Summary
This study demonstrates that Epstein-Barr virus type 2 (EBV-2) infects T cells in healthy Kenyan infants, persists in some infants for at least 2 years, and is shed in mothers’ breast milk and saliva, suggesting that EBV-2 infection of T cells is part of the persistence strategy.
Keywords: Epstein-Barr virus, T lymphocytes, cellular tropism, Burkitt lymphoma
Abstract
Background
The 2 strains of Epstein-Barr virus (EBV), EBV type 1 (EBV-1) and EBV-2, differ in latency genes, suggesting that they use distinct mechanisms to establish latency. We previously reported that EBV-2 infects T cells in vitro. In this study, we tested the possibility that EBV-2 infects T cells in vivo.
Methods
Purified T-cell fractions isolated from children positive for EBV-1 or EBV-2 and their mothers were examined for the presence of EBV and for EBV type.
Results
We detected EBV-2 in all T-cell samples obtained from EBV-2–infected children at 12 months of age, with some children retaining EBV-2–positive T cells through 24 months of age, suggesting that EBV-2 persists in T cells. We were unable to detect EBV-2 in T-cell samples from mothers but could detect EBV-2 in samples of their breast milk and saliva.
Conclusions
These data suggest that EBV-2 uses T cells as an additional latency reservoir but that, over time, the frequency of infected T cells may drop below detectable levels. Alternatively, EBV-2 may establish a prolonged transient infection in the T-cell compartment. Collectively, these novel findings demonstrate that EBV-2 infects T cells in vivo and suggest EBV-2 may use the T-cell compartment to establish latency.
Epstein-Barr virus (EBV) is a gammaherpesvirus that, following primary infection, persists for the life of the human host. The immortalization of B cells in vitro by EBV is thought to be a model for the first step in establishing latency in vivo. There are 2 strains of EBV, EBV type 1 (EBV-1) and EBV-2. Most studies of EBV infection of B cells use the EBV-1 strain that readily immortalizes B cells upon primary infection. EBV-2 differs genotypically from EBV-1 in key latency genes (eg, those encoding EBV nuclear antigen 2 [EBNA2], EBNA3a, and EBNA3c) and poorly immortalizes B cells in vitro [1–6]. This suggests that EBV-2 is impaired in the ability to independently drive B-cell differentiation and establish latency in the B-cell compartment in vivo. However, in Kenya, EBV-2 is prevalent in healthy adults [3, 7] and children (N. A. S., unpublished observation) and is present in almost half of all Burkitt lymphoma tumors [3]. The contradictory observations that EBV-2 weakly immortalizes B cells yet is able to persist in the human population led us to hypothesize that EBV-2 uses mechanisms distinct from EBV-1 to establish latency.
Previously, we reported that EBV-2 has a cell tropism for T cells [8]. In contrast to EBV-1, EBV-2 infects primary T cells, resulting in T-cell activation and proliferation, as well as alteration of cytokine expression profile [8]. These findings demonstrated that EBV-2 has the ability to modulate normal T-cell processes and suggest that EBV-2 may use the T-cell population to enhance the establishment of latency and persist in vivo. Thus, the goal of this study was to determine whether EBV-2 infects T cells in vivo, using samples from a population in Kenya, where the Burkitt lymphoma risk is high.
MATERIALS AND METHODS
Study Participants
Participants for this study were infants and mothers from Chulaimbo Subdistrict, a rural area in Kisumu County, Kenya. Infants in this study were born to human immunodeficiency virus (HIV)–negative mothers. Written informed consent was obtained for all study participants before any sample collection. Approval was obtained from the Kenya Medical Research Institute, SUNY Upstate Medical University, and University of Colorado ethical review boards. The recruitment process and study population has been previously described [7, 9]. Mothers were enrolled at 20–24 weeks of pregnancy, at which time a venous blood specimen was collected. Saliva and breast milk specimens were collected during postpartum week 6. Infants were followed from delivery through 24 months of age. Blood specimens collected by finger prick were obtained at ages 6, 10, 14, and 18 weeks and 9, 15, and 21 months, and from venous blood specimens were collected at 6, 12, 18 and 24 months of age. The infants in this study had 0–2 diagnosed cases of malaria through 24 months of age, with the exception of infant 3, who had 4 cases. Only 1 blood sample used for this study was collected from an infant with P. falciparum–positive blood smear (infant 7, collected at age 24 months).
Breast Milk and Saliva Specimen Collection and DNA Isolation
Breast milk and saliva specimens were collected from mothers at postpartum week 6, using an aseptic technique, and stored at −80°C. Whole milk was centrifuged to separate the lipid, supernatant, and cell debris, and supernatant was collected for DNA extraction. DNA was extracted from breast milk supernatant and saliva samples by use of a QiaAmp DNA mini kit (Qiagen), using the manufacturer’s protocol.
T-Cell Purification
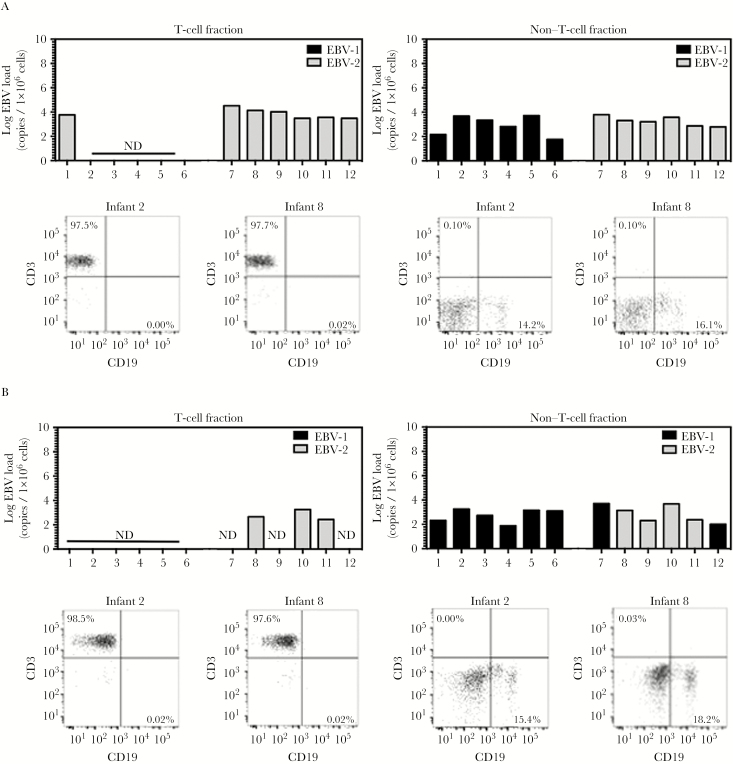
Peripheral blood mononuclear cells (PBMCs), collected via Ficoll-Paque purification, were used to isolate T-cell and non–T-cell fractions. PBMCs were stored in freezing medium (10% dimethyl sulfoxide and 90% fetal calf serum) in liquid nitrogen until analysis. T cells were isolated from thawed PBMCs by negative enrichment, using the human Pan T-Cell Isolation kit (Miltenyi-Biotec). After isolation of CD3+ T cells, magnetic columns were washed to collect the non–T-cell fraction. Purity analysis of all cell fractions was performed with anti–CD3-APC and anti-CD19-PerCp-Cy5.5 antibodies (BD Biosciences). The purity of T-cell fractions was as follows: >97% CD3+ T cells with ≤0.02% CD19+ B cells among infants aged 12 months of age, >96% with ≤0.04% CD19+ B cells among infants aged 24 months, and >95% with ≤0.04% CD19+ B cells among mothers at enrollment (Figures 2 and 3 and Supplementary Figure 1). DNA and RNA was extracted from cell fractions, using an AllPrep DNA/RNA kit (Qiagen).
Figure 2.
Epstein-Barr virus type 2 (EBV-2) infects T cells in vivo. Peripheral blood mononuclear cells (PBMCs) were collected from infant donors at 1, 2, and 24 months of age. Non–T-cell and T-cell fractions were isolated from the total PBMC population via magnetic columns, and DNA was subsequently isolated. Multiplex quantitative polymerase chain reaction (PCR) analysis of BALF5 in the EBV genome was performed to determine the log EBV copy number per 1 × 106 cells for each cellular fraction collected at ages 12 months (A) and 24 months (B). Additionally, a multiplex reverse-transcription PCR to detect EBNA3c was performed to identify the EBV type infecting each cell fraction. Post sort purity analysis for CD3 and CD19 was performed on all non–T-cell and T-cell fractions. Representative flow plots for infants 2 and 8 are shown under the corresponding bar graphs. Abbreviation: ND, not detected.
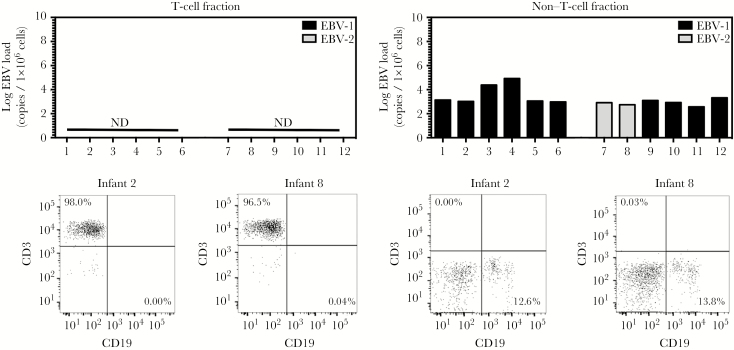
Figure 3.
Epstein-Barr virus type 2 (EBV-2) is not detected in the T-cell compartment of adults. Peripheral blood mononuclear cells (PBMCs) were collected from mothers at the time of enrollment into the study. Non–T-cell and T-cell fractions were isolated from the total PBMC population via magnetic columns, and DNA was subsequently isolated. Multiplex quantitative polymerase chain reaction (PCR) analysis of BALF5 in the EBV genome was performed to determine the log number of EBV copies per 1 × 106 cells for each cellular fraction. Additionally, a multiplex reverse-transcription PCR analysis to detect EBNA3c was performed to identify the EBV type infecting each cell fraction. Postsorting purity analysis for CD3 and CD19 was performed on all non–T-cell and T-cell fractions. Representative flow plots for infants 2 and 8 are shown under the corresponding bar graphs. Abbreviation: ND, not detected.
Polymerase Chain Reaction (PCR) Analysis of EBV Copy Number and Type
EBV DNA levels were quantified using primers and probes designed to detect a 70-bp region of the EBV BALF5 gene and the β-actin gene as a control for DNA input. EBV type was determined using primers to amplify a region of the EBNA-3C gene and probes to distinguish between EBV-1 and EBV-2 in this EBNA-3C region [7, 10, 11]. PCR analyses were run using an iCycler thermocycler equipped with an iCycler iQ real-time PCR detection system (Bio-Rad).
Reverse-Transcription PCR Analysis to Detect EBV Latent Gene Transcripts
Purified RNA was reverse transcribed using the QuantiTect reverse transcription kit (Qiagen). Primers and probes were obtained as previously described [12], and Biplex PCR analysis was performed to detect EBV latent transcripts and β-2-microglobulin, using the iTaq Universal Probes mix (BioRad). An EBV-2 lymphoblastoid cell line was used as a positive control for all amplicons studied [13].
RESULTS
EBV-2 Prevalence Among Healthy Kenyan Children
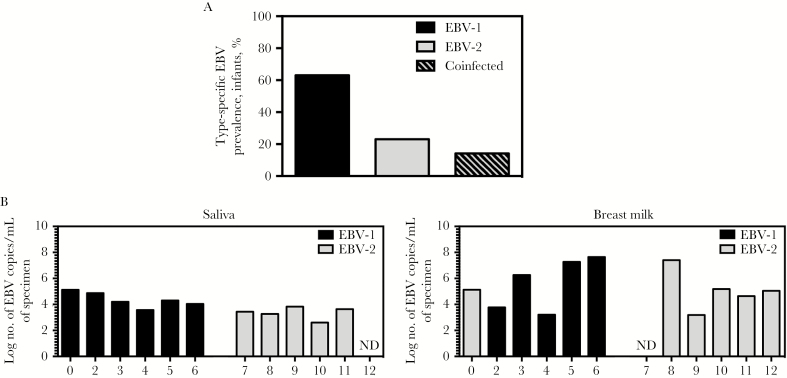
Burkitt lymphoma is associated with EBV infection and is a common pediatric cancer in sub-Saharan Africa [14]. EBV-2 was detected in Burkitt lymphoma tumors from this region >25 years ago [3]. However, the incidence of EBV-2 in healthy children from this region has not previously been published. Thus, we began by determining the EBV-2 prevalence in an infant cohort from a region in Western Kenya with a high risk for Burkitt lymphoma [15]. Blood samples collected from 85 healthy Kenyan children at 6 weeks through 12 months of age were analyzed for the presence of EBV DNA. All EBV-positive samples (84 collected at age 6 weeks, 62 collected at 10 weeks, 37 collected at 14 weeks, 23 collected at 18 weeks, 28 collected at 6 months, 28 collected at 9 months, and 32 collected at 12 months) were analyzed to determine EBV type. Infants in whom we were able to determine the EBV type in the blood for at least 1 time point through the first year of life (n = 35) were used to determine the prevalence of EBV-2. Through the first year of life, 23% of infants had only EBV-2 detected, and an additional 14% of infants were coinfected by both EBV types (Figure 1A). These findings demonstrate that EBV-2 was prevalent in healthy children from this region of Kenya, making this population relevant for studies of EBV-2 persistence.
Figure1.
Prevalence of Epstein-Barr virus (EBV) types in the Kisumu, Kenya, infant population and EBV types detectable in saliva and breast milk specimens from the infants’ mothers. A, Blood specimens were collected from infant donors at 6 weeks, 10 weeks, 14 weeks, 18 weeks, 6 months, 9 months, and 12 months of age. DNA was isolated from blood specimens and used to perform quantitative polymerase chain reaction (qPCR) analysis to detect BALF5 in the EBV genome (and thereby identify EBV-positive samples (84 collected at age 6 weeks, 62 collected at 10 weeks, 37 collected at 14 weeks, 23 collected at 18 weeks, 28 collected at 6 months, 28 collected at 9 months, and 32 collected at 12 months). DNA samples that were positive for EBV genome were further analyzed to determine the EBV type present in each sample, via a multiplex reverse-transcription PCR for EBNA3c that distinguishes EBV type 1 (EBV-1) and EBV-2; for 3 children, we determined the EBV type for samples collected ≥1 time point. DNA samples that were positive for EBV-1 or EBV-2 through the first year of life were used to determine the type-specific EBV prevalence in this population. Infants were considered coinfected if both EBV-1 and EBV-2 were detected at any time point tested. Prevalence data for samples in which the EBV type could be determined are shown in bar graph (22 of 35 were positive for EBV-1, 8 of 35 were positive for EBV-2, and 5 of 35 were positive for both types). B, Saliva and breast milk specimens were collected from infants’ mothers at postpartum week 6, and DNA was isolated. Multiplex quantitative PCR analysis of BALF5 in the EBV genome was performed to determine the log number of EBV copies per milliliter of saliva and breast milk. Additionally, multiplex reverse-transcription PCR analysis to detect EBNA3c was performed to identify the EBV type present in each sample. Abbreviation: ND, not detected.
EBV Type Detected in Infants and Mothers Correspond
To further evaluate EBV-2 in the Kenyan population, we focused our study on a subset of 12 infants and their mothers. Study participants were chosen on the basis of the availability of samples and the detection of a single EBV strain at 12 months of age. We selected 6 infants who were positive for EBV-1 (infants 1–6) and 6 who were positive for EBV-2 (infants 7–12; Table 1). We observed that all 12 infants were infected with EBV by 14 weeks of age (Table 1), consistent with our previous studies, in which we found EBV infection in infants in this region [16]. A likely source of EBV transmission is from the mother via saliva and/or breast milk [7, 17, 18]. To determine the EBV type present in saliva and breast milk specimens obtained from mothers at postpartum week 6, PCR analysis was performed. EBV was detected in 11 of 12 saliva samples (from mothers of infants 1–11; median, 3.82 log copies/mL; range, 2.60–5.12 log copies/mL) and 11 of 12 breast milk samples (from mothers of infants 1–6 and 8–12; median, 5.12 log copies/mL; range, 3.18–7.64 log copies/mL). The EBV type detected in saliva and/or breast milk specimens corresponded to the EBV type detected in blood specimens obtained from infant at 12 months of age (Figure 1B and Table 1), with the exception of infant 1. Different EBV strains were detected in the saliva (EBV-1) and breast milk (EBV-2) specimens collected from the mother of infant 1, and when we examined specimens obtained at early time points, we found that this infant was coinfected with both EBV strains at <12 months of age (Figure 1B and Table 1), supporting the idea that both saliva and breast milk were a source of transmissible virus. Based on these results and our previous finding that EBV-2 infects T cells in vitro [8], we next wanted to determine whether EBV-2 infects T cells in this population.
Table 1.
Infant Sex, Age at Initial Epstein-Barr Virus (EBV) Detection, and EBV Type Detected Before and After Age 12 Months
| Identifier | Sex | Age at First EBV Detection, wk | EBV Type(s) Detected, by Age | |
|---|---|---|---|---|
| <12 mo | 12 mo | |||
| 1 | Male | 14 | 1, 2 | 1 |
| 2 | Male | 6 | 1 | 1 |
| 3 | Female | 6 | 1 | 1 |
| 4 | Female | 6 | 1 | 1 |
| 5 | Male | 6 | 1 | 1 |
| 6 | Male | 6 | 1 | 1 |
| 7 | Female | 6 | 2 | 2 |
| 8 | Female | 6 | 2 | 2 |
| 9 | Male | 6 | 2 | 2 |
| 10 | Female | 10 | 1, 2 | 2 |
| 11 | Male | 6 | 2 | 2 |
| 12 | Male | 6 | 2 | 2 |
EBV-2 Infects T Cells in Healthy Children
To determine whether EBV-2 infects T cells in vivo, venous blood samples from infants at 12 month of age were used because there were sufficient numbers of PBMCs to perform cell fractionation. T-cell and non–T-cell fractions were isolated from PBMCs, using magnetic bead separation. The purity of CD3+ T-cell fractions in all samples collected at month 12 was >97%, with ≤0.02% CD19+ B cells, as determined by flow cytometry. The percentage of CD19+ B cells in the non–T-cell fraction ranged from 13.6% to 19.3%, with ≤0.12% CD3+ T cells (Figure 2A and Supplementary Figure 1). DNA and RNA were extracted from the purified fractions, and quantitative PCR (qPCR) was done to quantify EBV in each fraction.
EBV DNA was detected in the T-cell fraction of all 6 EBV-2–positive children (infants 7–12; median, 3.79 log copies/1 × 106 cells; range, 3.48–4.52 log copies/1 × 106 cells). Conversely, EBV was not detected in T cells from the majority of the children who were positive for EBV-1 at 12 months of age (infants 2–6). However, EBV was detected in the T-cell fraction of 1 EBV-1–positive child (infant 1; 3.77 log copies/1 × 106 cells; Figure 2A). Notably, infant 1 was found to be positive for EBV-2 at a younger age (Table 1), making it possible that EBV-2 established a persistent infection in the T-cell population at a level below the threshold of detection when whole PBMCs were analyzed. EBV DNA was found in the B-cell–containing non–T-cell fractions of all 12 children (median, 3.26 log copies/1 × 106 cells; range, 1.76–3.79 log copies/1 × 106 cells; Figure 2A), suggesting that EBV-2 persists in both T and B cells.
We confirmed the EBV type present in each cell fraction to determine whether, as observed in vitro, T-cell tropism is unique to EBV-2 [8]. Typing of the non–T-cell fractions demonstrated that the samples from infants 1–6 were EBV-1 positive and that those from infants 7–12 were EBV-2 positive (Figure 2A), corresponding to the type detected in the blood at 12 months of age (Table 1). Typing of the T-cell fractions revealed that, in all EBV-positive samples, including those from infant 1, EBV-2 but not EBV-1 could be detected (Figure 2A). This suggests that, in healthy children, EBV-2 can persist in both T cells and B cells, whereas EBV-1 persists only in B cells.
In vivo, EBV is known to persist in B cells in a latent state, in which viral gene expression is restricted to the latency genes, or in a latency zero state, in which no viral genes are expressed [4]. To assess the possibility that EBV-2 was latently infecting T cells, we performed gene expression analysis for the EBV latency genes LMP1 (encoding latent membrane protein 1), LMP2, EBNA1, and EBNA2. In healthy hosts, EBV primarily resides in the memory B-cell compartment in the latency zero state, with no detectable latent gene expression [4, 19]. Consistent with this, no latent gene transcripts were detected in the non–T-cell fractions in all samples (data not shown). Similarly, no EBV transcripts were detected in the T-cell fractions (data not shown), suggesting that EBV-2–infected T cells may be in a latency zero state. However, the limited amount of RNA acquired from these samples prevented us from performing additional analysis for lytic transcripts, leaving open the possibility that EBV-2 established a lytic infection in T cells. Notably, we were able to detect β-2-microglobulin mRNA in all non–T-cell and T-cell fractions, indicating that the samples were of sufficient quality to undergo PCR amplification (data not shown).
EBV-2 Persistence in T-Cell Compartment of Children
Collectively, data from the analysis of PBMCs obtained at 12 months of age demonstrated the presence of EBV-2–infected T cells but not EBV-1–infected T cells in healthy infants from Kenya. This suggests that EBV-2 could use the T-cell compartment, in addition to the B-cell compartment, as a latency reservoir. Thus, to better understand whether EBV-2 infection of T cells is a persistent or transient event, we next determined whether EBV-2 could be detected in T cells at 24 months of age in these same infants. T-cell and non–T-cell fractions were isolated from PBMCs collected from infants in our cohort at 24 months of age, and PCR assays to quantify the EBV load and determine the EBV type were performed as described above. The purity of CD3+ T-cell fractions in all 24-month samples was >96%, with ≤0.04% CD19+ B cells, as determined by flow cytometry. The percentage of CD19+ B cells in the non–T-cell fraction ranged between 14.8% and 20.5%, with ≤0.08% CD3+ T cells (Figure 2B and Supplementary Figure 1).
EBV DNA was detected in the B-cell–containing non–T-cell fractions of all 12 children at 24 months of age (median, 2.92 log copies/1 × 106 cells; range, 1.88–3.71 log copies/1 × 106 cells; Figure 2B). Typing of the non–T-cell fraction revealed that the majority of samples were EBV-1 positive (Figure 2B), suggesting that the predominate EBV type in circulation shifted from EBV-2 to EBV-1 in infants 7 and 12 by age 24 months. However, EBV-2 was still detectable in the non–T-cell fraction of 4 infants (infants 8–11), with EBV-2 also detected in the T-cell fraction of 3 of these infants (infants 8, 10, and 11; median, 2.65 log copies/1 × 106 cells; range, 2.43–3.25 log copies/1 × 106 cells; Figure 2B). These data demonstrate that EBV-2 is able to persist in the T-cell compartment of healthy children for at least 12 months.
EBV-2 Absence From the T-Cell Compartment of Adults
By adulthood, EBV has substantial time to establish a stable infection in the cellular compartments that serve as long-term latency reservoirs. Thus, to further decipher whether EBV-2 infection of T cells is persistent or transient, we next determined whether we could detect EBV-2 in T cells isolated from the mothers of the 12 infants included in this study. T-cell and non–T-cell fractions were isolated from PBMCs collected at 20–24 weeks of pregnancy and, as described above, qPCR assays to quantify EBV load and determine EBV type were performed. The purity of CD3+ T-cell fractions in these samples was >95%, with ≤0.04% CD19+ B cells. The percentage of CD19+ B cells in the non–T-cell fraction ranged from 11.3% to 15.7%, with ≤0.16% CD3+ T cells (Figure 3 and Supplementary Figure 1). Although EBV was detected in all of the B-cell–containing non–T-cell fractions isolated from the mothers (median, 3.04 log copies/1 × 106 cells; range, 2.56–4.92 log copies/1 × 106 cells), viral genome was not detectable in the T-cell fractions (Figure 3). Typing of the non–T-cell fractions demonstrated that EBV-1 was the predominate EBV type in circulation for 10 of 12 mothers (those of infants 1–6 and 9–12), with EBV-2 only detected in non–T-cell fractions from 2 mothers (those of infants 7 and 8; Figure 3). Notably, for some mothers, the EBV type detected in circulation did not correspond to the EBV type detected in the saliva (those of infants 9–11) and breast milk (those of infants 1 and 9–12; Figures 1B and 3), suggesting that EBV-2 may predominantly persist outside of peripheral circulation. These data leave open the question of whether EBV-2 establishes a long-term persistent infection in the T-cell compartment.
DISCUSSION
It is known that, in some disease states, EBV infects T cells in vivo, as evidenced by its detection in people with chronic active EBV infection [20], individuals with EBV-associated T-cell lymphomas [21], and children with HIV type 1 infection [22]. However, it had not been shown that EBV can infect T cells in healthy individuals. In this study, we demonstrated that EBV can infect CD3+ T cells in a population of healthy infants from Kenya, a region where the prevalence of EBV-2 is high. This finding suggests that T-cell infection is part of the normal life cycle of EBV-2 and not an aberration due to underlying disease processes. Consistent with our observations following EBV infection of T cells ex vivo [8], we detected EBV-2 but not EBV-1 in T cells. The ability of EBV-2 to infect T cells in healthy Kenyan infants is striking and suggests that the genetic variations unique to EBV-2 [2] promote T-cell infection.
The infants in this study are from a region of Kenya where malaria transmission is holoendemic [7, 9]. We have previously shown that children exposed to holoendemic malaria have suppressed EBV-specific immunity and an elevated viral load [10, 11], suggesting malaria could disturb the virus-host balance to favor T-cell infection. However, all EBV-2–positive infants had similar viral loads in their T-cell fractions regardless of the frequency of malarial parasite infection. Additionally, in the sample collected from an EBV-2–positive infant with malaria, the T-cell fraction was negative for EBV, suggesting that malarial parasite infection is not a prerequisite for EBV-2 infection of T cells.
A critical challenge studying EBV persistence in children is the limited amount of PBMCs available to analyze EBV persistence as compared to studies done in healthy US adults, from whom blood samples can be obtained through apheresis [23]. Because we had limited cell numbers from which to isolate T cells from infants, we used magnetic bead separation to isolate CD3+ T cells from the non–T-cell fractions. Thus, we did not have sufficient cells to isolate purified CD19+ B cells. So, while it is likely we detected EBV in B cells in the non–T-cell fraction, we cannot rule out EBV infection of other cell types. In addition, although EBV-2 preferentially infects CD8+ T cells ex vivo, because of limited cell numbers, we could not distinguish whether EBV-2 infected CD8+ or CD4+ T cells in vivo. We also could not perform limiting-dilution PCR analysis to determine the frequency of infected T cells. Importantly, B-cell contamination of the T-cell fractions was ≤0.04%, and infants who had only EBV-1 detected at any age had a T-cell fraction negative for EBV. Thus, the possibility that B-cell contamination could explain our results is unlikely. The EBNA3c qPCR used to identify EBV type is less sensitive (10 EBV copies/reaction) than the Balf5 qPCR used to quantify EBV load (4 EBV copies/reaction). Therefore, we selected infants for this study with comparatively higher EBV loads in whole blood at 12 months of age, to increase the chance we would be able to identify the EBV type in cell fractions. Notably, the EBV loads we detected in the Kenyan mothers and infants were comparable to EBV loads previously observed in PBMCs of healthy Gambian adults and children [24].
The observation that EBV-2 persists in the human population is paradoxical to the impaired ability of EBV-2 to immortalize B cells in culture, yet we found EBV-2 in B cells in vivo. This raises the possibility that EBV-2 could use a mechanism distinct from EBV-1 to establish a persistent infection. One possible mechanism would be that EBV-2 uses the T-cell compartment to establish persistence. EBV-2 could accomplish this by altering T-cell cytokine production that could dampen the immune response, recruit additional target cells to the infection site, and enhance lymphocyte activation and survival. Another, non–mutually exclusive possibility is that T cells serve as an additional latency reservoir for EBV-2. This idea is supported by our data demonstrating that EBV-2 could be detected in T cells isolated from all EBV-2–positive infants at 12 months of age and from 3 of these same infants at 24 months of age. However, EBV-2 was no longer detectable in 4 infants whose T-cell fractions were positive for EBV-2 at age 12 months. Additionally, while EBV-2 was readily detected in saliva and breast milk samples from mothers, EBV-2 was not detected in any maternal T-cell fractions. Thus, over time the frequency of EBV-2–infected T cells may drop to a concentration below the level of detection for our PCR protocol. Alternatively, T cells may not serve as a lifelong latency reservoir, but instead EBV-2 may establish a prolonged transient infection in the T cells subsequent to primary infection. The decrease or loss of EBV-2 in T cells with age could result from changes in the overall composition of peripheral T-cell subsets, as T cells at specific developmental stages may be more susceptible to infection. Along these lines, it has been shown that an EBV laboratory strain can infect immature human thymocytes in vitro [25]. Work to identify the receptors used by EBV-2 to infect T cells and the T-cell subsets that are susceptible to infection are currently ongoing. Comparison of the EBV type detected in the mother’s peripheral cell fractions verses saliva and breast milk also presents the possibility that EBV-2 may predominantly persist outside of circulation. The compartmentalization of EBV strains has previously been shown by Sitki-Green et al, demonstrating differences in EBV LMP1 variant profiles within the oral cavity and peripheral blood [26, 27]. Although EBV variants likely pass between compartments, these studies suggest that different compartments may provide specific EBV variants a more fit environment to persist. Additional studies focused on the healthy Kenyan population are required to determine whether EBV-2 infection of T cells is persistent or transient and to clarify the cellular compartment of long-term EBV-2 persistence.
In sum, we found that EBV-2 infects T cells in healthy Kenyan infants, persists in some infants infected in infancy through 2 years of age, and is shed in breast milk and saliva of their mothers. Our results indicate that EBV-2 infection of T cells is not an aberration but likely part of the strategy of EBV-2 to establish lifelong latency.
Supplementary Material
Notes
Acknowledgments. We thank the Chulaimbo Sub-District Hospital, for allowing us to use their facilities to perform this study; our clinical officer, medical officer, and data entry and field staff, for their involvement in the project; and the mothers and children, for their participation in this study.
The manuscript was approved by the director of the Kenya Medical Research Institute.
Financial support. This work was supported by the National Cancer Institute (grant CA102667 to R. R. and C. C., grant AI122670 to C. C., and D43 training grant [no 153707] to I. D. and S. O.), the National Institutes of Health, and Burroughs Welcome (Career Award for Medical Scientists [no. 1006818] to A. D.).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Rowe M, Young LS, Cadwallader K, Petti L, Kieff E, Rickinson AB. Distinction between Epstein-Barr virus type A (EBNA 2A) and type B (EBNA 2B) isolates extends to the EBNA 3 family of nuclear proteins. J Virol 1989; 63:1031–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Palser AL, Grayson NE, White RE et al. . Genome diversity of Epstein-Barr virus from multiple tumor types and normal infection. J Virol 2015; 89:5222–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Young LS, Yao QY, Rooney CM et al. . New type B isolates of Epstein-Barr virus from Burkitt’s lymphoma and from normal individuals in endemic areas. J Gen Virol 1987; 68 (Pt 11):2853–62. [DOI] [PubMed] [Google Scholar]
- 4. Babcock GJ, Hochberg D, Thorley-Lawson AD. The expression pattern of Epstein-Barr virus latent genes in vivo is dependent upon the differentiation stage of the infected B cell. Immunity 2000; 13:497–506. [DOI] [PubMed] [Google Scholar]
- 5. Tzellos S, Correia PB, Karstegl CE et al. . A single amino acid in EBNA-2 determines superior B lymphoblastoid cell line growth maintenance by Epstein-Barr virus type 1 EBNA-2. J Virol 2014; 88:8743–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Correia S, Palser A, Elgueta Karstegl C et al. . Natural variation of Epstein-Barr virus genes, proteins and pri-miRNA (revised). J Virol 2017; 91. doi: 10.1128/JVI.00375-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Daud II, Coleman CB, Smith NA et al. . Breast milk as a potential source of Epstein-Barr virus transmission among infants living in a malaria-endemic region of Kenya. J Infect Dis 2015; 212:1735–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Coleman CB, Wohlford EM, Smith NA et al. . Epstein-Barr virus type 2 latently infects T cells, inducing an atypical activation characterized by expression of lymphotactic cytokines. J Virol 2015; 89:2301–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Daud II, Ogolla S, Amolo AS et al. . Plasmodium falciparum infection is associated with Epstein-Barr virus reactivation in pregnant women living in malaria holoendemic area of Western Kenya. Matern Child Health J 2015; 19:606–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moormann AM, Chelimo K, Sumba PO, Tisch DJ, Rochford R, Kazura JW. Exposure to holoendemic malaria results in suppression of Epstein-Barr virus-specific T cell immunosurveillance in Kenyan children. J Infect Dis 2007; 195:799–808. [DOI] [PubMed] [Google Scholar]
- 11. Moormann AM, Chelimo K, Sumba OP et al. . Exposure to holoendemic malaria results in elevated Epstein-Barr virus loads in children. J Infect Dis 2005; 191:1233–8. [DOI] [PubMed] [Google Scholar]
- 12. Kubota N, Wada K, Ito Y et al. . One-step multiplex real-time PCR assay to analyse the latency patterns of Epstein-Barr virus infection. J Virol Methods 2008; 147:26–36. [DOI] [PubMed] [Google Scholar]
- 13. Simbiri KO, Smith NA, Otieno R et al. . Epstein-Barr virus genetic variation in lymphoblastoid cell lines derived from Kenyan pediatric population. PLoS One 2015; 10:e0125420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rochford R, Moormann AM. Burkitt’s Lymphoma. Curr Top Microbiol Immunol 2015; 390:267–85. [DOI] [PubMed] [Google Scholar]
- 15. Rainey JJ, Mwanda WO, Wairiumu P, Moormann AM, Wilson ML, Rochford R. Spatial distribution of Burkitt’s lymphoma in Kenya and association with malaria risk. Trop Med Int Health 2007; 12:936–43. [DOI] [PubMed] [Google Scholar]
- 16. Piriou E, Asito AS, Sumba PO et al. . Early age at time of primary Epstein-Barr virus infection results in poorly controlled viral infection in infants from Western Kenya: clues to the etiology of endemic Burkitt lymphoma. J Infect Dis 2012; 205:906–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Niederman JC, Miller G, Pearson HA, Pagano JS, Dowaliby JM. Infectious mononucleosis. Epstein-Barr-virus shedding in saliva and the oropharynx. N Engl J Med 1976; 294:1355–9. [DOI] [PubMed] [Google Scholar]
- 18. Mbulaiteye SM, Walters M, Engels EA et al. . High levels of Epstein-Barr virus DNA in saliva and peripheral blood from Ugandan mother-child pairs. J Infect Dis 2006; 193:422–6. [DOI] [PubMed] [Google Scholar]
- 19. Babcock GJ, Decker LL, Volk M, Thorley-Lawson DA. EBV persistence in memory B cells in vivo. Immunity 1998; 9:395–404. [DOI] [PubMed] [Google Scholar]
- 20. Fujiwara S, Kimura H, Imadome K et al. . Current research on chronic active Epstein-Barr virus infection in Japan. Pediatr Int 2014; 56:159–66. [DOI] [PubMed] [Google Scholar]
- 21. Gru AA, Haverkos BH, Freud AG et al. . The Epstein-Barr Virus (EBV) in T Cell and NK Cell Lymphomas: Time for a Reassessment. Curr Hematol Malig Rep 2015; 10:456–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bekker V, Scherpbier H, Beld M et al. . Epstein-Barr virus infects B and non-B lymphocytes in HIV-1-infected children and adolescents. J Infect Dis 2006; 194:1323–30. [DOI] [PubMed] [Google Scholar]
- 23. Khan G, Miyashita EM, Yang B, Babcock GJ, Thorley-Lawson DA. Is EBV persistence in vivo a model for B cell homeostasis? Immunity 1996; 5:173–9. [DOI] [PubMed] [Google Scholar]
- 24. Njie R, Bell AI, Jia H et al. . The effects of acute malaria on Epstein-Barr virus (EBV) load and EBV-specific T cell immunity in Gambian children. J Infect Dis 2009; 199:31–8. [DOI] [PubMed] [Google Scholar]
- 25. Watry D, Hedrick JA, Siervo S et al. . Infection of human thymocytes by Epstein-Barr virus. J Exp Med 1991; 173:971–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sitki-Green DL, Edwards RH, Covington MM, Raab-Traub N. Biology of Epstein-Barr virus during infectious mononucleosis. J Infect Dis 2004; 189:483–92. [DOI] [PubMed] [Google Scholar]
- 27. Sitki-Green D, Covington M, Raab-Traub N. Compartmentalization and transmission of multiple epstein-barr virus strains in asymptomatic carriers. J Virol 2003; 77:1840–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.