1, which confers distinctly different flowering time phenotypes when fused with VP64 (ABF1V) or 4EAR (ABF1E) (Zhang et al., 2016). Our previous study indicated that OsABF1 acts as a suppressor of floral transition in a photoperiod-independent manner. Simultaneous knockdown of both OsABF1 and its closest homologous gene, OsbZIP40, in rice by RNA interference results in a significantly earlier flowering phenotype. In this study, we further show that the OsABF1 RNAi and overexpression transgenic lines display distinctly different drought-tolerance phenotypes in response to osmotic stress. We performed RNA sequencing (RNA-seq) and chromatin immunoprecipitation sequencing (ChIP-seq) to determine the OsABF1 binding sequence and target genes, and to investigate its regulatory network in response to drought stress. Our results demonstrate that OsABF1-mediated up-regulation of COR413-TM1, an OsABF1 target gene that encodes a putative thylakoid membrane protein, may contribute to drought tolerance in rice. Moreover, OsABF1 regulates a variety of independent plant developmental processes that form a complex feedback circuit in the drought/abscisic acid signaling pathway.
Materials and methods
Plant material
The ABF1V, ABF1E, ABF1F, and OsABF1-RNAi transgenic plants were described in our previous study (Zhang et al., 2016). To generate the COR413-TM1 overexpression lines, COR413-TM1 cDNA was inserted into the pHCF vector at the PstI site using the Infusion system (Clontech). To generate the COR413-TM1-RNAi plants, a 272-bp fragment of the COR413-TM1 gene (from 284 to 555 bp) was inserted into the pANDA vector using the Gateway cloning system (Miki and Shimamoto, 2004). Each construct was introduced into Agrobacterium tumefaciens strain EHA105 and then transformed into rice cv. Kita-ake. The rice T-DNA insertion mutants osabf1-2 and osabf1-3 were obtained from the Salk Institute Genomic Analysis Laboratory (http://signal.salk.edu/cgi-bin/RiceGE) (Jeong et al., 2006).
Growth conditions and stress treatments
Seeds of the transgenic lines and the wild-type (WT) control were soaked in Petri dishes at 37 °C for 2 d. Uniformly germinated seeds were selected for hydroponic culture in bottomless 96-well plates in 1/10 Murashige and Skoog (MS) culture solution at 28 °C with a long-day (14 h light, 28 °C; 10 h dark, 24 °C) photoperiod. For the polyethylene glycol (PEG) osmotic treatment, 3-week-old seedlings were transferred to 20% PEG 4000 solution for the days indicated and then returned to the culture solution for recovery. For the drought treatment, the indicated rice genotypes were sown and cultured with 1/10 MS culture solution in transparent boxes for 2 weeks and then transferred to boxes containing wet soil. When plants were 4 weeks old, irrigation was stopped for the indicated number of days, and then water was added to the boxes for recovery.
To analyse the expression of OsABF1 and COR413-TM1 under different abiotic stresses and hormone treatments, seedlings of each genotype were cultivated under continuous light at 28 °C in a plant growth chamber. At 3 weeks old, seedlings were subjected to different abiotic treatments including: 20% PEG 4000, 200 mM NaCl, 1% H2O2, cold at 4 °C, or heat at 42 °C. Hormone treatments (0.1 mM ABA, gibberellic acid, 6-benzylaminopurine, jasmonic acid, 2,4-D, or Kinetin) were performed by addition into the culture solution and spraying on the leaves (Tang et al., 2012). The latest fully expanded leaves of the plants for each treatment were harvested in a time-course and used for RNA analysis.
ChIP assay
ABF1V-1 and WT plants grown under continuous light (CL) were used for ChIP assays, which were performed as described previously (Zhang et al., 2016). Briefly, 3 g of leaves from 4-week-old seedlings were cross-linked twice by 1% formaldehyde under vacuum for 15 min and stopped using 100 mM glycine. Then the samples were ground to powder in liquid nitrogen prior to isolating chromatin. After sonication, the chromatin complexes were incubated with anti-VP16 or anti-OsABF1 antibody (Zhang et al., 2016). The precipitated DNA was recovered in water for quantitative real-time PCR (qPCR) or ChIP-seq. For ChIP-qPCR, the enrichment value was normalized to that of input DNA (% of input).
ChIP-seq data analysis
ChIP-seq clean reads were mapped to the O. sativa ssp. japonica reference genome (Oryza sativa MSU 6.19), which was downloaded from the ENSEMBL site (http://www.ensembl.org/index.html) after removing adaptor and low-quality nucleotides using the Bowtie2 program with parameters -x Genome.fa-U single.fq-S sample.sam-N1 (Yates et al., 2016). Only the unique map reads were used for peak identification, and the ABF1V transgenic line and WT peaks were obtained using the model-based analysis software MACS with parameters -t sample.bam -f BAM -g 370000000 -n sample.name -w -p 1e-5 (Zhang et al., 2008). ABF1V-associated genes were defined by the following criteria: (a) the overlapped length of peaks with the 1000 bp upstream of the initiation codon and coding sequence region was larger than 50% compared with peak length; (b) the value of -10[log10(P-value)] was larger than 80; (c) fold enrichment (FE) was larger than 5; and (d) if a peak was shared between the ABF1V transgenic line and the WT, the FE ratio between the ABF1V line and the WT should be larger than 2. To determine the consensus DNA-binding motif, peaks in the ABF1V line and the WT were analysed using the motif-based sequence analysis tool MEME-ChIP (Machanick and Bailey, 2011). The candidate motifs were defined by the following criteria: (a) the size of the candidate motif was set from 5 to 30 bp; (b) a motif in a similar sequence was counted once at most; (c) the e-value of the candidate motif was lower than 1.0E–005; and (d) a candidate motif could be present on both sense and anti-sense strands.
DNA binding assay
The qPCR-based in vitro protein and DNA interaction assay was performed according to a previous method with minor modifications (Meng et al., 2013). Briefly, the OsABF1 coding DNA sequence (CDS) was cloned into the pCold TF plasmid at the HindIII site and transformed into the E. coli host strain (BL21) for the expression of recombinant protein according to the manual (TaKaRa, Cat.# 3365, v.0708). The E. coli cells were collected and suspended with lysis buffer [50 mM Tris (pH 8.0), 500 mM NaCl, 1 mM PMSF, and 20 mM β-mercaptoethanol], sonicated, and centrifuged at maximum speed for 1 h at 4 °C. The supernatant was diluted with lysis buffer at the designated amount and 700 μl of diluted supernatant was mixed with 50 μl of Dynabeads His-Tag beads (Life technologies, Cat. #10104D), incubated on a rotator for 5 min at room temperature, and washed five times with 1 ml washing buffer [500 mM NaCl, 50 mM Tris (pH 8.0), and 20 mM imidazole]. The beads were suspended in 50 μl of washing buffer for later use and the amount of bound protein was measured by the Bradford method. DNA fragments containing the indicated elements or non-correlated DNA fragments were synthesized and diluted to the designated concentrations. The DNA and protein binding reaction was conducted by mixing 10 μl of DNA fragments, 10 μl of beads, and 5 μl of 5×DNA binding buffer [20% glycerol, 2.5 mM DTT, 250 mM KCl, 1 mg ml–1 BSA, 50 mM Tris (pH 7.5), and 5 mM MgCl2] in 1.5-ml microcentrifuge tubes and was incubated at room temperature for 10 min. Then the beads were washed 10 times with 2×DNA binding buffer. The DNA–protein complex was eluted with elution buffer [500 mM NaCl, 50 mM Tris (pH 8.0), and 20 mM imidazole] and diluted 100-fold prior to qPCR analysis.
RNA-seq and data analysis
The WT plants and the ABF1V-1 and ABF1E-1 transgenic lines were cultivated under continuous light at 28 °C for 4 weeks in plant growth chambers. The WT drought (WT-D) plants were prepared by submerging the plants in 20% PEG solution for 4 h. Ten most-recently emerged and fully expanded leaves of each genotype were collected for RNA extraction. The sequencing library was constructed following the manufacturer’s instructions (Illumina Inc.). Paired-end sequencing libraries with an insert size of approximately 200 bp were sequenced on an Illumina HiSeq 2000 sequencer at the ANOROAD Company in Beijing. RNA-seq clean reads of three biological replicates were mapped to the O. sativa ssp. japonica reference genome after removing adaptor and low-quality nucleotides by TopHat (Trapnell et al., 2009). The expression value was calculated in FPKM (fragments per kilobase of exon model per million mapped fragments) and the differentially expressed genes were further analysed by Cuffdiff (q<0.05) (Trapnell et al., 2009, 2010). Differentially expressed genes were defined as those with fold changes ≥2, or ≤2/3.
A list of all primers used in this study is provided in Supplementary Table S1 at JXB online.
Accession Numbers
Sequence data from this article can be found in the MSU Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/analyses_search_locus.shtml) databases (Kawahara et al., 2013), under the following accession numbers: OsABF1 (LOC_Os01g64730), COR413-TM1 (LOC_Os05g49170), HOX24 (LOC_Os02g43330), AUMO1 (LOC_Os03g05880), OsEGY3 (LOC_Os03g51920), OsPP48 (LOC_Os03g16170), OsPP108 (LOC_Os09g15670), LEA14 (LOC_Os01g12580), OsbZIP23 (LOC_Os02g52780), OsbZIP46 (LOC_ Os06g10880), and OsbZIP72 (LOC_ Os09g28310). The RNA-seq and ChIP-seq data were deposited in the Gene Expression Omnibus with accession number SRP057432.
Results
Overexpression of OsABF1 enhances drought tolerance in rice
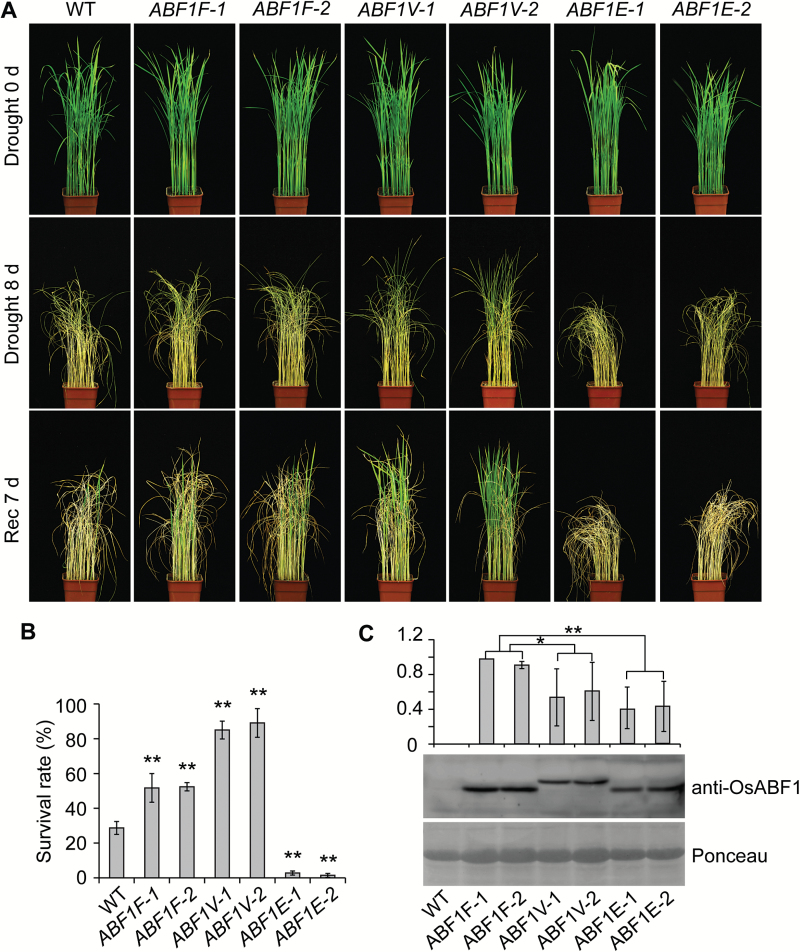
It has been reported that two T-DNA insertion mutants in the OsABF1 gene, osabf1-1 (in the O. sativa cv. Hwayong background) and osabf1-2 (in the O. sativa cv. Dongjin background; see Supplementary Fig. S1A), are more vulnerable to salinity and drought treatments, compared with wild-type rice (Amir Hossain et al., 2010b). Here, we showed that knockdown of OsABF1 in the OsABF1-RNAi transgenic lines (Zhang et al., 2016) caused hypersensitivity to drought treatment, compared to the wild-type (WT) plants (O. sativa cv. Kita-ake; Supplementary Fig. S1B, C). Our results suggest that OsABF1 is a universal positive regulator of drought tolerance in rice. To test this hypothesis, we examined the performance of transgenic lines expressing Flag-tagged OsABF1 and OsABF1 fused to either a transcriptional activation domain or a repression domain: Pubi:OsABF1-3Flag (ABF1F), Pubi:OsABF1-VP64 (ABF1V) (transcriptional activation of OsABF1), or Pubi:OsABF1-4EAR (ABF1E) (transcriptional repression of OsABF1) (see Supplementary Fig. S2A). We compared these transgenic lines with WT Kita-ake rice plants under water-deprivation conditions (Huang et al., 2009). The ABF1F and ABF1V lines were significantly more tolerant to the treatment, with survival rates of about 1.8-fold and 3.0-fold higher than that of the WT, respectively. By contrast, the ABF1E lines were more vulnerable to drought stress than the WT (Fig. 1A, B and Supplementary Fig. S2B, C). We also used the PEG treatment to simulate drought conditions and got similar results to the water-deprivation treatment (Supplementary Fig. S3). Interestingly, although ABF1F proteins were more abundant in the ABF1F lines than ABF1V proteins in the ABF1V lines (Fig. 1C), the ABF1F lines were more sensitive to drought stress than the ABF1V lines, suggesting that the increase of the transcriptional activation activity of OsABF1 in the ABF1V transgenic line enhanced the performance of rice under drought stress. We also compared the OsABF1 protein level of the WT under water deprivation conditions to the OsABF1 transgenic lines. However, due to the low sensitivity of anti-OsABF1, OsABF1 protein was not detectable in the WT, which indicated that the protein level in the OsABF1 transgenic lines was much higher than that in the WT under stressed conditions.
Fig. 1.
Overexpression of ABF1F, ABF1V, or ABF1E confers different degrees of osmotic-stress sensitivity in rice. (A) Representative images of indicated genotypes subjected to drought treatment. Plants were grown under long days (LDs, 14 h light/10 h dark) for 3 weeks before water deprivation. The images were taken before the water deprivation treatment (drought 0 d), after 8 d of water deprivation (drought 8 d), and then after water had been restored for 7 d (Rec 7 d). (B) Survival rate of each genotype subjected to water deprivation treatments as described in (A). Mean values ±SD are shown. The values of the indicated genotypes were compared to that of the WT (Student’s t-tests, **P<0.01, n=3). (C) Protein expression analysis of ABF1V, ABF1E, and ABF1F in the indicated lines. The immunoblot was probed with anti-OsABF1 antibody. The relative abundance of OsABF1 in each genotype was calculated by the formula: (OsABF1/Ponceau [each genotype])/(OsABF1/Ponceau [ABF1F-1]). The relative abundance of OsABF1 in ABF1F-1 was arbitrarily set to 1. Mean values ±s.e.m. (standard error of the mean) are shown (Student’s t-tests, *P<0.05, **P<0.01, n=3).
Genome-wide analysis of ABF1V-associated genes by ChIP-seq
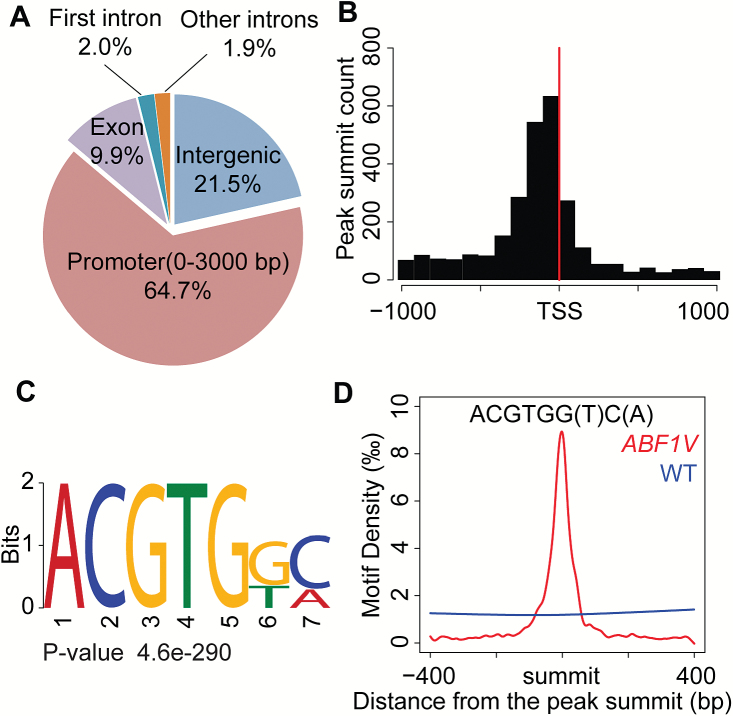
To determine the direct target genes of OsABF1, we used the ABF1V-1 transgenic line and anti-VP16 antibodies to perform chromatin immunoprecipitation and high-throughput sequencing (ChIP-seq) (see Supplementary Fig. S4). The WT Kita-ake was used as the negative control. In total, we identified 3882 genes with binding peaks localized in the promoter or CDS regions (see Supplementary Table S2). Analysis of the distribution of the sites in the rice genome demonstrated that 64.7% of the OsABF1 binding sites are located in the promoter regions, and only 35.3% are in the gene body and intergenic regions (Fig. 2A). The distance from each peak to the nearest transcription start site (TSS) was calculated and is depicted in a histogram in Fig. 2B, revealing the enrichment of association sites within 200 bp upstream of the TSSs. Furthermore, our analysis of the OsABF1 binding sequences detected an over-representation of the ACGTG(G/T)(C/A) consensus sequence (Fig. 2C). A density-plot analysis revealed that this motif was strictly enriched in the peak summits in the ABF1V line and the enrichment measurements returned rapidly to background levels outside ±200 bp of the peak summits. In contrast, the density was evenly distributed below 0.2% in the WT control (Fig. 2D). The ACGTG(G/T)(C/A) motif identified here is similar to the previously identified ABRE motif (T/G/C/)ACGT(G/T)GC, in that both contain an ACGT core (Ono et al., 1996; Hobo et al., 1999; Nijhawan et al., 2008). The variation of bases surrounding the ACGT core may determine the binding specificity of individual bZIP TFs.
Fig. 2.
Identification of the ABF1V binding sites by analysis of the ChIP-seq data. (A) Overview of ABF1V binding peak distribution in the rice genome. (B) Enrichment of ABF1V binding peaks in the promoter region. TSS, transcriptional start site. (C) Consensus sequence identified in the ABF1V binding peaks by the MEME-ChIP program. (D) Density plot of the ACGTG(G/T)(C/A) motif around the summits of overlapping peaks in the ChIP-seq data of the ABF1V line drawn by the R program. The density curve of WT ChIP-seq data was used as a negative control.
Dynamic affinity of OsABF1 with its target sequences
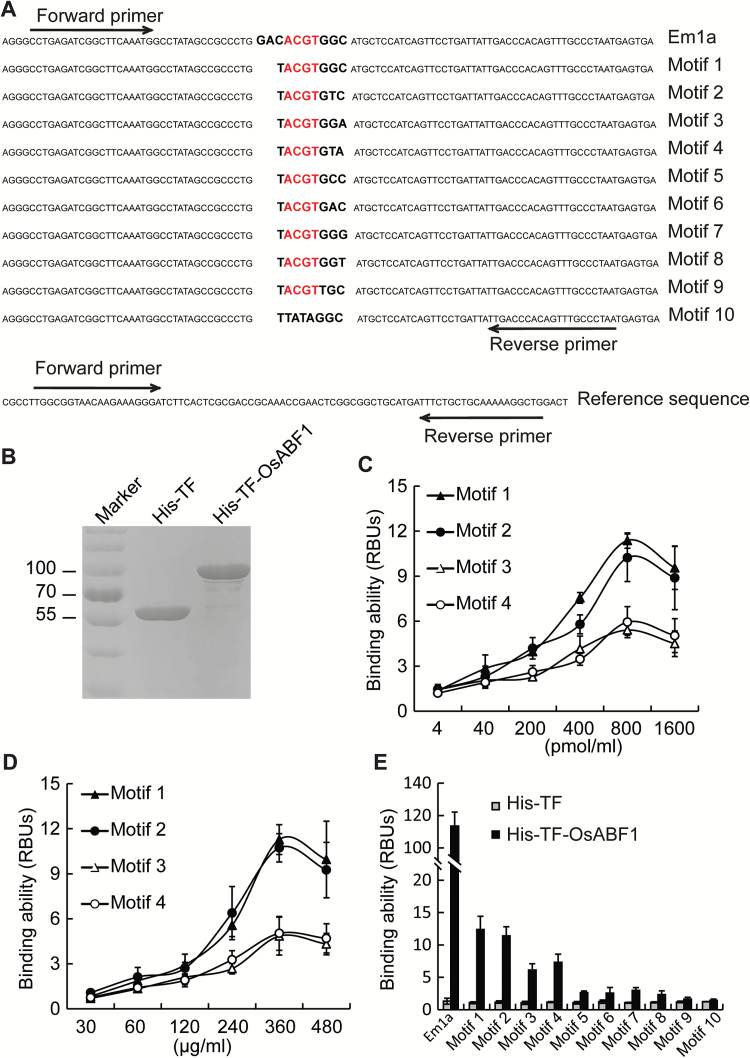
To test if the bases immediately flanking the ACGT core affect the binding specificity of OsABF1, we analysed the binding ability of OsABF1 to different sequences containing an ACGT core via a qPCR-based protein-DNA binding assay (Meng et al., 2013). DNA sequences containing the indicated motifs in conjunction with a non-specific reference sequence were synthesized commercially (Fig. 3A). The OsABF1 CDS was cloned into the pCold TF plasmid. The His-TF (His-tagged Trigger Factor Chaperone) control protein and the His-TF-OsABF1 recombinant protein were expressed and purified from the E. coli host strain BL21 (Fig. 3B). To find an optimal DNA concentration for the assay, we tested the binding ability of His-TF-OsABF1 with different concentrations of motif 1 (ACGTGGC), motif 2 (ACGTGTC), motif 3 (ACGTGGA), and motif 4 (ACGTGTA) (Fig. 3C). The relative binding units (RBUs) gradually reached a peak with an increase in DNA concentration from 4 to 800 pmol ml–1, but then slightly decreased with a higher DNA concentration of 1600 pmol ml–1 (Fig. 3C). To find the appropriate protein concentration for the assay, we tested the binding ability of His-TF-OsABF1 to the indicated motif with increasing concentrations of protein (Fig. 3D). The RBUs increased gradually as the protein concentrations increased from 30 to 360 μg ml–1, but decreased when 480 μg ml–1 of protein was used. Both assays in Fig. 3C and D demonstrated that His-TF-OsABF1 has a greater ability to bind motif 1 and 2, compared with motif 3 and 4 (Factorial analysis of variance, P<0.001, n=3). When either nucleotide G or T is in the second position following the ACGT core (ACGTGGX and ACGTGTX), the OsABF1 binding ability remained the same; however, its binding ability was enhanced when C versus A is in the third position following the ACGT core (ACGTGXC vs. ACGTGXA). We investigated the binding ability of OsABF1 to 11 motif variations under the experimentally determined optimal conditions described above, using 800 pmol ml–1 of the DNA probe and 360 μg ml–1 of His-TF-OsABF1 protein (Fig. 3E). The results indicated that OsABF1 had the highest binding ability to the previously identified ABRE complex sequence named Em1a (Hobo et al., 1999), which contains an ACGTGGC motif in the sense strand and an ACGTGTC motif in the antisense strand, suggesting that OsABF1 strongly prefers the site containing the ACGTG(G/T)C sequence. Moreover, the replacement of G/T with C/A (in motif 5 or 6) two nucleotides after the ACGT core significantly reduced the binding ability (Student’s t-test, P<0.001, n=3). The same reduction in binding ability was observed with the replacement of C/A with G/T (in motif 7 or 8) three nucleotides after the ACGT core. Furthermore, the replacement of G with T (in motif 9) immediately after the ACGT core completely stopped the interaction. Taken together, these results demonstrated that the bases immediately flanking the ACGT core are critical for the binding specificity of OsABF1 in vitro.
Fig. 3.
Quantitative PCR (qPCR)-based in vitro DNA binding assay. (A) DNA sequences containing the indicated motifs and an unrelated DNA sequence (reference sequence) used as an internal control are shown. Forward and reverse arrows indicate the primers used for qPCR. (B) The purified His-TF and His-TF-OsABF1 recombinant proteins were separated on an SDS-PAGE gel and stained with Coomassie Brilliant Blue. His-TF indicates the His-tagged trigger factor chaperone, which facilitates co-translational folding of newly expressed polypeptides. (C) Binding ability of His-TF-OsABF1 to increasing concentrations of motif 1, 2, 3, or 4. The relative binding units (RBUs) were calculated using the formula: [RBU=2–△Ct, △Ct=Ct(Motif)–Ct(Reference); Ct (cycle threshold) represents the number of cycles required for the fluorescence signal to exceed the background level]. Mean values ±s.e.m. are shown (n=3). (D) Binding ability of each motif to increasing concentrations of His-TF-OsABF1 protein. RBUs were calculated as in (C). Mean values ±s.e.m. (standard error of the mean) are shown (n=3). € Binding ability of His-TF-OsABF1 to each motif. His-TF was used as a negative control.
OsABF1 regulates the transcription of genes associated with a variety of independent plant developmental processes
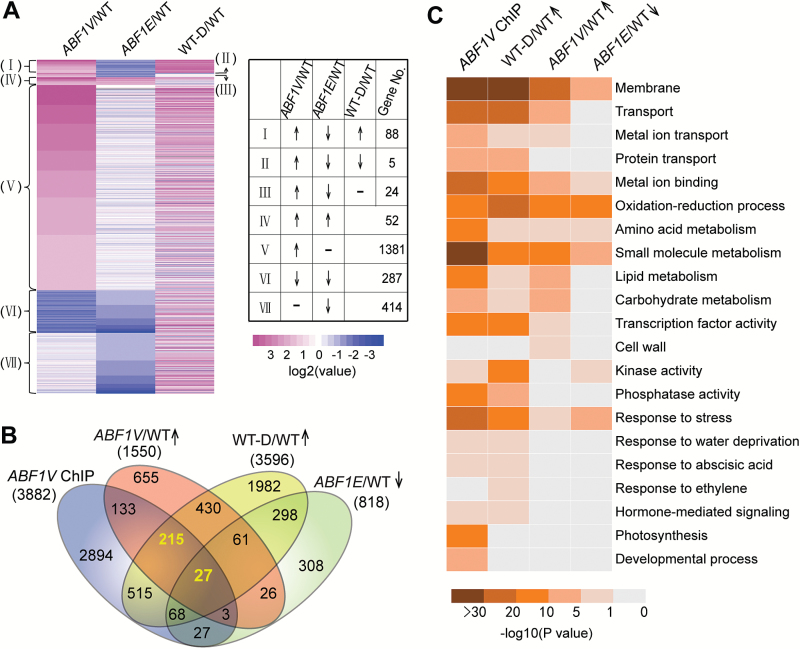
Next, we sought to identify genes under the regulation of OsABF1 (or its HTFs) using the RNA-seq assay of the WT and the ABF1V-1 and ABF1E-1 transgenic plants grown under normal conditions, along with the WT plants treated with PEG to simulate drought conditions (WT-D) (Fig. 4 and Supplementary Fig. S5). The transcript levels from each sample were normalized to the WT expression levels. The numbers of up- and down-regulated genes were, respectively, 171 and 818 in the ABF1E line, 1550 and 2255 in the ABF1V line, and 3596 and 4089 in the WT-D (see Supplementary Tables S3, S4, and S5). The number of differentially expressed genes (DEGs) in the ABF1E line was much lower than that in the ABF1V line, suggesting that the transcriptional activation activity of OsABF1 was largely suppressed or even reversed by fusion with the 4EAR effector. To find genes associated with the abnormal phenotypes of the OsABF1 HTF transgenic lines, we classified the DEGs into seven groups (Fig. 4A). Among the 1550 up-regulated genes in the ABF1V line (Groups I–V), 117 were down-regulated in the ABF1E line (Groups I–III), which may account for the opposite phenotypes between the ABF1V and ABF1E lines (Fig. 1). More than 75% of these 117 genes were also up-regulated in the WT-D (Group I). In addition, cluster analysis of gene expression profiles demonstrated that the ABF1E line grouped together with the WT, but the ABF1V line was closer to the WT-D (see Supplementary Fig. S5). These results support the hypothesis that a major function of OsABF1 is to mediate drought responses through transcriptional activation.
Fig. 4.
Identification of OsABF1-regulated genes by analysis of RNA-seq and ChIP-seq data. (A) Heatmap of differentially expressed genes. The ABF1V-1 and ABF1E-1 transgenic lines and the WT were grown under continuous light for 28 d. WT-D represents the WT sample grown under continuous light for 28 d and then treated with 20% PEG for 4 h. The color scale indicates the log-ratio calculated by normalizing the expression value of the gene in each sample to that in the WT. The differentially expressed genes were classified into seven groups from I to VII. ↑, up-regulated genes; ↓, down-regulated genes; –, unchanged genes. (B) Venn diagram showing the overlapping profiles among ABF1V-associated genes by ChIP-seq, up-regulated genes in the ABF1V transgenic line, up-regulated genes in the WT-D treatment, and down-regulated genes in the ABF1E line in comparison to that of the WT by RNA-seq. (C) Enriched functional categories in ABF1V-bound genes and in up-regulated genes in the WT-D treatment and ABF1V transgenic line, and down-regulated genes in the ABF1E transgenic line.
Analyses of the overlap between the RNA-seq and ChIP-seq data revealed that 242 OsABF1 binding genes were up-regulated in both the ABF1V transgenic line and the WT-D (Fig. 4B). These genes were regarded as candidate target genes under the direct regulation of OsABF1 in response to drought stress. Ontology analysis revealed that genes bound by OsABF1 and genes up-regulated by drought stress are both enriched in the pathways related to membrane, transport, oxidation–reduction processes, metabolism, transcription factor activity, kinase activity, phosphatase activity, and stress response (Fig. 4C), suggesting that OsABF1 regulates the transcription of genes associated with a variety of independent plant developmental processes. We selected seven candidate target genes based on their association with these pathways to examine their transcription profiles and in vivo binding with OsABF1 by quantitative reverse transcription PCR (qRT-PCR) and ChIP-qPCR. The transcription of all seven candidate genes was up-regulated in the ABF1V line and the WT-D, but down-regulated in the ABF1E line (Fig. 5A and Supplementary Figs S6A–S11A). Moreover, OsABF1 physically interacted with at least one site containing the ACGTG(G/T)(C/A) motif localized within the promoter region of each of the seven candidate genes (Fig. 5B–D and Supplementary Figs S6B–D to S11B–D).
Fig. 5.
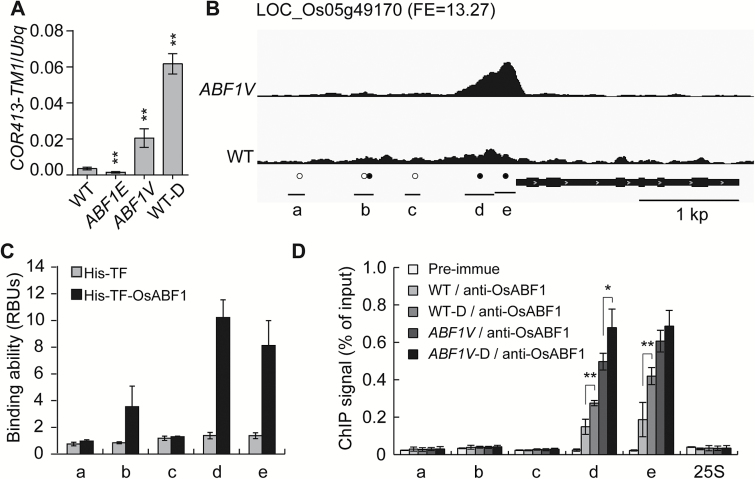
Identification of COR413-TM1 as a direct target of OsABF1. (A) Transcriptional analysis of COR413-TM1 in the WT, the ABF1E and ABF1V transgenic lines, and the WT-D treatment by quantitative reverse transcription PCR (qRT-PCR) with normalization to Ubiquitin-1 (Ubq). Mean values ±s.e.m. (standard error of the mean) are shown (Student’s t-tests, **P<0.01, n=3). (B) ABF1V binding profile in the promoter of COR413-TM1 in the ABF1V line and the WT ChIP-seq data. The bars at the bottom represent the distribution of DNA fragments containing the ACGTGG(T)C(A) motif or ACGT core, as indicated by black or white dots. (C) In vitro binding assay of His-TF-OsABF1 recombinant protein to the indicated DNA fragments, as shown in (B). His-TF recombinant protein was used as a negative control. (D) Verification of the OsABF1 direct binding sites in the COR413-TM1 promoter by ChIP-qPCR analysis. ChIP samples were prepared using the ABF1V line and the WT plants under normal conditions or under drought stress, and precipitated with anti-OsABF1 antibodies. The pre-immune serum was used as a negative control. Results of ChIP-qPCR were quantified by normalization of the immunoprecipitation signal with the corresponding input signal. The binding to 25S rDNA was used as a negative control. The means ±SD (n=3) are shown.
COR413-TM1 is a direct target of OsABF1
Membrane and stress response functional categories were highly enriched in genes bound by OsABF1, up-regulated in drought conditions, up-regulated in the ABF1V line, and down-regulated in the ABF1E line (Fig. 4C). Therefore, we further investigated one of the seven experimentally verified genes, COR413-TM1, which encodes a cold-inducible thylakoid membrane protein (Breton et al., 2003). Its mRNA level decreased about 60% in the ABF1E line, increased about 4-fold in the ABF1V line, and increased 10-fold in the WT-D compared with the WT (Fig. 5A). ChIP-seq data showed a sharp binding peak at its promoter region, which has five sites containing an ACGTG(G/T)(C/A) motif or an ACGT core sequence (Fig. 5B). In vitro qPCR-based DNA binding assays indicated that recombinant His-TF-ABF1 protein efficiently binds to the b, d, and e DNA fragments of COR413-TM1 (Fig. 5C). In vivo ChIP experiments using an ABF1V transgenic plant with anti-OsABF1 antibodies showed that ABF1V robustly binds to the d and e sites, which are immediately upstream of the TSS (Fig. 5D). Furthermore, we performed ChIP experiments using WT plants with anti-OsABF1 antibodies. Similar robust binding signals were also observed at the d and e sites, providing further validation that COR413-TM1 is a direct target gene under the regulation of OsABF1 in vivo. The observation that OsABF1 binds to the b site in vitro (Fig. 5C) but not in vivo (Fig. 5D) suggested that other factors may co-operate with OsABF1 in determining the target genes in plants. Consistent with this hypothesis, the finding that COR413-TM1 is a direct target gene of OsbZIP71, which preferentially binds to G-box (CACGTG) motifs (Liu et al., 2014), further suggests that co-regulation of COR413-TM1 expression by bZIP transcription factors is integral to the drought signaling cascade.
OsABF1-mediated up-regulation of COR413-TM1 in response to osmotic stress
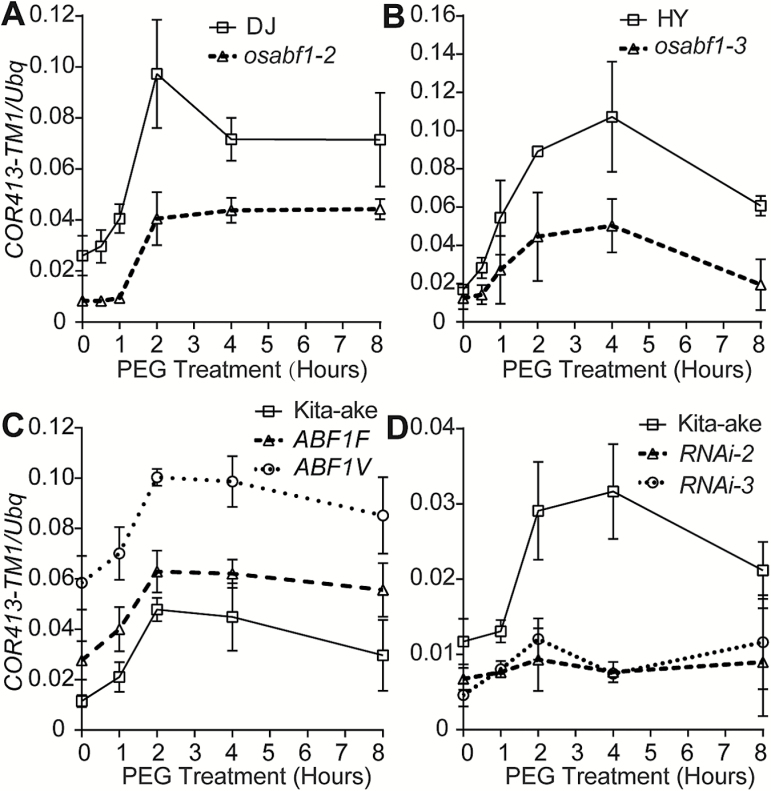
To investigate the role of OsABF1 in the regulation of COR413-TM1 expression in response to osmotic stress, we compared the COR413-TM1 mRNA levels in the OsABF1 loss-of-function mutants osabf1-2 and osabf1-3, and their respective WT control under PEG treatment (Fig. 6). The expression of COR413-TM1 in the WT control was robustly induced by PEG treatment within 4 h. In contrast, the osmotic stress-induced expression of COR413-TM1 was partially impaired in the osabf1-2 and osabf1-3 mutants (Fig. 6A, B), suggesting that OsABF1 functions redundantly with other genes in activating the expression of COR413-TM1. Consistent with this hypothesis, we were not able to induce COR413-TM1 mRNA levels by PEG treatment in the OsABF1 RNAi lines (RNAi-2 and RNAi-3) (Fig. 6D), in which both OsABF1 and its homolog OsbZIP40 (Zhang et al., 2016), are knocked out. Taken together, these results suggest that OsABF1 is required for full activation of COR413-TM1 transcription in response to osmotic stress.
Fig. 6.
Loss of function of OsABF1 impaired induction of COR413-TM1 expression in response to osmotic stress. (A) Dynamic changes of COR413-TM1 mRNA levels in the osabf1-2 mutant and wild-type Dongjin (DJ) rice. (B) Dynamic changes of COR413-TM1 mRNA levels in the osabf1-3 mutant and wild-type Hwayoung (HY) rice. (C) Dynamic changes of COR413-TM1 mRNA levels in the OsABF1F and OsABF1V lines and wild-type Kita-ake rice. (D) Dynamic changes of COR413-TM1 mRNA levels in the OsABF1 RNAi lines and wild-type Kita-ake rice. Mean values ±SD of three replicates are shown.
Overexpression of COR413-TM1 confers a drought-tolerant phenotype
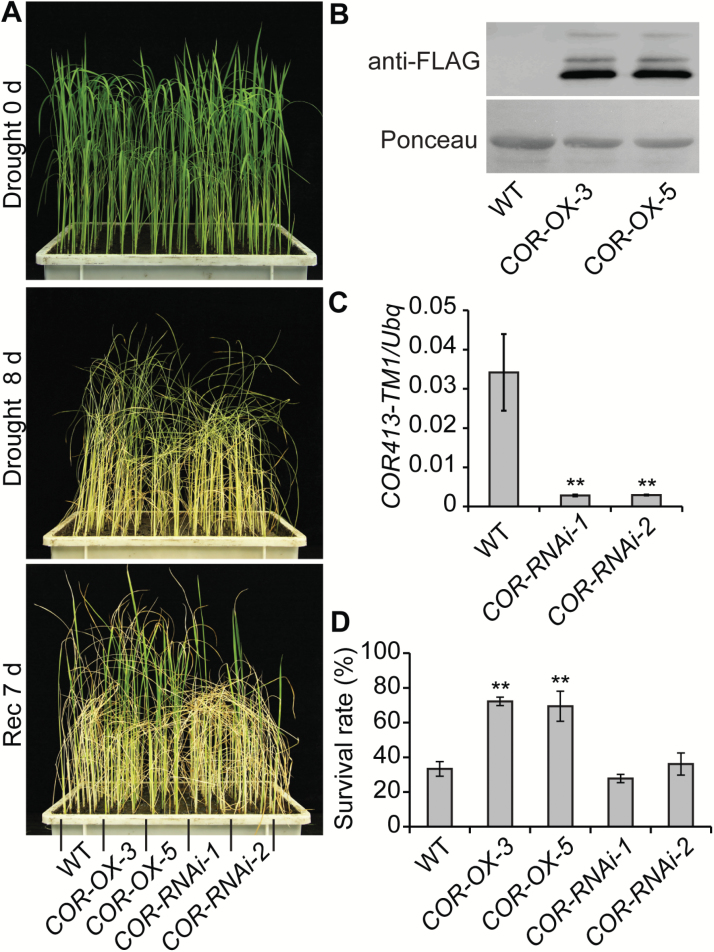
To test if COR413-TM1 could play a role in the response to drought in rice, we obtained 28 overexpression transgenic lines (COR-OXs), carrying the construct Pubi:COR413-TM1-Flag. The osmotic stress assays demonstrated that 25 out of the 28 lines were more tolerant to both drought and PEG treatments than the WT (two representative lines are shown in Fig. 7A and Supplementary Fig. S12A). Overexpression of COR413-TM1-Flag protein in the transgenic lines was verified by western blot assays using anti-Flag antibodies (Fig. 7B). The survival rates of the transgenic lines were significantly higher than that of the WT under osmotic stress (Fig. 7D and Supplementary Fig. S12B). In addition, the transgenic lines showed no other obvious abnormal phenotypes, underscoring the potential utility of COR413-TM1 to improve crop performance in drought conditions (Supplementary Fig. S13). The RNA interference (RNAi) approach was also applied to knockdown COR413-TM1. qRT-PCR analysis revealed that the expression level of COR413-TM1 significantly decreased in the COR413-TM1 RNAi lines (Fig. 7C); however, the performance of the RNAi lines under osmotic stress was similar to that of the WT (Fig. 7A, D), suggesting that functional redundancy exists between COR413-TM1 and other genes. Taken together, these results support the hypothesis that the up-regulation of COR413-TM1 at least partially contributes to drought tolerance mediated by OsABF1 in rice.
Fig. 7.
Overexpression of COR413-TM1 confers a drought-tolerant phenotype. (A) Images of the WT, the COR413-TM1 overexpression lines (COR-OX-3 and COR-OX-5), and the RNAi lines (COR-RNAi-1 and COR-RNAi-2) subjected to drought conditions. The 28-d-old seedlings were treated under drought conditions for 8 d (Drought 8 d) and then recovered by watering for 7 d (Rec 7 d). (B) Protein expression analysis of COR413-TM1-Flag in the indicated genotypes by immunoblot probed with anti-Flag antibodies. (C) Transcriptional analysis of COR413-TM1 by qRT-PCR. Mean values ±s.e.m. (standard error of the mean) are shown (Student’s t-tests, **P<0.01, n=3). (D) Survival rate of the indicated genotypes subjected to drought treatment as in (A). Mean values ±SD are shown. The value of the indicated genotype was compared to that of the WT (Student’s t-tests, **P<0.01, n=3).
Discussion
Improving drought tolerance in crops is important for sustainability in agriculture. The utilization of TFs to enhance drought tolerance in crops often results in unfavorable side effects because TFs can target thousands of downstream genes. For example, overexpression of OsABF1 significantly improved rice performance under drought conditions but also led to a severe late-flowering phenotype. Therefore, it may be more desirable to identify the OsABF1 target genes that are specifically responsible for drought tolerance. In this study, we used ABF1V and ABF1E transgenic plants, in which the OsABF1 targets show opposite expression patterns, to identify the genes directly associated with osmotic and drought stress responses in rice. Through bioinformatics and genetic analyses, we identified a direct target gene of OsABF1, COR413-TM1, which encodes a putative thylakoid membrane protein specific to the plant kingdom (Breton et al., 2003). In cereals and A. thaliana, the expression levels of several members of this gene family are regulated by cold, water stress, light, and ABA, and they may be associated with membrane structure reinforcement or environmental stress signaling (Seki et al., 2002; Nijhawan et al., 2008). Our data also showed that the expression of COR413-TM1 is induced by PEG, NaCl, cold, heat, ABA, and others hormone treatments, but not by H2O2 and 6-benzylaminopurine treatments (Supplementary Fig. S14). Furthermore, we demonstrated that OsABF1 mediates drought-induced up-regulation of COR413-TM1 (Figs 5 and 6), and overexpression of COR413-TM1 enhanced drought tolerance in transgenic rice without other side effects (Fig. 7 and Supplementary Fig. S13), suggesting its potential utility in developing drought-tolerant crops. It is interesting to note that the drought tolerance of COR413-TM1 was not as strong as that of the ABF1V line (Figs 1B and 7B), suggesting that an additive effect with other target genes may exist. LEA14 (encoding late embryo abundant protein 14) is also a target of OsABF1 (Supplementary Table S6 and Supplementary Fig. S9), and overexpression of LEA proteins has been shown to enhance drought tolerance in transgenic rice (Xiao et al., 2007; Duan and Cai, 2012). Therefore, OsABF1 appears to mediate drought tolerance by modulating the expression of multiple downstream genes, including COR413-TM1 and LEA14.
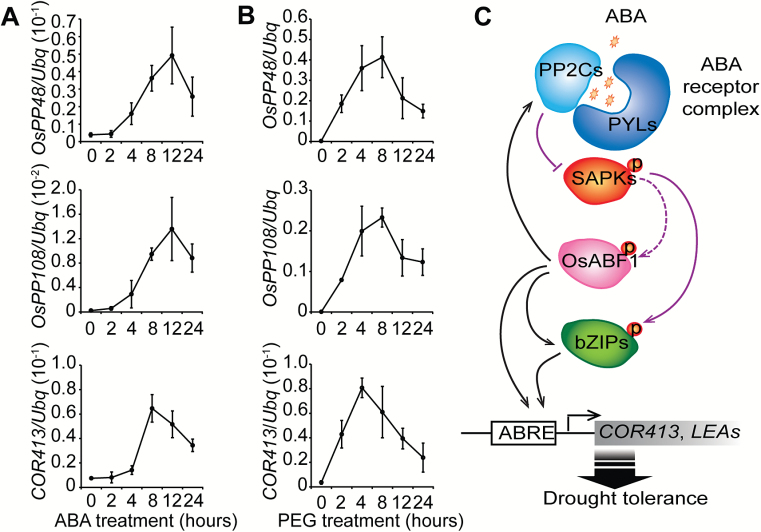
Interestingly, we found two clade-A protein phosphatase 2C (PP2C) genes, OsPP48 and OsPP108, that are under the direct regulation of OsABF1 (Supplementary Table S6 and Supplementary Figs S10, S11). The PP2C and Pyrabactin resistance1 (PYR)/PYR1-like (PYL)/RACR proteins are the components of the ABA receptor discovered in A. thaliana (Ma et al., 2009; Park et al., 2009). The phosphatase activities of PP2Cs are inhibited by ABA-bound PYR/PYL/RACR, releasing the activation of the subclass III SnRK2s (SAPKs), which further phosphorylate TFs including ABFs and AREBs (Furihata et al., 2006; Fujii and Zhu, 2009; Fujita et al., 2009; Yoshida et al., 2010). In rice, OsPP48/OsPP2C30, OsPYL/RACR5, and SAPK2 (a homolog of the SnRK2 protein kinase) were reported to form a complex to mediate ABA signaling (Kim et al., 2012). Because both drought and ABA treatments increase the expression of OsABF1, which further up-regulates the expression of OsPP48 and OsPP108, it may be that PYR/PYLs/RACR, SAPKs, PP2Cs, and OsABF1 form a negative feedback regulatory loop in the ABA signaling pathway (Fig. 8). We also found that OsABF1 directly regulates the expression of OsbZIP23, OsbZIP46, and OsbZIP72, which are associated with osmotic stress tolerance in rice (Hobo et al., 1999; Xiang et al., 2008; Lu et al., 2009; Tang et al., 2012) (Supplementary Fig. S15). OsbZIP23 can activate the expression of OsPP76/OsPP2C49, which revealed feedback regulation of ABA signaling (Zong et al., 2016). Our finding that OsABF1 directly targets both bZIPs (OsbZIP23, OsbZIP46, and OsbZIP72) and PP2Cs (OsPP48 and OsPP108) adds further insight into the complex drought–ABA signaling network in rice (Fig. 8).
Fig. 8.
OsABF1 mediates a feedback regulation pathway in ABA signaling and osmotic stress/drought responses. (A) Dynamic changes of the mRNA levels of each gene in response to ABA treatment. (B) Dynamic changes of the mRNA levels of each gene in response to PEG treatment. (C) A working model showing the crucial role of OsABF1 in the ABA-dependent pathway in osmotic/drought stress regulation. Regulation at the transcriptional level is depicted by black arrows and at the post-transcriptional level by purple arrows. The dashed line indicates predicted regulation.
Supplementary Data
Supplementary data are available at JXB online.
Fig. S1. Diagrams of T-DNA insertion positions and the performance of OsABF1-RNAi lines under osmotic stress.
Fig. S2. Diagram of the OsABF1 HTF constructions and the performance of OsABF1 transgenic lines under water deprivation conditions.
Fig. S3. Performance of OsABF1V, OsABF1E, and OsABF1F lines under polyethylene glycol (PEG) treatment.
Fig. S4. Genome-wide distribution of ABF1V-associated sites.
Fig. S5. Heat map of RNA-seq data.
Fig. S6. Identification of HOX24 as a direct target of OsABF1.
Fig. S7. Identification of AUMO1 as a direct target of OsABF1.
Fig. S8. Identification of OsEGY3 as a direct target of OsABF1.
Fig. S9. Identification of LEA14 as a direct target of OsABF1.
Fig. S10. Identification of OsPP48 as a direct target of OsABF1.
Fig. S11. Identification of OsPP108 as a direct target of OsABF1.
Fig. S12. Performance of COR413-TM1 overexpression lines under osmotic stress.
Fig. S13. Yield traits of COR413-TM1 overexpression lines.
Fig. S14. Dynamic transcription of OsCOR413-TM1 under abiotic stress or hormone treatments.
Fig. S15. Identification of OsbZIP23, OsbZIP46, and OsbZIP72 as direct targets of OsABF1.
Table S1. Oligonucleotide primers used in this study.
Table S2. ABF1V-associated genes.
Table S3. Differentially expressed genes between the WT and the ABF1V transgenic line.
Table S4. Differentially expressed genes between the WT and the ABF1E transgenic line.
Table S5. Differentially expressed genes between the WT and the WT-D treatment.
Table S6. Candidate genes directly regulated by OsABF1.
Author contributions
BL and ZT conceived this project and designed the research; CZ, CL, JL, YL, CY, and HL performed the research; BL and ZT wrote the article with contributions from all the authors.
Supplementary Material
Acknowledgments
This work is supported in part by the National Transgenic Science and Technology Program (2016ZX08010-002), National Natural Science Foundation of China (31422041 to B.L., 31371649 to H.L., 3157101834 to T.Z.), and CAAS Innovation Program.
References
- Amir Hossain Md, Cho JI, Han M, Ahn CH, Jeon JS, An G, Park PB. 2010a. The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. Journal of Plant Physiology 167, 1512–1520. [DOI] [PubMed] [Google Scholar]
- Amir Hossain Md, Lee Y, Cho JI et al. 2010b. The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Molecular Biology 72, 557–566. [DOI] [PubMed] [Google Scholar]
- Beerli RR, Segal DJ, Dreier B, Barbas CF 3rd. 1998. Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proceedings of the National Academy of Sciences, USA 95, 14628–14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton G, Danyluk J, Charron JB, Sarhan F. 2003. Expression profiling and bioinformatic analyses of a novel stress-regulated multispanning transmembrane protein family from cereals and Arabidopsis. Plant Physiology 132, 64–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Cai W. 2012. OsLEA3-2, an abiotic stress induced gene of rice plays a key role in salt and drought tolerance. PLoS ONE 7, e45117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstein M. 2013. Plant breeding: discovery in a dry spell. Nature 501, S7–S9. [DOI] [PubMed] [Google Scholar]
- Fujii H, Zhu JK. 2009. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proceedings of the National Academy of Sciences, USA 106, 8380–8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Nakashima K, Yoshida T et al. 2009. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant & Cell Physiology 50, 2123–2132. [DOI] [PubMed] [Google Scholar]
- Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. 2006. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proceedings of the National Academy of Sciences, USA 103, 1988–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobo T, Kowyama Y, Hattori T. 1999. A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proceedings of the National Academy of Sciences, USA 96, 15348–15353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XY, Chao DY, Gao JP, Zhu MZ, Shi M, Lin HX. 2009. A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes & Development 23, 1805–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA. 2005. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309, 293–297. [DOI] [PubMed] [Google Scholar]
- Izawa T, Foster R, Chua NH. 1993. Plant bZIP protein DNA binding specificity. Journal of Molecular Biology 230, 1131–1144. [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. 2002. bZIP transcription factors in Arabidopsis. Trends in Plant Science 7, 106–111. [DOI] [PubMed] [Google Scholar]
- Jeong DH, An S, Park S et al. 2006. Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. The Plant Journal 45, 123–132. [DOI] [PubMed] [Google Scholar]
- Kagale S, Rozwadowski K. 2011. EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6, 141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Choi HI, Im MY, Kim SY. 2002. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. The Plant Cell 14, 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, de la Bastide M, Hamilton JP et al. 2013. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Hwang H, Hong JW et al. 2012. A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. Journal of Experimental Botany 63, 1013–1024. [DOI] [PubMed] [Google Scholar]
- Liu C, Mao B, Ou S, Wang W, Liu L, Wu Y, Chu C, Wang X. 2014. OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Molecular Biology 84, 19–36. [DOI] [PubMed] [Google Scholar]
- Lu G, Gao C, Zheng X, Han B. 2009. Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 229, 605–615. [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. 2009. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. [DOI] [PubMed] [Google Scholar]
- Machanick P, Bailey TL. 2011. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27, 1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Li H, Wang Q, Liu B, Lin C. 2013. Blue light-dependent interaction between cryptochrome2 and CIB1 regulates transcription and leaf senescence in soybean. The Plant Cell 25, 4405–4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki D, Shimamoto K. 2004. Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant & Cell Physiology 45, 490–495. [DOI] [PubMed] [Google Scholar]
- Nijhawan A, Jain M, Tyagi AK, Khurana JP. 2008. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiology 146, 333–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SJ, Song SI, Kim YS, Jang HJ, Kim SY, Kim M, Kim YK, Nahm BH, Kim JK. 2005. Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiology 138, 341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Izawa T, Chua NH, Shimamoto K. 1996. The rab16B promoter of rice contains two distinct abscisic acid-responsive elements. Plant Physiology 112, 483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N et al. 2009. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L, Barbero GF, Geerinck J et al. 2010. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski I, Ma J, Triezenberg S, Ptashne M. 1988. GAL4-VP16 is an unusually potent transcriptional activator. Nature 335, 563–564. [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J et al. 2002. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. The Plant Journal 31, 279–292. [DOI] [PubMed] [Google Scholar]
- Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang P, Zhu JK. 2005. Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. The Plant Cell 17, 2384–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CP, Galbraith DW. 2006. AtSAP18, an orthologue of human SAP18, is involved in the regulation of salt stress and mediates transcriptional repression in Arabidopsis. Plant Molecular Biology 60, 241–257. [DOI] [PubMed] [Google Scholar]
- Song L, Huang SC, Wise A et al. 2016. A transcription factor hierarchy defines an environmental stress response network. Science 354, aag1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA. 2008. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319, 1384–1386. [DOI] [PubMed] [Google Scholar]
- Tang N, Zhang H, Li X, Xiao J, Xiong L. 2012. Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiology 158, 1755–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Herr W. 2003. The herpes simplex virus VP16-induced complex: the makings of a regulatory switch. Trends in Biochemical Sciences 28, 294–304. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Tang N, Du H, Ye H, Xiong L. 2008. Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiology 148, 1938–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Huang Y, Tang N, Xiong L. 2007. Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theoretical and Applied Genetics 115, 35–46. [DOI] [PubMed] [Google Scholar]
- Yates A, Akanni W, Amode MR et al. 2016. Ensembl 2016. Nucleic Acids Research 44, D710–D716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. 2010. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. The Plant Journal 61, 672–685. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biology 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Liu J, Zhao T et al. 2016. A drought-inducible transcription factor delays reproductive timing in rice. Plant Physiology 171, 334–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Liu J, Li HY et al. 2015. Using hybrid transcription factors to study gene function in rice. Science China Life sciences 58, 1160–1162. [DOI] [PubMed] [Google Scholar]
- Zong W, Tang N, Yang J, Peng L, Ma S, Xu Y, Li G, Xiong L. 2016. Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought-resistance-related genes. Plant Physiology 171, 2810–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.