Summary
In this study, we report the development of a serologically based immunophenotyping approach to the study of Onchocerca volvulus population diversity. Differences in immunoreactivity measured by this assay may be used to characterize O. volvulus populations from different geographic locations.
Keywords: Onchocerca volvulus, population biology, immunophenotype, single nucleotide polymorphism, immunoreactivity
Abstract
We have developed a serologically based immunophenotyping approach to study Onchocerca volvulus (Ov) population diversity. Using genomic sequence data and polymerase chain reaction–based genotyping, we identified nonsynonymous single-nucleotide polymorphisms (SNPs) in the genes of 16 major immunogenic Ov proteins: Ov-CHI-1/Ov-CHI-2, Ov16, Ov-FAR-1, Ov-CPI-1, Ov-B20, Ov-ASP-1, Ov-TMY-1, OvSOD1, OvGST1, Ov-CAL-1, M3/M4, Ov-RAL-1, Ov-RAL-2, Ov-ALT-1, Ov-FBA-1, and Ov-B8. We assessed the immunoreactivity of onchocerciasis patient sera (n = 152) from the Americas, West Africa, Central Africa, and East Africa against peptides derived from 10 of these proteins containing SNPs. Statistically significant variation in immunoreactivity among the regions was seen in SNP-containing peptides derived from 8 of 10 proteins tested: OVOC1192(1–15), OVOC9988(28–42), OVOC9225(320–334), OVOC7453(22–36), OVOC11517(14–28), OVOC3177(283–297), OVOC7911(594–608), and OVOC12628(174–188). Our data show that differences in immunoreactivity to variant antigenic peptides may be used to characterize Ov populations, thereby elucidating features of Ov population biology previously inaccessible because of the limited availability of parasite material.
Onchocerciasis—also known as “river blindness”—is a neglected tropical disease caused by the parasitic filarial nematode Onchocerca volvulus (Ov). The parasite is transmitted by blackflies of the genus Simulium. The disease is characterized by a papular dermatitis with associated pruritus, subcutaneous nodules containing adult-stage worms, lymphadenitis, and, most notably, ocular lesions leading to impaired vision or blindness [1–3]. Ov is endemic in 31 countries in sub-Saharan Africa, Venezuela, Brazil, and Yemen, with an estimated 37 million people infected [4–6]. It has been estimated that onchocerciasis is responsible for the annual loss of a half-million disability-adjusted life-years [7, 8].
The ongoing public health and socioeconomic impact of onchocerciasis requires a better understanding of Ov biology. In particular, studies of Ov population biology may help elucidate its spread, emergence of drug resistance, and persistence despite control measures. There is still much to be learned about Ov population biology, although a reference sequence of the Ov genome is available [9] and sequencing technologies have made large-scale analysis of allelic differences possible [10].
Early Ov population biology studies focused on differences in O-150 repetitive DNA sequences between “rainforest” (nonblinding) and “savanna” (blinding) strains. Members of the O-150 DNA interspersed family of repeats were shown to cluster differentially in parasites from rainforest and savanna strains, to be geographically distributed, and to be dispersed to the Americas during the trans-Atlantic slave trade [11, 12]. Analysis of O-150 repeat sequences in Ov parasites from Sudan revealed unique/discrete sequence clusters in populations from the northern, southern, and eastern foci of Sudan, with parasites from the southern and eastern foci being more closely related to those endemic to West African savanna rather than to northern Sudan [13]. Subsequent studies showed that the frequencies of particular genetic polymorphisms in the P-glycoprotein and β-tubulin genes differed between ivermectin-naive and ivermectin-exposed populations of Ov [14]. In contrast, a study of the nuclear internal transcribed spacer 2 rDNA sequences in Brazilian as well as African savanna and forest Ov strains showed low levels of genetic differentiation and a lack of allele clustering by geography or strain type [15].
A recent study of Ov population genomics assessing global diversity in nuclear, mitochondrial, and Wolbachia endosymbiont genomes from West Africa, Uganda, and Ecuador demonstrated gene flow and genetic admixture among West African forest and savanna populations [10]. A set of ancestry informative markers was identified and found to be useful in estimating the relative proportion of forest and savanna ancestry in Ov isolates. In addition, the usage of single loci such as the O-150 sequence to determine Ov phylogeny and ancestry was shown to provide limited information in comparison to multilocus genotype data. These very informative studies relied, however, on archived samples of adult parasites obtained following surgical removal of subcutaneous nodules (onchocercomata), a resource not easily available.
Because a serologically based method to identify strain differences among Toxoplasma gondii parasites [16] had been previously developed, we employed such a strategy for the characterization of Ov populations. To this end, we were able to develop a novel serologically based method to immunophenotype Ov populations using serum from Ov-infected patients from geographically disparate parts of the world.
METHODS
Patients and Sera
Sera from 152 Ov-infected individuals from the Americas (Ecuador and Guatemala), West Africa (Nigeria and Togo), Central Africa (Cameroon), and East Africa (Uganda), as well as 3 uninfected nonendemic volunteers, was used and obtained with informed consent using protocol O9-I-N178 approved by the National Institute of Allergy and Infectious Diseases Institutional Review Board (Table 1). The diagnosis of Ov infection was based on the identification of Ov microfilariae in skin snips or skin biopsy material, or by positive polymerase chain reaction (PCR) of the O-150 repeat sequence as previously described [17].
Table 1.
General Characteristics of Studied Population
| Characteristic | Total (N = 152) |
The Americas (n = 48) | West Africa (n = 42) | Central Africa (n = 42) | East Africa (n = 20) |
|---|---|---|---|---|---|
| Sex, No. (%) | |||||
| Male | 84 (55.3) | 25 (52.1) | 26 (61.9) | 24 (57.1) | 9 (45) |
| Female | 61 (40.1) | 22 (45.8) | 10 (23.8) | 18 (42.9) | 11 (55) |
| Unspecified | 7 (4.6) | 1 (2.1) | 6 (14.3) | 0 (0) | 0 (0) |
| Age, y | |||||
| Median | 35 | 28.5 | 39 | 50.5 | 12 |
| Min, Max | (8, 80) | (11, 70) | (14, 70) | (22, 80) | (8, 14) |
| Race, No. | |||||
| Amerindian | 20 | 20 | 0 | 0 | 0 |
| Black | 127 | 23 | 42 | 42 | 20 |
| Mixed | 5 | 5 | 0 | 0 | 0 |
Onchocerca volvulus Genomic DNA Extraction
Genomic DNA (gDNA) was extracted from Ov skin nodules and skin snips obtained from Ecuadorian and Ugandan Ov-infected patients. Skin nodules containing adult female O. volvulus worms were surgically excised from individuals in the Zapallo River Basin, Ecuador, as previously described [10]. The sera of these Ov-infected individuals were not available for enzyme-linked immunosorbent assays (ELISAs) in this study. Twenty adult female worms were freed from surrounding host tissue and processed as previously described with modifications [18]. In brief, nodules were incubated in 5 mL Hank’s Balanced Salt Solution/penicillin-streptomycin-amphotericin B plus 3mM calicum chloride (CaCl2) and 200 μL Liberase Thermolysin Low (Roche, Indianapolis, Indiana) for 48–72 hours, followed by dissection of parasite tissue from host tissue. Parasite material was freeze-thawed 3 times and homogenized. Genomic DNA was extracted from homogenates using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, California).
Genomic DNA was also extracted from skin snips derived from Ugandan Ov-infected patients using methods previously described [17]. The sera of these patients were used for ELISAs in this study.
Onchocerca volvulus Genomic DNA Sequencing
Genomic DNA extracted from the 20 adult female worms contained in subcutaneous nodules was pooled and submitted to the North Carolina State Genomic Sciences Laboratory (Raleigh, North Carolina) for Illumina library construction and sequencing. Prior to library preparation, the skin nodule gDNA template was quantified by a Qubit 2.0 fluorometer (Invitrogen, Carlsbad, California). Library construction was performed using an Illumina TruSeq Nano Library kit. In brief, the gDNA was fragmented using a Covaris S2 ultrasonicator (Covaris, Woburn, Massachusetts) and purified using AMPure XP beads (Beckman Coulter, Brea, California). The fragments were then end-repaired, followed by 550-bp insert size-selection using sequential AMPure XP bead isolation steps. After adapter ligation, the library was enriched by PCR amplification. The amplified library was checked for quality and final concentration using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, California) with a high-sensitivity DNA chip before sequencing on an Illumina HiSeq 2500 DNA sequencer, utilizing a 100-bp paired-end sequencing flow cell with a HiSeq Reagent Kit v4 (Illumina, San Diego, California). Flow cell cluster generation for the HiSeq2500 was performed using an automated cBot system (Illumina, San Diego, California). The software package Real Time Analysis, was used to generate raw bcl, or base call files, which were then de-multiplexed by sample into fastq files for data submission using bcl2fastq2 software.
Identification of Single-Nucleotide Polymorphisms
Single nucleotide polymorphisms (SNPs) were identified by analysis of published immunogenic protein sequence data and whole-genome sequencing (WGS) data. The SNPs identified by these methods are listed in Supplementary Table 1.
Using the amino acid sequences of previously identified Ov-specific immunogenic proteins [19], the National Center for Biotechnology Information (NCBI) translated nucleotide database and Wellcome Trust Sanger Institute BLAST server were queried for related expressed sequence tags (ESTs) and gene sequences. Sequences of each query and its matches were aligned with MegAlign in Lasergene version 12 (DNASTAR, Madison, Wisconsin). Nonsynonymous mutations were identified in the alignments and designated as putative SNPs if present at least 2 times.
Using the gDNA of the 20 adult female worms from skin nodules of Ecuadorian patients and skin snips from Ugandan patients, we performed allelic discrimination qPCR (TaqMan SNP Genotyping Assays, ThermoFisher Scientific) to confirm the presence of some of the SNPs identified by EST alignments. Seven of the 9 nonsynonymous SNPs identified by EST alignments were verified by allelic discrimination qPCR: OVOC7453 S29F, OVOC7453 K93N, OVOC12871.3 P151R, OVOC12871.3 R180P, OVOC12769.2 I86M, OVOC8754 A165T, OVOC3177 A288T. The remaining 2 were confirmed by analysis of WGS data as described below.
WGS data from the aforementioned 20 adult female worms were mined for SNPs in immunogenic proteins genes using methods previously described [20]. SNPs in immunogenic protein genes were also identified by analysis of WGS data that were derived 10 of the same 20 adult female worms as part of a larger study of Ov genomic diversity [10].
Synthetic Peptides
Pairs of 15-mer peptides derived from 16 major immunogenic Ov proteins (Ov-CHI-1/Ov-CHI-2, Ov16, Ov-FAR-1, Ov-CPI-1, Ov-B20, Ov-ASP-1, Ov-TMY-1, OvSOD1, OvGST1, Ov-CAL-1, M3/M4, Ov-RAL-1, Ov-RAL-2, Ov-ALT-1, Ov-FBA-1, and Ov-B8), and each containing one of the SNP variant amino acids identified above, were synthesized and conjugated at their C-terminal cysteine residues to keyhole limpet hemocyanin (KLH) (Atlantic Peptides, LLC, Lewisburg, Pennsylvania). The peptides were synthesized on a Rainin Symphony using Fmoc chemistry. The coupling solution used contained 2-(6-Chloro-1H-benzotriazol-1-yl)-N,N,N’,N’-tetramethylaminium hexafluorophosphate (Chemical Abstract Service [CAS] number 330645-87-9) and diisopropylethylamine (CAS number 7087-68-5), and the deprotection solution contained piperidine (CAS number 110-89-4). The peptides were cleaved from the resin with a trifluoroacetic acid cleavage solution. The sequences and names of the synthetic peptides are listed in Table 2.
Table 2.
Synthetic Variant Peptides Derived From Immunogenic Proteins
| Peptide Sequencea | Positionb | Protein Name | Gene Number | Peptide Name |
|---|---|---|---|---|
| NKFASQKGM[T/I]GFGTSC | 204–218 | Ov-CAL-1 | OVOC860 | OVOC860(204–218) |
| DYIGSGP[P/R]KGTGLHRC | 144–158 | Ov16 | OVOC12871.3 | OVOC12871.3(144–158) |
| MSKPEK[T/A]GAQQTSLLC | 1–15 | Ov-TMY-1 | OVOC1192 | OVOC1192(1–15) |
| QQQQQRDERE[I/T]PPFLC | 28–42 | Ov-RAL-2 | OVOC9988 | OVOC9988(28–42) |
| VTTRREKD[G/S]RYHDDGC | 320–334 | Ov-B20 | OVOC9225 | OVOC9225(320–334) |
| VQLQGAK[S/F]ARAKNPSC | 22–36 | Ov-CPI-1 | OVOC7453 | OVOC7453(22–36) |
| VSGIIRFKQ[D/V]KEGLPC | 14–28 | OvSOD1 | OVOC11517 | OVOC11517(14–28) |
| LENLV[A/T]ELKNRLASKC | 283–297 | Ov-B8 | OVOC3177 | OVOC3177(283–297) |
| DHYDFLK[P/S]RIILDPNC | 594–608 | Ov-RAL-1 | OVOC7911 | OVOC7911(594–608) |
| FMVTTQN[I/M]RSTAVTLC | 174–188 | M3/M4 | OVOC12628 | OVOC12628(174–188) |
| INHEL[E/K]GEATATTRSC | 384–398 | Ov-CHI-1/ -2 | OVOC12569 | OVOC12569(384–398) |
| NEETKEELK[A/T]TFPNTC | 156–170 | Ov-FAR-1 | OVOC8754 | OVOC8754(156–170) |
| KYRSDLI[H/N]GKLKNRNC | 38–52 | Ov-ASP-1 | OVOC9575 | OVOC9575(38–52) |
| SENSIKPK[G/E]KLQPQMC | 55–69 | OvGST1 | OVOC7314 | OVOC7314(55–69) |
| EEGEGEE[M/T]PEDNDGGC | 28–42 | Ov-ALT-1 | OVOC12769.2 | OVOC12769.2(28–42) |
| DATLNLN[A/T]INQFPGKC | 332–346 | Ov-FBA-1 | OVOC7786 | OVOC7786(332–346) |
aThe cysteine (C) at the C terminus was added for coupling purposes. Bold type indicates polymorphic sites.
bThese numbers indicate the position of each peptide in the protein coding sequence.
ELISA to Test Immunoreactivity Against Peptides
Measurements of patient immunoglobulin G (IgG) immunoreactivity against SNP-variant peptides were performed by ELISA. Flat-bottom Immulon 4 plates (Dynatech Laboratories, Chantilly, Virginia) were coated overnight at 4°C with 1 μg/mL [OVOC3177(283–297), OVOC7314(55–69), OVOC12628(174–188), OVOC7453(22–36), OVOC12871.3(144–158), OVOC1192(1–15), OVOC860(204–218), OVOC7911(594–608), OVOC9988(28–42), OVOC9575(38–52)], 5 μg/mL [OVOC9225(320–334), OVOC7786(332–346), OVOC8754(156–170), OVOC12569(384–398)], or 10 μg/mL [OVOC11517(14–28), OVOC12769.2(28–42)] of KLH-conjugated synthetic peptides in phosphate-buffered saline (PBS). Higher concentrations of peptide (5 μg/mL and 10 μg/mL) were needed to perform ELISAs with peptides that elicited relatively weak signals. After incubation with peptide, plates were washed with PBS and 0.05% Tween 20 (Sigma-Aldrich, St Louis, Missouri), followed by blocking with PBS and 5% bovine serum albumin (BSA)/0.05% Tween 20 for 2 hours at 37°C. Duplicate serum samples were diluted 1:100 in PBS and 1% BSA/0.05% Tween 20 and incubated overnight at 4°C. Plates were then washed and incubated with alkaline phosphatase–conjugated goat antihuman IgG (Jackson ImmunoResearch Laboratories, West Grove, Pennsylvania) for 2 hours at 37°C. Plates were again washed, and 1 mg/mL p-nitrophenyl phosphate, disodium salt (Sigma-Aldrich) in sodium carbonate buffer (KD Medical, Columbia, Maryland) was added. Colorimetric development was detected at 405 nm using a microplate reader (Molecular Devices, Sunnyvale, California), and optical density (OD) was used as a surrogate measure of antibody levels.
Immunoreactivity to all 32 SNP-variant peptides was tested using a representative set of hypothesis-generating sera from the 4 geographic areas (data not shown). No substantial differences were found in the immunoreactivity against 1 SNP variant compared to its cognate. Thus, in scaling up the analysis to include a large set of hypothesis-confirming sera, only 1 of any given 2 SNP variants were used in the ELISA assays. The number of peptides ultimately assayed was further reduced to 10 due to availability of material.
Statistical Analysis
To compare immunoreactivity of hypothesis-generating sera against 1 SNP variant compared to its cognate, 2-tailed t tests were performed.
For the hypothesis-confirming tests, we used a log transformation of the ELISA values for the 10 peptides. Because graphically there were no large deviations from normality of homoscedasticity (Figure 1B–K), we compare the 4 geographic regions with analysis of variance (ANOVA) and compared pairs of regions with standard 2-sided t tests. We performed linear model likelihood ratio tests to adjust for the effects of age group and sex on ELISA values. We defined “immunoreactive” patients for each of the 10 values as ELISA values exceeding the cutoffs determined as the minimum values that ensure 100% specificity in the receiver operating characteristic (ROC) curves. Two-sided Fisher exact tests (Fisher-Irwin version) were used to compare the resulting binary responses between geographic regions. We present false discovery rate [21] P values adjusted (Padj) for the fact that we tested ELISAs on 10 peptides. The pairwise comparisons present unadjusted P values. All P values are 2-sided and P < .05 is considered significant (and the use of adjusted or unadjusted P values is clear from the context). All statistical analyses and graphs were performed using Prism 6.0 (GraphPad Software, San Diego, California) or R version 3.3.1 software.
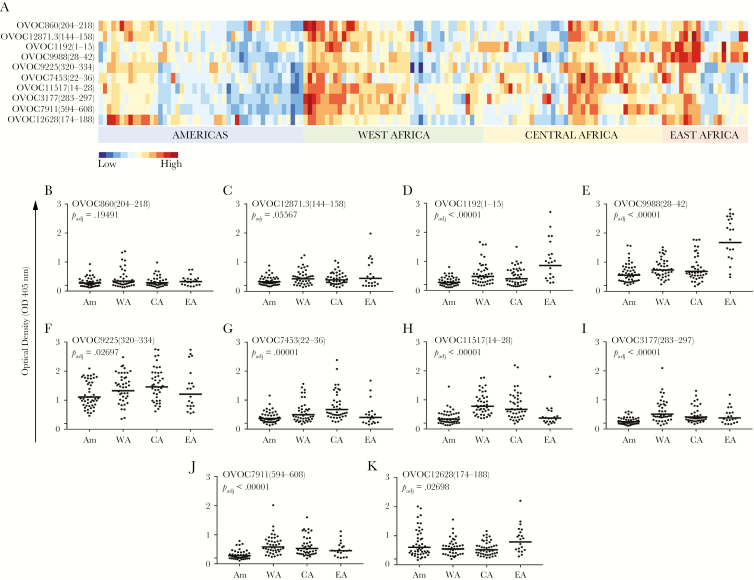
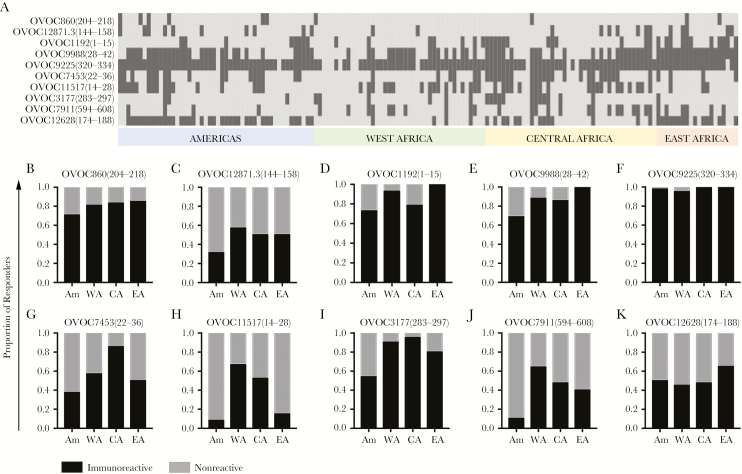
Figure 1.
Immunoreactivity against variant peptides significantly differs by region. A, Heatmap of semisupervised clustering of the normalized optical density (OD) values, with a color scale ranging from low (dark blue) to high (dark red). B–K, Scatterplot graphs of OD for each variant peptide, with geometric means indicated by the horizontal lines. The geographic regions are indicated on the x-axis; the OD values are indicated on the y-axis. The false discovery rate–adjusted P values from analysis of variance are listed on each graph. Abbreviations: Am, Americas; CA, Central Africa; EA, East Africa; WA, West Africa.
RESULTS
General Characteristics of Patient Population
Sera from 152 patients were used in this study. The patients’ geographic origins included the Americas (Ecuador and Guatemala), West Africa (Nigeria and Togo), Central Africa (Cameroon), and East Africa (Uganda). General characteristics including sex, age, and race are listed in Table 1.
Identification of SNPs in Immunogenic Proteins
Single-nucleotide polymorphisms in immunogenic Ov proteins were identified by in silico alignments of sequences in the NCBI translated nucleotide database and Wellcome Trust Sanger Institute BLAST server, as well as by Illumina sequencing and TaqMan PCR of Ov gDNA as described in the Methods. Supplementary Table 1 lists the identified nonsynonymous SNPs and the means by which they were identified. These SNPs are distributed along the entire lengths of the proteins. Certain immunogenic proteins were noted to be relatively rich in SNPs, including Ov-CHI-1/Ov-CHI-2, Ov-ASP-1, and Ov-ALT-1.
Immunoreactivity Against Variant Peptides Significantly Differs by Region
To investigate whether immunoreactivity of patient sera from different geographic regions (Americas, West Africa, Central Africa, and East Africa) could be used to distinguish parasite populations, we developed an ELISA to test IgG reactivity against SNP-variant peptides derived from immunogenic Ov proteins (OD values are listed in Supplementary Table 2). As shown in Figure 1, immunoreactivity of patient sera against 8 of 10 SNP-variant peptides differed significantly by region. The spectrum of OD values is shown in the heatmap (Figure 1A). As can be seen, a range of reactivity to the peptides was detected for the different geographic regions and peptides. For example, Ov-infected sera from East Africa showed relatively high reactivity to OVOC9988(28–42) and OVOC1192(1–15), whereas sera from West and Central Africa showed relatively low reactivity to OVOC12628(174–188). Sera from Central Africa showed relatively high reactivity to OVOC7453(22–36).
Geographic variations in immunoreactivity against each peptide are also illustrated in the scatterplot graphs shown in Figure 1B–K. ANOVA revealed statistical differences in immunoreactivity against 8 of 10 variant peptides across the 4 geographic regions: OVOC1192(1–15) (Padj < .00001), OVOC9988(28–42) (Padj < .00001), OVOC9225(320–334) (Padj = .02697), OVOC7453(22–36) (Padj = .00001), OVOC11517(14–28) (Padj < .00001), OVOC3177(283–297) (Padj < .00001), OVOC7911(594–608) (Padj < .00001), and OVOC12628(174–188) (Padj = .02698). Reactivity against OVOC860(204–218) and OVOC12871.3(144–158) was not significantly different among the 4 regions (Padj = .19491 and Padj = .05567). Significant differences in 7 of the 8 peptides found significant were maintained after adjusting for age group and sex (the adjusted effect for OVOC12628[174–188] became borderline significant, Padj = .07715; see Supplementary Table 5).
Pairwise t test results comparing OD values between geographic regions revealed statistical differences in immunoreactivity between regions, as shown in Supplementary Table 3. The most significant differences (unadjusted P < .05) in immunoreactivity against the variant peptides were found between the Americas and West Africa. The least number of differences were found between samples from the West Africa and Central Africa.
Binary Response Analysis of Immunoreactive and Nonreactive Patients
To explore the geographic differences in immunoreactivity by binary response analysis, patients were categorized as “immunoreactive” if their ELISA values exceeded the cutoffs determined as the minimum values that ensure 100% specificity in the ROC curves. The overall distribution of the immunoreactive and nonreactive patients identified for each synthetic peptide is pictured in Figure 2A. Geographic variations in the distributions of immunoreactive and nonreactive patients were again seen. All regions had a high number of immunoreactive patients to OVOC9225(320–334), but had about half nonreactive patients to OVOC12628(174–188), for example.
Figure 2.
Binary responses of immunoreactivity against variant peptides significantly differs by region. A, Binary responses for each peptide with immunoreactive (dark gray) and nonreactive (light gray) patients. B–K, Normalized proportions of binary responses for each variant peptide. Geographic regions are indicated on the x-axis, and the proportions of immunoreactive (black) and nonreactive (gray) patients are indicated on the y-axis. Abbreviations: Am, Americas; CA, Central Africa; EA, East Africa; WA, West Africa.
Proportions of binary responses for each variant peptide by geographic area are illustrated in Figure 2B–K. Not unlike the quantitative data presented in Figure 1, the number of immunoreactive patients to OVOC9225(320–334) in all geographic regions was higher overall as compared to other peptides. In contrast, there were more nonreactive patients to OVOC12628(174–188) and OVOC11517(14–28).
Pairwise Fisher exact test results comparing binary responses between geographic regions demonstrated differences as shown in Supplementary Table 4. The most significant differences (unadjusted P < .05) in immunoreactivity against the variant peptides were found between Americas and West Africa. Regions that had the least number of significant differences between them were the Central Africa and East Africa, as well as West Africa and Central Africa.
DISCUSSION
The study of Ov population biology has been limited because extraction of parasite material from hosts requires invasive procedures. Here, we report the development of a novel and noninvasive serologically based approach to the study of Ov population biology. SNP variant Ov antigen-derived peptides were used in immunoassays to investigate whether the immunoreactivity of patient sera from the Americas, West Africa, Central Africa, and East Africa could differentiate parasite populations in each region. Our immunogenic Ov peptide-based ELISA method elucidated differential antibody responses by geographic region. We conclude that the differential immunoreactivity elicited by the ELISA reflects differences among Ov populations in each region.
The geographic variations in antibody responses in this study are consistent with the recent finding that Ov populations are biogeographically structured on the basis of multilocus genomic data [10]. The clear distinctions between Ugandan, West African, and Ecuadorian isolates seen by Choi and colleagues were also observed in the present study.
The suggestion that the divergent selection of Ov in response to different vector species may play an important role in generating population structure is also relevant to our findings. For example, molecular phylogenetic analysis of the Simulium damnosum complex revealed the existence of East, West, and Central African clades [22]. The geographic distribution of Ov vector species along these lines correlates well with the differences in Ov populations that we inferred from immunoreactivity of sera obtained from these regions against Ov antigens.
This present study does have several limitations to consider. First, in a subset of patients, we were unable to relate the differences in immunoreactivity with differences in the antigen SNP allele sequences of parasites extracted from the patients. Therefore, some of the variability in antibody responses could be due to host factors. A second limitation was our assumption that Ov populations are delineated by the studied geographic regions. It could be that worms in Nigeria (West Africa) and in Cameroon (Central Africa) comprise a population distinct from worms in Togo (West Africa); moreover, forest, and savanna Ov populations were likely grouped together by our regional classifications. Another limitation is that we were unable to control for the influence of other infections or diseases on the antibody responses of the populations studied.
The characterization of Ov populations by this novel serologically based immunotyping approach has important implications for elimination efforts. The diversity of antibody responses against various Ov peptides could inform development of Ov vaccines that are currently under way [23, 24]. The correlation of Ov population immunotypes with drug resistance could provide a noninvasive means of monitoring the effects of interventions in areas where there is persistent microfilaridermia despite multiple rounds of mass drug administration [25].
Overall, our results show that the population biology of Ov can be characterized by patterns of host immunoreactivity to Ov antigenic peptides in immunoassay-based formats using archived serum or plasma. Data obtained by this noninvasive approach may contribute to not only an understanding of Ov population biology but could also inform Ov elimination efforts. Further characterization of Ov populations using additional immunogenic protein-derived peptides may certainly increase the utility of this multilocus immunophenotyping strategy in clinical applications.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Acknowledgments. The authors wish to acknowledge Rocky Mountain Laboratories Research Technologies Section, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH).
Financial support. This research was supported by the Division of Intramural Research, NIAID, NIH.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Budden FH. The natural history of ocular onchocerciasis over a period of 14–15 years and the effect on this of a single course of suramin therapy. Trans R Soc Trop Med Hyg 1976; 70:484–91. [DOI] [PubMed] [Google Scholar]
- 2. Murdoch ME, Hay RJ, Mackenzie CD et al. A clinical classification and grading system of the cutaneous changes in onchocerciasis. Br J Dermatol 1993; 129:260–9. [DOI] [PubMed] [Google Scholar]
- 3. Abdel-Hameed AA, Noah MS, Schacher JF, Taher SA. Lymphadenitis in Sowda. Trop Geogr Med 1987; 39:73–6. [PubMed] [Google Scholar]
- 4. Hotez PJ, Kamath A. Neglected tropical diseases in sub-Saharan Africa: review of their prevalence, distribution, and disease burden. PLoS Negl Trop Dis 2009; 3:e412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. World Health Organization. Onchocerciasis fact sheet http://www.who.int/mediacentre/factsheets/fs374/en/. Accessed 4 December 2016.
- 6. Dunn C, Callahan K, Katabarwa M et al. The contributions of onchocerciasis control and elimination programs toward the achievement of the Millennium Development Goals. PLoS Negl Trop Dis 2015; 9:e0003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Remme JH. Research for control: the onchocerciasis experience. Trop Med Int Health 2004; 9:243–54. [DOI] [PubMed] [Google Scholar]
- 8. Coffeng LE, Stolk WA, Zoure HG et al. African programme for onchocerciasis control 1995–2015: updated health impact estimates based on new disability weights. PLoS Negl Trop Dis 2014; 8:e2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cotton JA, Bennuru S, Grote A et al. The genome of Onchocerca volvulus, agent of river blindness. Nat Microbiol 2016; 2:16216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Choi YJ, Tyagi R, McNulty SN et al. Genomic diversity in Onchocerca volvulus and its Wolbachia endosymbiont. Nat Microbiol 2016; 2:16207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ogunrinade A, Boakye D, Merriweather A, Unnasch TR. Distribution of the blinding and nonblinding strains of Onchocerca volvulus in Nigeria. J Infect Dis 1999; 179:1577–9. [DOI] [PubMed] [Google Scholar]
- 12. Zimmerman PA, Katholi CR, Wooten MC, Lang-Unnasch N, Unnasch TR. Recent evolutionary history of American Onchocerca volvulus, based on analysis of a tandemly repeated DNA sequence family. Mol Biol Evol 1994; 11:384–92. [DOI] [PubMed] [Google Scholar]
- 13. Higazi TB, Katholi CR, Mahmoud BM et al. Onchocerca volvulus: genetic diversity of parasite isolates from Sudan. Exp Parasitol 2001; 97:24–34. [DOI] [PubMed] [Google Scholar]
- 14. Eng JK, Prichard RK. A comparison of genetic polymorphism in populations of Onchocerca volvulus from untreated- and ivermectin-treated patients. Mol Biochem Parasitol 2005; 142:193–202. [DOI] [PubMed] [Google Scholar]
- 15. Morales-Hojas R, Cheke RA, Post RJ. A preliminary analysis of the population genetics and molecular phylogenetics of Onchocerca volvulus (Nematoda: Filarioidea) using nuclear ribosomal second internal transcribed spacer sequences. Mem Inst Oswaldo Cruz 2007; 102:879–82. [DOI] [PubMed] [Google Scholar]
- 16. Kong JT, Grigg ME, Uyetake L, Parmley S, Boothroyd JC. Serotyping of Toxoplasma gondii infections in humans using synthetic peptides. J Infect Dis 2003; 187:1484–95. [DOI] [PubMed] [Google Scholar]
- 17. Zimmerman PA, Guderian RH, Aruajo E et al. Polymerase chain reaction-based diagnosis of Onchocerca volvulus infection: improved detection of patients with onchocerciasis. J Infect Dis 1994; 169:686–9. [DOI] [PubMed] [Google Scholar]
- 18. Schulz-Key H, Albiez EJ, Büttner DW. Isolation of living adult Onchocerca volvulus from nodules. Tropenmed Parasitol 1977; 28:428–30. [PubMed] [Google Scholar]
- 19. Lustigman S, James ER, Tawe W, Abraham D. Towards a recombinant antigen vaccine against Onchocerca volvulus. Trends Parasitol 2002; 18:135–41. [DOI] [PubMed] [Google Scholar]
- 20. Neafsey DE, Waterhouse RM, Abai MR et al. Mosquito genomics. Highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science 2015; 347:1258522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 1995; 57:289–300. [Google Scholar]
- 22. Krueger A, Hennings IC. Molecular phylogenetics of blackflies of the Simulium damnosum complex and cytophylogenetic implications. Mol Phylogenet Evol 2006; 39:83–90. [DOI] [PubMed] [Google Scholar]
- 23. Hess JA, Zhan B, Bonne-Année S et al. Vaccines to combat river blindness: expression, selection and formulation of vaccines against infection with Onchocerca volvulus in a mouse model. Int J Parasitol 2014; 44:637–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Steisslinger V, Korten S, Brattig NW, Erttmann KD. DNA vaccine encoding the moonlighting protein Onchocerca volvulus glyceraldehyde-3-phosphate dehydrogenase (Ov-GAPDH) leads to partial protection in a mouse model of human filariasis. Vaccine 2015; 33:5861–7. [DOI] [PubMed] [Google Scholar]
- 25. Awadzi K, Boakye DA, Edwards G et al. An investigation of persistent microfilaridermias despite multiple treatments with ivermectin, in two onchocerciasis-endemic foci in Ghana. Ann Trop Med Parasitol 2004; 98:231–49. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.